Background: Reasons for the differences in activity between the peroxisomal and mitochondrial DCRs are unknown.
Results: Structure-function studies on peroxisomal DCR (pDCR) and comparison with its mitochondrial counterpart reveal differences in catalytic residues and hinge movements.
Conclusion: Catalysis by pDCR is unique and involves aspartate.
Significance: Our studies explain why human peroxisomal DCR, but not mitochondrial DCR, can process very long chain fatty acids.
Keywords: Enzyme Catalysis, Molecular Biology, Peroxisomes, Protein Crystallization, Structural Biology, β-Oxidation, Peroxisome
Abstract
Peroxisomes play an essential role in maintaining fatty acid homeostasis. Although mitochondria are also known to participate in the catabolism of fatty acids via β-oxidation, differences exist between the peroxisomal and mitochondrial β-oxidation. Only peroxisomes, but not mitochondrion, can shorten very long chain fatty acids. Here, we describe the crystal structure of a ternary complex of peroxisomal 2,4-dienoyl CoA reductases (pDCR) with hexadienoyl CoA and NADP, as a prototype for comparison with the mitochondrial 2,4-dienoyl CoA reductase (mDCR) to shed light on the differences between the enzymes from the two organelles at the molecular level. Unexpectedly, the structure of pDCR refined to 1.84 Å resolution reveals the absence of the tyrosine-serine pair seen in the active site of mDCR, which together with a lysine and an asparagine have been deemed a hallmark of the SDR family of enzymes. Instead, aspartate hydrogen-bonded to the Cα hydroxyl via a water molecule seems to perturb the water molecule for protonation of the substrate. Our studies provide the first structural evidence for participation of water in the DCR-catalyzed reactions. Biochemical studies and structural analysis suggest that pDCRs can catalyze the shortening of six-carbon-long substrates in vitro. However, the Km values of pDCR for short chain acyl CoAs are at least 6-fold higher than those for substrates with 10 or more aliphatic carbons. Unlike mDCR, hinge movements permit pDCR to process very long chain polyunsaturated fatty acids.
Introduction
The past two decades have seen a firm link being established between obesity and disease (1). A decrease in the ability of the body to utilize fat or intake of excessive dietary fats beyond the body's capacity to metabolize them can lead to obesity and has been shown to be detrimental to health (2). Obesity has been associated with increased risk of cardiovascular diseases, diabetes, osteoarthritis, cancer, and other physiological disorders (1). Although the harmful effects of accumulation of saturated fats are well documented, comparatively little is known about the physiological effects of excess polyunsaturated fats. Unsaturated fats play a role in the vital functions of the body like cognition, memory, inflammation, aging, cardiovascular processes, etc. (3–5). Although some studies have demonstrated that unsaturated fatty acids can harm the body by modulating immune responses (6) and cell growth and multiplication (7), there is no clear understanding of the effects of accumulation of unsaturated fats.
Saturated and unsaturated fats are catabolized by the mitochondrion and peroxisomes (8). Both the organelles employ similar machineries for processing fats, and so far the only major difference known is that the peroxisomes can process linear as well as branched fatty acids with much longer chains (C > 20) when compared with the mitochondrion (9). Further, some studies have indicated that the degradation of fatty acids is incomplete in peroxisomes (10). Catabolic processing of very long chain fatty acids by peroxisomes results in medium chain acyl CoAs like octanoyl CoA that are conjugated to carnitine and shuttled to the mitochondrion for further degradation (11). So far, the reasons underlying these differences in β-oxidation in humans are not known. In contrast to humans, β-oxidation in peroxisomes from yeast and plants proceeds to completion (12). The reason for this difference has been attributed to the composition of the active site: in particular, the ability of the enzyme to orient the shorter substrate correctly for catalysis (13).
Fatty acids are converted to acetyl or propionyl CoA, depending on whether the aliphatic chain has even or odd number of carbons. The complete degradation of fatty acids requires the concerted activity of four enzymes, each catalyzing a discrete reaction. Acyl CoA dehydrogenase catalyzes the first dehydrogenation step, resulting in a double bond between C2 and C3 of the fatty acid CoA conjugate (14). In the next step, enoyl CoA hydratase adds a water molecule across the double bond, resulting in the formation of 3-hydroxyacyl CoA, which is further oxidized by 3-hydroxyacyl CoA dehydrogenase to its keto form 3-ketoacyl CoA. In the final step, β-ketoacyl CoA thiolase inserts a thiol group from an incoming CoA between the C2 and C3 carbons. This releases a molecule of acetyl CoA and results in a fatty acid CoA conjugate shorter by two carbons (14, 15). Polyunsaturated fatty acids contain pre-existing double bonds at odd or even positions that are cis in nature as opposed to the trans conformation of the double bond in trans-2-enoyl CoA, which is formed during the first step of β-oxidation. Therefore, to continue the β-oxidation stalled at the cis double bonds, three additional enzyme activities are required to produce the trans-2-enoyl CoA derivative that can further undergo the chain shortening (16). First, an isomerase Δ3,5,Δ2,4-enoyl CoA isomerase converts 3,5-dienoyl CoA to 2,4-dienoyl CoA. Next, 2,4-dienoyl CoA reductase produces trans-3-enoyl CoA, which is finally isomerized by Δ3,Δ2-enoyl CoA isomerase to trans-2-enoyl CoA. Among all of these enzymatic steps, the reaction catalyzed by 2,4-dienoyl CoA reductase is considered to be the rate-limiting step in the β-oxidation of polyunsaturated fatty acids (16).
Structurally, 2,4-dienoyl CoA reductases (hereafter referred to as DCRs)4 belong to the short chain dehydrogenase/reductase (SDR) superfamily (17). Members belonging to this family share a highly conserved Rossmann fold used for binding cofactors like NADH or NADPH. In addition, catalysis involves the participation of an invariant lysine residue (18). Interestingly, a serine hydrogen-bonded to a tyrosine, together with the lysine and an asparagine, have been shown to be critical for reactions catalyzed by the SDR family (19). Although mitochondrial DCRs (mDCR) and their counterparts in the peroxisome (pDCR) catalyze similar reactions, they share only 35% sequence identity. More important, two of the residues (tyrosine and serine) of the catalytic tetrad, a hallmark of the SDR family of enzymes, including mDCR (20), are missing in the pDCR (see Fig. 1A). This gives rise to an intriguing question: how does pDCR catalyze the reaction in spite of not carrying the conserved catalytic tetrad? Further, what structural features of pDCR permit it to process very long chain polyunsaturated fatty acids? How are these features different from those of the mDCR, which cannot process very long chain poly unsaturated fatty acids? To answer these questions, we decided to determine the crystal structure of the ternary complex of pDCR and perform structure-based mutagenesis and biochemical studies. Here, we report the structure-function analysis of pDCR bound with NADP and 2,4-hexadienoyl CoA (HXC) and discuss the unique features of the enzymatic mechanism. Because the conversion of 2,4-dienoyl CoA to trans-3-enoyl CoA is the rate-limiting step in the catabolism of poly unsaturated fatty acids, the mechanistic insights gained in this study have profound implications for treatment of diseases characterized by accumulation of polyunsaturated fatty acids.
