Background: The function of KaiB remains to be solved.
Results: Dimeric KaiB1–94 generated circadian oscillation in vitro, but it did not in cells.
Conclusion: KaiB tetramer-dimer transformation is responsible for the regulation of the SasA-mediated clock output pathway.
Significance: We demonstrated the role of KaiB in the regulation of the SasA-KaiC interaction, involved in the transmission of time-information from KaiABC-machinery to transcription apparatus.
Keywords: Bioluminescence, Circadian, Circadian Clock, Circadian Rhythms, Cyanobacteria, Fluorescence Correlation Spectroscopy, KaiB, SasA, Thermosynechococcus elongatus, Oligomeric Structure
Abstract
The molecular machinery of the cyanobacterial circadian clock consists of three proteins, KaiA, KaiB, and KaiC. The three Kai proteins interact with each other and generate circadian oscillations in vitro in the presence of ATP (an in vitro KaiABC clock system). KaiB consists of four subunits organized as a dimer of dimers, and its overall shape is that of an elongated hexagonal plate with a positively charged cleft flanked by two negatively charged ridges. We found that a mutant KaiB with a C-terminal deletion (KaiB1–94), which lacks the negatively charged ridges, was a dimer. Despite its dimeric structure, KaiB1–94 interacted with KaiC and generated normal circadian oscillations in the in vitro KaiABC clock system. KaiB1–94 also generated circadian oscillations in cyanobacterial cells, but they were weak, indicating that the C-terminal region and tetrameric structure of KaiB are necessary for the generation of normal gene expression rhythms in vivo. KaiB1–94 showed the highest affinity for KaiC among the KaiC-binding proteins we examined and inhibited KaiC from forming a complex with SasA, which is involved in the main output pathway from the KaiABC clock oscillator in transcription regulation. This defect explains the mechanism underlying the lack of normal gene expression rhythms in cells expressing KaiB1–94.
Introduction
The circadian clock is an endogenous biological mechanism that generates daily cycles in physiological activity (circadian rhythms) (1, 2). Cyanobacteria are the simplest organisms that exhibit circadian rhythms (2), and the circadian clock gene cluster kaiABC is essential for circadian rhythms in cyanobacteria (3). The rhythms involve circadian oscillations in the phosphorylation level (4) and ATPase activity (5) of KaiC and complex formation among KaiA, KaiB, and KaiC (6, 7). KaiC has a duplicated RecA/DnaB structure. Its N-terminal domain shows ATPase activity (8), whereas its C-terminal domain shows both ATPase activity (5, 8) and intersubunit phosphorylation activity (9, 10). KaiA and KaiB have opposite effects on KaiC ATPase activity (5, 8) and phosphorylation level; KaiA increases them (10–13), and KaiB decreases them (5, 12, 14). KaiB is composed of two asymmetric dimers (15, 16) that form a tetramer. KaiB directly associates with the C-terminal clock oscillator domain (13) of KaiA (17). Its overall shape is that of an elongated hexagonal plate with a positively charged cleft (PC)7 that is flanked by two negatively charged ridges (NRs) (16). Rhythm analysis of cyanobacterial cells carrying a mutant kaiB gene show that the PC is necessary to KaiB clock function (16). Deletion of the C-terminal residues from amino acids 95–108 of KaiB (yielding mutant KaiB1–94) derived from the cyanobacteria Synechococcus sp. strain PCC 7942 (hereafter Synechococcus) and Thermosynechococcus elongatus results in loss of the NRs and extensively weakens in vivo circadian rhythms (16). Here we examine the structure of KaiB1–94 and the role it plays in the generation circadian oscillations.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
KaiA, KaiB, KaiC, and SasA derived from T. elongatus (18) were expressed in Escherichia coli BL21 cells and purified as previously described (16, 17, 19–21). We generated gene constructs for a truncated mutant KaiB with a C-terminal deletion from residues 95–108 (KaiB1–94) and a mutant KaiC with alanine (KaiCAA) or aspartate (KaiCDD) substitutions at the two KaiC phosphorylation sites using PCR-mediated site-directed mutagenesis and cloned them in the pGEX-6P-1 vector (GE Healthcare) as described previously (20).
To determine their purity, we subjected the purified proteins to SDS-PAGE on 15% gels (22) and stained the gels with Coomassie Brilliant Blue (CBB). We estimated protein concentrations using the Bio-Rad Protein Assay with BSA as the standard as previously described (20). Unless otherwise stated, KaiA, KaiB, KaiC, and SasA refer to the KaiA dimer, KaiB tetramer, KaiC hexamer, and SasA trimer (21), respectively, and their concentrations are expressed in terms of their oligomeric status.
Estimation of the Molecular Mass of KaiB by Gel Filtration Chromatography
We estimated the molecular mass of KaiB using analytical gel filtration chromatography on a Superdex 75 5/150GL column (GE Healthcare) equilibrated with 20 mm Tris-HCl buffer (pH 7.5) containing 150 mm NaCl (gel filtration buffer) at 4 °C using an ÄKTA explorer (GE Healthcare) and a Gel Filtration Calibration Kit LMW (GE Healthcare) for the molecular mass standards. We monitored the elution profiles of proteins by absorbance at 280 nm (A280) and performed all analyses in 20 mm Tris-HCl buffer (pH 7.5) containing 50 mm NaCl, 20 mm Tris-HCl buffer (pH 7.5) containing 500 mm NaCl, 20 mm MES-NaOH buffer (pH 6.5) containing 150 mm NaCl, and 20 mm Tris-HCl buffer (pH 8.5) containing 150 mm NaCl.
Sedimentation Equilibrium by Analytical Ultracentrifugation
We performed sedimentation equilibrium analytical ultracentrifugation using a Beckman Optima XL-A analytical ultracentrifuge with an An60Ti rotor. Samples were dialyzed against 20 mm HEPES-NaOH buffer (pH 7.5) containing 150 mm NaCl (sedimentation equilibrium buffer), which was used as the blank. We performed measurements at 20 °C at 36,000, 38,000, and 40,000 rpm for KaiB1–94 and at 20,000, 22,000, and 24,000 rpm for wild-type KaiB (KaiBWT). We monitored concentration profiles of the samples by A280 and recorded them at a spacing of 0.001 cm in step mode, with 20 averages per step. We analyzed equilibrium data with Beckman Optima XL-A/XL-I data analyses software, Version 6.04, which was provided as an add-on to Version 6.0 (Microcal Inc.), and we calculated global, single-species fits using different loading absorbance values at 280 nm (0.2, 0.25, and 0.3 for KaiB1–94 and 0.18 for KaiBWT). Based on the amino acid compositions of the proteins, we used the partial specific volumes 0.767 ml/g for KaiB1–94 and 0.759 ml/g for KaiBWT for the analyses.
Estimation of the Molecular Mass and Stoichiometry of the KaiB-KaiC Complex by Gel Filtration Chromatography
Reaction mixtures containing 5 μm KaiB1–94 or 2.5 μm KaiBWT and 1 μm KaiCDD in 20 mm HEPES-NaOH buffer (pH 7.5) containing 1 mm ATP, 5 mm MgCl2, and 150 mm NaCl (HEPES reaction buffer) were incubated at 40 °C for 18 h and analyzed by gel filtration chromatography on a Superdex 200/HR 10/300 column (GE Healthcare) equilibrated with HEPES reaction buffer containing 0.1 mm ATP and 5 mm MgCl2 at 4 °C. We used thyroglobulin (670 kDa), apoferritin (440 kDa), β-amylase (200 kDa), and BSA (66 kDa) as molecular mass standards. Fractions containing the KaiB-KaiCDD complex were subjected to SDS-PAGE on 18% gels, and then the gels were stained with CBB. We determined the amount of KaiB and KaiCDD contained in the complex by densitometry using a CS analyzer (ATTO) to determine the stoichiometry of the complex.
Labeling of KaiB with Cy3 and Fluorescence Correlation Spectroscopy (FCS) Measurements
Cy3-NHS fluorescent dye esters (GE Healthcare) were covalently coupled to the amines of lysine residues and the N-terminal amino acid residue of KaiB. KaiB (8 μm KaiB1–94 or 4 μm KaiBWT) was incubated with 80 μm Cy3-NHS ester in gel filtration buffer at 4 °C for 2 h. After labeling, the reaction mixtures were loaded on a PD MidiTrapTM G-25 column (GE Healthcare) for removal of the remaining unbound dye from the labeled proteins. We calculated the amounts of Cy3 introduced onto KaiB from the absorbance of the labeled proteins at 552 nm (A552). Under these conditions, 0.85 ± 0.10 and 0.64 ± 0.01 molecules of Cy3 (n = 3) per subunit of KaiB1–94 and KaiBWT were introduced into KaiB1–94 and KaiBWT, respectively.
