Background: C. jejuni produces an N-linked heptasaccharide that is attached to multiple proteins and has been linked with full virulence.
Results: A modified N-glycan displaying a phosphoethanolamine (pEtN) moiety linked to the terminal GalNAc was identified attached to 9 proteins.
Conclusion: The addition of pEtN to the N-glycan is mediated by the pEtN transferase EptC.
Significance: Modification of the N-glycan by pEtN confirms that EptC targets multiple substrates in C. jejuni.
Keywords: Campylobacter, Glycoprotein, Mass Spectrometry (MS), Membrane Proteins, Proteomics, Phosphoethanolamine
Abstract
Campylobacter jejuni is the major worldwide cause of bacterial gastroenteritis. C. jejuni possesses an extensive repertoire of carbohydrate structures that decorate both protein and non-protein surface-exposed structures. An N-linked glycosylation system encoded by the pgl gene cluster mediates the synthesis of a rigidly conserved heptasaccharide that is attached to protein substrates or released as free oligosaccharide in the periplasm. Removal of N-glycosylation results in reduced virulence and impeded host cell attachment. Since the N-glycan is conserved, the N-glycosylation system is also an attractive option for glycoengineering recombinant vaccines in Escherichia coli. To determine whether non-canonical N-glycans are present in C. jejuni, we utilized high throughput glycoproteomics to characterize C. jejuni JHH1 and identified 93 glycosylation sites, including 34 not previously reported. Interrogation of these data allowed the identification of a phosphoethanolamine (pEtN)-modified variant of the N-glycan that was attached to multiple proteins. The pEtN moiety was attached to the terminal GalNAc of the canonical N-glycan. Deletion of the pEtN transferase eptC removed all evidence of the pEtN-glycan but did not globally influence protein reactivity to patient sera, whereas deletion of the pglB oligosaccharyltransferase significantly reduced reactivity. Transfer of eptC and the pgl gene cluster to E. coli confirmed the addition of the pEtN-glycan to a target C. jejuni protein. Significantly reduced, yet above background levels of pEtN-glycan were also observed in E. coli not expressing eptC, suggesting that endogenous E. coli pEtN transferases can mediate the addition of pEtN to N-glycans. The addition of pEtN must be considered in the context of glycoengineering and may alter C. jejuni glycan-mediated structure-function interactions.
Introduction
Campylobacter jejuni is a Gram-negative pathogen responsible for a major proportion of food (typically poultry-derived)- and water-borne diarrheal illness worldwide, with an estimated 1% of the population in the United Kingdom and United States infected annually (1, 2). Prior infection with C. jejuni has also been associated with development of immune-mediated sequelae such as Reiter's syndrome (3), Guillain-Barré Syndrome (4), and immunoproliferative small intestinal disease (5). Despite studies to identify factors responsible for C. jejuni virulence in humans and colonization of the poultry host, our understanding of these processes remains incomplete (6, 7). Virulence-associated factors include cell-surface structures such as carbohydrates (8–10) and post-translationally modified flagellar and membrane proteins (11–15). These components generate significant variability that manifests in both the derived chemical structures and between C. jejuni isolates. Variations in lipooligosaccharide (LOS),3 capsule polysaccharide, and O-linked glycosylation of the flagella structural subunit flagellin are thus a quintessential feature of C. jejuni biology (16–21).
C. jejuni contains a conserved N-glycosylation system responsible for the modification of membrane-associated, periplasmic, and secreted proteins (22) and for the generation of a “free” oligosaccharide (fOS) involved in osmotic stability (23). The N-linked glycan is a heptasaccharide (GalNAc-α1,4-GalNAc-α1,4-[Glcβ1,3]-GalNAc-α1,4-GalNAc-α1,4-GalNAc-α1,3-Bac-β1, where Bac is bacillosamine (2,4-diacetamido-2,4,6 trideoxyglucopyranose)) encoded by the pgl gene cluster (24–26) and is attached to proteins (22, 27) in the periplasm by the PglB oligosaccharyltransferase (28) at the consensus sequon (D/E)XNX(S/T) (where X ≠ proline (29)). Removal of components of the N-glycosylation pathway results in reduced adherence to and invasion of gut epithelial cells and lowered colonization of the chicken gastrointestinal tract (11–12). Analysis of fOS (30), N-linked glycan (22, 30), and pathway intermediates (e.g. lipid-bound glycan (31)) suggest that the heptasaccharide is the sole N-glycan formed, although enzymes within the pgl biosynthetic pathway appear to have broader specificity than the substrates used to construct the canonical glycan (25, 32–33). Monoacetylated Bac in the laboratory passaged NCTC 11168 strain (32) remains the only glycan variant identified to-date. As passaged C. jejuni isolates differ in the manifestation of phenotypes associated with motility, morphology, and virulence (34, 35), it is unclear if the utilization of monoacetylated Bac is an authentic process common to all C. jejuni.
In contrast to the apparently conserved nature of the N-glycan, other carbohydrate structures such as capsule polysaccharide and LOS are subject to phase variation and can also be modified by phosphate-containing moieties, including phosphoramidate (36–37) and phosphoethanolamine (pEtN (19, 38)). The addition of similar moieties to protein substrates has been documented in other pathogens, most notably the decoration of Neisseria gonorrhoeae pilin with pEtN, phosphocholine (39, 40), and phosphoglycerol (41, 42). C. jejuni also modifies the flagellar rod protein FlgG with pEtN, and this addition is mediated by the pEtN transferase Cj0256 (Ref. 15; recently named EptC (43)). Deletion of eptC resulted in a decrease in motility and increased sensitivity to polymyxin B (15). Modification of FlgG with pEtN is required for full motility (43), whereas pEtN modification of lipid A contributes to polymyxin B resistance (15).
Here we report the identification of multiple proteins modified with an N-linked glycan displaying an additional moiety of +123.01 Da, consistent with the presence of pEtN. High resolution tandem mass spectrometry (MS/MS) confirmed that pEtN was attached to the terminal GalNAc of the canonical N-glycan rather than as a direct modification of the protein substrates. Deletion of eptC (cj0256) from C. jejuni JHH1 confirmed that EptC generates the pEtN-glycan. Heterologous expression of EptC in combination with the pgl cluster in E. coli led to the production of a pEtN-glycan-modified C. jejuni substrate protein (AcrA (44)). Modification with pEtN may influence C. jejuni and host-pathogen structure-function relationships mediated by the N-linked glycan.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Bacterial strains used in this study are provided in supplemental Table S1. C. jejuni were cultured in parallel on 100 individual Skirrow's agar plates in a micro-aerophilic environment of 5% O2, 5% CO2, and 90% N2 at 37 °C for 48 h. Collection of cells and generation of protein extracts for glycoproteomics were as previously described (27). Growth of C. jejuni wild-type, and deletion/complementation mutants was performed in Mueller-Hinton (MH) or brain heart infusion (BHI) medium supplemented with 1% yeast extract and 7% blood (BHI+). Escherichia coli were grown in Luria-Bertani (LB) broth or on agar at 37 °C under ambient oxygen conditions. Where required for selection, 30 μg/ml kanamycin, 25 μg/ml chloramphenicol, 25 μg/ml trimethoprim, and 100 μg/ml ampicillin were used.
Protease Digestion for Glycopeptide Enrichment
Dried proteins were resuspended in 6 m urea, 2 m thiourea, 40 mm NH4HCO3, and reduced/alkylated before digestion with Lys-C (1/200 w/w) and then trypsin (1/50 w/w) as previously described (27). For pepsin and thermolysin digestion, proteins were reduced/alkylated and processed according to Chen et al. (45). Briefly, for pepsin digests, samples were diluted 1:4 with 0.1% trifluoroacetic acid (TFA) and adjusted to a pH of ∼2.5 with 10% TFA. 1:25 (w/w) of pepsin to protein was added, and digestion was allowed to proceed for 24 h at 25 °C. For thermolysin digestion, samples were adjusted to a dilution of 1:4 with 100 mm NH4HCO3, and 1:25 (w/w) thermolysin to protein added. Samples were incubated for 24 h at 25 °C. All peptide digests were dialyzed against ultrapure water overnight using a Mini Dialysis kit with a molecular mass cutoff of 1000 Da (Amersham Biosciences) and on completion were collected and lyophilized.
