Background: TDP2 is the only known tyrosine 5′-phosphodiesterase.
Results: Recombinant human TDP2 preferentially processes single-stranded DNA ends. Its catalytic site is similar to APE-1 and consists of at least four essential residues Asn-120, Glu-152, Asp-262, and His-351 and two divalent metals.
Conclusion: We elucidated the catalytic mechanisms of TDP2 and its substrate preference.
Significance: TDP2 is critical for the repair of trapped topoisomerase complexes.
Keywords: Anticancer Drug, DNA Repair, DNA Topoisomerase, DNase, Phosphodiesterases, APE1, EAPII, TTRAP, Topoisomerase II, Topoisomerase III
Abstract
TDP2 is a multifunctional enzyme previously known for its role in signal transduction as TRAF and TNF receptor-associated protein (TTRAP) and ETS1-associated protein 2 (EAPII). The gene has recently been renamed TDP2 because it plays a critical role for the repair of topoisomerase II cleavage complexes (Top2cc) and encodes an enzyme that hydrolyzes 5′-tyrosine-DNA adducts that mimic abortive Top2cc. Here we further elucidate the DNA-processing activities of human recombinant TDP2 and its biochemical characteristics. The preferred substrate for TDP2 is single-stranded DNA or duplex DNA with a four-base pair overhang, which is consistent with the known structure of Top2cc or Top3cc. The kcat/Km of TDP1 and TDP2 was determined. It was found to be 4 × 105 s−1m−1 for TDP2 using single-stranded 5′-tyrosyl-DNA. The processing of substrates as short as five nucleotides long suggests that TDP2 can directly bind DNA ends. 5′-Phosphodiesterase activity requires a phosphotyrosyl linkage and tolerates an extended group attached to the tyrosine. TDP2 requires Mg2+ or Mn2+ for efficient catalysis but is weakly active with Ca2+ or Zn2+. Titration with Ca2+ demonstrates a two-metal binding site in TDP2. Sequence alignment suggests that TDP2 contains four conserved catalytic motifs shared by Mg2+-dependent endonucleases, such as APE1. Substitutions at each of the four catalytic motifs identified key residues Asn-120, Glu-152, Asp-262, and His-351, whose mutation to alanine significantly reduced or completely abolished enzymatic activity. Our study characterizes the substrate specificity and kinetic parameters of TDP2. In addition, a two-metal catalytic mechanism is proposed.
Introduction
DNA topoisomerases are ubiquitous enzymes that regulate DNA supercoiling and resolve DNA entanglements (knots and catenanes) by forming reversible DNA cleavage complexes. DNA cleavage results from the formation of covalent tyrosyl-phosphoribose bonds between the enzymes and the DNA backbone. With the exception of type IB topoisomerases (Top1 and Top1mt), which forms 3′-phosphotyrosyl bonds, all other mammalian topoisomerases (Top3α, Top3β, Top2α, and Top2β) form 5′-tyrosyl-DNA linkages (1–5). Topoisomerase cleavage complexes are normally very short lived (milliseconds) and reverse upon nucleophilic attack from the sugar hydroxyl end of the broken DNA after completion of the DNA topoisomerization reactions. Thus, reversal reactions are critically dependent upon the proper alignment of the broken DNA ends. This explains why drugs that bind at the enzyme-DNA interface trap topoisomerase cleavage complexes (6) and why DNA alterations (mismatches and base lesions) also generate persistent cleavage complexes (7–10). An important class of anti-cancer drugs, the Top2 inhibitors, which include etoposide, doxorubicin, daunorubicin, and mitoxantrone, target and stabilize Top2 cleavage complexes (11).
TDP2 (also known as TTRAP and EAP II) was discovered as a human 5′-tyrosyl-DNA phosphodiesterase that specifically cleaves off tyrosine from the 5′-end of DNA, leaving a phosphate group ready for religation (12). It was named following the prior discovery by Nash and coworkers (14) of TDP1, which repairs Top1 cleavage complexes (Top1cc) by cleaving 3′-tyrosyl-DNA bonds (14). TDP2 was initially named TTRAP and EAPII because of its role in NF-κB (15) and MAPK-ERK (16) signaling transduction, in transcription regulation (17), in zebrafish development (18), and in HIV-1 integration (19). It also promotes cancer cell proliferation (20) and regulates rRNA biogenesis (21). The 5′-tyrosyl-DNA phosphodiesterase activity of TDP2 and its function in the repair of trapped topoisomerase II cleavage complexes (12, 22) suggests the importance of TDP2 as a target for anticancer therapy. It was recently shown that TDP2 is up-regulated in p53 mutant tumor cells (23).
In this study we investigated the biochemical and substrate characteristics of TDP2 and generated point mutants to elucidate the catalytic residues involved in the 5′-tyrosyl-phosphodiesterase activity.