We performed FCS measurements using a multiphoton FCS/fluorescence cross-correlation spectroscopy system using Fluctuation Dual Emission Uni-laser eXcitation (Fluc DEUXTM) (MBL, Nagoya, Japan). Cy3 was excited by the 445-nm laser, and its emission was detected at 615–690 nm. Cy3-KaiB1–94 and Cy3-KaiBWT (0.1 μm) were separately incubated with 0, 0.03, 0.05, 0.10, and 0.30 μm KaiCDD in the presence of 0, 0.05, 0.10, or 0.30 μm SasA in HEPES reaction buffer at 25 °C for 18 h, and the reaction mixtures were subjected to FCS measurements at 25 °C for 5 s (5 times consecutively). The diffusion time defined as the average time required for the diffusion of fluorescent particles across the detection area reflects the size of the particles. If the effect of the molecular shape on its diffusion is negligible, diffusion time is proportional to the cubic root of the molecular mass, as described in Equation 1 (23, 24),
 |
where DTa and DTb are the diffusion times of molecules a (Cy3-KaiB1–94 or Cy3-KaiBWT) and b (Cy3-KaiB1–94-KaiCDD complex or Cy3-KaiBWT-KaiCDD), respectively, and MMa and MMb are the molecular masses of molecules a and b, respectively. Accordingly, we calculated the size of the Cy3-KaiB1–94-KaiCDD and Cy3-KaiBWT-KaiCDD complexes to be 320 ± 70 and 370 ± 90 kDa, respectively, using the molecular masses of Cy3-KaiBWT (∼50 kDa) and Cy3-KaiB1–94 (∼25 kDa). These values are consistent with the molecular masses determined by gel filtration chromatography (see Fig. 2A).
FIGURE 2.
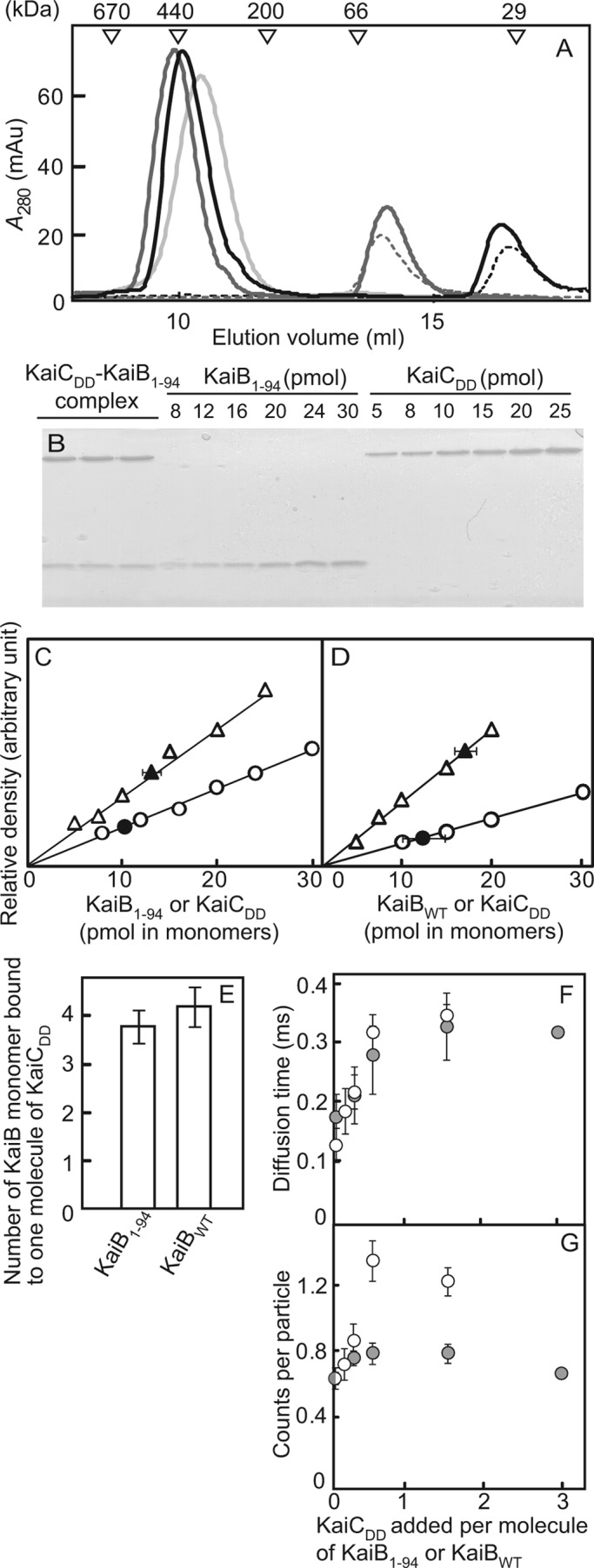
Stoichiometry of the KaiB-KaiC complex and the formation of the KaiB-KaiC complex analyzed by FCS analysis. A, shown are elution profiles. Reaction mixtures containing 5 μm KaiB1–94 or 2.5 μm KaiBWT and 1 μm KaiCDD were incubated at 40 °C for 18 h in HEPES reaction buffer and then subjected to gel filtration chromatography. Black line, KaiB1–94 + KaiCDD; gray line, KaiBWT + KaiCDD; light gray line, KaiCDD; black dotted line, KaiB1–94; gray dotted line, KaiBWT. mAu, milliabsorbance units. B, shown is SDS-PAGE. Aliquots of a fraction containing the KaiB1–94-KaiCDD complex or the KaiBWT-KaiCDD complex were subjected to SDS-PAGE. Standard amounts of KaiB1–94, KaiBWT, and KaiCDD were also applied to the gels. Only the SDS-PAGE gel of the KaiB1–94-KaiCDD complex is shown. Lanes 1–3, triplicate samples from the fraction containing the KaiB1–94-KaiCDD complex; lanes 4–9 contain 5–40 pmol of KaiB1–94; lanes 10–15 contain 5–40 pmol of KaiCDD. C, shown are calibration curves for the KaiB1–94-KaiCDD complex. The relative intensities of the bands were plotted against known amounts of KaiB1–94 and KaiCDD, and the KaiB1–94 and KaiCDD bands from the KaiB1–94-KaiCDD complex were plotted. Typical plots from three independent experiments are shown. Open circles, known amounts of KaiB1–94; open triangles, known amounts of KaiCDD; closed circles and closed triangles, KaiB1–94 and KaiCDD in the complex, respectively. D, calibration curves for the KaiBWT-KaiCDD complex are shown. The relative intensities of the bands were plotted against known amounts of KaiBWT and KaiCDD, and the KaiBWT and KaiCDD bands from the KaiBWT-KaiCDD complex were plotted. Typical plots from three independent experiments are shown. Open circles, known amounts of KaiBWT; open triangles, known amounts of KaiCDD; closed circles and closed triangles, KaiBWT and KaiCDD in the complex, respectively. E, stoichiometry of the KaiB-KaiCDD complex is shown. The number of molecules of KaiB1–94 or KaiBWT monomer per molecule of KaiCDD contained in the KaiB-KaiCDD complex was estimated from the plots shown in A–D. Values shown are the means ± S.D. F, KaiCDD-induced changes in the diffusion times of Cy3-KaiB1–94 and Cy3-KaiBWT molecules are shown. Cy3-KaiB1–94 (0.2 μm) and Cy3-KaiBWT (0.1 μm) were separately incubated with 0, 0.025, 0.05, 0.15, and 0.3 μm KaiCDD, and their diffusion times were measured by FCS analysis. Values shown are the means ± S.D. from quadruple measurements. Open circles, Cy3-KaiB1–94; gray circles, Cy3-KaiBWT. G, KaiCDD-induced changes in the counts per particle for the Cy3-KaiB1–94 and Cy3-KaiBWT molecules are shown. The counts per particle were calculated from the data shown in F.
Surface Plasmon Resonance Analysis of Interaction between KaiB and KaiC
We analyzed the interaction between KaiB1–94 and KaiCDD and between KaiBWT and KaiCDD at 25 °C by surface plasmon resonance (25) using a Biacore and its associated software (GE Healthcare). KaiCDD (10 μm) was immobilized on a CM5 sensor chip using an amine coupling kit (GE Healthcare) in HEPES reaction buffer containing 0.1 mm ATP, 0.1 mm dithiothreitol (DTT), and 50 mm NaCl. We monitored the association to, and dissociation from, the immobilized KaiCDD of KaiB1–94, KaiBWT, KaiA, and SasA continuously at stepwise concentrations of 0.1, 0.25, 0.5, 1.0, and 1.5 μm in HEPES reaction buffer. We computed the dissociation (kd) and association (ka) rates by fitting a 1:1 binding model to the experimental data. We calculated the value for apparent equilibrium dissociation constant (KD) as kd/ka.
Measurement of the Phosphorylation Levels of KaiC
Reaction mixtures of KaiA and KaiC (each 0.5 μm) were incubated in the presence or absence of 1.0 μm KaiB1–94 or 0.5 μm KaiBWT in gel filtration buffer containing 1 mm ATP and 5 mm MgCl2 at 25 or 40 °C, and 10-μl aliquots of the reaction mixtures were removed to stop the reaction at specific time intervals. The aliquots were subjected to SDS-PAGE on 12.5% gels (acrylamide:bisacrylamide = 144:1), and the gels were stained with CBB. The intensities of bands were measured by densitometry using a CS Analyzer. KaiC showed a triplet band on SDS-PAGE (4, 9); the two upper bands correspond to phosphorylated KaiC (p-KaiC), and the lowest band corresponds to nonphosphorylated KaiC (np-KaiC). We calculated the relative amount of p-KaiC to total KaiC (the level of p-KaiC) from band intensities and analyzed the circadian oscillations of the level of p-KaiC using the program RAP (26).
