Background: The role of miRNA-mediated regulation of RECK in keratinized tumors is unclear.
Results: miRNAs express differentially in subtypes of OSCCs, and keratinization-associated miRNAs inversely correlate with RECK in oral cancer cells.
Conclusion: miR-7 and miR-21 negatively regulate the tumor suppressor gene RECK.
Significance: Keratinization-associated miRNAs may serve as novel targets to reduce tumor aggressiveness.
Keywords: Gene Regulation, Microarray, MicroRNA, RNA Interference (RNAi), Tumor Cell Biology, RECK, Keratinization, miR-21, miR-7, Oral Squamous Cell Carcinoma
Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that posttranscriptionally regulate gene expression during many biological processes. Recently, the aberrant expressions of miRNAs have become a major focus in cancer research. The purpose of this study was to identify deregulated miRNAs in oral cancer and further focus on specific miRNAs that were related to patient survival. Here, we report that miRNA expression profiling provided more precise information when oral squamous cell carcinomas were subcategorized on the basis of clinicopathological parameters (tumor primary site, histological subtype, tumor stage, and HPV16 status). An innovative radar chart analysis method was developed to depict subcategories of cancers taking into consideration the expression patterns of multiple miRNAs combined with the clinicopathological parameters. Keratinization of tumors and the high expression of miR-21 were the major factors related to the poor prognosis of patients. Interestingly, a majority of the keratinized tumors expressed high levels of miR-21. Further investigations demonstrated the regulation of the tumor suppressor gene reversion-inducing cysteine-rich protein with kazal motifs (RECK) by two keratinization-associated miRNAs, miR-7 and miR-21. Transfection of miR-7 and miR-21-mimics reduced the expression of RECK through direct miRNA-mediated regulation, and these miRNAs were inversely correlated with RECK in CAL 27 orthotopic xenograft tumors. Furthermore, a similar inverse correlation was demonstrated in CAL 27 cells treated in vitro by different external stimuli such as trypsinization, cell density, and serum concentration. Taken together, our data show that keratinization is associated with poor prognosis of oral cancer patients and keratinization-associated miRNAs mediate deregulation of RECK which may contribute to the aggressiveness of tumors.
Introduction
Oral cancer is one of the most prevalent cancers worldwide, with squamous cell carcinomas being the most common type, accounting for ∼90% of all oral cancers (1). Approximately 300,000 new cases of oral cancers (amounting to 3% of total cancers) are anticipated annually (2). Although many studies have demonstrated how clinical and histological staging may explain why some cancers, but not others, behave aggressively, these stagings do not always properly reflect the extent of disease (3). A better prediction for the prognosis of patients has been proposed using gene expression data in combination with OSCC4 tumor stage information rather than tumor stage information alone (4, 5). Despite the medical advances and new treatments for oral cancer, the average five-year survival rate of 50% has not improved for decades (6). Hence, more molecular insights into oral cancer pathogenesis are needed to develop proper diagnostics and therapeutics.
MicroRNAs (miRNAs) are endogenous small non-coding RNAs, ∼18–25 nucleotides in length, that act as posttranscriptional regulators of gene expression in diverse cellular processes such as proliferation, differentiation, development, and cell death (7). Over 60% of all mammalian mRNAs are predicted targets of miRNAs, indicating their extensive roles in the regulation of numerous cellular processes (8). The discovery of miRNAs and the elucidation of their function in regulating gene expression levels demonstrates their increasing importance in cancer genetics (9). In cancer, some miRNAs have been found to play roles as regulators for tumor suppressors or oncogenes, depending on the tumor microenvironment and/or tissue type.
When epithelial cells keratinize, keratins and keratin filament-associated proteins are synthesized and assembled to form keratin filaments (10). During these processes, signaling molecules such as matrix metalloproteinases (MMPs) modulate the process of keratinization (10, 11). In particular, MMP-9 has been implicated in the process of keratinization (11). MMPs are well known to play crucial roles for tumor progression by degrading the extracellular matrix barriers encompassing the tumor and permitting invasion into surrounding connective tissues (12). At least three cancer-associated MMPs (MMP-2, MMP-9, and MT1-MMP) are regulated by RECK (13, 14). Reversion-inducing cysteine-rich protein with kazal motifs (RECK) is a membrane-anchored glycoprotein detected in a variety of normal human tissues (14). The reduced expression of RECK has been observed in various types of tumor tissues and is frequently associated with poor prognosis (15–19). Therefore, studies of the relation between RECK and keratinization may yield important insights into the mechanisms of carcinogenesis and the prognosis of oral cancer patients.
Although the majority of oral cancers are OSCCs, the heterogeneity within OSCCs and the limited molecular data regarding carcinogenesis hamper our understanding of the biological differences in OSCC subclasses. Therefore, the aim of this study was to identify prognostic indicators for oral cancer and to understand how aberrantly expressed miRNAs and their targets are involved in the pathogenesis of this disease.
EXPERIMENTAL PROCEDURES
Patients and Tissue Samples
All human OSCCs and normal tongue tissues were collected from the Tissue Bank at the Moffitt Cancer Center (Protocol no. MCC-15370) and approved by the Institutional Review Board of the University of South Florida (no. 106444). Written consent was given by the patients for their information to be stored in the Moffitt Tissue Bank and used for research. Clinicopathological characteristics of these 17 patients are shown in supplemental Table S1. Tissues were snap-frozen and stored at −80 °C until further use.
Cell Culture and Transfection
Seven head and neck cancer cells (CAL 27, SCC-25, HN, BHY, FaDu, RPMI 2650, and OQ01), four cervical cancer cells (HeLa, SiHa, Ca Ski, and End1/E6E7), and HEK293 cells were used in this study. CAL 27, SCC-25, FaDu, RPMI 2650, HeLa, SiHa, Ca Ski, End1/E6E7, and HEK293 cells were purchased from the ATCC. HN and BHY cells were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen, GmbH (DSMZ) (Braunschweig, Germany). OQ01 was a primary cultured head and neck cancer cell line provided by Dr. Lung-Ji Chang, University of Florida. CAL 27 cells were cultured in DMEM supplemented with 1.5 g/liter sodium bicarbonate; SCC-25 in DMEM/F12 supplemented with 15 mm HEPES, 1.2 g/liter sodium bicarbonate, 0.5 mm sodium pyruvate, 400 ng/ml hydrocortisone; FaDu and RPMI 2650 in minimum essential medium supplemented with 1.5 g/liter sodium bicarbonate, 1.0 mm sodium pyruvate, 0.1 mm non-essential amino acids; HN, BHY, and OQ01 in DMEM/F12; HeLa, SiHa, and HEK293 in DMEM; Ca Ski in RPMI1640 medium; End1/E6E7 in Keratinocyte medium (ScienCell, San Diego, CA). All the media except Keratinocyte medium were supplemented with 10% FBS, 100 μg/ml streptomycin, and 100 units/ml penicillin. Keratinocyte medium was prepared according to the protocol of the manufacturer. All cell lines were incubated in a humidified atmosphere of 37 °C with 5% CO2. All transfections were performed using Lipofectamine 2000 (Invitrogen) according to the protocol of the manufacturer. miRNA mimics and nonspecific negative control molecules were purchased from Ambion (Austin, TX). Transfected cells were incubated in a 37 °C incubator with 5% CO2 for 72 h.
RNA Isolation
All the RNA used in this study was isolated using the mirVanaTM miRNA isolation kit (Ambion). Human tissues measuring ∼60 mm3 in volume were homogenized using the PowerGen 125 (Fisher Scientific, Pittsburgh, PA) in 600 μl of the lysis/binding buffer, after which total RNA was isolated according to the instructions of the manufacturer. A NanoDrop ND-100 spectrophotometer (Nanodrop Technology, Inc., Wilmington, DE) was used to quantify the isolated RNA. Additionally, an Agilent 2100 bioanalyzer from the Interdisciplinary Center for Biotechnology Research at the University of Florida was used to monitor the size distribution and overall quality of total RNA prior to miRNA profiling.
