Abstract
Although sequencing of Mycobacterium tuberculosis genome lead to better understanding of transcription units and gene functions, interactions occurring during transcription initiation between RNA polymerase and promoters is yet to be elucidated. Different stages of transcription initiation include promoter specific binding of RNAP, isomerization, abortive initiation and promoter clearance. We have now analyzed these events with four promoters of M. tuberculosis viz . PgyrB1, PgyrR, PrrnPCL1 and PmetU. The promoters differed from each other in their rates of open complex formation, decay, promoter clearance and abortive transcription. The equilibrium binding and kinetic studies of various steps revealed distinct rate limiting events for each of the promoter, which also differed markedly in their characteristics from the respective promoters of Mycobacterium smegmatis. Surprisingly, the transcription at gyr promoter was enhanced in the presence of initiating nucleotides and decreased in the presence of alarmone, pppGpp, a pattern typically seen with rRNA promoters studied so far. The gyr promoter of M. smegmatis, on the other hand, was not subjected to pppGpp mediated regulation. The marked differences in the transcription initiation pathway seen with rrn and gyr promoters of M. smegmatis and M. tuberculosis suggest that such species specific differences in the regulation of expression of the crucial housekeeping genes could be one of the key determinants contributing to the differences in growth rate and lifestyle of the two organisms. Moreover, the distinct rate limiting steps during transcription initiation of each one of the promoters studied point at variations in their intracellular regulation.
Introduction
M. tuberculosis is one of the most formidable pathogens known to mankind. The resurgence of the pathogen, its alliance with HIV infection and emergence of the drug resistant strains has resulted in a global challenge to combat tuberculosis [1]. The distinctive features of the bacterium such as slow growth rate, dormancy, unique cell wall composition, resistance towards phagocytosis by macrophages etc demand a thorough investigation of its biology at the molecular level. A number of studies carried out so far reveal significant differences in the transcription process in mycobacteria when compared to E. coli and other bacteria [2], [3]. Presence of as many as 13 sigma factors for transcription from promoters with diverse architecture [2], [4], [5] and inability of the mycobacterial promoters to function in E. coli [5]–[7] are some of the key features warranting a detailed study of the transcription process in the pathogen.
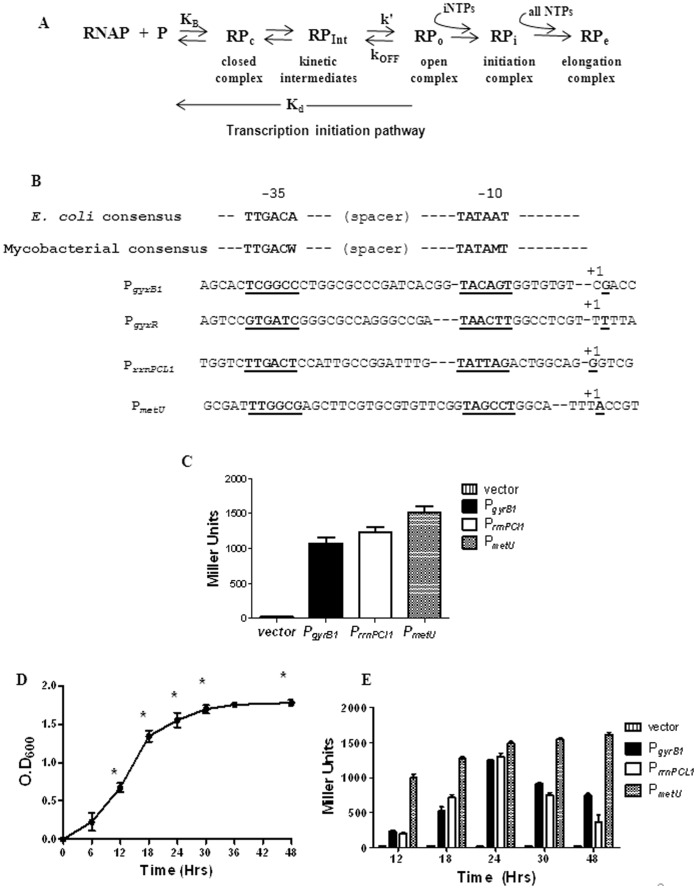
Transcription constitutes the first stage in gene expression and comprises of multiple steps viz initiation, elongation and termination ( Figure 1A ). During the initiation, RNA polymerase (RNAP) binds to the promoter, leading to the formation of several intermediates which differ from each other in their kinetic properties [8]–[11]. After initial binding of RNAP to the promoter to form the closed complex, the DNA strands unwind to form a catalytically competent open complex, associated with a series of conformational changes in the enzyme as well as DNA [8]–[11]. Binding of the initial ribonucleotides (iNTPs) to the RNAP results in the formation of ternary complex, poised to enter into the elongation mode [8]–[11]. After the synthesis of abortive transcripts of 2–15 nucleotides in most of the promoters studied, the RNAP leaves the promoter to enter into the elongation phase of transcription [12]. Because of these elaborate orchestrated steps, the initiation pathway is also fine-tuned by a number of regulatory mechanisms to meet the requirements posed by various physiological conditions of the cell [11], [13]. Typically, a few promoters (rrn, initiator tRNA) achieve higher promoter strength in exponential phase due to the stabilization of open complex by initiating nucleotides (iNTPs). In contrast, during the stationary phase, increase in the concentration of guanosine tetra/penta phosphate ((p)ppGpp), leads to inhibition of transcription from these promoters [14], [15]. Thus the strength of these promoters varies with the growth phase as they are subjected to growth phase dependent regulation.
Figure 1. Transcription with M. tuberculosis promoters. A Scheme of transcription initiation. B Sequences of promoters used in this study.
The -35, -10 elements and the transcription start sites are underlined. Sequences are aligned with E. coli σ70 and mycobacterial σA dependent promoter consensus. C in vivo reporter assays. Transcriptional activities of the promoters were determined by β-galactosidase reporter assays from the early exponential phase cultures of M. smegmatis. The promoter activities are represented in Miller units on Y axis. D Growth curve of M. smegmatis . The OD600 of the culture was recorded at 0, 6, 12, 24, 30, 36 and 48 Hrs. The time points at which the β-galactosidase reporter assays were carried out are indicated with asterisks. E in vivo reporter assays in different phases of growth. M. smegmatis cells harbouring pSD5B promoter constructs were grown upto 48 hours and promoter activity was determined at different times of growth by β-galactosidase assay as mentioned above.
The present work is the first detailed kinetic analysis of the events during transcription initiation in M. tuberculosis. We have carried out promoter-polymerase interaction studies using a few of the house-keeping promoters to characterize the mechanisms of transcription initiation. The kinetics of RNAP-DNA interactions was measured in different promoter sequence contexts to determine their key rate–limiting steps. Further, the role of iNTPs and pppGpp in regulating transcription initiation was studied. While the M. tuberculosis ribosomal RNA promoter exhibited the characteristics seen with E.coli and other bacteria, the promoters for the gyrase operon showed unusual and hitherto unknown pattern of transcription initiation. Most significantly, the promoters for the same genes from M. smegmatis and M. tuberculosis differed markedly in their kinetic properties and response to the effectors.
