Abstract
The maize genome is replete with chromosomal duplications and repetitive DNA. The duplications resulted from an ancient polyploid event that occurred over 11 million years ago. Based on DNA sequence data, the polyploid event occurred after the divergence between sorghum and maize, and hence the polyploid event explains some of the difference in DNA content between these two species. Genomic rearrangement and diploidization followed the polyploid event. Most of the repetitive DNA in the maize genome is retrotransposable elements, and they comprise 50% of the genome. Retrotransposon multiplication has been relatively recent—within the last 5–6 million years—suggesting that the proliferation of retrotransposons has also contributed to differences in DNA content between sorghum and maize. There are still unanswered questions about repetitive DNA, including the distribution of repetitive DNA throughout the genome, the relative impacts of retrotransposons and chromosomal duplication in plant genome evolution, and the hypothesized correlation of duplication events with transposition. Population genetic processes also affect the evolution of genomes. We discuss how centromeric genes should, in theory, contain less genetic diversity than noncentromeric genes. In addition, studies of diversity in the wild relatives of maize indicate that different genes have different histories and also show that domestication and intensive breeding have had heterogeneous effects on genetic diversity across genes.
Genomic technologies have produced a wealth of data on the organization and structure of genomes. These data range from extensive marker-based genetic maps to “chromosome paintings” based on fluorescent in situ hybridization to complete genomic DNA sequences. Although genomic approaches have changed the amount and type of data, the challenges of interpreting genomic data in an evolutionary context have changed little from the challenges faced by Stebbins (1) and the coauthors of the evolutionary synthesis. The challenges are to infer the mechanisms of evolution and to construct a comprehensive picture of evolutionary change.
In this paper, we will focus on the processes that contribute to the evolution of plant nuclear genomes by using maize (Zea mays) as a model system. In some respects, it is premature to discuss the evolution of plant genomes, because the pending completion of the Arabidopsis (Arabidopsis thaliana) genome, with rice (Oryza sativa) following, is sure to unlock many mysteries about plant genome evolution. However, it must be remembered that Arabidopsis and rice are being sequenced, precisely because their genomes are atypically small and streamlined. Even after these genomes are sequenced, it will still be a tremendous challenge to understand the evolution of plant nuclear genomes, like the maize genome, for which entire DNA sequences will not be readily available.
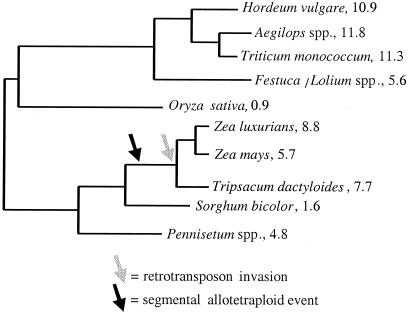
Maize is a member of the grass family (Poaceae). The grasses represent a range of genome size and structural complexity, with rice on one extreme. A diploid with 12 chromosomes (2n = 24), rice has one of the smallest plant genomes, with only 0.9 pg of DNA per 2C nucleus (Fig. 1). Other grass species exhibit far larger genomes. Wheat, for example, is a hexaploid with 21 chromosomes (2n = 42) and a haploid DNA content of 33.1 pg (2). Genera like Saccharum (sugarcane) and Festuca are even more complicated, displaying wide variation in ploidy level and over 100 chromosomes in some species. As a diploid with 10 chromosomes (2n = 20) and a 2C genome content roughly 6-fold larger than rice, maize lies somewhere in the middle of grass genome size and structural complexity (Fig. 1).
Figure 1.
A phylogeny of diploid grass species. Numerical values next to species names represent the 2C genome content of the species, measured in picograms. The phylogeny and genome content information is taken from figure 1 of ref. 51. The arrows represent the hypothesized timing of evolutionary events.
This paper focuses on the impact of chromosomal duplication, transposition, and nucleotide substitution on the evolution of the maize genome. We will discuss chromosomal duplication and transposition separately and will pay particular attention to their effects on DNA content. Nucleotide substitution will be discussed in the context of genetic diversity. Patterns of genetic diversity provide insight into the population genetic processes that act on different regions of the genome and thus uncover the evolutionary forces that act on genomes. We focus on maize throughout the paper but also generalize to other species when appropriate.
Polyploidy and Chromosomal Duplication
An Ancient Polyploid Origin.
The first hints of the complex organization of the maize genome came from cytological studies. Although maize is diploid, early studies by McClintock (3, 4) demonstrated the association of nonhomologous chromosomes during meiosis. Later studies documented the formation of bivalents and multivalents in maize haploids (5, 6). Altogether, cytological observations suggested that the maize genome contains extensive regions of homology, probably reflecting chromosomal duplications.
Evidence for chromosomal duplication also came from linkage information. In 1951, Rhoades (7, 8) noted that some regions of linkage maps did not contain mutants, and he proposed that the lack of mutants reflected genetic redundancy caused by chromosomal duplication. Rhoades' proposal has since been supported by molecular data. For example, isozyme studies have documented the presence of duplicated, linked loci in maize (9–12), and restriction fragment length polymorphism mapping studies have shown that many markers map to two or more chromosomal locations (13, 14). These mapping studies have established that some chromosomes—e.g., chromosomes 1 and 5 and chromosomes 2 and 7—share duplicated segments. Perhaps the most surprising information about the extent of gene duplication in maize is that 72% of single-copy rice genes are duplicated in maize (15).
Extensive chromosomal duplication in maize has been interpreted as evidence for a polyploid origin of the genome (7, 16), but until recently, there had been no estimation of the timing and mode of this polyploid event. In 1997, Gaut and Doebley (17) inferred the timing and mode of the polyploid event by studying DNA sequences from maize duplicated genes. To infer the mode of origin, Gaut and Doebley first modeled patterns of genetic divergence under three different types of polyploid formation: autopolyploidy, genomic allopolyploidy, and segmental allopolyploidy. (Briefly, allopolyploids are created by hybridization between species, with a genomic allopolyploid based on species that have fully differentiated chromosomes and a segmental allopolyploid based on species that have only partially differentiated chromosomes. Autopolyploidy refers to a polyploid event based on an intraspecific event. Stebbins contributed a great deal toward the definition and use of these terms, and precise definitions can be found in ref. 1.) The models' predictions were then compared with patterns of DNA sequence divergence in 14 pairs of maize duplicated genes. The sequence data were consistent with a segmental allotetraploid model of origin but inconsistent with the other two models of polyploid formation. Hence, the authors concluded that the maize genome was the product of a segmental allotetraploid event. They estimated the timing of the event by applying a molecular clock to the sequence data.
