Background: Biochemical studies indicate the presence of an ADP-inhibited state in V1-ATPase.
Results: Single-molecule analysis showed two types of pauses during rotation, a second-scale pause and the other an irreversible long pause.
Conclusion: Long pause corresponds to the ADP-inhibited state of V1-ATPase.
Significance: This is the first time to show that V1-ATPase has a second-scale pause and also that an ADP-inhibited V1-ATPase could resume active rotation under external manipulation.
Keywords: ATP Synthase, Enzyme Kinetics, H+-ATPase, Molecular Motors, Single-molecule Biophysics, Vacuolar ATPase, Long and Short Pauses, Mechanical Manipulation by Magnetic Tweezers, Single-molecule Experiment
Abstract
V1-ATPase, the hydrophilic V-ATPase domain, is a rotary motor fueled by ATP hydrolysis. Here, we found that Thermus thermophilus V1-ATPase shows two types of inhibitory pauses interrupting continuous rotation: a short pause (SP, 4.2 s) that occurred frequently during rotation, and a long inhibitory pause (LP, >30 min) that terminated all active rotations. Both pauses occurred at the same angle for ATP binding and hydrolysis. Kinetic analysis revealed that the time constants of inactivation into and activation from the SP were too short to represent biochemically predicted ADP inhibition, suggesting that SP is a newly identified inhibitory state of V1-ATPase. The time constant of inactivation into LP was 17 min, consistent with one of the two time constants governing the inactivation process observed in bulk ATPase assay. When forcibly rotated in the forward direction, V1 in LP resumed active rotation. Solution ADP suppressed the probability of mechanical activation, suggesting that mechanical rotation enhanced inhibitory ADP release. These features were highly consistent with mechanical activation of ADP-inhibited F1, suggesting that LP represents the ADP-inhibited state of V1-ATPase. Mechanical activation largely depended on the direction and angular displacement of forced rotation, implying that V1-ATPase rotation modulates the off rate of ADP.
Introduction
Two types of rotary ATPase/synthase are found in nature as follows: vacuolar type ATPase/synthase (VoV1) and F-type ATPase/synthase (FoF1) (1–3). Eukaryotic VoV1 counterparts are also found in some eubacteria, including Thermus thermophilus and Enterococcus hirae, and archaea (1). These counterparts are sometimes referred as A-type ATPase or AoA1 (4). Similar to FoF1, VoV1 consists of two distinct motor domains as follows: hydrophilic V1, which catalyzes either ATP hydrolysis or synthesis, and hydrophobic Vo, which facilitates proton translocation across membranes. These domains are coupled by rotation of a central rotor complex against a surrounding stator apparatus (5, 6). Although VoV1 principally acts as an H+-pump in the vacuoles of eukaryotes, archaea employ VoV1 as an ATP synthase as well as some eubacteria such as T. thermophilus of which VoV1 was investigated in this study.
For T. thermophilus VoV1, the V1 motor domain is called V1-ATPase and exhibits ATP hydrolysis activity when isolated from the Vo sector. T. thermophilus V1-ATPase has the subunit composition of A3B3DF. A3B3 forms a hexameric ring in which three A and B subunits are alternately arranged. The catalytic reaction center resides at the A-B interface, mainly on the A subunit. The D and F subunits form the rotary shaft; the D subunit penetrates the central cavity of the A3B3 ring, and the F subunit binds to the protruding part of subunit D (7).
T. thermophilus V1-ATPase has been well characterized in both biochemical studies and single-molecule experiments (8–10). V1-ATPase has common features of rotation with F1-ATPase. V1 demonstrates counterclockwise rotation when viewed from the membrane side, as does F1. The elementary step size of the rotation is 120°, each probably coupled with a single turnover of ATP hydrolysis. The torque of V1-ATPase is ∼35 piconewtons·nm, which is slightly smaller than that of F1-ATPase (40 piconewtons·nm) (11, 12).
The coupling reaction scheme of V1-ATPase is distinct from that of F1-ATPase. Although the 120° step of F1 is composed of 80° and 40° substeps, isolated V1 does not show any substeps. In V1-ATPase, ATP hydrolysis takes place at the same angle of ATP binding (12), and F1 executes ATP hydrolysis and release of inorganic phosphate at +80° from the ATP-binding angle, termed the catalytic angle (13–15). Furthermore, the latest single-molecule experiment using a drag-free probe suggested that V1-ATPase releases reaction products at the same angle as hydrolysis and ATP binding (9). Thus, the reaction scheme of V1 is likely to differ from that of F1. Comparative research on V1-ATPase and F1-ATPase would provide important implications about the common working principles and unique mechanisms of these motors.
The inhibitory effects of regulatory proteins or chemical reagents on enzymes are generally observed as a pause or stall of catalytic turnover at the single-molecule level. In the case of F1-ATPase, the ADP-inhibited form is the most common inhibitory state found in various types of F1-ATPases and is thought to play a regulatory role (16). During catalytic turnover of ATP hydrolysis, F1 stochastically fails to release ADP during turnover and pauses catalysis by tightly binding to ADP. In single-molecule rotation assays, the ADP-inhibited form of F1 is observed as a pausing state lasting 30 s that intervenes in continuous rotation (17). The ϵ subunit, the inhibitory subunit of F1 and tentoxin, a cyclic tetrapeptide, has also been reported to cause frequent and stable pauses of rotation, respectively (18, 19). RNA polymerase is also known to demonstrate transiently pausing states, which have been discussed in relation to the regulatory mechanism for translation (20). Thus, kinetic and thermodynamic analyses of the pausing state and catalytically active state reveal important implications concerning the regulatory mechanism of enzymes.
In contrast to the basic features of the rotation, the slow transition between the actively rotating state and the inhibitory pausing state of V1-ATPase has not been well characterized in single-molecule rotation assays. Biochemical studies of T. thermophilus V1-ATPases showed that ADP inhibition occurs by the same mechanism as the ADP inhibition of F1-ATPase (10). In this study, we characterized the transition between the rotating state and pausing state of T. thermophilus V1-ATPase in the single-molecule rotation assay. We found two types of V1-ATPase pausing states. One is a frequently occurring and second-scale reversible “short pause.” The other type of inhibitory state, which always terminated the rotation, was an extremely stable “long pause” with a long lifetime, so we were unable to determine the mean time of the long pause under the present experimental conditions. We analyzed kinetic features of these two types of pauses and discuss them in relation to the ADP inhibition of V1-ATPase observed in bulk ATPase assays. We also attempted to rescue V1-ATPase from the long pause state with external force. Like the ADP-inhibited form of F1-ATPase, forcible rotation induced the reproducible activation of V1-ATPase from the long pause. The observed angular dependence of the activation suggests that V1-ATPase modulates the off rate of inhibitory ADP upon rotation.
EXPERIMENTAL PROCEDURES
Preparation of His-tagged Wild-type V1
His-tagged wild-type V1, A(His-10/C28S/C508S)3B(C264S)3D(E48C/Q55C)F, was expressed in Escherichia coli and purified as described previously (8). E. coli cells were suspended in buffer A, composed of 100 mm sodium phosphate (NaPi) at pH 8.0, 300 mm NaCl, and 20 mm imidazole (pH 8.0). Cell lysate was prepared by sonication, followed by heat treatment at 65 °C for 30 min. Denatured proteins were removed by centrifugation at 10,000 rpm for 90 min. Solution was applied to a nickel-nitrilotriacetic acid column equilibrated with buffer A. V1 was eluted with buffer B (buffer A with 200 mm imidazole-HCl at pH 8.0). The buffer was exchanged for 20 mm Tris-HCl (pH 8.0) containing 1 mm EDTA with a centrifugal filter (Amicon ultrafiltration unit, Millipore) and then applied to an ion exchange column (UNO-Q, Bio-Rad). Protein was eluted with a linear gradient of NaCl (0–400 mm). After collecting eluted fractions of V1, the buffer was exchanged for 100 mm NaPi (pH 8.0) containing 10 mm EDTA with an Amicon ultrafiltration unit.
Removal of Bound ADP
Purified V1 contained bound nucleotides and had low ATPase activity. Tightly bound ADP was removed as described previously (8, 21). Enzyme solution was heated at 65 °C for 10 min, followed by cooling on ice for 30 min. This process was repeated five times, and the solution was then applied to a desalting column (PD-10 column, Amersham Biosciences) equilibrated with ADP removal buffer consisting of 100 mm NaPi (pH 8.0) and 10 mm EDTA (8). After the nucleotide-depletion procedure, V1 was incubated with DTT at 25 °C for 2 h to reduce the cysteine residues for the subsequent biotinylation procedure. To remove excess DTT, the sample was subjected to a gel filtration column (Superdex 75HR, GE Healthcare) equilibrated with 20 mm MOPS-KOH (pH 7.0) containing 150 mm NaCl. The eluted sample was concentrated with a centrifugal filter. Purified enzyme was biotinylated with maleimide-PEG2-biotin (Pierce) at a 1:5 ratio at room temperature for 2 h. Free biotin was removed with a desalting column (NAP10, GE Healthcare). Labeled V1 was flash-frozen in liquid N2 and stored at −80 °C until used.