Results
Promoter Characteristics and Activities
For comparison of the promoter-RNAP interactions, two stable RNA promoters viz. ribosomal RNA, initiator tRNA (PrrnPCL1, PmetU) and another house-keeping promoter (PgyrB1 ) were chosen in addition to a weak promoter (PgyrR) ( Figure 1B ). M. tuberculosis has only a single operon for rRNA transcription driven by two promoters [16]–[18]. Amongst the two promoters, PrrnPCL1 is the major house-keeping promoter and is stronger than PrrnP1 [16]. Moreover, PrrnPCL1 is found in the genome of every sequenced species of mycobacteria and appears to be conserved across the genus [18]. PmetU is the only promoter driving the transcription of the single initiator tRNA gene in M. tuberculosis [19]. The -10 and -35 elements of these two stable RNA promoters resemble the mycobacterial σA consensus sequence [6]. Two promoters from the gyr operon of M. tuberculosis included in the study are illustrated in Figure 1B [6]. PgyrB1 is the major promoter of the gyr operon that directs the high levels of transcription from the gyrB - gyrA dicistron [6]. The -10 element of the promoter is similar to the σA dependent promoter consensus sequence. PgyrR is an overlapping and divergently organized promoter to PgyrB1 whose activity is appoximately 13 times weaker than PgyrB1 and hence is a representative weak promoter in this study [6].
The relative in vivo activities of the promoters subjected to the present analysis were determined using the promoter-lacZ transcriptional fusion constructs transformed to M. smegmatis mc2 155. PmetU showed the highest activity followed by PgyrB1 and PrrnPCL1 ( Figure 1C ). The activities were also measured at different times of growth ( Figure 1D ). The promoter strength of PmetU did not vary significantly at different growth phases, while the activities of both PrrnPCL1 and PgyrB1 decreased as the cells entered the stationary phase ( Figure 1E ) suggesting the regulation of these promoters is growth phase dependent. To understand the mechanism of differential regulation of the promoters, equilibrium and kinetic analysis of transcription initiation was carried out.
Promoter–polymerase Interaction
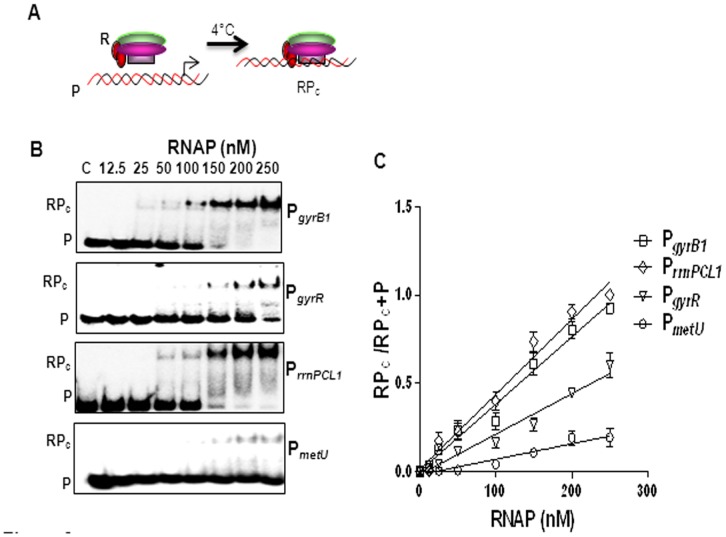
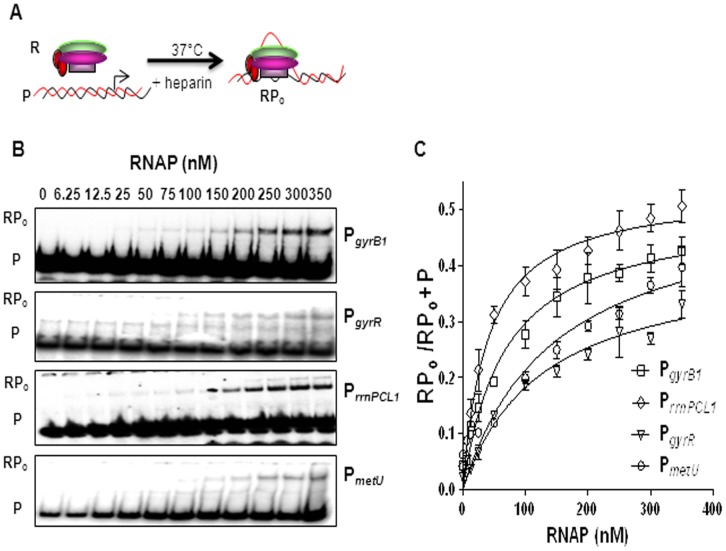
For the equilibrium binding analysis of M. tuberculosis promoters with RNAP, binding constant (KB) for closed complex formation was measured. KB is the determinant of the strength of the closed complex, which is obtained by titration of the promoters with the RNAP and plotting the ratios of the bound fraction to total DNA (RPc/RPc+P) against the RNAP concentrations ( Figure 2A, B ). The affinity of RNAP for PrrnPCL1 and PgyrB1 during closed complex formation was 5 and 3.3 fold higher respectively compared to the other stable RNA promoter, PmetU. Kd which is the determinant of the strength of the open complex, was obtained by plotting the ratio of RPo/RPo+P (fraction DNA bound) against RNAP concentrations ( Figure 3A, B , Table 1 ). From the Table 1 it is apparent that PrrnPCL1 and PgyrB1 have comparable Kd values (lower than PgyrR and PmetU). Thus, although the affinity of RNAP for both PrrnPCL1 and PgyrB1 was higher than other two promoters studied, (see above) the overall extent of open complex formation was low, which could be either due to slow rate of formation or reduced longevity of open complex. Therefore, next,the rates of formation as well as stability of open complexes were measured for all the promoters. The rates of formation of open complex for the promoters were determined by measuring the rate constants of isomerization (see next paragraph). The stability of the open complexes was determined by monitoring the decay of the complexes with time.
Figure 2. Determination of KB. A.
Scheme of assay, R represents RNAP, P represents promoter fragment and RPc represents closed complex. B Promoter fragments were incubated with different RNAP concentrations for 20 min and the complexes formed were resolved using 4% native-PAGE. C The amount of radioactivity in bound and free fragments was measured by densitometry and indicated as RPc and P respectively. RPc/RPc+P ratios were plotted as function of RNAP concentrations. KB was calculated from the slope of the graph. The values obtained are mean of three independent experiments (Table 1).
Figure 3. Determination of Kd. A.
Scheme of assay, R represents RNAP, P represents promoter fragment and RPo represents open complex. B Promoter fragments were titrated using a range of RNAP concentrations, incubated at 37°C for 10 min, challenged with heparin and analyzed using 4% native-PAGE. C The amount of radioactivity in bound and free fragments was measured by densitometry and indicated as RPo and P respectively. RPo/RPo+P ratios were plotted as a function of RNAP concentrations to obtain the hyperbolic graph. The values (Table 1) are mean of three independent experiments.