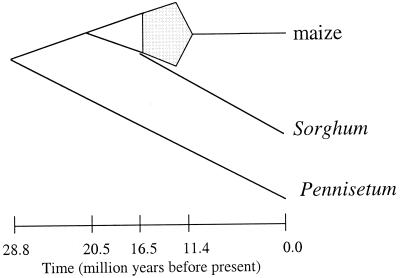
The hypothesized origin of the maize genome is detailed in Fig. 2 (17). Briefly, this hypothesis states that (i) maize is the product of a segmental allotetraploid event, (ii) the two diploid progenitors (or “parents”) of maize diverged ≈20.5 mya, (iii) the tetraploid event occurred between 16.5 and 11.4 mya, sometime after the divergence of Sorghum from one of the progenitor lineages, and (iv) the genome “rediploidized” before 11.4 mya. Although valuable, there are at least three reasons to be cautious about the hypothesis. The first reason is that the hypothesis is based on a relatively small number of DNA sequences—i.e., only 14 pairs of duplicated sequences. The second reason is that some of the sequences were not mapped to a chromosomal location. Ideally, these analyses should be based on a far greater number of sequences, all of which are known to reside in regions of known chromosomal duplication. Finally, it was not possible to test molecular clock assumptions rigorously for all of the sequence data, and thus some of the clock-based time estimates are subject to an unknown amount of error. Despite the need for caution, the study of Gaut and Doebley (17) provides the first glimpse into the mode and timing of an ancient plant polyploid event, and it also proposes a hypothesis that is testable with additional data.
Figure 2.
A hypothesis for the origin of the maize genome (17). Under this hypothesis, Pennisetum and maize diverged ≈29 million years ago (mya), followed ≈9 million years later by the divergence of the two diploid progenitors of maize. Sorghum diverged from one of these progenitor lineages (≈16.5 mya) before the two diploid progenitors united to form allopolyploid maize. The polyploid event occurred sometime between 16.5 mya and 11.4 mya, with subsequent diploidization completed by 11.4 mya. Gray shading represents the period in which allotetraploidy and diploidization occurred.
The Polyploid Event and the Divergence of Maize and Sorghum.
Fig. 1 places the segmental allotetraploid event in a phylogenetic context, and this context raises three important points about the comparison of maize to sorghum. First, if the allotetraploid event occurred after maize and sorghum diverged, then the maize genome should be duplicated more extensively than the sorghum genome. A corollary prediction is that maize and sorghum should not share common chromosomal duplications. Ultimately, these predictions can be tested with comparative genetic maps. At this point, however, it is unclear from comparative genetic maps as to whether the two genomes share extensive duplications in common, largely because published sorghum maps lack sufficient coverage (18–21). However, mapping information indicates that a higher proportion of markers is duplicated in maize than in sorghum. For example, Pereira et al. (19) found that 44% of restriction fragment length polymorphism markers detected more bands in maize than in sorghum; conversely, only 7% of markers detected more bands in sorghum than in maize. This information is consistent with the phylogenetic placement of the allotetraploid event (Fig. 1).
The second point centers on chromosome number. Maize and sorghum (Sorghum bicolor) have the same number of chromosomes (2n = 20). If maize underwent an allotetraploid event after the divergence of maize from sorghum, why do these plants have an identical number of chromosomes? At present, there is no suitable answer to this question, but there has been discussion about the evolution of chromosome number. Traditionally, it has been assumed that the basal haploid chromosome number of the tribe Andropogoneae, which encompasses maize, sorghum, and Tripsacum, was n = 5 (22, 23). More recently, it has been suggested that the basal haploid chromosome of the tribe was n = 10 (24). If the basal number was 10, one can hypothesize both that the chromosome number of S. bicolor has remained unchanged and that maize was the product of an allopolyploid event between two species with a reduced number of chromosomes (n = 5). This scenario is plausible, because the tribe contains diploid taxa with n = 5 (e.g., Elionurus and Sorghum species; ref. 24) and because comparative maps provide support that maize consists of two n = 5 subgenomes (25, 26).
Wilson et al. (27) have asserted that maize came from an ancestor with neither 5 nor 10 chromosomes. Based on genetic map data, they argued that the chromosome number of maize before the allotetraploid event was n = 8. The chromosome number was doubled subsequently to n = 16 (2n = 32) during the maize allotetraploid event and then reduced further by diploidization and fusion to the current number (n = 10; 2n = 20). Unfortunately, however, the argument of Wilson et al. contains errors regarding the timing and phylogenetic context of the allotetraploid event. For example, they suggest that the allotetraploid event occurred after the divergence of maize and Tripsacum, whereas most evidence suggests that the allotetraploid event occurred before the divergence of maize and Tripsacum. When these errors are taken into account, their arguments for the evolution of chromosome number seem unlikely. In short, there are no definitive answers either as to the evolution of chromosome number in this group or as to why S. bicolor and maize have the same number of chromosomes.
The third and final point about maize and sorghum centers on the difference in genome content between the two species. The segmental allotetraploid event predicts 2-fold variation in DNA content between sorghum and maize, but it does not account for the actual 3.5-fold variation in DNA content (Fig. 1). Based on this information, differences in DNA content probably reflect the allopolyploid event and additional evolutionary changes, such as the accumulation of repetitive DNA.
Genome Rearrangement After an Allopolyploid Event.
It must be remembered that extant maize is a diploid, and thus the segmental allotetraploid hypothesis presumes that the maize genome rearranged and diploidized. Is this presumption reasonable? Is genome rearrangement common after allopolyploid events?
Thus far, studies of synthetic plant polyploids suggest that genomes rearrange rapidly after allopolyploid events (reviewed in ref. 28). In one study, Song et al. (29) created four synthetic allopolyploids. After recovery of F2 polyploids, each line was selfed until the F5 generation. Plants from the F2 and each subsequent generation were subjected to Southern hybridization with a panel of 89 probes. Southern blotting revealed remarkable differences in fragment profiles from generation to generation. In one synthetic polyploid, 66% of the probes detected fragment loss, fragment gain, or a change in fragment size, demonstrating that extensive rearrangement can occur rapidly after allopolyploid formation. Feldman and coworkers (30–32) performed similar studies in Triticum and Aegilops. Their results suggest that allopolyploids lose noncoding sequences in a directed, nonrandom fashion and that coding sequences are modified extensively (30–32).
