Abstract
The PML–RARA fusion protein is found in approximately 97% of patients with acute promyelocytic leukemia (APL). APL can be associated with life-threatening bleeding complications when undiagnosed and not treated expeditiously. The PML–RARA fusion protein arrests maturation of myeloid cells at the promyelocytic stage, leading to the accumulation of neoplastic promyelocytes. Complete remission can be obtained by treatment with all-trans-retinoic acid (ATRA) in combination with chemotherapy. Diagnosis of APL is based on the detection of t(15;17) by karyotyping, fluorescence in situ hybridization or PCR. These techniques are laborious and demand specialized laboratories. We developed a fast (performed within 4–5 h) and sensitive (detection of at least 10% malignant cells in normal background) flow cytometric immunobead assay for the detection of PML–RARA fusion proteins in cell lysates using a bead-bound anti-RARA capture antibody and a phycoerythrin-conjugated anti-PML detection antibody. Testing of 163 newly diagnosed patients (including 46 APL cases) with the PML–RARA immunobead assay showed full concordance with the PML–RARA PCR results. As the applied antibodies recognize outer domains of the fusion protein, the assay appeared to work independently of the PML gene break point region. Importantly, the assay can be used in parallel with routine immunophenotyping for fast and easy diagnosis of APL.
Keywords: PML–RARA protein, t(15;17), APL, immunobead, flow cytometry
Introduction
The presence of t(15;17) with the PML–RARA fusion gene is considered to be the hallmark of acute promyelocytic leukemia (APL), also known as acute myeloid leukemia (AML) type M3.1 Initially t(15;17) was assumed to be present in all APL patients. However, over the last 10 years a sizeable minority of APL cases (∼5%) has been identified as lacking this classical translocation, but containing the PML–RARA fusion gene. Such nonclassical PML–RARA fusion genes can be caused by chromosomal aberrations, such as insertions or complex chromosomal aberrations, which may be missed by cytogenetics and fluorescence in situ hybridization (FISH), but are detected by PCR.2, 3, 4
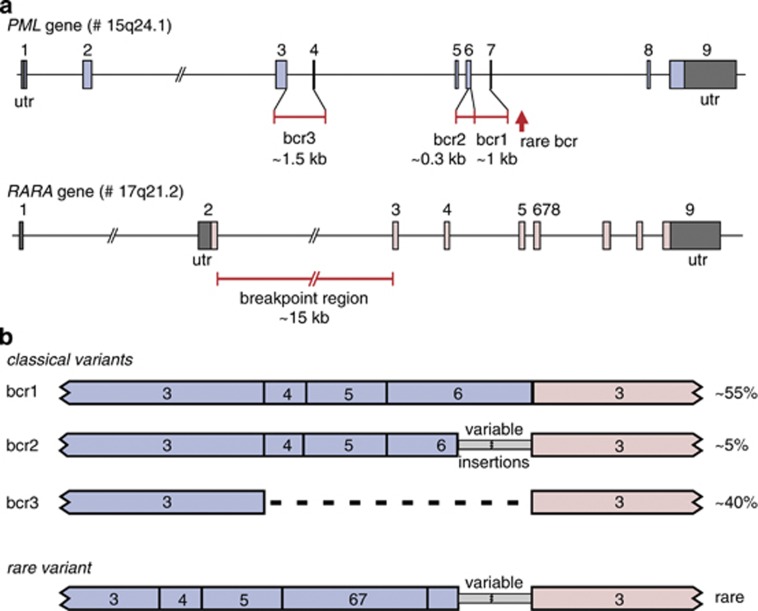
The break points in the RARA gene are all located in intron 2 (∼15 kb), whereas the vast majority of break points in the PML gene cluster in the break point cluster region (bcr) of intron 6 (bcr1;∼55% of cases), exon 6 (bcr2; ∼5% of cases) or intron 3 (bcr3; ∼40% of cases).3 Two APL cases with t(15;17) have been reported with breaks in PML exon 7 (Figure 1).5 A rare subgroup of APL cases (2–3%) has variant RARA gene translocations with a non-PML fusion partner. So far seven alternative RARA gene partners have been identified in APL cases: PLZF, NPM, NUMA, STAT5B, FIP1L1, PRKAR1A and BCOR.6, 7, 8, 9, 10, 11, 12, 13, 14 The PLZF gene appears to be the most frequent alternative partner and is present in ∼1% of all APL cases (Table 1).7, 10
Figure 1.
Structure of the PML and RARA genes with the break point regions and the corresponding fusion gene transcripts. (a) The PML gene contains three well-defined small break point cluster regions (bcr's): bcr1 in intron 6, bcr2 in the downstream part of exon 6 and bcr3 in intron 3. In addition two rare break points in intron 7 have been reported. In the RARA gene the break points cluster in intron 2 (∼15 kb). (b) The three well-defined bcr's and the rare intron 7 breaks in the PML gene result in four different PML–RARA fusion transcripts. Percentages refer to the relative frequency of the various PML–RARA variants.
Table 1. RARA fusion genes in APL.
| Chromosome aberration | Fusion gene | Relative frequency in APL |
|---|---|---|
| t(15;17) (q24;q21) (classical) | PML–RARA | ∼92% |
| Other 15q24 and 17q21 aberrationsa | PML–RARAa | ∼5% |
| t(11;17) (q23;q21) | PLZF-RARA | ∼1% |
| t(5;17) (q35;q21) | NPM-RARA | ∼0.5% |
| t(11;17) (q13;q21) | NUMA-RARA | Rare |
| Der(17) | STAT5B-RARA | Rare |
| t(4:17) (q12;q21) | FIP1L1-RARA | Two cases |
| Complex der(17) | PRKAR1A-RARA | Single case |
| t(x;17) (p11;q21) | BCOR-RARA | Single case |
| No aberration detectable | No RARA gene rearrangement | ∼1% |
Finally, in ∼1% of APL molecular techniques have not identified a RARA gene rearrangement.2 Further studies are needed to assess what genetic event causes APL characteristics in these patients. In some of these cases the PCR technique may have given false-negative results due to unusual positioning of the PML or RARA gene break points, not covered by the applied primers.5
APL patients with the PML–RARA fusion gene aberration have been shown to be highly sensitive to retinoid differentiating agents, such as all-trans-retinoic acid (ATRA). Combination therapy (ATRA and chemotherapy) has significantly improved treatment outcome in APL patients. The long-term outcome is now favorable because of the low risk of relapse and prevention of life-threatening coagulopathy at diagnosis. Also in recent years, As2O3 was introduced as a new and efficient treatment alternative for APL patients. Consequently, stem cell transplantation in first remission is no longer recommended.15, 16
It should be noted that APL patients with the most frequent variant fusion gene, the PLZF-RARA fusion gene (Table 1), are not sensitive to ATRA treatment.10, 16, 17 However, other rare variant APL cases with NPM-RARA and NUMA-RARA fusion genes are sensitive to ATRA treatment, comparable to PML–RARA positive cases.11
As the first description of APL, life-threatening bleeding problems have been identified as the most notorious manifestation of the disease. This coagulopathy in many APL patients leads to pulmonary and cerebral hemorrhages, if the appropriate treatment regimen is not initiated instantly.15, 16, 18 Even upon treatment, APL-related coagulopathy takes 5–8 days to improve. Many hematologists consider the correct diagnosis in a patient with APL a medical emergency, because of the coagulopathy manifestations, which continue to be a major cause of death in APL patients. Consequently, a positive diagnosis of APL is of utmost importance for patient care in leukemia treatment. Such diagnosis preferably should be provided within hours and not within days.
