Background: Yca1 is a metacaspase and plays an important role in cell death of the yeast Saccharomyces cerevisiae.
Results: Crystal structure of Yca1 reveals a monomeric architecture that differs significantly from other canonical caspases.
Conclusion: Both catalytic mechanism and function of Yca1 may be distinct from canonical caspases.
Significance: This structure offers insights into metacaspase function.
Keywords: Apoptosis, Caspase, Cysteine Protease, Signal Transduction, Structural Biology, X-ray Crystallography, metacaspase
Abstract
Yca1, the only metacaspase in Saccharomyces cerevisiae, is thought to be a clan CD cysteine protease that includes the caspase subfamily. Although yeast is a single cell eukaryote, it can undergo a cell death process reminiscent of apoptosis. Yca1 has been reported to play an important role in the regulation of such apoptotic process. However, the structure and functional mechanism of Yca1 remain largely enigmatic. In this study, we report the crystal structure of the Yca1 metacaspase at 1.7 Å resolution, confirming a caspase-like fold. In sharp contrast to canonical caspases, however, Yca1 exists as a monomer both in solution and in the crystals. Canonical caspase contains six β-strands, with strand β6 pairing up with β6 of another caspase molecule to form a homodimerization interface. In Yca1, an extra pair of antiparallel β-strands forms a continuous β-sheet with the six caspase-common β-strands, blocking potential dimerization. Yca1 was reported to undergo autocatalytic processing in yeast; overexpression in bacteria also led to autoprocessing of Yca1 into two fragments. Unexpectedly, we found that both the autocatalytic processing and the proteolytic activity of Yca1 are greatly facilitated by the presence of calcium (Ca2+), but not other divalent cations. Our structural and biochemical characterization identifies Yca1 as a Ca2+-activated cysteine protease that may cleave specific substrates during stress response in yeast.
Introduction
Yeast apoptosis was first reported in a Saccharomyces cerevisiae Δcdc48 mutant strain in 1997 (1). Since then, a number of apoptosis-related genes have been discovered and characterized in yeast, including the caspase-like protease Yca1 (2), the inhibitor of apoptosis (IAP)2 Bir1 (3, 4), the apoptosis-inducing factor Aif1 (5, 6), the BH3-containing Bcl-2 family homologue YBP (7), and others (8). To date, Yca1, also known as Mca1 (2) and Yor197w (9), remains the only identified metacaspase in S. cerevisiae. Yca1 is a clan CD cysteine protease (10), which has a protein fold similar to that of the canonical caspases. Specifically, Yca1 is classified as a type I metacaspase (11), with a characteristic N-terminal prodomain that is thought to be cleaved off upon activation. Although yeast is a single cell eukaryote, it is widely believed to undergo programmed cell death, or apoptosis, in response to detrimental environmental cues (6, 12–15). Consequently, early skepticism on yeast apoptosis has now been replaced by systematic investigation on the genetic and biochemical pathways that control the onset of apoptosis in yeast.
Yca1 appears to be a positive regulator of apoptosis upon stress induction by hydrogen peroxide, acetic acid, and toxin (6, 12–15). Nuclear localization of poly(Q) expansions in S. cerevisiae was abolished inΔyca1-null mutant strain, blocking apoptosis (16). Despite these advances, how Yca1 facilitates apoptosis remains largely unknown. Yca1 is thought to be a functional protease (17); however, its substrate proteins are yet to be identified. It is also unclear what triggers the protease activity of Yca1 and whether it can be inhibited by other cellular factors. Yca1 has also been reported to play a role in apoptosis-unrelated cellular processes. For instance, inactivation of Yca1 was shown to result in longer G1/S transition and down-regulation of Yca1 bypassed the G2/M checkpoint upon nocodazole treatment (18). In addition, the N-terminal poly(Q/N) prodomain of Yca1 was found to be critical for clearance of insoluble protein aggregates (19).
The N-terminal prodomain of Yca1, containing Asn/Gln-rich sequences, was reported to exhibit a biochemical property of self-aggregation (20). The catalytic His-Cys dyad required for proteolytic activity is predicted to be highly conserved among metacaspases and canonical caspases in metazoan. Supporting this notion, Yca1 was reported to undergo autoproteolytic processing to yield a large subunit and a C-terminal small subunit of ∼12 kDa (9). In sharp contrast to canonical caspases, but similar to other metacaspases, Yca1 appears to cleave artificial peptide substrates after positively charged amino acids Arg and Lys (21). Nevertheless, no physiologically relevant substrate of Yca1 has been identified. Using bacterial cell lysate that contained overexpressed Yca1 protein, Silva et al. (22) identified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a putative Yca1 substrate. Yca1 does not appear to directly interact with Bir1 or the yeast Omi/HtrA homologue Nma111 (4).
In this study, we report the crystal structure of the full-length yeast metacaspase Yca1 at 1.7 Å resolution. The overall structure is quite similar to canonical caspases. However, unlike canonical caspases, Yca1 exists as a monomer both in solution and in the crystals. Structural analysis provides a satisfying explanation to this observation. We serendipitously found that the autoproteolytic processing of Yca1 is greatly facilitated by the presence of Ca2+. We reconstituted an in vitro proteolysis assay using the Bir1p protein as an artificial substrate and showed that the proteolytic activity of Yca1 is also markedly stimulated by Ca2+. These biochemical observations, together with structural analysis, provide a framework for deciphering the roles of Yca1 in yeast apoptosis and other cellular processes.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The full-length, wild-type Yca1 (GenBankTM ID: 854372) and substrate Bir1 fragments (GenBank ID: 853551) were subcloned from S. cerevisiae S288C genomic DNA into pET15b and pET21b vectors (Novagen), respectively, using standard PCR-based protocols. Identities of individual clones were verified by double-strand plasmid sequencing. Mutagenesis of Yca1 was performed using the two-step PCR method and verified by plasmid sequencing. All proteins were overexpressed in Escherichia coli BL21 (DE3) at 22 or 30 °C. The soluble fraction of bacterial lysate was purified by nickel-nitrilotriacetic acid affinity chromatography (Qiagen) followed by ion exchange chromatography (Source-15Q or Source-15S, GE Healthcare) and gel filtration (Superdex-200, GE Healthcare). Protein concentrations were determined by UV spectroscopic measurement at 280 nm.