EXPERIMENTAL PROCEDURES
DNA Constructs
Full-length TDP2 open-reading frame was cloned into pET151/D-TOPO (Invitrogen) after PCR amplification from pCMV6-TDP2-Entry (OriGene, Rockville, MD) using the following primers: TDP2-Fwd, 5′-CAC CAT GGA GTT GGG GAG TTG CCT GGA GGG CGG GAG GGA G-3′, and TDP2_Rvs, 5′-GTA CGT TAC AAT ATT ATA TCT AAG TTG CAC AGA-3′. Mutations were introduced into pET151/D-TDP2 with the QuikChange Lightning site-directed mutagenesis kit (Stratagene, Santa Clara, CA). The primers used for site-directed mutagenesis are listed in supplemental Table S1. Primers from Cortes Ledesma et. al (12) were used for E152A and D262A.
Purification of Recombinant Human Proteins
N-terminal histidine-tagged recombinant human TDP2 and mutants were expressed from pET151/D-TOPO in BL21 (DE3) cells and purified by gravity-flow chromatography on nickel-nitrilotriacetic acid columns (Qiagen, Valencia, CA). Briefly, Escherichia coli BL21(DE3) carrying the constructs with TDP2 or mutants was grown in Terrific Broth at 37 °C until the absorbance at 600 nm reached 0.2. The culture was cooled to room temperature and, after the addition of IPTG to 1 mm, was grown at 25 °C for 16 h. The bacteria were harvested by centrifugation at 4000 rpm for 30 min at 4 °C and resuspended in binding buffer (40 mm Tris-HCl, pH 7.5, 10% glycerol, 300 mm NaCl, 0.05% Tween-20, 1 mm dithiothreitol (DTT(, 30 mm imidazole) with the addition of protease inhibitor and lysozyme. The cells were lysed by incubating on ice for 30 min with occasional stirring until the suspension becomes viscous. Then the lysate was clarified by sonication on ice for 2 min with 50% power and centrifugation at 16,000 rpm at 4 °C for 30 min. The supernatant was applied to a 1 ml nickel-nitrilotriacetic acid column, which was pre-equilibrated with binding buffer. The column was washed with 10 ml of binding buffer and eluted with elution buffer (binding buffer plus 250 mm imidazole). The peak fractions containing the recombinant protein, tested by SDS-PAGE, were pooled and dialyzed against storage buffer (25 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm DTT, 50% glycerol). Protein concentration was measured by NanoDrop using the extinction coefficient of 43 × 103 cm−1m−1 (Thermo Scientific, Wilmington, DE). The protein was stored at −80 °C in aliquots for future use. Human Tdp1 was expressed in E. coli BL21 (DE3) cells and purified as described (24).
Preparation of DNA Substrates and in Vitro Repair Reactions
Oligonucleotides with 5′-phosphotyrosine linkage were synthesized by Midland (Midland, TX). All other oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). Terminal deoxynucleotidyltransferase (Invitrogen) and [α-32P]cordycepin-5′-triphosphate (PerkinElmer Life Sciences) were used for 3′-end labeling. The sequences of the oligonucleotides used in experiments are listed in supplemental Table S1.
One nanomolar labeled DNA substrates in a 10-μl reaction volume was incubated with indicated concentrations of recombinant human TDP2 for 30 min at 25 °C in TDP2 reaction buffer containing 80 mm KCl, 5 mm MgCl2, 0.1 mm EDTA, 1 mm DTT, 40 μg/ml bovine serum albumin, 50 mm Tris-HCl, pH 7.5, and 0.01% Tween 20. Reactions were terminated by adding 1 volume of gel loading buffer (formamide containing 5 mm EDTA). The oligonucleotide (N14Y) and conditions used for TDP1 reaction are described in Dexheimer et al. (25). Samples were subjected to 16% denaturing PAGE. Gels were dried and exposed on phosphorimaging screens. Imaging and quantification were done using a Typhoon 8600 imager (GE Healthcare) and ImageJ software (National Institutes of Health, Bethesda, MD).
Metal titration of TDP2 activity was performed under assay conditions mentioned above. The ratios of MgCl2 and CaCl2 were varied as described in the legend of Fig. 4C. The combined divalent metal ion concentration was 1 mm in the mixed metal reactions. Visualization and quantification of substrates and products were carried out as above.
FIGURE 4.
Metal selectivity of human recombinant TDP2. A, representative denaturing PAGE gels show the processing of the ssY19 with the presence of 5 mm Mg2+, Co2+, Mn2+, Ca2+, Zn2+, or without metal ions. B, dosimetric quantification of 5′-end processing for the different substrates is shown. C, TDP2 processing activity in the presence of mixed metals is shown. 5′-Tyrosyl-DNA phosphodiesterase activity of TDP2 was assayed in the presence of varying concentrations of Mg2+ in the absence or presence of Ca2+. The reactions were carried out using 7.9 nm TDP2 and stopped at 2 min. In the mixed metal reactions, the total amount of metals was 1 mm.
Determination of Kinetic Parameters
The initial velocity was determined by incubating 0.79 nm TDP2 with various concentrations of single-stranded Y19 (ssY19)2 substrate under standard conditions. Briefly, 1 nm radiolabeled ssY19 plus 500, 1,000, 5,000, or 10,000 nm unlabeled ssTY19 for up to 120 min. Reactions were stopped at different time points and quantitated as described above. Kinetic parameters, such as Vmax (m/s), kcat (s−1), Km (m), and kcat/Km (s−1m−1) for substrates were obtained using Lineweaver-Burk plot.