Protein Quantification and ATPase Assay
The protein concentration of V1 was determined from UV absorbance using a molar extinction coefficient of 360,000 m−1 cm−1 that was calibrated from quantitative amino acid analysis. ATPase activity was measured with an ATP-regenerating system containing 50 mm Tris-HCl (pH 8.0), 100 mm KCl, 2 mm MgCl2, 2 mm phosphoenol pyruvate, 0.2 mg/ml NADH, 0.1 mg/ml pyruvate kinase, 0.1 mg/ml lactate dehydrogenase, and the indicated amount of Mg-ATP. The rate of ATP hydrolysis was monitored from the rate of NADH oxidation, determined by the absorbance decrease at 340 nm.
Rotation Assay
Coverslips coated with nickel-nitrilotriacetic acid were prepared as described previously (22). Streptavidin-coated colloidal gold beads were prepared as follows: colloidal gold of 40 nm diameter (BB International, UK) was mixed with 10 mm boric acid/NaOH buffer containing 3 mg/ml Tween 20 and incubated for 30 min. Biotin-EG3-undecanethiol (SensoPath) was mixed with carboxy-EG6-undecanethiol (Dojindo, Japan) and hydroxy-EG6-undecanethiol (Dojindo, Japan). The solution was then mixed with biotin mixture and incubated at 70 °C overnight. Then solution was washed several times with 10 mm boric acid/NaOH buffer containing 3 mg/ml Tween 20 by centrifugation (15,000 rpm, 5 min) and suspended in this buffer. Next, streptavidin-containing solution (1 mg of streptavidin in boric acid/NaOH buffer containing Tween 20) was mixed with the colloid solution at a 1:1 ratio and incubated for 3 h at room temperature with gentle mixing. The solution was washed several times with the boric acid buffer by centrifugation at 15,000 rpm for 5 min. The sediment of the colloid was suspended in the boric acid buffer and stored at 4 °C.
A flow cell of 5–10 μl in volume was made of two coverslips (bottom, 24 × 36 mm2, and top, 24 × 24 mm2) separated by two spacers of 50-μm thickness. Biotinylated V1 in buffer C (50 mm Tris-HCl (pH 8.0), 100 mm KCl) was infused into a flow cell and incubated for 5 min. Unbound V1 was washed out with 50 μl of buffer C. Then buffer C containing 1% BSA was infused into the flow cell to reduce nonspecific binding of the beads or colloid particles. After incubation for several minutes, the solution of a rotation marker particle, magnetic beads (Seradyn, Thermo Scientific), or the custom colloidal gold beads was infused into the flow cell. After incubation for 20 min or more, unbound beads were washed out with 70 μl of buffer C. Observation of rotation was initiated after infusion of 140 μl of buffer D (50 mm Tris-HCl (pH 8.0), 100 mm KCl, 2 mm MgCl2) containing an indicated amount of Mg-ATP. For ADP-free experiments, the ATP-regenerating system (50 μg/ml pyruvate kinase, 1 mm phosphoenolpyruvate) was added to buffer D. Rotation of the bead was observed under phase-contrast microscopy (IX70, Olympus) using a ×100 objective lens. Images were captured with a charge-coupled device camera (FC300M; Takenaka) and recorded with a DV-CAM device (DSR-11; Sony) at 30 frames/s. Analysis of rotation was performed using custom software (Digimo). Time-averaged rotation speed was calculated over five consecutive revolutions. All experiments were carried out at 23–25 °C.
RESULTS
Two Types of Inhibitory Pauses, Short Pause (SP)2 and Long Pause (LP)
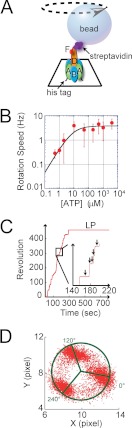
Rotation of V1 molecules was observed at the single-molecule level by attaching a magnetic bead of 200–500 nm in diameter as a rotation marker to the D subunit constituting the rotary shaft with the F subunit (Fig. 1A). The A3B3 ring was immobilized on the glass surface via His tags introduced at the N termini of the A subunits. The rotary motion of the rotor complex was visualized as the rotation of the magnetic beads under an optical microscopic field. Because of the high viscous friction against the rotating bead, the rotation rate was limited to ∼8 rev/s. The rotational velocity was determined at ATP concentrations from 0.5 μm to 4 mm to obtain the Michaelis-Menten curve (Fig 1B). The Vmax and Km values were determined to be 3.8 rev/s and 8.1 μm, giving the apparent rate constant of ATP binding (konATP) as 1.39 × 106 s−1 m−1, which was consistent with the konATP values determined with a 40-nm gold colloid in our previous study (9) and ATP hydrolysis activity (8). Fig. 1C shows a typical time course of V1 rotation at 4 mm ATP, where time constants of ATP binding dwell and ATP cleavage dwell are 0.3 and 2.5 ms, respectively (9), both of which are negligible compared with the time for 120° rotation (∼42 ms). Therefore, V1 molecules generally demonstrated smooth rotation. However, rotation of V1 frequently stopped for several seconds, which is too long to be attributed to the catalytic or ATP-waiting pause of 0.3–2.5 ms (insets of Fig. 1C). This inhibitory pause was transient; after a few seconds, V1 spontaneously resumed to rotate again. The positions of the pause were separated by a 120° interval, suggesting that the pause was due to a slow transition to a catalytically inactive state (Fig. 1D). The rotation and the second-scale pause were repeatedly observed until V1 completely stopped rotation. The angular position of this final long pause corresponded to one of the three positions of the second-scale pause (see below). The coincidence of the pausing angles suggests that the final long pause is also caused by an intrinsic inactivation process. This long pause was practically irreversible; once V1 lapsed into the long pausing state, V1 never resumed rotation even after 30 min. In the longest observation period, the paused V1 was observed for more than 5 h. The molecule did not resume active rotation. Hereafter, we refer to the final pause as LP and to the reversible second-scale pause as SP.
FIGURE 1.
Two types of pauses found in rotation assay of V1-ATPase. A, experimental setup for the single-molecule experiment (not to scale). A single molecule of V1 was attached to a glass surface through His10 tags at each A subunit. A streptavidin-coated magnetic bead with a diameter of ∼500 nm was attached to the D subunit that, along with the F subunit, constitutes the rotary shaft. V1 rotates counterclockwise. B, rotational rate versus [ATP]. By fitting with Michaelis-Menten equation, the maximum rate (Vmax) and Michaelis-Menten constant (Km) were determined as 3.8 rev/s and 8.1 μm. The apparent rate constant of ATP binding (konATP) was estimated as 1.39 × 10−6 s−1 m1 from Vmax/Km. C, rotation trajectory of a single V1 molecule. The molecule showed frequent reversible pauses, which we called the short pause. After ∼5 min of rotation, the molecule lapsed into an irreversible long pause state, from which spontaneous recovery was never observed unless forcibly rotated. D, X-Y trajectory of the molecule shown in C before lapsing into the LP state. The pausing positions of the SP states were separated by 120° intervals.