Crystallization, Data Collection, and Structure Determination
The full-length, wild-type Yca1 was concentrated to ∼7 mg/ml after gel filtration. Limited proteolysis was performed before crystals were grown at 18 °C using the hanging-drop vapor diffusion method. Diamond-shaped wild-type Yca1 protein crystals appeared after 30 days in buffers containing 0.8 m potassium phosphate monobasic and 0.8 m sodium phosphate monobasic. Crystals were harvested and flash-frozen in a well buffer containing 20% glycerol.
Diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) and integrated and scaled with the HKL-2000 package (23). The structures were determined by molecular replacement using PHASER (24) with the atomic coordinates of GSU0716 (Protein Data Bank (PDB) code 3BIJ (www.pdb.org)) as the initial search model. Model building and structure refinement were performed using COOT (25) and Phenix (26), respectively.
Enzymatic Assays
The full-length, wild-type Yca1 or mutant C276A was incubated with or without substrate at 18 °C overnight or otherwise indicated in a buffer containing 20 mm Tris (pH 8.0) and 150 mm NaCl in the presence or absence of CaCl2. Proteins were resolved on 16% SDS-PAGE gel and stained by Coomassie Blue G250.
RESULTS
Structure of Yca1
To decipher the function and mechanism of Yca1, we overexpressed the full-length (residues 1–432), wild-type (WT) Yca1 in E. coli and biochemically purified the protein to homogeneity. The elution volume of Yca1 corresponds to an apparent molecular mass of ∼35 kDa (supplemental Fig. 1A), suggesting that Yca1 may exist in solution as a monomer. Confirming a previous study (9), the recombinant Yca1 protein contains a large subunit and a small subunit (supplemental Fig. 1A). Presumably, Yca1 had been autocatalytically processed. To determine the site(s) of autoprocessing, we subjected the small subunit to N-terminal peptide sequencing analysis, which yielded two sequences, NH2-Thr-Val-Lys-Gly-Gly-COOH and NH2-Gly-Gly-Met-Gly-Asn-COOH. This analysis suggests that autoprocessing of Yca1 may occur at the carboxyl end of Lys331 and Lys334 (supplemental Fig. 1B).
The N-terminal sequences of Yca1 are quite hydrophilic and presumably flexible, with 22 Gln, 12 Asn, 11 Gly, and 16 Pro residues in the N-terminal 100 amino acids. Prior to crystallization, the two-subunit Yca1 was treated with a trace amount of V8 protease, which readily removed the N-terminal 83 amino acids in Yca1. The resulting Yca1 protein was crystallized in the space group H3, with one molecule of Yca1 in each asymmetric unit. Sequence analysis indicated sequence homology between Yca1 and an uncharacterized protein GSU0716 from Geobacter sulfurreducens for which an atomic structure is available (PDB code 3BIJ (www.pdb.org)). The structure was hence determined by molecular replacement and refined at 1.68 Å resolution (Table 1).
TABLE 1.
Data collection and refinement statistics
One crystal was used for this structure. Values in parentheses are for the highest resolution shell.
| Data collection | |
| Source | SSRF BL17U |
| Wavelength (Å) | 0.99582 |
| Space group | H3 |
| cell dimensions | |
| a, b, c (Å) | 114.60, 114.60, 62.11 |
| α, β, γ (°) | 90.00, 90.00, 120.00 |
| Resolution (Å) | 50–1.68 (1.74–1.68) |
| Mosaicity | 0.19–0.46 |
| Rmergea (%) | 5.5 (26.8) |
| I/σI | 32.9 (8.2) |
| Completeness (%) | 99.2 (93.6) |
| Redundancy | 5.5 (5.6) |
| Refinement | |
| Resolution (Å) | 33–1.68 |
| No. of reflections | 34,430 |
| Rwork/Rfreeb (%) | 17.04/18.72 |
| No. of atoms | 2414 |
| Protein | 2131 |
| Ligand/ion | 15 |
| Water | 268 |
| B-factors | 23.52 |
| Protein | 22.68 |
| Ligand/ion | 13.83 |
| Water | 30.70 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.197 |
| Ramachandran plot statisticsc (%) | |
| Most favored | 91.1 |
| Additional allowed | 8.9 |
| Generously allowed | 0.0 |
| Disallowed | 0.0 |
a Rmerge = ΣhΣi|Ih,i − Ih|/ΣhΣiIh,i, where Ih is the mean intensity of the i observations of symmetry related reflections of h.
b R = Σ|Fobs − Fcalc|/ΣFobs, where Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 5% of the reflections selected randomly).
c Ramachandran plot was performed by Procheck.
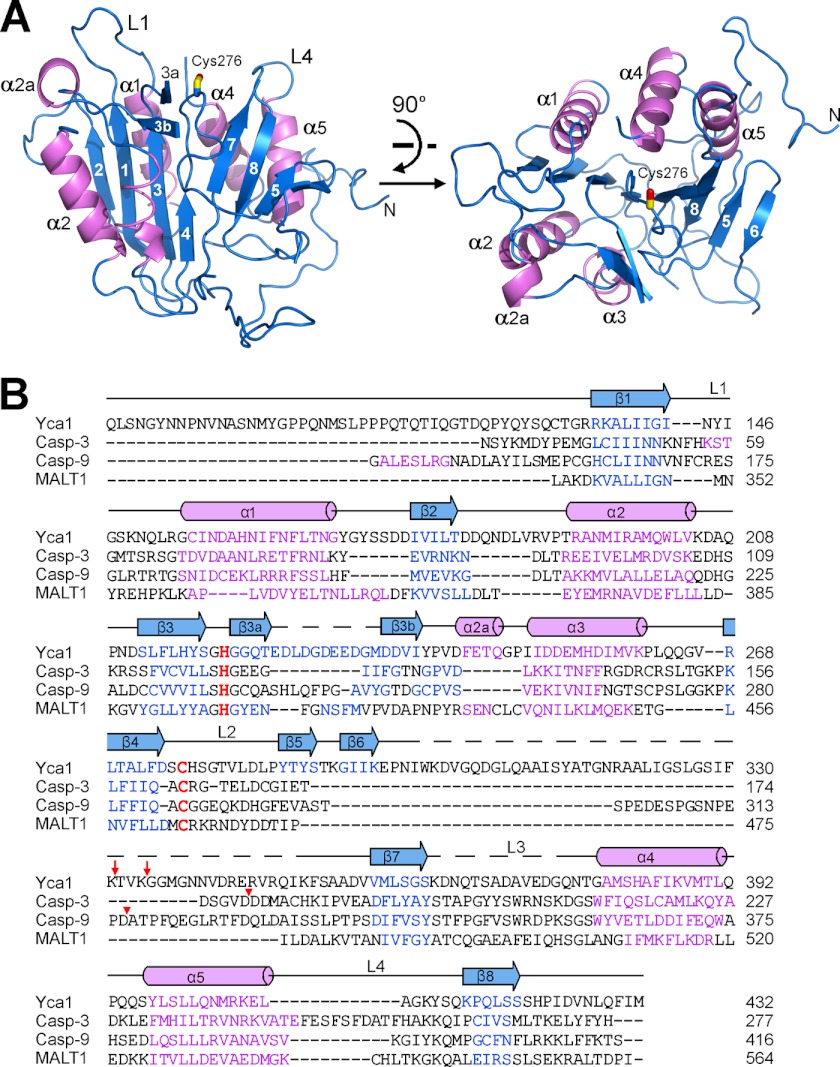
The structure of Yca1 comprises a centrally located eight-stranded β-sheet, with three α-helices (α1, α4, and α5) on one side and two (α2 and α3) on the other side (Fig. 1). A β-hairpin, comprising β3a and β3b, and a short α-helix α2a are located between strand β3 and helix α3, close to the active site of Yca1 (Fig. 1A). By convention of the caspase active site definition (27), Yca1 contains three well ordered loops L1, L2, and L4. L1 and L4 constitute two opposing sides of a putative substrate-binding groove. The putative catalytic residue Cys276 is located on the L2 loop, spatially between L1 and L4. The L3 loop, which serves as the base of the substrate-binding groove, is disordered in the structure.