Identification of Glycopeptides Using Zwitterionic–Hydrophilic Interaction Liquid Chromatography (ZIC-HILIC) Enrichment and Reversed Phase LC-MS/MS
ZIC-HILIC enrichment was performed according to Scott et al. (27) with minor modification. Essentially, 100 μg of starting peptide was used to improve detection of the 204 m/z GalNAc oxonium ion based on extracted ion chromatograms (XIC) compared with the previously reported 20 μg enrichment (27). ZIC-HILIC fractions were resuspended in 0.1% formic acid and separated using a trapless EASY-nLC system (Proxeon, Odense Denmark) coupled to an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose CA) as in Scott et al. (27). The instrument was operated using Xcalibur v2.2 (Thermo Scientific) with a capillary temperature of 200 °C in a data-dependent mode automatically switching between MS and higher energy collisional dissociation (HCD) MS/MS. The rationale for employing HCD fragmentation alone (rather than collision-induced dissociation (CID) and HCD (27) was that HCD spectra contain both glycan diagnostic ions (e.g. 204.08 m/z; GalNAc) and peptide fragment ions allowing glycopeptide identification. For each MS scan, the three most abundant precursor ions were selected for HCD fragmentation (normalized collision energy 45). MS resolution was set to 60,000 with an AGC of 1e6, maximum fill time of 500 ms, and a mass window of 600–2000 m/z. HCD fragmentation was carried out with an AGC of 2e5, maximum fill time of 500 ms, and mass window 170–2000 m/z. For definition of unusual glycan structures on glycopeptides, CID was employed using an LTQ-Orbitrap Velos mass spectrometer with automatic switching between MS and CID MS/MS. For each MS scan, the 10 most abundant precursor ions were selected for fragmentation with normalized collision energy 35. MS parameters were set as above, whereas CID fragmentation was carried out with an AGC of 2e4 and maximum fill time of 100 ms. The mass window for CID was dynamically defined by the selected ion m/z with an upper m/z limit of 2000 and lower m/z of 28% of the selected m/z.
Data Base Interrogation of Identified Glycopeptides
Raw files were processed in Proteome Discover v1.0 Build 43 (Thermo Scientific) to generate .mgf files and searched using MASCOT against a composite data base composed of the HB93-13, RM1221, 81-176, NCTC11168, and 81116 genomes in FASTA format. Scan events that did not result in peptide identification were exported to GPMAW 8.2 (Lighthouse Data, Odense Denmark) “mgf graph,” which generated .mgf files for MS/MS spectra containing the GalNAc diagnostic oxonium 204.086 m/z ion. These scans were manually annotated based on the presence of the deglycosylated peptide ion (parent mass minus 1405.561 Da and corresponding to the elemental composition C56H91N7O34 of the C. jejuni glycan) within a tolerance of 20 ppm. To facilitate glycopeptide assignments from HCD scan events, ions below the mass of the predicted deglycosylated peptides were extracted with Xcalibur v2.2 using the Spectrum list function. Ions with a deconvoluted mass above the deglycosylated peptide mass and ions corresponding to known carbohydrate oxonium ions such as 204.08, 366.14, and 407.16 m/z were removed by post-spectral processing. MASCOT v2.2 searches were conducted via the Australasian Proteomics Computational Facility with the C. jejuni taxonomy selected. Searches were carried out with a parent ion mass accuracy of 20 ppm and a product ion accuracy of 0.02 Da with no protease specificity and instrument selected as MALDI-QIT-TOF (use of this setting was due to the observation of multiple internal cleavage products and extensive NH3 and H2O loss from a, b, y ions, which are all included within this scoring setting) as well as the fixed modification carbamidomethyl (C) and variable modifications, oxidation (M), deamidation (N), and formylation (N-term). A MASCOT ion score cut-off of 20 was accepted for positive identifications, and all data were searched with the decoy setting activated to generate a zero false positive rate against a decoy data base. XIC corresponding to ±0.025 m/z of the monoisotopic peak of identified glycopeptides were generated within Xcalibur. Peaks were processed with a 15-point gaussian smooth and the corresponding ions inspected to ensure correct charge state.
Construction of C. jejuni JHH1 Chromosomal Mutants
To create the C. jejuni JHH1 ΔpglB mutant, primers PglBF and PglBR (supplemental Table S1) were used to amplify pglB::kan from genomic NCTC 11168 ΔpglB DNA (22). The resulting PCR product was inserted into the pGEM-T easy vector. Positive insertions were confirmed by PCR, and the resulting plasmid was named pNS1 (supplemental Table S1). To create the ΔeptC mutant, up- and downstream regions of eptC were amplified using 0256-1-PstI-F, 0256-2-HincII-R, 0256-3-HincII-F, and 0256-4-XhoI-R from JHH1 genomic DNA. PCR amplicons digested with HincII and PstI or XhoI, dependent on the amplicon, were introduced into PCR script digested with PstI and XhoI generating PCRscript-cj0256up/down. The APH(3′)-III phosphotransferase gene was gel-isolated after digestion of plasmid pWM10 with SmaI. The resulting product was inserted into PCRscript-cj0256up/down digested with HincII. The candidates were antibiotic-selected, and the resulting plasmid was named pNS2. Plasmids (pNS1 and pNS2) were used to create disruptions in the JHH1 background as described (46). C. jejuni JHH1 from overnight confluent plates of MH or BHI+ were resuspended in 15% glycerol, 9% sucrose, and washed 5 times before electroporation using a Micropulser™ (Bio-Rad) at 2.48 kV uniformly achieving ∼5–6-ms electroporation events. Cells were diluted with prewarmed BHI and recovered on BHI+ plates for 5 h. Cells were then collected by centrifugation at 8000 × g for 2 min at 25 °C and plated on BHI+ plates containing 30 μg/ml kanamycin. Plates were incubated for up to 4 days at 37 °C, 5% O2, 5% CO2, and 90% N2. The resulting colonies were evaluated for pglB and eptC disruption by PCR.
Western Blotting
Proteins were separated by SDS-PAGE gels and transferred to polyvinylidene difluoride membrane. Membranes were blocked overnight in 5% bovine serum albumin (Sigma) and probed with either α-N-linked glycan (1/10,000, (23)), α-His (1/3000; Rockland, Gilbertsville PA), α-HA (1/4000, Santa Cruz, Santa Cruz, CA), anti-JlpA (1/1000, (47)), or patient sera samples (1/2000, kindly provided by Dr. S. Cawthraw (48)).
Induction of eptC and Isolation of pEtN-glycan-modified AcrA from E. coli
E. coli CLM24 cells containing pACYC and pWA2 and either pMLBAD or pNS3 from overnight growth were used to inoculate 100-ml cultures. On achieving an A600 of 0.7, cells were induced by the addition of arabinose to 0.2% (w/v). After induction at 37 °C for 5 h, arabinose was added again to ensure expression during overnight cultivation. Cells were harvested by centrifugation at 6000 × g, 4 °C, stored at −80 °C, and freeze-dried. Cells were resuspended in 10 mm imidazole PBS and sonicated for 4 × 1min with 1 min on ice between rounds. Cellular debris was removed by centrifugation, and soluble AcrA was purified by nickel-nitrilotriacetic acid as previously described (44). Purified AcrA was separated using 10% SDS-PAGE and subjected to in-gel tryptic digestion (49).
Motility Assay
Motility assays were conducted using semi-solid MH medium supplemented with 0.4% agar. Plates were inoculated using 1 μl of overnight biphasic culture (A600 of ∼0.5). Plates were incubated for 24 h at 37 °C under microaerophilic conditions, and the diameter of motility was measured.
Determination of Polymyxin B Resistance
Minimum inhibitory concentrations were determined using polymyxin B Etest® strips (Biomérieux, France). Confluent overnight plates of C. jejuni were harvested and normalized to A600 0.35, and a 1:10 dilution of culture was then incubated in pre-warmed BHI (with 1% yeast extract) at 37 °C under microaerophilic conditions for 4 h. This diluted culture was then mixed 1:10 with 50 °C NZCYM medium (Oxoid, Basingstoke UK) supplemented with 0.6% agar. NZCYM top agar was gently poured onto MH plates and allowed to dry. An Etest® strip was placed on the center, and the plates were incubated at 37 °C for 24 h.
RESULTS
Identification of a pEtN-modified N-Linked Glycan in C. jejuni
To determine whether C. jejuni is capable of synthesizing novel N-glycans and attaching them to proteins, whole cell lysates from C. jejuni JHH1 were proteolyzed in parallel with thermolysin, pepsin, and trypsin, and the resulting digests were subjected to glycoproteomics analysis (27). A total of 12 ZIC-HILIC enrichments (biological replicates for each digest and protein lysate) were subjected to LC-MS, and 263 unique glycopeptides representing 93 glycosylation sites from 58 glycoproteins were identified (supplemental Table S2). These data contained 34 previously uncharacterized glycosylation sites and 15 novel glycoproteins (supplemental Table S3).