Table 1. Summary of equilibrium binding constants and kinetic parameters.
| Constant/Property | PgyrB1 | PrrnPCL1 | PgyrR | PmetU |
| KB (x 107 M−1) | 2.1±0.03 | 3.1±0.16 | 0.27±.001 | 0.7±0.08 |
| Kd (x 107 M) | 68.05±12.16 | 40.14±6.09 | 146.2±44.58 | 127.6±27.19 |
| k' (fast) (min−1) | 1.4±0.7 | 1.2±0.08 | 0.54±0.20 | 0.24±0.15 |
| k' (slow) (x 10−2 min−1) | 0.24±0.4 | 0.21±0.74 | 0.61±0.24 | 0.04±0.04 |
| KB x k' (fast) | 2.1×107 | 2.7×107 | 1.3×106 | 1.35×106 |
| koff (fast) (min−1) | 0.74±0.27 | 2.63±1.41 | 1.35±0.92 | 0.48±0.16 |
| koff (slow) (x10−1 min−1) | 0.5±0.17 | 0.4±0.29 | 0.63±0.16 | 0.16±0.09 |
| t1/2 (fast) (min) | 5.17 | 1.51 | 7.75 | 4.4 |
| t1/2 (slow) (min) | 17.78 | 6.12 | 13.32 | 34.84 |
| Abortivetranscription | ++++ | – | +++ | – |
| Promoter clearance (min) | 23 | 2.3 | 18 | 8.93 |
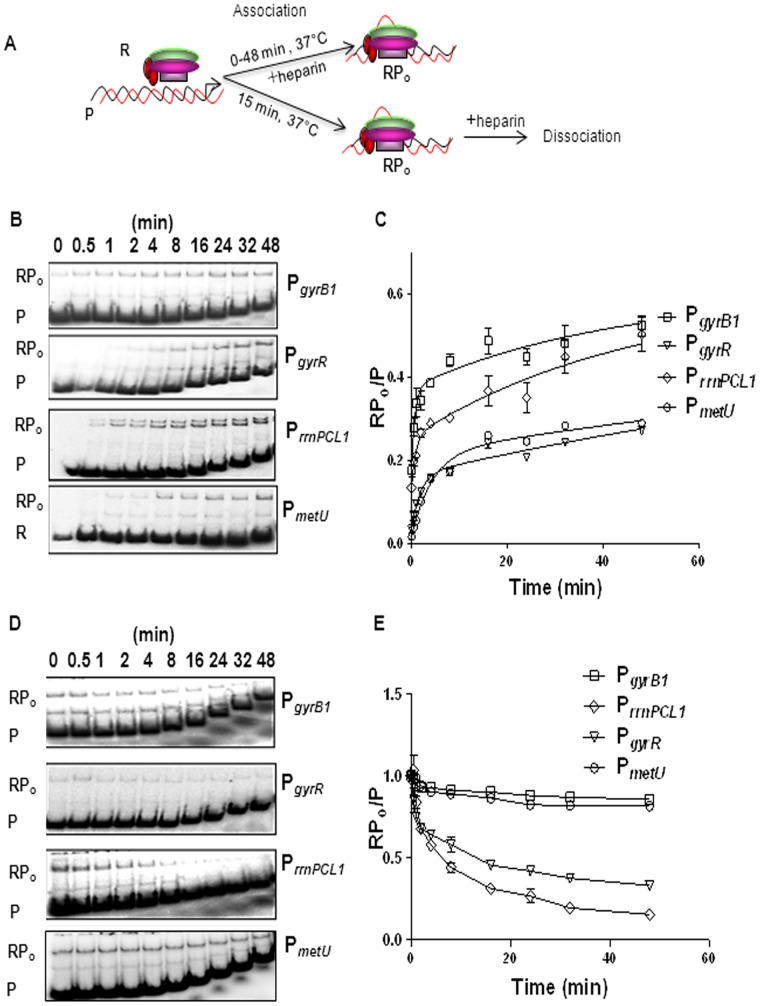
Pseudo first order rate constant (k') which is the measure of isomerization rate was calculated by monitoring the time dependent interaction of RNAP and promoters. From the results of association kinetics, ( Figure 4A, B , Table 1 ) it is evident that the rate of open complex formation was highest for the PgyrB1 followed by PrrnPCL1, thus providing an explaination for the higher strength of these two promoters. Notably, the rate of isomerization at PmetU was slow, although the in vivo activity of this promoter was high (see Figure 1 E), suggesting the possibility that the downstream kinetic events after isomerization step could be the key determinants of the high strength of the promoter. Indeed, when the rate of the dissociation of RNAP from the promoter was monitored, RNAP dissociated at a very low rate ( Figure 4C and D ). PgyrB1 also showed slower dissociation rate indicating another contributing feature for its strength. As expected, like other well studied rRNA promoters, higher dissociation rate was seen at PrrnPCL1, compared to the more stable open complexes at PgyrB1 and PmetU ( Table 1 ). Biphasic curve obtained in all these cases could be an indication of an initial faster dissociation of closed complex and a slower dissociation of a comparatively more stable open complex.
Figure 4. Kinetics of association and dissociation. A.
Scheme of assay, R represents RNAP, P represents promoter fragment and RPo represents open complex. B The association of RNAP and promoter DNA to form open complex was monitored over time intervals ranging from 0 to 48 min, as indicated. C The amount of radioactivity in bound and free fragments was measured by densitometry and indicated as RPo and P respectively. RPo/P ratios were plotted against time and k′ was measured by double exponential association analysis. The values of k′ are mean of three independent experiments and are shown in Table 1. D Open complex at each promoter fragment was formed by incubating RNAP and promoter fragments for 15 min at 37°C. Dissociation of RNAP was monitored by challenging the pre-formed open complex with heparin for time intervals ranging from 0 to 48 min, as indicated. E The data was fit into the double exponential decay equation to measure koff. The biphasic nature of the double exponential decay curve is suggestive of the existence of two complexes decaying at different rates. The steeper and the trailing parts of the curve represent the faster and slower decaying phases respectively. The values of koff are mean of three independent experiments (Table 1).