FIGURE 1.
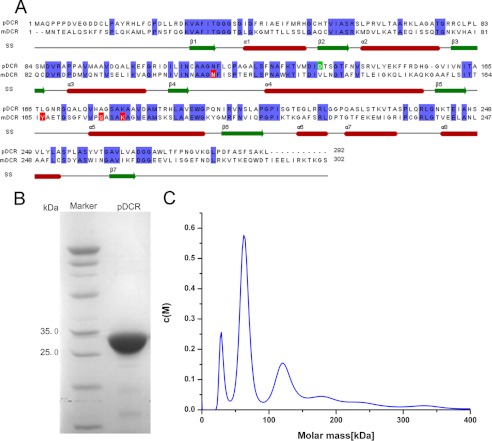
Characterization of pDCR. A, sequence alignment of pDCR with mDCR. Conserved residues are highlighted in blue. Positions of the catalytic tyrosine, serine, lysine, and asparagine of mDCR are highlighted in red. The catalytically critical aspartate of pDCR is highlighted in green. The secondary structural elements of pDCR are shown. B, purity of pDCR before crystallization. Coomassie Blue-stained gel of 1 μl of pure pDCR separated by SDS-PAGE. C, analytical ultracentrifugation analysis of pDCR. Sedimentation velocity experiments reveal that a majority of pure pDCR exists as a mixture of monomers and dimers.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
A 878-base pair DNA fragment coding pDCR was amplified using the forward primer 5′-TACTTCCAATCCAATGCATGGCCCAGCCGCCGC-3′and reverse primer 5′-TTATCCACTTCCAATGCATGAGCTTAGCAGAG-3′. The PCR product was ligated into vector pMCSG7 (21) and transformed into Escherichia coli BL21 (DE3) cells. The cells were cultured in LB medium containing ampicillin (100 μg/ml) at 37 °C until the A600 nm reached 0.8. The culture was then induced with 0.2 mm isopropyl-β-d-thiogalactoside for 20 h at 16 °C. The cells were harvested by centrifugation, lysed by sonication, and clarified by centrifugation; the supernatant was then applied to a nickel-nitrilotriacetic acid resin gravity column (Qiagen) that had been previously equilibrated with PBS (137 mm NaCl, 2.7 mm KCl, 50 mm NaHPO4, 10 mm KH2PO4, pH 7.4). The column was first washed with 50 ml of PBS, followed by a wash with 50 ml of PBS containing 20 mm imidazole, and finally eluted with PBS containing 500 mm imidazole (see Fig. 1B). Fractions were analyzed by SDS-PAGE, and those fractions containing pDCR were pooled and dialyzed overnight against 50 mm PBS, pH 7.4, 10% glycerol. The His tag was cleaved by tobacco etch virus treatment. Uncut protein was separated by nickel affinity chromatography. pDCR was again pooled and dialyzed overnight against 50 mm PBS, pH 7.4, 10% glycerol, 1 mm EDTA, 1 mm DTT, and then concentrated to ∼15 mg/ml for crystallization (see Fig. 1C).
Site-directed Mutagenesis
Site-directed mutagenesis of pDCR was carried out using the QuikChange® site-directed mutagenesis kit (Stratagene, La Jolla, CA). The following primers and their antisense primers (not shown) were used to introduce the mutations: 1) ctc cct ctc tct atg gcc gtc cga gcg ccc cca (D86A); 2) acc gtg atg gac atc gct acc agc ggc acc ttc (D137A); 3) gag aag ttc ttc agg gcc cac gga ggg gtg atc (D155A); 4) gcc aag gcc gct gtg gcc gcg atg acg cgg cac (D186A); 5) ccc atc agt ggc aca gcg ggg ctc cgg cga ctg (E215A); and 6) gcc gtg ctg gtg gcc gct ggc ggg gca tgg ttg (D268A). The reactions were set up according to the manufacturer's instructions. The entire lengths of the genes were sequenced to verify the mutations. The mutants were expressed and purified the same way as the wild-type enzyme and assayed for activity under identical conditions.
Synthesis of Substrate
First, the trans-2,trans-4-decadienoic acid was prepared from trans-2,trans-4-decadienal (Aldrich) by Ag2O oxidation (22). The substrate trans-2,trans-4-decadienoyl CoA thiolester and the trans-2,trans-4-hexadienoyl CoA thiolester were synthesized from the corresponding free acid and CoA by mixed anhydride method (23) and purified by reverse phase high performance liquid chromatography.
Activity Assay and Kinetic Studies
Reductase activity was determined kinetically by following the substrate-dependent decrease in absorbance of NADPH at 340 nm at room temperature (20). The assay mixture contained 50 mm PBS, pH 7.4, 100 μm EDTA, 125 μm NADPH, and 10 μg of enzyme in a final volume of 1.0 ml. The mixtures were preincubated for 20 min at room temperature, after which the reactions were initiated by the addition of 40 μm trans-2,trans-4-hexadienoyl CoA or trans-2,trans-4-decadienoyl CoA, and the decrease in absorbance at 340 nm was monitored for 90 s. Estimation of Km and Vmax values was performed using the same assay buffer with substrate concentrations ranging from 1 to 100 μm (25). Measurements were made at five substrate concentrations, and the averages of two assays were used for each point.
Crystallization and Structure Determination
Crystallization screening was done using commercially available sparse matrix screens. pDCR in ternary complex with NADP+ and CoA and in complex with substrate grew under similar conditions. A ternary complex of pDCR with the oxidized cofactor and CoA or substrate was prepared by incubating the protein solution with 6 mm NADP+ and 6 mm CoA or 50 mm HXC for several hours prior to crystallization. Diffraction quality crystals appeared overnight at 22 °C in a mother liquor solution containing 0.1 m MES, pH 6.0, and 8% PEG 6000. Single crystals were frozen in liquid nitrogen prior to x-ray diffraction testing and data collection. Diffraction data were collected at a wavelength 0.9795 Å at Southeast Regional Collaborative Access Team 22-ID Beamline at Advanced Photon Source, Argonne National Lab. All of the data sets were collected at cryogenic temperature. The data were indexed, integrated, and scaled using HKL 2000 (24). The structure of pDCR in ternary complex with NADP+ and CoA was solved by molecular replacement using MolRep with structure of mDCER (Protein Data Bank code 1W6U, chain A) as a search template (25). The asymmetric unit consists of four molecules of pDCR based on the calculated Matthews coefficient value of 2.20 Å3/Da and a solvent content of 44.22%. Electron density for most of the peptide backbone was of good quality. Rounds of refinement with REFMAC5 (26) combined with inspection of map and manual adjustments, the addition of water molecules, NADP+ and CoA or HXC completed the ternary complex. Five percent of unique reflections were assigned, in a random fashion, to the “free” set, for calculation of the free R factor (Rfree), and the remaining 95% of the reflections constituted the “working” set, for calculation of R (27). Manual adjustments to the model were done using COOT (28). Details of data collection and refinement statistics are listed in Table 1.
