Abstract
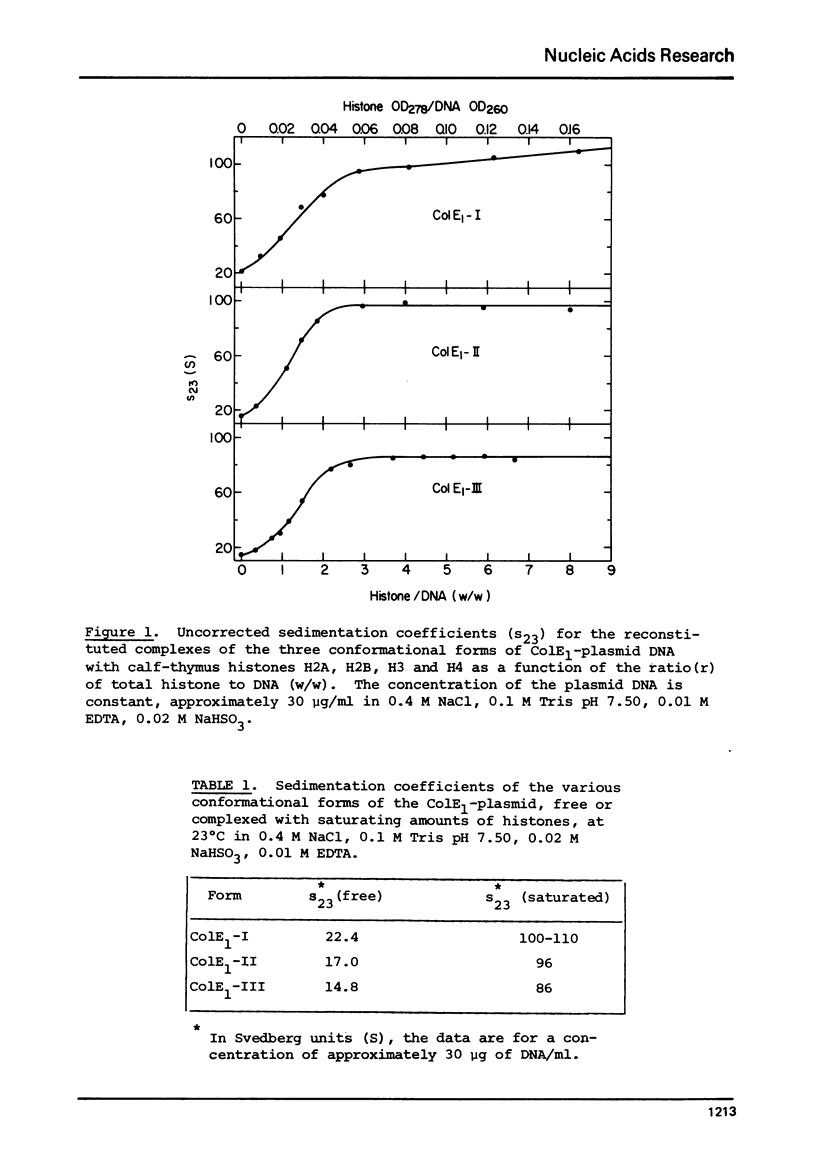
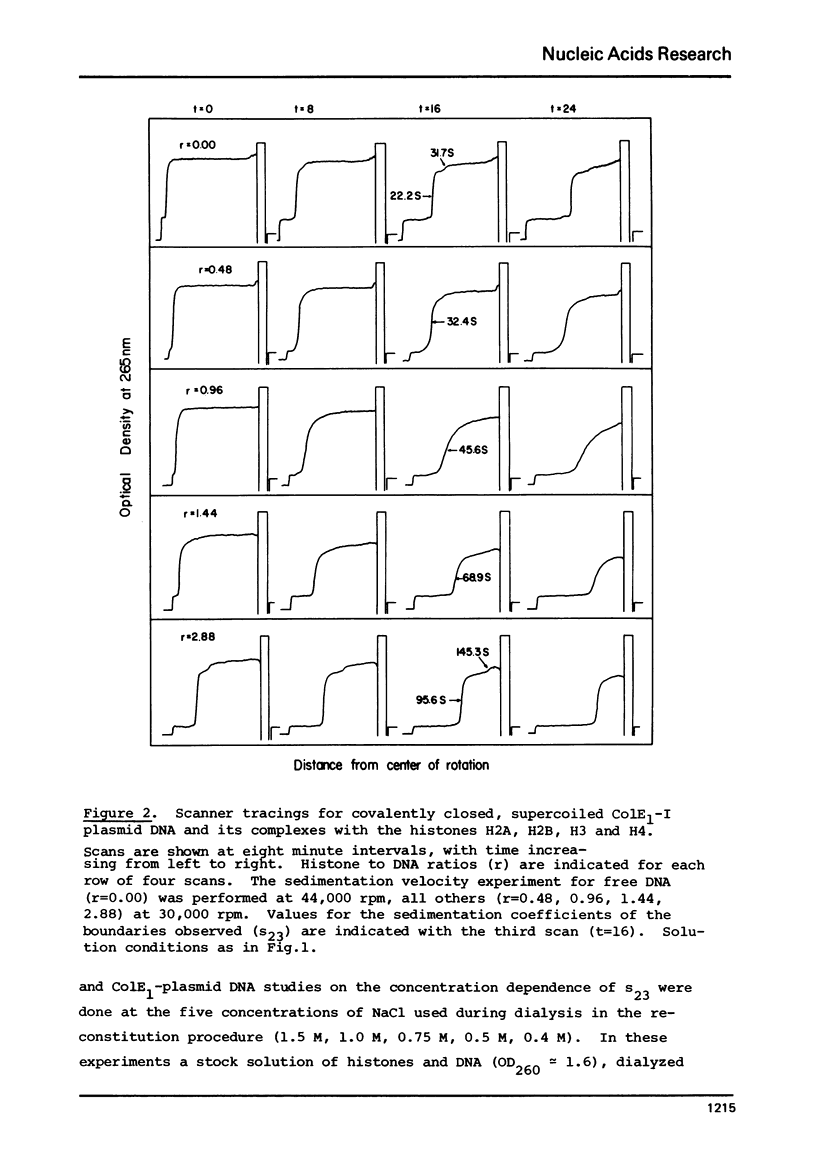
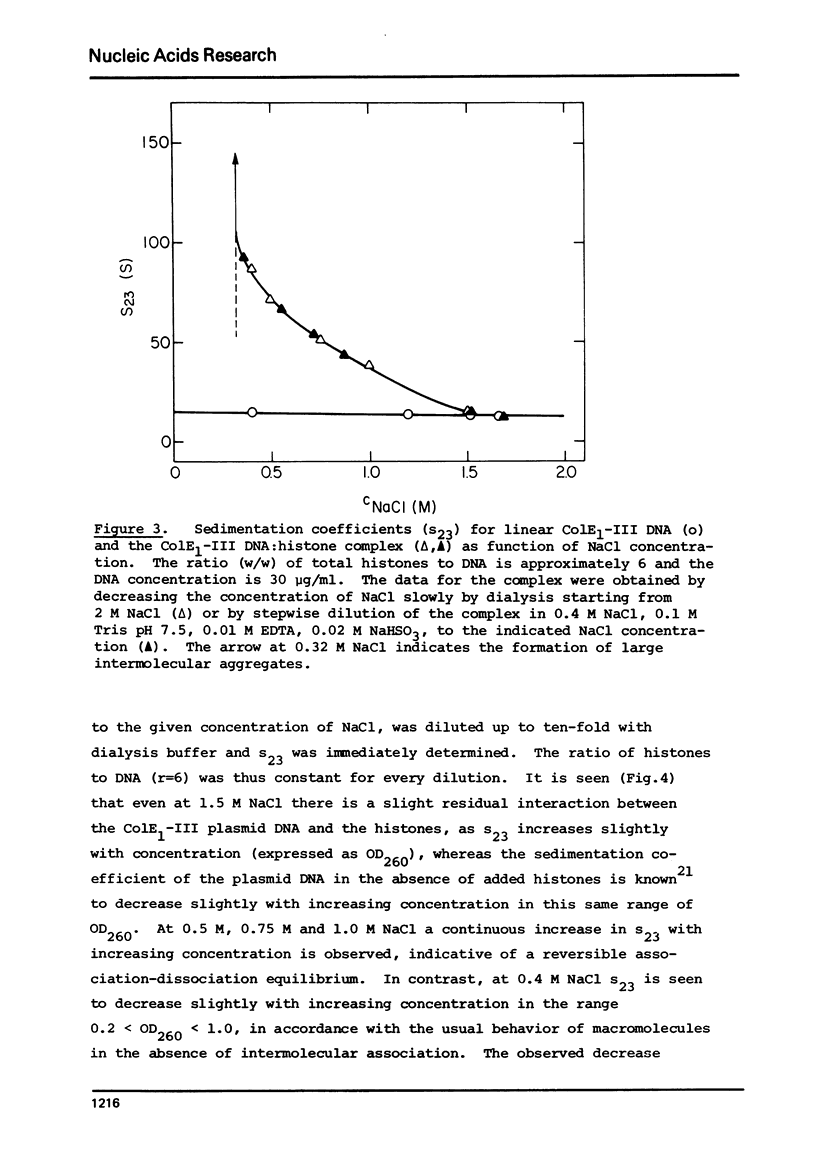
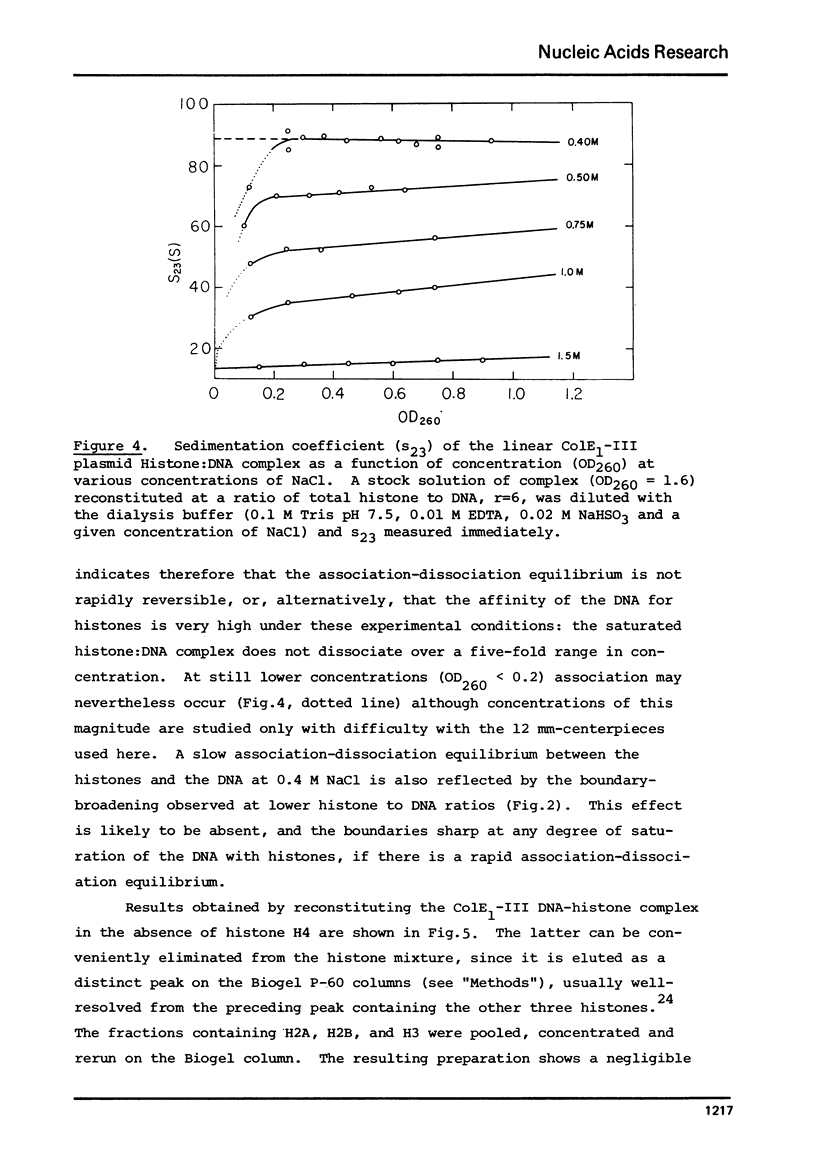
Complexes between the four calf-thymus histones (H2A, H2B, H3 and H4) and colE1-plasmid DNA have been reconstituted using the procedure of Oudet et al. ((1975), Cell 4, 281-300). The sedimentation rates of the complexes formed were studied under a variety of conditions. In 0.4 MNaCL, 0.1 M Tris pH 7.50, 0.01 M EDTA and 0.02 M NaHSO3, the final dialy-sis-solvent in the reconstitution procedure, the sedimentation coefficients s23 were found to increase when the complexes were reconstituted at increasing histone to DNA ratios. True plateau regions were reached in the case of the relaxed circular and linear forms of the plasmid DNA. The sedimenting boundaries observed for the complexes at saturation are sharp, reflecting a narrow distribution of sedimentation coefficients and a homogeneity of the complex comparable to that of the uncomplexed DNA. Studies of the dependence of s 23 on the concentration of the complex at constant DNA to histones ratio have been undertaken at salt concentrations