Effect of iNTPs on Open Complex Formation
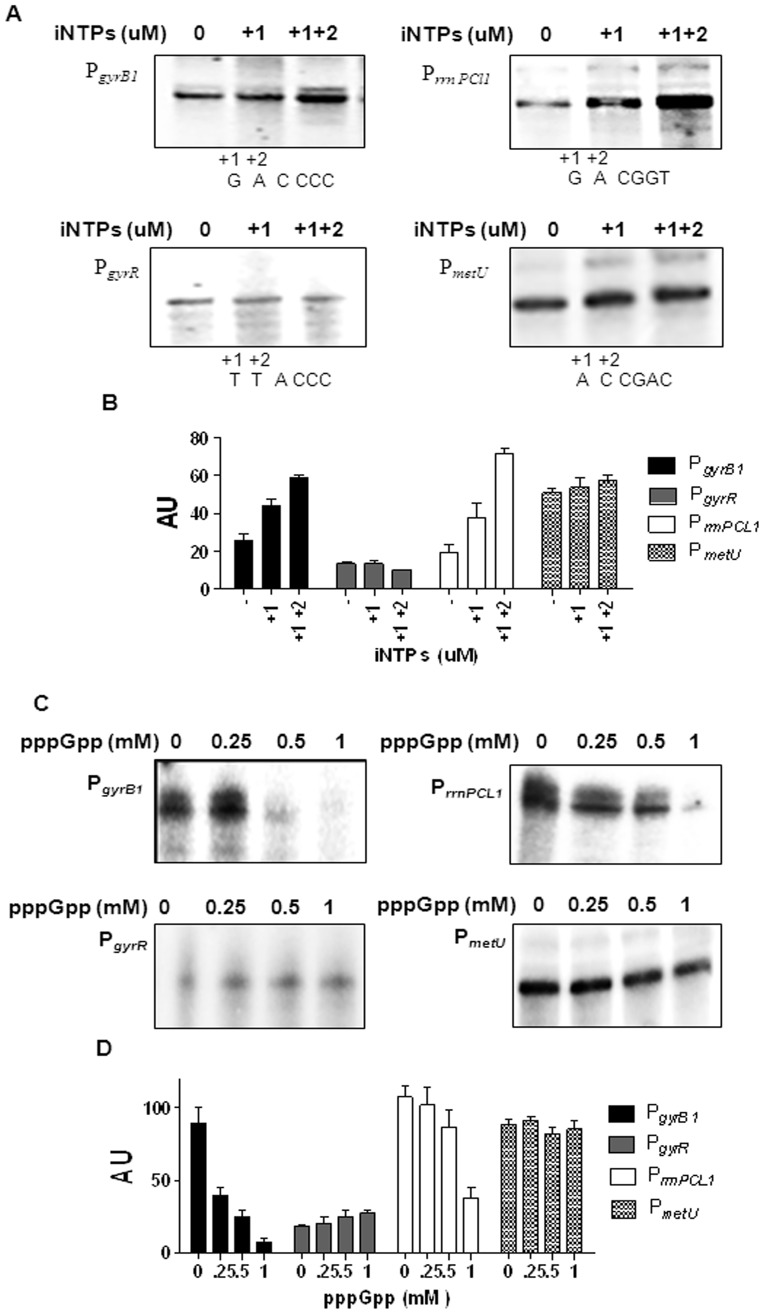
rRNA promoters from diverse organisms show enhanced promoter activity in the exponential phase. Since the promoter strength of PrrnPCL1 and also PgyrB1 varied with the growth phase (see Figure 1E ), we next examined the effect of iNTPs on the isomerization and the open complex stability of the PrrnPCL1, PgyrB1 and the other two promoters. The open complex at PrrnPCL1 increased on addition of +1 iNTP, and in presence of both +1 and +2 iNTPs ( Figure 5A, B , Figure S1A, B). In addition, the longevity of the open complex at PrrnPCL1 was increased in the presence of the first two NTPs (not shown). In contrast to the rRNA promoter, the other stable RNA promoter, PmetU, did not respond in a similar fashion when the +1 iNTP was added. Notably, the isomerization was stimulated at PgyrB1 in the presence of +1 iNTP with the further increase in the presence of both +1 and +2 iNTPs, similar to the pattern seen with PrrnPCL1 ( Figure 5A, B , Fig. S1A, B). However, unlike PrrnPCL1 the initiating nucleotides did not influence the longevity of the open complex at PmetU and PgyrB1 to a significant extent (data not shown), which could be attributed to the intrinsic stability of the complexes at these promoters in the absence of initiating nucleotides. In vitro assays carried out in the presence of pppGpp showed that transcription at PrrnPCL1 and PgyrB1 was inhibited ( Figure 5C, D ). There was no significant effect of pppGpp on transcription at PmetU ( Figure 5C, D ), revealing that the promoter is not subjected to a similar kind of regulation.
Figure 5. Effect of iNTPs on open complex. A.
Open complex was formed in the presence of 100 µM of iNTPs (+1 and +1, +2) and the transcription was initiated by adding heparin and all the four NTPs. The run-off transcripts were resolved on 8% urea–PAGE. The initial transcribed sequence (ITS) for all the promoters is shown. B The graph shows the quantification of transcripts formed in the absence and presence of +1 iNTP and +1, +2 iNTPs. The amount of run-off transcripts formed was measured by densitometry and indicated as AU on Y axis. C Effect of pppGpp on open complex. Open complex was formed in the presence of increasing concentrations of pppGpp as indicated and the transcription was initiated as described before. The run-off transcripts were resolved on 8% urea–PAGE. D The graph shows the quantification of transcripts formed in the presence of increasing concentrations of pppGpp. The amount of run-off transcripts formed was measured by densitometry and indicated as AU on Y axis.
Inefficient Promoter Clearance at Gyr Promoters
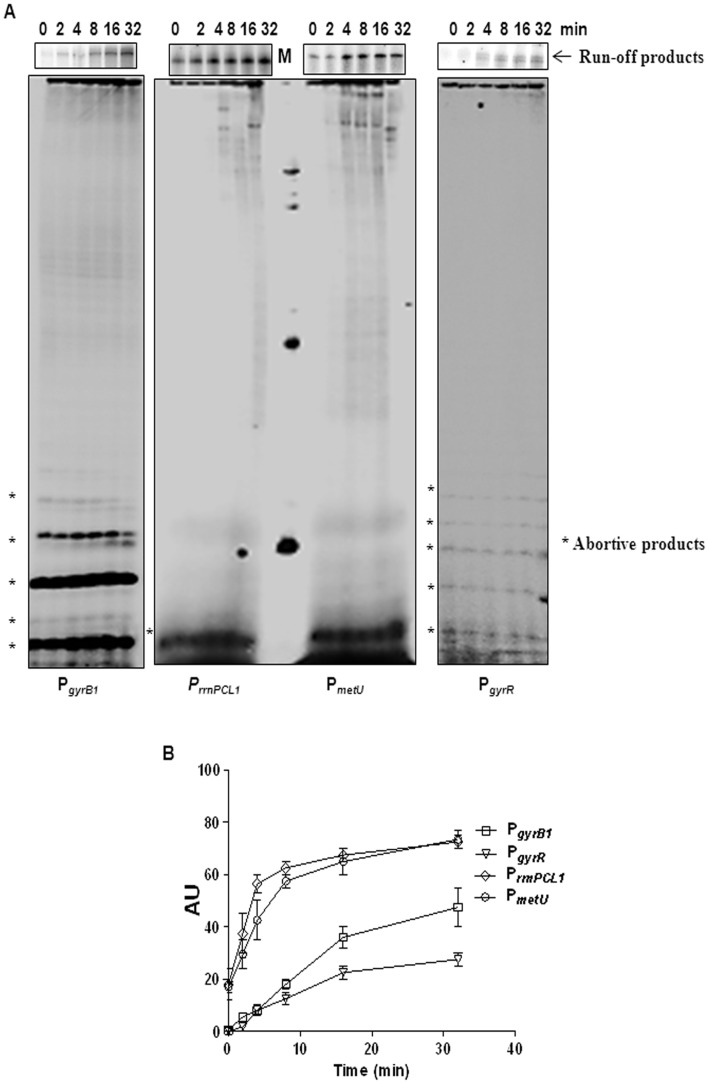
In addition to the efficiency in DNA binding and melting, overall promoter strength also depends on the rate of promoter clearance by RNAP. Thus the extent of abortive initiation during the transition from the initiation to elongation also has an important bearing on transcription initiation [20]. To determine the contribution of the post DNA-melting steps in overall transcription efficiency, the rate of promoter clearance and formation of abortive as well as run-off transcripts were measured ( Figure 6A, B , Table 1 ). The rate of promoter clearance was faster at two stable RNA promoters in contrast to the gyr operon promoters. The promoter which drives the dicistron transcription (PgyrB1) had 2.5 and 10 times slower clearance rate compared to PmetU and PrrnPCL1 respectively. The lower clearance rate seen with the gyr promoters seems to be resulting out of higher abortive transcription delaying the escape of RNAP from these promoters ( Figure 6 ). When the amount of run-off transcripts synthesized at these promoters were compared in the single and multiple round conditions, fewer run-off transcripts were synthesized at PmetU in the single round transcription compared to the PgyrB1 and PrrnPCL1 (data not shown). However, after multiple rounds of transcription, the accumulation of run-off transcripts at PmetU was comparable to that of PgyrB1 and PrrnPCL1.