Angular Position of the Pauses
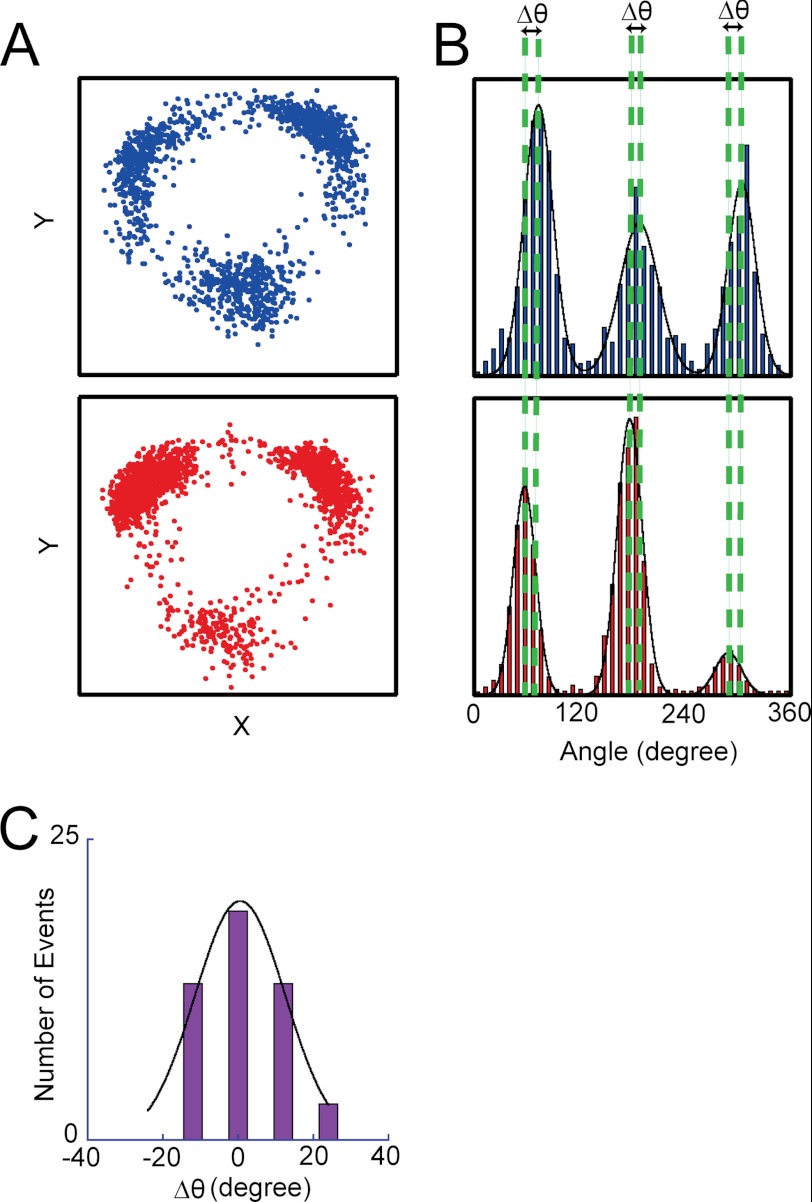
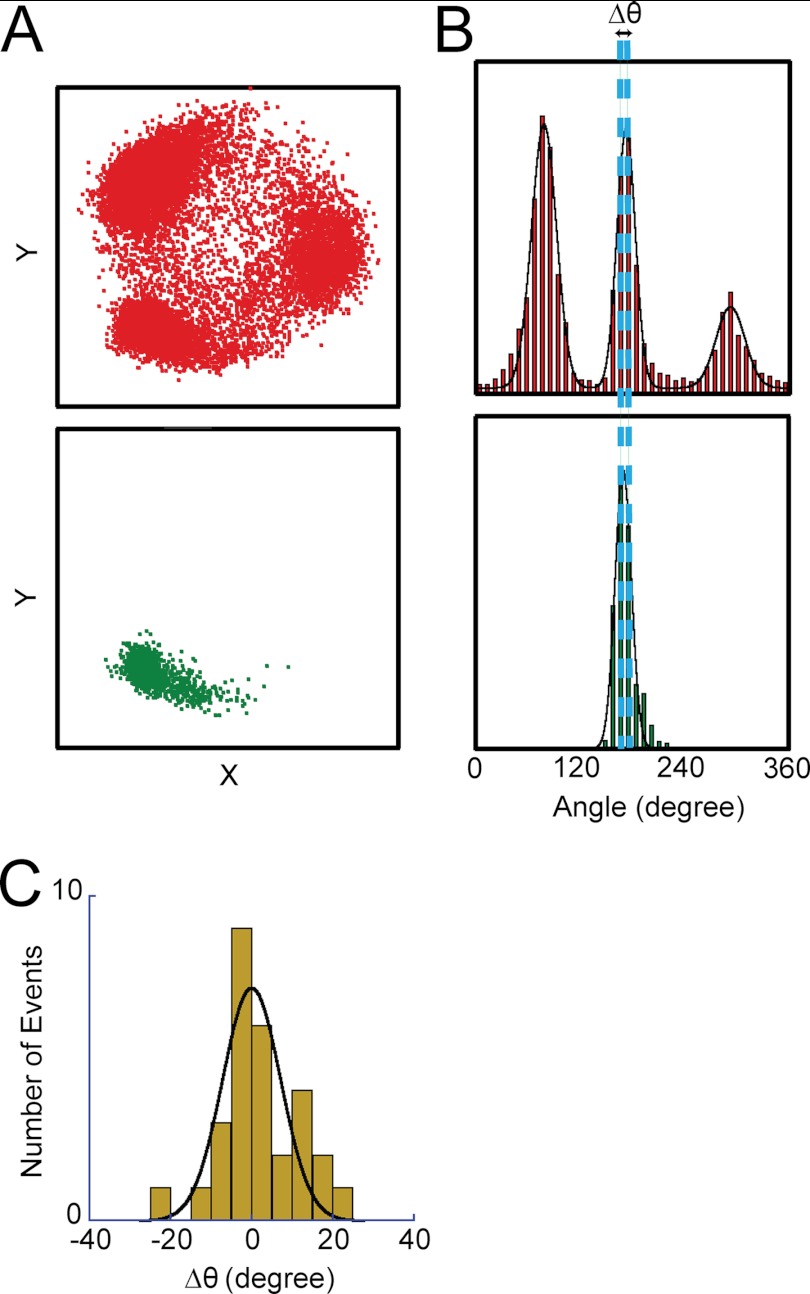
To characterize SP and LP states, we determined the angular positions of SP and LP in relation to the ATP-waiting angle. First, rotation was observed under ATP-limiting conditions to determine the ATP-waiting angles for individual molecules. At 4 μm ATP, which is well below the Km value, 8.1 μm, the overall reaction rate is determined by the ATP-binding step, and V1 mostly spends time in an ATP-waiting pause. Note that under ATP-limiting conditions, time constants obtained from fitting well correlate with theoretical time constants calculated by konATP (Figs. 2 and 3). This implies the occurrence of SP was much lower than that of the ATP-waiting pause at nonsaturating [ATP]. Therefore, ATP-waiting angles (Fig. 4A) were readily determined as three peaks found in the histogram of the angular position during the rotation (Fig. 4B, top panel). After identifying molecules that showed 3-fold symmetry pausing equally at three ATP-waiting angles, the buffer was exchanged with buffer containing 4 mm ATP, where the ATP-waiting pause diminished and SP became prominent. The histogram of the angle during rotation at 4 mm showed the three positions of SP (Fig. 4B, bottom panel). The angular positions of SP coincided well with those of the ATP-waiting angles. The histogram of the angular difference of SP from the ATP-waiting angle was only 0.6 ± 12° (mean ± S.D.; n = 48) (Fig. 4C). Thus, the angular positions for SP were found to be the same as the ATP-waiting angles.
FIGURE 2.
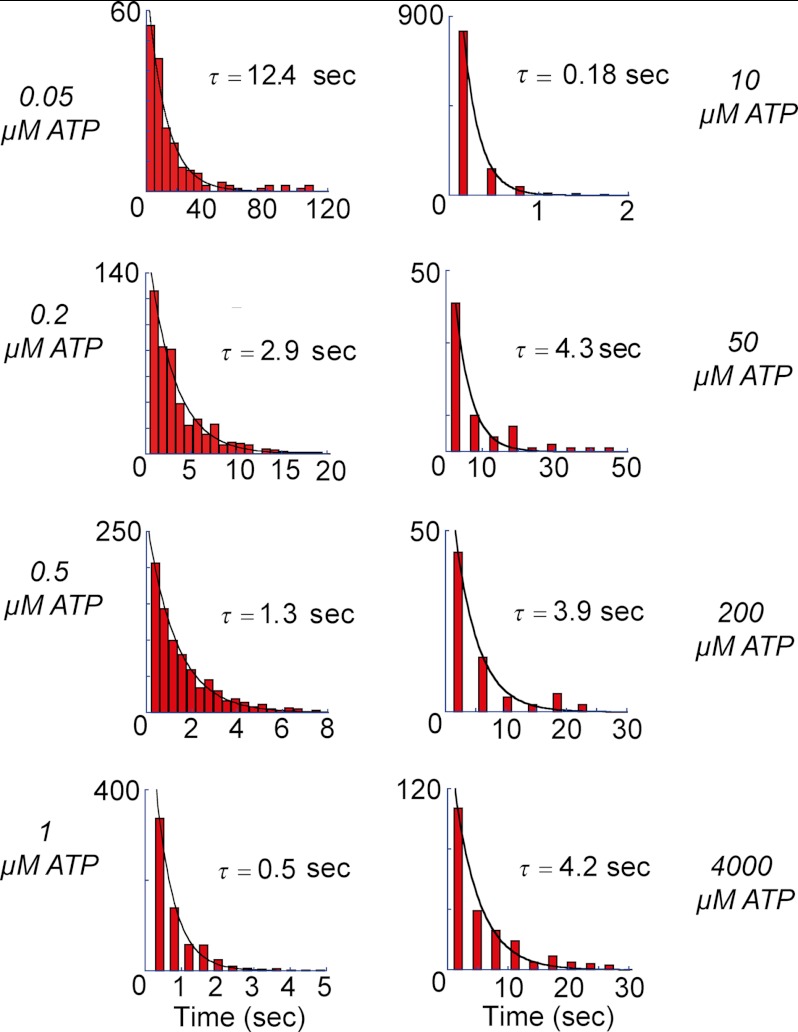
Dwell time histograms of temporary pauses, including both SP and ATP-waiting pause at varying [ATP] that are indicated at the side of the figures. Data were fitted with single exponential, displaying the derived time constants. As observed, time constants at low [ATP], including 10 μm, showed a gradual decrease, and even though at higher [ATP], they did not really change.