MicroRNA Microarray Profiling
Total RNAs isolated from three normal tongues and 17 OSCCs were processed by Asuragen Services (Austin, TX) according to standard operating procedures. The Agilent human miRNA microarrays Rel12.0 (Agilent, Santa Clara, CA) according to the Agilent miRNA protocol v2.1. Cluster 3.0 (for dendrograms) (20) and Java TreeView (for heat map) (21) software programs were used to visualize the hierarchical clustering of miRNA expression in 17 OSCCs.
Quantitative Real-time PCR (qRT-PCR)
Reverse transcription was performed using the TaqMan MicroRNA reverse transcription kit or TaqMan high-capacity cDNA reverse transcription kit for miRNA and mRNA, respectively (Applied Biosystems, Foster City, CA). Quantification of miRNA and mRNA expression was performed using TaqMan MicroRNA assays, TaqMan gene expression assays, and TaqMan Fast Universal PCR Master Mix (Applied Biosystems). The experiments were processed and analyzed using the Applied Biosystems StepOne real-time PCR machine. Fold change values were calculated using the 2−ΔΔCt method. In addition to the three normal tissues used for the microarray analyses, two normal tongue tissues were added in the qRT-PCR analyses to enhance statistical computation. U6 snRNA and 18S rRNA were used as internal controls to normalize all the miRNA or mRNA expressions. TaqMan primer sets were used to measure mRNA levels of HPV16-E6 and HPV16-E7 using real-time PCR (22), which are more reliable assays compared with the oversensitive DNA PCR assays (22, 23). The degree of HPV status was determined by the corresponding cycle threshold (Ct) values: -, undetectable; +, Ct > 32; ++, 25 < Ct < 32; +++, Ct < 25.
Combinatorial Radar Chart Analyses
OSCCs were separated into subgroups according to their clinicopathological characteristics (supplemental Table S1). The tumors were separated on the basis of the primary site (oropharynx or oral cavity), histological subtype (non-keratinized or keratinized), tumor stage (early stage or advanced stage), and HPV16 status (negative or positive). The TNM (tumor size (T), regional nodal involvement (N), presence or absence of distant metastasis (M)) classification system was used to determine tumor stage. Pathological TNM staging was primarily used, but clinical TNM staging was used for three tumor samples (T1715B, T1231D, and T470A), for which pathological TNM was unspecified. Radar charts generated by Microsoft Excel 2010 software were used to visualize the expression profiles of 10 miRNAs in 17 OSCCs. The relative expression values in OSCCs compared with those of normal controls obtained by qRT-PCR were adjusted so that the 10 miRNAs analyzed were represented on a similar scale. All values were converted into log units, and then overexpressed miRNAs were divided by the highest value obtained for the corresponding individual miRNA. For miR-486-5p, which was an underexpressed miRNA, the values were divided by the lowest expressed value among the samples to generate an opposite effect for convenient visualization.
Western Blot Analysis
NET/0.3%Nonidet P-40 buffer (150 mm NaCl, 5 mm EDTA, 50 mm Tris (pH 7.5), 0.3% Nonidet P-40) with Complete EDTA-free protease inhibitor (Roche) was used to prepare cell lysates. Proteins quantitated using BCA (Thermo Scientific, Rockford, IL) were separated on 7.5% polyacrylamide gel and transferred to a nitrocellulose membrane. The dilutions of primary antibodies were: 1:250 for mouse anti-RECK (BD Biosciences) and 1:5000 for mouse anti-tubulin antibodies (Sigma-Aldrich, St. Louis, MO). Secondary goat anti-mouse antibodies conjugated to horseradish peroxidase were used at 1:10000 dilutions (Southern Biotech, Birmingham, AL). Immunoreactive bands were detected by the SuperSignal chemiluminescent system (Thermo Scientific, Rockford, IL). Densitometric analyses for the developed films were performed using Image J software (National Institutes of Health). RECK protein expression levels were normalized to tubulin, and the percentage changes were calculated compared to the mock control. Diluted cell lysates of the untreated samples (25, 50, and 100%) were used to document the semiquantitative measurement of the Western blot analysis results.
Dual Luciferase Assay
The 3′ UTR of RECK was PCR-amplified and cloned between EcoRI and NotI of the pMiR-Target vector (OriGene Technologies, Rockville, MD). Mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the mutated sequences were confirmed by DNA sequencing. All the primers used for generating luciferase constructs are listed in supplemental Table S2. HEK293 cells transfected with luciferase reporters and miRNA mimics were harvested after 48 h. The luciferase activities were measured using a dual luciferase reporter assay system (Promega, Madison, WI) and FLUOstar OPTIMA (BMG Labtech, Germany). Renilla luciferase (pRL-CMV, Promega) expression levels were used as an internal control to normalize the relative expressions of firefly luciferase (24, 25).
Differential Trypsinization
CAL 27 cells were washed with PBS and treated with trypsin solution (0.25% trypsin/2.21 mm EDTA in Hank's buffered salt solution (CellGro, Manassas, VA)) in a 37 °C incubator with 5% CO2. Detached cells were collected in a 15-ml conical tube 5 min after treatment, and 3 ml of fresh trypsin solution was added back to the flask. The same procedure was performed to collect the detached cells after 10- and 15-min treatments. Cells collected after 5, 10, and 15 min were washed with PBS and subjected to the RNA isolation process and further experiments.
Tumor Xenografts
Tumor samples from xenografts were obtained in a previous study (26). In brief, CAL 27 cells at 500,000 cells/50 μl were injected submucosally in the floor of the mouth of anesthetized eight-week-old NOD-SCID mice. Oral tumors were grown for 2 weeks, after which the animals were sacrificed. The tumor tissues were then harvested, and RNA was extracted as described above for human tumors.
Statistical Analyses
For the microarray analysis, one-way analysis of variance was performed across all samples for statistical hypothesis testing, and two-tailed Student's t tests were performed for all pairwise comparisons. Significance was assigned to probes demonstrating a false discovery rate of corrected p values < 0.05, as described by Benjamini et al. (27). All experiments were repeated at least three times, and the statistical analyses were performed using GraphPad Prism 4.0 (Graph Pad Software, La Jolla, CA).
RESULTS
Differential miRNA Expression in OSCC Subtypes
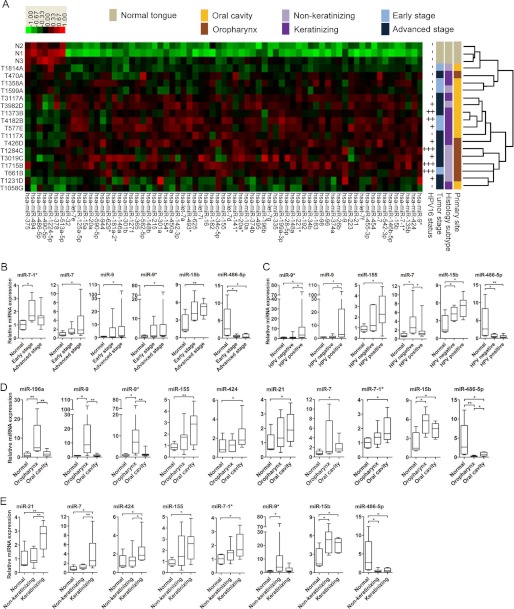
Microarray expression profiling of 17 OSCCs compared with three normal tongue tissues identified significant (p < 0.05) differences in the expression levels of 134 miRNAs. In particular, a heat map using supervised hierarchical clustering analyses with criteria of p < 0.05 and a Log2 difference over 1.5 demonstrated that OSCCs had seven underexpressed and 62 overexpressed miRNAs when compared with normal tissues (Fig. 1A). Moreover, the tumors formed a hierarchical clustering of groups (a distinct cluster indicates the differential expression between normal (N1-N3, gray) and OSCC samples (other colors)) depending on their clinicopathological parameters (Fig. 1A and supplemental Table S1). For example, six of seven oropharynx samples (brown) were clustered into a group in which five of the six were recorded as tumors originating at the base of the tongue. All of these tumors (T1284C, T3019C, T1715B, T661B, and T1231D) were non-keratinizing tumors (light purple). In addition, 10 HPV16-positive tumors formed two clusters: four (T3982D, T1373B, T4182B, and T577E) keratinizing and six (T426D, T1284C, T3019C, T1715B, T661B, and T1231D) non-keratinizing tumors all derived from oropharynx regions (Fig. 1A). Of the 69 aberrantly expressed miRNAs identified in the microarray analysis, nine up-regulated miRNAs (miR-9*, miR-424, miR-7–1*, miR-15b, miR-7, miR-21, miR-9, miR-155, and miR-196a) and one down-regulated miRNA (miR-486-5p) were selected for further verification by Taqman qRT-PCR. Selection of miRNAs was performed on the basis of the fold changes and the p values from our microarray data, although some miRNAs were chosen because of their potential involvement in oncogenesis (e.g. miR-196a (28), miR-155 (29), and miR-21 (30–32)).