TABLE 1.
Data collection and refinement statistics
The numbers in parentheses are statistics for the highest resolution shell.
| Data set | pDCR + NADP + CoA | pDCR + NADP + HXC |
|---|---|---|
| Data collection | ||
| Beam | 22-ID | 22-ID |
| Wavelength (Å) | 0.9795 | 0.9795 |
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 88.10, 94.50, 130.16 | 88.18, 95.44, 133.60 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50.00–1.81 (1.85–1.81) | 50.00–2.09 (2.22–2.09) |
| Rsym (%) | 10.1 (43.6) | 10.1 (20.6) |
| Completeness (%) | 92.1 (68.3) | 95.9 (77.9) |
| Redundancy | 6.0 (2.8) | 5.3 (3.2) |
| Refinement | ||
| Resolution (Å) | 39.43–1.84 | 20.00–2.10 |
| No. reflections | 88210 | 60105 |
| Rwork/Rfree (%) | 16.5/20.8 | 15.4/21.6 |
| No. atoms | 9379 | 9384 |
| Protein | 8167 | 8167 |
| Water | 828 | 805 |
| NADP | 192 | 192 |
| CoA/HXC | 192 | 220 |
| Root mean square deviations | ||
| Bond lengths(Å) | 0.023 | 0.020 |
| Bond angles(Å) | 2.374 | 2.386 |
| Mean B value (Å2) | 18.85 | 19.36 |
| Ramachandran analysis | ||
| Favored region (%) | 97.14 | 97.42 |
| Allowed region (%) | 2.12 | 1.84 |
| Outliers (%) | 0.74 | 0.74 |
Sedimentation Velocity Experiments
Sedimentation velocity experiments were performed using a Beckman XL-I analytical ultracentrifuge. Protein samples were diluted with PBS to 400 μl at a concentration of ∼1.0 mg/ml. The samples were loaded into a conventional double sector quartz cell and mounted in a Beckman four-hole An-60 Ti rotor. The data were collected at 60,000 rpm at a wavelength of 280 nm. Interference sedimentation coefficient distributions, c(M), were calculated from the sedimentation velocity data by using SEDFIT software.
RESULTS
An invariant lysine, a tyrosine, a serine, and an asparagine have been shown to participate in reactions catalyzed by enzymes belonging to the SDR superfamily, including mDCR (20). Analysis of the primary amino acid sequence of the pDCR by aligning it with that of its counterpart from mitochondrion revealed that pDCR does not contain two of the four residues important for catalysis (Fig. 1A). Tyr-199 and Ser-210, essential for catalysis in mDCR, are replaced by a leucine and an alanine residue, respectively, in pDCR. Therefore, to test whether human pDCR expressed and purified from E. coli cultures was active, we estimated its activity using 2,4-hexadienoyl CoA as a substrate. Interestingly, pDCR showed activity, suggesting that it can perform the catalysis without the tryrosine-serine pair (Table 2). Next, we retrieved and aligned sequences of pDCR homologs from the Protein Data Bank to find out whether any of them were lacking the catalytically critical amino acids. The position of the lysine is invariant in the primary sequence of all the species examined (supplemental Fig. S1). Surprisingly, although the tyrosine-serine pair does not seem to be conserved in the primary sequence alignment, inspection of the active site of the homologs reveals the presence of such a pair in all of the enzymes examined. For example, in the structure of the pDCR homolog, α-hydroxysteroid dehydrogenase in complex with NADH and glycochenodeoxycholic (Protein Data Bank code 1AHI) (29, 30), a Tyr-159 and Ser-146 pair is observed in the active site. Similarly, a Tyr-142 and Ser-153 pair is located in the active site of an SDR from Bacillus anthracis (Protein Data Bank code 3IMF), although these positions do not appear as conserved in the primary sequence alignment (supplemental Fig. S1).
TABLE 2.
Kinetic parameters of human peroxisomal 2,4-dienoyal CoA reductase
| Enzyme | 2,4-Hexadienoyl CoA |
2,4-Decadienoyl CoA |
||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| μm | μmol/min/mg | μm | μmol/min/mg | |
| pDCR | 71.6 ± 0.27 | 1.75 ± 0.34 | 12.7 ± 4.70 | 1.37 ± 0.15 |
pDCR has four tyrosine and 16 serine residues. Tyr-251 and Tyr-259 are highly conserved. In fact, all the homologs examined show the presence of an aromatic amino acid at this position (supplemental Fig. S1). Further, except for Ser-254, which is absolutely conserved, none of the other serine residues are conserved. The positions of none of the conserved tyrosines or the conserved Ser-254 match with the position of the tyrosine-serine pair of mDCR (Fig. 1A). Thus, the primary sequence analysis of pDCR was inconclusive, and therefore a structural view of pDCR was essential to verify whether pDCR also employs a tyrosine-serine pair in conjunction with a lysine and an asparagine, like other members of the SDR family, for catalysis.
Structure of pDCR
Large amounts of recombinant human pDCR could be produced in E. coli. Analytical ultracentrifugation analysis of purified pDCR (Fig. 1B) revealed that the protein exists as a mixture of monomers, dimers, and tetramers in solution (Fig. 1C). This is in contrast to the tetramer observed for mDCR packed in the crystal lattice (20). Such a pure (>95% purity as judged by SDS-PAGE analysis) but heterogeneous pDCR was active against the substrate HXC when tested for activity (Table 2). Thus, human pDCR produced and purified from E. coli was active. Whether monomeric pDCR can catalyze the reaction is still not known. However, pDCR could use a six carbon acyl CoA conjugated fatty acid as a substrate. This was intriguing because previous functional studies have suggested that human pDCR breaks down fatty acids to octanoyl CoA, which are then shuttled to the mitochondrion for further chain shortening (11). Thus, to find out whether pDCR crystallizes as a tetramer and whether a tyrosine-serine pair together with a lysine and an asparagine is involved in catalysis, it was essential to solve the structure of the ternary complex of the enzyme. The structure was also expected to shed light on substrate specificities of pDCR.