FIGURE 1.
Structure of Yca1. A, graphic representation of the Yca1 structure in two perpendicular views. The core structure contains five α-helices (colored magenta) and eight β-strands (blue), which form a central β-sheet sandwiched by the α-helices. Secondary structural elements, active site loops L1 and L4, and the catalytic residue Cys276 are labeled. B, sequence alignment among Yca1, canonical caspases (caspase-3 and caspase-9), and the paracaspase MALT1 (30). Color-coded secondary structural elements for Yca1 are indicated above the sequences. The putative catalytic dyad residues Cys-His are colored red. Sites of activation cleavage in caspases-3 and -9 are indicated by red triangles. Sites of autoprocessing in Yca1 are marked by red arrows. Figs. 1A, 2, and 4A were prepared using PyMOL (36).
Structural Comparison with Caspases
The overall structure of Yca1 conforms to the caspase fold. Specifically, the contiguous, six-stranded β-sheet (comprising β1–β4, β7, and β8) and the helices α1–α5 represent exactly the core caspase fold that is present in every caspase structure (27). However, there are important structural differences between Yca1 and caspases. The most prominent difference is the presence of two extra β-strands, β5 and β6, in Yca1, but not in canonical caspases (Fig. 1B). Strands β5 and β6 in Yca1 occupy the same general spatial location as that of the adjacent caspase molecule in the case of a caspase homodimer (supplemental Fig. 2). This analysis nicely explains why Yca1 cannot form a homodimer similar to that of canonical caspases.
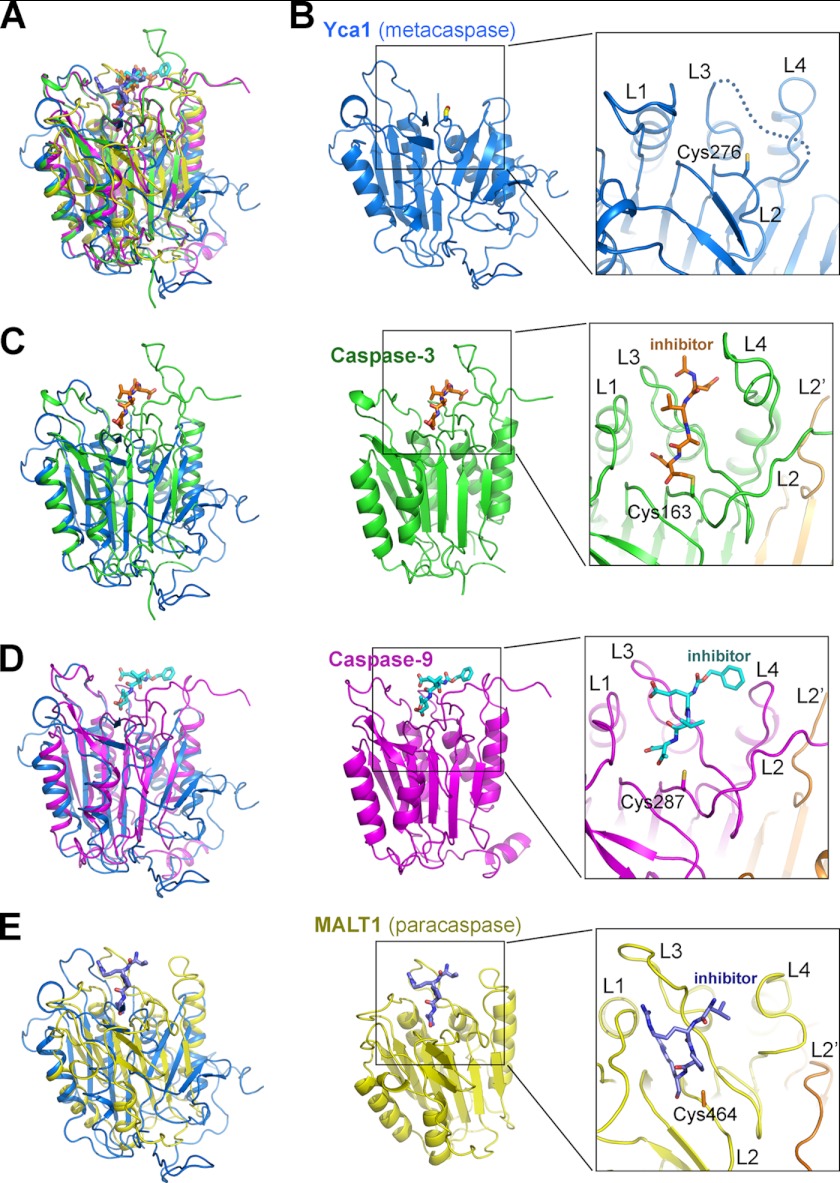
Yca1 can be superimposed reasonably well with representative caspases, such as caspase-3 (28) and caspase-9 (29), and the paracaspase MALT1 (30) (Fig. 2A). The active site is bound by a covalent inhibitor in the vast majority of caspase structures; in Yca1, however, the active site remains unoccupied (Fig. 2B). Notably, the sequence homology between Yca1 and other caspases is quite low, with 10.2%/11.4% identity between caspase-3/caspase-9 and Yca1 and 10.1% between Yca1 and MALT1. Due to these large extents of sequence divergence, there are important structural variations between Yca1 and other caspases. One important measurement of structural variation is root mean square deviation (r.m.s.d.). The r.m.s.d. is 7.45 Å between Yca1 and caspase-3 over only 62 aligned Cα atoms (Fig. 2C, left panel), whereas the value is 7.61 Å between Yca1 and caspase-9 for 127 Cα atoms (Fig. 2D, left panel). The structural variation between Yca1 and the paracaspase MALT1 is even greater, with an r.m.s.d. of 10.5 Å over 58 aligned Cα atoms (Fig. 2E, left panel). These structural differences are considerably larger than those between different classes of canonical caspases. For example, human caspase-3 can be superimposed with caspase-9 with an r.m.s.d. of 0.764 Å over 200 aligned Cα atoms.