Figure 6. In vitro transcription assays. A.
Promoter clearance assay was carried out as described in Materials and Methods. (α-32P) +2 nucleotide of ITS was used to label the transcripts. A 10 µl aliquot from the same assay mix was loaded onto 8% urea–PAGE (19∶1) to resolve the run-off transcripts and onto 23% urea-PAGE (10∶1) to resolve abortive transcripts. Arrow on the right side shows the run-off transcripts, while the asterisks show the abortive transcripts synthesized from the promoters. M on the top of the gel corresponds to marker. The bands from top to bottom in this lane corresponds to 91, 75, 32, and 11 nucleotides. B The amount of run-off transcripts was quantified (AU) and plotted against time. The promoter clearance rate was measured from the graph as the time required to form 90% of maximum transcript formed at each promter and is denoted as PC90%. The clearance rate for each promoter is the mean of three values (Table 1).
Discussion
Kinetics and equilibrium binding studies provide an insight into the strength and the mechanism of transcription initiation at the promoters. In addition to the sequence of promoter elements and overall promoter architecture, the strength of a given promoter is governed by events occurring at various stages of the transcription initiation process and the in vivo strength is the net result of cumulative effect of all the steps. In the present study with the four promoters of M. tuberculosis, we have dissected the individual steps in the transcription initiation to understand their characteristic rate limiting steps.
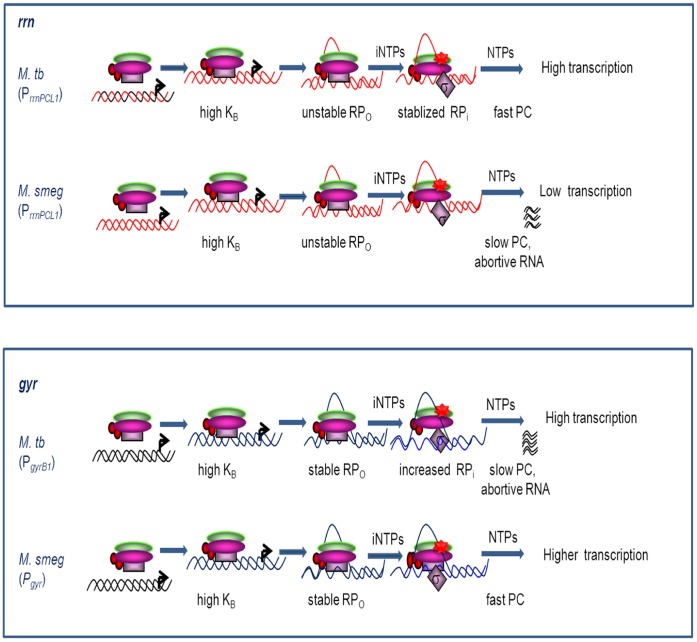
Generally, in every organism as if by a rule, the rrn operons are transcribed by the strongest house-keeping promoters. Very high frequency of initiation at rrn promoters is a characteristic feature that contributes to the abundance in rRNA transcripts [21] and the PrrnPCL1 of M. tuberculosis is no exception to this paradigm. The high strength of the promoter can be attributed to its −10 and −35 elements, which closely resemble to the σA consensus sequence [6], [16]. The instability of the open complex and increase in half-life in the presence of iNTPs seen with the promoter is a characteristic property of any typical rRNA promoter analyzed so far including promoters from M. smegmatis [15], [22]–[24]. However, the comparison of kinetics at PrrnPCL1 of M. tuberculosis with that of M. smegmatis also revealed interesting differences ( Figure 7 ). PrrnPCL1 of M. smegmatis showed slower promoter clearance and greater amount of abortive transcription in addition to an intrinsically unstable open complex [24]. The cumulative effect of these kinetic events results in (10 fold) lower transcriptional activity of M. smegmatis PrrnPCL1 in comparison to M. tuberculosis (24, Figure 7 , Figure S2). The two promoters also differed significantly in their response to iNTPs and pppGpp (24 and this work); the stimulation and inhibition by the two effectors was much more pronounced at PrrnPCL1 of M. tuberculosis. However, inadequacy of the PrrnPCL1 of M. smegmatis appears to be compensated by the very strong PrrnB , which appears to be one of the strongest promoter in the organism. Moreover, the presence of a second functional rRNA operon also ensures adequate rRNA transcription. All these observations indicate the importance of species specific variations in promoters to meet the cellular requirements. The constitutive high level transcripts synthesized from the single rRNA operon of M. tuberculosis seem to fulfill the need of the metabolic machinery of the cell possibly due to the slow growth characteristics of the organism. As a consequence, the present day M. tuberculosis strains and other closely related pathogens seem to have lost the second rRNA operon (rrnB) found in the fast growing species of the genus [16], [17]. The down-regulation of the operon by the action of the pppGpp during transcription initiation would ensure the fine tuning of rRNA expression to lower levels sufficient in the stationary phase. One would expect a very low level of rRNA expression in the dormant state of the organism during its intracellular survival. The positive regulation of the PrrnPCL1 by iNTPs and inhibition of its transcription initiation by pppGpp point out at the remarkable conservation of rRNA transcription regulation across the diverse bacterial species and this sensing mechanism appears to provide a unified theme for the growth phase dependent regulation of rRNA transcription. However, as summarized in Table S1, the rRNA operons are subjected to diverse controls, in addition to conserved features, adding another level of complexity.
Figure 7. Summary of transcription initiation at gyr and rrn promoters from M. tuberculosis and M. smegmatis.
Individual equilibrium and kinetic events occurring at M. tuberculosis and M. smegmatis gyr and rrn promoters are compared. RPc and RPo refer to closed complex and open complex respectively, RPI refers to initiation complex, iNTPs refers to initiating nucleotides, PC refers to promoter clearance. iNTPs enhance the formation of RPI at PgyrB1 and stability of open complex at rrn promoters.