Empirical studies detect rapid rearrangement of allopolyploid genomes, but rapid rearrangement is not equivalent to a complete diploidization. However, there is growing evidence that many plant, animal, and fungal genomes are the products of ancient polyploid events that were followed by rearrangement and a reduction in ploidy level. Yeast is one example. The DNA sequence of the yeast genome contains numerous blocks of duplicated genes. The phase (or direction) of the blocks are nonrandomly associated with centromeres, suggesting that the blocks were produced by the process of chromosomal duplication (33). Altogether, the data suggest that the yeast genome is the product of an ancient tetraploid event followed by rearrangement and diploidization (34). Vertebrates are another example of diploidized ancient polyploids; it is believed that vertebrates are degenerate polyploids owing to two polyploid events before the radiation of fish and mammals (35). Similar examples come from plants; for example, both Glycine (soybean) (36) and Brassica species (37, 38) seem to be degenerate polyploids. Based on this information, one can conclude that diploidization after polyploidy is evolutionarily common.
For maize, it should be possible to garner insights into the processes of rearrangement and diploidization from extant patterns of chromosomal duplication. Mapping studies have documented regions of chromosomal duplication in maize (Table 1). (It is important to note that Table 1 includes only those chromosomes that were explicitly defined as duplicated by the authors; Table 1 does not include all of the chromosome pairs on which markers are known to crosshybridize.) As Table 1 demonstrates, there is some disagreement among studies about chromosomal duplications, for two reasons. First, different studies use different data, leading to different conclusions. Second, and perhaps more importantly, researchers rarely denote their criteria for defining chromosomal duplications, and thus criteria likely differ among studies. Ultimately, chromosomal duplications should be defined by objective statistical criteria.
Table 1.
Duplicated chromosomes in maize and the studies that identified them
| Duplicated chromosomes | Reference nos. |
|---|---|
| 1–5 | 14, 27, 84 |
| 1–9 | 14, 27, 84 |
| 2–4 | 14 |
| 2–7 | 14, 27, 84 |
| 2–10 | 14, 15, 27, 84 |
| 3–8 | 14, 15, 27, 84 |
| 3–10 | 84 |
| 4–5 | 27, 84 |
| 6–8 | 14, 27, 84 |
| 6–9 | 27, 84 |
Nonetheless, there is a consensus about some chromosomal pairs. For example, it is now well established that portions of chromosome 1 are duplicated on chromosomes 5 and 9 (Table 1). The evolutionary implication for these pairings is that the process of diploidization rearranged one copy of chromosome 1. (Alternatively, chromosome 1 could be an amalgamation of regions from different parental chromosomes.) Chromosome 2 had a similar fate in that portions of chromosome 2 are also found on chromosomes 7, 10, and perhaps 4 (Table 1). More extensive evaluation of these duplications will provide an indication as to whether there has been any bias in rearrangements. For example, there is a strong bias for paracentric inversions, as opposed to translocations and pericentric inversions, between potato and tomato. It was reasoned that the bias toward paracentric inversions reflects the relatively low effect of paracentric inversions on fitness (39). Additional studies of chromosomal duplications in maize could provide additional insights into the kind of rearrangements that are most evolutionarily stable.
The Importance of Chromosomal Duplication in Genome Evolution.
Is maize typical with regard to its polyploid history and prevalent chromosomal duplication? There is no doubt that polyploidy is common in plants, with up to 70% of angiosperms owing their history to polyploidy (1, 40). Furthermore, genetic maps demonstrate that a great number of species contain chromosomal duplications. Even species with streamlined genomes contain chromosomal duplications; for example, rice has a large duplication between chromosomes 11 and 12 (41) and Arabidopsis also has at least one large chromosomal duplication (42). Other plant genomes with chromosomal duplications include sorghum (21), cotton (43), soybean (36), and Brassica species (37, 38). Some of these genomes are degenerate polyploids like maize, but others may owe their chromosomal duplications to independent segmental events.
It is important to note that chromosomal duplications are usually inferred from genetic maps, but most (if not all) genetic maps are based on low copy-number markers. Low copy-number markers are systematically biased against detecting duplicated chromosomal segments, and hence the extent of chromosomal duplication is likely grossly underestimated for most plant taxa. In addition, the resolution of most genetic maps is low, such that relatively small areas of chromosomal duplication cannot be detected. The result is that we do not have a realistic understanding of either the extent to which chromosomes are duplicated or the extent to which genomes contain functional redundancies. We can, however, look to Arabidopsis sequence data as preliminary examples of the extent of chromosomal duplication. Based on the sequences of chromosomes 2 and 4 (42, 44), it is estimated that 10–20% of the low-copy regions of the Arabidopsis genome lie within duplicated chromosomal regions (42). Given that the Arabidopsis genome is streamlined, this percentage is undoubtedly much higher in complex genomes. It is possible that most genes in most plant genomes reside in duplicated chromosomal regions.
Multiplication of Repeat Sequences
Extent and Identification of Repetitive DNA.
Repetitive DNA constitutes a high proportion of plant genomes. This fact has been confirmed experimentally by reassociation (or C0t) kinetics. For example, Flavell et al. (45) found that repetitive DNA (defined, in this case, as DNA with more than 100 copies per genome) constitutes ≈80% of genomes with a haploid DNA content >5 pg. In contrast, small genomes of <5 pg contain 62% repetitive DNA on average. Maize falls into this range; reassociation experiments indicate that the genome contains from 60% to 80% repetitive DNA (45, 46). The repetitive DNA of maize can be categorized further as 20% highly repetitive (over 800,000 copies per genome) and 40% middle repetitive (over 1,000 copies per genome; ref. 46).
It is obvious that repetitive DNA is a large component of the maize genome, and thus the proliferation of repeat sequences has had important evolutionary implications. However, reassociation studies alone cannot answer two important questions about repetitive DNA in maize: what is the repetitive DNA, and when did it arise?