RESULTS
TDP2 Prefers Single-stranded DNA Substrates
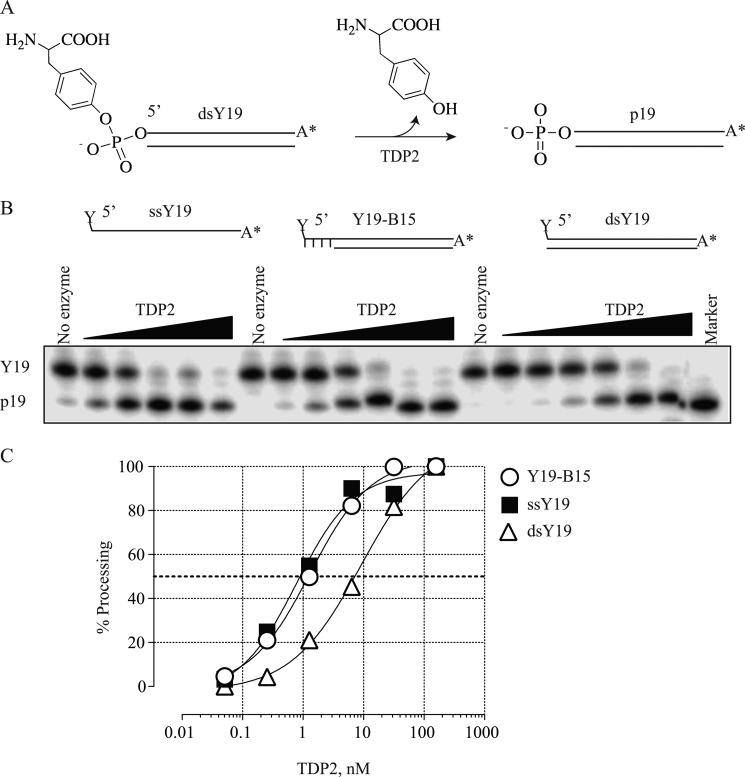
An illustration of TDP2 reactions is shown in Fig. 1A. Because TDP2 is active over a wide range of temperature (supplemental Fig. S1), all the following reactions were carried out at 25 °C.
FIGURE 1.
Processing of single-stranded DNA, DNA duplex with a 4-base overhang on the 5′-terminus, or blunt end DNA duplex by recombinant human TDP2. Increasing concentrations of rTDP2 were incubated with 1 nm 32P-labeled DNA substrates in TDP2 reaction buffer containing 5 mm Mg2+ at 25 °C for 30 min. A, shown is a schematic illustration of the TDP2 reaction. B, a representative denaturing PAGE gel shows TDP2-mediated removal of the tyrosine attached to the 5′-end of a single-stranded 19-mer oligonucleotide (ssY19), a duplex oligonucleotide with a 4-base overhang at the 5′-end (Y19-B15), or a 19-mer double-stranded oligonucleotide (dsY19). Cleavage leaves a phosphate group at the 5′-end (p-19). C, dosimetric quantification of 5′-end processing for the different substrates is shown. The dotted line indicates 50% processing.
Because Top2 dimers cleave double-stranded DNA with a 5′-overhang of 4 bases (1, 3, 4, 11), we reasoned that the physiological substrate for TDP2 would be a tyrosine linked to a protruding 5′-DNA end. The catalytic efficiency of TDP2 was examined on the following DNA substrates: ssY19, duplex with a 4-base overhang (Y19-B15), and duplex with blunt-end (dsY19), all bearing a 5′-phosphotyrosine terminus. TDP2 processed ssY19 and Y19-B15 with nearly 8-fold higher efficiencies than dsY19 (Fig. 1, B and C). These results show that TDP2 clearly prefers DNA substrates with single-stranded characteristics on the 5′-end. This is consistent with the fact that Top2 generates cleaved DNA molecules with 4-base overhangs on the 5′-ends (26). All substrates used in the rest of the study were single-stranded oligonucleotides unless otherwise stated.
TDP2 Substrate Specificity
First, we tested oligonucleotides of different lengths, Y5, Y11, Y19, and Y37, all sharing the same DNA sequence in the vicinity of the 5′-tyrosine. TDP2 processed all substrates but was more efficient with the longer substrates, suggesting its binding to single-stranded DNA (Fig. 2).
FIGURE 2.
Effect of oligonucleotide length on the 5′-tyrosyl-DNA phosphodiesterase activity of human recombinant TDP2. A, processing of ssY5, ssY11, ssY19, and ssY37 by rTDP2 is shown. The 5-, 11-, 19-, and 37-base oligonucleotides bearing a tyrosine at the 5′-end are shown above each reaction. Reactions were run on the same gel but exposed differently for ssY5 due to the impurity and low labeling efficiency for this substrate. The nonspecific band is indicated by an asterisk (*). B, dosimetric quantification of the 5-end processing for the different substrates is shown.