In Vivo Bioluminescence Rhythm Assay
We measured the bioluminescence rhythms of the T. elongatus strains carrying a PpsbA1::Xl luxAB reporter gene at 41 °C using a newly developed bioluminescence monitoring apparatus with a robotic plate conveyor. The apparatus was 1.3 times as sensitive to luminescence as our previous apparatuses (27). We analyzed bioluminescence data by a modified cosiner method of RAP as previously described (26, 28). We examined the wild-type strain, a kaiB-null strain carrying a mutant kaiB gene with a nonsense mutation downstream of its initiation codon, and a strain carrying an additional mutant kaiB gene encoding KaiB1–94, kaiB1–94, in the kaiB-null genetic background (16, 29).
Assay for the Complex Formation of SasA with KaiC by Gel Filtration Chromatography
Reaction mixtures containing 1.5 μm SasA and 0.6 μm KaiCDD were incubated in the presence or absence of 3.0 μm KaiBWT or 6.0 μm KaiB1–94 in HEPES reaction buffer containing 0.1 mm DTT at 25 °C for 18 h and analyzed by gel filtration chromatography on a Superdex 200/HR 10/300 column (GE Healthcare) equilibrated with HEPES reaction buffer containing 0.1 mm ATP, 5 mm MgCl2, and 0.1 mm DTT at 4 °C. Aliquots of the chromatography fractions were subjected to SDS-PAGE on 15% gels, and the gels were stained with CBB. We determined the amount of SasA in each fraction by densitometry.
RESULTS
Oligomeric Structures of KaiB1–94 and KaiBWT in Solution
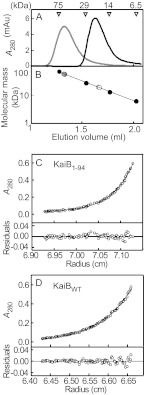
To determine the oligomeric structure of KaiB1–94 in solution, we estimated the molecular mass of KaiB1–94 by gel filtration chromatography in gel filtration buffer. KaiB1–94 (subunit molecular mass, 10.9 kDa) eluted as a single peak (Fig. 1A) corresponding to a 23.4 ± 0.4-kDa protein (Fig. 1B and Table 1) that corresponds to the approximate molecular mass of a dimer, whereas KaiBWT (subunit molecular mass, 12.4 kDa) eluted as a single peak (Fig. 1A) corresponding to a 55.4 ± 0.6-kDa protein (Fig. 1B and Table 1) that corresponds to the approximate molecular mass of a tetramer. We obtained similar results under varying buffer conditions (Table 1). In sedimentation equilibrium analysis, KaiB1–94 sedimented as a dimer with a molecular mass of 23 kDa (Fig. 1C and Table 1), and KaiBWT sedimented as a tetramer with a molecular mass of 48 kDa (Fig. 1D and Table 1), confirming the gel filtration chromatography results. Hereafter, when their concentrations are expressed, KaiBWT is considered a tetramer and KaiB1–94 a dimer.
FIGURE 1.
Determination of the oligomeric state of KaiB1–94 and KaiBWT. A, elution profiles of KaiB1–94 and KaiBWT by gel filtration chromatography are shown. KaiB1–94 and KaiBWT (the control) were subjected to analytical gel filtration chromatography at 4 °C on a Superdex75 5/150GL column equilibrated with 20 mm Tris-HCl buffer (pH 7.5) containing 150 mm NaCl. The elution positions of the molecular mass standards, conalbumin (75 kDa), carbonic anhydrase (29 kDa), ribonuclease (14 kDa), and aprotinin (6.5 kDa) are indicated above the elution profiles. Typical elution profiles from three independent experiments are shown. Black line, KaiB1–94; gray line, KaiBWT. mAu, milliabsorbance units. B, molecular masses of KaiB1–94 and KaiBWT were determined by gel filtration chromatography. Open circles, KaiB1–94; gray circles, KaiBWT; closed circles, the molecular mass standards. C and D, shown are sedimentation equilibrium analyses of KaiB1–94 and KaiBWT. The upper panels show the equilibrium profiles (○) displayed as the A280 (Au) versus the radial distance and theoretical concentration profiles (solid line) for a single molecular species with a molecular mass of 22 and 48 kDa, respectively. The lower panels show the residual plots from the curve fitting. C, KaiB1–94; D, KaiBWT.
TABLE 1.
Molecular mass of KaiB1–94
GF, gel filtration chromatography; SE, sedimentation equilibrium analysis; GFB, gel filtration buffer; SEB, sedimentation equilibrium buffer. Values for gel filtration chromatography are the means ± S.D. from triplicate measurements.
| KaiB | Method | Buffer | Molecular mass (n = 3) |
|---|---|---|---|
| kDa | |||
| KaiBWT | GF | GFB | 55.4 ± 0.6 |
| GF | SEB | 56.0 ± 1.0 | |
| KaiB1–94 | GF | GFB | 23.4 ± 0.4 |
| GF | SEB | 22.1 ± 0.4 | |
| GF | 20 mm Tris-HCl (pH 7.5), 50 mm NaCl | 22.5 ± 0.1 | |
| GF | 20 mm Tris-HCl (pH 7.5), 500 mm NaCl | 25.6 ± 0 | |
| GF | 20 mm MES-NaOH (pH 6.5), 150 mm NaCl | 24.0 ± 0.5 | |
| GF | 20 mm Tris-HCl (pH 8.5), 150 mm NaCl | 22.5 ± 0.5 | |
| KaiBWT | SE | SEB | 49.8 |
| KaiB1–94 | SE | SEB | 23.0 |
Complex Formation of KaiB1–94 and KaiBWT with KaiC and the Stoichiometry of the KaiB1–94-KaiCDD and KaiBWT-KaiCDD Complexes
First, we examined the complex formation of KaiB1–94 with KaiCWT, KaiCDD, and KaiCAA by FCS analysis (supplemental Fig. S1). The diffusion time of Cy3-KaiB1–94 incubated alone at 25 °C for 18 h was 0.13 ± 0.04 ms, and it increased in the presence of KaiCDD (0.37 ± 0.09 ms). On the other hand, the diffusion time scarcely increased in the presence of KaiCAA (0.16 ± 0.03 ms). These results indicate that KaiB1–94 formed a complex with KaiCDD, whereas it scarcely formed a complex with KaiCAA. Furthermore, the diffusion time of Cy3-KaiB1–94 increased in the presence of KaiCWT when the phosphorylation level of KaiCWT had been elevated to about 70% by KaiA (0.26 ± 0.03 ms), whereas it increased only slightly in the presence of KaiCWT when the phosphorylation level of KaiCWT (about 30%) had not been elevated by KaiA (0.18 ± 0.03 ms). Therefore, the complex formation of KaiB1–94 with KaiC depends on the phosphorylation level of KaiC; the higher the phosphorylation level, the more efficient complex formation occurred. Probably, KaiB1–94 associates with p-KaiC more strongly than with np-KaiC. These results are consistent with a previous observation that KaiBWT associates with p-KaiC more strongly than with np-KaiC (30, 31). Hereafter, we used KaiCDD to examine the complex formation of KaiB with KaiC because the phosphorylation level of KaiCWT changes during incubation, especially in the presence of KaiB and/or KaiA (Ref. 4 and see Fig. 3A).
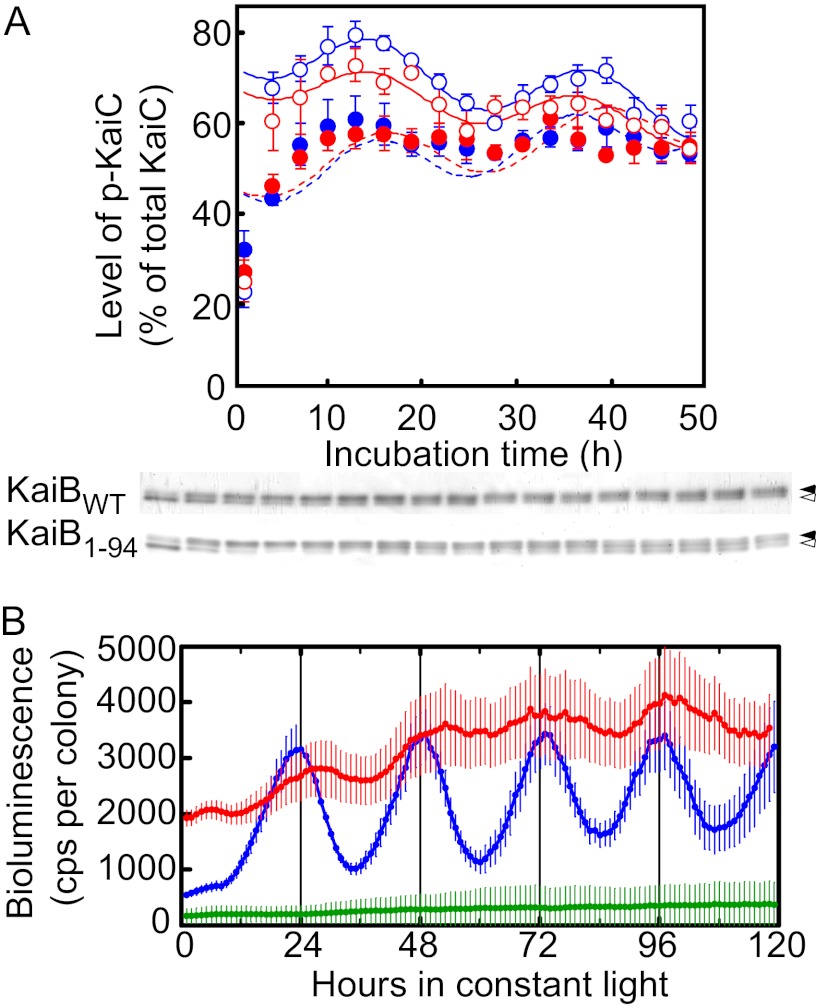
FIGURE 3.