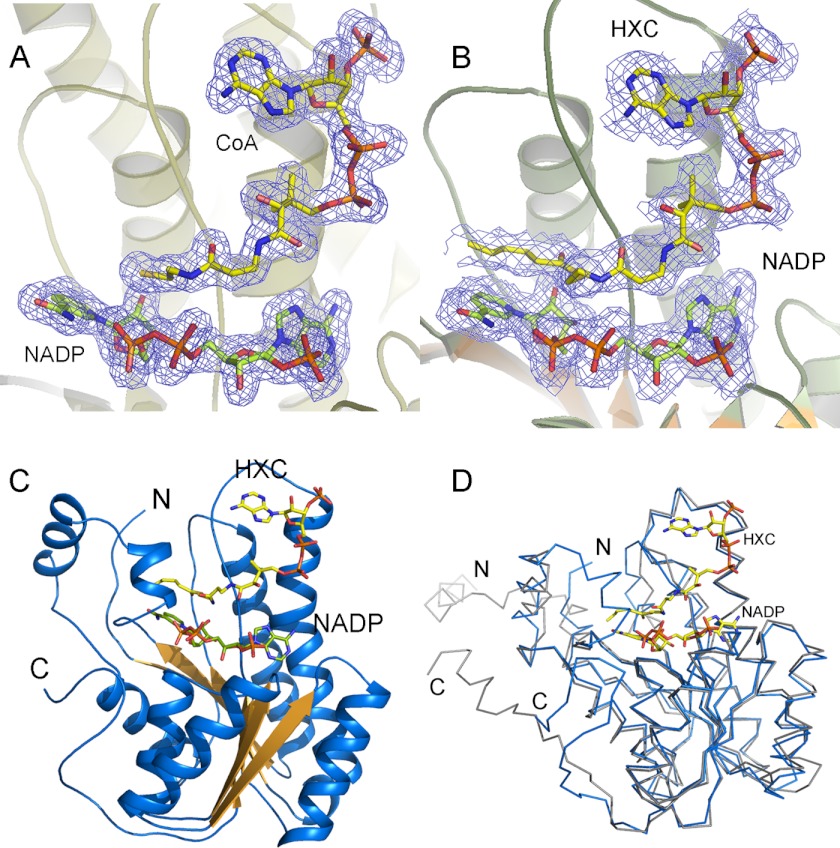
The halo-enzyme could not be crystallized. Protein mixed with the CoA or substrate and cofactor NADP+ crystallized in P212121 space group with four molecules of pDCR packed in the asymmetric unit. The structure could be solved by molecular replacement using the structure of mDCR (Protein Data Bank code 1W6U) as a search template. Residues 4–278 could be traced without ambiguity and refined to 1.84 and 2.10 Å resolutions with an R value of 16.5% (Rfree 20.8%) and 15.4% (Rfree 21.6%) for the ternary complex with CoA and HXC, respectively. Additional statistics for the binary complexes are listed in Table 1. The last 15 amino acids of pDCR are disordered in all the chains of both the ternary complexes. The four chains of pDCR within a tetramer are identical, and superimposition of the chains revealed that there are no obvious differences between them. Electron density for the cofactor NADP was clear in all the chains of both the binary complexes (Fig. 2A). Similarly, the CoA could be modeled easily into the characteristic shape of the difference density calculated before inclusion of the CoA in the model. There is no change in the position of the NADP or CoA in any of the chains of the ternary complexes. However, differences are observed for the substrate HXC. Except for chain D, electron density for the aliphatic tail of the substrate HXC (Fig. 2B) was not clear, suggesting that either short chain acyl CoAs are poor substrates or, conversely, the active site has a large cavity that can accommodate aliphatic chains larger than the C6 used in the present study. A sharp increase in B-factor for positions after the sulfur atom (average B factor of ∼40) is observed. The position of the CPA group after the P2 phosphate of HXC is different in chain D than that observed in other chains. This movement has implications for the mechanism of action. The movement of the CPA group is relayed to the rest of the aliphatic tail, which translates into folding of the tail and positioning of the delta carbon within the range for a hydride transfer from the C4 carbonyl carbon (Fig. 2B). Because the structural elements of all the chains are identical and because chain D has well defined electron density for the substrate, further description and analysis of the pDCR structure are based on chain D.
FIGURE 2.
Structure of pDCR. A and B, electron density for NADP with CoA (A) and NADP with HXC (B) from the two ternary complexes of pDCR contoured at 1.5 σ. C, overall structure of pDCR shown in cartoon representation. The ligands are depicted as sticks. D, superimposition of pDCR (shown in blue) over mDCR shown in gray. N and C termini of both the enzymes are marked.
The structure of pDCR reveals an α/β architecture with a typical Rossman fold. A central twisted sheet made up of seven parallel strands flanked by helices on either side forms the core of the protein (Fig. 2C). Often secondary structural elements seen inserted in this core impart substrate specificities for a wide array of proteins catalyzing diverse reaction using a similar fold (31). In DCRs, helix α4 and the loop connecting strand β4 with helix α4 are extended to form the substrate-binding site. Similarly, a large loop is inserted between strand β6 and helix α6 probably for regulating access to the active site (20).
An InterProScan scan of the pDCR structure by the ProFunc server (32) identified sequence motifs belonging to the SDR superfamily and the glucose/ribitol dehydrogenase family of enzymes. A search of the Protein Data Bank (33) for structural homologs retrieved the structure of a short chain dehydrogenase from a bacterium, B. anthracis, as the best match (40% sequence identity, E value 2.9 × 10−31 with 255 overlapping residues) and the mitochondrial DCR (35% sequence identity, E value 1.2 × 10−29 with 258 overlapping residues) among the top matches. Because the DCR from the mitochondrion and the peroxisome are related functionally, the pDCR structure is analyzed in context with the already reported structure of mDCR. Although the sequence identity between the enzymes from the two organelles is very low, the overall structure is conserved (Fig. 2D). Superimposition studies reveal that there is hardly any difference between the positions of the C α atoms of the two enzymes. Because the overall topology of both the enzymes is similar, the differences in the properties of the two DCRs could be attributed to the low primary sequence identity between the two enzymes. Both the DCRs pack as a tetramer in the crystal lattice (Fig. 3A). The loop connecting helix α4 with strand β4 that is disordered in all the chains of mDCR is ordered in pDCR. On the other hand, the last 15 amino acids missing in the pDCR structure are well ordered in the mDCR. However, these amino acids are located far away from the active site.
FIGURE 3.
