Abstract
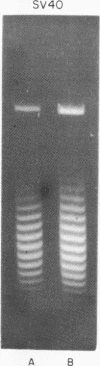
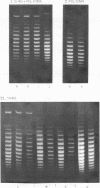
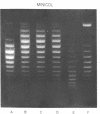
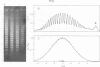
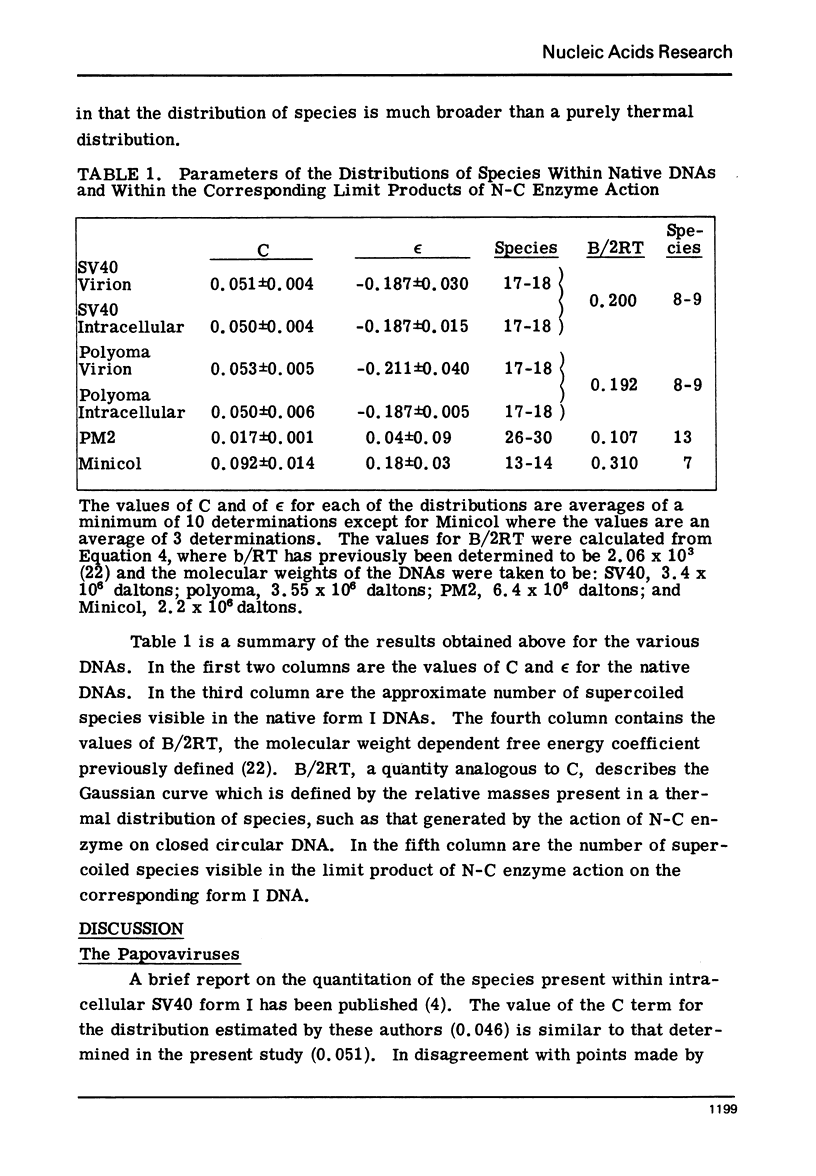
Systems for gel electrophoresis in the presence of one of the intercalative unwinding ligands, ethidium or chloroquine, have been developed which permit the resolution of highly supercoiled closed circular DNA molecules differing by unit values of the topological winding number, α. All native closed circular DNAs examined, including the viral and intracellular forms of SV40 and polyoma DNA, bacterial plasmid DNAs, and the double stranded closed circular DNA genome of the marine bacteriophage, PM2, are more heterogeneous with respect to the number of superhelical turns present than are the thermal distributions observed in the limit products of the action of nicking-closing (N-C) enzyme on the respective DNAs. In the cases of SV40 and polyoma, where it has been shown that the supercoiling is a combined consequence of the binding of the four nucleosomal histones, H2a, H2b, H3 and H4, and the action of N-C enzyme, the breadth of the distributions within the form I DNAs poses specific problems since the work of other laboratories indicates that the number of nucleosomes on the respective minichromosomes falls within a narrow distribution of 21. If it is assumed that all nucleosomes have identical structures, and that the DNA within a nucleosome is not free to rotate, the native DNA would be anticipated to be less heterogeneous than the thermal equilibrium mixtures present in N-C enzyme relaxed SV40 and polyoma DNAs.
The absolute number of superhelical turns (at 37°C in 0.2 M NaCl) in virion polyoma DNA has been determined to be 26 ± 1, which is the same value obtained for virion SV40 DNA. This is consistent with the observations that polyoma DNA has a higher molecular weight, a lower superhelix density, but the same number of nucleosomes as SV40 DNA. In addition, the distributions within the virion and intracellular form I DNAs of both SV40 and polyoma were found to be indistinguishable.