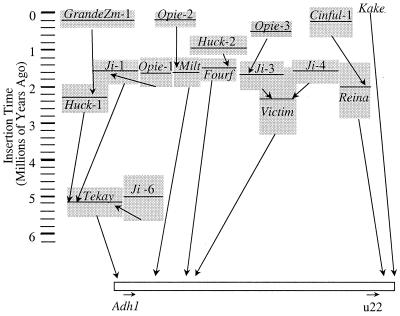
To date, the most complete answers to these two questions come from studies of the maize Adh1 region by Bennetzen and coworkers (47–50). They isolated a 280-kilobase yeast artificial chromosome clone of the Adh1 region and characterized the composition of the repetitive intergenic DNA. Retrotransposons comprise roughly 62% of the 240 kilobases analyzed, with an additional 6% of the clone consisting of miniature inverted-repeat transposable elements, remnants of DNA transposons, and other low-copy repeats. In total, the region contained 23 retrotransposons representing 10 distinct families. Of the 23 retroelements, 10 inserted within another element, resulting in a nested or “layered” structure of intergenic DNA within maize (Fig. 3). The architecture of this region suggests that retrotransposons preferentially target other retroelements for insertion.
Figure 3.
The estimated insertion times of retrotransposons in the Adh1 region (49). Each gray box represents a retrotransposon. The horizontal line through the box is the estimate of insertion time, and the height of the box represents the standard deviation of the estimate. Arrows between boxes indicate the order of insertion. For example, Huck-2 inserted into Fourf ≈1 mya.
Perhaps the most interesting feature of the Adh1 region is that it seems to be a representative region of the maize genome. Three observations support this contention. First, Southern blot and other analyses suggest that the retrotransposon families in the Adh1 region comprise at least 50% of the maize genome; altogether, just three of the retroelement families found in the Adh1 region constitute a full 25% of the genome (48). Second, 85% of repetitive DNAs from other regions were also present in the Adh1 region (although it should be noted that the sample of repetitive DNAs from other regions was small and thus this estimate may not be robust). Finally, a more recent study suggests that retrotransposons hybridize fairly uniformly to maize bacterial artificial chromosome clones, suggesting that the distribution of retrotransposons is reasonably homogeneous throughout the genome (B. Meyers, personal communication).
The Timing of Retrotransposon Multiplication.
Maize repetitive DNA seems to be primarily retrotransposons, but the second question remains: when did these retroelements multiply? To answer this question, SanMiguel et al. (49) sequenced the long terminal repeat (LTR) of retrotransposons in the Adh1 region. The rationale was as follows: when a single retrotransposon inserts into genomic DNA, both copies of the LTR are identical. Over time, the LTRs accumulate nucleotide substitutions and diverge in sequence. If the accumulation of nucleotide substitutions occurs at a regular pace, the number of nucleotide differences between the two LTRs provide insight into the date of LTR divergence and hence the date of retrotransposon insertion.
SanMiguel et al. (49) applied this approach to estimate the insertion time for 17 LTRs from the Adh1 region (Fig. 3). The results show that the oldest retrotransposon insertion is ≈5.2 mya and that most (15 of 17) retrotransposons inserted within the last 3.0 million years. The question arises as to whether these time estimates are reasonable. One feature that supports the results is that the time estimates correspond to the layering of retrotransposons (Fig. 3). In other words, in most cases (10 of 11) the insertion date for a retrotransposon is less than the insertion date for the retrotransposon into which it inserted. (The one exception is an instance in which the insertion dates are statistically indistinguishable.) Another observation that supports these results is that the sorghum Adh1 region lacks retrotransposons (50). Based on this information and ignoring the possibility of extensive retrotransposon loss in sorghum (51), retrotransposons in the maize Adh1 region must have amassed in the ≈16 million years since the divergence of sorghum and maize.
The implications of the study are important. If the Adh1 region is representative and the retrotransposons in this region constitute 50% of the genome, the maize genome has doubled in size in the last 5–6 million years. Like the polyploid event, retrotransposon proliferation represents a doubling of genome content over a relatively short evolutionary time scale.
Fig. 1 indicates that retrotransposon multiplication likely began in the evolutionary lineage leading to maize and Tripsacum, which diverged roughly ≈4.5–4.8 mya (52). Thus, most maize retrotransposon activity postdates the divergence of genera, but the oldest retrotransposons in the maize Adh1 region likely predate the split between Zea and Tripsacum. This discussion underscores the importance of studying Tripsacum to understand evolutionary events in maize better; if Fig. 1 is accurate, Tripsacum should share both chromosomal duplications and some retrotransposon activity in common with maize. It is known that Zea and Tripsacum share at least one low-copy retrotransposon that is absent from other closely related genera (53), but there is generally little information about chromosomal duplications or retrotransposons in Tripsacum.
Based on the available information, two large events differentiate the maize lineage from the sorghum lineage. The first event, segmental allotetraploidy, resulted in a 2-fold increase in maize DNA content. The second event, retrotransposon proliferation, produced another 2-fold increase in maize DNA content. Together, these events adequately explain the 3.5-fold difference in DNA content between maize and sorghum. However, it should be noted that there is also substantial variation in genomic DNA content among Zea and Tripsacum species (Fig. 1) (2, 54); this variation may reflect different amounts of retrotransposon proliferation or independent chromosomal duplications.
Remaining Questions.
Studies of the Adh1 region by Bennetzen and coworkers (47–50) have provided invaluable insight into the structure and dynamics of maize intergenic DNA, but at least three important questions remain.
Question 1.
Are retrotransposons distributed homogeneously among genomic regions? The Adh1 studies, as well as other studies (B. Meyers, personal communication), suggest that retrotransposon distribution may be roughly homogenous among regions of the maize genome. However, other lines of evidence suggest that such homogeneity is unlikely. For example, evolutionary theory predicts that transposable elements should gather in regions of low recombination, such as centromeres (55, 56). This prediction holds in Arabidopsis, where sequence data from chromosomes 2 and 4 indicate an increase in the frequency of transposable elements near centromeres (57).
There are other reasons to suggest that retrotransposon distribution may not be homogeneous throughout the maize genome. One obvious reason is that there are heterogeneities in chromosomal structure, such as euchromatin, heterochromatin, nucleolus organizing regions, telomeres, centromeres, and knobs. Nonetheless, recent research indicates that retrotransposons constitute a substantial fraction of both heterochromatic centromeres and heterochromatic knobs (58, 59); for one chromosome 9 knob, retroelements comprise roughly one-third of knob-specific clones (60). Many of the retrotransposons in knob and centromeric DNA belong to the element families found in the Adh1 region. Despite these commonalties, there are also substantive differences among knobs, centromeres, and the Adh1 region. For example, centromeres contain a centromere-specific retrotransposon (CentA; ref. 59). Similarly, chromosomal knobs associate with 180-bp and 350-bp repeat elements that are otherwise sparse in the genome (58). Altogether, the emerging picture is one in which some retroelement families are fairly ubiquitous, and other repetitive DNAs are heterogeneous in their distribution (e.g., ref. 61).