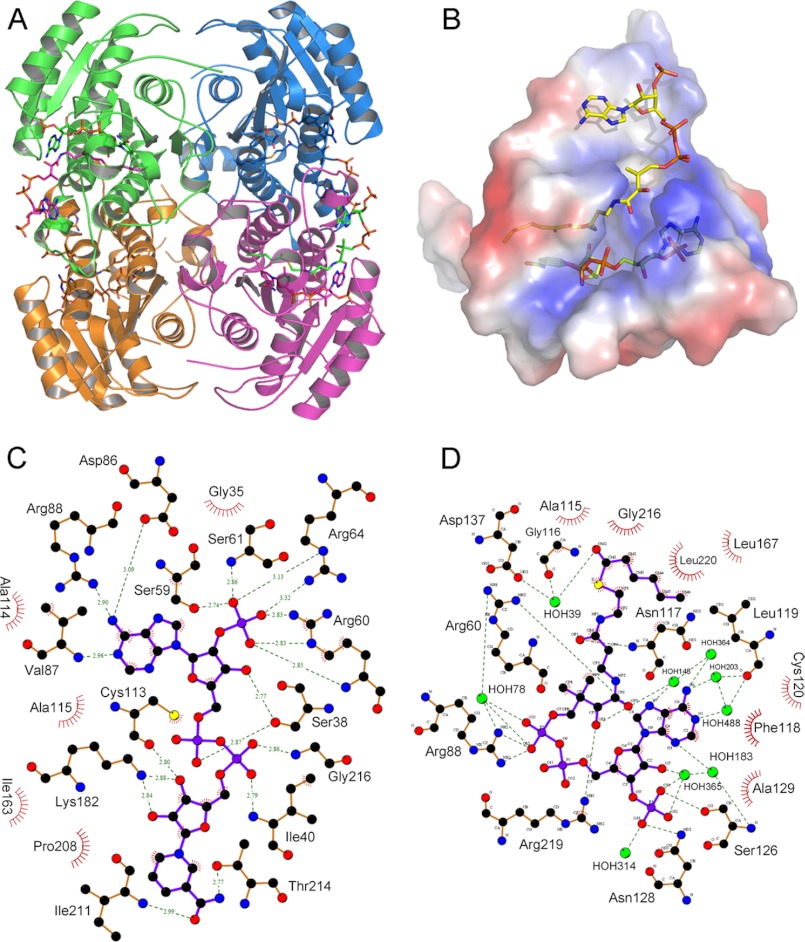
Cofactor and substrate-binding sites. A, cartoon representation of the tetramer of pDCR. Ligands are shown as sticks. B, surface electrostatic potential representation of region around NADP and HXC. Blue and red represent positive and negative potential, respectively. C and D, interactions of NADP (C) and HXC (D) with protein plotted using LigPlot. For clarity, water molecules have been removed from C. Hydrogen bonding distances have been removed from D. Ligands are shown as purple sticks, and water is shown as cyan spheres.
NADP-binding Site
As observed for other members of the SDR family, NADP binds just above the β-sheet in an elongated cavity near the center of the protein (Fig. 3B). This location closely mirrors the location of NADP in mDCR (20). NADP is docked in the active site using a combination of highly conserved side chain, backbone, and water-mediated contacts that hold and orient the NADP for delivery of the hydride to the substrate (Fig. 3C). Several hydrogen bonds tether the adenine moiety into the binding site. The amine group of the adenine ring is hydrogen-bonded to the carboxyl oxygen of Asp-86, whereas the side chains of two arginine residues hold the 2′ phosphate group of the adenine moiety in place. Except for Arg-64 being replaced by Lys-92, similar interactions hold the adenine moiety in mDCR. The backbone carbonyl of Met-85 makes a hydrogen bond to the C2 carbon of the adenine ring, and the hydroxyl oxygen of Ser-59 forms a hydrogen bond with the 2′ phosphate group of the adenine moiety. The hydroxyl group at the C3 position of the ribose ring forms two hydrogen bonds: a 2.5 Å hydrogen bond with Ser-38 (replaced by Thr-69 in mDCR) and a water-mediated hydrogen bond with Ser-59. Interestingly, the α-phosphate that has dual conformations in mDCR assumes only one conformation in pDCR, which is identical in all the chains of pDCR. It is hydrogen-bonded to Ser-38 and Glu-215 (Fig. 3C). On the other hand, the β-phosphate is hydrogen-bonded to the backbone amide nitrogen of Ile-40 and the amine nitrogen of the nicotamide moiety. The invariant Lys-182 donates a hydrogen bond to both the hydroxyl oxygens of the ribose ring of the nicotamide. Thus, the NADP is engaged and positioned by the protein for catalysis via numerous hydrogen bonding interactions.
Substrate-binding Site
2,4-hexadienoyl CoA, 2,4-decadienoyl CoA, and CoA were used individually as substrates for cocrystallization with the cofactor NADP. Interpretable electron density for only 2,4-hexadienoyl CoA, CoA and NADP was observed. The reason for the lack of density for 2,4-decadienoyl CoA is currently unknown. Thus, two ternary complexes of pDCR, 2,4-hexadienoyl CoA/NADP, and CoA/NADP containing the CoA and NADP were used to gain mechanistic insights.
Not surprisingly, the CoA from both the ternary complexes overlaps upon superimposition the two structures. The CoA binds on the surface in proximity to helix α4 and the loop connecting strand β4 with helix α4 (Fig. 2C). The location and nature of the substrate-binding site is reminiscent of mDCR, where HXC is docked using conserved interactions (20). The adenine ring is stacked against a catalytically critical phenylalanine. Interestingly, the amine group of adenine does not participate in any interactions of the CoA with the protein. Instead, the N1 nitrogen interacts directly with the side chain of Cys-120 (Fig. 3D). Further, N-1, N-3, and N-7 nitrogens of the adenine moiety are seen linked to a group of water molecules forming an intricate network, probably acting as a conduit for the relay of charge. The only interaction of the ribose ring is intramolecular interaction with a hydrogen bond between the C2 hydroxyl group and the oxygen atom of the 3′ phosphate group on the ribose. This 3′ phosphate is hydrogen-bonded to Ser-126, Asn-128, and a number of water molecules. Although the α-phosphate does not contact the protein, the β-phosphate is localized in space by the side chains of Arg-60 and Arg-88 (Fig. 3D). After the β-phosphate, the position of the rest of the CoA and the acyl chain is different in chain D of pDCR, which happens to have interpretable density for the acyl chain when compared with other chains of pDCR or mDCR. A kink is observed between the β-phosphate and the pantothenate moiety, which pushes the CDP and CEP atoms inwards, resulting in the hydroxyl oxygen O3P being pushed toward the interior of the protein. The side chain of Arg-219 is hydrogen-bonded to hydroxyls O3P and O1P of CoA, whereas O2P is hydrogen-bonded to Arg-60. Similarly, the amide N1 of CoA is hydrogen-bonded to β-phosphate of NADP. Interestingly, an aspartate, Asp-137, is hydrogen-bonded to the O2M hydroxyl oxygen of the substrate via a water molecule. This has implications for the mechanism of catalysis of pDCR. The aspartate is replaced by a valine in mDCR. The aliphatic chain of the substrate is not linear; it is bent between consecutive carbon atoms, giving an appearance of a folded chain. This folding of the aliphatic tail probably helps in accommodating longer acyl chains into the active site. The substrate HXC in chain D appears more folded than rest of the substrates modeled in DCRs. Such a folding brings the δ carbon of HXC in proximity of the carbonyl carbon of the NADP for a hydride transfer.