Full text
PDF

















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cremisi C., Pignatti P. F., Croissant O., Yaniv M. Chromatin-like structures in polyoma virus and simian virus 10 lytic cycle. J Virol. 1975 Jan;17(1):204–211. doi: 10.1128/jvi.17.1.204-211.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeys R. J., Jackson D. A. Dye titrations of covalently closed supercoiled DNA analyzed by agarose gel electrophoresis. Biochem Biophys Res Commun. 1976 Mar 22;69(2):446–454. doi: 10.1016/0006-291x(76)90542-8. [DOI] [PubMed] [Google Scholar]
- DeLeys R. J., Jackson D. A. Electrophoretic analysis of covalently closed SV40 DNA: Boltzmann distributions of DNA species. Nucleic Acids Res. 1976 Mar;3(3):641–652. doi: 10.1093/nar/3.3.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejo R. T., Canelo E. S., Sinsheimer R. L. DNA of bacteriophage PM2: a closed circular double-stranded molecule. Proc Natl Acad Sci U S A. 1969 Aug;63(4):1164–1168. doi: 10.1073/pnas.63.4.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejo R. T., Lebowitz J. A simple electrophoretic method for the determination of superhelix density of closed circular DNAs and for observation of their superhelix density heterogeneity. Anal Biochem. 1976 May 7;72:95–103. doi: 10.1016/0003-2697(76)90510-8. [DOI] [PubMed] [Google Scholar]
- Frearson P. M., Crawford L. V. Polyoma virus basic proteins. J Gen Virol. 1972 Feb;14(2):141–155. doi: 10.1099/0022-1317-14-2-141. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray H. B., Jr, Upholt W. B., Vinograd J. A buoyant method for the determination of the superhelix density of closed circular DNA. J Mol Biol. 1971 Nov 28;62(1):1–19. doi: 10.1016/0022-2836(71)90127-6. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Visualization of prokaryotic DNA in a regularly condensed chromatin-like fiber. Proc Natl Acad Sci U S A. 1976 Feb;73(2):563–567. doi: 10.1073/pnas.73.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselkorn R., Rouvière-Yaniv J. Cyanobacterial DNA-binding protein related to Escherichia coli HU. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1917–1920. doi: 10.1073/pnas.73.6.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Kasamatsu H., Wu M. Protein-SV40 DNA complex stable in high salt and sodium dodecyl sulfate. Biochem Biophys Res Commun. 1976 Feb 9;68(3):927–936. doi: 10.1016/0006-291x(76)91234-1. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W., Wendel I. Stepwise relaxation of supercoiled SV40 DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):199–208. doi: 10.1101/sqb.1974.039.01.026. [DOI] [PubMed] [Google Scholar]
- McMillen J., Consigli R. A. Characterization of polyoma DNA-protein complexes. I. Electrophoretic identification of the proteins in a nucleoprotein complex isolated from polyoma-infected cells. J Virol. 1974 Dec;14(6):1326–1336. doi: 10.1128/jvi.14.6.1326-1336.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Pettijohn D. E., Hecht R. RNA molecules bound to the folded bacterial genome stabilize DNA folds and segregate domains of supercoiling. Cold Spring Harb Symp Quant Biol. 1974;38:31–41. doi: 10.1101/sqb.1974.038.01.006. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Gros F. Characterization of a novel, low-molecular-weight DNA-binding protein from Escherichia coli. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3428–3432. doi: 10.1073/pnas.72.9.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shure M., Vinograd J. The number of superhelical turns in native virion SV40 DNA and minicol DNA determined by the band counting method. Cell. 1976 Jun;8(2):215–226. doi: 10.1016/0092-8674(76)90005-2. [DOI] [PubMed] [Google Scholar]
- Tai H. T., Smith C. A., Sharp P. A., Vinograd J. Sequence heterogeneity in closed simian virus 40 deoxyribonucleic acid. J Virol. 1972 Feb;9(2):317–325. doi: 10.1128/jvi.9.2.317-325.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne H. V. Electrophoretic separation of polyoma virus DNA from host cell DNA. Virology. 1966 Jun;29(2):234–239. doi: 10.1016/0042-6822(66)90029-8. [DOI] [PubMed] [Google Scholar]
- Timmis K., Cabello F., Cohen S. N. Covalently closed circular DNA molecules of low superhelix density as intermediate forms in plasmid replication. Nature. 1976 Jun 10;261(5560):512–516. doi: 10.1038/261512a0. [DOI] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Radloff R., Watson R., Laipis P. The twisted circular form of polyoma viral DNA. Proc Natl Acad Sci U S A. 1965 May;53(5):1104–1111. doi: 10.1073/pnas.53.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring M. Variation of the supercoils in closed circular DNA by binding of antibiotics and drugs: evidence for molecular models involving intercalation. J Mol Biol. 1970 Dec 14;54(2):247–279. doi: 10.1016/0022-2836(70)90429-8. [DOI] [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]