FIGURE 2.
Structural comparison of Yca1 with caspases and the paracaspase MALT1. A, structural overlay of Yca1 (blue), caspase-3 (green), caspase-9 (magenta), and MALT1 (yellow). B, a close-up view of the active site of Yca1. Among the four active site loops, L1, L2, and L4 exhibit well defined conformation. A large portion of the L3 loop (dotted line), however, is disordered, likely due to its flexible nature in the absence of substrate binding. C, comparison of Yca1 (PDB code 4F6O) with caspase-3 (PDB code 1CP3 (28)). A close-up view on the caspase-3 active site is shown. The covalently bound inhibitor acetyl-Asp-Val-Ala-Asp-fluoromethyl ketone (Ac-DVAD-fmk) is highlighted in orange. D, comparison of Yca1 with caspase-9 (PDB code 1JXQ (29)). A close-up view on the caspase-9 active site is shown. The covalently bound inhibitor benzyloxycarbonyl-Glu-Val-Asp-dichlorobenzylmethyl ketone (z-EVD-dcbmk) is highlighted in cyan. E, comparison of Yca1 with the paracaspase MALT1 (PDB code 3UOA (30)). A close-up view on the MALT1 active site is shown. The covalently bound inhibitor benzyloxycarbonyl-Val-Arg-Pro-Arg (z-VRPR-fmk) is highlighted in purple.
When compared with canonical caspases, Yca1 also exhibits a distinct conformation at the active site. Most notably, the L2 loop of Yca1, which harbors the catalytic residue Cys276, turns inward, becoming part of the core structure (Fig. 2B); L2 is followed by an extended sequence and a pair of β-strands (Fig. 1). By sharp contrast, the L2 loop of caspase-3 or caspase-9 extends outward, away from the core structure (Fig. 2, C and D, right panels; supplemental Fig. 3). Consequently, the L2 loop in caspase-3 or -9 is involved in the formation of the loop-bundle interactions, which play an essential role for sustaining the robust proteolytic activity of caspase-3 or -9 (27, 31, 32). The inward-turning feature of the L2 loop in Yca1 is shared by the paracaspase MALT1 (Fig. 2E, right panel; supplemental Fig. 3). However, unlike MALT1, which forms a stable homodimer in the presence of peptide inhibitor (30), Yca1 is unable to form the same type of homodimer due to the presence of two extra β-strands (supplemental Fig. 2).
Calcium-dependent Autoprocessing and Activity
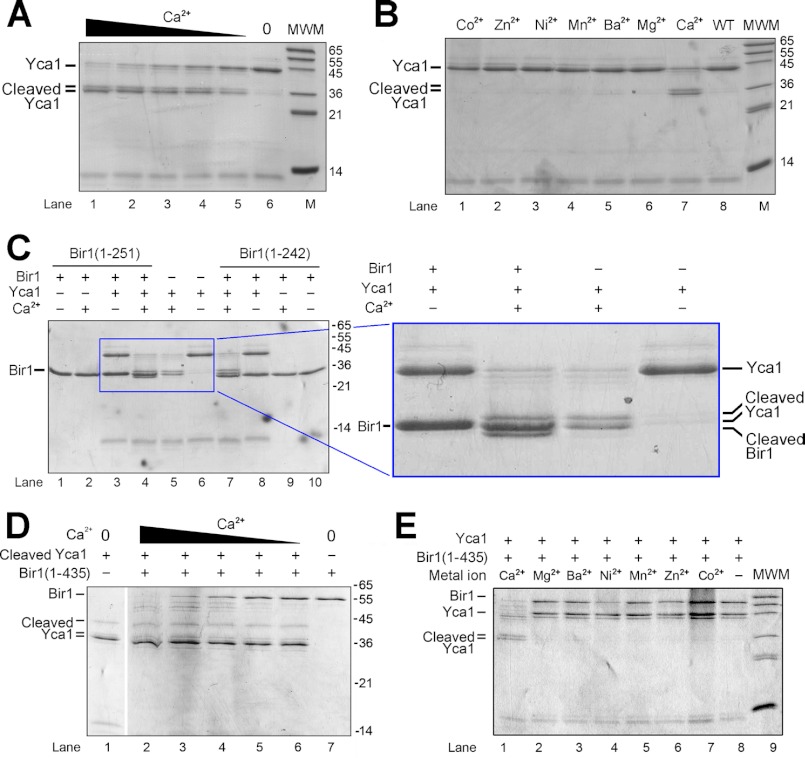
A recent study showed that the Arabidopsis metacaspase AtMCP2d strictly requires Ca2+ for its proteolytic activity in vitro (33), suggesting a critical role for Ca2+ in the catalytic mechanism. Recombinant WT Yca1 is already autocatalytically processed (supplemental Fig. 1). We serendipitously discovered that upon incubation with Ca2+, the large subunit of Yca1 was further processed into two smaller, 36-kDa fragments (Fig. 3A). N-terminal peptide sequencing of these two fragments revealed that the autoprocessing occurred after Arg72 and Lys86 (data not shown). Supporting this conclusion, single mutation of Arg72 or Lys86 reduced the doublet to a single band, whereas simultaneous mutation of Arg72 and Lys86 led to complete absence of the doublet (supplemental Fig. 4). The conversion of the Yca1 large subunit to the two smaller fragments becomes increasingly more complete at higher concentrations of Ca2+ (Fig. 3A). To examine whether other divalent cations may have a similar effect, we tested six additional metal ions: Mg2+, Zn2+, Mn2+, Ni2+, Ba2+, and Co2+. The results unambiguously showed that only Ca2+ specifically enhanced the autocatalytic processing of Yca1 (Fig. 3B).