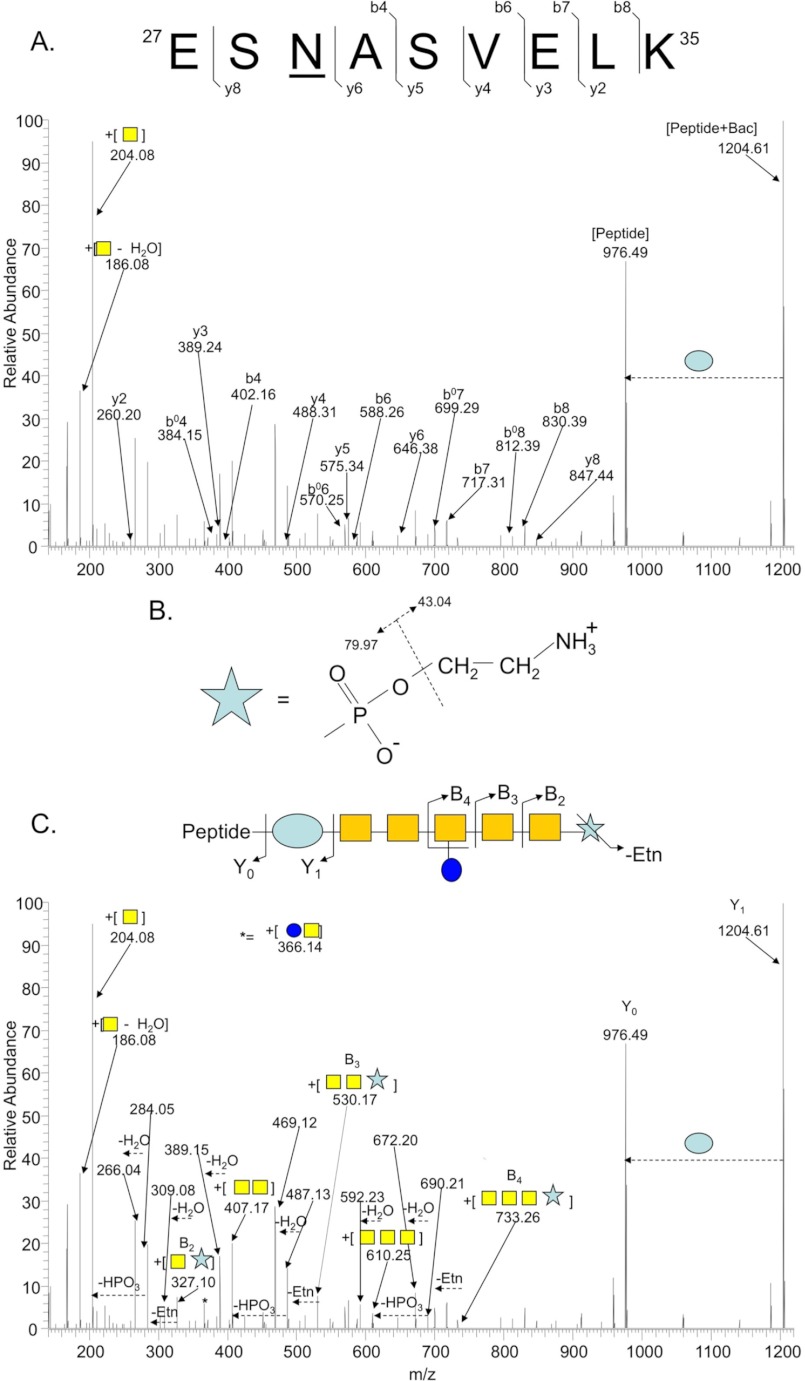
We next interrogated the dataset to determine whether modifications to the canonical N-linked glycan could be identified. Manual inspection of HCD MS/MS spectra was undertaken, with an emphasis on high quality spectra that did not generate an acceptable MASCOT score and that contained the diagnostic GalNAc oxonium ion, 204.08 m/z. Such spectra are more likely to contain a modified N-glycan as we relied on high mass accuracy within MS scans and included the predicted mass of the canonical heptasaccharide (1405.561 Da) as a fixed modification. Approximately 5000 MS/MS spectra were examined. We observed no evidence of monoacetylated Bac-containing N-linked glycans. We detected several highly charged (z ≥ +3) parent ions that generated fragment ions consistent with the canonical heptasaccharide, but with an overall increased glycan mass of ∼123.01 Da, as determined by Y0 (deglycosylated peptide) and Y1 (peptide + bacillosamine) fragment ions compared with the parent ion mass. None of these spectra generated MASCOT identifications of the glycopeptide. Manual definition of the y- and b-ion series generated by fragmentation of these parent ions confirmed peptide sequences (Fig. 1A). As the mass of 123.01 Da does not correspond to a known sugar, we searched the small molecule METLIN data base to identify possible candidates (mass of 142.03 ± 0.02 Da; corresponding to 123.01 + 19.018 [H2O and H+] Da). METLIN retrieved pEtN (MH+ = 142.0264) as the most likely candidate.
FIGURE 1.
Identification of a pEtN-modified N-linked glycan attached to antigenic protein HisJ (Cj0734c). A, shown is peptide fragmentation (lowercase y and b) by HCD MS/MS supporting the assignment of the peptide sequence 27ESNASVELK35. B, fragmentation of pEtN leads to the loss of ethanolamine (43.04 Da) followed by phosphate (79.97 Da). C, shown is pEtN and N-glycan fragmentation (uppercase Y and B) supporting the terminal location of pEtN attached to GalNAc. Oxonium ions of N-glycan sugars and pEtN are also annotated.
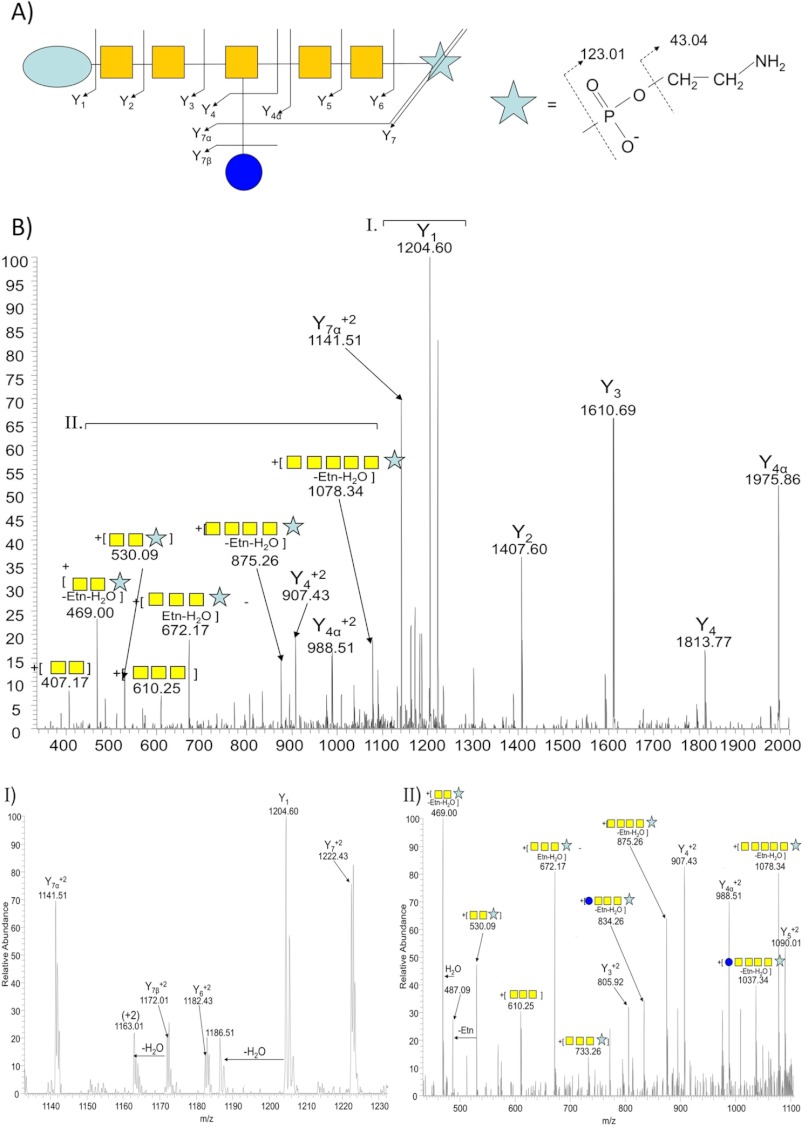
In MS/MS (CID or HCD), pEtN generates internal fragment ions resulting from the loss of ethanolamine followed by phosphate (Fig. 1B). We manually inspected the +123.01 Da-containing glycopeptide dataset and in each case we observed fragment ions corresponding to those predicted for pEtN as well as internal fragment ions showing a pEtN-GalNAc linkage (Fig. 1C). To define the location of pEtN in the N-glycan, we performed CID MS/MS, which predominantly generates glycan-associated fragment ions (27). This confirmed the location of pEtN attached to the terminal GalNAc in the modified N-glycan (Fig. 2, A–C).