FIGURE 1.
Differential miRNA expression in OSCC subtypes. A, 17 OSCC tumors form clusters on the basis of miRNA expression patterns and clinicopathological parameters. Heat maps were created by supervised hierarchical clustering analysis of 69 miRNAs (p < 0.05) with Log2 difference of > 1.5 between 17 OSCCs and three normal tongue samples. miRNA expression (columns) is shown for individual human tissue samples (rows). Four different clinicopathological classifications were applied to characterize the tumors: primary site (yellow/brown), histological subtype (light purple/dark purple), tumor stage (light blue/dark blue), and HPV16 status (−, +, ++, or +++). Gray represents normal tongue samples (N1-N3). Red and green colors in the heat map indicate overexpression and underexpression, respectively. Differential expressions of 10 miRNAs were verified in subtypes of OSCCs using Taqman qRT-PCR. Tumor samples were separated into subtypes on the basis of clinicopathological parameters: early (n = 6) versus advanced stage (n = 11) (B); HPV-negative (n = 7) versus HPV-positive (n = 10) (C); oropharynx (n = 7) versus oral cavity (n = 10) tumors (D); non-keratinizing (n = 8) versus keratinizing (n = 9) tumors (E). In addition to the three normal tissues used for the microarray analyses, two normal tongue tissues were added in the qRT-PCR analyses to enhance statistical computation. All qRT-PCR results are expressed as mean ± S.E. from at least three independent experiments. The results were analyzed using Mann-Whitney U test. *, p < 0.05, **, p < 0.01.
Combined Clinicopathological Parameters and miRNA Expression Signatures for OSCC Subtypes
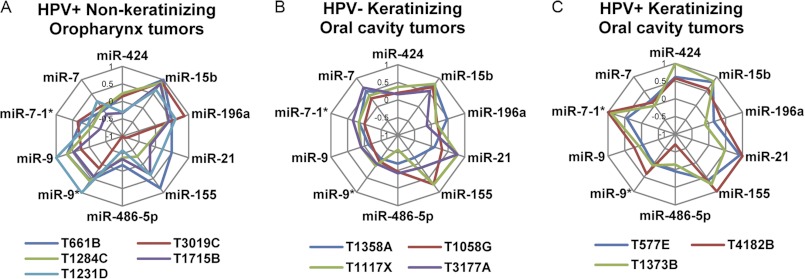
Because the tumors formed distinct clusters on the basis of our microarray data (Fig. 1A), the results obtained from the qRT-PCR experiments were further analyzed for the differences among the subtypes of OSCCs instead of considering all 17 tumors as a single homogenous group. Interestingly, significantly different miRNA expression levels were observed when subtypes of OSCCs were taken into consideration, as shown in Fig. 1, B–E. These observations indicated that the heterogeneous expression of miRNAs in OSCCs depends on their individual characteristics. Radar chart analyses were used to visualize the expression patterns of the 10 miRNAs selected for verification in individual OSCCs. When all 17 tumors were analyzed together, a complex heterogeneity was observed (supplemental Fig. S1A). In contrast, normal tissues exhibited similar contours, with the exception of one sample (supplemental Fig. S1B, N2). Interestingly, when using different combinations of these clinicopathological parameters, unique miRNA expression patterns were generated for specific subtypes of OSCCs (Fig. 2). Each subtype of tumor formed a contour that was easily distinguishable from that of the overall tumor population (Fig. 2 and supplemental Fig. S1A). Thus, these data demonstrated that combinatorial interpretations of clinicopathological features and miRNA expression profiles could be used as specific signatures of individual subtypes of oral tumors.
FIGURE 2.
Combined clinicopathological parameters and miRNA expression signatures for OSCC subtypes. Similar contours of miRNA expression patterns are formed distinguishing HPV-positive non-keratinizing oropharynx tumors (A), HPV-negative keratinizing oral cavity tumors (B), and HPV-positive keratinizing oral cavity tumors (C).
Advanced Tumor Stage, Keratinization State, and High Expression of miR-21 Are Indicators of Poor Prognosis for Oral Cancer Patients
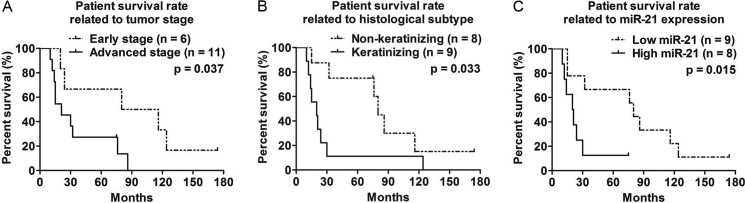
The overall 5-year survival rate for the patients in this study was 41.2% (7 of 17, Supplemental Table S1). Clinicopathological parameters and miRNA expression patterns of OSCCs were examined to evaluate their correlation to the survival rate of the patients. Kaplan-Meier survival analyses demonstrated that patients with advanced stage tumors (5-year survival, 27.3% (3/11); hazard ratio, 3.36; 95% confidence interval, 1.08–10.49; p = 0.037) and/or keratinizing tumors (5-year survival, 11.1% (1/9); hazard ratio, 3.34; 95% confidence interval, 1.10–10.12; p = 0.033) were significantly associated with a poor survival prognosis (Fig. 3, A and B). From our clinical data, we found that 89% of patients (eight of nine) with keratinized tumors survived less than 3 years (36 months) compared with 75% of patients (six of eight) with non-keratinized tumors who survived more than 5 years (60 months) (supplemental Table S1). Primary site (p = 0.66) and HPV status (p = 0.80) were not associated with the prognosis of patients (supplemental Fig. S2A). Patients with tumors expressing high levels of miR-21, however, displayed a significant correlation (5-year survival, 12.5% (1/8); hazard ratio, 5.31; 95% confidence interval, 1.39–20.38; p = 0.015) with poorer survival rates than those with lower levels of miR-21 (Fig. 3C). Tumors with miR-21 expression levels higher than the median value were considered “high miR-21-expressing tumors”. Other miRNAs examined in survival rate analyses were determined to be unrelated with the prognosis of patients (supplemental Fig. S2B). Interestingly, seven (T4182B, T577E, T3117A, T426D, T1117X, T470A, and T1373B) of the eight tumors that expressed high levels of miR-21 were classified as keratinizing tumors. The only non-keratinizing tumor (T3982D) in the “high miR-21-expressing tumor” group had the lowest miR-21 expression of the eight tumors. Thus, these results suggested that the expression of miR-21 was associated with the keratinization of tumors and that this association could potentially contribute to the poorer survival of the patients.
FIGURE 3.
Advanced tumor stage, keratinization state, and high expression of miR-21 as indicators of poor prognosis for oral cancer patients. Kaplan-Meier curves with Log-rank test were applied to identify the factors related to prognosis. A, advanced stage (p = 0.037). B, keratinization (p = 0.033). C, high expression of miR-21 (p = 0.015) were associated with poor survival rate.