Although the clinical and cytomorphological picture of APL seems relatively clear, the leukemic cells in a subset of patients (5–10%) do not exhibit the typical APL morphology.19 Consequently, it is strongly recommended to make a fast and accurate diagnosis of the PML–RARA aberration. Because of the risk of early death due to bleeding, best clinical practice recommends that APL should be excluded in each patient with newly diagnosed AML.15, 16
At present, the diagnosis of PML–RARA positive APL cases is based on the result of cytomorphology and immunophenotyping combined with karyotyping, FISH, and/or PCR. Immunophenotyping results are typically completed on the same day, but the other techniques are time consuming (usually 1–2 days for FISH and PCR techniques and 1–2 weeks for karyotyping) and are restricted to specialized laboratories with well-trained personnel. In addition some laboratories use the anti-PML protein immunofluorescence test, which is claimed to result in a macrogranular nuclear staining pattern in case of a wild-type (nontranslocated) PML gene, but gives multiple smaller dots in case of a PML–RARA fusion gene.15, 20, 21 This assay is fast, but requires sufficient fluorescence microscopy experience for reliable reading and false-negativity cannot be excluded.20, 21
Therefore, novel methods are needed for fast and easy diagnosis of PML–RARA positive APL, without subjective interpretation steps. We have developed a new antibody-based flow cytometric immunobead assay for specific detection of the PML–RARA fusion protein, comparable to our BCR-ABL immunobead assay.22 To allow detection of all known PML–RARA fusion proteins independent of the break point (bcr1, bcr2, bcr3, and the rare bcr in exon 7), antibodies were generated that recognize immunogenic epitopes located upstream of the bcr's (upstream of bcr3) for PML and downstream of the break point region in intron 2 known for RARA. The antibodies were combined into a prototype immunobead assay in which the anti-RARA antibody was used as a capture antibody and the anti-PML antibody as a phycoerythrin (PE)-conjugated detection antibody. After initial testing on cell lines and a small series of patient samples, the EuroFlow Consortium (EU-FP6, LSHB-CT-2006-018708) evaluated 163 acute leukemias at diagnosis, which showed full concordance between PCR and immunobead results.
Materials and Methods
Cell lines
Several leukemic cell lines were used to investigate the specificity of the PML–RARA immunobead assay. The NB4 cell line with t(15;17) and PML–RARA transcripts was used as positive control; the PML break point in this cell line is located in bcr1 (intron 6).23 Several other leukemic cell lines were used as negative controls: ME-1 with inv(16) and CBFB-MYH11 transcripts, MV4-11 with t(4;11) and MLL-AF4 transcripts, K562 with t(9;22) and BCR-ABL transcripts, 697 with t(1;19) and E2A-PBX1 transcripts, RCH-ACV with t(1;19) and E2A-PBX1 transcripts, KASUMI-1 with t(8;21) and AML1-ETO transcripts, and REH with t(12;21) and TEL-AML1 transcripts.3 All leukemic cell lines used in this study are available from the German Collection of Microorganisms and Cell cultures (DSMZ GmbH, Braunschweig, Germany).
Healthy controls
Peripheral blood (PB) samples were obtained from healthy donors after informed consent according to the local Medical Ethics Committee guidelines. Total white blood cells (WBC) were obtained by NH4Cl lysis of the erythrocytes. PB mononuclear cells (PBMC) were obtained by ficoll separation. To mimic leukemic samples with varying tumor loads, cells of the PML–RARA expressing cell line NB4 were mixed at different concentrations in either PML–RARA negative cell lines or in normal PBMC or WBC.
Patient samples
Freshly collected or frozen bone marrow (BM) or PB samples were obtained from patients in accordance with the Declaration of Helsinki. All participants in the EuroFlow Consortium (EU-FP6 grant LSHB-CT-2006-018708) obtained approval from the local Medical Ethics Committee for secondary use of remaining diagnostic material of patients suspected to have a hematological malignancy. Cell samples were processed by either NH4Cl lysis or ficoll separation.
Antibodies for specific detection of PML and RARA domains
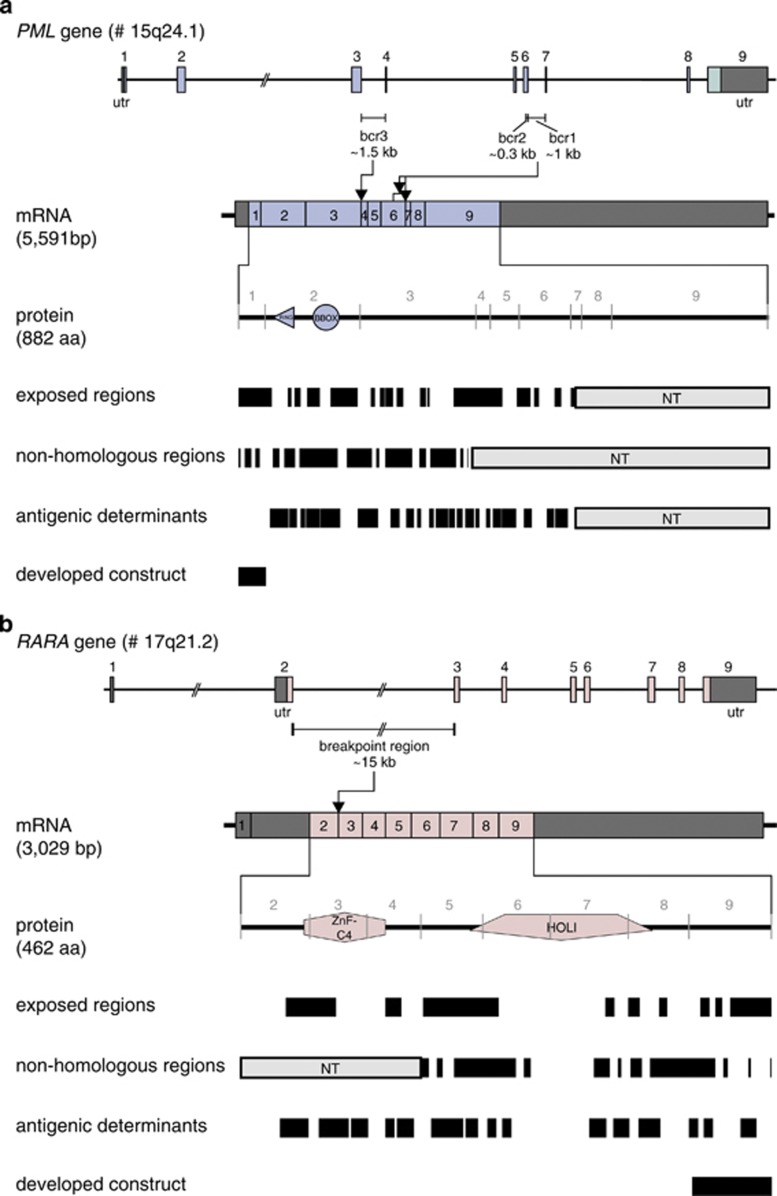
Antibodies were raised against recombinantly expressed epitopes of PML and RARA domains that are present in all known fusion protein variants, which result from various break point regions. The first PML domain (first 46 amino acids) located before the first Zinc finger domain was used for immunization (Figure 2a). For the generation of anti-RARA antibodies, the C-terminal domain of RARA (last 70 amino acids) was recombinantly expressed and used for immunization (Figure 2b).
Figure 2.
Selection of the PML and RARA constructs for raising anti-PML and anti-RARA antibodies. (a) To increase the chance of making an anti-PML antibody that worked in the immunobead assay, the PML immunogen had to meet specific criteria. The PML protein domains encoded by exon 1–3 (upstream of all known break points) were analyzed with various web-based programs to identify exposed regions (accessibility for the antibody), nonhomologous regions (no cross-reactivity with other human proteins) and antigenic determinants (difference between human and mouse). Accordingly, the first PML domain (first 46 amino acids) before the ring-type zinc finger domain was selected for cloning, protein expression, protein purification and subsequent immunization. The anti-PML antibody 5D8 was used in the immunobead assay. (b) Using the same selection criteria, the C-terminal domain of RARA (last 70 amino acids) were used for cloning, protein production and subsequent immunization. The anti-RARA antibody 9G4.16 was used in the immunobead assay.
The recombinant proteins were expressed in HEK 293FS cells and after purification were used for immunization of SJL (anti-PML) or Balb/c (anti-RARA) mice using subcutaneous immunization techniques optimized at BD Diagnostics (Durham, NC, USA), followed by fusion of spleen cells with the P3X,653 cell line for hybridoma formation. This resulted in the generation of the anti-PML 5D8 and the anti-RARA 9G4.16 antibodies.