FIGURE 2.
CID MS/MS of pEtN-glycan-modified glycopeptide 27ESNASVELK35. A, a fragmentation map shows complete coverage of the pEtN-glycan. B, shown is fragmentation of the doubly-charged ion of 27ESNASVELK35 (m/z = 1253.03597). Highlighted regions of both 1120–1240 m/z (I) and 450–1100 m/z (II) are provided to enable complete glycan annotation.
We next refined our spectral processing approach to enhance data base searching by removing pEtN-glycan fragment ions before analysis. MS/MS spectra were then reanalyzed, and a total of 8 pEtN-glycan-modified peptides were confidently identified (minimum MASCOT ion score of 20) from 7 glycoproteins (Table 1 and supplemental Fig. S1). Further support for these pEtN-glycan-modified glycopeptides was provided by the LC elution profiles of pEtN-glycan-modified glycopeptides compared with their canonical N-glycan-modified forms (supplemental Fig. S2). Glycopeptide species were chromatographically related, with pEtN-glycan-modified peptides typically eluting ∼1–2 min before their canonical glycopeptide forms.
TABLE 1.
pEtN-glycan modified glycopeptides identified in C. jejuni JHH1 and NCTC11168 O. 9 modified peptides from 8 proteins were identified
Corresponding MS/MS spectra are provided in supplemental Figs. S1 and S3.
| Cj # | Protein Name | Precursor mass (charge) | Precursor MH+ | Peptide mass (MH+) | Peptide sequence | MASCOT score |
|---|---|---|---|---|---|---|
| Da | ||||||
| Cj0131 | Putative peptidase M23 family protein | 979.74768 (+3) | 2937.22715 | 1408.6261 | 73DDNTSAMVIADEK85 | 34 |
| Cj0131 | Putative peptidase M23 family protein | 962.76045 (+3) | 2886.26548 | 1357.6780 | 68IILHKDDNTSAM79 | 48 |
| Cj0168c | Putative periplasmic protein | 938.38813 (+3) | 2813.14853 | 1284.5815 | 21ANTPSDVNQTHT32 | 34 |
| Cj0289c | Major antigenic peptide Peb3 | 746.64479 (+3) | 2237.91849 | 709.3515 | 88DFNVSK93 | 20 |
| Cj0399 | Colicin V production protein homolog | 1006.11781 (+3) | 3016.33756 | 1487.7700 | 173LQDIVSDLNNTQK179 | 45 |
| Cj0734c | Histidine-binding protein precursor HisJ/CjaC | 835.69240 (+3) | 2505.06131 | 976.4946 | 27ESNASVELK35 | 31a |
| Cj0982c | Putative amino acid transporter periplasmic solute-binding protein | 917.06178 (+3) | 2749.16947 | 1220.6005 | 137DSNITSVEDLK147 | 31 |
| Cj0983 | Surface-exposed lipoprotein JlpA | 840.03658 (+3) | 2518.09385 | 989.5262 | 104GEANASISIK113 | 21 |
| Cj1013cb | Putative cytochrome c biogenesis protein | 962.76045 (+3) | 3279.48667 | 1750.9082 | 529QDLNSTLPVVNTNHAK544 | 36 |
a MASCOT score given for C. jejuni JHH1 identification. MASCOT score = 20 for identical peptide in C. jejuni NCTC 11168 O.
b Peptide identified in C. jejuni NCTC 11168 O alone.
An N-Linked Glycan-modified with pEtN Is Present in C. jejuni NCTC 11168 O
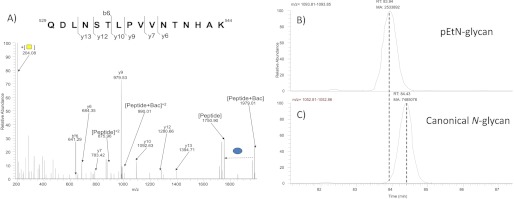
To examine whether pEtN-glycan modification was unique to C. jejuni JHH1, we subjected peptides from the NCTC 11168 O strain (34) to ZIC-HILIC enrichment and glycopeptide analysis. On this occasion we searched the data to only extract MS/MS spectra containing diagnostic pEtN-glycan fragment ions (pEtN-GalNAc, 327.09; pEtN-GalNAc minus ethanolamine (Etn), 284.05). We observed two glycopeptides with pEtN-glycan modification. These were 27ESNASVELK35 (the underline highlights the site of glycan attachment) from HisJ (CjaC or Cj0734c; MASCOT score = 20), which was also observed in the C. jejuni JHH1 dataset, and a novel glycopeptide 529QDLNSTLPVVNTNHAK544 derived from Cj1013c (Fig. 3A; MASCOT score = 36). Consistent with the behavior of glycopeptides from C. jejuni JHH1, pEtN-glycan-modified glycopeptides from NCTC 11168 eluted ∼1–2 min before the canonical glycopeptide forms (Fig. 3, B and C).
FIGURE 3.
A pEtN-glycan-modified glycopeptide (529QDLNSTLPVVNTNHAK544) from NCTC 11168 O. A, HCD MS/MS denoting peptide fragment ions confirming the identity (MASCOT ion score, 36) of XIC of pEtN-glycan-modified (B) and canonical N-glycan-modified (C) 529QDLNSTLPVVNTNHAK544.
EptC (Cj0256) Is the C. jejuni N-Glycan pEtN Transferase
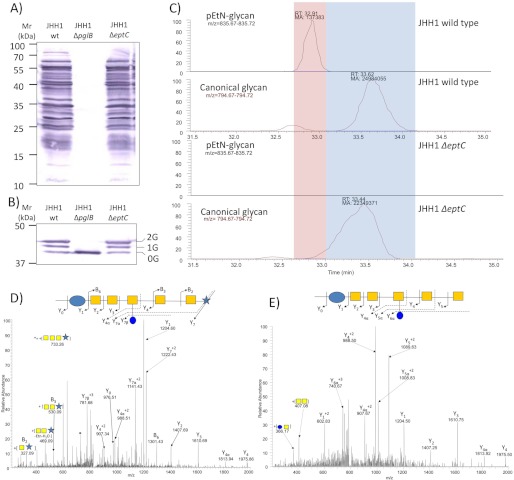
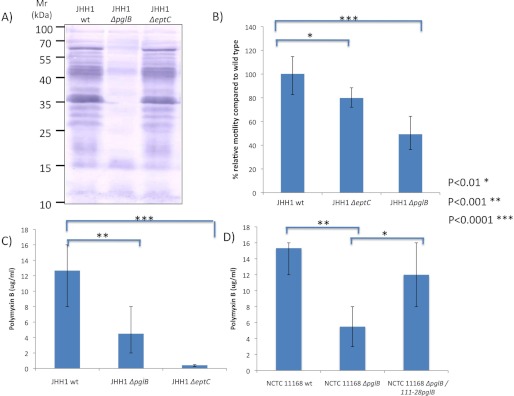
EptC (Cj0256) is the sole pEtN transferase predicted in C. jejuni 81-176 (15), and we confirmed this by in silico analysis of the C. jejuni 81116, ICDCCJ07001, BAA-1458, NCTC 11168, IA3902, M1, RM1221, and S3 genomes. Furthermore, no strains are predicted to contain eptC paralogs. Since EptC demonstrates substrate promiscuity (15) by modifying both lipid A and FlgG, we next tested whether EptC is responsible for pEtN modification of the N-glycan. A chromosomal disruption of eptC was created in the C. jejuni JHH1 background. We also deleted the pglB gene to compare the effects of loss of the canonical N-glycan with loss of the pEtN modification alone. Western blotting of protein lysates from C. jejuni JHH1 wild-type and ΔeptC with N-glycan-specific antibody showed no major differences in the N-glycoproteome (Fig. 4A), and anti-JlpA antibody revealed no differences in the pEtN-glycan-modified protein JlpA (Table 1) after eptC deletion (Fig. 4B). This is most likely due to the low mass of pEtN (123.01 Da), which cannot be differentiated from non-pEtN modified forms on SDS-PAGE gels. C. jejuni JHH1 ΔpglB showed a complete loss of glycosylated proteins.
FIGURE 4.
Comparison of glycoproteomes from C. jejuni JHH1 wild type and ΔpglB and ΔeptC deletion mutants. Western blotting with anti-N-linked glycan antibodies (A) and anti-JlpA (0G, 1G, and 2G refer to the number of occupied N-glycosites (47) (B) is shown. C, shown is XIC of pEtN-glycan and canonical N-glycan attached to peptide 27ESNASVELK35. CID MS/MS spectra of pEtN- glycan-modified (D) and canonical N-glycan-modified (E) forms of peptide 27ESNASVELK35, taken from wild-type and ΔeptC C. jejuni JHH1, respectively.