Coordinated Expression of Keratinization-associated miRNAs in Human Tissues and Cell Lines
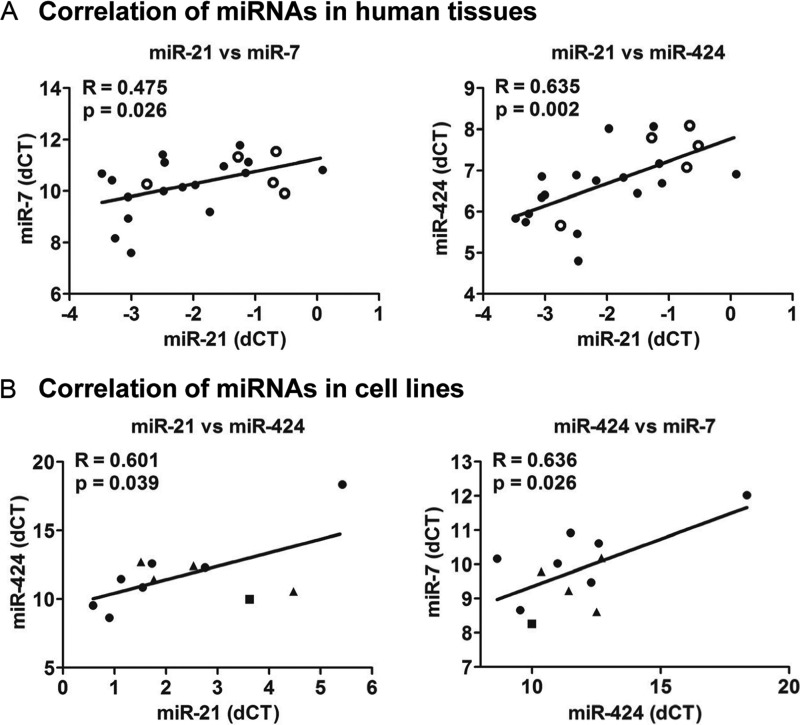
Our findings that both high miR-21 levels and the keratinization status were related to the survival rate of oral cancer patients made us pay particular attention to the expression levels of three miRNAs (miR-21, miR-7, and miR-424) that were found to be up-regulated in keratinizing tumors compared with either normal controls or non-keratinizing tumors (Fig. 1E). Using linear regression analysis, we observed a significant correlation when the expression levels of these miRNAs were examined together (Fig. 4). In human tissues (five normal and 17 OSCCs), miR-21 expression was directly correlated with both miR-7 and mir-424 expression levels (Fig. 4A). In human cell lines (seven head and neck cancer cells, four cervical cancer cells, and HEK293 cells), miR-21 and miR-7 expression directly correlated with miR-424 expression levels but not to each other (Fig. 4B). Together, these data suggested that the keratinization-associated miRNAs may have redundant regulatory effects for a common target.
FIGURE 4.
Coordinated expression of keratinization-associated miRNAs in human tissues and cell lines. A, linear regression analyses of miRNAs in human oral tissues. Positive correlation was observed between miR-21 and miR-7 (r = 0.475, p = 0.026) and miR-21 and miR-424 (r = 0.635, p = 0.002). ● and ○ indicate 17 OSCCs and 5 normal tongue samples, respectively. B, linear regression analyses of miRNAs in human cancer cell lines. Positive correlation was observed between miR-21 and miR-424 (r = 0.601, p = 0.039) and miR-424 and miR-7 (r = 0.636, p = 0.026). ●, ▴, and ■ indicate head and neck cancer, cervical cancer, and HEK293 cells, respectively.
RECK as a Common Target of Keratinization-associated miRNAs
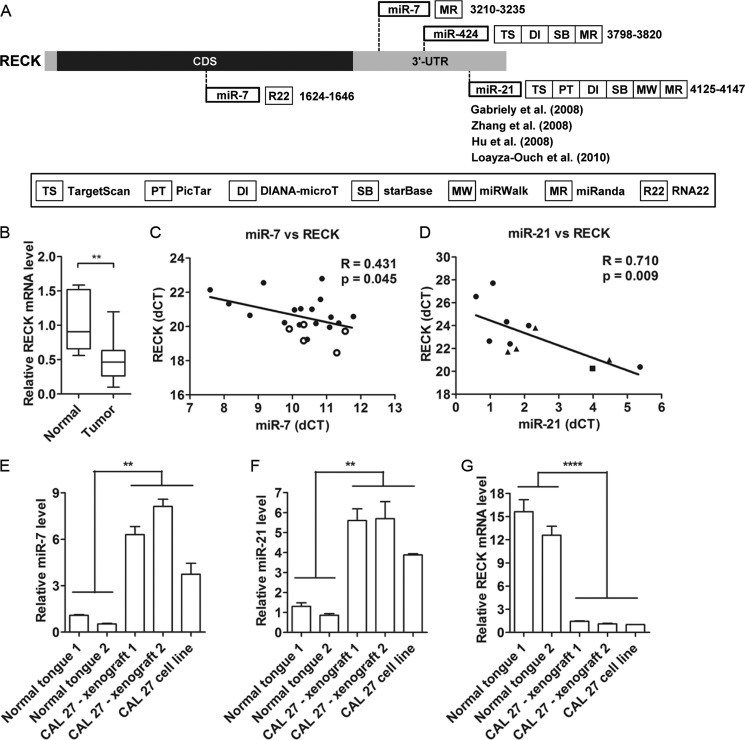
In silico analysis was performed to predict candidate targets coregulated by the three keratinization-associated miRNAs. Using several different prediction algorithms (supplemental Fig. S3), RECK was selected for further validation because RECK is a known tumor suppressor underexpressed in cancers. Putative binding sites for miR-7, miR-21, and miR-424 were detected, mostly on the 3′ UTR of RECK, with the exception of a second site for miR-7, which was also found within the coding region (Fig. 5A). The mRNA level of RECK was determined to be significantly underexpressed in the 17 OSCCs compared with the normal tissues (Fig. 5B). Next, the expression of RECK and miRNAs were analyzed by linear regression analysis, and an inverse correlation was observed between miR-7 and RECK in human tissues (Fig. 5C). When the same strategy was applied to human cancer cell lines, miR-21 and RECK were inversely correlated with each other (Fig. 5D). No correlation was observed, however, between miR-424 and RECK in either tissues or cell lines, indicating that miR-424 may not be a key regulator of RECK expression. Thus, these results showed that two keratinization-associated miRNAs, miR-7 and miR-21, could be important for regulating the expression of RECK.
FIGURE 5.
RECK is a common target of keratinization-associated miRNAs. A, in silico analysis identified a candidate target for miR-7, miR-21, and miR-424. Seven different miRNA target prediction programs were used in this analysis. B, RECK mRNA expression was measured in human oral tissues using Taqman qPCR. C, expression of miR-7 was inversely correlated with RECK in human oral tissues (r = 0.431, p = 0.045). ● and ○ indicate 17 OSCCs and five normal samples, respectively. D, expression of miR-21 was inversely correlated with RECK in human cancer cell lines. ●, ▴, and ■ indicate head and neck cancer cells, cervical cancer cells, and HEK293 cells, respectively. miR-7 (E) and miR-21 (F) expression levels were elevated whereas RECK expression levels (G) were reduced in CAL 27 xenograft mouse tumors. All results are expressed as mean ± S.E. from at least three independent experiments. Student's t test was used for the analyses. **, p < 0.01; ****, p < 0.0001.
A similar inverse correlation in the expression of miR-7, miR-21, and RECK were observed in CAL 27 orthotopic xenograft tumors. These were keratinizing tumors on the basis of H&E staining as confirmed by oral pathologists in the Department of Oral and Maxillofacial Diagnostic Sciences and documented in our earlier study (26). In particular, the expression of miR-7 and miR-21 were increased in the CAL 27 xenograft tumors (Fig. 5, E and F, respectively) compared with normal human tongue tissues. Meanwhile, RECK expression was reduced in these tumors (Fig. 5G).