Human TDP1, the counterpart of TDP2, hydrolyzes the phosphodiester bond between a DNA 3′-end and a tyrosyl moiety but has also been shown to remove a broad spectrum of 3′ adducts from DNA, including 3′-tetrahydrofuran and 3′-biotin adducts (25, 27, 28). Therefore, we asked whether TDP2 could remove 5′ adducts other than a tyrosine. To test this possibility, TDP2 was incubated with single-stranded 19-mer oligonucleotides bearing a 5′-fluorescein, 5′-biotin, or 5′-digoxigenin. TDP2 failed to show any activity on these adducts (Fig. 3, A and B). This is in an agreement with a previous study (12) employing abasic site or 5′-AMP substrates, which could arise in vivo. However, TDP2 lacked AP endonuclease or 5′-deadenylase activity.
FIGURE 3.
Specificity of human recombinant TDP2 for the 5′-phosphotyrosine group. A, shown is a schematic illustration of fluorescein, biotin, or tyrosine linked to the 5′-end of the 19-mer oligonucleotide DNA substrate and representative denaturing PAGE gel of 5′-end processing reactions. FAM, 6-carboxyfluorescein. B, shown is differential processing of substrates containing a digoxigenin (Digo) directly attached to the 5′-end of the oligonucleotide or through a tyrosine.
TDP2 efficiently cleaved the 5′-digoxigenin moiety when it was linked via phosphotyrosyl linkage (Fig. 3B), indicating its selectivity for 5′-phosphotyrosyl linkage. Because the labeling reaction of Y19 with digoxigenin was partial, the substrate actually contained a mixture of Y19 and 5′-digoxigenin tyrosine. The size difference allowed direct comparison of TDP2 processing efficiency for the two substrates. TDP2 processed the 5′-digoxigenin-tyrosine substrate more efficiently than Y19, suggesting that TDP2 could potentially remove longer peptides attached to the tyrosine, as the proteolytic processing of abortive Top2 (or Top3) complexes is unlikely to cleave down to the last amino acid residue.
Metal Selectivity and Two Metal Binding Mechanisms of TDP2
Caldecott and co-workers (12) showed in their landmark publication that TDP2 5′-phosphodiesterase activity requires Mg2+. To understand the specificities of TDP2 for metals, substrate-processing efficiencies with different metal ions and with combinations of metal ions were assessed. Our results show that TDP2 is inactive in the absence of divalent metal, and that its activity is maximal in the presence of Mg2+ and Co2+ (Fig. 4, A and B). Mn2+ was also an effective cofactor, whereas Ca2+ or Zn2+ enabled less than 50% activity even at enzyme concentrations almost 2 orders of magnitude higher (Fig. 4, A and B). Mg2+ concentration as low as 0.1 mm was sufficient for efficient cleavage of ssY19 substrate (Fig. 4C) (29).
Metal ion titration assays have been used to distinguish between one and two metal ion involvement in catalysis (30, 31). For an enzyme that has two metal binding sites, the affinity for metals at each site may differ. Presumably only one of the two metal binding sites can be substituted with a non-productive metal. Therefore, titration with a non-productive metal should yield a biphasic response with stimulation at low concentrations of the productive metal and inhibition at high concentrations. To determine whether a second metal is involved in TDP2 catalysis, we tested TDP2 activity in the presence of Mg2+ alone or varied ratios of Mg2+ and Ca2+. Titration with non-productive calcium ions yielded a dose-response curve characteristic of a two-metal-ion mechanism (Fig. 4C). At higher ratios of Ca2+/Mg2+ (0.0001–0.08 mm Mg2+), Ca2+ led to stimulation of activity relative to the Mg2+ alone controls. However, with increasing Mg2+, Ca2+ exerted an inhibitory effect. The biphasic model by nonproductive calcium ions provides biochemical evidence for a two-metal-ion mechanism for TDP2.
Kinetic Characterization of the Tyrosyl Phosphodiesterase Activities of Human TDP2 and TDP1
To determine the kinetic parameters of TDP2, increasing concentrations of ssY19 substrate were incubated with TDP2, and the rate of processing was measured for each substrate concentration. The kinetic parameters, including Km (m), kcat (s−1), and kcat/Km (m−1s−1), were obtained from the double reciprocal Lineweaver-Burk plot (Fig. 5). The kcat of TDP2 for Y19 was 3 s−1 with a Km of 8.18 × 10−6 m. The resulting specificity constant, kcat/Km, was calculated as 4 × 105 m−1s−1. In comparison, the kcat of TDP1 for a 14-mer single-stranded oligo with a 3′-phosphotyrosine (N14Y) under comparable conditions was 7 s−1 with a Km of 8 × 10−8 m, giving rise to a kcat/Km of 9 × 107 m−1s−1 (Table 1, supplemental Fig. S2).
FIGURE 5.
Lineweaver-Burk plot of the 5′-tyrosyl-DNA phosphodiesterase activity of human recombinant TDP2. TDP2 (0.79 nm) was incubated in the presence of 1 nm radiolabeled ssY19 substrate plus the indicated concentrations of unlabeled ssTY19 for up to 2 h. kcat, Km, and kcat/Km were calculated from Lineweaver-Burk plots. The catalytic parameters are summarized beneath the plot.
TABLE 1.