FIGURE 3.
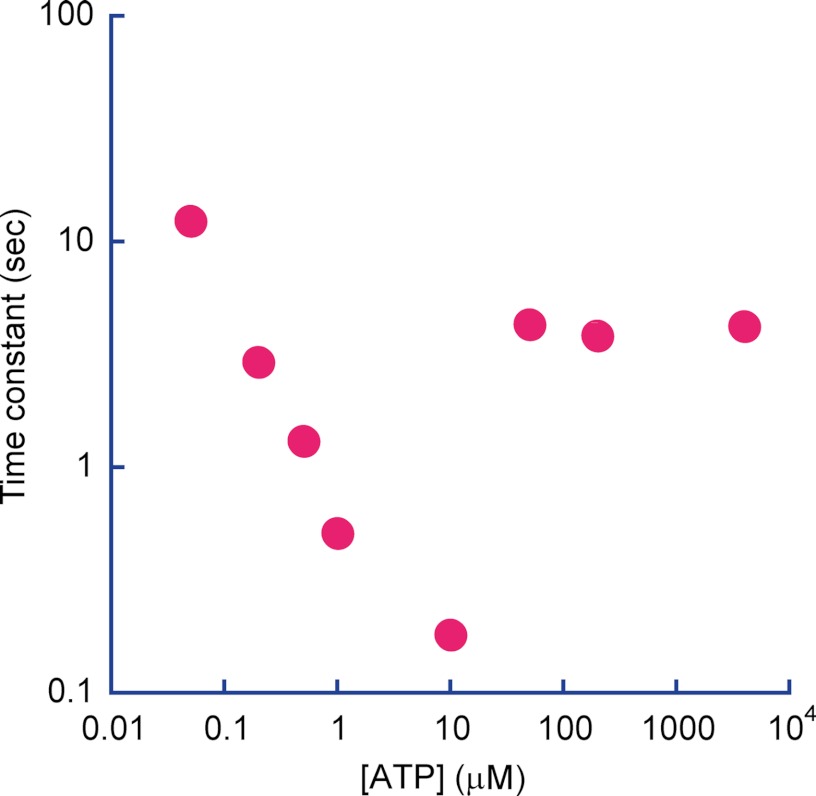
Time constants from Fig. 2 were plotted against corresponding [ATP]. As seen from 0.05 to 10 μm ATP, time constants decreased by matching with the kon for ATP binding at those [ATP]. However, at higher [ATP], the time constants were almost same around 4 s, indicating the dominant rate-limiting step is a short pause.
FIGURE 4.
Angular positions of SP and ATP binding. A, X-Y trajectories of a single V1 molecule at 4 μm ATP (top panel) and at 4 mm ATP (bottom panel). After observing a rotation at 4 μm ATP in which V1 demonstrated ATP-waiting pauses, the ATP concentration was increased by buffer exchange to 4 mm, at which V1 demonstrated the SPs. Therefore, three positions in each trajectory represent the ATP-waiting angles and the SP angles, respectively. B, angle histograms of the single molecule of V1 is shown in A at 4 μm ATP (top panel) and at 4 mm (bottom panel). Dotted lines show the mean positions of ATP-waiting angles and the SP angles. C, angular deviation between ATP binding and SP dwell determined from 48 observations. Mean is 0.6 ± 12°.
The angular position of LP was also determined. Because, as mentioned above, V1 lapses into the final LP state after repeated SPs under saturating ATP conditions, the final pausing angle of LP was determined from comparison with those of SP for individual molecules at 4 mm ATP. As shown in Fig. 5, A and B, the angular position of LP was approximately identical to one of the three angles of SP. The angular deviation determined from statistical analysis (Fig. 5C) was −0.1 ± 7.2° (mean ± S.D.; n = 29). Thus, the angular position of LP is also the same as the ATP-waiting angle.
FIGURE 5.
Angular positions of SP and LP. A, X-Y trajectory of a single V1 molecule before entering LP (top panel) and after LP (bottom panel). Rotation was observed at 4 mm ATP. B, angle histograms of SP (top panel) and LP (bottom panel) dwells from X-Y trajectories in A. C, angular deviation between SP and LP determined from 29 observations. The angular position of LP was compared with that of the nearest SP. Mean was −0.1 ± 7.2°.
Kinetic Analysis of Short Pause and Long Pause
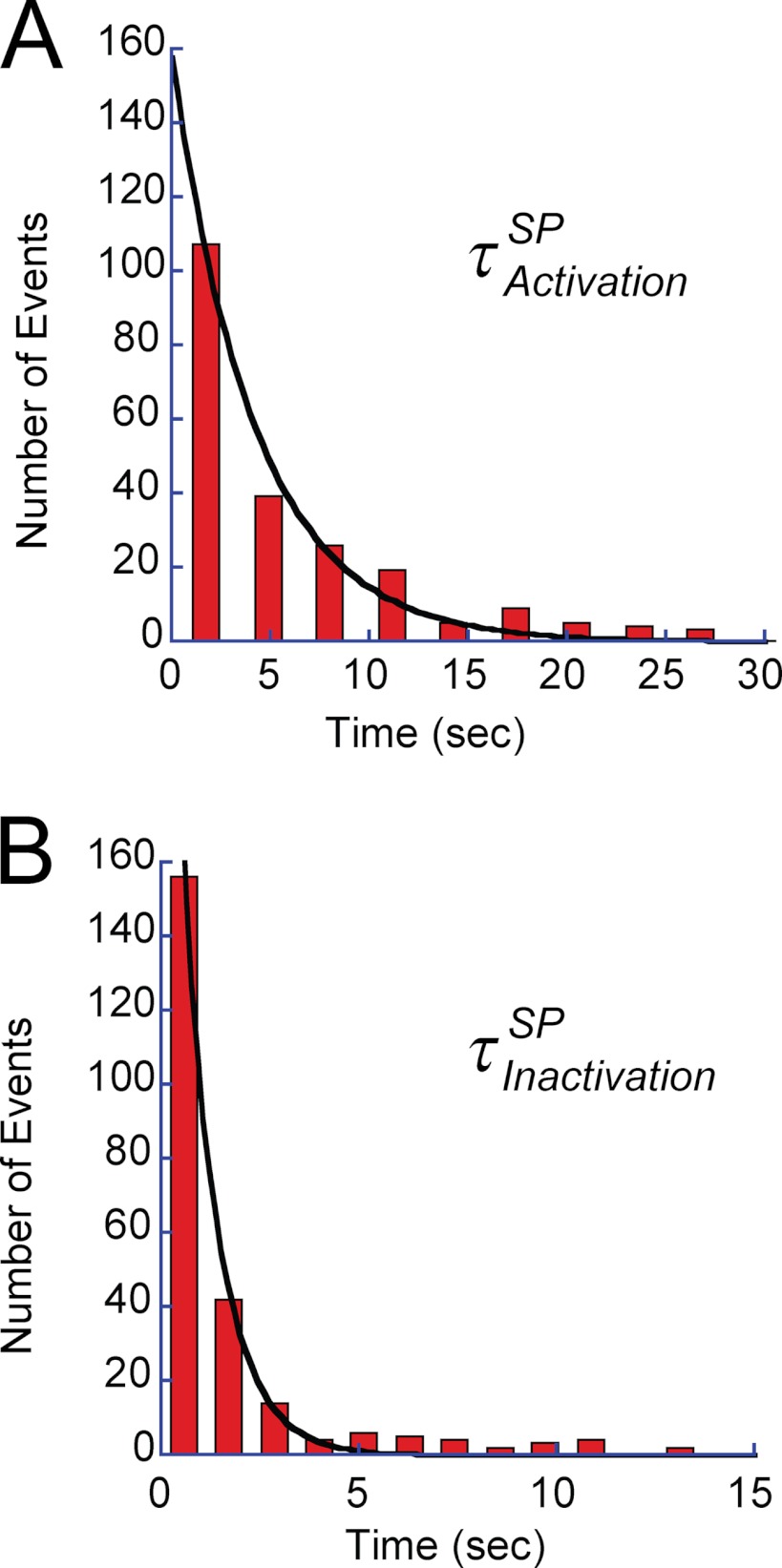
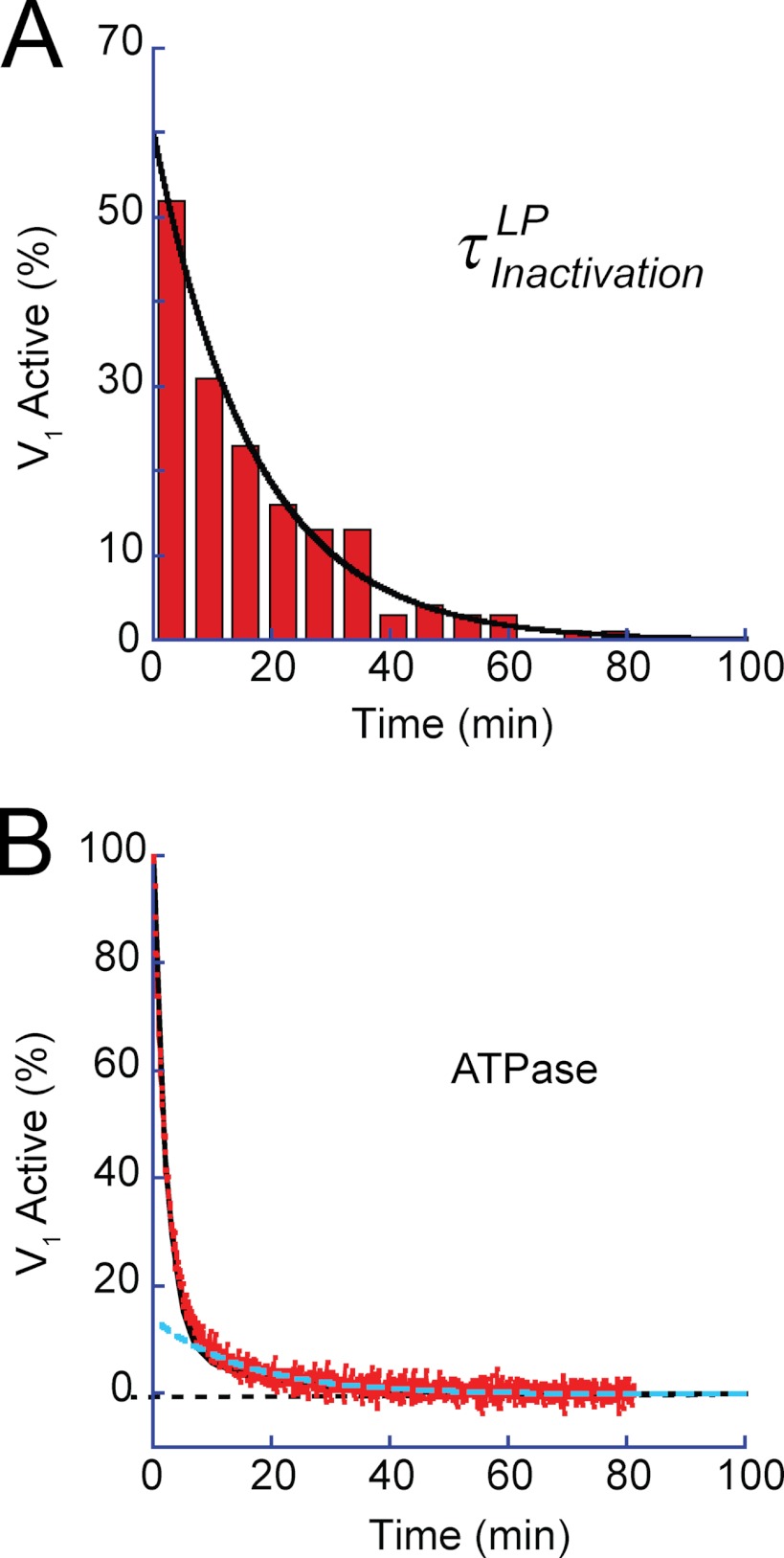
Fig. 6, A and B, shows the histograms of the duration time of SP and the rotation time between successive SPs, respectively. Both demonstrated exponential decay, giving time constants of the inactivation into SP and activation from SP as follows: τinactivationSP (0.9 s) and τactivationSP (4.2 s), respectively. Thus, V1 remained in the active state (rotation state) for only 18% of the observation time. The free energy difference of the inactive state (SP state) from the active state (rotation state) was estimated from the equilibrium constant (0.21) of the active state to be −1.5 kBT, where kBT represents the thermal energy. Although biochemical studies have reported that T. thermophilus V1 lapses into the ADP-inhibited form during ATP hydrolysis (26), the time constant of the inactivation in the literature (∼3 min) is too long to reflect SP. However, the literature value is rather close to the rotation time before lapsing into LP. Fig. 7A shows the histogram of the rotation time before LP, including both the rotation time and the duration time of SP. The obtained histogram demonstrated an exponential decay, giving the time constant of the inactivation into LP state, τinactivationLP, of 17.6 min. For comparison, the time-dependent inactivation during bulk ATPase assay with the ATP-regenerating system was measured (Fig. 7B). As reported previously (8, 10), the ATPase activity was gradually inactivated and finally reached nearly zero in 