Circadian oscillations in vitro and in vivo. A, shown are circadian oscillations in vitro. Generation of temperature-compensated circadian oscillations in vitro in the level of p-KaiC by KaiB1–94 in an in vitro KaiABC clock system is shown. Reaction mixtures containing KaiA, KaiBWT or KaiB1–94, and KaiCWT (0.5 μm each) were incubated in 20 mm Tris-HCl buffer (pH 7.5) containing 1 mm ATP, 5 mm MgCl2, and 150 mm NaCl at 25 °C or 40 °C for the periods indicated and were subjected to SDS-PAGE on 12.5% gels. The gels were stained with CBB, and the relative amounts of np-KaiC and p-KaiC were estimated by densitometry. Typical photographs from three independent experiments are shown. Circadian oscillations in the level of p-KaiC were analyzed by RAP, and the simulated curves are shown. Values shown are the means ± S.D. from triplicate measurements. KaiBs added: open red circles, KaiB1–94 (40 °C); open blue circles, KaiBWT (40 °C); closed red circles, KaiB1–94 (25 °C); closed blue circles, KaiBWT (25 °C). Closed arrows, p-KaiC; open arrows, np-KaiC. B, shown are circadian bioluminescence rhythms of a T. elongatus PpsbA1::Xl luxAB reporter strain carrying kaiBWT or kaiB1–94 in a kaiB-null genetic background. Bioluminescence rhythms of T. elongatus PpsbA1::Xl luxAB reporter strains carrying kaiBWT (WT, blue, n = 10), kaiB1–94 (KaiB1–94, red, n = 13), and no additional kaiB gene (KaiB-null, green, n = 75) in a kaiB-null background (the kaiB gene had a nonsense codon just downstream of its initiation codon) were measured at 41 °C. Data points and error bars indicate the mean ± S.D. from n independent samples. cps, counts per s.
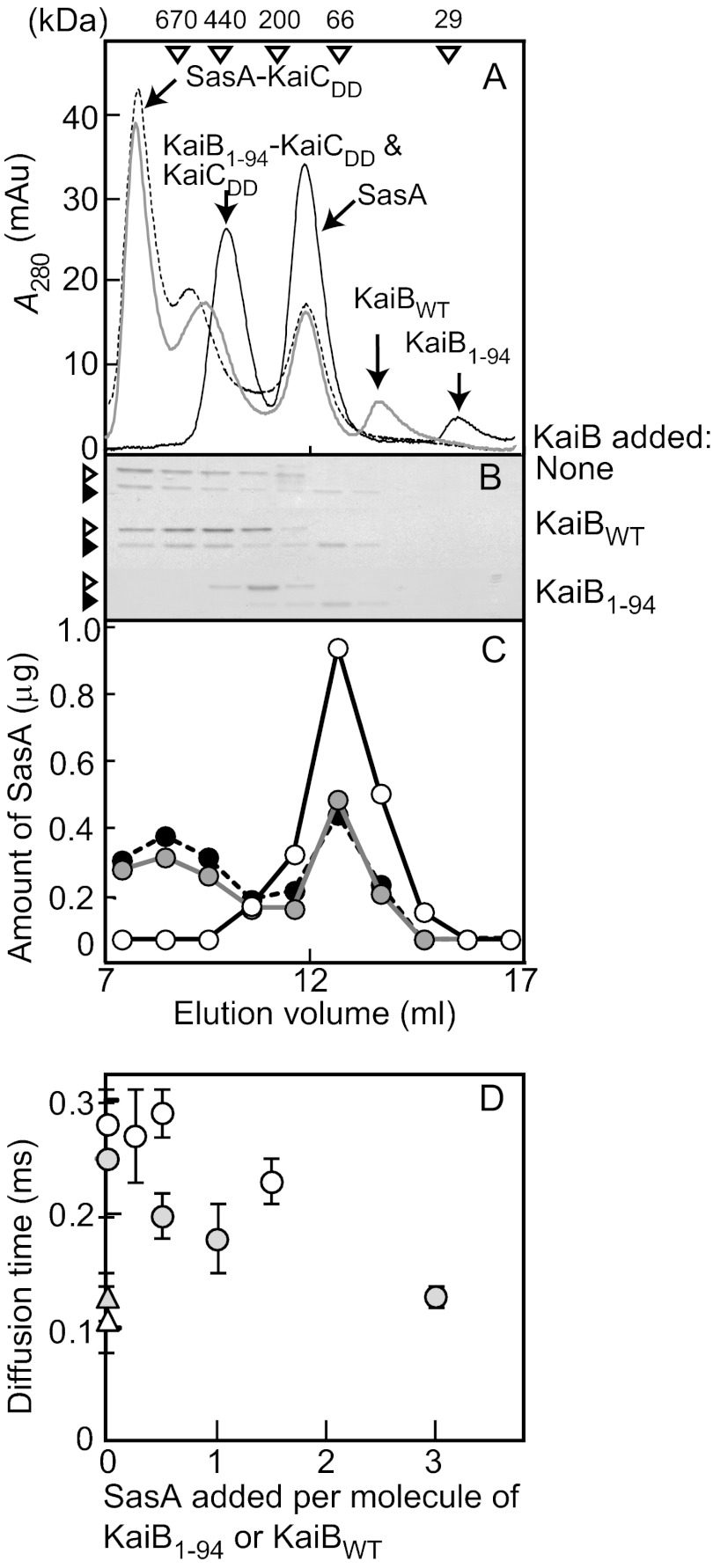
Next we examined the KaiB1–94-KaiCDD complex by gel filtration chromatography followed by SDS-PAGE. KaiB1–94 and KaiCDD each eluted as a single peak, KaiB1–94 with an apparent molecular mass of 25 ± 2 kDa and KaiCDD with an apparent molecular mass of 319 ± 7 kDa (n = 3) (Fig. 2A). When KaiCDD (1 μm) was incubated with an excess of KaiB1–94 (5 μm) at 40 °C for 18 h (n = 3), the reaction products eluted as two peaks, one with a molecular mass of 26 ± 1 kDa (KaiB1–94) and the other with a molecular mass of 350 ± 20 kDa (Fig. 2A), suggesting the formation of a KaiB1–94-KaiCDD complex. We also confirmed the formation of a KaiBWT-KaiCDD complex (366 ± 20 kDa) by gel filtration chromatography (Fig. 2A). The apparent molecular masses of the KaiB1–94-KaiCDD and KaiBWT-KaiCDD complexes were slightly higher than that of the KaiCDD hexamer (Fig. 2A).
To determine the stoichiometry of the KaiB1–94-KaiCDD complex, we collected it by gel filtration chromatography (Fig. 2A, 9.0–9.5 ml) and estimated the amount of KaiB1–94 and KaiCDD it contained by SDS-PAGE (Fig. 2B) followed by densitometry (Fig. 2, C and D). When KaiCDD (1 μm) was incubated with an excess of KaiB1–94 (5 μm) at 40 °C for 18 h, the fraction containing the complex contained 3.8 ± 0.3 subunits of KaiB1–94 per molecule of hexameric KaiCDD (Fig. 2E). When we applied the same procedure to the KaiBWT-KaiCDD complex, the stoichiometry showed that it contained 4.2 ± 0.4 subunits of KaiBWT per molecule of hexameric KaiCDD (Fig. 2E), indicating that the KaiB1–94-KaiCDD complex consisted of two molecules of KaiB1–94 (a dimer) and one molecule of hexameric KaiCDD, whereas the KaiBWT-KaiCDD complex consisted of one molecule of KaiBWT (a tetramer) and one molecule of hexameric KaiCDD.
We confirmed these results using FCS with Cy3-NHS fluorescent dye ester (Cy3)-labeled KaiB proteins (Cy3-KaiB1–94 and Cy3-KaiBWT). When 0.2 μm Cy3-KaiB1–94 and 0.1 μm Cy3-KaiBWT were incubated in the presence of 0, 0.025, 0.05, 0.15, or 0.3 μm KaiCDD, the diffusion time varied directly with the KaiCDD concentration (Fig. 2F), indicating complex formation between KaiCDD and Cy3-KaiB1–94 or Cy3-KaiBWT.