Surprisingly, in contrast to the rrn promoter, the other stable RNA promoter, PmetU, which drives the transcription from a single initiator tRNA gene in M. tuberculosis, does not appear to be influenced by iNTPs and pppGpp. In this context, studies with E.coli tRNA promoters provide interesting parallels. E. coli has two initiator tRNA promoters, PmetY and PmetZ, transcribing the single initiator tRNA and an operon containing three tandemly repeated tRNAf1 Met respectively [25]. However, they respond differently to these regulators. While the transcription from PmetY is unaffected by pppGpp, the PmetZ is subjected to inhibition [26]. Thus, it is apparent that unlike the rrn operons, regulation of promoters of initiator tRNA is not universally conserved. Also in contrast to the PrrnPCL1, the isomerization step was found to be the rate limiting step at PmetU in M. tuberculosis. The lower amounts of run-off transcripts from the promoter in single round transcription assays, compared to the gyr and rrn promoters could be due to the slower rate of formation of the open complex. However, the high promoter strength of the PmetU under in vivo conditions could be accounted by its higher open complex stability, faster promoter clearance and low levels of abortive transcription. The accumulation of large amounts of transcripts after multiple rounds of transcription at PmetU ( Figure 4D ) could be explained not only because of its promoter strength but also possibly due to frequent recycling of RNAP likely to occur at the tRNA genes. The process of re-initiation of transcription could be facilitated because of the presence of an intrinsic terminator at the end of the short gene to allow RNAP to fall off at a distance not far away from the promoter [19], [27]. Notably, high levels of transcripts seen with class III transcripts in eukaryotes is attributed to efficient recycling of the Pol III on tRNA genes [28].
The extent of closed complex formation and rate of isomerization are the major determinants of promoter strength of PgyrB1, the major promoter transcribing the gyr operon of the organism. The rate limiting step at PgyrB1 seems to be the promoter clearance by RNAP. The promoters efficient at early steps of promoter-polymerase interaction tend to be inefficient in promoter clearance [29]. The present data with PgyrB1 supports this hypothesis. The stimulation of open complex formation upon addition of the iNTPs and the opposing effect of inhibition by pppGpp is an unusual property of the promoter, a feature distinct from Pgyr of M. smegmatis, which do not appear to respond in a similar fashion [24]. To our knowledge, this is the first ever description of nucleotide mediated activation and pppGpp mediated inhibition of transcription initiation of gyr promoter in any organism or for that matter promoter of any topoisomerase gene.
Surprisingly, the present studies reveal that the process of transcription initiation at M. tuberculosis PgyrB1 is markedly distinct from the Pgyr, a single promoter transcribing gyrase operon of M. smegmatis, which is a non- pathogenic member of the same genus often used as surrogate host for a variety of studies (Table S2). Slower isomerization rates, faster promoter clearance, lower abortive initiation are the characteristic features of the transcription initiation at Pgyr from M. smegmatis [24]. In contrast, the principle gyr promoter of M. tuberculosis, the subject of the present analysis, exhibited entirely opposite effects viz. faster rate of open complex formation, slower promoter clearance and higher abortive transcription. Notably, the two promoters exhibit markedly distinct mode of ‘Relaxation Stimulated Transcription’ (RST), a homeostatic control employed by cells to regulate gyrase activity and topological status of the genome [6], [30], [31]. In addition, distinct influence exerted by iNTPs and pppGpp on the promoter strength of PgyrB1 could ensure control of the promoter linked to the growth phase. These differences in the regulation of the gyr operons between the two different species may indeed reflect their growth rates, physiology and contrastingly different life-style.
To conclude, during initiation of transcription, each of the M. tuberculosis promoters studied is subjected to different rate-limiting steps and regulation. While unstable open complexes appear to serve as the sensors of initiating nucleotide concentration in rRNA promoter, distinctly, tRNA promoter is rate-limited at open complex formation and not subjected to growth phase dependent control. The opposing effects of the regulatory effectors, on the principle promoter of the gyr operon of the organism indicate the fine control connecting growth phase to supercoiling homeostasis of the genome, a mechanism probably required for metabolic shut down.
Materials and Methods
Promoter DNA, Transcription Templates and RNAP
The strains, plasmids and the sequences of the promoter fragments used for this study are listed in the Table S3. Since promoters of gyr operon are divergent and overlap, the sequences of these promoters were altered such that only one of the two promoters was functional. The sequence of -10 element of PgyrB1 was rearranged from TACAGT to ACTTAG in the fragment containing PgyrR and the sequence corresponding to -10 element of PgyrR was changed from TCTTCT to CTCGTG in fragment containing PgyrB1 ( Figure 1B ).
pARN104, a derivative of pUC18 was used as a vector to clone the promoter fragments amplified from M. tuberculosis H37Ra genomic DNA with specific primers. For in vitro transcription assays, templates were prepared by PCR amplification from the constructs using a set of vector specific primers followed by gel purification. The primers used in this study are listed in Table S4. RNAP was isolated from M. smegmatis SM07 [32] by a modified procedure involving in vivo reconstitution of the enzyme with σA [33]. The σA content in the RNAP preparation was 95% stoichiometric to the β, β' subunits. The specific activity of the purified RNAP was determined both by the standard method of 3[H]-UTP incorporation and by titrating the promoter fragment with a range of RNAP concentrations as described [34], [35]. pppGpp was synthesized as described [36].
β-galactosidase Reporter Assays
The cells were grown in MB7H9 (Difco) medium supplemented with 2% glucose (Sigma) and 0.05% Tween80 (Sigma). Promoter strength was measured by β-galactosidase reporter assay and the activity represented in Miller units (Miller units = 1,000×A 420/(time (min) x volume of culture (ml) x optical density at 600 nm] [37]. M. smegmatis mc2155 transformed with the vector pSD5B [38] was used as the negative control. To determine the in vivo promoter strength in different growth phases, the cultures of M. smegmatis mc2155 transformed with the promoter fusion constructs were grown for 12, 18, 24 30, 48 hours and the β-galactosidase reporter assay was carried out as described before.
Electrophoretic Mobility Shift Assay (EMSA)
For EMSA, oligonucleotides having the individual promoter sequences were used. The 5' promoter fragments were end labeled at their 5' ends of one of the strands with (γ -32P) ATP and T4 polynucleotide kinase (New England Biolabs) at 37°C for 30 min. The labeled strand was annealed with two molar excess of complementary strand. The binding reactions were carried out in transcription buffer containing 50 mM Tris HCl, (pH-8.0 at 25°C), 3 mM magnesium acetate, 100 µM EDTA, 100 µM DTT, 50 mM KCl, 50 µg ml−1 BSA, 5% glycerol [27]. The buffer used for pppGpp assays also included 35 mM of potassium glutamate. The electrophoresis was carried out either at 4°C or room temperature on a 4% native-PAGE. The amount of radioactivity in bound and free promoter fragments was measured by phosphorimager (Fujifilm) and densitometry analysis by Image Guage ver. 2.54.
Determination of Equilibrium Constants (KB and Kd)
To study the RNAP-DNA closed complexes, 1 nM of promoter fragments were titrated with varied amounts of RNAP. The incubation was carried out at ice for 20 min and the fractions were resolved on 4% native-PAGE at 4°C. The KB was determined by Prism software from three independent sets of experiments as described [39]. For determination of the Kd of the open complex, different concentrations of RNAP and 1 nM of promoters were incubated at 37°C for 10 min followed by heparin (50 µg ml−1) challenge for 1 min.The fractions were resolved on 4% native-PAGE at 37°C. The equilibrium dissociation constant for the heparin resistant complexes (Kd) was measured by the equation Y = Ymax[RNAP]/Kd+[RNAP], where Ymax corresponds to binding maximum [40], [41].