We next compared the C. jejuni JHH1 wild-type and ΔeptC strains by examining XIC from ZIC-HILIC-enriched glycopeptide fractions for all glycopeptides identified in Table 1. In the wild-type, we were readily able to detect both the canonical and pEtN-glycan-modified forms of each glycopeptide, whereas in JHH1 ΔeptC, only the canonical modification could be observed (Fig. 4C). MS/MS confirmed the presence of the pEtN-glycan on glycopeptides from wild-type (Fig. 4, D and E) that corresponded to peaks not observed in the ΔeptC mutant.
Generation of a pEtN-glycan in E. coli
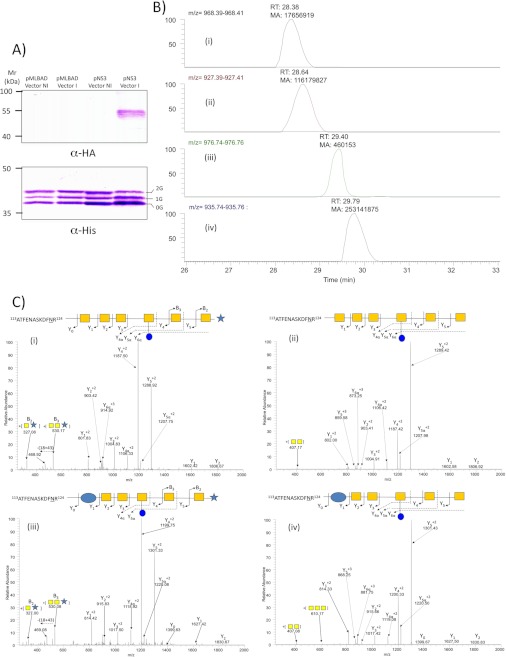
To further demonstrate the involvement of EptC in pEtN transfer to the N-glycan, we employed the E. coli CLM24 expression system containing pACYC (encoding the pgl gene cluster) and pWA2 (containing the glycosylation substrate protein, AcrA) (44, 50). Introduction of EptC with a C-terminal hemagglutinin (HA) tag in the arabinose-inducible system pMLBAD (pNS3; supplemental Table S1) enabled the selective induction of eptC in E. coli (Fig. 5A). Expression of EptC did not lead to gross changes in AcrA levels (Fig. 5B). The glycoforms of His-tagged AcrA were isolated from induced pMLBAD (empty vector control; no eptC) and induced pNS3 (expressing eptC) and subjected to tryptic digestion followed by ZIC-HILIC enrichment. In the presence of EptC, the AcrA peptide 113ATFENASKDFNR124 was clearly glycosylated both with and without pEtN modification on the canonical C. jejuni glycan (Fig. 5C-D and supplemental Fig. S4) as well as on the E. coli-specific glycan variant (Hex6HexNAc, Refs. 31 and 51); Fig. 5, C and D).
FIGURE 5.
Expression of the C. jejuni N-linked glycosylation system, EptC-HA, and AcrA-His in E. coli. A, upper, induction (I) of EptC with arabinose from pNS3 compared with non-induced controls (NI) and empty vector control pMLBAD is shown. Lower, comparison of AcrA glycosylation (0G, 1G, and 2G refer to the number of occupied N-glycosites). B, XIC of pEtN- glycan-modified (i and iii), E. coli heptasaccharide-modified (i and ii) and canonical C. jejuni N- glycan-modified (iii and iv) glycopeptides from AcrA 113ATFENASKDFNR124 are shown. C, confirmation of glycan structures by CID MS/MS (i–iv).
Influence of pEtN Modification on the N-Linked Glycoproteome
The observation of pEtN on multiple N-linked glycoproteins, several of which are known targets of the human humoral immune response (47, 49, 52), suggested a possible association with protein immunogenicity. This would also be consistent with studies showing that pEtN is an immunodominant modification of Neisseria meningitidis LOS (53). Coupled with the association of the C. jejuni N-linked glycan itself in reactivity to patient sera (54), we attempted to determine whether pEtN modification influenced immunoreactivity to C. jejuni proteins. Examination of reactivity with eight patient sera from the long term humoral response study of Cawthraw et al. (48) revealed that deletion of pglB resulted in loss of reactivity; however, no discernible changes in reactivity were observed against proteins from C. jejuni JHH1 ΔeptC (Fig. 6A and supplemental Fig. S5). The substrate promiscuity of EptC also lead us to examine the known phenotypes of eptC deletion in comparison with a C. jejuni JHH1 background deficient in N-linked glycosylation. Consistent with the phenotypes reported by Cullen and Trent (15), deletion of eptC in JHH1 resulted in an ∼25% decrease in motility (Fig. 6B) compared with wild-type and an increase in polymyxin B sensitivity (Fig. 6C). Interestingly, C. jejuni JHH1 ΔpglB also demonstrated an ∼3-fold increase in polymyxin B sensitivity (Fig. 6C) and ∼50% reduction in motility. To assess whether these phenotypes are consistent across N-linked glycosylation-deficient C. jejuni subspecies, we tested an NCTC 11168 ΔpglB mutant, which also demonstrated an ∼3-fold increase in polymyxin B sensitivity. Restoration of pglB by complementation resulted in a reversion to near wild-type levels of polymyxin B resistance (Fig. 6D).
FIGURE 6.
The effect of ΔpglB and ΔeptC on C. jejuni phenotypes. A, shown is a Western blot using patient serum. B, shown are motility assays of C. jejuni JHH1 wild type, ΔeptC, and ΔpglB. C, polymyxin B sensitivity of C. jejuni JHH1 wild-type, ΔpglB, and ΔeptC. D, shown is restoration of polymyxin B resistance in C. jejuni NCTC 11168 ΔpglB::pglB.
DISCUSSION
N-linked glycosylation has been associated with virulence in C. jejuni (11, 12), and protein targets have now been identified across several strains (Refs. 22, 26–27, 29, and 32 and this study). Unlike the complex branching, trimming, and additional sugar subunits associated with glycan diversity in eukaryotic N-glycans, all evidence until now has suggested that the C. jejuni N-glycan is rigidly conserved as a heptasaccharide consisting of an Asn-linked bacillosamine, five HexNAc (GalNAc), and a Hex branch (glucose) from the fourth position (3rd GalNAc). Our glycopeptide ZIC-HILIC enrichment approach (27) combined with the use of multiple, parallel proteolytic digests provided enhanced glycoproteome coverage that enabled the identification of 34 novel glycosylation sites from 15 previously uncharacterized C. jejuni glycoproteins. The size of the C. jejuni glycoproteome is considered to be as low as 150 protein targets (55) and no more than 260 proteins (predicted periplasmic, secreted, and membrane-associated proteins containing an N-linked sequon (27)). Our work combined with that of others in different C. jejuni strains has brought the number of verified C. jejuni glycoproteins to 70, making this the most comprehensive N-glycoproteome characterized for a free-living organism.
Our ability to mine deeply into the C. jejuni glycoproteome allowed us to search for evidence of non-canonical N-linked glycans. Manual interrogation of MS/MS spectra concentrated on high quality scans, with clear glycan fragment ions but no positive assignment by MASCOT. These spectra contained no evidence for the monoacetylated Bac linked to Asn previously observed in the laboratory passaged NCTC 11168 strain (32). Furthermore, we found no examples of truncated or elongated N-glycans modified by the addition or subtraction of GalNAc residues. Multiple glycopeptides containing a modified N-linked glycan consistent with the addition of a terminal pEtN were identified. The pEtN-glycan was not restricted to C. jejuni JHH1 as pEtN-modified glycopeptides were also identified in C. jejuni NCTC 11168 O. Extensive internal fragmentation of the pEtN-glycan and a predominant charge state that is non-ideal for HCD fragmentation (z ≥ +3 (27)) meant that pEtN-glycan-modified peptide sequences were difficult to identify even within this optimized workflow. Several MS/MS spectra containing clear pEtN-glycan fragment ions did not generate discernible peptide-related fragment ions, suggesting that additional sites of modification are likely to exist.
C. jejuni contains only a single protein, EptC (Cj0256), that is predicted to possess pEtN transferase activity (15), in contrast to other bacteria (e.g. E. coli) that contain multiple pEtN transferases (43). Neisseria sp. also encode multiple pEtN transferases that target specific substrates, including LptA, which modifies lipid A (56), Lpt3 and Lpt6, which target heptose within the lipopolysaccharide inner core (57), and phospho-form transferase A that can modify protein substrates (58, 59). In C. jejuni, EptC modifies both lipid A and FlgG and thus demonstrates broader substrate specificity than pEtN transferases from other organisms. It is currently unknown if protein targets in addition to FlgG are modified directly by EptC, although considering the broad range of substrates, this appears probable. Disruption of eptC resulted in the loss of the pEtN-glycan modification from all glycopeptides identified in this study, confirming that EptC also acts as an N-glycan pEtN transferase.