Prototype immunobead assay
The here described and tested prototype immunobead assay for detection of the PML–RARA fusion protein was essentially developed as described for the BCR-ABL immunobead assay.22 Small adaptations were made in the pretreatment and lysis reagents to improve the detection of the PML–RARA fusion protein, which has a nuclear localization. This is in contrast to the cytoplasmically expressed BCR-ABL fusion protein. In short, to inhibit protease activity before cells lysis, intact cells were pretreated for 10 min on ice with the protease inhibitors 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, (AEBSF, Sigma, St Louis, MO, USA; 15 mℳ AEBSF, 15 μl of 1 ℳ stock in phosphate-buffered saline (PBS)) and phenylmethanesulfonyl fluoride (PMSF, Sigma; 1 mℳ, 5 μl of 200 mℳ stock in ethanol) in PBS at a concentration of 107 cells/ml. Next, cells were gently spun down (5 m at 500 g at 4 °C) and washed with 1 ml of 5% fetal bovine serum in PBS. After removal of the supernatant the cells were resuspended in BD Pharmingen cell lysis buffer supplemented with 0.1% sodium dodecyl sulfate, 2 mM MgCl, 50 U/ml of Benzonase, and the BD Baculogold protease inhibitor cocktail solution diluted as described by the manufacturer (BD Biosciences, San José, CA, USA). After 15 min incubation on ice, the lysate was spun down at 14 000 r.p.m. (20 000 g) for 10 min at 4 °C to remove cell debris, and the supernatant was transferred to a fresh Eppendorf tube then used in the cytometric bead assay.
In all, 50 μl of cell lysate was incubated with 50 μl of bead-conjugated anti-RARA (9G4.16) antibody (6000 beads) and 50 μl of PE-conjugated anti-PML (5D8) antibody for 2 h at room temperature while being shaken. Incubation was performed in either a 96-well filter plate (Multiscreen HTS filter plates, MSBVN1250, Millipore, Tullagreen, Ireland) or in BD Falcon 12 × 75 mm tubes. After incubation, the filter plate was washed twice with 200 μl of BD CBA wash buffer and after the final drain, the beads were resuspended in the plate with 200 μl of BD CBA wash buffer. The beads incubated in BD Falcon 12 × 75mm tubes were washed one time by the addition of 1 ml of BD CBA wash buffer, and after pelleting the beads for 7 min at 500 g at 4 °C, the beads were resuspended in 300 μl of BD CBA wash buffer. Analysis was performed on the results obtained from at least 1000 beads that were acquired on a BD FACS Canto II or BD LSR II flow cytometer using BD FACSDiva software, version 6.1 (BD Biosciences). Instruments were setup according to the BD Cytometer Setup and Tracking module and subsequently photo multipliers were adjusted using capture beads according to the manufacturer's recommendations, resulting in mean fluorescence intensity (MFI) values of 20 000, 40 000 and 100 for the APC, APC-Cy7 and PE channels, respectively.
Specificity and sensitivity testing of the PML–RARA prototype immunobead assay
The specificity of the PML–RARA prototype kit was confirmed by the analysis of cell lysates from leukemic cell lines with different types of fusion proteins (see above).
The sensitivity of the PML–RARA prototype assay was determined by analyzing serial dilutions of the NB4 cell line into both PBMC and WBC. All analyses were performed in duplicate. MFI values detected in 100% PBMC and 100% WBC were used as negative control values.
Testing of the PML–RARA prototype immunobead assay by the EuroFlow Consortium
The PML–RARA prototype immunobead assay was tested in nine EuroFlow laboratories on BM and/or PB samples of 163 leukemia patients at initial diagnosis, including 46 APL patients (according to morphology and/or immunophenotyping; 9 PB and 37 BM samples). Because of the rarity of APL samples, a large fraction of the patient samples was used from local cell banks. Only six of the 46 APL samples were fresh cell samples. To facilitate the standardized collection of patient data with accompanying results from PML–RARA testing, a Case Report Form was developed for each sample with drop-down menus for all relevant information. This case report form was electronically linked to a corresponding Microsoft Excel file to guarantee transfer of data without typing errors.
The PML–RARA prototype immunobead assay was performed according to recommendations developed at BD Biosciences (see above). However, in three laboratories without a refrigerated microcentrifuge, centrifugation of lysates was performed at 20 000 g for 2 min at room temperature instead of 10 min at 4 °C. Storage of the recovered lysates at 80 °C was allowed in order to analyze multiple samples collectively for better comparison with several positive and negative controls. Controls were provided by Dynomics (lysates from 10% or 20% NB4 in PBMC as positive controls and negative control lysates derived from PBMC).
PCR analysis of PML–RARA transcripts
PCR analysis of the different types of PML–RARA fusion transcripts was performed with the standardized Europe Against Cancer (EAC) protocols using TaqMan based real-time quantitative PCR (RQ-PCR; see Gabert et al., 2003 for details).24 The ABL, GUS or B2M internal control genes for the real-time quantitative-PCR studies were selected from the EAC study by Beillard et al.25 According to the EAC guidelines, if too few control gene copies were detected in patients without PML–RARA transcripts, those patient samples were excluded from the study because of the low quality of the RNA.25
Statistical analysis
Statistical analyses using the Mann–Whitney U-test were performed to compare mean MFI values between different PML–RARA positive samples, between different PML break point types (bcr1, bcr2, bcr3) and between different experimental conditions, for example, microcentrifugation at room temperature for 2 min versus at 4 oC for 10 min and sample analysis on the same day versus overnight. The relation between MFI values and age of the samples was evaluated by Spearman's rank correlation coefficient. A value of P<0.05 was regarded to be statistically significant.
Results
Testing of the PML–RARA immunobead assay on cell lines and a small number of patient samples
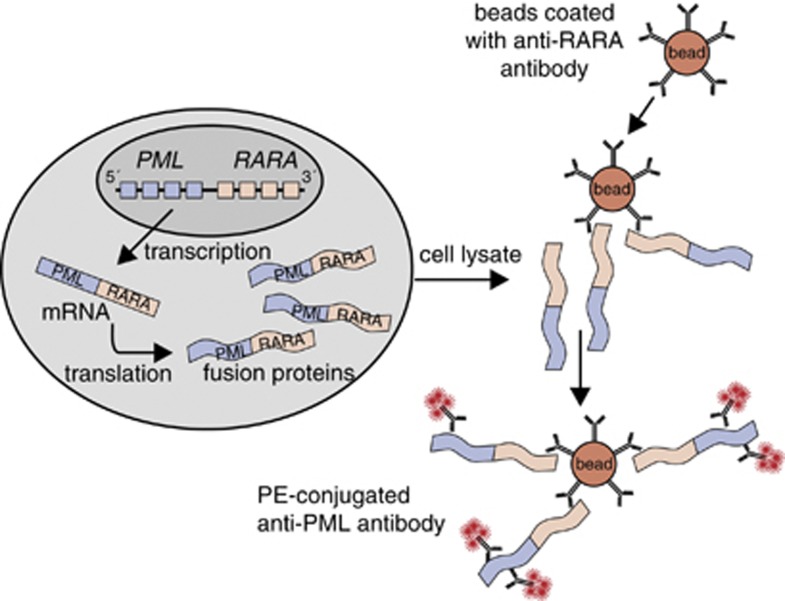
Antibodies were developed against regions within PML and RARA proteins, that are present in all described PML–RARA fusion proteins (Figure 2). The anti-PML antibody 5D8 was raised against exposed, nonhomologous amino-acid sequences. The involved region is located upstream of the bcr's (Figure 2a). Using the same criteria, the anti-RARA antibody was raised against sequences downstream of the RARA break point region (Figure 2b). The selected anti-PML and anti-RARA antibodies were tested for their suitability as bead-bound capture antibody and PE-conjugated detection antibody. Best results were obtained when the anti-RARA antibody was used as bead-bound capture antibody and the anti-PML antibody as PE-conjugated detection antibody; this configuration was used in the prototype PML–RARA immunobead assay (Figure 3).