Direct Regulation of RECK by miR-7 and miR-21
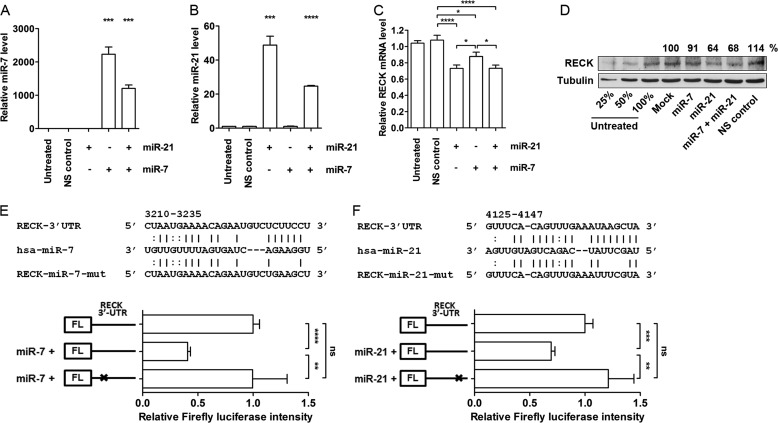
To investigate the effects of miR-7 and miR-21 on the regulation of RECK in oral cancer cells, CAL 27 cells were transfected with either a miR-7-mimic, miR-21-mimic, or both. Although the transfection of miR-7- and miR-21-mimics successfully increased the level of each specific miRNA, the relative fold increase was different for each even though the same concentrations of miRNA-mimics were used (Fig. 6, A and B). RECK mRNA levels were measured 72 h post-transfection. Despite the different degree of increases in miRNA levels upon transfection, miR-21-mediated regulation showed a stronger repression (∼ 30%) effect on RECK mRNA level compared with miR-7-mediated suppression (10–20%, Fig. 6C). Cotransfection of the two miRNA-mimics at the same final concentrations (each 12.5 nm used) suppressed RECK mRNA levels similar to the level shown by miR-21 single transfection (25 nm) (Fig. 6C). The regulation of RECK by miR-7 and miR-21 was not limited to CAL 27 cells. A similar reduction was observed in HeLa cells 48 h post-transfection with the miRNA-mimics (supplemental Fig. S4). RECK protein levels were monitored in HEK293 cells transfected with miRNA mimics. Strong repression (30–40%) of RECK protein was observed when transfected with miR-21-mimic alone, whereas miR-7 alone generated a mild repression (∼10%, Fig. 6D). The data from HEK293 cells were presented because the protein level of RECK was barely detectable in untransfected CAL 27 cells. The direct regulation of RECK by miR-7 and miR-21 was further evaluated by luciferase reporter assays. The wild-type RECK 3′ UTR containing both putative miRNA binding sites were cloned downstream of firefly luciferase reporter. Mutated versions of the RECK 3′ UTR were generated for the putative binding sites for miR-7 or miR-21. Each mutants contained four altered nucleotides on the miRNA seed region binding sequence on RECK (Fig. 6, E and F). Cotransfection of the reporters with miR-7 or miR-21 significantly reduced the firefly luciferase activity. Mutating the miRNA binding sites on RECK abrogated the miRNA-mediated regulation and rescued the luciferase activity. This demonstrates that both miR-7 and miR-21 directly interact with RECK transcript and that the miRNA binding regions are responsible for miRNA-mediated regulation of RECK (Fig. 6, E and F). Together, these data demonstrated that RECK could be directly coregulated by miR-7 and miR-21 and that different endogenous levels of these miRNAs in cells could contribute to the down-regulation of RECK transcript.
FIGURE 6.
Direct regulation of RECK by miR-7 and miR-21. miR-7 (A), miR-21 (B), and RECK mRNA (C) expression levels were measured 72 h post-transfection of miRNA-mimics used at a final concentration of 25 nm. D, under same experimental conditions, cells were harvested and protein levels of RECK were assessed by Western blot assay. Nonspecific (NS) control is the mirVana miRNA mimic negative control (Ambion/Applied Biosystems, Austin, TX). E and F, luciferase reporter analyses of RECK 3′ UTR and miR-7/miR-21. The top panels show the predicted miR-7 and miR-21 binding sites within RECK 3′ UTR and corresponding seed region mutants. Lines (I) between RECK and miRNA complementary nucleotides are the typical Watson-Crick interactions (A–U and G–C, respectively) and colons are the weak non-typical base pair interactions. Four nucleotide mutations were generated on each miRNA seed region binding sequences. A dual luciferase assay was used to determine the direct regulation of RECK by miR-7 and miR-21. Renilla luciferase was used as an internal control to normalize the expression of firefly luciferase activity. All results are expressed as mean ± S.E. from at least three independent experiments. Student's t test was used for the analyses. ns, not significant. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Inverse Correlation of RECK and Keratinization-associated miRNAs Depending on Serum Concentration, Cell Density, and Adherence of CAL 27 Cells
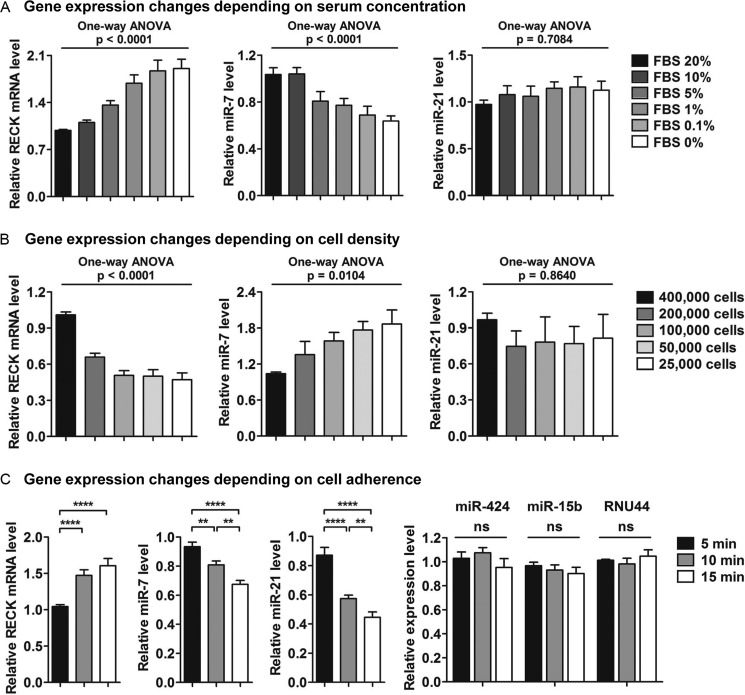
RECK is required at the cellular level for stable cell substrate adhesion, and its expression is affected by external physiological stimuli such as cell density and serum concentration (33, 34). Thus, we investigated the expression of RECK and the keratinization-associated miRNAs in CAL 27 cells seeded in varying conditions of nutrient supply and confluency. Different concentrations of FBS ranging from 20% to 0% inversely affected the expression of both RECK and miR-7 (Fig. 7A). RECK expression gradually increased when serum was increasingly deprived, whereas miR-7 expression decreased. In contrast, miR-21 expression levels remained constant despite different concentrations of FBS, suggesting that RECK was not regulated by miR-21 under these conditions.
FIGURE 7.
The inverse correlation of RECK and keratinization-associated miRNAs is dependent on serum concentration, cell density, and adherence of CAL 27 cells. A, RECK and miR-7 expression levels are inversely correlated in a serum-dependent manner. One-way analysis of variance was used to analyze the increasing or decreasing trends of gene expression. B, the inverse correlation of RECK and miR-7 expression levels are dependent on the density of cells. One-way analysis of variance was used to analyze the increasing or decreasing trends of gene expression. C, a gradual reduction of miR-7 and miR-21 was observed in CAL 27 cells that had stronger cell surface adherent properties. Conversely, RECK mRNA levels increased as cells exhibited more adherent properties. miR-424, miR-15b, and RNU44 are representative negative controls showing that gene level changes were not a common phenomenon during the differential trypsinization process. All results are expressed as mean ± S.E. from at least three independent experiments. Student's t tests were used for the statistical analyses. ns, not significant. **, p < 0.01; ****, p < 0.0001.