The counts per particle of Cy3-KaiB1–94 (0.2 μm) also varied with KaiCDD concentration (Fig. 2G). The counts per particle values of Cy3-KaiB1–94 in the presence of KaiCDD at molar ratios greater than 0.5 were approximately twice those in its absence, which suggested that one molecule of the Cy3-KaiB1–94-KaiCDD complex contained two molecules of the Cy3-KaiB1–94 dimer (Fig. 2G). In contrast, counts per particle did not significantly change when Cy3-KaiBWT (0.1 μm) was incubated with an excess of KaiCDD (0.3 μm), which suggests that one molecule of the KaiBWT-KaiCDD complex contained one molecule of the KaiBWT tetramer (Fig. 2G).
Interaction of KaiB1–94 Versus KaiBWT with KaiCDD
Surface plasmon resonance analysis showed that the association rate (ka) of KaiCDD was 8 times as high with KaiB1–94 as with KaiBWT, and its dissociation rate (kd) was 0.6 times as high (Table 2). Thus, the binding affinity of KaiB1–94 for KaiCDD was 15 times the binding affinity of KaiBWT. We also examined the binding affinities of KaiA and SasA, other known KaiC-binding proteins, and determined that KaiB1–94 had the highest binding affinity in the group (Table 2).
TABLE 2.
Kinetic and steady state parameters for the interaction of KaiBWT, KaiB1–94, KaiA, and SasA with KaiCDD obtained by surface plasmon resonance analysis
Values are the means ± S.D. from triplicate measurements.
| Protein | ka | kd | KD |
|---|---|---|---|
| m−1s−1 (×104) | s−1 (×10−3) | m (×10−8) | |
| KaiBWT | 1.3 ± 0.057 | 0.46 ± 0.058 | 3.5 ± 0.38 |
| KaiB1–94 | 10.4 ± 0.61 | 0.26 ± 0.040 | 0.24 ± 0.029 |
| KaiA | 3.9 ± 1.0 | 2.5 ± 0.10 | 6.8 ± 1.4 |
| SasA | 7.7 ± 1.1 | 1.2 ± 0.066 | 1.5 ± 0.24 |
Circadian Oscillations in the Level of p-KaiC in an in Vitro KaiABC Clock System and Circadian Gene Expression Rhythms in T. elongatus Cells
When 0.5 μm KaiA, KaiB (KaiB1–94 or KaiBWT), KaiC, and ATP were incubated together at 25 or 40 °C in an in vitro KaiABC clock system (4), the level of p-KaiC oscillated in the presence of KaiB1–94 and KaiBWT (Fig. 3A). At 40 °C, the period length was 21.7 ± 0.9 h for the oscillations driven by KaiB1–94 and 22.6 ± 0.2 h for those driven by KaiBWT (Fig. 3A), which was consistent with the circadian oscillations observed in T. elongatus cells (24.4 ± 0.3 h) (Ref. 16 and Fig. 3B). Thus, KaiB1–94, like KaiBWT, can generate circadian oscillations in the in vitro KaiABC clock system. At 25 °C, the period length was 21.7 ± 1.3 h for the oscillations driven by KaiB1–94 and 23.3 ± 0.5 h for those driven by KaiBWT (Fig. 3A), indicating that the period lengths were temperature-compensated in our system.
Circadian rhythms were normal in the kaiB-null host cells carrying kaiBWT but showed a greatly reduced amplitude in those carrying kaiB1–94 (Fig. 3B).
Inhibition of SasA-KaiCDD Complex Formation by KaiB1–94 and KaiBWT
To determine why cyanobacterial cells expressing KaiB1–94 showed weakened circadian gene expression rhythms even though KaiB1–94 is able to generate normal circadian oscillations in vitro, we examined the effects of KaiB on the SasA-mediated clock output pathway, that is, on the formation of the SasA-KaiC complex.
Gel filtration chromatography yielded only a single peak for both KaiCDD and SasA corresponding to a 340 ± 20-kDa protein and a 140 ± 10-kDa protein, respectively. When reaction mixtures containing 1.5 μm SasA and 0.6 μm KaiCDD were incubated in the absence or presence of 3 μm KaiBWT or 6 μm KaiB1–94 at 25 °C for 18 h, the elution profile of the sample without KaiB showed 3 peaks (Fig. 4A) that we analyzed by SDS-PAGE (Fig. 4B). Peak 1 (>670 kDa, the molecular mass of the largest standard used) corresponded to the SasA-KaiCDD complex, peak 2 (440 ± 50 kDa) corresponded to the other form of the SasA-KaiCDD complex, and peak 3 (130 ± 5 kDa) corresponded to SasA. The amount of SasA contained in each fraction estimated by SDS-PAGE followed by densitometry showed that 34 ± 10% (n = 3) of the total SasA was unbound (Fig. 4C).
FIGURE 4.
Inhibition of the complex formation of SasA with KaiCDD by KaiB1–94 and KaiBWT. A, elution profiles of gel filtration chromatography are shown. Reaction mixtures containing 1.5 μm SasA and 0.6 μm KaiCDD in HEPES reaction buffer containing 0.1 mm DTT were incubated in the presence or absence of 6.0 μm KaiB1–94 or 3.0 μm KaiBWT at 25 °C for 18 h and were analyzed by gel filtration chromatography. Other conditions were as described in the Fig. 2 legend. Typical plots from three independent experiments are shown. KaiBs added: black solid line, KaiB1–94; gray solid line, KaiBWT; black dotted line, none. mAu, milliabsorbance units. B, shown is SDS-PAGE. Aliquots of the fractions (elution volumes, 7–17 ml) were subjected to SDS-PAGE on 15% gels, and the gels were visualized by CBB staining. Open arrowheads indicate KaiCDD, and closed arrowheads indicate SasA. C, shown is the amount of SasA. The amount of SasA in each fraction was estimated with SDS-PAGE (Fig. 4B) using densitometry. KaiBs added: closed circles and black dotted line, none; open circles and black solid line, KaiB1–94; gray circles and gray solid line, KaiBWT. D, shown are SasA-induced changes in the diffusion times of the Cy3-KaiB1–94 molecule and the Cy3-KaiBWT molecule in the presence of KaiCDD. Reaction mixtures containing Cy3-KaiB (0.2 μm Cy3-KaiB1–94 or 0.1 μm Cy3-KaiBWT) and 0.1 μm KaiCDD were incubated in the presence or absence of the indicated amounts of SasA in HEPES reaction buffer containing 0.1 mm DTT at 25 °C for 18 h, and then the diffusion time was measured. The diffusion times of Cy3-KaiB1–94 and Cy3-KaiBWT in the absence of KaiCDD and SasA were measured as controls. Values shown are the means ± S.D. from quadruple measurements. Other conditions were the same as described in the Fig. 2F legend. Open circles, Cy3-KaiB1–94; open triangles, Cy3-KaiB1–94 without KaiCDD and SasA; gray circles, Cy3-KaiBWT; gray triangles, Cy3-KaiBWT without KaiCDD and SasA.
The elution profile of the sample with 6 μm KaiB1–94 showed three peaks (Fig. 4A); peak 1 (360 ± 10 kDa) corresponded to KaiCDD and the KaiB1–94-KaiCDD complex, peak 2 (140 ± 10 kDa) corresponded to SasA, and peak 3 (20 ± 0 kDa) corresponded to KaiB1–94. The presence of KaiB1–94 all but extinguished the SasA-KaiCDD complex and greatly increased free SasA (Fig. 4A). The free SasA fractions contained 92 ± 10% (n = 3) of the total SasA added (Fig. 4, B and C), which indicated nearly complete inhibition of SasA-KaiCDD complex formation by KaiB1–94. In the presence of 3 μm KaiBWT, the elution profile of the sample showed four peaks (Fig. 4A) corresponding to the SasA-KaiCDD complex (>670 kDa), the other form of the SasA-KaiCDD complex, KaiCDD and KaiBWT-KaiCDD complex (420 ± 40 kDa), SasA (140 ± 10 kDa), and KaiBWT (50 ± 0 kDa). The presence of KaiBWT slightly reduced the SasA-KaiCDD complex peak and slightly increased the free SasA peak; the free SasA fractions contained 43 ± 5% (n = 3) of the total SasA added (control 34 ± 10%) (Fig. 4C), which suggests that KaiBWT inhibited formation of the SasA-KaiCDD complex, albeit very slightly. Thus, KaiB1–94 was a stronger inhibitor of SasA-KaiCDD complex formation than KaiBWT.
We also examined KaiB inhibition of SasA-KaiCWT complex formation by FCS analysis with Cy3-SasA (supplemental Fig. S2). The diffusion time of Cy3-SasA (0.2 μm) incubated alone at 25 °C for 18 h was 0.14 ± 0.00 ms. On the other hand, when Cy3-SasA was incubated with 0.2 μm KaiCWT, its diffusion time increased to 0.30 ± 0.06 ms, indicating formation of a Cy3-SasA-KaiCWT complex. The addition of 2.0 μm KaiB1–94 almost completely canceled this diffusion time increase (0.16 ± 0.03 ms), indicating that KaiB1–94 inhibited SasA-KaiCWT complex formation as well as SasA-KaiCDD complex formation (see above). However, the addition of KaiBWT did not cancel the diffusion time increase under the conditions examined (diffusion time: 0.35 ± 0.03 and 0.33 ± 0.01 ms in the presence of 1.0 μm KaiBWT and 2.0 μm KaiBWT, respectively) (supplemental Fig. S2).