Figure 3.
Configuration of the prototype PML–RARA immunobead assay. The bead-bound anti-RARA antibody 9G4.16 serves as the capture antibody, whereas the PE-conjugated anti-PML antibody 5D8 serves as the detection antibody. The leukemia cells are lysed and incubated with beads and the detection antibody, then washed and evaluated by flow cytometry for PE positivity of the immunobeads.
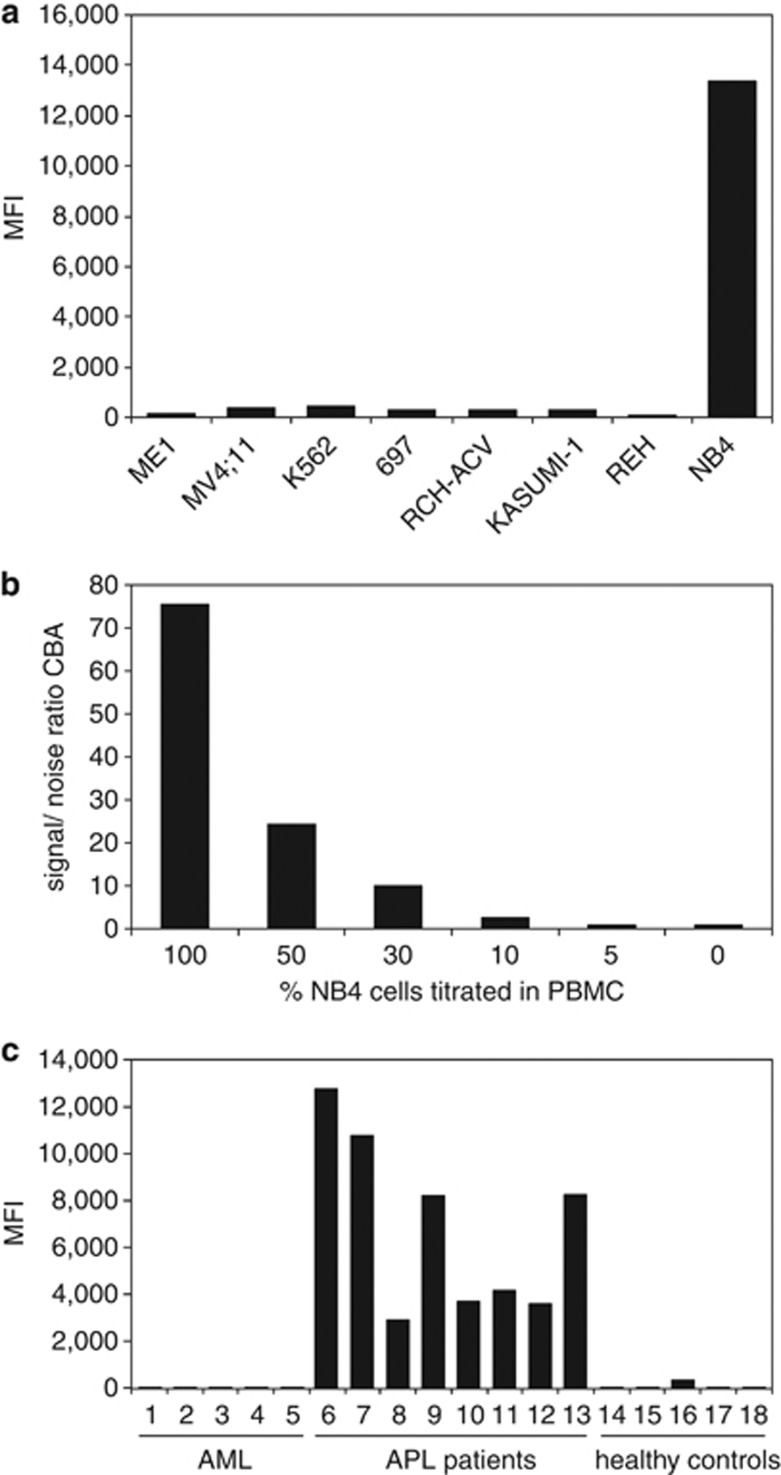
The specificity of the assay was determined by testing leukemic cell lines with t(15;17) or other translocations. Only the t(15;17) positive NB4 cell line gave high PE-fluorescent signals, whereas the other cell lines tested exhibited low background signals (Figure 4a). The sensitivity of the PML–RARA immunobead assay was determined by dilution of the t(15;17) positive NB4 cell line in both WBC and PBMC. The PML–RARA specific fluorescent signal was still detectable at concentrations as low as 10% of the NB4 cell line (Figure 4b). This sensitivity is sufficient, as PB and BM samples of most APL patients at diagnosis contain more than 70% of leukemic cells. Testing of a small series of APL patients with bcr1 or bcr3 PML–RARA variants showed that the immunobead assay was positive in all APL patients and the results were independent of the PML break point position (Figure 4c).
Figure 4.
Detection of PML–RARA fusion protein in cell lines and patient samples with the prototype PML–RARA immunobead assay. (a) Cell lysates from cell lines with different translocations expressing various fusion proteins: ME-1 with inv(16) expressing CBFβ-MYH11, MV4;11 with t(4;11) expressing MLL-AF4, K562 with t(9;22) expressing BCR-ABL, 697 with t(1;19) expressing E2A-PBX1, RCH-ACV with t(1;19) expressing E2A-PBX1, KASUMI-1 with t(8;21) expressing AML1-ETO, REH with t(12;21) expressing TEL-AML1 did not give a PE fluorescence signal in the PML–RARA immunobead assay, whereas the cell line NB4 with t(15;17) expressing PML–RARA (bcr1) gave a strong PE signal. (b) The PML–RARA positive NB4 cell line was diluted into PBMC of a healthy individual and cell lysates were prepared. The immunobead assay was carried out in triplicate for every diluted sample. At a concentration of 10% NB4 cells a signal-to-noise (s/n) ratio compared with the PBMC background of 2.7 was obtained. (c) The novel PML–RARA immunobead assay was carried out on a series of cell lysates of AML patients without the t(15;17) translocation (samples 1–5), APL patients with translocations in the PML gene involving both the bcr1 and bcr3 region and of two APL patients in which the break point region involved in the t(15;17) translocation was not determined (samples 6–13), and of healthy individuals (samples 14–19). Strong positive PE signals were only obtained in the cell lysates of APL patients independent of the bcr involved.
Testing of the PML–RARA prototype immunobead assay by the EuroFlow Consortium
The PML–RARA prototype immunobead assay was distributed to nine EuroFlow laboratories. A series of 163 leukemia patients was evaluated, consisting of 112 AML patients (including 46 APL patients), one chronic myeloid leukemia (CML) patient in accelerated phase, 34 B-cell precursor acute lymphoblastic leukemia (BCP-ALL) patients, and 16 T-ALL patients. For each analysis of patient samples, both a positive and negative control lysate were used to verify the proper performance of the immunobead assay.