FIGURE 3.
Calcium greatly facilitates the autocatalytic processing and the proteolytic activity of Yca1. A, calcium facilitates the autocatalytic processing of Yca1 in a concentration-dependent manner. Shown here is an SDS-PAGE gel stained by Coomassie Blue. The Ca2+ concentrations in lanes 1–5 are 25, 12.5, 6.25, 3.13, and 1.57 mm, respectively. MWM indicates molecular mass markers. B, only Ca2+, but not other divalent metal ions, facilitated autoprocessing of Yca1. The divalent metal ions tested in lanes 1–6 are Co2+, Zn2+, Ni2+, Mn2+, Ba2+, and Mg2+, respectively. The final concentration of the metal ions was uniformly 10 mm in the reaction. C, Yca1 cleaves the substrate protein Bir1p (residues 1–251) in a Ca2+-dependent manner. The substrate protein Bir1p was cleaved only in the presence of Ca2+ (lane 4), producing a slightly smaller fragment. D, calcium facilitates the proteolytic activity of autoprocessed Yca1 in a concentration-dependent manner. The Ca2+ concentrations in lanes 2–6 are 25, 12.5, 6.25, 3.13, and 1.57 mm, respectively. The Bir1p fragment contains residues 1–435. E, only Ca2+, but not other divalent metal ions, facilitated proteolytic activity of Yca1. The divalent metal ions tested in lanes 4–9 are Mg2+, Ba2+, Ni2+, Mn2+, Zn2+, and Co2+, respectively. The final concentration of the metal ions was uniformly 10 mm in the reaction.
In fruit flies, cleavage of the inhibitor of apoptosis protein DIAP1 by activated caspases constitutes an important regulatory mechanism for cell death (34). Because Bir1p contains two baculovirus IAP repeat (BIR) domains and might be an IAP homolog, we examined whether Bir1p could be a substrate of the Yca1 metacaspase. In the absence of Yca1 or Ca2+, the Bir1p fragment 1–251 remained intact (Fig. 3C, lanes 1–3). In the presence of 10–50 mm Ca2+, Bir1p (residues 1–251) was cleaved into a fragment of slightly smaller molecular mass (Fig. 3C, lane 4). The extra fragment is different from the doublet of 36 kDa derived from the Yca1 large subunit (Fig. 3C, right panel). The C-terminal sequences of Bir1p (1–251) 243RNRFERIKN251 contain four Arg and Lys residues, which represent potential cleavage sites for Yca1. In particular, Ile249–Lys250 of Bir1p is similar to the Yca1 sequences Phe330–Lys331 and Val333–Lys334, with a hydrophobic residue preceding Lys. This analysis suggests that the cleavage site of Bir1p is likely to be at its C terminus. Supporting this prediction, Bir1p (residues 1–242) was no longer cleaved by Yca1 (Fig. 3C, lanes 7–10).
These results strongly suggest that calcium may be required not only for the autocatalytic processing of Yca1 (Fig. 3A), but also for the maintenance of its catalytic activity. To examine the latter scenario, we obtained the fully autoprocessed Yca1 by incubating Yca1 with 10 mm Ca2+ for 12 h. Then we removed Ca2+ and examined the catalytic activity of the autoprocessed, Ca2+-free Yca1. Supporting our conclusion, cleavage of the Bir1p fragment (residues 1–435) by the autoprocessed Yca1 was greatly accelerated by increasing concentrations of Ca2+ (Fig. 3D, lanes 2–6). When compared with the fragment 1–251, the larger Bir1p fragment 1–435 allows better separation of the cleavage products from the uncleaved fragment. In addition, only Ca2+, but not any other divalent cation tested, was able to enhance substrate cleavage by Yca1 (Fig. 3E).
Catalytic Residues
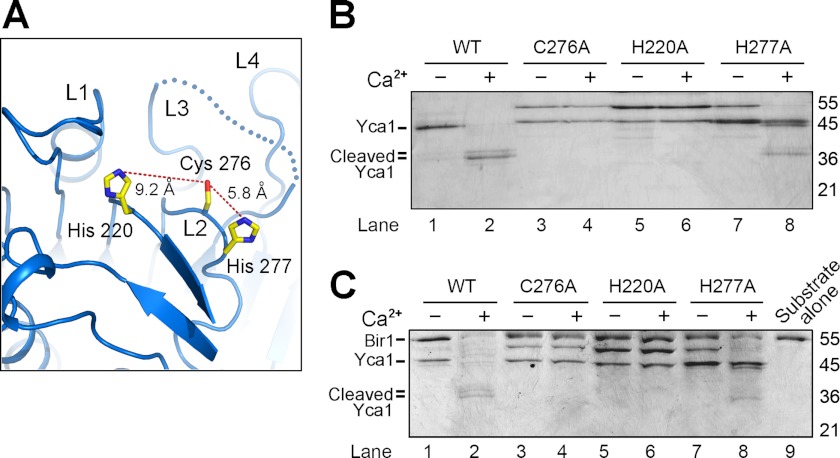
Sequence alignment with other caspases reveals two invariant amino acids, Cys276 and His220, in Yca1, which may constitute the catalytic dyad residues (Fig. 1B). Interestingly, however, the Nϵ atom of the His220 imidazole ring is ∼9.2 Å away from the Sγ atom of Cys276, whereas the distance between His277-Nϵ and Cys276-Sγ is 5.8 Å (Fig. 4A). To clearly identify the catalytic residues, we generated three missense mutations, each targeting a candidate amino acid in the active site, and purified these three mutant proteins to homogeneity. In sharp contrast to the WT Yca1, the mutants C276A and H220A failed to undergo Ca2+-stimulated autoprocessing (Fig. 4B, lanes 1–6). The mutant H277A, however, was further processed in the presence of Ca2+ (Fig. 4B, lanes 7 and 8). These results indicate that Cys276 and His220 constitute the catalytic dyad residues. When compared with a hydrogen bond distance required for catalysis, the longer distance between Cys276 and His220 might be circumvented by a local conformational change induced by substrate binding.