We examined SasA inhibition of KaiB-KaiCDD complex formation by FCS analysis with Cy3-KaiB1–94 and Cy3-KaiBWT (Fig. 4D). In the absence of KaiCDD, the diffusion time was 0.13 ± 0.02 ms for 0.2 μm Cy3-KaiB1–94 and 0.11 ± 0.03 ms for 0.1 μm Cy3-KaiBWT. In the presence of 0.1 μm KaiCDD, it increased to 0.28 ± 0.03 ms for Cy3-KaiB1–94 and 0.25 ± 0.05 ms for Cy3-KaiBWT (Fig. 4D), indicating formation of the Cy3-KaiB1–94-KaiCDD and Cy3-KaiBWT-KaiCDD complexes. The diffusion time of 0.2 μm Cy3-KaiB1–94 was slightly shorter in the presence of 0.3 μm SasA (0.23 ± 0.02 ms) than in its absence (0.28 ± 0.03) (Fig. 4D), which indicates that SasA weakly inhibited formation of the KaiB1–94-KaiCDD complex. In the presence of KaiCDD, on the other hand, SasA reduced the diffusion time of 0.1 μm Cy3-KaiBWT from 0.25 ± 0.05 to 0.13 ± 0.01 ms, the level it was in the absence of KaiCDD (0.11 ± 0.03 ms) (Fig. 4D), which indicates that SasA almost completely inhibited formation of the KaiBWT-KaiCDD complex. Thus, SasA inhibited KaiBWT-KaiCDD complex formation more strongly than KaiB1–94-KaiCDD complex formation.
DISCUSSION
That KaiB1–94 is a dimer (Fig. 1 and Table 1) but interacts with KaiCDD in much the same way as tetrameric KaiBWT (Fig. 2) and can generate normal circadian oscillations in vitro (Fig. 3A) indicates that KaiB tetramericity is not required for the generation of circadian oscillations per se. The tetrameric structure of KaiB, however, and its negatively charged C-terminal region seem to play an important role in the generation of circadian gene expression rhythms in vivo because the circadian bioluminescence rhythms of cyanobacterial cells expressing KaiB1–94 in a ΔkaiB genetic background are greatly weakened (Ref. 16 and Fig. 3B). We demonstrated that whereas KaiBWT inhibits the formation of the SasA-KaiC complex slightly, KaiB1–94 inhibits it strongly (Fig. 4 and supplemental Fig. S2), probably because of the high affinity of KaiB1–94 for KaiCDD (Table 2).
SasA is a KaiC binding histidine kinase in the SasA-RpaA two-component regulatory system (32, 33) and is involved in the main clock output pathway. Ultimately, the time information of the KaiABC clock oscillator is transmitted to the transcription apparatus and generates genome-wide circadian transcription rhythms in cyanobacteria. Disruption of the sasA gene greatly lowers the level of kaiBC expression and the amplitude of kaiA and kaiBC expression rhythms (32). KaiB1–94 competitively inhibits the SasA-KaiC interaction more strongly than KaiBWT does due to its higher affinity for KaiC (Table 2); it is likely, therefore, that KaiB1–94 hinders the SasA-KaiC interaction that is necessary for the transmission of the time information from the KaiABC clock oscillator to the transcription apparatus through the SasA-mediated clock output pathway (32, 33). We hypothesize that the defective rhythm phenotypes of cyanobacterial cells expressing KaiB1–94 in a ΔkaiB genetic background follow from a KaiB1–94-induced defect in the SasA-KaiC interaction involved in the SasA-mediated clock output pathway.
KaiB (KaiB1–94 and KaiBWT) inhibited formation of the SasA-KaiC (KaiCDD and KaiCWT) complex (Fig. 4, A–C, and supplemental Fig. S2), and SasA inhibited formation of the KaiB (KaiB1–94 and KaiBWT)-KaiCDD complex (Fig. 4D). It is unknown whether the two proteins compete for an identical binding site on KaiC because the binding sites of KaiB and SasA on KaiC are not determined. Previously, the N-terminal domain of SasA was suggested to be homologous to KaiB (32). However, comparison between the NMR structure of SasA N-terminal domain and the x-ray crystal structure of KaiB demonstrated that KaiB and SasA N-terminal domain have different folds (34). Therefore, it is not likely that SasA and KaiB bind to the same site on KaiC. We have demonstrated by surface plasmon resonance analysis and gel filtration chromatography that the SasA binding site of KaiC is located on its N-terminal domain (21), as suggested by NMR analysis (35). The SasA inhibition of KaiBWT-KaiCWT complex formation has been demonstrated by Native-PAGE using proteins derived from Synechococcus (36).
Our demonstration that two molecules of KaiB1–94 bind to one molecule of KaiCDD (Fig. 2) suggests that two molecules of KaiB1–94 dimer bind to one molecule of a KaiC hexamer. Because one molecule of KaiBWT binds to one molecule of KaiCDD (Fig. 2), it is likely that upon interaction with KaiC, the KaiB molecule changes from a tetramer to a dimer. This is consistent with the cryo-electron microscopy analysis of the KaiBWT-KaiCWT complex that revealed the possibility of KaiB binding to KaiC as two dimers (37). Based on the crystal structure of KaiB, we have proposed that the interdimer interface of the KaiB tetrameric molecule could be destabilized by changes in pH or ionic strength and dissociate into two dimers (16).
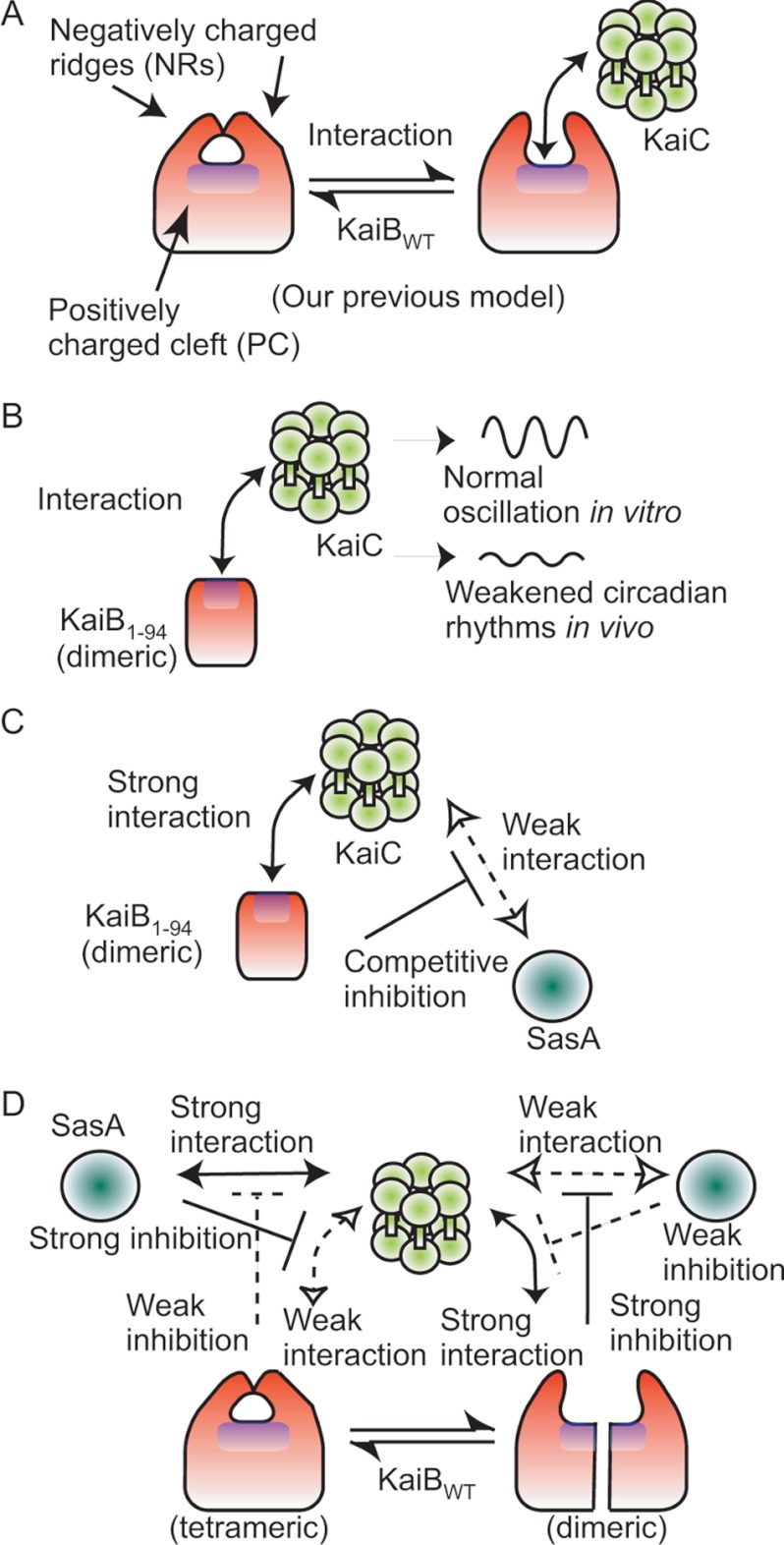
On one surface of the KaiB tetramer, a PC, which is the active site, is covered with two NRs, and the PC may be exposed for ligand binding by NR movement (Fig. 5A) (16). In agreement with this model, KaiB1–94, which lacks NRs and has a continuously exposed PC, is a more stable binding partner for KaiCDD than KaiBWT (Table 2). It is likely that PC exposure by NR movement may play an important role in the interaction of KaiB with KaiC (Fig. 5, A and B). An alternative model is that KaiB associates with KaiC using a surface, which is exposed by the deletion of NRs on KaiB or the dimerization of KaiB. There are two areas on KaiC that are highly negatively charged, one around the pore opening and inside the pore of KaiC N-terminal domain and the other around the intersubunit interface of one of two adjacent KaiC N-terminal domains (Ref. 38 and supplemental Fig. S3). Possible electrostatic interaction between the PC on KaiB and the negatively charged surfaces of KaiC might occur.