Evidence from previous studies suggests that the EptC-mediated addition of pEtN to the N-glycan occurs after the PglK flippase translocation of the complete canonical N-glycan into the periplasm. EptC has already been shown to modify lipid A and FlgG, both of which are periplasmic substrates (15), and EptC itself is N-glycosylated at Asn-215 (27), confirming that at least this region of the protein is exposed to the periplasm. We have thus far been unable to identify a pEtN-glycan fOS (data not shown), suggesting modification occurs after attachment of the canonical glycan to protein substrates; however, we cannot state unequivocally that such a fOS does not exist. Finally, the exact position of pEtN on the terminal GalNAc remains unresolved, as it is not yet possible to produce sufficient amounts of modified N-glycan for structural analyses.
Expression of EptC in E. coli containing the pgl gene cluster resulted in the addition of the pEtN-glycan to the target protein, AcrA. Intriguingly, we also found evidence of low level, yet above background, addition of pEtN to the N-linked glycan (∼20% of that seen in E. coli expressing the pgl gene cluster and eptC, based on comparison of area under the curve for XIC from E. coli expressing eptC or empty vector control) in the absence of eptC (supplemental Fig. S6). Non-EptC pEtN modification of the C. jejuni N-linked glycan in E. coli at even these low levels demonstrates that at least one of the five predicted E. coli pEtN transferases also possesses this function. When expressed in E. coli, we were also able to identify glycopeptides from AcrA that were modified with an extended C. jejuni N-glycan containing an additional hexose (supplemental Fig. S6) as well as an N-glycan containing an unusual Asn-linked carbohydrate of mass 244.12 Da (supplemental Fig. S6). These findings confirm that further N-glycan variations exist within the E. coli expression system than previously reported (31, 51) and that endogenous processes within E. coli can generate unexpected glycoforms.
Detection of even very low levels of endogenous pEtN modification on N-linked glycans in E. coli CLM24, the favored strain used for glycoprotein expression studies (60), may be a concern with respect to the homogeneity of glyco-conjugated vaccines expressed in this heterologous system. The addition of pEtN to C. jejuni substrates by E. coli pEtN transferases does have precedent, as E. coli EptA is capable of modifying C. jejuni lipid A (15). It is unclear whether the addition of pEtN will have an influence on vaccine reactivity or efficiency; however, pEtN has been noted as an immunodominant epitope (53), and therefore, care must be taken to ensure glycoprotein vaccines derived from this system either do not display the pEtN moiety or that the pEtN-glycan is harmless to recipients, and immunological reactivity is minimal compared with the intended epitope. Alternatively, these concerns may be overcome by the generation of an E. coli CLM24 derivative, in which the genes encoding endogenous pEtN transferases have been removed to limit background addition of pEtN to glyco-conjugates. Determination of the relative quantities of peptides modified with the canonical and pEtN-modified N-glycans is not feasible with currently available technology. It is, therefore, impossible to state unequivocally the ratio in which the two forms co-exist in C. jejuni; however, the enrichment, separation, and MS resolution and sensitivity required to detect the pEtN-glycan suggest it is present at relatively low levels, although factors beyond abundance and including the higher charge state, lack of peptide fragment ions, and change in LC retention time may account for reduced detection of pEtN-glycan-modified peptides.
Direct attachment of pEtN to FlgG by EptC (15) and to the pilin (40) of Neisseria sp. by phospho-form transferase A (39) have previously been observed, and pEtN modification of LOS has also been documented in a number of C. jejuni strains (37, 38). In N. gonorrhoeae there is recognition of the interplay between glycosylation and pEtN modification (39, 40), with overlapping site occupation observed between the O-linked glycosylation system and phospho-form transferase A -mediated phospho-form modifications (59). Extensive heterogeneity of pEtN and glycan modifications have been recognized in at least three proteins: PilE (39, 40) NGO1043, and NGO1237 (59), with two of these, PilE and NGO1043, recognized as major immunogens (40, 59). The link between pEtN and immunogenicity in Neisseria proteins and our identification of pEtN-glycan modification on previously identified C. jejuni immunogens, such as JlpA (47), Peb3 (61), and HisJ (62), provided further circumstantial evidence regarding a possible role in immunogenicity. We were, however, unable to detect a global effect of eptC deletion on reactivity of proteins with patient sera. In contrast, deletion of pglB, which results in loss of N-glycan attachment to proteins, significantly reduced protein reactivity. Disruption of eptC in the JHH1 background resulted in phenotypes that were reported previously (15); however, similar effects were also observed for pglB deletion mutants. Others have reported that deletion of genes within the pgl cluster, such as pglE, can result in motility defects (63); however, alterations in other genetic backgrounds have not displayed this phenotype (11). Increased sensitivity of ΔpglB mutants to polymyxin B was unexpected and suggests modification with the N-linked glycan enhances resistance; however, whether this is mediated globally via modification of a specific protein or at the level of the fOS is currently unknown.
Since EptC is multifunctional and targets several different substrates, it is difficult to assess the functional implications of the pEtN-glycan. Although no clear differences in reactivity to patient sera were observed, this does not preclude a role for the pEtN-glycan in innate immunity or in modifying host cell interactions. For example, previous studies have shown that C. jejuni N-linked glycoproteins and LOS displaying terminal GalNAc can bind the human macrophage galactose-type lectin (MGL) (64). Although speculative, the addition of pEtN to the N-glycan may block interactions between the previously terminal GalNAc of the canonical glycan and MGL, thus disrupting adherence between C. jejuni and MGL-expressing dendritic and macrophage cells. To assess this possibility, large-scale production of the pEtN-glycan will be required. Additionally, deletion of eptC results in loss of motility and is, therefore, likely to have a significant impact on the ability of C. jejuni to colonize both human and chicken hosts. Furthermore, a motility-reduced transposon mutagenesis library identified eptC (referred to as cj0256) leading to a 35% reduction in motility that was also associated with reduced invasion of INT-407 cells 65). Again, however, the multifunctional nature of EptC makes interpretation of such results problematic, as the individual contributions of pEtN modification of lipid A, FlgG, and the N-glycan cannot be determined. The motility defect observed in strain JHH1 ΔeptC is consistent with strain 81-176 (15); however, the magnitude of this reduction (25%) is less than observed for 81-176 (45%) and more consistent with published reports for strain 480 (35%) (65). The strain variations observed in these studies may be due to a multitude of currently unknown factors that influence motility in C. jejuni, including expression in clinical environments and unique genetic composition.
In conclusion, the depth of our C. jejuni glycoproteome dataset enabled us to identify a novel pEtN modification of the canonical N-linked glycan, which is attached to nine glycosylation sites in eight glycoproteins derived from two different C. jejuni strains. The addition of pEtN to the N-linked glycan is mediated by EptC and was transferable into E. coli. Although the biological role of the N-glycan and pEtN-glycan are currently unknown, the loss of the pEtN modification does not appear to have the same global effect on patient sera reactivity as seen for the N-linked glycan alone. The addition of pEtN, however, is likely to influence any glycan-mediated charge-based interactions and thus alter glycan-associated structure/function relationships. Further studies are required to fully understand N-glycan diversity and to discern the role of these glycans in C. jejuni virulence.
Supplementary Material
This work was supported by Australian Research Council Discovery Project Grant ARC DP110103573 (to S. J. C.).

This article contains supplemental Table S1-S3 and Figs. S1–S6.
- LOS
- lipooligosaccharide
- CID
- collision-induced dissociation
- fOS
- free oligosaccharide
- HCD
- higher energy collisional dissociation
- XIC
- extracted ion chromatogram
- ZIC-HILIC
- zwitterionic hydrophilic interaction liquid chromatography
- MH
- Mueller-Hinton
- BHI
- brain heart infusion
- pEtN
- phosphoethanolamine.