FIGURE 4.
Characterization of active site residues in Yca1. A, a close-up view on the putative catalytic residues Cys276, His220, and His277. B, the mutations C276A and H220A in Yca1 resulted in complete abrogation of autocatalytic processing in the presence of Ca2+. By contrast, the Yca1 mutant H277A retained the ability to undergo Ca2+-stimulated autoprocessing (lanes 7 and 8). C, only the WT Yca1, but not the mutant C276A or H220A, was able to cleave the substrate protein Bir1p (residues 1–435) in a Ca2+-dependent manner. The mutant Yca1-H277A retained a markedly reduced level of catalytic activity in the presence of calcium.
Curiously, despite mutation of the candidate catalytic residues Cys276 and His220, the purified mutants each contained a fraction of autoprocessed large fragment (Fig. 4B, lanes 3 and 5). At present, it remains unclear what caused the processing of these Yca1 mutants. It is possible that processing of Yca1 mutants might be carried out by other proteases in cells; in this case, such cleavage likely occurred during cell growth because use of multiple protease inhibitors during Yca1 purification failed to reduce such processing. Nonetheless, neither mutant Yca1-C276A nor mutant Yca1-H220A was able to cleave the Bir1p substrate (residues 1–435), regardless of the absence or presence of calcium (Fig. 4C, lanes 3–6). By contrast, the mutant Yca1-H277A retained the ability to cleave the Bir1p substrate (Fig. 4C, lanes 7 and 8).
Structural Comparison with MCA2
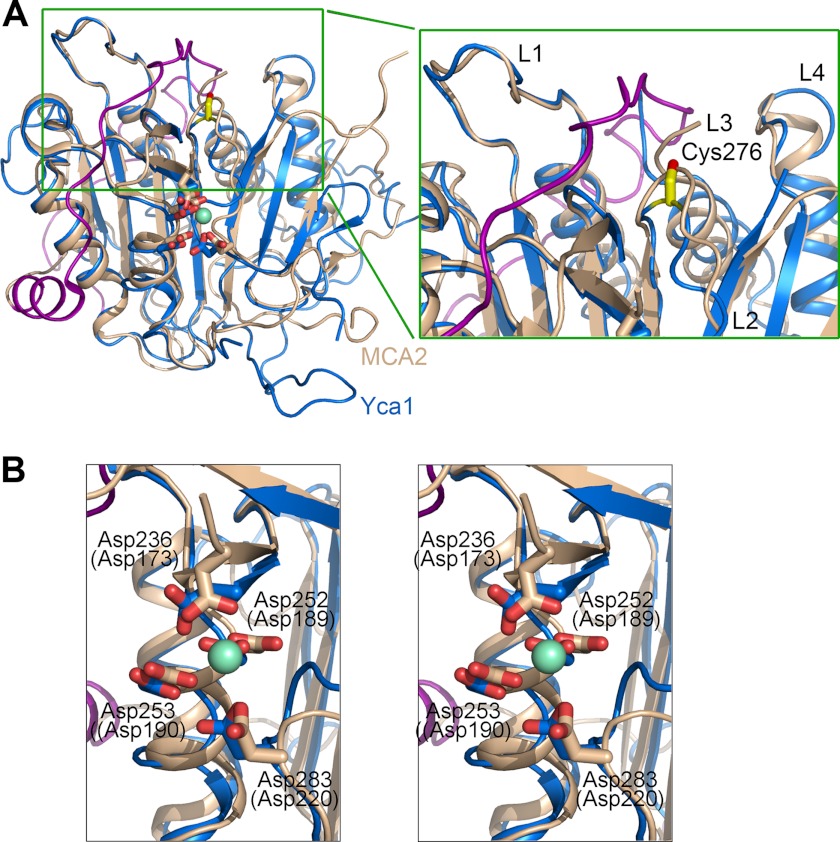
In the final stage of our manuscript preparation, we noted the online publication of a similar study (35), which reported the crystal structures of a metacaspase MCA2 in Trypanosoma brucei. In that study, the structure of the MCA2 catalytic mutant, C213G or C213A, was captured in either free or samarium-bound conformation (35), where the samarium-binding site was proposed to be occupied by calcium. The amino acid sequences of Yca1 are 24.9% identical to those of MCA2. The core structures of Yca1 and MCA2 are very similar, with an r.m.s.d. of 2.3 Å over 177 aligned Cα atoms between Yca1 (PDB code 4F6O) and the free MCA2-C213G (PDB code 4AF8). Yca1 can be superimposed to samarium-bound MCA2 (PDB code 4AFP) with an r.m.s.d. of 2.6 Å over 196 aligned Cα atoms (Fig. 5A).
FIGURE 5.
Structural comparison between Yca1 and MCA2 (35). A, structural superposition of Yca1 and MCA2 reveals a conserved overall fold with a similar active site conformation. Yca1 (PDB code 4F6O) and MCA2 (4AFP (35)) are colored blue and beige, respectively. The N-terminal fragment of MCA2, which was thought to inhibit its protease activity (35), is highlighted in magenta. A close-up view of the active site loops is shown on the right. B, a stereo view of the structural overlay around the samarium-binding site in MCA2. The four acidic, samarium-binding residues in MCA2 (in brackets) are invariant in Yca1. The four acidic residues in Yca1 are labeled.
The structural similarity is exemplified in two aspects. First, in both structures, the active site loops L1 and L4 exhibit nearly identical conformations (Fig. 5A, right panel). The L3 loop, which constitutes the base of the substrate-binding groove, is disordered in both structures. Second, the four samarium-binding, acidic amino acids in MCA2, Asp173, Asp189, Asp190, and Asp220, are invariant in Yca1, corresponding to Asp236, Asp252, Asp253, and Asp283, respectively. In the structure overlay, these amino acids occupy nearly identical positions (Fig. 5B). This analysis suggests that the calcium/samarium-binding site is conserved between MCA2 and Yca1.