80 min. This time-dependent inactivation is explained by ADP inhibition; V1 fails to release ADP produced upon hydrolysis and transforms into the stable inhibitory state. Fitting the time course with a single exponential function gave the time constant of 3.4 min. Although the obtained time constant is in the range of minutes, similar to τinactivationLP (17.6 min), it is evidently faster than τinactivationLP. However, a closer look at the time course of inactivation in the bulk ATPase assay indicated a slow inactivation process (cyan dotted line in Fig. 7B) that followed the fast inactivation of 3.4 min. This finding suggests that there are two independent pathways for ADP inhibition. The double exponential fitting provided two time constants, 2.13 and 15.7 min. From the equilibrium level of the fitting, the equilibrium constant of inactivation was determined to be 0.018, giving the energy difference between active state and LP as −4.0 kBT. The later time constant is essentially consistent with τinactivationLP, suggesting that the slow inactivation process leading to the LP state corresponds to the slow inactivation observed in the bulk ATPase assay. One feasible explanation for why the fast-inactivating molecule was not observed is that the molecules inactivated via the fast pathway stopped rotation before being identified under the optical microscope; because the probability of finding actively rotating wild-type V1 particles is very low compared with the mutant V1 that we have often used for kinetic analysis of V1 rotation (1, 8, 21, 23–26), over 5 min was usually required to find the first rotating particle after ATP infusion into the flow cell.
FIGURE 6.
Kinetic analysis of SP. A, dwell time histogram of SP at 4 mm ATP. The histogram was fitted with a single exponential function of 4.2 ± 0.3 s. B, histogram of rotation time between SPs. Single exponential fitting gave the time constant of inactivation into the SP state as 0.9 ± 0.04 s. The number of molecules and trials were 7 and 250, respectively.
FIGURE 7.
Kinetic analysis of LP and inactivation in ATPase assay. A, dwell time histogram of rotation time until lapsing into the LP state. The rotation time includes the SPs. Single exponential fitting gave a time constant of 17.6 ± 1.5 min. B, time course of spontaneous inactivation observed in bulk ATPase assay. Activity was monitored with an ATP-regenerating system. The inactivation proceeded in two phases as follows: a fast phase (τfast = 2.13 min) and a slow phase (τslow = 15.7 min). Black and dotted cyan lines represent the fractions of the fast and slow phases.
Pauses under Non-load Conditions
To test the possible effect of the large viscous drag of the magnetic beads on kinetic properties of SP and LP, we observed the rotation of V1 using 40-nm gold colloid, which demonstrates negligible viscous drag much lower than that of the magnetic beads. As shown in Fig. 8A, the drag-free rotation assay also showed the frequent and transient pauses lasting seconds during continuous 120° steps, and each intervened with a catalytic dwell of 2.5 ms (9). The angular positions of the catalytic dwell (Fig. 8B, top panel) were consistent with the pausing angles of SP (Fig. 8B, bottom panel). The angular position of the catalytic dwell was reported to correspond to the ATP-binding angle (12), which again supports the finding that SP occurs at the same angle of ATP binding. Dwell time analysis determined the time constants of activation and inactivation as 3.50 ± 0.54 s (48 events) for τactivationSP and 0.55 ± 0.24 s (36 events) for τinactivationSP, which are essentially consistent with the values determined in the rotation assay with magnetic beads.
FIGURE 8.