The work of Bernardi and coworkers (62, 63) is an intriguing addition to this picture. They fractionated DNA by G:C content and hybridized each G:C fraction to 38 coding-region probes. The coding genes hybridize almost exclusively to a DNA fraction of very narrow G:C content (1% of the total range), and this narrow fraction corresponds to 17% of the DNA content of the genome. To explain this hybridization pattern, Bernardi and coworkers (62, 63) reasoned that maize coding genes must be located in “gene-rich” regions and that these gene-rich regions must be flanked by DNA with highly homogeneous G:C contents. They proposed that this flanking DNA could consist of retrotransposons like those flanking the Adh1 gene (48).
The results from G:C fractionation experiments and studies of the Adh1 region are inconsistent. On the one hand, the study of the Adh1 region, coupled with studies of centromeres and knobs, suggest that retrotransposon distribution is widespread, representing 50% of the genome. On the other hand, Bernardi and coworkers' work implicitly suggests that retrotransposon distributions are heterogeneous, with a higher concentration of retroelements in the 17% of the genome that represents coding DNA. Ultimately, there may be a resolution to differences implied by different studies, but such a resolution will require more sequencing of large chromosomal clones representing diverse genomic regions.
Question 2.
What contributes more to the evolution of DNA content: multiplication of repetitive DNA or chromosomal duplication? The evolutionary history of maize suggests that retrotransposon multiplication and chromosomal duplication (by way of polyploidy) each have generated a 2-fold increase in DNA content within the last 16 million years. Hence, the net effect of these two evolutionary processes is similar in maize. In contrast, it seems that the multiplication of repeat sequences is the primary contributor to differences in DNA content between many taxa (45). For example, barley and rice have similar complements of low-copy genes (64) but a 12-fold difference in DNA content (Fig. 1). The difference in DNA content is thus probably attributable to differences in the amount of repetitive DNA (64).
It is premature to make the general statement that repeat proliferation contributes more to the evolution of DNA content than chromosomal duplications for two reasons. First, as mentioned previously, mapping studies are biased against the discovery of duplications, and for this reason, there is as yet no accurate indication of the extent of chromosomal duplication in complex genomes. Second, duplication and repeat proliferation are not independent. Duplication plays a role in repeat proliferation, because duplication doubles repetitive DNA as well as low-copy DNA.
Question 3.
Are chromosomal duplication events correlated with an increase in the rate of transposition? This question originates from the work of Matzke, Matzke, and colleague (65, 66). They argue that polyploid genomes contain duplications of all genes and thus are relatively well buffered against mutations caused by transposon insertion. As a consequence, transposable elements multiply and are maintained in polyploid genomes. For maize, the fact that two major events (polyploidy and retrotransposon multiplication) are located on the same phylogenetic lineage gives credence to the idea that these phenomena are biologically correlated (Fig. 1), but it is not yet known whether this correlation is widely observed.
Genetic Variation in Genes Along Chromosomes
Genetic Diversity as a Function of Recombination, Natural Selection, and Chromosomal Position.
Genomes are dynamic entities that can be modified extensively by polyploidy and transposon multiplication. However, ongoing evolutionary processes like mutation, recombination, natural selection, and migration also shape the genome. The effect of these extant processes on the genome can be inferred from careful study of genetic diversity.
Diversity throughout the genome is affected strongly by the interplay of recombination and natural selection. In Drosophila, for example, genetic diversity varies along the chromosome as a function of recombination rate (67, 68). Loci near centromeres tend to have low recombination rates and also tend to have low levels of genetic diversity, but both recombination rate and genetic diversity increase toward the tip of chromosomes. This relationship is not because recombination is mutagenic; rather, it reflects an interdependence between natural selection and recombination (67, 69). In regions of low recombination, for example, linkage between nucleotide sites ensures that selection for or against a single nucleotide substitution will affect a large region of the genome. In regions of high recombination, nucleotide sites are nearly independent; thus, selection on a single site affects a much smaller region of the genome. The result of the interdependence between selection and recombination is that (i) levels of genetic diversity can be a function of chromosomal position and (ii) large chromosomal regions can be depauperate of genetic diversity.
The correlation between chromosomal position and genetic diversity has been confirmed in plants (70, 71), but it is not yet clear whether recombination in maize follows a simple pattern along chromosomes. For example, it has been documented that maize single-copy regions act as recombination hot spots, but recombination rates also vary among single-copy regions (72–74). Altogether, these studies suggest that the relationship between chromosomal position and recombination rate may not be as straightforward in maize as in Drosophila. More thorough elucidation of recombination rates in maize requires comparisons between genetic and physical maps; such physical maps are being produced but are not yet completed.
Nonetheless, we have a goal to quantify patterns of genetic diversity more accurately in the maize genome. To make this quantification, we have begun a long-term study of 100 maize genes along chromosomes 1 and 3. To measure genetic diversity in each gene, we will sample DNA sequences from ≈70 individuals representing maize, its progenitor, and two other wild Zea taxa. The project has many long-term goals, including (i) to investigate the relationship between chromosomal position and genetic diversity, (ii) to examine the impact of domestication on genetic diversity in maize, (iii) to compare the evolutionary history among species across genes, and (iv) to create a public single-nucleotide-polymorphism database.
The first stage of this ongoing project is to measure genetic diversity in 25 chromosome 1 genes from 16 maize individuals representing Mexican and South American land races and 9 individuals representing U.S. inbred lines. The results of this first stage will be reported in detail elsewhere, but we can make a preliminary contrast of diversity in centromeric vs. noncentromeric genes. Average diversity per base pair in four genes within 5 centimorgans of the centromere is θ = 0.0144, as determined by using Watterson's estimator (75). This level of diversity is slightly lower than average diversity in 11 noncentromeric genes (average Watterson's θ = 0.0170), but the centromeric genes do not have extremely low levels of diversity. For example, all four centromeric genes contain more diversity than 3 of the 11 noncentromeric genes. Thus, we report that there is as yet no clear evidence for a strong reduction in genetic diversity near the centromere of chromosome 1.