The main structural difference between Yca1 and MCA2 lies in the N-terminal fragment, which encircles MCA2 and inhibits the protease activity of MCA2 (35). There are numerous interactions between residues in the N-terminal fragment and the caspase domain of MCA2. In Yca1, however, the N-terminal fragment does not bind to the caspase domain detectably and can be readily removed by limited proteolysis. In addition, there is no detectable sequence homology between the N-terminal sequences of Yca1 and MCA2. Neither the key amino acid Tyr31 nor the sequence motif Pro30-Tyr31-Leu32 in MCA2, which both play an essential role in the proposed autoinhibition, is conserved in Yca1. Thus it is unlikely that the N-terminal fragment of Yca1 also regulates its protease activity through autoinhibition. To examine this scenario experimentally, we generated a truncated variant Yca1-Δ86, which had the N-terminal 86 amino acids deleted, and a double mutant Yca1-R72A/K86A. Subsequent protease activity assay reveals that both Yca1-Δ86 and Yca1-R72A/K86A exhibited a similar level of protease activity when compared with the WT Yca1 protein (supplemental Fig. 5).
DISCUSSION
In this study, we report the first crystal structure of a yeast metacaspase, Yca1 from S. cerevisiae, and its biochemical characterization. Our results demonstrate that as anticipated, the metacaspase adopts a caspase fold, with active site loops arranged similarly as other caspases. However, important structural differences exist between Yca1 and other caspases. The most prominent and unique feature of Yca1 is an eight-stranded β-sheet, which presumably precludes the possibility of homodimerization via the same type of interface as observed in other caspases (supplemental Fig. 2).
At present, there is insufficient information to determine whether the autocatalytic processing of Yca1 occurs in cis or in trans. Yca1 is processed at a minimum of four sites, namely Arg72, Lys86, Lys331, and Lys334. Understandably, all four amino acids are located on flexible surface loops that are disordered in the crystals. The amino acid that is closest to Lys86 and visible in the crystal structure of Yca1 is Leu89. However, the distance between Leu89 and the catalytic residue Cys276 is ∼35 Å, which is beyond the distance spanned by three consecutive amino acids (Lys86–Leu89). This analysis suggests that unless the structure after Leu89 unfolds, autocleavage after Lys86 is unlikely to be mediated in cis. By a similar analysis, the amino acid that is closest to Lys331/Lys334 and structured in the Yca1 crystal is Phe351; the distance between Phe351 and the catalytic residue Cys276 is ∼26 Å, which is less than that spanned maximally by 17 consecutive amino acids (Lys334–Phe351) and thus in principle may allow cis cleavage to occur. Our experimental evidence indicates that autocleavage at Arg72/Lys86 can occur in trans, at least in vitro using purified recombinant proteins (supplemental Fig. 6).
In contrast to most other caspases, Yca1 was crystallized in the absence of an inhibitor. Consequently, the active site loops exhibit considerably higher temperature factors (B factors), and the bulk of the L3 loop is disordered. We suggest that the inhibitor-bound conformation of the Yca1 active site might be quite similar to that of the paracaspase MALT1 and further speculate that a different kind of homodimerization might be utilized for the autocatalytic processing and perhaps proteolytic activity of Yca1. Conclusive answers to these questions await future experiments.
Our biochemical characterization identifies Yca1 to be a Ca2+-activated protease. 1.5 mm Ca2+ induced autocatalytic processing of Yca1 (Fig. 3A), whereas the Ca2+ concentration range of 2–5 mm was sufficient to support a weak level of the Yca1 protease activity. To make the visual effect more apparent, we used 10–25 mm Ca2+ in our protease assays. We speculate that the requirement for increased Ca2+ concentrations might be due to low binding affinity for Ca2+. Notably, a related study showed that 50 mm Ca2+ is required for activation of metacaspase in Arabidopsis (33). Local high Ca2+ concentrations might offer tight regulation of Yca1 protease activity in vivo to prevent undesirable cell death under normal cell growth conditions or under slightly increased intracellular calcium concentrations.
Although the structure of a samarium-bound metacaspase has been elucidated (35), how calcium facilitates the autocatalytic processing of Yca1 remains unclear. Our in vitro observation strongly suggests that activation of Yca1 in S. cerevisiae may also be calcium-dependent. In this case, release of calcium into the cytoplasm might be an important trigger for the activation of Yca1 and its associated biological consequences, including, perhaps, cell death.
Supplementary Material
Acknowledgments
We thank J. He and Q. Wang at Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U for on-site assistance.
This work was supported by funds from the Ministry of Science and Technology (Grant 2009CB918801), National Natural Science Foundation (Project 30888001), and Beijing Municipal Commissions of Education and Science and Technology.
This article was selected as a Paper of the Week.

This article contains supplemental Figs. 1–6.
The atomic coordinates and structure factors (code 4F6O) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- IAP
- inhibitor of apoptosis
- r.m.s.d.
- root mean square deviation
- ac
- acetyl
- fmk
- fluoromethyl ketone
- z
- benzyloxycarbonyl
- dcbmk
- dichlorobenzylmethyl ketone.