Kinetic parameters of TDP2 and TDP1
| Enzyme | Substrate | kcat | Km | kcat/Km |
|---|---|---|---|---|
| s−1 | m | m−1s−1 | ||
| TDP2 | ssY19 | 3 | 8 × 10−6 | 4 × 105 |
| TDP1 | N14Y | 7 | 8 × 10−8 | 9 × 107 |
TDP2 Catalytic Site Residues
Because TDP2/TTRAP has been related to the cation-dependent endonuclease family (32), we aligned the amino acid sequence of TDP2 with APE1 and exonuclease III (ExoIII) (Fig. 6). Sequence alignment of human TDP2, APE1, and thermophilic exonuclease III shows that TDP2 contains all four conserved catalytic motifs that are characteristic of these nucleases (Fig. 6, boxes). We mutated one conserved residue in each motif (Fig. 6, arrowheads) and tested the activity of the mutants. Changing Glu-152, Asp-262, or His-351 to alanine completely abolished TDP2 activity, whereas the N120A mutant retained some activity, albeit 6000-fold reduced compared with wild type TDP2 (Fig. 7). On the other hand, mutating the less conserved aspartic acid residue Asp-122 near box I (Fig. 6) only reduced TDP2 activity by 4-fold (Fig. 7C). These results suggested that Asn-120, Glu-152, Asp-262, and His-351 likely form the catalytic active sites of TDP2.
FIGURE 6.
Alignment of human TDP2, APE1 and thermophilic exonuclease III (ExoIII) amino acid sequences. The conserved motifs characteristic of the Mg2+/Mn2+- dependent phosphodiesterase superfamily are indicated in the boxes. The four conserved putative catalytic residues (Asn-120, Glu-152, Asp-262, and His-351) mutated in this study are indicated by arrows and summarized in the table with the corresponding residues in APE1 and exonuclease III.
FIGURE 7.
Putative catalytic residues of human TDP2. Shown is a representative denaturing PAGE gel (A) and dosimetric quantification (B) of 5′-tyrosyl-DNA phosphodiesterase activity by wild type TDP2 and catalytic inactive mutants. C, numbers correspond to the TDP2 concentrations needed to reach 50% processing of the wild type enzyme.
DISCUSSION
Top2 can relax superhelical DNA either positively or negatively coiled, and unknot and decatenate DNA via concerted breakage and rejoining of both strands of the DNA double helix simultaneously (1, 3–5, 33). The critical roles that Top2 plays in DNA metabolism and genomic integrity make it an important target for cancer therapy. Drugs targeting Top2 can be divided into two classes: Top2 inhibitors that eliminate Top2 catalytic activities and Top2 poisons that increase the level of Top2-DNA covalent complexes (4, 11). The latter includes most of the clinically active drugs, such as etoposide, doxorubicin, and mitoxantrone. Proficient repair of the lesions caused by Top2 poisons may result in drug resistance. There are two major enzymatic pathways to remove the topoisomerase-linked DNA damage: nucleases and phosphodiesterases. Work in Schizosaccharomyces pombe suggested a role for Mre11 and CtIP in removal of trapped Top2 from DNA (34). Nucleases remove a stretch of DNA containing the trapped topoisomerase, whereas TDP2 can specifically cleave the phosphotyrosyl linkage initially formed during Top2-mediated cleavage of DNA. Our data suggest that the 5′-tyrosyl-DNA phosphodiesterase activity of TDP2 is robust under various conditions including broad pH range and low Mg2+ or Mn2+ concentrations (29).
The recently resolved crystal structure of trapped Top2 has shown that a single etoposide molecule intercalated at the cleavage sites of Top2 dislodges the two cleaved DNA ends, thereby disfavoring religation (35). Products of such disturbed reactions are DNA double-strand breaks with protruding 4-base 5′-overhangs attached to the catalytic active residue tyrosine 821 of Top2 (1, 3–5, 33, 35). This should be the physiological substrate for TDP2. Our data are consistent with this possibility, as we find that TDP2 prefers a single-stranded DNA end for 5′-tyrosyl processing as well as the 5′-staggered overhang substrate over the blunt end duplex (see Fig. 1). Furthermore, the fact that TDP2 can process an oligonucleotide substrate as short as 5 nucleotides (see Fig. 2) and a one-base surrogate substrate (36) also indicates that a long DNA fragment is not required for TDP2 recognition. Although the Km of TDP2 (8 × 10−6 m) for its substrate appears large, it is possible that ssY19 is not the optimal substrate. Alternatively, the presence of cofactors could also improve the Km value of TDP2, thus enhancing its in vivo efficiency. Additionally, a kcat/Km of 4 × 105 s−1m−1 also suggests that ssY19 may not be the optimal substrate for TDP2. An example of a highly specific enzyme is APE1, the closest relative to TDP2, which yields a kcat/Km value on the order of 107–108 s−1m−1 on AP site-containing oligonucleotide substrates (37, 38).