between 0.4 and 1.5 M NaCL in the above solvent. The formation of the complexes is reversible, at least at the higher ionic strengths. At salt concentrations below 0.36 M the complex precipitates from solution. Omission of histone H4 from the reconstitution mixture abolishes complex formation.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Axel R., Melchior W., Jr, Sollner-Webb B., Felsenfeld G. Specific sites of interaction between histones and DNA in chromatin. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4101–4105. doi: 10.1073/pnas.71.10.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm E. L., Strickland W. N., Strickland M., Thwaits B. H., van der Westhuizen D. R., von Holt C. Purification of the five main calf thymus histone fractions by gel exclusion chromatography. FEBS Lett. 1973 Aug 15;34(2):217–221. doi: 10.1016/0014-5793(73)80797-5. [DOI] [PubMed] [Google Scholar]
- Böttger M., Scherneck S., Fenske H. A sedimentation study of the interaction of superhelical SV40 DNA with H1 histone. Nucleic Acids Res. 1976 Feb;3(2):419–429. doi: 10.1093/nar/3.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Noll M., Kornberg R. D. Electron microscopy of defined lengths of chromatin. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3320–3322. doi: 10.1073/pnas.72.9.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Iwai K. Carboxymethylcellulose chromatography in 6M urea and zone electrophoresis of calf thymus histone. J Biochem. 1970 Mar;67(3):465–472. doi: 10.1093/oxfordjournals.jbchem.a129269. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Noll M. Subunit structure of chromatin. Nature. 1974 Sep 20;251(5472):249–251. doi: 10.1038/251249a0. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Renz M., Day L. A. Transition from noncooperative to cooperative and selective binding of histone H1 to DNA. Biochemistry. 1976 Jul 27;15(15):3220–3228. doi: 10.1021/bi00660a010. [DOI] [PubMed] [Google Scholar]
- Richards B. M., Pardon J. F. The molecular structure of nucleohistone (DNH). Exp Cell Res. 1970 Sep;62(1):184–196. doi: 10.1016/0014-4827(79)90519-6. [DOI] [PubMed] [Google Scholar]
- SCHUMAKER V. N., SCHACHMAN H. K. Ultracentrifugal analysis of dilute solutions. Biochim Biophys Acta. 1957 Mar;23(3):628–639. doi: 10.1016/0006-3002(57)90386-4. [DOI] [PubMed] [Google Scholar]