Discordant Evolutionary Histories Among Genes.
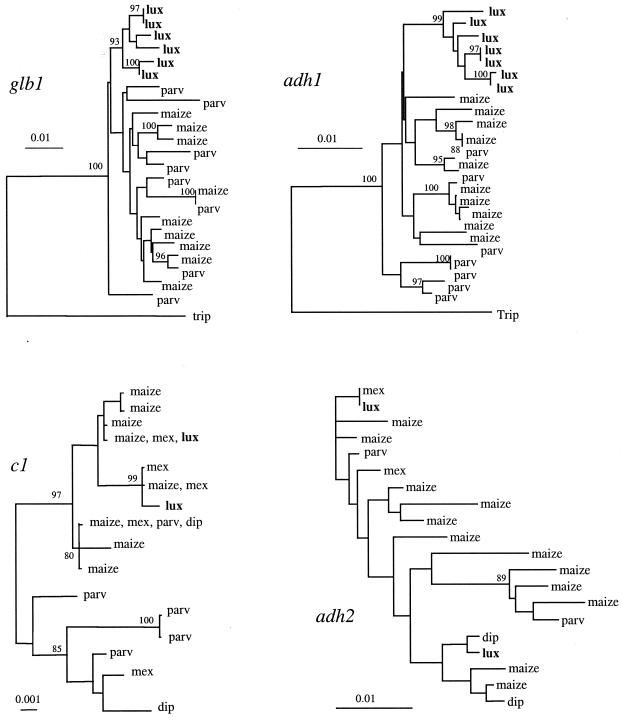
One interesting feature of genetic diversity studies of maize and its wild relatives is that evolutionary histories differ among loci. As an example, consider Fig. 4, which summarizes sequence data from four genes. The genes Adh1 and Glb1 provide very similar pictures of the relationship of the wild species Z. luxurians to other members of the genus Zea (52, 76); in short, for both of these genes, Z. luxurians sequences comprise a separate, well defined clade. In contrast, Z. luxurians individuals contain sequences that are very similar (or even identical) to sequences from other Zea taxa for Adh2 (77) and c1 (78). Thus, the picture of evolutionary history from Adh1 and Glb1 is not consistent with information from c1 and Adh2. (Fig. 4 focuses on genealogical or phylogenetic information for ease of presentation, but sequence statistics also suggest that these genes have different evolutionary histories.) One interesting feature of Fig. 4 is that Adh1 and Glb1 are located within a 12-centimorgan region of chromosome 1; Adh2 and c1 are found on chromosomes 4 and 9, respectively.
Figure 4.
Genealogies of four genes, based on the neighbor-joining method (82) with Kimura 2-parameter distances (83). Taxa are abbreviated as follows: maize, domesticated maize; parv, ancestor of domesticated maize (Z. mays subsp. parviglumis); mex, Z. mays subsp. mexicana; lux, Zea luxurians; dip, Zea diploperennis; trip, Tripsacum dactyloides. Sequences from Z. luxurians are shown in bold. The data are from refs. 52 and 76–78. Scale bars indicate level of divergence among sequences; bootstrap values >80% are shown.
We have sampled extensively from the wild relatives of maize for only a handful of genes, but discordant patterns, such as those demonstrated in Fig. 4, continue to be identified. The challenge of these data will be to infer the evolutionary processes that contribute to discordant evolutionary histories among genes. Several possibilities exist, including differences in nucleotide substitution rates, introgression (migration) rates, and natural selection among genes. One interesting possibility is that genealogical patterns among genes may correlate with chromosomal location.
In this context, it is worth noting that studies of Drosophila species have also demonstrated discordant patterns of genetic diversity among loci. For example, Wang et al. (79) studied three loci in three Drosophila species. Two of the loci (Hsp82 and period) yielded very similar pictures of genetic divergence among taxa. At these two loci, sequences were well differentiated among taxa. However, the pattern of genetic diversity in the third Drosophila locus (Adh) was incongruent with data from the first two loci. In this last locus, DNA sequences from different taxa were not highly diverged. Wang et al. (79) used population genetic tools to contrast genealogical information among Drosophila loci, and they concluded that introgression among species has occurred at a much higher rate at one locus (Adh) than at the other two loci (Hsp82 and period). In short, Drosophila studies strongly suggest that the processes affecting genetic diversity can vary among loci and also demonstrate the importance of comparing genealogical information across species and across loci.
In crops, artificial selection can cause discordant patterns of genetic diversity among loci. Thus far, levels of nucleotide sequence diversity have been measured in maize and its wild progenitor (Z. mays subsp. parviglumis) for six genes (summarized in ref. 80). All six genes indicate that maize has reduced genetic diversity relative to its wild progenitor, probably reflecting a genetic bottleneck during domestication (52, 76). However, the level of reduction in genetic diversity varies substantially among genes. For four of the six genes, maize retains at least half of the genetic diversity of its wild progenitor. For the remaining two genes (c1 and tb1), maize contains less than 20% of the level of diversity of its wild progenitor (78, 81). Low diversity in c1 and tb1 likely reflects artificial selection by the early domesticators of maize. The tb1 gene was probably selected to affect morphological changes in branching pattern (81), and c1 may have been selected for production of purple pigment in maize kernels (78).
Just as domestication has had a heterogeneous effect across loci, so has the process of maize breeding. For nine genes that we have sampled extensively thus far, U.S. inbred lines average roughly 65% the level of genetic diversity of the broader sample of maize. This level of reduction from maize land races to U.S. maize is commensurate with the original reduction in genetic diversity from wild progenitor to domesticated maize (52). Altogether, owing to reductions in diversity caused by initial domestication and subsequent intensive breeding, our initial estimates indicate that U.S. inbreds contain only ≈40% of the level of genetic diversity of the wild ancestor of maize.
Thus far, studies of genetic diversity have shown that maize genes have different levels of genetic diversity, and diversity in some genes has been affected strongly by artificial selection. In addition, studies of wild Zea taxa indicate that genes differ in their evolutionary histories among taxa. Our ongoing study of 100 genes will help determine whether patterns of evolutionary history among genes are, in fact, correlated with chromosomal location and will also contribute to the overall understanding of the evolutionary forces acting on plant genomes.
Acknowledgments
The authors acknowledge National Science Foundation Grants DBI-9872631 and DEB-9815855 and U.S. Department of Agriculture Grant 98-35301-6153.