REFERENCES
- 1. Butzler J. P. (2004) Campylobacter, from obscurity to celebrity. Clin. Microbiol. Infect. 10, 868–876 [DOI] [PubMed] [Google Scholar]
- 2. Snelling W. J., Matsuda M., Moore J. E., Dooley J. S. (2005) Campylobacter jejuni. Lett. Appl. Microbiol. 41, 297–302 [DOI] [PubMed] [Google Scholar]
- 3. Hannu T., Kauppi M., Tuomala M., Laaksonen I., Klemets P., Kuusi M. (2004) Reactive arthritis following an outbreak of Campylobacter jejuni infection. J. Rheumatol. 31, 528–530 [PubMed] [Google Scholar]
- 4. Hughes R. A., Cornblath D. R. (2005) Guillain-Barré syndrome. Lancet 366, 1653–1666 [DOI] [PubMed] [Google Scholar]
- 5. Lecuit M., Abachin E., Martin A., Poyart C., Pochart P., Suarez F., Bengoufa D., Feuillard J., Lavergne A., Gordon J. I., Berche P., Guillevin L., Lortholary O. (2004) Immunoproliferative small intestinal disease associated with Campylobacter jejuni. N. Engl. J. Med. 350, 239–248 [DOI] [PubMed] [Google Scholar]
- 6. Konkel M. E., Monteville M. R., Rivera-Amill V., Joens L. A. (2001) The pathogenesis of Campylobacter jejuni-mediated enteritis. Curr. Issues Intest. Microbiol. 2, 55–71 [PubMed] [Google Scholar]
- 7. Young K. T., Davis L. M., Dirita V. J. (2007) Campylobacter jejuni. Molecular biology and pathogenesis. Nat. Rev. Microbiol. 5, 665–679 [DOI] [PubMed] [Google Scholar]
- 8. Bacon D. J., Szymanski C. M., Burr D. H., Silver R. P., Alm R. A., Guerry P. (2001) A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40, 769–777 [DOI] [PubMed] [Google Scholar]
- 9. Heikema A. P., Bergman M. P., Richards H., Crocker P. R., Gilbert M., Samsom J. N., van Wamel W. J., Endtz H. P., van Belkum A. (2010) Characterization of the specific interaction between sialoadhesin and sialylated Campylobacter jejuni lipooligosaccharides. Infect. Immun. 78, 3237–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Naito M., Frirdich E., Fields J. A., Pryjma M., Li J., Cameron A., Gilbert M., Thompson S. A., Gaynor E. C. (2010) Effects of sequential Campylobacter jejuni 81-176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J. Bacteriol. 192, 2182–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Szymanski C. M., Burr D. H., Guerry P. (2002) Campylobacter protein glycosylation affects host cell interactions. Infect. Immun. 70, 2242–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karlyshev A. V., Everest P., Linton D., Cawthraw S., Newell D. G., Wren B. W. (2004) The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology 150, 1957–1964 [DOI] [PubMed] [Google Scholar]
- 13. Ewing C. P., Andreishcheva E., Guerry P. (2009) Functional characterization of flagellin glycosylation in Campylobacter jejuni 81-176. J. Bacteriol. 191, 7086–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goon S., Kelly J. F., Logan S. M., Ewing C. P., Guerry P. (2003) Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50, 659–671 [DOI] [PubMed] [Google Scholar]
- 15. Cullen T. W., Trent M. S. (2010) A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc. Natl. Acad. Sci. U.S.A. 107, 5160–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karlyshev A. V., Champion O. L., Churcher C., Brisson J. R., Jarrell H. C., Gilbert M., Brochu D., St Michael F., Li J., Wakarchuk W. W., Goodhead I., Sanders M., Stevens K., White B., Parkhill J., Wren B. W., Szymanski C. M. (2005) Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol. 55, 90–103 [DOI] [PubMed] [Google Scholar]
- 17. Hanniffy O. M., Shashkov A. S., Moran A. P., Senchenkova S. N., Savage A. V. (2001) Chemical structure of the core oligosaccharide of aerotolerant Campylobacter jejuni O:2 lipopolysaccharide. Carbohydr. Res. 330, 223–229 [DOI] [PubMed] [Google Scholar]
- 18. Hanniffy O. M., Shashkov A. S., Moran A. P., Prendergast M. M., Senchenkova S. N., Knirel Y. A., Savage A. V. (1999) Chemical structure of a polysaccharide from Campylobacter jejuni 176.83 (serotype O:41) containing only furanose sugars. Carbohydr. Res. 319, 124–132 [DOI] [PubMed] [Google Scholar]
- 19. Gilbert M., Parker C. T., Moran A. P. (2008) in Campylobacter (Nachamkin I., Szymanski C. M., Blaser M. J., eds) 3rd Ed., pp. 483–504, American Society for Microbiology, Washington, D. C [Google Scholar]
- 20. Karlyshev A. V., Wren B. W., Moran A. P. (2008) in Campylobacter (Nachamkin I., Szymanski C. M., Blaser M. J., eds) 3rd Ed., pp. 505–521, American Society for Microbiology, Washington, D. C., [Google Scholar]
- 21. Howard S. L., Jagannathan A., Soo E. C., Hui J. P., Aubry A. J., Ahmed I., Karlyshev A., Kelly J. F., Jones M. A., Stevens M. P., Logan S. M., Wren B. W. (2009) Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infect. Immun. 77, 2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Young N. M., Brisson J. R., Kelly J., Watson D. C., Tessier L., Lanthier P. H., Jarrell H. C., Cadotte N., St Michael F., Aberg E., Szymanski C. M. (2002) Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277, 42530–42539 [DOI] [PubMed] [Google Scholar]
- 23. Nothaft H., Liu X., McNally D. J., Li J., Szymanski C. M. (2009) Study of free oligosaccharides derived from the bacterial N-glycosylation pathway. Proc. Natl. Acad. Sci. U.S.A. 106, 15019–15024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Glover K. J., Weerapana E., Imperiali B. (2005) In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc. Natl. Acad. Sci. U.S.A. 102, 14255–14259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Glover K. J., Weerapana E., Chen M. M., Imperiali B. (2006) Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphate transferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry 45, 5343–5350 [DOI] [PubMed] [Google Scholar]
- 26. Kelly J., Jarrell H., Millar L., Tessier L., Fiori L. M., Lau P. C., Allan B., Szymanski C. M. (2006) Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J. Bacteriol. 188, 2427–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scott N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., Falconer L., Kolarich D., Djordjevic S. P., Højrup P., Packer N. H., Larsen M. R., Cordwell S. J. (2011) Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, M000031-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kowarik M., Numao S., Feldman M. F., Schulz B. L., Callewaert N., Kiermaier E., Catrein I., Aebi M. (2006) N-Linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science 314, 1148–1150 [DOI] [PubMed] [Google Scholar]
- 29. Kowarik M., Young N. M., Numao S., Schulz B. L., Hug I., Callewaert N., Mills D. C., Watson D. C., Hernandez M., Kelly J. F., Wacker M., Aebi M. (2006) Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25, 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu X., McNally D. J., Nothaft H., Szymanski C. M., Brisson J. R., Li J. (2006) Mass spectrometry-based glycomics strategy for exploring N-linked glycosylation in eukaryotes and bacteria. Anal. Chem. 78, 6081–6087 [DOI] [PubMed] [Google Scholar]
- 31. Reid C. W., Stupak J., Chen M. M., Imperiali B., Li J., Szymanski C. M. (2008) Affinity-capture tandem mass spectrometric characterization of polyprenyl-linked oligosaccharides. Tool to study protein N-glycosylation pathways. Anal. Chem. 80, 5468–5475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ding W., Nothaft H., Szymanski C. M., Kelly J. (2009) Identification and quantification of glycoproteins using ion-pairing normal-phase liquid chromatography and mass spectrometry. Mol. Cell. Proteomics 8, 2170–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wacker M., Feldman M. F., Callewaert N., Kowarik M., Clarke B. R., Pohl N. L., Hernandez M., Vines E. D., Valvano M. A., Whitfield C., Aebi M. (2006) Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc. Natl. Acad. Sci. U.S.A. 103, 7088–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gaynor E. C., Cawthraw S., Manning G., MacKichan J. K., Falkow S., Newell D. G. (2004) The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J. Bacteriol. 186, 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carrillo C. D., Taboada E., Nash J. H., Lanthier P., Kelly J., Lau P. C., Verhulp R., Mykytczuk O., Sy J., Findlay W. A., Amoako K., Gomis S., Willson P., Austin J. W., Potter A., Babiuk L., Allan B., Szymanski C. M. (2004) Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 279, 20327–20338 [DOI] [PubMed] [Google Scholar]