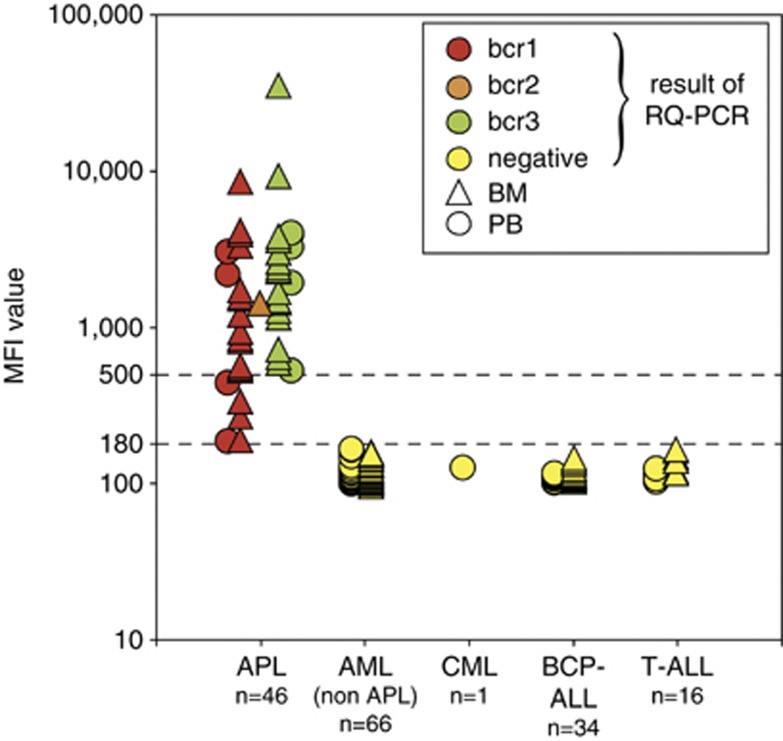
MFI values of the non-APL patient samples had a median value of 114 (range 100–173) and a mean of 119 (s.d. 15.4). The MFI values of the AML (non-APL) samples were slightly higher than the MFI values of the B-cell precursor acute lymphoblastic leukemia samples and T-ALL samples: mean of 121 (s.d. 18.3) versus 118 (s.d. 10.4) and 113 (s.d. 8.7), respectively. The cutoff between negativity and positivity was based on the MFI values of negative AML (non-APL) samples, using 3-s.d. values, resulting in an MFI value of 176. Consequently, a safe MFI cutoff value of 180 was defined. All tested PCR-negative samples and negative controls had an MFI below this cutoff value. Based on the MFI cutoff value and the values of the positive samples, several ranges of positive MFI values were arbitrarily defined to cluster the leukemias according to the expression level of PML–RARA protein: high-level positivity: MFI ⩾1000; medium-level positivity: MFI ⩾500, but <1000; low-level positivity: MFI ⩾180, but <500; negativity: MFI <180.
Full concordance (163/163; 100%) was observed between the PML–RARA PCR results for fusion transcript detection and the prototype PML–RARA immunobead results for fusion protein detection (Tables 2 and 3 and Figure 5). A total of 46 APL patients (22 children and 24 adults) were positive for PML–RARA with both methods. All other 117 leukemia patients including 66 AML patients (18 children and 48 adults), 1 CML patient (accelerated phase), 34 BCP-ALL patients and 16 T-ALL patients, were PML–RARA negative in both the assays (Table 2).
Table 2. Results of PML–RARA detection by PCR and the immunobead assay performed in 163 patient samples.
| Newly diagnosed patients | No. of patients | PML–RARA mRNA positive |
PML–RARA immunobead assay |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Positive |
MFI positive |
MFI negative |
Mean | 3-s.d. | |||||
| Median | Range | Median | Range | ||||||
| APL (AML-M3) | 46 | 46 | 46 | 1514 | 192–32.160 | NA | NA | NA | NA |
| AML, non-APL | 66 | 0 | 0 | NA | NA | 114 | 100–173 | 121 | 55 |
| CML accelerated phase | 1 | 0 | 0 | NA | NA | 131 | NA | NA | NA |
| BCP-ALL | 34 | 0 | 0 | NA | NA | 115 | 106–151 | 118 | 31 |
| T-ALL | 16 | 0 | 0 | NA | NA | 113 | 113–130 | 113 | 26 |
| Total | 163 | 46 | 46 | 1514 | 192–32.160 | 114 | 100–173 | 119 | 46 |
Abbreviations: AML-M3, acute myeloid leukemia type M3; APL, acute promyelocytic leukemia; BCP-ALL, B-cell precursor acute lymphoblastic leukemia; CML, chronic myeloid leukemia; NA, not applicable.
Table 3. Results of PML–RARA detection by PCR and the novel immunobead assay in AML patients according to age, PML break point and MFI value.
| Leukemia type | No. of samples |
PML–RARA mRNA positivity |
MFI of PML–RARA protein positivity |
||||
|---|---|---|---|---|---|---|---|
| bcr1 | bcr2 | bcr3 | Low (180–500) | Medium (500–1000) | High (>1000) | ||
| APL | |||||||
| Childhood | 22 | 11 | 1 | 10 | 1 | 7 | 14 |
| Adult | 24 | 10 | 0 | 14 | 4 | 4 | 16 |
| Other AML | |||||||
| Childhood | 18 | 0 | 0 | 0 | 0 | 0 | 0 |
| Adult | 48 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 112 | 21 | 1 | 24 | 5 | 11 | 30 |
| (46%) | (2%) | (52%) | (11%) | (24%) | (65%) | ||
Abbreviations: AML, acute myeloid leukemia; APL, acute promyelocytic leukemia.
Figure 5.
Results of the EuroFlow testing of the prototype PML–RARA immunobead assay. MFI values were plotted per patient group. The results of the RQ-PCR analyses are indicated with symbols (bcr1, bcr2, bcr3 and negative). The lower MFI values in APL patients with bcr1 breaks as compared with APL patients with bcr3 breaks reached statistical significance (Mann–Whitney U-test: P=0.04), if all samples (PB and BM) were analyzed. As PB samples tend to have lower percentages of leukemic cells (particularly in bcr1 cases), we repeated the Mann–Whitney U-test for the BM samples only (17 bcr1 and 18 bcr3), which showed no significant difference between bcr1 and bcr3 cases (P= 0.06). Further details about the PML–RARA positive samples are given in Figures 6, 7, 8.
MFI values in patients with positivity for the PML–RARA immunobead assay
The vast majority of PML–RARA positive patients had high MFI values (65%) or medium MFI values (24%). Low MFI values (above the cutoff value of MFI 180) were found in only 11% of the positive patients (five cases): 192, 195, 273, 339 and 444 (Figure 5). The five cases consisted of four adult patients and one child (Table 3). The sample with the lowest MFI value of 192 concerned a PB sample with only 37% leukemic cells, which was substantially less than in most of the other APL samples (Figure 6). The samples with the MFI values of 195 and 273 concerned 2 BM samples, which had a long time lapse between sampling and processing of 3 days (65 and 60 h, respectively), which most likely has resulted in protein degradation (see below). Finally, the samples with the MFI values of 339 and 444 concerned a BM sample with 80% blast cells and a PB sample with 42% blast cells, respectively.
Figure 6.
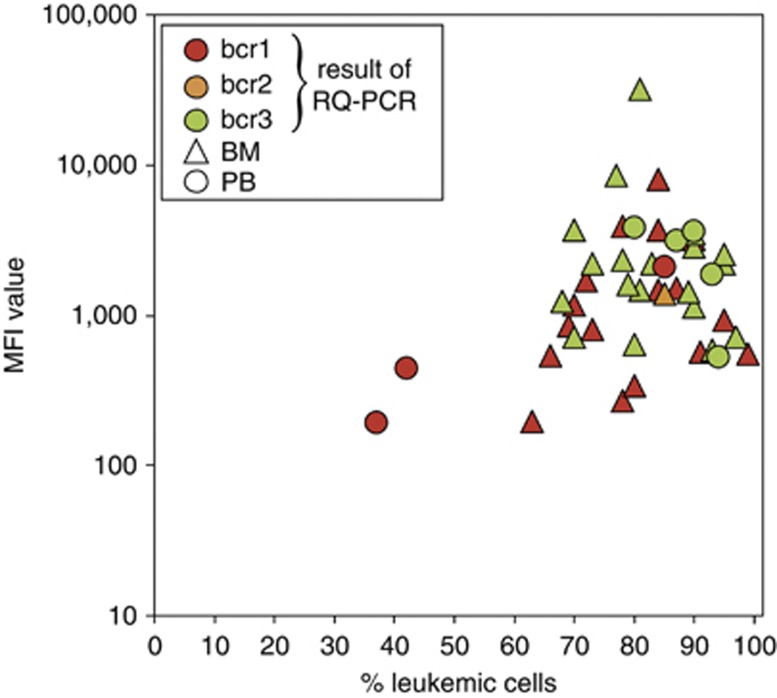
Level of PML–RARA fusion protein in relation to percentage of leukemic cells. No significant relationship was found between MFI values and percentages of leukemic cells. MFI values of the APL patients with bcr1 breaks had some correlation with percentage of leukemic cells (Spearman's rho coefficient 0.45; P=0.048), whereas such correlation was not observed for patients with bcr3 breaks (Spearman's rho coefficient −0.21; P=0.32).