Abbreviations
- mya
million years ago
- LTR
long terminal repeat
Footnotes
This paper was presented at the National Academy of Sciences colloquium “Variation and Evolution in Plants and Microorganisms: Toward a New Synthesis 50 Years After Stebbins,” held January 27–29, 2000, at the Arnold and Mabel Beckman Center in Irvine, CA.
References
- 1.Stebbins G L. Variation and Evolution in Plants. New York: Columbia Univ. Press; 1950. [Google Scholar]
- 2.Bennett M D, Leitch I J. Ann Bot. 1995;76:113–176. [Google Scholar]
- 3.McClintock B. Proc Natl Acad Sci USA. 1930;16:791–796. doi: 10.1073/pnas.16.12.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McClintock B. Z Zellforsch Mikrosk Anat. 1933;19:191–237. [Google Scholar]
- 5.Ting Y C. Cytologia. 1966;31:324–329. [Google Scholar]
- 6.Snope A J. Chromosoma. 1967;21:243–349. [Google Scholar]
- 7.Rhoades M M. Am Nat. 1951;85:105–110. [Google Scholar]
- 8.Rhoades M M. In: Corn and Corn Improvement. Sprague G F, editor. New York: Academic; 1955. pp. 123–219. [Google Scholar]
- 9.Goodman M M, Stuber C W, Newton K, Weissinger H H. Genetics. 1980;96:697–710. doi: 10.1093/genetics/96.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wendel J F, Stuber C W, Goodman M M, Beckett J B. J Hered. 1989;80:218–228. doi: 10.1093/oxfordjournals.jhered.a110839. [DOI] [PubMed] [Google Scholar]
- 11.Wendel J F, Stuber C W, Edwards M D, Goodman M M. Theor Appl Genet. 1986;72:178–185. doi: 10.1007/BF00266990. [DOI] [PubMed] [Google Scholar]
- 12.McMillin D E, Scandalios J G. Proc Natl Acad Sci USA. 1980;77:4866–4870. doi: 10.1073/pnas.77.8.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis G M, McMullen M D, Baysdorfer C, Musket T, Grant D, Staebell M, Xu G, Polacco M, Koster L, Melia-Hancock S, et al. Genetics. 1999;152:1137–1172. doi: 10.1093/genetics/152.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helentjaris T, Weber D, Wright S. Genetics. 1988;118:353–363. doi: 10.1093/genetics/118.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahn S, Tankley S D. Proc Natl Acad Sci USA. 1993;90:7980–7984. doi: 10.1073/pnas.90.17.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson E. Chron Bot. 1945;9:88–92. [Google Scholar]
- 17.Gaut B S, Doebley J F. Proc Natl Acad USA. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitkus R, Doebley J, Lee M. Genetics. 1992;132:1119–1130. doi: 10.1093/genetics/132.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereira M G, Lee M, Bramel-Cox P, Woodman W, Doebley J, Whitkus R. Genome. 1994;37:236–243. doi: 10.1139/g94-033. [DOI] [PubMed] [Google Scholar]
- 20.Berhan A M, Hulbert S H, Butler L G, Bennetzen J L. Theor Appl Genet. 1993;86:598–604. doi: 10.1007/BF00838715. [DOI] [PubMed] [Google Scholar]
- 21.Chittenden L M, Schertz K F, Lin Y R, Wing R A, Paterson A H. Theor Appl Genet. 1994;87:925–933. doi: 10.1007/BF00225786. [DOI] [PubMed] [Google Scholar]
- 22.Celarier R P. Rhodora. 1956;58:135–143. [Google Scholar]
- 23.Molina M D, Naranjo C A. Theor Appl Genet. 1987;73:542–550. doi: 10.1007/BF00289192. [DOI] [PubMed] [Google Scholar]
- 24.Spangler R, Zaitchik B, Russo E, Kellogg E A. Syst Bot. 1999;24:267–281. [Google Scholar]
- 25.Moore G, Devos K M, Wang Z, Gale M D. Curr Biol. 1995;5:737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- 26.Devos K M, Gale M D. Plant Mol Biol. 1997;35:3–15. [PubMed] [Google Scholar]
- 27.Wilson W A, Harrington S E, Woodman W L, Lee M, Sorrells M E, McCouch S R. Genetics. 1999;153:453–473. doi: 10.1093/genetics/153.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wendel J F. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- 29.Song K, Lu P, Tang K, Osborn T C. Proc Natl Acad Sci USA. 1995;92:7719–7723. doi: 10.1073/pnas.92.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B, Vega J M, Segal G, Abbo S, Rodova H, Feldman M. Genome. 1998;41:272–277. [Google Scholar]
- 31.Liu B, Vega J M, Feldman M. Genome. 1998;41:535–542. doi: 10.1139/g98-052. [DOI] [PubMed] [Google Scholar]
- 32.Feldman M, Liu B, Segal G, Abbo S, Levy A A, Vega J M. Genetics. 1997;147:1381–1387. doi: 10.1093/genetics/147.3.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfe K H, Shields D C. Nature (London) 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 34.Seoighe C, Wolfe K H. Proc Natl Acad Sci USA. 1998;95:4447–4452. doi: 10.1073/pnas.95.8.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Postlethwait J H, Yan Y-L, Gates M A, Horne S, Arnores A, Brownlie A, Donovan A, Egan E S, Force A, Gong Z Y, et al. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- 36.Shoemaker R C, Polzin K, Labate J, Specht J, Brummer E C, Olson T, Young N, Concibido V, Wilcox J, Tamulonis J P, et al. Genetics. 1996;144:329–338. doi: 10.1093/genetics/144.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bohuon E J R, Keith D J, Parkin I A P, Sharpe A G, Lydiate D J. Theor Appl Genet. 1996;93:833–839. doi: 10.1007/BF00224083. [DOI] [PubMed] [Google Scholar]
- 38.Cavell A C, Lydiate D J, Parkin I A P, Dean C, Trick M. Genome. 1998;41:62–69. [PubMed] [Google Scholar]
- 39.Bonierbale M W, Plaisted R L, Tanksley S D. Genetics. 1988;120:1095–1103. doi: 10.1093/genetics/120.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masterson J. Science. 1994;264:421–423. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- 41.Harushima Y, Yano M, Shomura A, Sato M, Shimano T, Kuboki Y, Yamamoto T, Lin S-Y, Antonio B A, Parco A, et al. Genetics. 1998;148:479–494. doi: 10.1093/genetics/148.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayer K S, Schuller C, Wambutt R, Murphy G, Volckaert G, Pohl T, Dusterhoft A, Stiekema W, Entian K D, Terryn N, et al. Nature (London) 1999;402:769–777. [Google Scholar]