- 36. McNally D. J., Lamoureux M. P., Karlyshev A. V., Fiori L. M., Li J., Thacker G., Coleman R. A., Khieu N. H., Wren B. W., Brisson J. R., Jarrell H. C., Szymanski C. M. (2007) Commonality and biosynthesis of the O-methyl phosphoramidate capsule modification in Campylobacter jejuni. J. Biol. Chem. 282, 28566–28576 [DOI] [PubMed] [Google Scholar]
- 37. Szymanski C. M., Michael F. S., Jarrell H. C., Li J., Gilbert M., Larocque S., Vinogradov E., Brisson J. R. (2003) Detection of conserved N-linked glycans and phase-variable lipooligosaccharides and capsules from Campylobacter cells by mass spectrometry and high resolution magic angle spinning NMR spectroscopy. J. Biol. Chem. 278, 24509–24520 [DOI] [PubMed] [Google Scholar]
- 38. St Michael F., Szymanski C. M., Li J., Chan K. H., Khieu N. H., Larocque S., Wakarchuk W. W., Brisson J. R., Monteiro M. A. (2002) The structures of the lipooligosaccharide and capsule polysaccharide of Campylobacter jejuni genome sequenced strain NCTC 11168. Eur. J. Biochem. 269, 5119–5136 [DOI] [PubMed] [Google Scholar]
- 39. Aas F. E., Egge-Jacobsen W., Winther-Larsen H. C., Løvold C., Hitchen P. G., Dell A., Koomey M. (2006) Neisseria gonorrhoeae type IV pili undergo multisite, hierarchical modifications with phosphoethanolamine and phosphocholine requiring an enzyme structurally related to lipopolysaccharide phosphoethanolamine transferases. J. Biol. Chem. 281, 27712–27723 [DOI] [PubMed] [Google Scholar]
- 40. Hegge F. T., Hitchen P. G., Aas F. E., Kristiansen H., Løvold C., Egge-Jacobsen W., Panico M., Leong W. Y., Bull V., Virji M., Morris H. R., Dell A., Koomey M. (2004) Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. Proc. Natl. Acad. Sci. U.S.A. 101, 10798–10803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stimson E., Virji M., Barker S., Panico M., Blench I., Saunders J., Payne G., Moxon E. R., Dell A., Morris H. R. (1996) Discovery of a novel protein modification. α-Glycerophosphate is a substituent of meningococcal pilin. Biochem. J. 316, 29–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chamot-Rooke J., Mikaty G., Malosse C., Soyer M., Dumont A., Gault J., Imhaus A. F., Martin P., Trellet M., Clary G., Chafey P., Camoin L., Nilges M., Nassif X., Duménil G. (2011) Posttranslational modification of pili upon cell contact triggers N. meningitidis dissemination. Science 331, 778–782 [DOI] [PubMed] [Google Scholar]
- 43. Cullen T. W., Madsen J. A., Ivanov P. L., Brodbelt J. S., Trent M. S. (2012) Characterization of a unique modification of the flagellar rod protein FlgG by the Campylobacter jejuni lipid A phosphoethanolamine transferase, linking bacterial locomotion and antimicrobial peptide resistance. J. Biol. Chem. 287, 3326–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Feldman M. F., Wacker M., Hernandez M., Hitchen P. G., Marolda C. L., Kowarik M., Morris H. R., Dell A., Valvano M. A., Aebi M. (2005) Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 102, 3016–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen R., Jiang X., Sun D., Han G., Wang F., Ye M., Wang L., Zou H. (2009) Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J. Proteome Res. 8, 651–661 [DOI] [PubMed] [Google Scholar]
- 46. Davis L., Young K., DiRita V. (2008) Genetic manipulation of Campylobacter jejuni. Curr. Protoc. Microbiol. Chapter 8, Unit 8A 2.1–8A 2.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scott N. E., Bogema D. R., Connolly A. M., Falconer L., Djordjevic S. P., Cordwell S. J. (2009) Mass spectrometric characterization of the surface-associated 42 kDa lipoprotein JlpA as a glycosylated antigen in strains of Campylobacter jejuni. J. Proteome Res. 8, 4654–4664 [DOI] [PubMed] [Google Scholar]
- 48. Cawthraw S. A., Feldman R. A., Sayers A. R., Newell D. G. (2002) Long term antibody responses following human infection with Campylobacter jejuni. Clin. Exp. Immunol. 130, 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cordwell S. J., Len A. C., Touma R. G., Scott N. E., Falconer L., Jones D., Connolly A., Crossett B., Djordjevic S. P. (2008) Identification of membrane-associated proteins from Campylobacter jejuni strains using complementary proteomics technologies. Proteomics 8, 122–139 [DOI] [PubMed] [Google Scholar]
- 50. Wacker M., Linton D., Hitchen P. G., Nita-Lazar M., Haslam S. M., North S. J., Panico M., Morris H. R., Dell A., Wren B. W., Aebi M. (2002) N-Linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298, 1790–1793 [DOI] [PubMed] [Google Scholar]
- 51. Linton D., Dorrell N., Hitchen P. G., Amber S., Karlyshev A. V., Morris H. R., Dell A., Valvano M. A., Aebi M., Wren B. W. (2005) Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 55, 1695–1703 [DOI] [PubMed] [Google Scholar]
- 52. Pawelec D., Jakubowska-Mróz J., Jagusztyn-Krynicka E. K. (1998) Campylobacter jejuni 72Dz/92 cjaC gene coding 28-kDa immunopositive protein, a homologue of the solute-binding components of the ABC transport system. Lett. Appl. Microbiol. 26, 69–76 [DOI] [PubMed] [Google Scholar]
- 53. Mackinnon F. G., Cox A. D., Plested J. S., Tang C. M., Makepeace K., Coull P. A., Wright J. C., Chalmers R., Hood D. W., Richards J. C., Moxon E. R. (2002) Identification of a gene (lpt-3) required for the addition of phosphoethanolamine to the lipopolysaccharide inner core of Neisseria meningitidis and its role in mediating susceptibility to bactericidal killing and opsonophagocytosis. Mol. Microbiol. 43, 931–943 [DOI] [PubMed] [Google Scholar]
- 54. Szymanski C. M., Yao R., Ewing C. P., Trust T. J., Guerry P. (1999) Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32, 1022–1030 [DOI] [PubMed] [Google Scholar]
- 55. Nothaft H., Amber S., Aebi M., Szymanski C. M. (2008) in Campylobacter (Nachamkin I., Szymanski C. M., Blaser M. J., eds) 3rd Ed., pp. 447–469, American Society for Microbiology, Washington, D. C [Google Scholar]
- 56. Cox A. D., Wright J. C., Li J., Hood D. W., Moxon E. R., Richards J. C. (2003) Phosphorylation of the lipid A region of meningococcal lipopolysaccharide: identification of a family of transferases that add phosphoethanolamine to lipopolysaccharide. J. Bacteriol. 185, 3270–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wenzel C. Q., St Michael F., Stupak J., Li J., Cox A. D., Richards J. C. (2010) Functional characterization of Lpt3 and Lpt6, the inner-core lipooligosaccharide phosphoethanolamine transferases from Neisseria meningitidis. J. Bacteriol. 192, 208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Naessan C. L., Egge-Jacobsen W., Heiniger R. W., Wolfgang M. C., Aas F. E., Røhr A., Winther-Larsen H. C., Koomey M. (2008) Genetic and functional analyses of PptA, a phospho-form transferase targeting type IV pili in Neisseria gonorrhoeae. J. Bacteriol. 190, 387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Anonsen J. H., Egge-Jacobsen W., Aas F. E., Børud B., Koomey M., Vik A. (2012) Novel protein substrates of the phospho-form modification system in Neisseria gonorrhoeae and their connection to O-linked protein glycosylation. Infect. Immun. 80, 22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ihssen J., Kowarik M., Dilettoso S., Tanner C., Wacker M., Thöny-Meyer L. (2010) Production of glycoprotein vaccines in Escherichia coli. Microb. Cell Fact. 9, 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pei Z. H., Ellison R. T., 3rd, Blaser M. J. (1991) Identification, purification, and characterization of major antigenic proteins of Campylobacter jejuni. J. Biol. Chem. 266, 16363–16369 [PubMed] [Google Scholar]
- 62. Wyszyńska A., Tomczyk K., Jagusztyn-Krynicka E. K. (2007) Comparison of the localization and post-translational modification of Campylobacter coli CjaC and its homolog from Campylobacter jejuni, Cj0734c/HisJ. Acta Biochim. Pol. 54, 143–150 [PubMed] [Google Scholar]
- 63. Vijayakumar S., Merkx-Jacques A., Ratnayake D. B., Gryski I., Obhi R. K., Houle S., Dozois C. M., Creuzenet C. (2006) Cj1121c, a novel UDP-4-keto-6-deoxy-GlcNAc C-4 aminotransferase essential for protein glycosylation and virulence in Campylobacter jejuni. J. Biol. Chem. 281, 27733–27743 [DOI] [PubMed] [Google Scholar]
- 64. van Sorge N. M., Bleumink N. M., van Vliet S. J., Saeland E., van der Pol W. L., van Kooyk Y., van Putten J. P. (2009) N-glycosylated proteins and distinct lipooligosaccharide glycoforms of Campylobacter jejuni target the human C-type lectin receptor MGL. Cell Microbiol. 11, 1768–1781 [DOI] [PubMed] [Google Scholar]
- 65. Golden N. J., Acheson D. W. (2002) Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect. Immun. 70, 1761–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.