Detection of PML–RARA proteins, derived from all three PML break points
The immunobead assay was designed to detect all PML–RARA fusion proteins that originate from the bcr1, bcr2 and bcr3 break points in the PML genes. As summarized in Table 3, all three types of break point regions in the PML gene were included in the 46 APL/AML cases with a positive PCR result for PML–RARA transcripts: 21 cases with bcr1 (46%), 1 case with bcr2 (2%), and 24 cases with bcr3 (52%). This implies that indeed the anti-PML and anti-RARA antibodies recognize epitopes that are sufficiently upstream and downstream of the PML and RARA break point cluster, respectively (Figure 5). The lower MFI values in APL patients with bcr1 breaks as compared with APL patients with bcr3 breaks just reached statistical significance (P=0.04 by Mann–Whitney U-test; Figure 5). We speculated that this difference might be caused by the fact that bcr1 PML–RARA proteins are 158 amino acids longer than bcr3 PML–RARA proteins and consequently might be more vulnerable for degradation. However, then we realized that by coincidence the two cases with the lowest percentages of leukemic cells (37 and 43%) belonged to the bcr1 PML–RARA patient group, both being blood samples. In fact, 5 of the 6 cases with less than 70% leukemic cells belonged to the bcr1 PML–RARA group (Figure 6). So, the seemingly lower MFI values of the APL patients with bcr1 breaks is merely related to lower percentages of leukemic cells in the tested samples. Interestingly, if the Mann–Whitney U-test was performed for BM samples only (17 bcr1 and 18 bcr3), no significant difference was found between the MFI values of patient samples with bcr1 versus bcr3 breaks (P=0.06).
Relation between PML–RARA protein level and percentage of leukemic cells
To evaluate whether differences in MFI levels between leukemias might be related to the percentage of leukemic cells present in the tested samples, MFI values were plotted against APL cell percentages (Figure 6). MFI values from APL patients with bcr1 breaks seemed to have some correlation with the percentage of leukemic cells (Spearman's rho coefficient =0.45; P= 0.048), whereas no significant relation was observed for patients with bcr3 breaks (Spearman's rho coefficient=−0.21; P= 0.32). It should be remarked that the percentages of leukemic cells in the bcr1 group varied over a broader range (37–99%) as compared with the bcr3 group (68–97%) (Figure 6).
Microcentrifugation of cell lysates and temperature
The 46 PML–RARA positive patient samples were evaluated for effects resulting from temperature differences during microcentrifugation. A total of 27 cases were in the 10 min 4 °C group and had a median MFI value of 1460 and a mean of 3042 (s.d. 6098). The high mean value and the high s.d. value appeared to be caused by the fact that this group contained 4 of the 5 low MFI values and the single very high MFI value of 32 160. The other 19 cases were in the 2 min room temperature group with a median MFI value of 1697 and a mean value of 2108 (s.d. 1769). The Mann–Whitney U-test did not show a significant difference (P=0.7). Consequently, the temperature of the microcentrifugation step does not seem to influence the results of the PML–RARA immunobead assay.
No effect of time lapse between sampling and processing of the patient material within the first 36 h
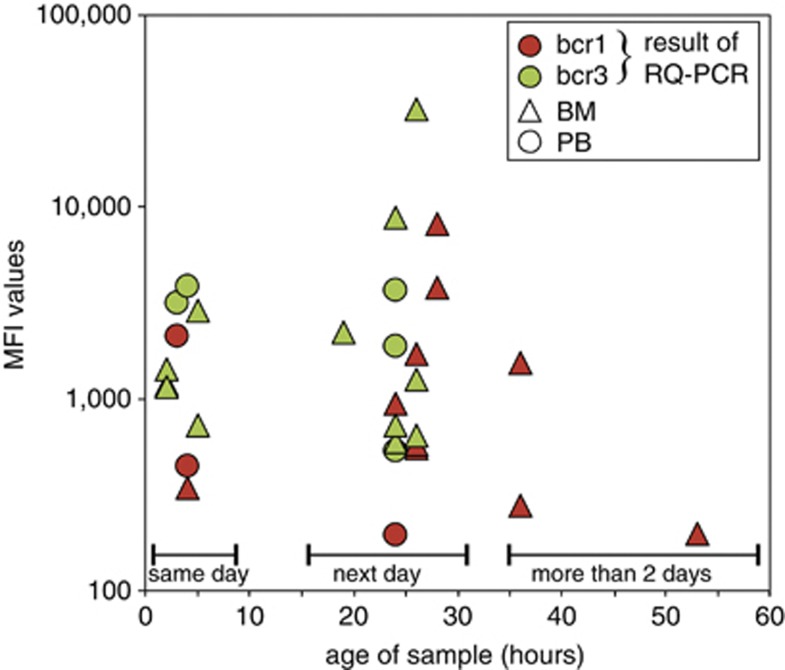
To understand whether longer transportation and/or processing times might have a negative impact on the quality and reliability of the PML–RARA immunobead assay, the EuroFlow laboratories were asked to make a fair estimate of the time lapse between sampling and processing of the patient material. These assessed and estimated time lapses were reported in the case report forms. The time lapse was reported in 30 of the 46 PML–RARA positive cases: 10 cases were processed on the same day (within 8 h), 17 cases on the next day (18–36 h), and 3 cases were processed on the third day or later (>48 h). Comparison of the MFI values did not show a clear decrease between cases processed within 8 h (same day) and 18–36 h (next day) (P=0.88 by Mann–Whitney U-test; Figure 7). We cannot draw conclusions from the three samples processed on the third day or later (>48 h), other than the fact that 2 of the 3 samples had a very low MFI value (MFI: 195 and 273).
Figure 7.
Level of PML–RARA fusion protein expression versus time lapse in sample processing.Comparison of the MFI values did not show a clear decrease between cases processed within 8 h (same day), and 18–36 h (next day) (Mann–Whitney U-test: P=0.88). Three samples were processed after more than 2 days; two of them had low MFI values.
These results indicate that processing within 36 h does not harm the performance of the PML–RARA prototype immunobead assay. The results of patient samples processed after 36 h (after 2 days) should be analyzed with caution.
Paired blood and BM samples
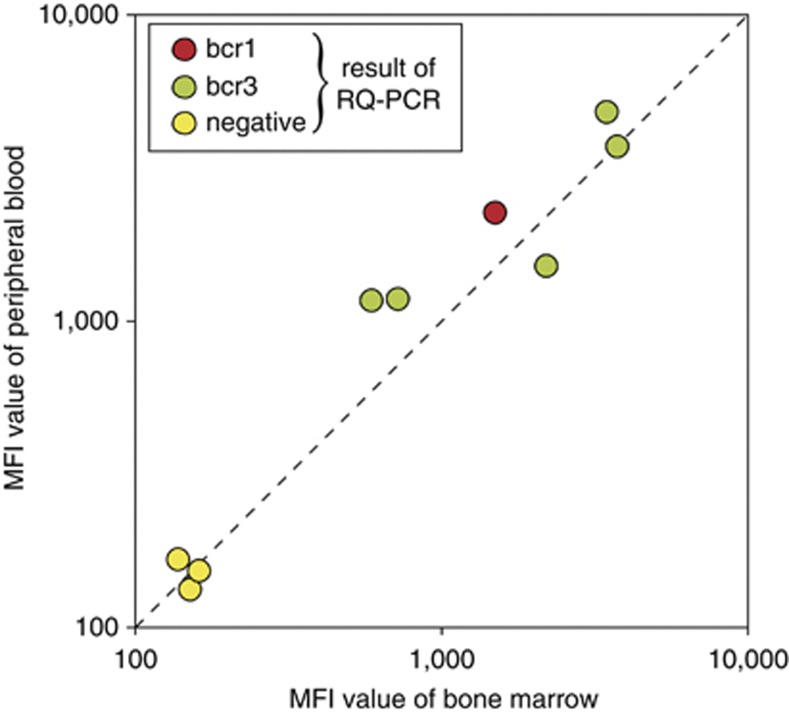
Nine paired fresh BM and PB samples were analyzed, six of which were derived from APL patients at the time of diagnosis and three from non-APL patients. The results of the six paired PB–BM APL samples showed that the intra-individual PML–RARA protein expression in leukemic cells from BM and PB is fully comparable (Figure 8).