REFERENCES
- 1. Madeo F., Fröhlich E., Fröhlich K. U. (1997) A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 139, 729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Uren A. G., O'Rourke K., Aravind L. A., Pisabarro M. T., Seshagiri S., Koonin E. V., Dixit V. M. (2000) Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 6, 961–967 [DOI] [PubMed] [Google Scholar]
- 3. Yoon H. J., Carbon J. (1999) Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc. Natl. Acad. Sci. 96, 13208–13213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walter D., Wissing S., Madeo F., Fahrenkrog B. (2006) The inhibitor-of-apoptosis protein Bir1p protects against apoptosis in S. cerevisiae and is a substrate for the yeast homologue of Omi/HtrA2. J. Cell Sci. 119, 1843–1851 [DOI] [PubMed] [Google Scholar]
- 5. Wissing S., Ludovico P., Herker E., Büttner S., Engelhardt S. M., Decker T., Link A., Proksch A., Rodrigues F., Corte-Real M., Fröhlich K. U., Manns J., Candé C., Sigrist S. J., Kroemer G., Madeo F. (2004) An AIF orthologue regulates apoptosis in yeast. J. Cell Biol. 166, 969–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khan M. A., Chock P. B., Stadtman E. R. (2005) Knockout of caspase-like gene, YCA1, abrogates apoptosis and elevates oxidized proteins in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 102, 17326–17331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Büttner S., Ruli D., Vögtle F. N., Galluzzi L., Moitzi B., Eisenberg T., Kepp O., Habernig L., Carmona-Gutierrez D., Rockenfeller P., Laun P., Breitenbach M., Khoury C., Fröhlich K. U., Rechberger G., Meisinger C., Kroemer G., Madeo F. (2011) A yeast BH3-only protein mediates the mitochondrial pathway of apoptosis. EMBO J. 30, 2779–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carmona-Gutierrez D., Ruckenstuhl C., Bauer M. A., Netzberger C., Eisenberg T., Braun R. J., Rockenfeller P., Khoury C. M., Moitzi B., Sommer C., Ring J., Schroeder S., Habernig L., Mazzoni C., Winderickx J., Gourlay C. W., Madeo F. (2011) A new Canterbury tale: the eighth International Meeting on Yeast Apoptosis in Canterbury, UK, 2–6 May 2011. Cell Death Differ. 18, 1948–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Madeo F., Herker E., Maldener C., Wissing S., Lächelt S., Herlan M., Fehr M., Lauber K., Sigrist S. J., Wesselborg S., Fröhlich K. U. (2002) A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9, 911–917 [DOI] [PubMed] [Google Scholar]
- 10. Rawlings N. D., Barrett A. J., Bateman A. (2012) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 40, D343–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsiatsiani L., Van Breusegem F., Gallois P., Zavialov A., Lam E., Bozhkov P. V. (2011) Metacaspases. Cell Death Differ. 18, 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bettiga M., Calzari L., Orlandi I., Alberghina L., Vai M. (2004) Involvement of the yeast metacaspase Yca1 in ubp10Δ-programmed cell death. FEMS Yeast Res. 5, 141–147 [DOI] [PubMed] [Google Scholar]
- 13. Guaragnella N., Pereira C., Sousa M. J., Antonacci L., Passarella S., Côrte-Real M., Marra E., Giannattasio S. (2006) YCA1 participates in the acetic acid-induced yeast programmed cell death also in a manner unrelated to its caspase-like activity. FEBS Lett. 580, 6880–6884 [DOI] [PubMed] [Google Scholar]
- 14. Guaragnella N., Bobba A., Passarella S., Marra E., Giannattasio S. (2010) Yeast acetic acid-induced programmed cell death can occur without cytochrome c release which requires metacaspase YCA1. FEBS Lett. 584, 224–228 [DOI] [PubMed] [Google Scholar]
- 15. Guaragnella N., Passarella S., Marra E., Giannattasio S. (2010) Knock-out of metacaspase and/or cytochrome c results in the activation of a ROS-independent acetic acid-induced programmed cell death pathway in yeast. FEBS Lett. 584, 3655–3660 [DOI] [PubMed] [Google Scholar]
- 16. Sokolov S., Pozniakovsky A., Bocharova N., Knorre D., Severin F. (2006) Expression of an expanded polyglutamine domain in yeast causes death with apoptotic markers. Biochim. Biophys. Acta 1757, 660–666 [DOI] [PubMed] [Google Scholar]
- 17. Watanabe N., Lam E. (2005) Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are arginine/lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J. Biol. Chem. 280, 14691–14699 [DOI] [PubMed] [Google Scholar]
- 18. Lee R. E., Puente L. G., Kaern M., Megeney L. A. (2008) A non-death role of the yeast metacaspase: Yca1p alters cell cycle dynamics. Plos One 3, e2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee R. E., Brunette S., Puente L. G., Megeney L. A. (2010) Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc. Natl. Acad. Sci. U.S.A. 107, 13348–13353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Erhardt M., Wegrzyn R. D., Deuerling E. (2010) Extra N-terminal residues have a profound effect on the aggregation properties of the potential yeast prion protein Mca1. Plos One 5, e9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vercammen D., van de Cotte B., De Jaeger G., Eeckhout D., Casteels P., Vandepoele K., Vandenberghe I., Van Beeumen J., Inzé D., Van Breusegem F. (2004) Type II metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana cleave substrates after arginine and lysine. J. Biol. Chem. 279, 45329–45336 [DOI] [PubMed] [Google Scholar]
- 22. Silva A., Almeida B., Sampaio-Marques B., Reis M. I., Ohlmeier S., Rodrigues F., Vale A., Ludovico P. (2011) Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a specific substrate of yeast metacaspase. Biochim. Biophys. Acta 1813, 2044–2049 [DOI] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26. Huesgen P. F., Miranda H., Lam X., Perthold M., Schuhmann H., Adamska I., Funk C. (2011) Recombinant Deg/HtrA proteases from Synechocystis sp. PCC 6803 differ in substrate specificity, biochemical characteristics, and mechanism. Biochem. J. 435, 733–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shiozaki E. N., Chai J., Shi Y. (2002) Oligomerization and activation of caspase-9, induced by Apaf-1 CARD. Proc. Natl. Acad. Sci. 99, 4197–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mittl P. R., Di Marco S., Krebs J. F., Bai X., Karanewsky D. S., Priestle J. P., Tomaselli K. J., Grütter M. G. (1997) Structure of recombinant human CPP32 in complex with the tetrapeptide acetyl-Asp-Val-Ala-Asp fluoromethyl ketone. J. Biol. Chem. 272, 6539–6547 [DOI] [PubMed] [Google Scholar]
- 29. Renatus M., Stennicke H. R., Scott F. L., Liddington R. C., Salvesen G. S. (2001) Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. 98, 14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu J. W., Jeffrey P. D., Ha J. Y., Yang X., Shi Y. (2011) Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region. Proc. Natl. Acad. Sci. U.S.A. 108, 21004–21009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chai J., Wu Q., Shiozaki E., Srinivasula S. M., Alnemri E. S., Shi Y. (2001) Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding. Cell 107, 399–407 [DOI] [PubMed] [Google Scholar]
- 32. Shiozaki E. N., Chai J., Rigotti D. J., Riedl S. J., Li P., Srinivasula S. M., Alnemri E. S., Fairman R., Shi Y. (2003) Mechanism of XIAP-mediated inhibition of caspase-9. Mol. Cell 11, 519–527 [DOI] [PubMed] [Google Scholar]
- 33. Watanabe N., Lam E. (2011) Calcium-dependent activation and autolysis of Arabidopsis metacaspase 2d. J. Biol. Chem. 286, 10027–10040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li X., Wang J., Shi Y. (2011) Structural mechanisms of DIAP1 autoinhibition and DIAP1-mediated inhibition of drICE. Nat. Commun. 2, 408. [DOI] [PubMed] [Google Scholar]
- 35. McLuskey K., Rudolf J., Proto W. R., Isaacs N. W., Coombs G. H., Moss C. X., Mottram J. C. (2012) Crystal structure of a Trypanosoma brucei metacaspase. Proc. Natl. Acad. Sci. U.S.A. 109, 7469–7474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 0.99, Schrödinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.