Cell density was another factor that inversely affected the expression of RECK and miR-7 (Fig. 7B). CAL 27 cells were plated at different densities by 2-fold serial dilutions ranging from 4 × 105 to 2.5 × 104 cells on a 24-well plate. Moving from high to low cell density, RECK expression decreased, whereas miR-7 levels increased. Again, the expression of miR-21 was unaffected by changes in cell density.
During routine cell passage, it was noted that CAL 27 cells generally had stronger adherence to the culture plate and that normal trypsin treatment conditions used for other cancer cell types (e.g. HeLa) would detach only a fraction of CAL 27 cells. Because RECK is required at the cellular level for stable cell substrate adhesion, an experiment was therefore designed to determine whether the collection of detached cells at different time points, (5, 10, and 15 min, during the trypsinization process) would inversely affect the levels of RECK and its regulatory miRNAs. Interestingly, cells that were easily detached at 5 min had significantly lower expression levels of RECK compared with cells that were more adherent and required longer trypsin treatment times (10 and 15 min) to detach. In contrast, the less adherent cells had significantly higher levels of both miR-7 and miR-21 compared with the more adherent cells (Fig. 7C). The expression levels of miR-424, miR-15b, and RNU44, normalized to U6 snRNA, remained unchanged during the differential trypsinization procedure, indicating that the expression changes observed for RECK and the keratinization-associated miRNAs during this process were specific to miR-7 and miR-21. Therefore, the treatment of CAL 27 cells to different physiological conditions further supported an inverse correlation between RECK and keratinization-associated miRNAs.
DISCUSSION
In this study, we have identified 69 aberrantly expressed miRNAs in OSCCs in comparison to normal tongue tissues. Because miRNA expressions are known to be tissue- and tumor-specific (35), using the appropriate subset of tumors with the corresponding normal controls is therefore important to reduce the potential complexities associated with analyzing heterogeneous tumor tissues. Thus, our miRNA profiling mainly focused on tongue cancer which is the most prevalent type of oral cancer (36, 37). To date, this is the largest miRNA expression profiling study on tongue cancers that takes into consideration multiple clinicopathological parameters that enable the interpretation of aberrant miRNA expression levels in subtypes of this disease.
Understanding the clinical relevance of miRNA expression patterns in OSCCs is a necessary requirement to better classify these heterogeneous tumors and circumvent the therapeutic challenges faced upon their clinical management. Our data have not only shown that the TNM system is a good prognostic parameter, as proposed by Hiratsuka et al. (38), but also illustrated that the keratinization status of OSCCs was closely associated with poor prognosis of patients. Consistent with our findings, several studies have also associated keratinization with carcinogenesis and survival of cancer patients. Keratinization has been associated with the increased incidence of neck metastases and the decreased survival rate of patients with tongue cancer (39). Another study also has demonstrated a significantly poorer survival rate of patients with keratinizing tumors than those with non-keratinizing squamous cell carcinomas, indicating the prognostic importance of keratinization status in tumors of the nasopharynx (40). Altered keratin expression has been observed in the carcinogen-induced hamster cheek pouch carcinogenesis model, which results in the keratinization of squamous cell carcinomas of the oral epithelium, including tongue (39, 41). In agreement with these studies, our findings also indicated that keratinization of tumors was highly associated with poorer survival of patients and that miR-21 could potentially be responsible for this keratinization process.
Our study mainly focused on two miRNAs, miR-7 and miR-21, which were up-regulated in keratinized tumors compared with normal or non-keratinized tumors. Although aberrant expressions of these miRNAs were reported in different types of cancer, this is the first report to identify the link between these miRNAs and keratinization. Overexpression of miR-21 has been associated with poorer survival of patients with tongue squamous cell carcinomas (42). The importance of miR-21 as a poor prognosis indicator, however, is not limited to oral cancer, as similar results have been observed for other types of cancers (30, 32). Recent mechanistic and functional studies focus on the tumor suppressive aspects of miR-7 in cancers (43–46). Although many aberrantly expressed miRNAs have been reported in diverse human cancers, it does not necessarily mean that all of them play a causative role for tumorigenesis. Although the oncogenic miRNAs may induce oncogenesis, some deregulated miRNAs may be the secondary consequence from the loss of normal cellular identity and further contribute to the phenotypic variability of tumors (47). The expression of miR-7 in cancers seems to vary depending on cell or tissue type. For example, miR-7 has been reported to be down-regulated in schwannoma tumors (43) and glioblastoma (48) but up-regulated in breast cancer (49) and lung cancer (50). It is possible that the increase of miR-7 level in tongue tumors examined was due to the consequence or a responsive event during oral carcinogenesis instead of being the cause of cancer development. These observations suggest that although miR-7 act as a tumor suppressive miRNA in normal physiological conditions, the elevation of miR-7 in such environment may contribute to the regulation of a tumor suppressor RECK.
By using in silico analyses, RECK was determined as the best candidate for further investigation among the keratinization-associated miRNA targeted genes because RECK is a key modulator for regulating the extracellular matrix integrity in physiological and pathological states by negatively regulating MMPs (13). In addition to miR-21-mediated regulation of RECK proposed in other studies (51–54), we identified miR-7 as another regulator for RECK and further confirmed that both miRNAs regulate RECK in oral cancer. Although these miRNAs can simultaneously target RECK, our data suggested that different amounts of these miRNAs were required to achieve similar efficacies in target regulation. It is intriguing that RECK has been reported to be down-regulated in tumors of the carcinogen-induced hamster cheek pouch carcinogenesis model (55), which has been known to form keratinized tumors (39, 41). Therefore, the increase in keratinization-associated miR-7 and miR-21 during carcinogenesis could lead to down-regulation of RECK. Together with the deregulated activation of MMPs during keratinization as discussed under “Introduction,” the keratinization-associated miR-7 and miR-21 can help facilitate the aggressiveness of tumors, leading to poor survival.
Our findings show additional evidence that the keratinization-associated miRNAs, miR-7 and miR-21, are inversely correlated with the expression of RECK. CAL 27 orthotopic xenograft tumors are keratinized tumors (26) and so it is a good model to compare the inverse expression patterns between RECK and the keratinization-associated miRNAs. Also, varying the cell density or nutrient supply was a good in vitro system to monitor the inverse correlation between RECK and miRNAs because RECK expression alters upon these changes (33, 34). In both altered conditions, miR-7 appears to act more dynamically to modulate RECK expression in response to environmental changes, and miR-21 is relatively stable in regulating RECK, regardless of the external stimuli. Similar observations were noted on the effects of miR-7 and miR-21 on RECK in CAL 27 or HEK293 cells. Although the regulation of RECK by miR-21 is more likely to be consistent throughout the experiments, miR-7-mediated regulation varies in different assays. For example, miR-7 overexpression mildly changed the endogenous RECK levels, but the repression effects in luciferase assays was stronger than that of miR-21-mediated repression. Western blot analysis and luciferase assays were performed in HEK293 cells to achieve a stronger sensitivity and better transfection efficiency. Undetectable RECK in CAL 27 may, in part, be due to the high expression of these keratinization-associated miRNAs. Together, these data suggest that the increase of keratinization-associated miRNAs posttranscriptionally repress RECK in cancers and that environmental changes may further modulate RECK mRNA levels via changes to specific miRNA levels. In particular, miR-21 may play a more central role in regulating RECK, whereas miR-7 may be more involved in regulating RECK levels on the basis of dynamic changes in tumor microenvironment.
In conclusion, our data imply that the interpretation of miRNA expression patterns can be better resolved when one takes into consideration clinicopathological data of OSCC subtypes. Moreover, our innovative approach of using radar chart analyses provides clearer visualizations of miRNA expression patterns in subtypes of OSCCs. Of significance, our patient survival analyses demonstrated that keratinization and high miR-21 levels were important indicators of oral cancer patient prognosis and that miR-7 and miR-21, two keratinization-associated miRNAs, could contribute to the regulation of the tumor suppressor gene RECK in oral cancers. By understanding the modulation kinetics between keratinization-associated miRNAs and RECK (which is also involved in the keratinization process), the stimuli that affect their expression levels and the mechanisms of how those molecular events are associated with poor prognosis could ultimately lead to improved therapeutics for oral cancer.