Figure 8.
Comparison of PML–RARA fusion protein expression levels between paired BM and PB samples of nine acute leukemias at diagnosis. In all paired BM and PB samples of the same patient, fully comparable MFI values were observed. Remarkably, the MFI values of the BM and PB levels in the six APL cases showed a wide variation between patients, but were comparable in each individual patient.
Discussion
The PML–RARA fusion protein is the hallmark of APL and is present in 92% of APL patients with the classical t(15;17) translocation and in 5% of APL patients with nonclassical genetic aberrations such as insertions or complex chromosomal aberrations. PML–RARA positive APL patients often present with coagulopathy and without the right treatment, 40% of the patients develop pulmonary and cerebral hemorrhages, which can be lethal. Therefore, APL is a medical emergency, and the diagnosis of PML–RARA positive APL should be triaged fast enough to enable patients to enter a correct treatment protocol as soon as possible.
In this study, we developed a simple and fast flow cytometric immunobead assay for the detection of PML–RARA fusion proteins based on the same principle as described for the BCR-ABL protein immunobead assay.22 For the detection of PML–RARA fusion proteins in cell lysates, antibodies were raised against the N-terminal domain of PML, upstream of the break point region BCR3 and present in all known PML–RARA fusion proteins, and against the C-terminal domain of RARA (downstream of the RARA break points in intron 2). The antibodies were either bead-coupled or labeled with a fluorochrome. PML–RARA proteins were detected in the immunobead assay using a bead-bound capture antibody against one side of the fusion protein (anti-RARA) and a fluorochrome-conjugated detection antibody against the other side of the fusion protein (anti-PML).
The prototype immunobead assay appeared to be specific for the detection of the PML–RARA fusion proteins as a positive signal was obtained only when the epitopes of both the anti-PML and anti-RARA antibodies were present in a protein lysate. No background signals were obtained in cell lines with wild-type proteins or other fusion proteins. The sensitivity of the PML–RARA prototype immunobead assay was at least 10% for the NB4 cell line. This sensitivity should be sufficient to successfully identify all APL patients with a PML–RARA fusion gene, as PB and BM samples of most APL patients at diagnosis contain more than 70% of leukemic cells; this was also the case in 87% of our APL patient samples. Nevertheless, it is recommended that the assay will be conducted on BM samples, as in APL patients with pancytopenia very few leukemic cells might be present in PB samples.15 The suitability of the novel immunobead assay was further shown by detection of PML–RARA fusion proteins originating from both the bcr1 (downstream) and bcr3 (upstream) break points in the PML gene.
After further optimization, the prototype immunobead assay was distributed for large-scale testing to nine diagnostic laboratories enrolled in the EuroFlow consortium. The results obtained with the PML–RARA immunobead assay were fully concordant with the PCR results in a series of 163 patient samples including 46 APL patients, 66 non-APL AML patients, one CML patient in accelerated phase of the disease, 34 BCP-ALL patients and 16 T-ALL patients.
In the series of APL patients included in this study, the adult and childhood cases were equally represented and they showed comparable levels of PML–RARA fusion protein. The detection of PML–RARA fusion proteins in the 46 APL cases studied at diagnosis was independent of the type of PML break point (bcr1, bcr2, or bcr3). This implies that the location of the epitopes used for generation of the anti-PML and anti-RARA antibodies were chosen sufficiently upstream and downstream of the known break point clusters. As the bead assay is designed to be independent of the PML break points, PCR analysis might be needed at a later stage for identification of the PML break point variant in order to use the appropriate PCR primers for disease monitoring in individual patients.3, 24, 26
Analysis of the MFI levels detected with the prototype immunobead assay did not show a clear correlation with the percentages of leukemic cells present in the samples. This could imply that different APL cases may express different levels of PML–RARA fusion proteins. This explanation is supported by the evaluation of PML–RARA expression levels in paired BM and PB samples, which showed clear patient-to-patient differences between the six APL patients analyzed. In contrast, comparable PML–RARA expression levels were detected within in the paired BM and PB samples of the same patient.
Processing of leukemia samples in the same day, or the first day after sampling gave comparable results. However, it is not clear from this study whether a longer time lapse between sampling and processing would lead to suboptimal results (too few samples tested), but it is advised to keep the time lapse between sampling and processing within 36 h. As it is not known whether protein integrity is affected during transportation and the time lapse between sampling and processing, it would be advantageous to supplement the immunobead assay with an internal control for the assessment of protein quality in the cell lysates, as also proposed for the previously reported study performed with the BCR-ABL protein kit.22
We conclude that the flow cytometric immunobead assay is a very powerful diagnostic tool for detection of PML–RARA fusion proteins in leukemic cells. Because of the severe course of APL, a clinical assay with high specificity and sensitivity is critically needed to positively identify APL patients within a total group of AML patients.15, 16 Comparable to the BCR-ABL immunobead assay described previously, the PML–RARA immunobead assay also exhibits several obvious advantages over classical PCR and FISH techniques. The immunobead assay is independent of the break point position in the fusion gene, there is no need for special laboratory facilities, the assay requires no specialized equipment other than a standard flow cytometer, the results can be obtained in approximately 4 h, and the assay can be run in parallel to routine immunophenotyping without the need for extra technician time. It should be noted that the sensitivity of the current prototype PML–RARA immunobead assay is at least 10%, which is less sensitive than PCR and also slightly less sensitive than FISH, but sufficiently sensitive for the formal diagnosis of acute leukemia, if BM samples are analyzed with sufficiently high percentages of leukemic cells. Consequently, the novel PML–RARA immunobead assay will contribute to an earlier diagnosis of APL and thereby will enable clinicians to start specific treatment as soon as possible. Use of the novel PML–RARA immunobead assay hopefully will prevent the patients from developing any life-threatening bleeding problems because of early stage treatment.
Acknowledgments
The research and development of the flow cytometric immunobead assay and the EuroFlow Consortium were supported by the European Commission (grant STREP EU-FP6, LSHB-CT-2006-018708; Flow cytometry for fast and sensitive diagnosis and follow-up of hematological malignancies). We are grateful to Dr Jean-Luc Sanne of the European Commission for his support and monitoring of the EuroFlow project. We thank Dr P Valk (Department of Hematology, Erasmus MC, Rotterdam) for providing extra APL samples. We thank Dr Charlene Bush-Donovan, Dr Frans Nauwelaers, Dr Mark Herberger and Dr Vera Imper (BD Biosciences) for their technical support in the design and production of the prototype PML–RARA immunobead assay.
EHA Dekking, AL Noordijk, PM Vermeulen and MT de Man are employees of Dynomics, a spin-off company of Erasmus MC, Rotterdam, The Netherlands; JJM van Dongen and VHJ van der Velden are inventors of the immunobead assay (patent PCT/NL01/00.945), which has been licensed by Erasmus MC to Dynomics; R Varro and H Wai are employees of BD Biosciences; EP Dixon is employee of BD Diagnostics; JJM van Dongen is stockholder of Dynomics and advised Dynomics in the development and optimization of the immunobead assay. It is thanks to this crucial combination of disciplines (the University setting of Erasmus MC, the spin-off company Dynomics, the Industry represented by BD Biosciences and the EuroFlow Consortium, where the clinic matches the diagnostic) that we have been provided with a complete chain of events, which has led us from invention through development and production to end up in a final clinical testing, which we present here, the description of a new diagnostic product for the diagnosis of APL.