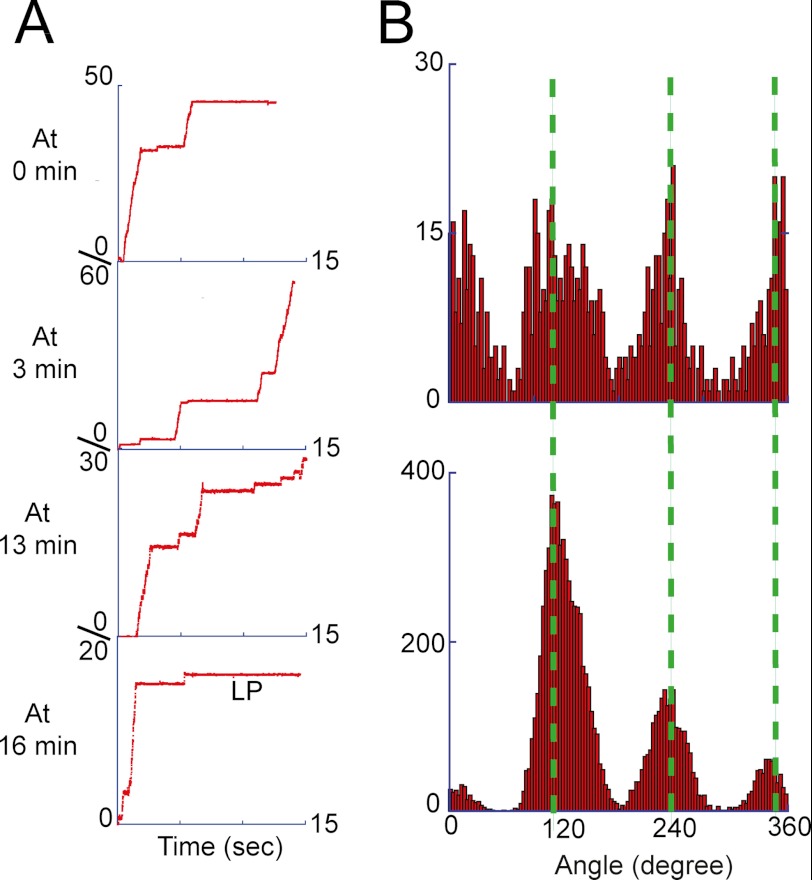
SP and LP under non-load conditions. The rotation assay was conducted using 40-nm gold colloid as the rotation probe at 4 mm ATP. The rotation was recorded every 3 or 4 min for a minute at maximum due to the data storage limitation of the fast-framing camera. A, sequential rotation trajectories of a single V1 molecule. The fourth trajectory shows that after ∼16 min this molecule stopped rotation with LP. The recording rate for this molecule was 250 frames/s. B, angle histogram from the time trajectories of continuous rotations, i.e. after the SPs were omitted (top), and that of the overall time trajectory before lapsing into the LP state (bottom), where SP dominated the data points, representing the angular positions of SP. Green lines show the mean positions of the SP.
We also attempted to determine the rotation time until LP. However, the fast-framing camera requisite for the rotation assay with 40-nm gold colloid did not allow continuous observation over 5 min due to the limitation of image data storage. Therefore, we conducted a time-lapse rotation assay; every 3–4 min, we recorded the rotary motion of the targeted molecule with a fast-framing camera for at least 10 s. With this method, the final long pause was observed. Although the time resolution was around 3 min, the average rotation time before LP was 16.7 ± 7.8 min. This observation is also essentially consistent with the τinactivationLP (17.6 min) determined in the rotation assay with magnetic beads. Thus, the viscous drag was confirmed not to affect the kinetics of pauses. In the following experiments, we again used the magnetic beads as the rotation marker in the single-molecule rotation assay.
Mechanical Activation from LP State
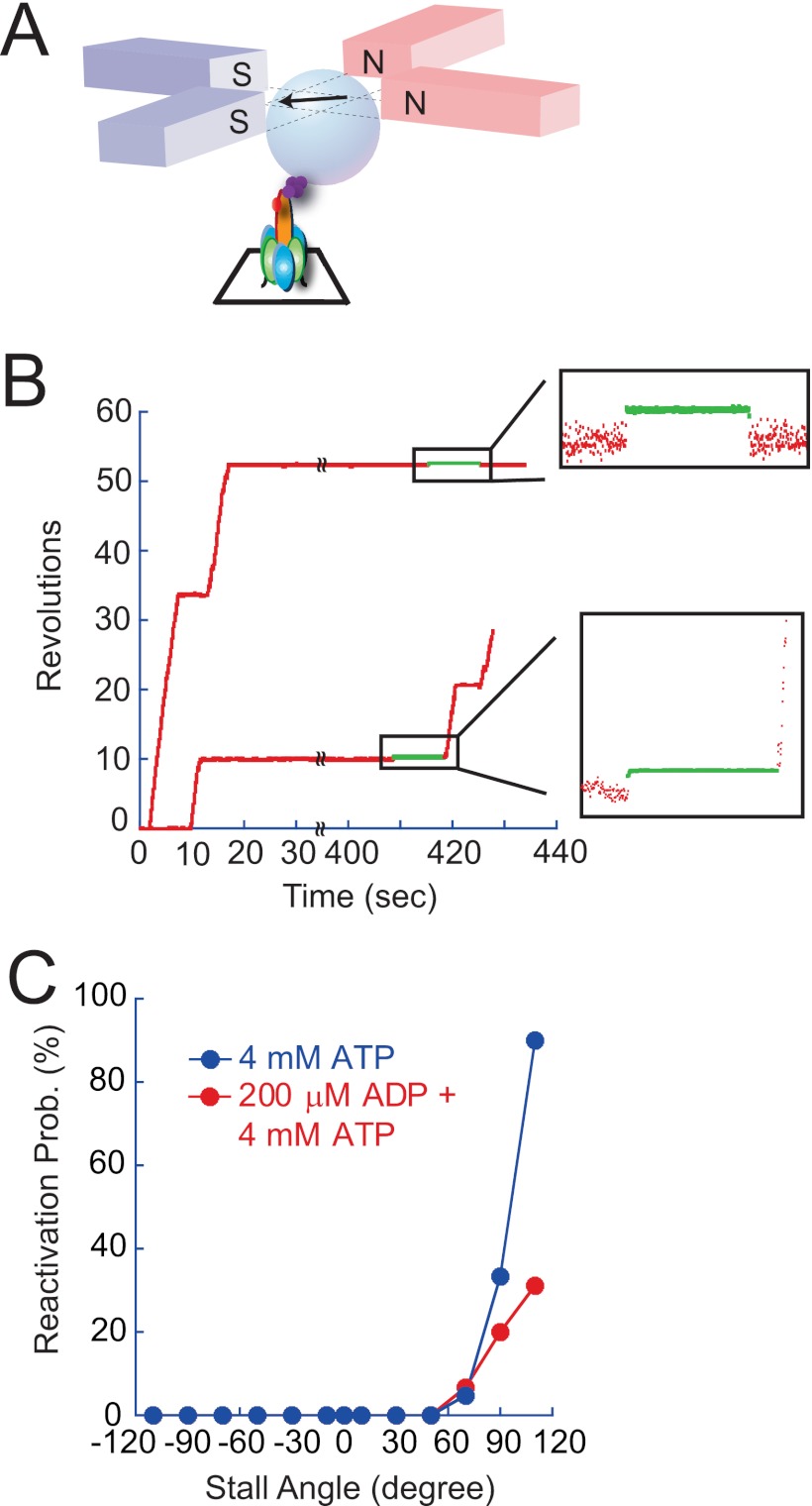
Although the kinetic features of LP do not match perfectly, the basic characteristics of LP are consistent with those expected for the ADP-inhibited form of V1 from bulk ATPase measurement in terms of time scale and apparent irreversibility. In the case of F1, external mechanical force has been reported to activate the ADP-inhibited form; when F1 in the ADP-inhibited form is forcibly rotated in the forward direction by 80°, F1 always resumes active rotation immediately after release from the external force (27). We investigated whether V1 in LP is also activated by external force similar to ADP-inhibited F1. Fig. 9A represents a schematic image of the experimental system; the magnetic bead was attached onto the D subunit as in the aforementioned rotation assay. In this experiment, the magnetic beads acted not only as the rotation marker but also as the handle to control the angular position of the rotary shaft of V1. The torque for external control was generated with magnetic tweezers composed of two pairs of electromagnets mounted 1 cm above the microscopic stage. The magnetic tweezers generated a magnetic field parallel to the stage, and the magnitude and orientation of the magnetic field was controlled by the electric current on each electromagnet.
FIGURE 9.
Mechanical activation from LP state. A, schematic image of manipulation with magnetic tweezers. A magnetic bead was attached onto the rotor as a handle for manipulation and as a rotation marker. Experiments were carried out at 4 mm ATP. B, typical time courses of the mechanical activation, a failed case (top) and a successful case (bottom). After ensuring that the molecule entered into the LP state by waiting more than 5 min, the manipulation was conducted with magnetic tweezers. In these cases, V1 in the LP state was forcibly rotated in the direction of rotation by +100° (top) and +120° (bottom). The green regions of the expanded time courses represent the time under manipulation. Symbols on the x axis and trajectories indicate the continuity of LP state until 400 s. C, probabilities of activation after manipulation at 4 mm ATP (blue) and at 4 mm ATP and 200 μm ADP (red).