- Sahasrabuddhe C. G., Van Holde K. E. The effect of trypsin on nuclease-resistant chromatin fragments. J Biol Chem. 1974 Jan 10;249(1):152–156. [PubMed] [Google Scholar]
- Sollner-Webb B., Camerini-Otero R. D., Felsenfeld G. Chromatin structure as probed by nucleases and proteases: evidence for the central role of histones H3 and H4. Cell. 1976 Sep;9(1):179–193. doi: 10.1016/0092-8674(76)90063-5. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Streeck R. E., Zachau H. G. Nucleosome formation abolishes base-specific binding of histones. Nature. 1975 Dec 4;258(5534):447–450. doi: 10.1038/258447a0. [DOI] [PubMed] [Google Scholar]
- Strätling W. H., Seidel I. Relaxation of chromatin structure by ethidium bromide binding: determined by viscometry and histone dissociation studies. Biochemistry. 1976 Nov 2;15(22):4803–4809. doi: 10.1021/bi00667a009. [DOI] [PubMed] [Google Scholar]
- de Pomerai D. I., Chesterton C. J., Butterworth P. H. Preparation of chromatin. Variation in the template properties of chromatin dependent on the method of perparation. Eur J Biochem. 1974 Aug 1;46(3):461–471. doi: 10.1111/j.1432-1033.1974.tb03639.x. [DOI] [PubMed] [Google Scholar]