References
- Pandolfi PP, Alcalay M, Fagioli M, Zangrilli D, Mencarelli A, Diverio D, et al. Genomic variability and alternative splicing generate multiple PML/RAR alpha transcripts that encode aberrant PML proteins and PML/RAR alpha isoforms in acute promyelocytic leukaemia. EMBO J. 1992;11:1397–1407. doi: 10.1002/j.1460-2075.1992.tb05185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwade D, Biondi A, Mozziconacci MJ, Hagemeijer A, Berger R, Neat M, et al. Characterization of acute promyelocytic leukemia cases lacking the classic t(15;17): results of the European Working Party. Groupe Français de Cytogénétique Hématologique, Groupe de Français d'Hematologie Cellulaire, UK Cancer Cytogenetics Group and BIOMED 1 European Community-Concerted Action ‘Molecular Cytogenetic Diagnosis in Haematological Malignancies'. Blood. 2000;96:1297–1308. [PubMed] [Google Scholar]
- Van Dongen JJM, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13:1901–1928. doi: 10.1038/sj.leu.2401592. [DOI] [PubMed] [Google Scholar]
- Walz C, Grimwade D, Saussele S, Lengfelder E, Haferlach C, Schnittger S, et al. Atypical mRNA fusions in PML–RARA positive, RARA-PML negative acute promyelocytic leukemia. Genes Chromosomes Cancer. 2010;49:471–479. doi: 10.1002/gcc.20757. [DOI] [PubMed] [Google Scholar]
- Chillón MC, González M, García-Sanz R, Balanzategui A, González D, López-Pérez R, et al. Two new 3' PML break points in t(15;17)(q22;q21)-positive acute promyelocytic leukemia. Genes Chromosomes Cancer. 2000;27:35–43. [PubMed] [Google Scholar]
- Redner RL, Rush EA, Faas S, Rudert WA, Corey SJ. The t(5;17) variant of acute promyelocytic leukemia expresses a nucleophosmin-retinoic acid receptor fusion. Blood. 1996;87:882–886. [PubMed] [Google Scholar]
- Grimwade D, Gorman P, Duprez E, Howe K, Langabeer S, Oliver F, et al. Characterization of cryptic rearrangements and variant translocations in acute promyelocytic leukemia. Blood. 1997;90:4876–4885. [PubMed] [Google Scholar]
- Hummel JL, Wells RA, Dubé ID, Licht JD, Kamel-Reid S. Deregulation of NPM and PLZF in a variant t(5;17) case of acute promyelocytic leukemia. Oncogene. 1999;18:633–641. doi: 10.1038/sj.onc.1202357. [DOI] [PubMed] [Google Scholar]
- Arnould C, Philippe C, Bourdon V, Gregoire MJ, Berger R, Jonveaux P. The signal transducer and activator of transcription STAT5b gene is a new partner of retinoic acid receptor alpha in acute promyelocytic-like leukaemia. Hum Mol Genet. 1999;8:1741–1749. doi: 10.1093/hmg/8.9.1741. [DOI] [PubMed] [Google Scholar]
- Zelent A, Guidez F, Melnick A, Waxman S, Licht JD. Translocations of the RARalpha gene in acute promyelocytic leukemia. Oncogene. 2001;20:7186–7203. doi: 10.1038/sj.onc.1204766. [DOI] [PubMed] [Google Scholar]
- Catalano A, Dawson MA, Somana K, Opat S, Schwarer A, Campbell LJ, et al. The PRKAR1A gene is fused to RARA in a new variant acute promyelocytic leukemia. Blood. 2007;110:4073–4076. doi: 10.1182/blood-2007-06-095554. [DOI] [PubMed] [Google Scholar]
- Kondo T, Mori A, Darmanin S, Hashino S, Tanaka J, Asaka M. The seventh pathogenic fusion gene FIP1L1-RARA was isolated from a t(4;17)-positive acute promyelocytic leukemia. Haematologica. 2008;93:1414–1416. doi: 10.3324/haematol.12854. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Tsuzuki S, Tsuzuki M, Handa K, Inaguma Y, Emi N. BCOR as a novel fusion partner of retinoic acid receptor alpha in a t(X;17)(p11;q12) variant of acute promyelocytic leukemia. Blood. 2010;116:4274–4283. doi: 10.1182/blood-2010-01-264432. [DOI] [PubMed] [Google Scholar]
- Menezes J, Acquadro F, Perez-Pons de la Villa C, García-Sánchez F, Álvarez S, Cigudosa JC. FIP1L1/RARA with break point at FIP1L1 intron 13: a variant translocation in acute promyelocytic leukemia. Haematologica. 2011;96:1565–1566. doi: 10.3324/haematol.2011.047134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MA, Grimwade D, Tallman MS, Lowenberg B, Fenaux P, Estey EH, et al. Management of acute promyelocytic leukemia: recommendations from an expert panel on behalf of the European LeukemiaNet. Blood. 2009;113:1875–1891. doi: 10.1182/blood-2008-04-150250. [DOI] [PubMed] [Google Scholar]
- Tallman MS, Altman JK. How I treat acute promyelocytic leukemia. Blood. 2009;114:5126–5135. doi: 10.1182/blood-2009-07-216457. [DOI] [PubMed] [Google Scholar]
- Guidez F, Parks S, Wong H, Jovanovic JV, Mays A, Gilkes AF, et al. RARalpha-PLZF overcomes PLZF-mediated repression of CRABPI, contributing to retinoid resistance in t(11;17) acute promyelocytic leukemia. Proc Natl Acad Sci USA. 2007;104:18694–18699. doi: 10.1073/pnas.0704433104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein E, McMahon B, Kwaan H, Altman JK. Frankfurt O, Tallman MS. The coagulopathy of acute promyelocytic leukaemia revisited. Best Pract Res Clin Haematol. 2009;22:153–163. doi: 10.1016/j.beha.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Liso V, Bennett J. Morphological and cytochemical characteristics of leukaemic promyelocytes. Best Pract Res Clin Haematol. 2003;16:349–355. doi: 10.1016/s1521-6926(03)00061-6. [DOI] [PubMed] [Google Scholar]
- Falini B, Flenghi L, Fagioli M, Lo Coco F, Cordone I, Diverio D, et al. Immunocytochemical diagnosis of acute promyelocytic leukemia (M3) with the monoclonal antibody PG-M3 (anti-PML) Blood. 1997;90:4046–4053. [PubMed] [Google Scholar]
- Villamor N, Costa D, Aymerich M, Esteve J, Carrió A, Rozman M, et al. Rapid diagnosis of acute promyelocytic leukemia by analyzing the immunocytochemical pattern of the PML protein with the monoclonal antibody PG-M3. Am J Clin Pathol. 2000;114:786–792. doi: 10.1309/J6PU-3XY6-R0C3-NW26. [DOI] [PubMed] [Google Scholar]
- Weerkamp F, Dekking E, Ng YY, Van der Velden VHJ, Wai H, Böttcher S, et al. Flow cytometric immunobead assay for the detection of BCR-ABL fusion proteins in leukemia patients. Leukemia. 2009;23:1106–1117. doi: 10.1038/leu.2009.93. [DOI] [PubMed] [Google Scholar]
- Lanotte M, Martin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R. NB4, a maturation inducible cell line with t(15:17) marker isolated from human acute promyelocytic leukemia (M3) Blood. 1991;77:1080–1086. [PubMed] [Google Scholar]
- Gabert J, Beillard E, Van der Velden VHJ, Bi W, Grimwade D, Pallisgaard N, et al. Standardization and quality control studies of 'real-time' quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia - a Europe Against Cancer program. Leukemia. 2003;17:2318–2357. doi: 10.1038/sj.leu.2403135. [DOI] [PubMed] [Google Scholar]
- Beillard E, Pallisgaard N, Van der Velden VHJ, Bi W, Dee R, Van der Schoot E, et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using 'real-time' quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR) - a Europe against cancer program. Leukemia. 2003;17:2474–2486. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- Grimwade D, Jovanovic JV, Hills RK, Nugent EA, Patel Y, Flora R, et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J Clin Oncol. 2009;27:3650–3658. doi: 10.1200/JCO.2008.20.1533. [DOI] [PubMed] [Google Scholar]