FIGURE 5.
Model for the conformational change of KaiB. On one surface of the KaiB tetramer, a PC is covered with two NRs, and the active sites (16) and KaiA-interacting site (17) are located on the PC. Therefore, we hypothesize that the PC is exposed for ligand binding by NR movement (Refs. 16 and 39; A). The PC on KaiB1–94, on the other hand, is always exposed, which allows it to interact with KaiC because it lacks the NRs and exists as a dimer (B). KaiB1–94 is more stable than KaiBWT as a binding partner for KaiC, which suggests that the KaiB binding site is located on the PC and that the PC may be exposed to KaiC by NR movement (A). Thus, KaiB1–94 competitively inhibits the interaction of SasA with KaiC by its strong interaction with KaiC, which is necessary for the generation of normal circadian gene expression rhythms in cyanobacterial cells (C). On interaction with KaiC, the KaiB tetramer molecule may dissociate into two dimeric molecules that bind to one hexameric KaiC molecule (D). The tetrameric structure and NRs of the KaiB molecule are necessary for normal circadian gene expression rhythms in cells (D) but not for circadian oscillations in the in vitro KaiABC clock system.
Our demonstration that the stable interaction of KaiB1–94 with KaiC inhibited the interaction of SasA with KaiC (Fig. 4) indicated that the C-terminal region of KaiB is necessary for driving normal circadian transcription rhythms through regulation of the KaiC-SasA interaction (Fig. 5, C and D). Furthermore, our recent observations are consistent with our model as follows. Electron spin resonance analysis demonstrated that the mobility of the spin labels introduced into the amino acid residues located on the PC greatly increased on incubation at 40 °C (39), which indicates that the local structure surrounding the spin labels on the PC is relaxed during incubation (39); the 64th residue located inside the KaiB molecule near the PC interacts with KaiA (17).
In the KaiABC clock oscillator, KaiA associates preferentially with np-KaiC (40) (KaiCAA mimics np-KaiC) to elevate the phosphorylation level of KaiC (11–13). Then, KaiA dissociates from the p-KaiC (KaiCDD mimics completely phosphorylated KaiC) (40). In contrast, KaiB associates preferentially with the p-KaiC (Ref. 6 and supplemental Fig. S1) and then reduces the phosphorylation level of KaiC (12, 14). A series of these reactions and interactions among Kai proteins generate circadian oscillations such as oscillations in the phosphorylation level (4) and ATPase activity (5) of KaiC and complex formation among Kai proteins (6, 7). SasA also associates preferentially with p-KaiC, which enhances the autophosphorylation of SasA (21). KaiB and SasA compete each other for KaiC (Fig. 4 and supplemental Fig. S2). Therefore, it is likely that KaiB regulates the normal SasA-KaiC interaction required for the transmission of time information from KaiC to the transcription apparatus, resulting in genome-wide transcription cycles in cyanobacterial cells (21, 32, 33). SasA trimer and KaiC hexamer associate each other at a 1:1 molar ratio at their N-terminal domains, and the phosphorylation states of their C-terminal domains affect the affinity of the interaction (21). Interestingly, the phosphorylation state of KaiC also affects the rigidity of the C-terminal domains of KaiC hexamer and the interaction between the N-terminal domains and C-terminal domains of the KaiC, and KaiC associates with KaiB when the structure of KaiC C-terminal domains is rigid (35).
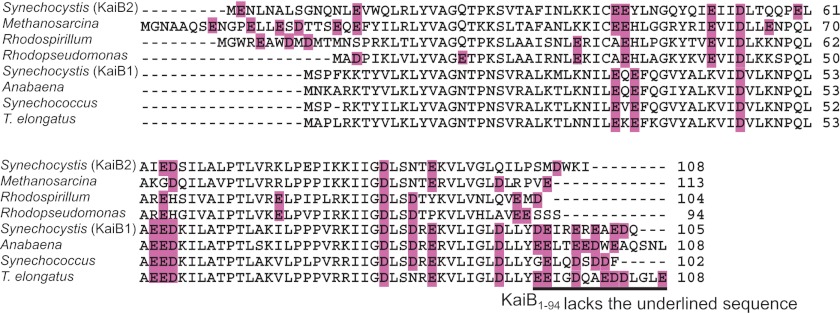
KaiB and KaiC occur in Archaea and Proteobacteria as well as in cyanobacteria (41). The C-terminal cluster of negatively charged residues is highly conserved in cyanobacterial clock KaiBs (Ref. 16 and Fig. 6) but not in cyanobacterial non-clock KaiBs (such as Synechocystis KaiB2 (41)) and not in KaiB homologues in Archaea and Proteobacteria (Fig. 6). The negatively charged C terminus of KaiB likely plays an important role in the SasA-mediated clock output pathway through KaiB-KaiC interactions affecting the SasA-KaiC interaction. SasA is present only in cyanobacteria, which suggests that the negatively charged C-terminal region of cyanobacterial KaiB may have co-evolved with SasA as a component of the cyanobacterial circadian system.
FIGURE 6.
Amino acid sequence alignment of KaiB. We aligned the amino acid sequences of KaiB from three strains of cyanobacteria, two strains of archaea, and two strains of green-sulfur bacteria using ClustalX (42). The negatively charged residues (glutamate and aspartate are shaded in purple). The C-terminal deletion in KaiB1–94 is indicated by underlining. Cyanobacterial strains: Synechococcus sp. strain PCC 7942; T. elongatus BP-1; Anabaena sp. strain PCC 7120; Synechocystis sp. PCC 6803 KaiB1; Synechocystis sp. PCC 6803 KaiB2. Archaeal strain: Methanosarcina mazei Goe1. Proteobacterial strains: Rhodospirillum rubrum ATCC 11170; Rhodopseudomonas palustris CGA009.
Supplementary Material
Acknowledgments
We thank Satoko Ogawa and Kumiko Tanaka for technical support and Miriam Bloom (SciWrite Biomedical Writing and Editing Services) for professional editing. Development of the automated bioluminescence monitoring apparatus CL96–4 and the plate conveyor robot CI-08L were supported by SENTAN, JST. The apparatuses CL96–4 and CI-08L are commercially available through Churitsu Electric Corp. (Nagoya, Japan).
This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to M. I. and K. O.).

This article contains supplemental Figs. S1–S3.
- PC
- the positively charged cleft of KaiB
- NRs
- the two negatively charged ridges of KaiB
- p-KaiC
- phosphorylated KaiC
- CBB
- Coomassie Brilliant Blue
- FCS
- fluorescence correlation spectroscopy
- KaiB1–94
- a truncated mutant KaiB with a C-terminal deletion from residues 95 to 108
- KaiBWT
- wild-type KaiB
- KaiCAA
- a mutant KaiC with alanine substitutions at the two KaiC phosphorylation sites
- KaiCDD
- a mutant KaiC with aspartate substitutions at the two KaiC phosphorylation sites
- np-KaiC
- nonphosphorylated KaiC
- KD
- apparent equilibrium dissociation constant
- Synechococcus
- Synechococcus sp. strain PCC 7942.