Determination of Association and Dissociation Rate Constants
For determination of association rate constants, closed complexes were pre-formed as described. The aliquots (9 µl) from the assay mixture were withdrawn at different time points (0 to 48 min) and challenged with heparin (50 µg ml−1) followed by immediate loading onto 4% native-PAGE electrophoresed at room temperature to analyze the bound fractions. For dissociation assays, open complexes were formed by incubating promoter fragments and RNAP for 15 min at 37°C and the assay mixtures were subjected to heparin challenge (50 µg ml−1). Aliquots (10 µl) were withdrawn at time intervals ranging from 0 to 48 min followed by loading onto 4% running native-PAGE electrophoresed at room temperature. The first order and dissociation rate constants were calculated by fitting the values as described earlier [34].
Assays to Determine the Effect of Ribonucleotides on Isomerization and Stability
Initially promoter DNA was incubated with RNAP (50 nM; 100 nM in case of PgyrR) in the presence of ribonucleotides. The ribonucleotides were added to a final concentration of 100 µM in different combinations (+1, +1+2). The reactions were incubated to form competitor resistant complex as described above and supplemented with NTP mix (100 µM), 1 µCi (α- P32) UTP and incubated at 37°C for 15 min. The reactions were terminated with 2x stop dye (95% formamide, 0.025% (w/v) bromophenol blue, 0.025% (w/v) xylene cyanol, 5 mM EDTA and 0.025% SDS and 8 M urea). The samples were kept at 95°C for 1 min and snap chilled before loading onto 8% urea-PAGE. The effect of iNTPs on open complex was also checked by EMSA. Briefly, the labeled promoter fragments were incubated with RNAP (50 nM; 100 nM in case of PgyrR) in the presence of ribonucleotides (100 µM). The reactions were incubated to form competitor resistant complex as described above.
In vitro Transcription Reactions
After RNAP [100 nM] and promoter DNA (50 nM) were incubated at 37°C for 10 min for open complex formation, RNA synthesis was initiated by the addition of NTP mix (100 µM), 1 µCi (α- P32) UTP and incubated at 37°C for 15 min and terminated with 2x stop dye (95% formamide, 0.025% (w/v) bromophenol blue, 0.025% (w/v) xylene cyanol, 5 mM EDTA and 0.025% SDS and 8 M urea). The samples were heated at 95°C for 1 min and snap chilled before loading onto 8% urea-PAGE. For single round transcription, 50 µg ml−1 heparin was added along with NTP mix (100 µM) and 1 µCi (α- P32) UTP. For promoter clearance analysis, promoter DNA (50 nM) and RNAP (100 nM) were incubated in transcription buffer and the reactions were carried out as described (13, 32). +2 NTP in the Initial Transcribed Sequence (ITS) of each promoter was used as the labeled nucleotide. (α- P32) ATP was used to label the transcripts in case of PgyrB1 , PmetU (α- P32) UTP for PrrnPCl1 and PgyrR. The samples were analyzed in 23% urea-PAGE (10∶1) to resolve abortive transcripts. A 10 µl aliquot from the same assay mix was loaded onto 8% urea – PAGE (19∶1) to resolve the run-off transcripts. The run-off transcripts were quantified as arbitrary units (AU) and plotted against time. The time corresponding to 90% of the maximum transcript formed, at each promoter, was calculated as the promoter clearance rate (PC90%).
Assays with pppGpp
For the assays with pppGpp, RNAP (100 nM) was incubated with pppGpp (1 mM) in transcription buffer (with 35 mM potassium glutamate) for 15 min. in vitro transcription assays to study the effect of pppGpp were carried out as described above.
Supporting Information
Effect of iNTPs on formation and dissociation of open complex. A iNTPs were incubated with promoter fragments and RNAP as described in Materials and Methods to determine their effect on isomerization. The initial transcribed sequence of each template is shown on the left side of the picture. B The amount of RNAP-promoter complex formed in the presence and absence of iNTPs was quantified (AU) and plotted. Slower moving complex was quantified in case of PrrnPCL1.
(TIF)
Iin vivo promoter activity of P rrnPCL1 from M.smegmatis and M.tuberculosis . in vivo activites of PrrnPCL1 from M. smegmatis (M.smeg) and M. tuberculosis (M.tb) was measured by β galactosidase assay and plotted on Y axis as Miller units.
(TIF)
Comparison of transcription at rRNA promoter s of E. coli, M. smegmatis and M. tuberculosis .
(PDF)
Comparison of transcription at gyr promoter s of E. coli, M. smegmatis and M. tuberculosis .
(PDF)
Strains, plasmids and oligonucleotides used in this study.
(PDF)
Sequence of primers used in this study.
(PDF)
Acknowledgments
We thank Dipankar Chatterji for M. smegmatis SM07, pET21b- relNTD construct and discussions. Thanks are due to Bianca Sclavi for the critical reading of the manuscript and valuable inputs. We acknowledge the Phosphorimager facility of IISc supported by the Department of Biotechnology, Government of India.
Funding Statement
PT and AC are recipients of Senior Research Fellowships from University Grant Commission and Council of Scientific and Industrial Research, Government of India, respectively. VN is a J.C. Bose Fellow of Department of Science and Technology, Government of India and is recipient of Centre of Excellence in tuberculosis research grant from Department of Biotechnology, Government of India. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Johnson R, Streicher EM, Louw GE, Warren RM, van Helden PD, et al. (2006) Drug resistance in Mycobacterium tuberculosis . Curr Issues Mol Biol 8: 97–111. [PubMed] [Google Scholar]
- 2.Smith I, Bishai WR, Nagaraja V (2005) Control of mycobacterial transcription. In: Tuberculosis and the Tubercle Bacillus. ST Cole, McMurray, D. N., and Jacobs, Jr., W. R. (ed). Washington, DC: ASM Press, 219–231.