Mutagenesis of pDCR
The absence of tyrosine-serine pair in pDCR prompted us to search for other amino acids critical for catalysis. Hydratases and isomerases functioning in the β-oxidation use acidic residues as proton donors for catalysis (34). We mutated several acidic residues around the active site to alanine and estimated their residual activities against 6- and 10-carbon-long unsaturated acyl chains of fatty acid CoA conjugates (Table 3). In the ternary complex of pDCR, Asp-86 is interacting with the adenine ring of the cofactor NADP. Although this residue appears conserved, a few homologs have an asparagine in place of the aspartate. D86A mutation almost completely abolished the activity of the enzyme against both the substrates. Another acidic residue, Asp-137, is seen interacting with the substrate via a water molecule. This aspartate is not conserved, and interestingly most of the homologs have an asparagine at this position. D137A mutation completely abolished the activity of pDCR. Mutating two other aspartates, Asp-186 and Asp-268, to alanine almost abolished the activity of the enzyme, whereas D155A and E215A mutations resulted in partial residual activity. None of these acidic residues are conserved among the homologs. Although these residues are located away from the ligands, they are likely to play a role in the relay of charge using water molecules as a conduit. Thus, mutagenesis studies on pDCR indicate that acidic residues are imported for catalysis
TABLE 3.
Relative activity of pDCR mutants
| Mutation | Relative activity |
|
|---|---|---|
| 2,4-decadienoyl CoA | 2,4-Hexadienoyl CoA | |
| % | ||
| Wild type | 100 | 100 |
| D86A | 1.70 | 1.76 |
| D137A | 3.32 | 1.27 |
| D155A | 36.3 | 59.6 |
| D186A | 6.21 | 1.86 |
| E215A | 28.9 | 7.59 |
| D268A | 2.75 | 1.14 |
DISCUSSION
Mechanism of Catalysis
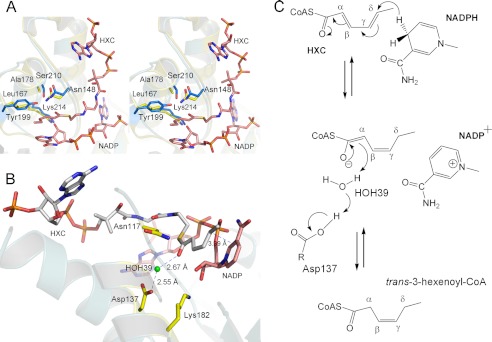
Although the conserved catalytic serine of the SDR family could not be identified by primary sequence alignment for mDCR, a structural view of mDCR confirmed the presence of Tyr-199 and Ser-210 at the catalytic center close to the strictly conserved Lys-214 and Asn-148 (Fig. 4A). Primary sequence analysis of pDCR also failed to identify a tyrosine-serine pair implicated in catalysis (Fig. 1A). Therefore, we searched the active site of pDCR for the tyrosine-serine pair, which together with a lysine and an asparagine residue has been shown to be critical for SDR catalyzed reactions. Surprisingly, superimposition of the active site of pDCR over mDCR revealed that the tyrosine and serine pair of mDCR is replaced by a leucine and an alanine, respectively, in pDCR (Fig. 4A). The invariant lysine occupies a similar position in both the structures. In addition, the position of the asparagine, Asn-117 (Asn-148 in mDCR), is conserved in both the structures. Thus, in the absence of a tyrosine-serine pair in the active site, pDCR might be using a different mechanism for catalysis than that reported for mDCR.
FIGURE 4.
Mechanism of catalysis. A, stereo view of catalytic amino acids of mDCR (blue sticks) superimposed over pDCR (yellow sticks). Tyr-199 is replaced by Leu-167, and Ser-210 is replaced by Ala-178 in pDCR. Lys-214 and Asn-148 of mDCR are conserved in pDCR. Ligands are shown as sticks. B, positions of the catalytically critical amino acids of pDCR and location of the potential proton donor, HOH39, shown as a green sphere. C, schematic diagram of the conversion of HXC to trans-3-hexenoyl CoA by pDCR.
Catalysis by DCRs is known to proceed via the formation of an enolate intermediate (35). Kinetics of isotope-labeled experiments indicate that although the order of binding of the substrate and cofactor to DCRs is random, the enzymatic reaction proceeds in sequential steps. The initial part of the catalysis involves the transfer of a hydride from NADPH to trans-2,trans-4-dienoyl CoA. In the structure of pDCR, the C4 carbon of the nicotamide moiety is 3.4 Å away from the δ carbon of HXC and well positioned to donate the pro-4S hydride, resulting in an enolate intermediate (Fig. 4B). Unlike mDCR, in pDCR there are no side chains of tyrosine and serine within hydrogen bonding distance of the Cα position that could stabilize the intermediate. The invariant lysine, critical for activity, is in the vicinity of Cα and is likely to play a role in the stabilization of the intermediate (Fig. 4A). An aspartate, Asp-137, is hydrogen-bonded via a solvent molecule to the Cα (Fig. 4B). The aspartate probably facilitates the transfer of proton from the solvent molecule to the Cα, resulting in the formation of the product (Fig. 4C). This is partly supported by the fact that the pKa of the aspartate is elevated to 6.5, and pDCR showed optimum activity at pH 6.5 when assayed under conditions mentioned under “Experimental Procedures” (data not shown). Further, a D137A mutation completely abolished the activity of pDCR (Table 3). Asn-117, Gln-175, and Lys-182 located close to Asp-137 probably increase the pKa of the critical aspartate and polarize the proton donor. Although there is a general consensus that the proton is donated by water molecules in DCR-catalyzed reactions, direct structural evidence was lacking. Our studies on pDCR, for the first time, provide this crucial evidence.
Substrate Specificities of DCRs
Very long chain and long chain fatty acids are shortened in peroxisomes and shuttled to the mitochondrion for complete degradation (10). Previous studies have suggested octanoyl CoA as the end product of human peroxisomal β-oxidation. However, our in vitro enzymatic studies reveal that pDCR can utilize HXC as a substrate. The ternary complex of pDCR with HXC and NADP support this biochemical evidence. Inspection of the environment around HXC in the active site of the ternary complex of pDCR revealed that pDCR should be able to utilize HXC as a substrate. Intriguingly, the Km for HXC was almost 6-fold higher than that for decadienoyl CoA (Table 2). Therefore, under physiological conditions, the processing of the substrate by pDCR may be a function of the Km for a particular substrate, and pDCR seems to have lower Km values for longer acyl CoAs. In addition, the inability to orient shorter substrates correctly for catalysis, as reported previously for other enzymes of the β-oxidation pathway, could be another reason why pDCRs cannot process short chain fatty acids (13).