Supplementary Material
Acknowledgments
We would like to thank members of the Chan laboratory for their technical assistance, discussion, and encouragement. We are grateful to Tissue Procurement at the H. Lee Moffitt Cancer Center for providing the specimens.
This work was supported in part by a UF/Moffitt Collaborative Initiative grant (to E. K. L. C. and J. Q. C.). This work was also supported by National Institute of Dental and Craniofacial Research Grants K99DE018191 (to A. J.) and T32 DE007200 (to R. P.), by an alumni graduate fellowship (to H. M. J.), and by the Andrew J. Semesco Foundation, Ocala, FL.
The normalized microarray data reported in this paper have been submitted to the Gene Expression Omnibus Repository with accession number GSE28100.

This article contains supplemental Tables S1 and S2.
- OSCC
- oral squamous cell carcinoma
- miRNA
- microRNA
- MMPs
- matrix metalloproteinases
- RECK
- reversion-inducing cysteine-rich protein with kazal motifs
- qRT-PCR
- quantitative PCR
- HPV
- human papillomavirus.
REFERENCES
- 1. Scully C., Felix D. H. (2006) Oral medicine. Update for the dental practitioner oral cancer. Br. Dent. J. 200, 13–17 [DOI] [PubMed] [Google Scholar]
- 2. Jemal A., Tiwari R. C., Murray T., Ghafoor A., Samuels A., Ward E., Feuer E. J., Thun M. J. (2004) Cancer statistics, 2004. CA. Cancer J. Clin. 54, 8–29 [DOI] [PubMed] [Google Scholar]
- 3. Lin W. H., Chen I. H., Wei F. C., Huang J. J., Kang C. J., Hsieh L. L., Wang H. M., Huang S. F. (2011) Clinical significance of preoperative squamous cell carcinoma antigen in oral-cavity squamous cell carcinoma. Laryngoscope 121, 971–977 [DOI] [PubMed] [Google Scholar]
- 4. Méndez E., Houck J. R., Doody D. R., Fan W., Lohavanichbutr P., Rue T. C., Yueh B., Futran N. D., Upton M. P., Farwell D. G., Heagerty P. J., Zhao L. P., Schwartz S. M., Chen C. (2009) A genetic expression profile associated with oral cancer identifies a group of patients at high risk of poor survival. Clin. Cancer Res. 15, 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chung C. H., Parker J. S., Karaca G., Wu J., Funkhouser W. K., Moore D., Butterfoss D., Xiang D., Zanation A., Yin X., Shockley W. W., Weissler M. C., Dressler L. G., Shores C. G., Yarbrough W. G., Perou C. M. (2004) Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell 5, 489–500 [DOI] [PubMed] [Google Scholar]
- 6. Casiglia J., Woo S. B. (2001) A comprehensive review of oral cancer. Gen. Dent. 49, 72–82 [PubMed] [Google Scholar]
- 7. Bartel D. P. (2009) MicroRNAs. Target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garzon R., Marcucci G., Croce C. M. (2010) Targeting microRNAs in cancer. Rationale, strategies and challenges. Nat. Rev. Drug Discov. 9, 775–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bragulla H. H., Homberger D. G. (2009) Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J. Anat. 214, 516–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kobayashi T., Kishimoto J., Ge Y., Jin W., Hudson D. L., Ouahes N., Ehama R., Shinkai H., Burgeson R. E. (2001) A novel mechanism of matrix metalloproteinase-9 gene expression implies a role for keratinization. EMBO Rep. 2, 604–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coussens L. M., Fingleton B., Matrisian L. M. (2002) Matrix metalloproteinase inhibitors and cancer. Trials and tribulations. Science 295, 2387–2392 [DOI] [PubMed] [Google Scholar]
- 13. Oh J., Takahashi R., Kondo S., Mizoguchi A., Adachi E., Sasahara R. M., Nishimura S., Imamura Y., Kitayama H., Alexander D. B., Ide C., Horan T. P., Arakawa T., Yoshida H., Nishikawa S., Itoh Y., Seiki M., Itohara S., Takahashi C., Noda M. (2001) The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell 107, 789–800 [DOI] [PubMed] [Google Scholar]
- 14. Takahashi C., Sheng Z., Horan T. P., Kitayama H., Maki M., Hitomi K., Kitaura Y., Takai S., Sasahara R. M., Horimoto A., Ikawa Y., Ratzkin B. J., Arakawa T., Noda M. (1998) Regulation of matrix metalloproteinase-9 and inhibition of tumor invasion by the membrane-anchored glycoprotein RECK. Proc. Natl. Acad. Sci. U.S.A. 95, 13221–13226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Furumoto K., Arii S., Mori A., Furuyama H., Gorrin Rivas M. J., Nakao T., Isobe N., Murata T., Takahashi C., Noda M., Imamura M. (2001) RECK gene expression in hepatocellular carcinoma: correlation with invasion-related clinicopathological factors and its clinical significance. Reverse-inducin, g-cysteine-rich protein with Kazal motifs. Hepatology 33, 189–195 [DOI] [PubMed] [Google Scholar]
- 16. Namwat N., Puetkasichonpasutha J., Loilome W., Yongvanit P., Techasen A., Puapairoj A., Sripa B., Tassaneeyakul W., Khuntikeo N., Wongkham S. (2011) Down-regulation of reversion-inducing, cysteine-rich protein with Kazal motifs (RECK) is associated with enhanced expression of matrix metalloproteinases and cholangiocarcinoma metastases. J. Gastroenterol. 46, 664–675 [DOI] [PubMed] [Google Scholar]
- 17. Song S. Y., Son H. J., Nam E., Rhee J. C., Park C. (2006) Expression of reversion-inducing, cysteine-rich protein with Kazal motifs (RECK) as a prognostic indicator in gastric cancer. Eur. J. Cancer 42, 101–108 [DOI] [PubMed] [Google Scholar]
- 18. Takeuchi T., Hisanaga M., Nagao M., Ikeda N., Fujii H., Koyama F., Mukogawa T., Matsumoto H., Kondo S., Takahashi C., Noda M., Nakajima Y. (2004) The membrane-anchored matrix metalloproteinase (MMP) regulator RECK in combination with MMP-9 serves as an informative prognostic indicator for colorectal cancer. Clin. Cancer Res. 10, 5572–5579 [DOI] [PubMed] [Google Scholar]
- 19. Masui T., Doi R., Koshiba T., Fujimoto K., Tsuji S., Nakajima S., Koizumi M., Toyoda E., Tulachan S., Ito D., Kami K., Mori T., Wada M., Noda M., Imamura M. (2003) RECK expression in pancreatic cancer. Its correlation with lower invasiveness and better prognosis. Clin. Cancer Res. 9, 1779–1784 [PubMed] [Google Scholar]
- 20. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saldanha A. J. (2004) Java Treeview. Extensible visualization of microarray data. Bioinformatics 20, 3246–3248 [DOI] [PubMed] [Google Scholar]
- 22. Cattani P., Siddu A., D'Onghia S., Marchetti S., Santangelo R., Vellone V. G., Zannoni G. F., Fadda G. (2009) RNA (E6 and E7) assays versus DNA (E6 and E7) assays for risk evaluation for women infected with human papillomavirus. J. Clin. Microbiol. 47, 2136–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cattani P., Zannoni G. F., Ricci C., D'Onghia S., Trivellizzi I. N., Di Franco A., Vellone V. G., Durante M., Fadda G., Scambia G., Capelli G., De Vincenzo R. (2009) Clinical performance of human papillomavirus E6 and E7 mRNA testing for high-grade lesions of the cervix. J. Clin. Microbiol. 47, 3895–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lian S. L., Li S., Abadal G. X., Pauley B. A., Fritzler M. J., Chan E. K. (2009) The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA 15, 804–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li S., Lian S. L., Moser J. J., Fritzler M. L., Fritzler M. J., Satoh M., Chan E. K. (2008) Identification of GW182 and its novel isoform TNGW1 as translational repressors in Ago2-mediated silencing. J. Cell Sci. 121, 4134–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jakymiw A., Patel R. S., Deming N., Bhattacharyya I., Shah P., Lamont R. J., Stewart C. M., Cohen D. M., Chan E. K. (2010) Overexpression of dicer as a result of reduced let-7 MicroRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer 49, 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 [Google Scholar]