Before applying a magnetic field to V1, we ascertained that the molecule lapsed into the LP state. Considering the time constant of SP (4.2 s), we set our criteria for LP as a pause longer than 5 min while dwelling at one of the three SP angles. After waiting for 5 min, a magnetic field was applied to forcibly rotate the V1 molecule in LP and stall it at a target angle. After the set time period lapsed, the molecule was released from the magnetic field. Similar to the mechanical activation of ADP-inhibited F1, V1 showed essentially two behaviors. One is the reactivation from LP state. V1 resumed active rotation immediately after release from the magnetic tweezers. Once reactivated, V1 made a continuous rotation until being trapped in the SP state. The rotation velocity was the same as that before entering the LP state. These findings imply that V1 reactivated from the LP state completely resumed the catalytic activity of ATP hydrolysis. The other type of response to the manipulation was the return to the original pausing position. In this case, V1 again showed LP unless activated by another manipulation. Fig. 9B shows typical time courses of the reactivation and the failure of reactivation. Unclassifiable behaviors were also observed (less than 1%). The typical behavior of the minor events was that the molecule returned to the original angle and resumed rotation spontaneously within 30 s, which is too short to be classified as LP. In these cases, V1 likely changed the pausing state from LP to SP upon the manipulation. This type of data were omitted from the data analysis.
Angle Dependence of Mechanical Reactivation
We stalled V1 in the LP state at angles ranging from −110 to +110° for 10 s at 4 mm ATP. The results are summarized in Fig. 9C, which shows the reactivation probability defined as the number of reactivation events against the total number of trials. In the range from −110 to +50°, no reactivation was observed. Manipulation over +50° induced reactivation. When rotated +110°, most molecules resumed active rotation immediately after being released. Thus, we confirmed that V1 paused in the LP state can be reactivated with forcible forward rotation similar to F1 under ADP inhibition. To explore further similarities with F1, we tested the effect of solution ADP on the reactivation of V1 in the LP state because solution ADP is known to suppress the reactivation of F1 under ADP inhibition (27). The probability of reactivation was determined in the presence of 200 μm ADP and 4 mm ATP. The suppressive effect of ADP was evident at +110 and +90°, but it was not observed at +70° (Fig. 9C). The reactivation probability at +110° decreased from 90 to 31%. Thus, reactivation from the LP state was also found to be sensitive to solution ADP, as expected from the contention that LP represents the ADP-inhibited form of V1.
DISCUSSION
In this study, we found that V1 makes two types of pauses during rotation as follows: a second-scale SP and an LP, which had a time constant that exceeded the observation time of the rotation assay, i.e. more than tens of minutes. Although V1 resumes active rotation after a SP of 4.2 s, the LP is so stable that V1 cannot recover its catalytic activity to resume ATP-driven rotation from the LP state. Only when forcibly rotated over +50° in the forward direction did V1 emerge from the LP state and resume active rotation.
Biochemical studies of T. thermophilus V1 have suggested that V1 decreases ATPase activity with time constant of 3.4 min until the activity was almost ceased (Fig. 7B). The inactivated V1 tightly binds ADP at the catalytic site, and V1 does not recover its catalytic activity unless the bound ADP is completely depleted by thorough treatment for nucleotide depletion. The SP state was the most frequently found pause observed in the rotation assay. However, SP is not attributable to the biochemically suggested ADP inhibition because the time constants of inactivation and activation (τinactivationSP = 0.9 s and τactivationSP = 4.2 s) are both too short. Therefore, the SP state does not represent the ADP-inhibited form, but rather a newly identified inhibitory state of V1.
This inhibitory state with a short time span is not a type of artifact because of the viscous drag of the rotation probe. The rotation assay with the essentially drag-free 40-nm gold colloid probe also showed SP, and its occurrence frequency and duration time were both the same as those obtained with the more viscous magnetic bead probe. Similar short pause of a second-scale dwell time was reported for F1-ATPase (17). However, the physiological role and the mechanism of the short pause in both motors are unknown. Further study is necessary.
Interestingly, bulk ATPase assay did not show clear second-scale inactivation or activation caused by the SP state in the present condition. It should be mentioned that slight activation in the time course of bulk ATPase assay was previously reported (10). However, the degree of the activation was still very small. Therefore, these findings imply that V1 is almost in equilibrium between the active state and the SP state before being injected into the assay mixture for bulk ATPase measurement. From the time constants of the SP state, the estimated fraction of active V1 is only 18%. This contention explains, at least partially, the apparent discrepancy of the bulk ATPase rate and the rotation speed; the rotation speed is evidently faster than the rotation rate expected from the ATPase rate. Whereas the rotational rate of V1 at a Vmax condition (4 mm ATP) was reported to be 64 turns/s (12), the rotational rate estimated from the bulk ATPase rate (⅓ of ATPase rate) was only 12 turns/s (9). When the bulk ATPase rate is corrected assuming that only 18% of V1 molecules are in the active state, the genuine rotational rate is estimated to be 67 turns/s, which is highly consistent with the actual rotational rate, supporting the above contention of pre-existing equilibrium between the active and inactive states. This observation suggests that the transition into the SP state involves some conformational rearrangement that is irrelevant to catalysis. In fact, when the rotation and the dwell time of the SP state were analyzed in the presence of solution ADP, the kinetics of SP was not affected (data not shown), suggesting that at least ADP binding is irrelevant to the SP state.
The bulk ATPase assay indicated that the LP state has an expected minute-scale inactivation time constant. The extremely long duration time of the LP state is consistent with the prediction from the bulk ATPase assay; the ADP-inhibited form is so stable that V1 almost completely abolished catalytic activity, suggesting that the time constant of reactivation is very big. This finding is in contrast to the ADP-inhibited form of F1-ATPase, which spontaneously resumes active rotation after 30 s under the assistance of thermal agitation (17). However, the time constant of inactivation into the LP state (17.6 min) does not fully agree with that predicted from ATPase assay, 3.4 min (Fig. 7B) (10). We found that the slow inactivation proceeded after the fast inactivation of 3.4 min in the bulk ATPase assay. The time constant of this slow inactivation, 15.7 min, was essentially consistent with that for the LP state. Therefore, we conclude that the LP state corresponds to the ADP-inhibited form of V1 formed via the slow inactivation process. It is likely that we lost a significant fraction of rotating V1 molecules that stopped rotation before being identified in the rotation assay, because the probability of finding a rotating particle of wild-type V1 was so low that we always required over 5 min to find the first rotating particle after infusing ATP into flow chambers.
The effect of solution ADP on the LP state was investigated. Mechanical activation from the LP state was significantly suppressed by 200 μm ADP. This observation coincided with the ADP-inhibited form of F1; the activation of ADP-inhibited F1 by external force was largely suppressed with solution ADP. Thus, the rebinding of ADP to V1 is likely to stabilize the ADP-inhibited form as in the case of F1. Furthermore, the mechanical activation of V1 also demonstrated significant angle dependence; unless rotated over +70°, V1 never resumed active rotation. The observed strong angle dependence suggests that the off rate of inhibitory ADP from V1 is highly enhanced upon forward rotation. This finding implies that V1 also modulates the catalytic power upon rotation, as observed in F1-ATPase for efficient mechanochemical coupling between rotation and ATP hydrolysis/synthesis (15). However, compared with F1, the ADP-inhibited form of V1 was very persistent against mechanical activation with the magnetic tweezers. Whereas F1 was fully activated upon stalling at +70° for 10 s, the activation probability of V1 by stalling at +70° was less than 10%. Thus, activation energy for the activation of ADP-inhibited V1 appears to be much larger than that for F1.
Interestingly, mechanical activation was not observed when stalled in the ATP synthesis direction. Mechanical activation of F1 in the backward direction required ADP and Pi in solution, probably because F1 is activated only after it follows the reaction pathway of ATP synthesis. Based on this knowledge, we attempted to activate V1 in the LP state by backward rotation in the presence of ADP and Pi. We observed some activation events only in the presence of ADP and Pi, as in the case of F1 activation. However, the probability of the mechanical activation was too low to permit analysis (data not shown). The reason for this low probability is unclear. Backward activation of V1 might require other conditions or other subunits. To clarify these points, further analysis is required.