TDP2 knockdown cells are hypersensitive to etoposide but not camptothecin or MMS. In addition, TDP2 is inactive on abasic sites or 5′-AMP in nicked DNA (12), suggesting it is specific for tyrosine. Results from our study show that TDP2 only recognizes the bulky adduct attached to a tyrosine but not those directly attached to DNA and cleaves from the phosphotyrosine bond (see Fig. 3). We believe this is physiologically important because it suggests that TDP2 is not only specific but also efficient in recognizing and processing trapped Top2 cleavage complexes even in the form of a longer peptide. This also implies that Tdp2 should be able to process a wide range of 5′-phosphotyrosyl-DNA substrates generated by topoisomerases including Top2α, Top2β, Top3α, and Top3β (4, 5). Further experiments are warranted to determine whether TDP2 is involved in Top3 functions.
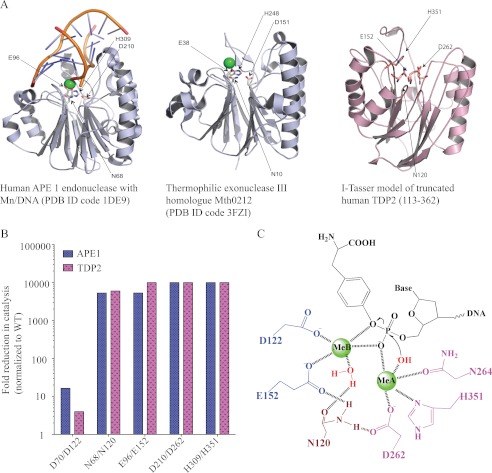
TDP2 shares 30% sequence similarity and 14% sequence identity with APE1 (32). The four catalytic motifs in APE1 are also conserved in TDP2; mutating one residue in each motif significantly impaired TDP2 activity (see Figs. 5 and 6, Asn-120/Glu-152/Asp-262/His-351 in TDP2 and Asn-68/Glu-96/Asp-210/His-309 in APE1). A predicted structure of truncated TDP2 (amino acid 113–362) is shown in Fig. 8A based on sequence and structure alignment to human APE1 (PDB ID 1DE9) and the thermophilic exonuclease III homologue Mth0212 (PDB ID 3FZI) by I-Tasser (39, 40), an online protein structure prediction program. The I-Tasser program predicted a largely disordered N terminus in TDP2 consisting of three helices (amino acids 20–35, 40–49, and 51–61). The predicted structure for the C-terminal core of TDP2 (amino acids 113–362) contains three β-sheets sandwiched by two α-helices on either side, forming a potential DNA binding groove. The four catalytic active residues form the putative catalytic site in the center of the core held by the β-sheets in the predicted structure.
FIGURE 8.
A, shown is the proposed structure of human TDP2 based on the I-Tasser program and sequence alignment with human APE1 (PDB ID 1DE9) and Thermophilic exonuclease III (PDB ID 3FZI). B, shown is a comparison of catalytic activities of APE1 and TDP2 mutants. The APE1 values were from published studies (37, 41, 42). C, shown is the proposed structure-based reaction mechanism for phosphodiester bond cleavage by TDP2.
Our mutational study shows that four residues Asn-120, Glu-152, Asp-262, and His-351 are critical for TDP2 catalytic activity (see Figs. 7 and 8). Mutations at residues Glu-152, Asp-262, or His-351 gave rise to catalytic inactive protein, and the N120A mutation reduced TDP2 efficiency by 6000-fold. Therefore, our study is in agreement and expands the initial results from Caldecott and co-workers (12) who had reported the effects of only two mutations, Glu-152 and Asp-262. The results from a mutational study of TDP2 mirror those of APE1 (see Fig. 8B) (37, 41, 42), further suggesting the two enzymes likely employ comparable catalytic mechanisms. In addition, the metal titration results clarify that TDP2 utilizes a two metal mechanism for catalysis (Fig. 4C). Therefore, based on its similarity to APE1 (30, 43) and our experimental evidence, we propose a catalytic mechanism for TDP2 with two metal binding sites (see Fig. 8C). In our proposed scheme, the metal A binding site is coordinated by Asp-262, His-351, and possibly Asn-264. The metal ion at site A would also coordinate a deprotonated water molecule for nucleophilic attack, assuming the orientation of DNA binding site is the same between APE1 and TDP2. Metal B is coordinated by the carboxylates of Asp-122 and Glu-152. Instead of having one acidic residue directly coordinating both metal ions, Asn-120 may indirectly bridge the two metal binding sites by hydrogen bonding to Glu-152 and Asp-262. In addition, TDP2 has an active profile from pH 7.5 to 10 (supplemental Fig. S3), indicating that the protonation states of the key residues in the active site are unperturbed in this range. The precise number of metal ion binding sites and the geometry of coordinating residues will only be elucidated by further structural studies.
In conclusion, our study shows the robust and specific activity of human TDP2 for 5′-phosphotyrosine residues covalently attached to single-stranded DNA ends and TDP2 activity upon substrates extended from the tyrosine. We also report the kinetic parameters for TDP2 in comparison with TDP1 and propose a two-metal catalytic mechanism based on biochemical titration experiments, sequence alignment, and mutational studies. Our study underlines the differences between TDP2 and TDP1, as TDP1 functions without metal cofactor and utilizes a transient covalent intermediate (13, 44). Together with the prior finding that TDP2 can also process 3′-tyrosyl substrates, albeit with much lower affinity (12, 29), our results suggest the broad range of activity of TDP2 and its preferential action at abortive Top2 and possibly Top3 cleavage complexes.