- 28. Chen C., Zhang Y., Zhang L., Weakley S. M., Yao Q. (2011) MicroRNA-196. Critical roles and clinical applications in development and cancer. J. Cell Mol. Med. 15, 14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R. M., Okamoto A., Yokota J., Tanaka T., Calin G. A., Liu C. G., Croce C. M., Harris C. C. (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198 [DOI] [PubMed] [Google Scholar]
- 30. Gao W., Shen H., Liu L., Xu J., Xu J., Shu Y. (2011) MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J. Cancer Res. Clin. Oncol. 137, 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chan S. H., Wu C. W., Li A. F., Chi C. W., Lin W. C. (2008) miR-21 microRNA expression in human gastric carcinomas and its clinical association. Anticancer Res. 28, 907–911 [PubMed] [Google Scholar]
- 32. Yan L. X., Huang X. F., Shao Q., Huang M. Y., Deng L., Wu Q. L., Zeng Y. X., Shao J. Y. (2008) MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14, 2348–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hatta M., Matsuzaki T., Morioka Y., Yoshida Y., Noda M. (2009) Density- and serum-dependent regulation of the RECK tumor suppressor in mouse embryo fibroblasts. Cell Signal. 21, 1885–1893 [DOI] [PubMed] [Google Scholar]
- 34. Morioka Y., Monypenny J., Matsuzaki T., Shi S., Alexander D. B., Kitayama H., Noda M. (2009) The membrane-anchored metalloproteinase regulator RECK stabilizes focal adhesions and anterior-posterior polarity in fibroblasts. Oncogene 28, 1454–1464 [DOI] [PubMed] [Google Scholar]
- 35. Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., Golub T. R. (2005) MicroRNA expression profiles classify human cancers. Nature 435, 834–838 [DOI] [PubMed] [Google Scholar]
- 36. Schantz S. P., Yu G. P. (2002) Head and neck cancer incidence trends in young Americans, 1973–1997, with a special analysis for tongue cancer. Arch. Otolaryngol. Head Neck Surg. 128, 268–274 [DOI] [PubMed] [Google Scholar]
- 37. Shiboski C. H., Shiboski S. C., Silverman S., Jr. (2000) Trends in oral cancer rates in the United States, 1973–1996. Community Dent. Oral Epidemiol. 28, 249–256 [DOI] [PubMed] [Google Scholar]
- 38. Hiratsuka H., Miyakawa A., Nakamori K., Kido Y., Sunakawa H., Kohama G. (1997) Multivariate analysis of occult lymph node metastasis as a prognostic indicator for patients with squamous cell carcinoma of the oral cavity. Cancer 80, 351–356 [PubMed] [Google Scholar]
- 39. Take Y., Umeda M., Teranobu O., Shimada K. (1999) Lymph node metastases in hamster tongue cancer induced with 9,10-dimethyl-1,2-benzanthracene. Association between histological findings and the incidence of neck metastases, and the clinical implications for patients with tongue cancer. Br. J. Oral Maxillofac. Surg. 37, 29–36 [DOI] [PubMed] [Google Scholar]
- 40. Reddy S. P., Raslan W. F., Gooneratne S., Kathuria S., Marks J. E. (1995) Prognostic significance of keratinization in nasopharyngeal carcinoma. Am. J. Otolaryngol. 16, 103–108 [DOI] [PubMed] [Google Scholar]
- 41. Gimenez-Conti I. B., Shin D. M., Bianchi A. B., Roop D. R., Hong W. K., Conti C. J., Slaga T. J. (1990) Changes in keratin expression during 7,12-dimethylbenz[a]anthracene-induced hamster cheek pouch carcinogenesis. Cancer Res. 50, 4441–4445 [PubMed] [Google Scholar]
- 42. Li J., Huang H., Sun L., Yang M., Pan C., Chen W., Wu D., Lin Z., Zeng C., Yao Y., Zhang P., Song E. (2009) MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin. Cancer Res. 15, 3998–4008 [DOI] [PubMed] [Google Scholar]
- 43. Saydam O., Senol O., Würdinger T., Mizrak A., Ozdener G. B., Stemmer-Rachamimov A. O., Yi M., Stephens R. M., Krichevsky A. M., Saydam N., Brenner G. J., Breakefield X. O. (2011) miRNA-7 attenuation in Schwannoma tumors stimulates growth by up-regulating three oncogenic signaling pathways. Cancer Res. 71, 852–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao X., Dou W., He L., Liang S., Tie J., Liu C., Li T., Lu Y., Mo P., Shi Y., Wu K., Nie Y., Fan D. (2012) MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene, in press [DOI] [PubMed] [Google Scholar]
- 45. Webster R. J., Giles K. M., Price K. J., Zhang P. M., Mattick J. S., Leedman P. J. (2009) Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J. Biol. Chem. 284, 5731–5741 [DOI] [PubMed] [Google Scholar]
- 46. Reddy S. D., Ohshiro K., Rayala S. K., Kumar R. (2008) MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 68, 8195–8200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kent O. A., Mendell J. T. (2006) A small piece in the cancer puzzle. MicroRNAs as tumor suppressors and oncogenes. Oncogene 25, 6188–6196 [DOI] [PubMed] [Google Scholar]
- 48. Kefas B., Godlewski J., Comeau L., Li Y., Abounader R., Hawkinson M., Lee J., Fine H., Chiocca E. A., Lawler S., Purow B. (2008) microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 68, 3566–3572 [DOI] [PubMed] [Google Scholar]
- 49. Foekens J. A., Sieuwerts A. M., Smid M., Look M. P., de Weerd V., Boersma A. W., Klijn J. G., Wiemer E. A., Martens J. W. (2008) Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc. Natl. Acad. Sci. U.S.A. 105, 13021–13026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chou Y. T., Lin H. H., Lien Y. C., Wang Y. H., Hong C. F., Kao Y. R., Lin S. C., Chang Y. C., Lin S. Y., Chen S. J., Chen H. C., Yeh S. D., Wu C. W. (2010) EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 70, 8822–8831 [DOI] [PubMed] [Google Scholar]
- 51. Hu S. J., Ren G., Liu J. L., Zhao Z. A., Yu Y. S., Su R. W., Ma X. H., Ni H., Lei W., Yang Z. M. (2008) MicroRNA expression and regulation in mouse uterus during embryo implantation. J. Biol. Chem. 283, 23473–23484 [DOI] [PubMed] [Google Scholar]
- 52. Gabriely G., Wurdinger T., Kesari S., Esau C. C., Burchard J., Linsley P. S., Krichevsky A. M. (2008) MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell Biol. 28, 5369–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Loayza-Puch F., Yoshida Y., Matsuzaki T., Takahashi C., Kitayama H., Noda M. (2010) Hypoxia and RAS-signaling pathways converge on, and cooperatively down-regulate, the RECK tumor-suppressor protein through microRNAs. Oncogene 29, 2638–2648 [DOI] [PubMed] [Google Scholar]
- 54. Zhang Z., Li Z., Gao C., Chen P., Chen J., Liu W., Xiao S., Lu H. (2008) miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab. Invest. 88, 1358–1366 [DOI] [PubMed] [Google Scholar]
- 55. Nagini S., Letchoumy P. V., A T., Cr R. (2009) Of humans and hamsters. A comparative evaluation of carcinogen activation, DNA damage, cell proliferation, apoptosis, invasion, and angiogenesis in oral cancer patients and hamster buccal pouch carcinomas. Oral Oncol. 45, e31–37 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.