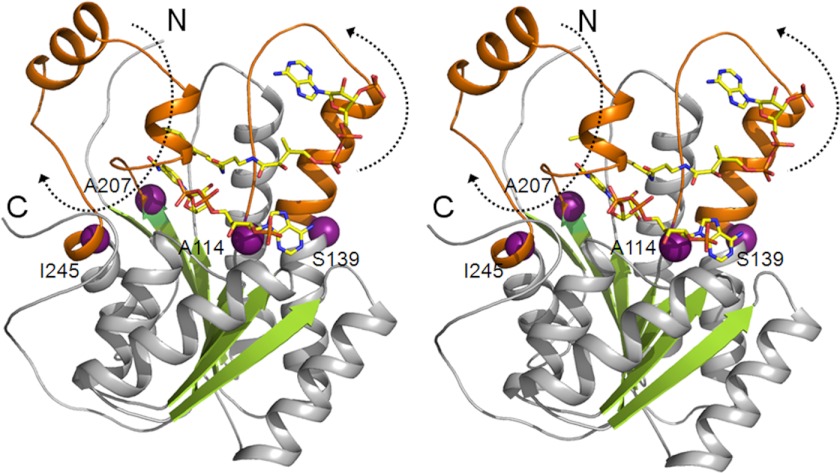
A key difference in the β-oxidation carried out by mitochondrion and peroxisomes lies in the length of the fatty acids processed by the organelles. Although peroxisome can shorten 26-carbon-long fatty acids like hexacosanoic acid, mitochondrion is known to shorten only long, medium, and short chain fatty acids like palmitic acid, which contains 16 carbons, and oleic, linoleic, and linolenic fatty acids that contain 18 carbons (9). The reasons for such differences in substrate specificities between the mitochondrion and peroxisome are unknown. We looked at the DCR structures from peroxisome and mitochondrion for clues to explain the substrate specificities. Analysis of the structures of DCRs from both the organelles in complex with the substrate HXC reveals that there is space around the substrate that can accommodate additional carbons. However, extending the aliphatic tail to a length similar to that of very long chain fatty acids could result in steric clashes. Therefore, to accommodate such very long chain fatty acids, some of the structural elements of the enzyme would have to move. To find out whether the structural elements of DCRs can move, we attempted to cocrystallize pDCR with a longer substrate: 2,4-decadienoyl CoA. However, no electron density could be observed for the longer substrate. We then analyzed the DCR structures from mitochondrion and peroxisome for hinge movements that could potentially open the active site for accommodating longer chains of fatty acids. Interestingly, a HingeProt analysis (36) revealed that the DCR from peroxisome is capable of very large hinge movements when compared with the DCR from mitochondrion, which did not show such movements of the structural elements. The extended portion of the helix α4 and the loop connecting strand β4 with helix α4 (where the adenine moiety of the substrate binds) swing back, opening up the active site. The structural elements swing around the residue Ala-114 of the loop and Ser-139 from the α4 helix. Further, the loop connecting strand β6 with helix α6, located close to the aliphatic tail of the substrate HXC and partially covering it, shows a large movement depicting opening and capping of the substrate-binding site (Fig. 5). Thus, the DCR from the peroxisome is able to accommodate and shorten very long chain fatty acids because of its inherent ability to widen the active site via hinge movement of structural elements. Such a movement of structural elements could explain why the peroxisomal enzymes are able to process very long chain fatty acids, whereas their mitochondrial counterparts cannot.
FIGURE 5.
Hinge movement of pDCR. Stereo view of the cartoon of pDCR depicting hinge movement. Large hinge movements of structural elements (orange) of pDCR were predicted by HingeProt server. Black dotted arrows indicate the direction of the movement. Such hinge movements are not observed for mitochondrial DCR (Protein Data Bank code 1W6U). The N and C termini of the protein are marked. The hinge residues are shown as spheres, whereas the ligands are shown as sticks.
In summary, the structure of the ternary complex of pDCR with NADP and its substrate provides essential and unique insights into the mechanism of catalysis. Unlike other members belonging to the SDR family, catalysis by pDCR does not involve a tyrosine-serine pair. Instead, a catalytically critical aspartate, together with an invariant lysine, polarizes a water molecule to donate a proton for the formation of the product. Our studies provide the first structural evidence for the participation of a water molecule in DCR-catalyzed reactions. We show that although pDCR can use 2,4-hexadienoyl CoA as a substrate, the affinities for short chain fatty acids are lower. Analysis of the hinge movement of DCRs from the mitochondrion and peroxisomes sheds light on the reason behind the unique ability of the peroxisome to shorten very long chain fatty acids. This information could be used to modulate activity of the enzymes involved in β-oxidation for adjusting the flux of fatty acids through the mitochondrion and peroxisomes.
Supplementary Material
Acknowledgments
We thank the Southeast Regional Collaborative Access Team 22-ID Beamline at the Advanced Photon Source, Argonne National Laboratory for data collection.
This work was supported by National Natural Science Foundation of China Grants 31070660, 31021062, and 31000334; Ministry of Science and Technology of China Grants 2009DFB30310, 2009CB918803, 2011CB911103, and 2009CB918803; and Chinese Academy of Sciences Research Grants YZ200839 and KSCX2-EW-J-3.

This article contains supplemental Fig. S1.
The atomic coordinates and structure factors (codes 4FC7 and 4FC6) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- DCR
- 2,4-dienoyl CoA reductase
- SDR
- short chain dehydrogenase
- mDCR
- mitochondrial DCR
- pDCR
- peroxisomal DCR
- HXC
- 2,4-hexadienoyl CoA
- NADP
- nicotinamide adenine dinucleotide phosphate.