Supplementary Material
This work was supported, in whole or in part, by the Center for Cancer Research, Intramural Program of the NCI, National Institutes of Health.

This article contains supplemental Table S1 and Figs. S1–S3.
- ssY19
- single-stranded Y19.
REFERENCES
- 1. Wang J. C. (1991) DNA topoisomerases. Why so many? J. Biol. Chem. 266, 6659–6662 [PubMed] [Google Scholar]
- 2. Schoeffler A. J., Berger J. M. (2008) DNA topoisomerases. Harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 41, 41–101 [DOI] [PubMed] [Google Scholar]
- 3. Champoux J. J. (2001) DNA topoisomerases. Structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 4. Pommier Y., Leo E., Zhang H., Marchand C. (2010) DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nitiss J. L. (2009) DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 9, 327–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pommier Y., Marchand C. (2012) Interfacial inhibitors. Targeting macromolecular complexes. Nat. Rev. Drug Discov. 11, 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sabourin M., Osheroff N. (2000) Sensitivity of human type II topoisomerases to DNA damage. Stimulation of enzyme-mediated DNA cleavage by abasic, oxidized. and alkylated lesions. Nucleic Acids Res. 28, 1947–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vélez-Cruz R., Riggins J. N., Daniels J. S., Cai H., Guengerich F. P., Marnett L. J., Osheroff N. (2005) Exocyclic DNA lesions stimulate DNA cleavage mediated by human topoisomerase II α in vitro and in cultured cells. Biochemistry 44, 3972–3981 [DOI] [PubMed] [Google Scholar]
- 9. Pourquier P., Pommier Y. (2001) Topoisomerase I-mediated DNA damage. Adv. Cancer Res. 80, 189–216 [DOI] [PubMed] [Google Scholar]
- 10. Dexheimer T. S., Kozekova A., Rizzo C. J., Stone M. P., Pommier Y. (2008) The modulation of topoisomerase I-mediated DNA cleavage and the induction of DNA-topoisomerase I crosslinks by crotonaldehyde-derived DNA adducts. Nucleic Acids Res. 36, 4128–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nitiss J. L. (2009) Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cortes Ledesma F., El Khamisy S. F., Zuma M. C., Osborn K., Caldecott K. W. (2009) A human 5'-tyrosyl-DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 461, 674–678 [DOI] [PubMed] [Google Scholar]
- 13. Dexheimer T. S., Antony S., Marchand C., Pommier Y. (2008) Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer Agents Med. Chem. 8, 381–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pouliot J. J., Yao K. C., Robertson C. A., Nash H. A. (1999) Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286, 552–555 [DOI] [PubMed] [Google Scholar]
- 15. Pype S., Declercq W., Ibrahimi A., Michiels C., Van Rietschoten J. G., Dewulf N., de Boer M., Vandenabeele P., Huylebroeck D., Remacle J. E. (2000) TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75, and TNF receptor-associated factors (TRAFs) and that inhibits nuclear factor-κB activation. J. Biol. Chem. 275, 18586–18593 [DOI] [PubMed] [Google Scholar]
- 16. Li C., Fan S., Owonikoko T. K., Khuri F. R., Sun S. Y., Li R. (2011) Oncogenic role of EAPII in lung cancer development and its activation of the MAPK-ERK pathway. Oncogene 30, 3802–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pei H., Yordy J. S., Leng Q., Zhao Q., Watson D. K., Li R. (2003) EAPII interacts with ETS1 and modulates its transcriptional function. Oncogene 22, 2699–2709 [DOI] [PubMed] [Google Scholar]
- 18. Esguerra C. V., Nelles L., Vermeire L., Ibrahimi A., Crawford A. D., Derua R., Janssens E., Waelkens E., Carmeliet P., Collen D., Huylebroeck D. (2007) Ttrap is an essential modulator of Smad3-dependent Nodal signaling during zebrafish gastrulation and left-right axis determination. Development 134, 4381–4393 [DOI] [PubMed] [Google Scholar]
- 19. Zhang J. Q., Wang J. J., Li W. J., Huang L., Tian L., Xue J. L., Chen J. Z., Jia W. (2009) Cellular protein TTRAP interacts with HIV-1 integrase to facilitate viral integration. Biochem. Biophys. Res. Commun. 387, 256–260 [DOI] [PubMed] [Google Scholar]
- 20. Xu G. L., Pan Y. K., Wang B. Y., Huang L., Tian L., Xue J. L., Chen J. Z., Jia W. (2008) TTRAP is a novel PML nuclear bodies-associated protein. Biochem. Biophys. Res. Commun. 375, 395–398 [DOI] [PubMed] [Google Scholar]