- 3. Jia Y, Patel SS (1997) Kinetic mechanism of transcription initiation by bacteriophage T7 RNA polymerase. Biochemistry 36: 4223–4232. [DOI] [PubMed] [Google Scholar]
- 4. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544. [DOI] [PubMed] [Google Scholar]
- 5.Gomez M, Smith I (2000) Determinants of mycobacterial gene expression, p.111–129. In H. F. Hatfull and W. R. Jacobs, Jr. (ed.), Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 6. Unniraman S, Chatterji M, Nagaraja V (2002) DNA gyrase genes in Mycobacterium tuberculosis: a single operon driven by multiple promoters. J Bacteriol 184: 5449–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manganelli R, Provvedi R, Rodrigue S, Beaucher J, Gaudreau L, et al. (2004) Sigma factors and global gene regulation in Mycobacterium tuberculosis . J Bacteriol 186: 895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saecker RM, Record MT Jr, Dehaseth PL (2011) Mechanism of bacterial transcription initiation: RNA polymerase-promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol 412: 754–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Saecker RM, Tsodikov OV, McQuade KL, Schlax PE Jr, Capp MW, et al. (2002) Kinetic studies and structural models of the association of E. coli sigma (70) RNA polymerase with the lambdaP(R) promoter: large scale conformational changes in forming the kinetically significant intermediates. J Mol Biol 319: 649–671. [DOI] [PubMed] [Google Scholar]
- 10. Helmann JD (2009) RNA polymerase: a nexus of gene regulation. Methods 47: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Browning DF, Busby SJ (2004) The regulation of bacterial transcription initiation. Nat Rev Microbiol 2: 57–65. [DOI] [PubMed] [Google Scholar]
- 12. Hsu LM (2009) Monitoring abortive initiation. Methods 47: 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. deHaseth PL, Zupancic ML, Record MT Jr (1998) RNA Polymerase-Promoter Interactions: the Comings and Goings of RNA Polymerase. J Bacteriol. 180: 3019–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Potrykus K, Cashel M (2008) (p)ppGpp: still magical? Annu Rev Microbiol 62: 35–51. [DOI] [PubMed] [Google Scholar]
- 15. Gaal T, Bartlett MS, Ross W, Turnbough CL Jr, Gourse RL (1997) Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278: 2092–2097. [DOI] [PubMed] [Google Scholar]
- 16. Gonzalez-y-Merchand JA, Garcia MJ, Gonzalez-Rico S, Colston MJ, Cox RA (1997) Strategies used by pathogenic and nonpathogenic mycobacteria to synthesize rRNA. J Bacteriol 179: 6949–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Verma A, Sampla AK, Tyagi JS (1999) Mycobacterium tuberculosis rrn promoters: differential usage and growth rate-dependent control. J Bacteriol 181: 4326–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stadthagen-Gomez G, Helguera-Repetto AC, Cerna-Cortes JF, Goldstein RA, Cox RA, et al. (2008) The organization of two rRNA (rrn) operons of the slow-growing pathogen Mycobacterium celatum provides key insights into mycobacterial evolution. FEMS Microbiol Lett 280: 102–112. [DOI] [PubMed] [Google Scholar]
- 19. Vasanthakrishna M, Kumar NV, Varshney U (1997) Characterization of the initiator tRNA gene locus and identification of a strong promoter from Mycobacterium tuberculosis . Microbiology 143 (Pt 11): 3591–3598. [DOI] [PubMed] [Google Scholar]
- 20. Hsu LM (2002) Promoter clearance and escape in prokaryotes. Biochim Biophys Acta 1577: 191–207. [DOI] [PubMed] [Google Scholar]
- 21. Dennis PP, Ehrenberg M, Bremer H (2004) Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol Mol Biol Rev 68: 639–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ohlsen KL, Gralla JD (1992) DNA melting within stable closed complexes at the Escherichia coli rrnB P1 promoter. J Biol Chem 267: 19813–19818. [PubMed] [Google Scholar]
- 23. Krasny L, Gourse RL (2004) An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. . 23 4473–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. China A, Tare P, Nagaraja V (2010) Comparison of promoter-specific events during transcription initiation in mycobacteria. Microbiology 156: 1942–1952. [DOI] [PubMed] [Google Scholar]
- 25. Kenri T, Imamoto F, Kano Y (1994) Three tandemly repeated structural genes encoding tRNA (f lMet) in the metZ operon of Escherichia coli K-12. Gene 138: 261–262. [DOI] [PubMed] [Google Scholar]
- 26. Nagase T, Ishii S, Imamoto F (1988) Differential transcriptional control of the two tRNA(fMet) genes of Escherichia coli K-12. Gene 67: 49–57. [DOI] [PubMed] [Google Scholar]
- 27. Unniraman S, Prakash R, Nagaraja V (2001) Alternate paradigm for intrinsic transcription termination in eubacteria. J Biol Chem 276: 41850–41855. [DOI] [PubMed] [Google Scholar]
- 28. Dieci G, Sentenac A (1996) Facilitated recycling pathway for RNA polymerase III. Cell 84: 245–252. [DOI] [PubMed] [Google Scholar]
- 29. Ellinger T, Behnke D, Bujard H, Gralla JD (1994) Stalling of Escherichia coli RNA polymerase in the +6 to +12 region in vivo is associated with tight binding to consensus promoter elements. J Mol Biol 239: 455–465. [DOI] [PubMed] [Google Scholar]
- 30. Menzel R, Gellert M (1987a) Fusions of the Escherichia coli gyrA and gyrB control regions to the galactokinase gene are inducible by coumermycin treatment. J Bacteriol 169: 1272–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Unniraman S, Nagaraja V (1999) Regulation of DNA gyrase operon in Mycobacterium smegmatis: a distinct mechanism of relaxation stimulated transcription. Genes to Cells 4: 697–706. [DOI] [PubMed] [Google Scholar]
- 32. Mukherjee R, Chatterji D (2008) Stationary phase induced alterations in mycobacterial RNA polymerase assembly: A cue to its phenotypic resistance towards rifampicin. Biochem Biophys Res Commun 369: 899–904. [DOI] [PubMed] [Google Scholar]
- 33. China A, Nagaraja V (2010) Purification of RNA polymerase from mycobacteria for optimized promoter-polymerase interactions. Protein Expr Purif 69: 235–242. [DOI] [PubMed] [Google Scholar]
- 34. Schroeder LA, deHaseth PL (2005) Mechanistic differences in promoter DNA melting by Thermus aquaticus and Escherichia coli RNA polymerases. J Biol Chem 280: 17422–17429. [DOI] [PubMed] [Google Scholar]
- 35. Sclavi B, Zaychikov E, Rogozina A, Walther F, Buckle M, et al. (2005) Real-time characterization of intermediates in the pathway to open complex formation by Escherichia coli RNA polymerase at the T7A1 promoter. Proc Natl Acad Sci U S A 102: 4706–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jain V, Saleem-Batcha R, China A, Chatterji D (2006) Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci 15: 1449–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller JH (1992) A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. In. NY: Cold Spring Harbor Laboratory Press, 71.
- 38. Jain S, Kaushal D, DasGupta SK, Tyagi AK (1997) Construction of shuttle vectors for genetic manipulation and molecular analysis of mycobacteria. Gene 190: 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chakraborty A, Nagaraja V (2006) Dual role for transactivator protein C in activation of mom promoter of bacteriophage Mu. J Biol Chem 281: 8511–8517. [DOI] [PubMed] [Google Scholar]
- 40. Tsujikawa L, Tsodikov OV, deHaseth PL (2002) Interaction of RNA polymerase with forked DNA: evidence for two kinetically significant intermediates on the pathway to the final complex. Proc Natl Acad Sci U S A 99: 3493–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schroeder LA, Choi AJ, DeHaseth PL (2007) The -11A of promoter DNA and two conserved amino acids in the melting region of sigma70 both directly affect the rate limiting step in formation of the stable RNA polymerase-promoter complex, but they do not necessarily interact. Nucleic Acids Res 35: 4141–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of iNTPs on formation and dissociation of open complex. A iNTPs were incubated with promoter fragments and RNAP as described in Materials and Methods to determine their effect on isomerization. The initial transcribed sequence of each template is shown on the left side of the picture. B The amount of RNAP-promoter complex formed in the presence and absence of iNTPs was quantified (AU) and plotted. Slower moving complex was quantified in case of PrrnPCL1.
(TIF)
Iin vivo promoter activity of P rrnPCL1 from M.smegmatis and M.tuberculosis . in vivo activites of PrrnPCL1 from M. smegmatis (M.smeg) and M. tuberculosis (M.tb) was measured by β galactosidase assay and plotted on Y axis as Miller units.
(TIF)
Comparison of transcription at rRNA promoter s of E. coli, M. smegmatis and M. tuberculosis .
(PDF)
Comparison of transcription at gyr promoter s of E. coli, M. smegmatis and M. tuberculosis .
(PDF)
Strains, plasmids and oligonucleotides used in this study.
(PDF)
Sequence of primers used in this study.
(PDF)