REFERENCES
- 1. Bünning E. (1973) The Physiological Clock. Circadian Rhythms and Biological Chronometry, 3rd Ed., Springer-Verlag, New York, NY [Google Scholar]
- 2. Sweeney B. M., Borgese M. B. (1989) A circadian rhythm in cell division in a prokaryote, the cyanobacterium Synechococcus WH7803. J. Phycol. 25, 183–186 [Google Scholar]
- 3. Ishiura M., Kutsuna S., Aoki S., Iwasaki H., Andersson C. R., Tanabe A., Golden S. S., Johnson C. H., Kondo T. (1998) Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523 [DOI] [PubMed] [Google Scholar]
- 4. Nakajima M., Imai K., Ito H., Nishiwaki T., Murayama Y., Iwasaki H., Oyama T., Kondo T. (2005) Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308, 414–415 [DOI] [PubMed] [Google Scholar]
- 5. Terauchi K., Kitayama Y., Nishiwaki T., Miwa K., Murayama Y., Oyama T., Kondo T. (2007) ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 104, 16377–16381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kageyama H., Nishiwaki T., Nakajima M., Iwasaki H., Oyama T., Kondo T. (2006) Cyanobacterial circadian pacemaker. Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol. Cell 23, 161–171 [DOI] [PubMed] [Google Scholar]
- 7. Akiyama S., Nohara A., Ito K., Maéda Y. (2008) Assembly and disassembly dynamics of the cyanobacterial periodosome. Mol. Cell 29, 703–716 [DOI] [PubMed] [Google Scholar]
- 8. Murakami R., Miyake A., Iwase R., Hayashi F., Uzumaki T., Ishiura M. (2008) ATPase activity and its temperature compensation of the cyanobacterial clock protein KaiC. Genes Cells 13, 387–395 [DOI] [PubMed] [Google Scholar]
- 9. Nishiwaki T., Iwasaki H., Ishiura M., Kondo T. (2000) Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 97, 495–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayashi F., Iwase R., Uzumaki T., Ishiura M. (2006) Hexamerization by the N-terminal domain and intersubunit phosphorylation by the C-terminal domain of cyanobacterial circadian clock protein KaiC. Biochem. Biophys. Res. Commun. 348, 864–872 [DOI] [PubMed] [Google Scholar]
- 11. Iwasaki H., Nishiwaki T., Kitayama Y., Nakajima M., Kondo T. (2002) KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 99, 15788–15793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Williams S. B., Vakonakis I., Golden S. S., LiWang A. C. (2002) Structure and function from the circadian clock protein KaiA of Synechococcus elongatus. A potential clock input mechanism. Proc. Natl. Acad. Sci. U.S.A. 99, 15357–15362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uzumaki T., Fujita M., Nakatsu T., Hayashi F., Shibata H., Itoh N., Kato H., Ishiura M. (2004) Crystal structure of the C-terminal clock-oscillator domain of the cyanobacterial KaiA protein. Nat. Struct. Mol. Biol. 11, 623–631 [DOI] [PubMed] [Google Scholar]
- 14. Kitayama Y., Iwasaki H., Nishiwaki T., Kondo T. (2003) KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 22, 2127–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hitomi K., Oyama T., Han S., Arvai A. S., Getzoff E. D. (2005) Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm. J. Biol. Chem. 280, 19127–19135 [DOI] [PubMed] [Google Scholar]
- 16. Iwase R., Imada K., Hayashi F., Uzumaki T., Morishita M., Onai K., Furukawa Y., Namba K., Ishiura M. (2005) Functionally important substructures of circadian clock protein KaiB in a unique tetramer complex. J. Biol. Chem. 280, 43141–43149 [DOI] [PubMed] [Google Scholar]
- 17. Mutoh R., Mino H., Murakami R., Uzumaki T., Takabayashi A., Ishii K., Ishiura M. (2010) Direct interaction between KaiA and KaiB revealed by a site-directed spin labeling electron spin resonance analysis. Genes Cells 15, 269–280 [DOI] [PubMed] [Google Scholar]
- 18. Yamaoka T., Satoh K., Katoh S. (1978) Photosynthetic activities of a thermophilic blue-green alga. Plant Cell Physiol. 19, 943–954 [Google Scholar]
- 19. Hayashi F., Suzuki H., Iwase R., Uzumaki T., Miyake A., Shen J. R., Imada K., Furukawa Y., Yonekura K., Namba K., Ishiura M. (2003) ATP-induced hexameric ring structure of the cyanobacterial circadian clock protein KaiC. Genes Cells 8, 287–296 [DOI] [PubMed] [Google Scholar]
- 20. Hayashi F., Itoh N., Uzumaki T., Iwase R., Tsuchiya Y., Yamakawa H., Morishita M., Onai K., Itoh S., Ishiura M. (2004) Roles of two ATPase-motif-containing domains in cyanobacterial circadian clock protein KaiC. J. Biol. Chem. 279, 52331–52337 [DOI] [PubMed] [Google Scholar]
- 21. Valencia S. J., Bitou K., Ishii K., Murakami R., Morishita M., Onai K., Furukawa Y., Imada K., Namba K., Ishiura M. (2012) Phase-dependent generation and transmission of time information by the KaiABC circadian clock oscillator through SasA-KaiC interaction in cyanobacteria. Genes Cells 17, 398–419 [DOI] [PubMed] [Google Scholar]
- 22. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 23. Eigen M., Rigler R. (1994) Sorting single molecules. Application to diagnostics and evolutionary biotechnology. Proc. Natl. Acad. Sci. U.S.A. 91, 5740–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Takahashi Y., Okamoto Y., Popiel H. A., Fujikake N., Toda T., Kinjo M., Nagai Y. (2007) Detection of polyglutamine protein oligomers in cells by fluorescence correlation spectroscopy. J. Biol. Chem. 282, 24039–24048 [DOI] [PubMed] [Google Scholar]
- 25. Karlsson R., Katsamba P. S., Nordin H., Pol E., Myszka D. G. (2006) Analyzing a kinetic titration series using affinity biosensors. Anal. Biochem. 349, 136–147 [DOI] [PubMed] [Google Scholar]
- 26. Okamoto K., Onai K., Ishiura M. (2005) RAP, an integrated program for monitoring bioluminescence and analyzing circadian rhythms in real time. Anal. Biochem. 340, 193–200 [DOI] [PubMed] [Google Scholar]
- 27. Okamoto K., Onai K., Furusawa T., Ishiura M. (2005) A portable integrated automatic apparatus for the real-time monitoring of bioluminescence in plants. Plant Cell Environ. 28, 1305–1315 [Google Scholar]
- 28. Onai K., Ishiura M. (2005) PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10, 963–972 [DOI] [PubMed] [Google Scholar]
- 29. Onai K., Morishita M., Itoh S., Okamoto K., Ishiura M. (2004) Circadian rhythms in the thermophilic cyanobacterium Thermosynechococcus elongatus. Compensation of period length over a wide temperature range. J. Bacteriol. 186, 4972–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nishiwaki T., Satomi Y., Nakajima M., Lee C., Kiyohara R., Kageyama H., Kitayama Y., Temamoto M., Yamaguchi A., Hijikata A., Go M., Iwasaki H., Takao T., Kondo T. (2004) Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc. Natl. Acad. Sci. U.S.A. 101, 13927–13932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu Y., Mori T., Pattanayek R., Pattanayek S., Egli M., Johnson C. H. (2004) Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc. Natl. Acad. Sci. U.S.A. 101, 13933–13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iwasaki H., Williams S. B., Kitayama Y., Ishiura M., Golden S., S., Kondo T. (2000) A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 101, 223–233 [DOI] [PubMed] [Google Scholar]
- 33. Takai N., Nakajima M., Oyama T., Kito R., Sugita C., Sugita M., Kondo T., Iwasaki H. (2006) A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 12109–12114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vakonakis I., Klewer D. A., Williams S. B., Golden S. S., LiWang A. C. (2004) Structure of the N-terminal domain of the circadian clock-associated histidine kinase SasA. J. Mol. Biol. 342, 9–17 [DOI] [PubMed] [Google Scholar]
- 35. Chang Y. G., Kuo N. W., Tseng R., LiWang A. (2011) Flexibility of the C-terminal, or CII, ring of KaiC governs the rhythm of the circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 14431–14436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pattanayek R., Williams D. R., Rossi G., Weigand S., Mori T., Johnson C. H., Stewart P. L., Egli M. (2011) Combined SAXS/EM-based models of the S. elongatus post-translational circadian oscillator and its interactions with the output His-kinase SasA. PLoS One 6, e23697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pattanayek R., Williams D. R., Pattanayek S., Mori T., Johnson C. H., Stewart P. L., Egli M. (2008) Structural model of the circadian clock KaiB-KaiC complex and mechanism for modulation of KaiC phosphorylation. EMBO J. 27, 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pattanayek R., Wang J., Mori T., Xu Y., Johnson C. H., Egli M. (2004) Visualizing a circadian clock protein. Crystal structure of KaiC and functional insights. Mol. Cell 15, 375–388 [DOI] [PubMed] [Google Scholar]
- 39. Mutoh R., Mino H., Murakami R., Uzumaki T., Ishiura M. (2011) Thermodynamically induced conformational changes of the cyanobacterial circadian clock protein KaiB. Appl. Magn. Reson. 40, 525–534 [Google Scholar]
- 40. Hayashi F., Ito H., Fujita M., Iwase R., Uzumaki T., Ishiura M. (2004) Stoichiometric interactions between cyanobacterial clock proteins KaiA and KaiC. Biochem. Biophys. Res. Commun. 316, 195–202 [DOI] [PubMed] [Google Scholar]
- 41. Aoki S., Onai K. (2009) Circadian clocks of Synechocystis sp. strain PCC 6803, Thermosynechococcus elongatus, Prochlorococcus spp., Trichodesmium spp., and other species. Bacterial Circadian Programs, pp. 259–282 (Ditty J. L., Mackey S. R., Johnson C. H., eds) Springer-Verlag, Berlin [Google Scholar]
- 42. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) The CLUSTAL_X windows interface. Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.