- 21. Vilotti S., Biagioli M., Foti R., Dal Ferro M., Lavina Z. S., Collavin L., Del Sal G., Zucchelli S., Gustincich S. (2012) The PML nuclear bodies-associated protein TTRAP regulates ribosome biogenesis in nucleolar cavities upon proteasome inhibition. Cell Death Differ 19, 488–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zeng Z., Cortés-Ledesma F., El Khamisy S. F., Caldecott K. W. (2011) TDP2/TTRAP is the major 5'-tyrosyl-DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage. J. Biol. Chem. 286, 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Do P. M., Varanasi L., Fan S., Li C., Kubacka I., Newman V., Chauhan K., Daniels S. R., Boccetta M., Garrett M. R., Li R., Martinez L. A. (2012) Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 26, 830–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Antony S., Marchand C., Stephen A. G., Thibaut L., Agama K. K., Fisher R. J., Pommier Y. (2007) Novel high throughput electrochemiluminescent assay for identification of human tyrosyl-DNA phosphodiesterase (Tdp1) inhibitors and characterization of furamidine (NSC 305831) as an inhibitor of Tdp1. Nucleic Acids Res. 35, 4474–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dexheimer T. S., Stephen A. G., Fivash M. J., Fisher R. J., Pommier Y. (2010) The DNA binding and 3'-end preferential activity of human tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 38, 2444–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deweese J. E., Osheroff N. (2009) The DNA cleavage reaction of topoisomerase II. Wolf in sheep's clothing. Nucleic Acids Res. 37, 738–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Interthal H., Chen H. J., Champoux J. J. (2005) Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J. Biol. Chem. 280, 36518–36528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murai J., Huang S. Y., Das B. B., Dexheimer T. S., Takeda S., Pommier Y. (2012) Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells. J. Biol. Chem. 287, 12848–12857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adhikari S., Karmahapatra S. K., Karve T. M., Bandyopadhyay S., Woodrick J., Manthena P. V., Glasgow E., Byers S., Saha T., Uren A. (2012) Characterization of magnesium requirement of human 5'-tyrosyl-DNA phosphodiesterase mediated reaction. BMC Res. Notes 5, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beernink P. T., Segelke B. W., Hadi M. Z., Erzberger J. P., Wilson D. M., 3rd, Rupp B. (2001) Two divalent metal ions in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1. Implications for the catalytic mechanism. J. Mol. Biol. 307, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 31. Vipond I. B., Baldwin G. S., Halford S. E. (1995) Divalent metal ions at the active sites of the EcoRV and EcoRI restriction endonucleases. Biochemistry 34, 697–704 [DOI] [PubMed] [Google Scholar]
- 32. Rodrigues-Lima F., Josephs M., Katan M., Cassinat B. (2001) Sequence analysis identifies TTRAP, a protein that associates with CD40 and TNF receptor-associated factors, as a member of a superfamily of divalent cation-dependent phosphodiesterases. Biochem. Biophys. Res. Commun. 285, 1274–1279 [DOI] [PubMed] [Google Scholar]
- 33. Liu L. F., Liu C. C., Alberts B. M. (1980) Type II DNA topoisomerases. Enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell 19, 697–707 [DOI] [PubMed] [Google Scholar]
- 34. Hartsuiker E., Neale M. J., Carr A. M. (2009) Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell 33, 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu C. C., Li T. K., Farh L., Lin L. Y., Lin T. S., Yu Y. J., Yen T. J., Chiang C. W., Chan N. L. (2011) Structural basis of type II topoisomerase inhibition by the anticancer drug etoposide. Science 333, 459–462 [DOI] [PubMed] [Google Scholar]
- 36. Adhikari S., Karmahapatra S. K., Elias H., Dhopeshwarkar P., Williams R. S., Byers S., Uren A., Roy R. (2011) Development of a novel assay for human tyrosyl-DNA phosphodiesterase 2. Anal. Biochem. 416, 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Erzberger J. P., Wilson D. M., 3rd (1999) The role of Mg2+ and specific amino acid residues in the catalytic reaction of the major human abasic endonuclease. New insights from EDTA-resistant incision of acyclic abasic site analogs and site-directed mutagenesis. J. Mol. Biol. 290, 447–457 [DOI] [PubMed] [Google Scholar]
- 38. Rothwell D. G., Hang B., Gorman M. A., Freemont P. S., Singer B., Hickson I. D. (2000) Substitution of Asp-210 in HAP1 (APE/Ref-1) eliminates endonuclease activity but stabilizes substrate binding. Nucleic Acids Res. 28, 2207–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER. A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim W. C., Berquist B. R., Chohan M., Uy C., Wilson D. M., 3rd, Lee C. H. (2011) Characterization of the endoribonuclease active site of human apurinic/apyrimidinic endonuclease 1. J. Mol. Biol. 411, 960–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chou K. M., Cheng Y. C. (2003) The exonuclease activity of human apurinic/apyrimidinic endonuclease (APE1). Biochemical properties and inhibition by the natural dinucleotide Gp4G. J. Biol. Chem. 278, 18289–18296 [DOI] [PubMed] [Google Scholar]
- 43. Mol C. D., Izumi T., Mitra S., Tainer J. A. (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature 403, 451–456 [DOI] [PubMed] [Google Scholar]
- 44. Interthal H., Chen H. J., Kehl-Fie T. E., Zotzmann J., Leppard J. B., Champoux J. J. (2005) SCAN1 mutant Tdp1 accumulates the enzyme-DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 24, 2224–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.