REFERENCES
- 1. Blomkalns A. L., Silver D. W. (2011) The obese patient, in Challenging and Emerging Conditions in Emergency Medicine, Wiley-Blackwell, New York [Google Scholar]
- 2. Flegal K. M., Graubard B. I., Williamson D. F., Gail M. H. (2005) Excess deaths associated with underweight, overweight, and obesity. J. Am. Med. Assoc. 293, 1861–1867 [DOI] [PubMed] [Google Scholar]
- 3. Kalmijn S., Feskens E. J., Launer L. J., Kromhout D. (1997) Polyunsaturated fatty acids, antioxidants, and cognitive function in very old men. Am. J. Epidemiol. 145, 33–41 [DOI] [PubMed] [Google Scholar]
- 4. Kenchaiah S., Evans J. C., Levy D., Wilson P. W., Benjamin E. J., Larson M. G., Kannel W. B., Vasan R. S. (2002) Obesity and the risk of heart failure. N. Engl. J. Med. 347, 305–313 [DOI] [PubMed] [Google Scholar]
- 5. Hotamisligil G. S., Shargill N. S., Spiegelman B. M. (1993) Adipose expression of tumor necrosis factor α. Direct role in obesity-linked insulin resistance. Science 259, 87–91 [DOI] [PubMed] [Google Scholar]
- 6. Stulnig T. M., Zeyda M. (2004) Immunomodulation by polyunsaturated fatty acids. Impact on T-cell signaling. Lipids 39, 1171–1175 [DOI] [PubMed] [Google Scholar]
- 7. Pearce M. L., Dayton S. (1971) Incidence of cancer in men on a diet high in polyunsaturated fat. Lancet 297, 455–504 [DOI] [PubMed] [Google Scholar]
- 8. Berg J. M., Tymoczko J. L., Styer L. (2002) Fatty Acid Metabolism, 5th Ed., W. H. Freeman and Company, New York [Google Scholar]
- 9. Reddy J. K., Hashimoto T. (2001) Peroxisomal β-oxidation and peroxisome proliferator activated receptor α. An adaptive metabolic system. Annu. Rev. Nutr. 21, 193–230 [DOI] [PubMed] [Google Scholar]
- 10. Fidaleo M. (2010) Peroxisomes and peroxisomal disorders. The main facts. Exp. Toxicol. Pathol. 62, 615–625 [DOI] [PubMed] [Google Scholar]
- 11. Ferdinandusse S., Mulders J., Ijlst L., Denis S., Dacremont G., Waterham H. R., Wanders R. J. (1999) Molecular cloning and expression of human carnitine octanoyltransferase. Evidence for its role in the peroxisomal β-oxidation of branched-chain fatty acids. Biochem. Biophys. Res. Commun. 263, 213–218 [DOI] [PubMed] [Google Scholar]
- 12. Hiltunen J. K., Qin Y. (2000) β-Oxidation strategies for the metabolism of a wide variety of acyl-CoA esters. Biochim. Biophys. Acta 1484, 117–128 [DOI] [PubMed] [Google Scholar]
- 13. Ylianttila M. S., Pursiainen N. V., Haapalainen A. M., Juffer A. H., Poirier Y., Hiltunen J. K., Glumoff T. (2006) Crystal structure of yeast peroxisomal multifunctional enzyme. Structural basis for substrate specificity of (3R)-hydroxyacyl CoA dehydrogenase units. J. Mol. Biol. 358, 1286–1295 [DOI] [PubMed] [Google Scholar]
- 14. Kerner J., Hoppel C. L. (2004) Carnitine and β-oxidation, in Encyclopedia of Biological Chemistry (William J. L., Lane M. D., eds) Elsevier, New York [Google Scholar]
- 15. Schulz H., Dennis E. Vance J. E. (2002) Oxidation of fatty acids in eukaryotes, in New Comprehensive Biochemistry, Elsevier, New York [Google Scholar]
- 16. Wanders R. J., Waterham H. R. (2006) Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 75, 295–332 [DOI] [PubMed] [Google Scholar]
- 17. Kavanagh K. L., Jörnvall H., Persson B., Oppermann U. (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families. Cell. Mol. Life Sci. 65, 3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fillgrove K. L., Anderson V. E. (2000) Orientation of coenzyme A substrates, nicotinamide and active site functional groups in (di)enoyl-coenzyme A reductases. Biochemistry 39, 7001–7011 [DOI] [PubMed] [Google Scholar]
- 19. Filling C., Berndt K. D., Benach J., Knapp S., Prozorovski T., Nordling E., Ladenstein R., Jörnvall H., Oppermann U. (2002) Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J. Biol. Chem. 277, 25677–25684 [DOI] [PubMed] [Google Scholar]
- 20. Alphey M. S., Yu W., Byres E., Li D., Hunter W. N. (2005) Structure and reactivity of human mitochondrial 2,4-dienoyl CoA reductase. J. Biol. Chem. 280, 3068–3077 [DOI] [PubMed] [Google Scholar]
- 21. Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., Donnelly M. I. (2002) A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 22. Thomason S. C., Kubler D. G. (1968) Acids as derivatives of aldehydes prepared with silver oxides. J. Chem. Ed. 45, 546 [Google Scholar]
- 23. Sobhi H. F., Minkler P. E., Hoppel C. L. (2010) Synthesis and characterization of cis-4-decenoyl CoA, 3-phenylpropionyl CoA, and 2,6-dimethylheptanoyl CoA. Anal. Biochem. 401, 114–124 [DOI] [PubMed] [Google Scholar]
- 24. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25. Lebedev A. A., Vagin A. A., Murshudov G. N. (2008) Model preparation in MolRep and examples of model improvement using x-ray data. Acta Crystallogr. D Biol. Crystallogr. 64, 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27. Brünger A. T. (1992) Free R-value. A novel statistical quantity for assessing the accuracy of crystal-structures. Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 28. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Penning T. M., Bennett M. J., Smith-Hoog S., Schlegel B. P., Jez J. M., Lewis M. (1997) Structure and function of 3-α-hydroxysteroid dehydrogenase. Steroids 62, 101–111 [DOI] [PubMed] [Google Scholar]
- 30. Tanaka N., Nonaka T., Tanabe T., Yoshimoto T., Tsuru D., Mitsui Y. (1996) Crystal structures of the binary and ternary complexes of 7α-hydroxysteroid dehydrogenase from Escherichia coli. Biochemistry 35, 7715–7730 [DOI] [PubMed] [Google Scholar]
- 31. Jörnvall H., Persson B., Krook M., Atrian S., Gonzàlez-Duarte R., Jeffery J., Ghosh D. (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34, 6003–6013 [DOI] [PubMed] [Google Scholar]
- 32. Laskowski R. A., Watson J. D., Thornton J. M. (2005) Profunc. A server for predicting protein function from 3d structure. Nucleic Acids Res. 33, W89–W93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr., Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. (1977) The Protein Data Bank. A computer-based archival file for macromolecular structures. J. Mol. Biol. 112, 535–542 [DOI] [PubMed] [Google Scholar]
- 34. Yu W., Chu X., Chen G., Li D. (2005) Studies of human mitochondrial 2,4-dienoyl CoA reductase. Arch. Biochem. Biophys. 434, 195–200 [DOI] [PubMed] [Google Scholar]
- 35. Fillgrove K. L., Anderson V. E. (2001) The mechanism of dienoyl CoA reduction by 2,4-dienoyl CoA reductase is stepwise. Observation of a dienolate intermediate. Biochemistry 40, 12412–12421 [DOI] [PubMed] [Google Scholar]
- 36. Emekli U., Schneidman-Duhovny D., Wolfson H. J., Nussinov R., Haliloglu T. (2008) HingeProt. Automated prediction of hinges in protein structures. Proteins 70, 1219–1227 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.