The energy differences of SP or LP state from active states are −1.5 and −4.0 kBT, respectively. These are significantly stable compared with that of the ADP inhibition of F1 (−0.6 kBT). The finding implies that the isolated V1 takes a longer rest than F1, pausing the catalytic turnover. This feature is physiologically advantageous to avoid futile ATP consumption in the cells because the isolated V1 is found in the T. thermophilus cytosolic fraction.3 Therefore, we assume that the SP and LP states are dual-suppression mechanisms to strictly prevent futile ATP consumption by the isolated V1 in T. thermophilus. It is well known that the yeast V1 domain is detached from vacuole membranes upon glucose starvation, and isolated V1 somehow lost the ATP hydrolysis activity (28). We do not know whether similar SP and LP states are found in eukaryotic V1. Further study for rotation mechanism in eukaryotic V1 is necessary.
Conclusions
Single-molecule rotation assay of V1-ATPase revealed that V1-ATPase has two types of inhibitory pausing states as follows: the short pause state is a newly found pausing state, and the long pause state was suggested to correspond to ADP inactivation that had been predicted in bulk ATPase analysis. V1-ATPase in the long pause state was forcibly activated by rotating in a forward direction by over +50°, although the long pause state was so stable that V1-ATPase never spontaneously resumed active rotation without external manipulation. The observed angle dependence of the mechanical activation was distinctive from that of F1-ATPase, suggesting that the energetic and kinetic features of the chemomechanical coupling of V1-ATPase is different from that of F1-ATPase. The single-molecule manipulation study on catalytically active V1-ATPase will elaborate the details of the common and distinctive features between the mechanical modulation of the catalytic power of V1-ATPase and F1-ATPase.
Acknowledgments
We thank all members of Noji Laboratory for critical discussions and technical advice. We thank H. Imamura for detailed and constructive comments during the preparation of this manuscript. We thank M. Kino-oka, R. Watanabe, and K. Hayashi for valuable comments.
This work was supported in part by Grant-in-aid for Scientific Research 18074005 (to H. N.) and by a Special Education and Research Expenses grant (to H. N.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
K. Yokoyama, unpublished data.
- SP
- short pause
- LP
- long pause.
REFERENCES
- 1. Yokoyama K., Imamura H. (2005) Rotation, structure, and classification of prokaryotic V-ATPase. J. Bioenerg. Biomembr. 37, 405–410 [DOI] [PubMed] [Google Scholar]
- 2. Nishi T., Forgac M. (2002) The vacuolar H+-ATPases. Nature's most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3, 94–103 [DOI] [PubMed] [Google Scholar]
- 3. Forgac M. (2007) Vacuolar ATPases. Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 4. Muench S. P., Trinick J., Harrison M. A. (2011) Structural divergence of the rotary ATPases. Q. Rev. Biophys. 44, 311–356 [DOI] [PubMed] [Google Scholar]
- 5. Lau W. C., Rubinstein J. L. (2012) Subnanometer-resolution structure of the intact Thermus thermophilus H+-driven ATP synthase. Nature 481, 214–218 [DOI] [PubMed] [Google Scholar]
- 6. Diab H., Ohira M., Liu M., Cobb E., Kane P. M. (2009) Subunit interactions and requirements for inhibition of the yeast V1-ATPase. J. Biol. Chem. 284, 13316–13325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saijo S., Arai S., Hossain K. M., Yamato I., Suzuki K., Kakinuma Y., Ishizuka-Katsura Y., Ohsawa N., Terada T., Shirouzu M., Yokoyama S., Iwata S., Murata T. (2011) Crystal structure of the central axis DF complex of the prokaryotic V-ATPase. Proc. Natl. Acad. Sci. U.S.A. 108, 19955–19960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakano M., Imamura H., Toei M., Tamakoshi M., Yoshida M., Yokoyama K. (2008) ATP hydrolysis and synthesis of a rotary motor V-ATPase from Thermus thermophilus. J. Biol. Chem. 283, 20789–20796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Furuike S., Nakano M., Adachi K., Noji H., Kinosita K., Jr., Yokoyama K. (2011) Resolving stepping rotation in Thermus thermophilus H+-ATPase/synthase with an essentially drag-free probe. Nat. Commun. 2, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yokoyama K., Muneyuki E., Amano T., Mizutani S., Yoshida M., Ishida M., Ohkuma S. (1998) V-ATPase of Thermus thermophilus is inactivated during ATP hydrolysis but can synthesize ATP. J. Biol. Chem. 273, 20504–20510 [DOI] [PubMed] [Google Scholar]
- 11. Hayashi K., Ueno H., Iino R., Noji H. (2010) Fluctuation theorem applied to F1-ATPase. Phys. Rev. Lett. 104, 218103. [DOI] [PubMed] [Google Scholar]
- 12. Imamura H., Takeda M., Funamoto S., Shimabukuro K., Yoshida M., Yokoyama K. (2005) Rotation scheme of V1-motor is different from that of F1-motor. Proc. Natl. Acad. Sci. U.S.A. 102, 17929–17933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adachi K., Oiwa K., Nishizaka T., Furuike S., Noji H., Itoh H., Yoshida M., Kinosita K., Jr. (2007) Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 130, 309–321 [DOI] [PubMed] [Google Scholar]
- 14. Shimabukuro K., Yasuda R., Muneyuki E., Hara K. Y., Kinosita K., Jr., Yoshida M. (2003) Catalysis and rotation of F1 motor. Cleavage of ATP at the catalytic site occurs in 1 ms before 40° substep rotation. Proc. Natl. Acad. Sci. U.S.A. 100, 14731–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Watanabe R., Okuno D., Sakakihara S., Shimabukuro K., Iino R., Yoshida M., Noji H. (2012) Mechanical modulation of catalytic power on F1-ATPase. Nat. Chem. Biol. 8, 86–92 [DOI] [PubMed] [Google Scholar]
- 16. Feniouk B. A., Yoshida M. (2008) Regulatory mechanisms of proton-translocating FoF1-ATP synthase. Results Probl. Cell Differ. 45, 279–308 [DOI] [PubMed] [Google Scholar]
- 17. Hirono-Hara Y., Noji H., Nishiura M., Muneyuki E., Hara K. Y., Yasuda R., Kinosita K., Jr., Yoshida M. (2001) Pause and rotation of F1-ATPase during catalysis. Proc. Natl. Acad. Sci. U.S.A. 98, 13649–13654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Imashimizu M., Bernát G., Sunamura E., Broekmans M., Konno H., Isato K., Rögner M., Hisabori T. (2011) Regulation of FoF1-ATPase from Synechocystis sp. PCC 6803 by γ and ϵ subunits is significant for light/dark adaptation. J. Biol. Chem. 286, 26595–26602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pavlova P., Shimabukuro K., Hisabori T., Groth G., Lill H., Bald D. (2004) Complete inhibition and partial re-activation of single F1-ATPase molecules by tentoxin. New properties of the re-activated enzyme. J. Biol. Chem. 279, 9685–9688 [DOI] [PubMed] [Google Scholar]
- 20. Herbert K. M., La Porta A., Wong B. J., Mooney R. A., Neuman K. C., Landick R., Block S. M. (2006) Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell 125, 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Imamura H., Ikeda C., Yoshida M., Yokoyama K. (2004) The F subunit of Thermus thermophilus V1-ATPase promotes ATPase activity but is not necessary for rotation. J. Biol. Chem. 279, 18085–18090 [DOI] [PubMed] [Google Scholar]
- 22. Itoh H., Takahashi A., Adachi K., Noji H., Yasuda R., Yoshida M., Kinosita K. (2004) Mechanically driven ATP synthesis by F1-ATPase. Nature 427, 465–468 [DOI] [PubMed] [Google Scholar]
- 23. Yokoyama K., Nakano M., Imamura H., Yoshida M., Tamakoshi M. (2003) Rotation of the proteolipid ring in the V-ATPase. J. Biol. Chem. 278, 24255–24258 [DOI] [PubMed] [Google Scholar]
- 24. Takeda M., Suno-Ikeda C., Shimabukuro K., Yoshida M., Yokoyama K. (2009) Mechanism of inhibition of the V-type molecular motor by tributyltin chloride. Biophys. J. 96, 1210–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Makyio H., Iino R., Ikeda C., Imamura H., Tamakoshi M., Iwata M., Stock D., Bernal R. A., Carpenter E. P., Yoshida M., Yokoyama K., Iwata S. (2005) Structure of a central stalk subunit F of prokaryotic V-type ATPase/synthase from Thermus thermophilus. EMBO J. 24, 3974–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Imamura H., Nakano M., Noji H., Muneyuki E., Ohkuma S., Yoshida M., Yokoyama K. (2003) Evidence for rotation of V1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 100, 2312–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hirono-Hara Y., Ishizuka K., Kinosita K., Jr., Yoshida M., Noji H. (2005) Activation of pausing F1 motor by external force. Proc. Natl. Acad. Sci. U.S.A. 102, 4288–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Parra K. J., Kane P. M. (1998) Reversible association between the V1 and Vo domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol. Cell. Biol. 18, 7064–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]