- 43.Reinisch A J, Dong J, Brubaker C L, Stelly D M, Wendel J F, Paterson A H. Genetics. 1994;138:829–847. doi: 10.1093/genetics/138.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin X Y, Kaul S S, Rounsley S, Shea T P, Benito M I, Town C D, Fujii C Y, Mason T, Bowman C L, Barnstead M, et al. Nature (London) 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 45.Flavell R B, Bennett M D, Smith J B, Smith D B. Biochem Genet. 1974;12:257–269. doi: 10.1007/BF00485947. [DOI] [PubMed] [Google Scholar]
- 46.Hake S, Walbot V. Chromosoma. 1980;79:251–270. [Google Scholar]
- 47.Springer P S, Edwards K J, Bennetzen J L. Proc Natl Acad Sci USA. 1994;91:863–867. doi: 10.1073/pnas.91.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.SanMiguel P, Tickhonov A, Jin Y-K, Melake-Berhan A, Springer P S, Edwards K J, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 49.SanMiguel P J, Gaut B S, Tikhonov A, Nakajima Y, Bennetzen J L. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 50.Tikhonov A P, SanMiguel P J, Nakajima Y, Gorenstein N M, Bennetzen J L, Avramova Z. Proc Natl Acad Sci USA. 1999;96:7409–7414. doi: 10.1073/pnas.96.13.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bennetzen J L, Kellogg E A. Plant Cell. 1997;9:1509–1514. doi: 10.1105/tpc.9.9.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hilton H, Gaut B S. Genetics. 1998;150:863–872. doi: 10.1093/genetics/150.2.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vicient C M, Martinez-Izquierdo J A. Gene. 1997;184:257–261. doi: 10.1016/s0378-1119(96)00610-5. [DOI] [PubMed] [Google Scholar]
- 54.Bennett M D, Smith J B. Philos Trans R Soc London Ser B. 1991;334:309–345. [Google Scholar]
- 55.Charlesworth B, Langley C H, Stephan W. Genetics. 1986;112:947–962. doi: 10.1093/genetics/112.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Charlesworth B, Sniegowski P, Stephan W. Nature (London) 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 57.Copenhaver G N, Nickel K, Kuromori T, Benito M I, Kaul S, Lin X Y, Bevan M, Murphy G, Harris B, Parnell L D, et al. Science. 1999;286:2468–2474. doi: 10.1126/science.286.5449.2468. [DOI] [PubMed] [Google Scholar]
- 58.Ananiev E V, Phillips R L, Rines H W. Proc Natl Acad Sci USA. 1998;95:10785–10790. doi: 10.1073/pnas.95.18.10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ananiev E V, Phillips R L, Rines H W. Proc Natl Acad Sci USA. 1998;95:13073–13078. doi: 10.1073/pnas.95.22.13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ananiev E V, Phillips R L, Rines H W. Genetics. 1998;149:2025–2037. doi: 10.1093/genetics/149.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Q, Arbuckle J, Wessler S R. Proc Natl Acad Sci USA. 2000;97:1160–1165. doi: 10.1073/pnas.97.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barakat A, Carels N, Bernardi G. Proc Natl Acad Sci USA. 1997;94:6857–6861. doi: 10.1073/pnas.94.13.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carels N, Barakat A, Bernardi G. Proc Natl Acad Sci USA. 1995;92:11057–11060. doi: 10.1073/pnas.92.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saghai-Maroof M A, Yang G P, Biyashev R M, Maughan P J, Zhang Q. Theor Appl Genet. 1996;92:541–551. doi: 10.1007/BF00224556. [DOI] [PubMed] [Google Scholar]
- 65.Matzke M A, Mittelsten-Scheid O, Matzke A J M. BioEssays. 1999;21:761–767. doi: 10.1002/(SICI)1521-1878(199909)21:9<761::AID-BIES7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 66.Matzke M A, Matzke A J M. Trends Ecol Evol. 1998;13:241. doi: 10.1016/s0169-5347(98)01390-1. [DOI] [PubMed] [Google Scholar]
- 67.Begun D J, Aquadro C F. Nature (London) 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- 68.Hamblin M T, Aquadro C F. Genetics. 1999;153:859–869. doi: 10.1093/genetics/153.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Charlesworth D, Charlesworth B, Morgan M T. Genetics. 1995;141:1619–1632. doi: 10.1093/genetics/141.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dvorak J, Luo M-C, Yang Z-L. Genetics. 1998;148:423–434. doi: 10.1093/genetics/148.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stephan W, Langley C H. Genetics. 1998;150:1585–1593. doi: 10.1093/genetics/150.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Civardi L, Xia Y, Edwards K J, Schnable P S, Nikolau B J. Proc Natl Acad Sci USA. 1994;91:8268–8272. doi: 10.1073/pnas.91.17.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okagaki R J, Weil C F. Genetics. 1997;147:815–821. doi: 10.1093/genetics/147.2.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Timmermans M C P, Das O P, Messing J. Genetics. 1996;143:1771–1783. doi: 10.1093/genetics/143.4.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watterson G A. Theor Popul Biol. 1975;7:188–193. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 76.Eyre-Walker A, Gaut R L, Hilton H, Feldman D L, Gaut B S. Proc Natl Acad Sci USA. 1998;95:4441–4446. doi: 10.1073/pnas.95.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goloubinoff P, Paabo S, Wilson A C. Proc Natl Acad Sci USA. 1993;90:1997–2001. doi: 10.1073/pnas.90.5.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hanson M A, Gaut B S, Stec A O, Fuerstenberg S I, Goodman M M, Coe E H, Doebley J. Genetics. 1996;143:1395–1407. doi: 10.1093/genetics/143.3.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang R L, Wakeley J, Hey J. Genetics. 1997;147:1091–1106. doi: 10.1093/genetics/147.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.White S E, Doebley J F. Genetics. 1999;153:1455–1462. doi: 10.1093/genetics/153.3.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang R L, Stec A, Hey J, Lukens L, Doebley J. Nature (London) 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- 82.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 83.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 84.Gale M D, Devos K M. Proc Natl Acad Sci USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]