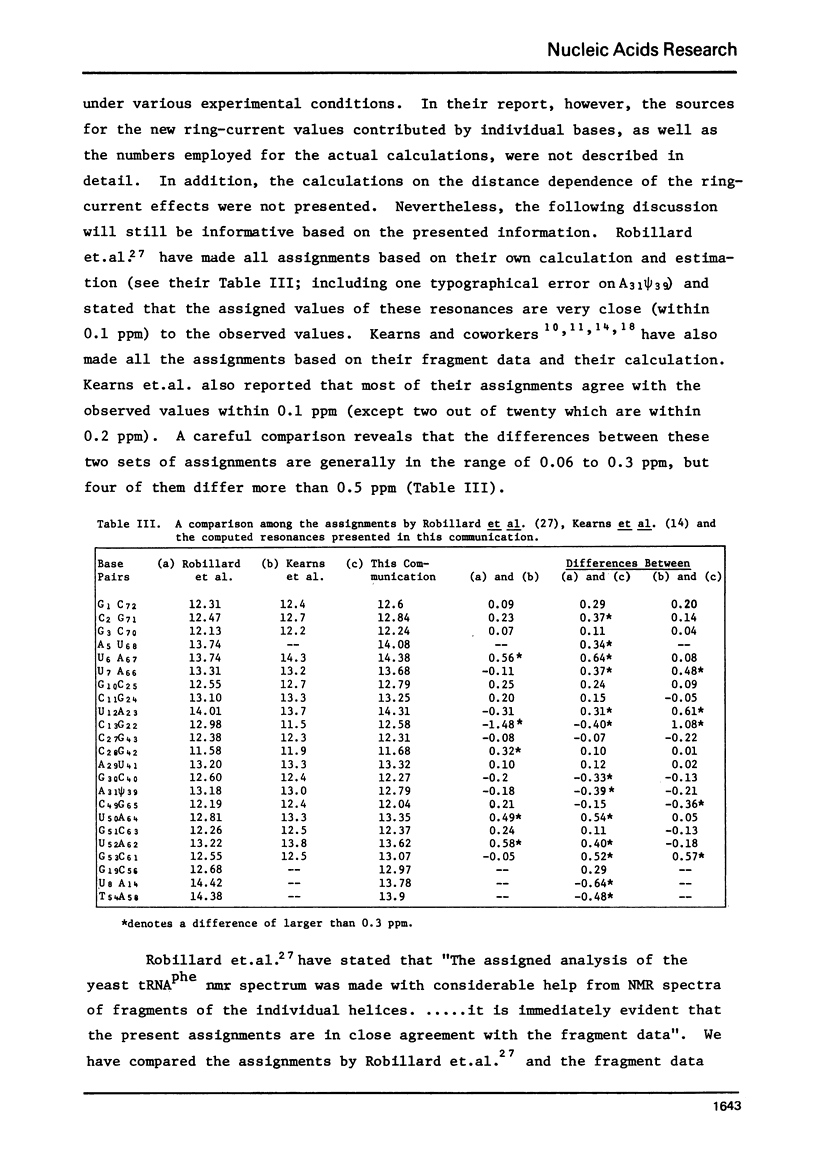
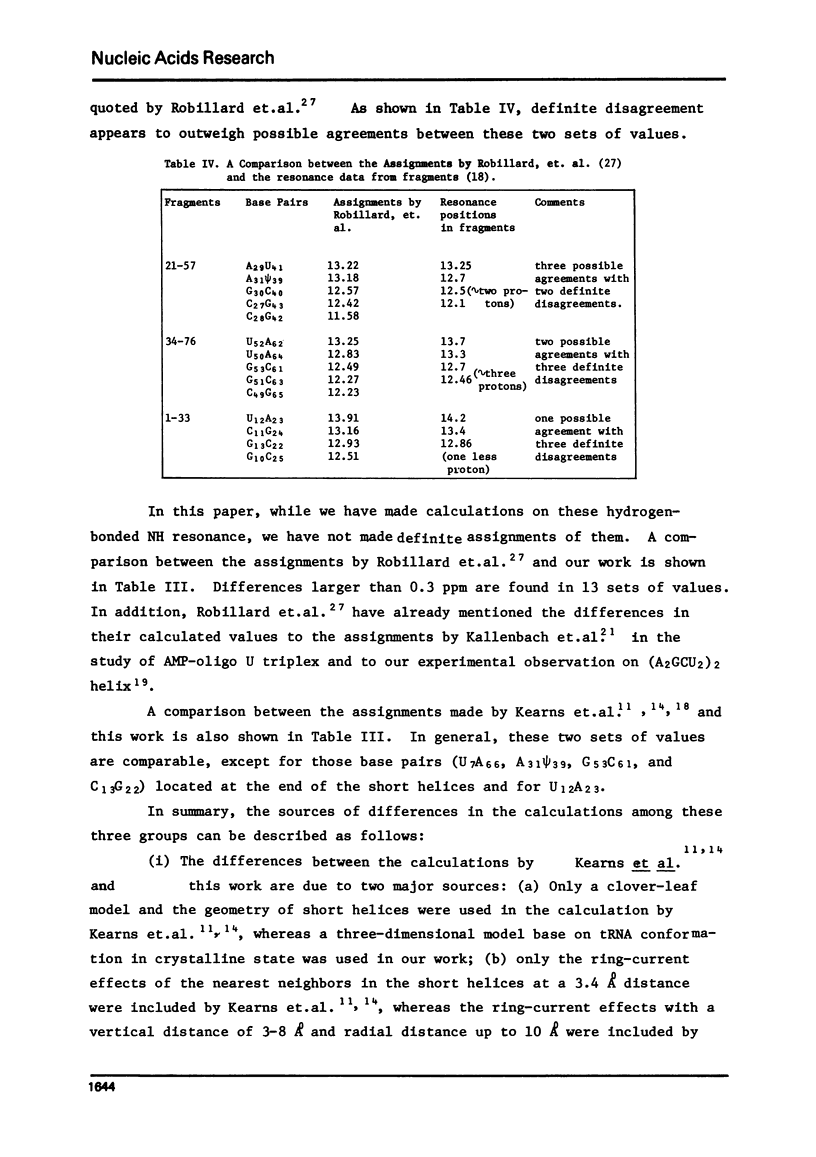
Abstract
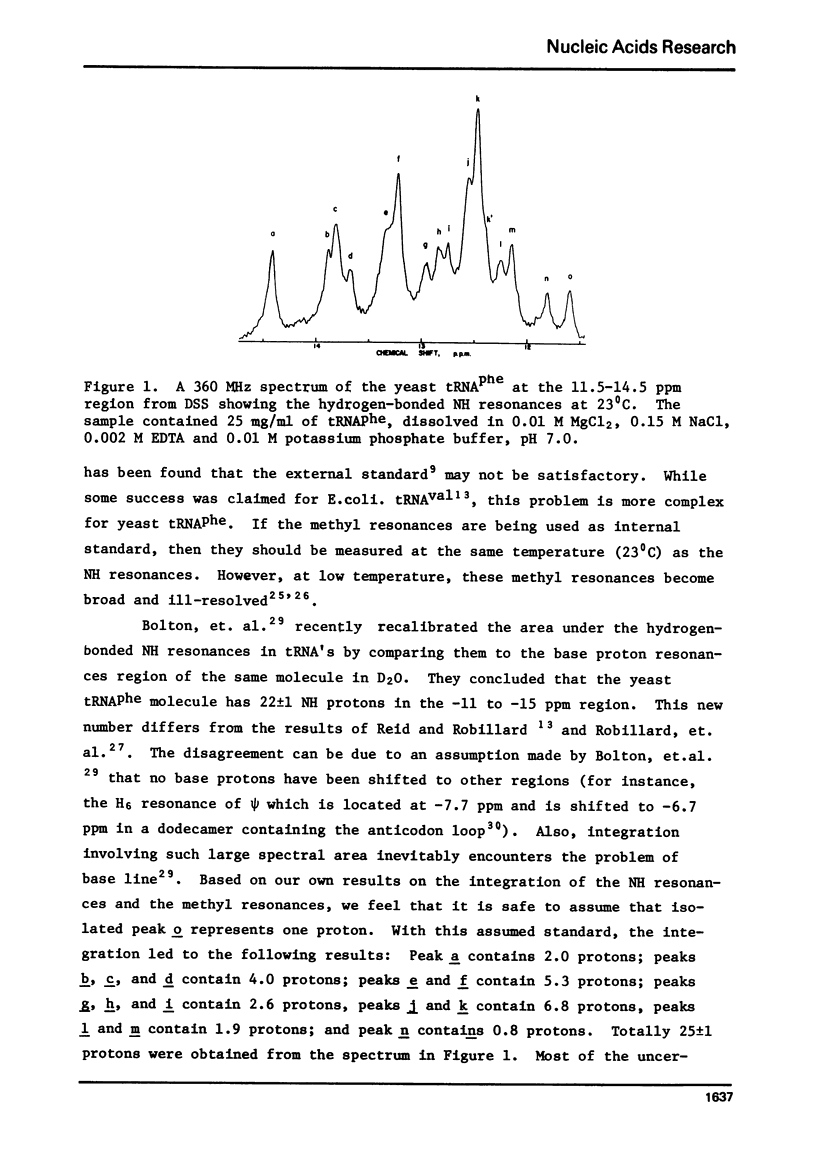
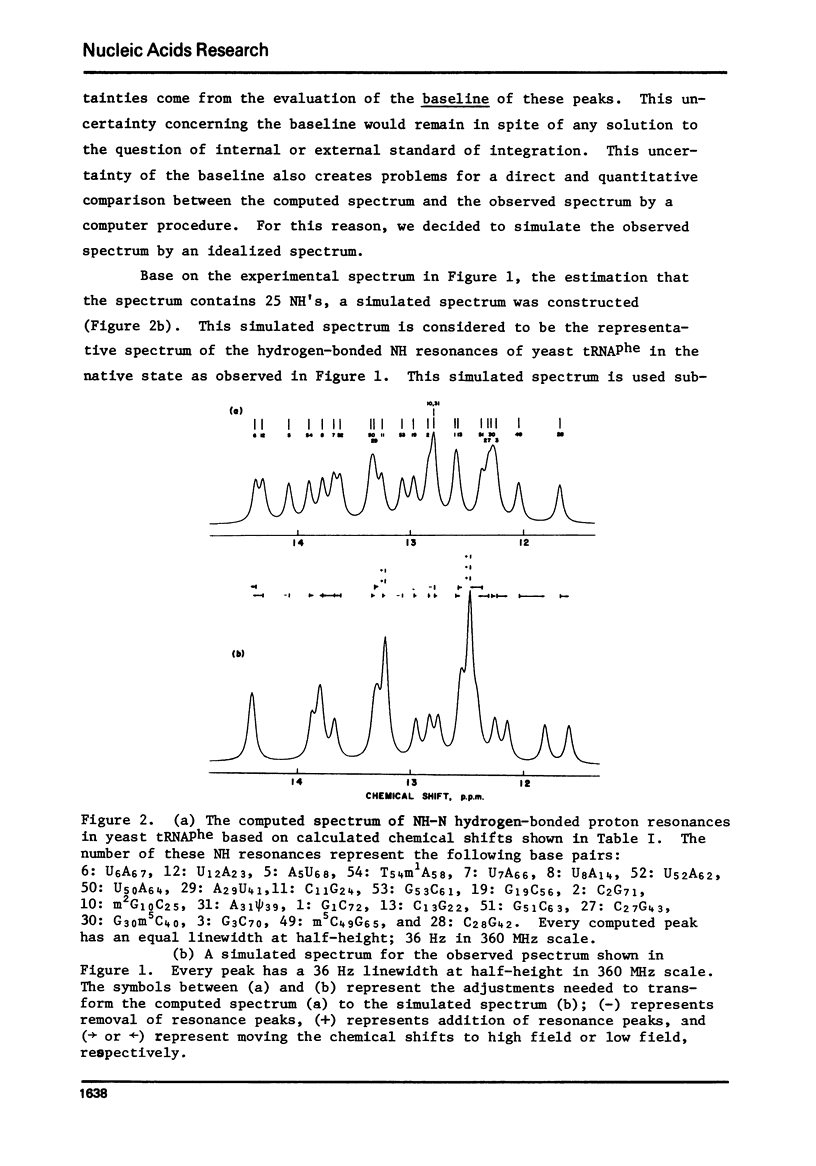
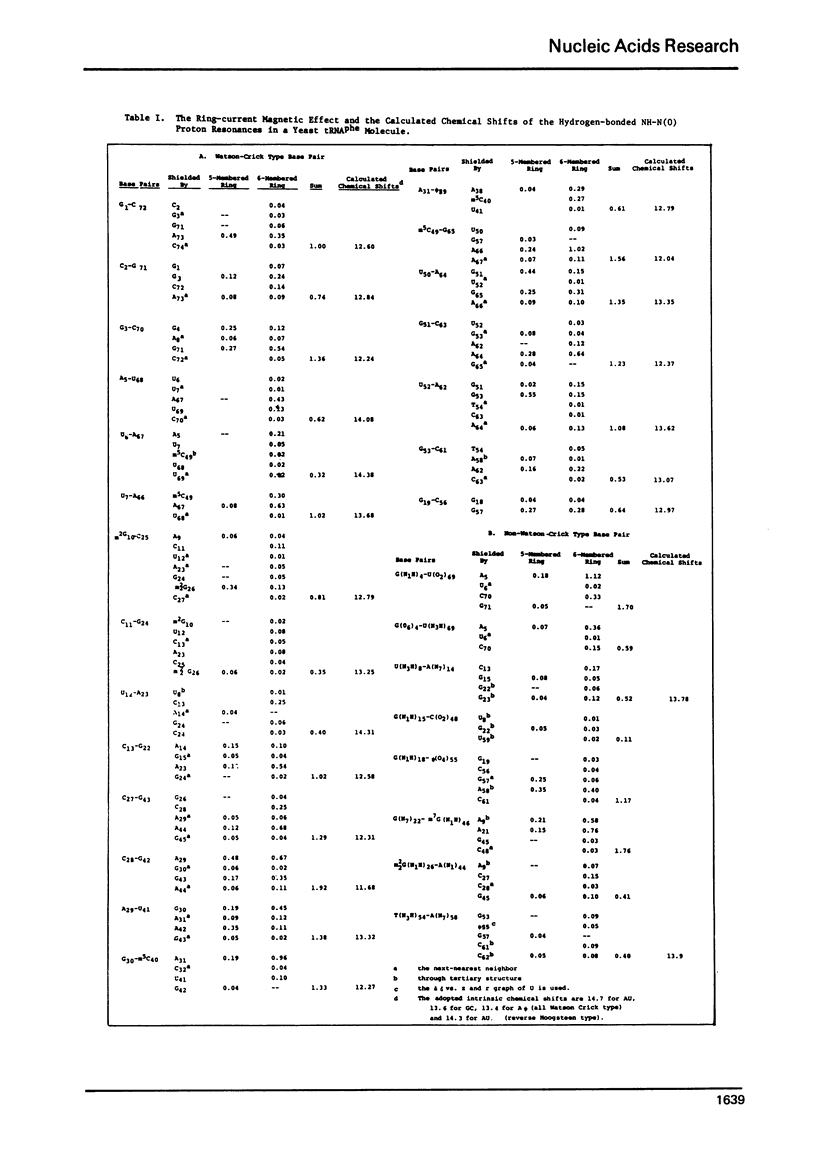
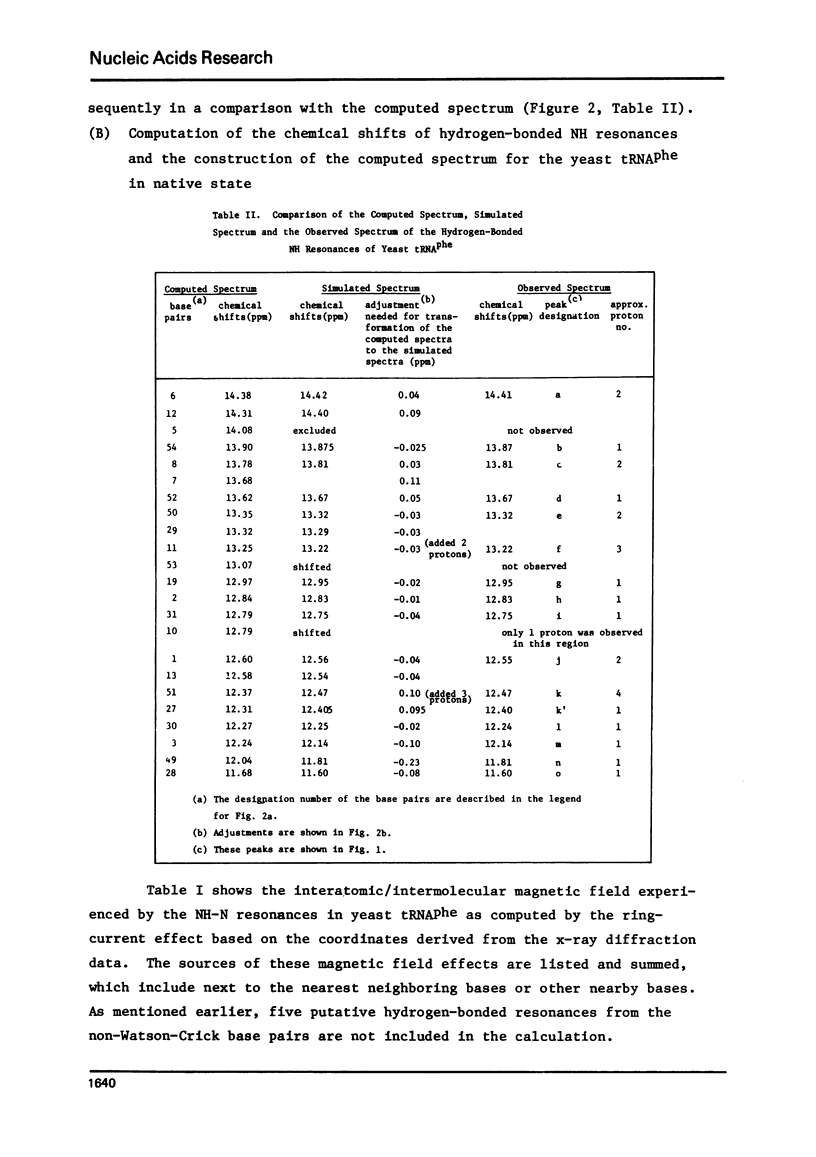
The hydrogen-bonded NH resonances of Baker's yeast tRNAphe in H2O solution with Mg++ have been measured by a 360 MHz spectrometer at 23 degrees C. Totally, fifteen peaks and one shoulder can be resolved which represent 25 +/- 1 protons. Based on the refined atomic coordinates of the tRNAphe in the orthorhombic crystal, on the recent advances in the distance dependence of the ring-current magnetic field effects and on the adopted values for the isolated hydrogen-bonded NH resonances, a computed spectrum consisting of 23 protons was constructed. A quantitative comparison by computer was made between the computed spectrum and the spectrum simulated from the observed spectrum. These two spectra are closely similar but not identical. We suggest that the conformation of yeast tRNAphe in aqueous solution is closely similar but not identical to that found in the crystal, especially in the T psi C region and D region. Also the NH resonances in 3-4 proposed hydrogen bonds (most likely for tertiary structure) may exchange very rapidly in aqueous solution.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Bond P. J. Structures for Poly(U)-poly(A)-poly(U)triple stranded polynucleotides. Nat New Biol. 1973 Jul 25;244(134):99–101. doi: 10.1038/newbio244099a0. [DOI] [PubMed] [Google Scholar]
- Arter D. B., Walker G. C., Uhlenbeck O. C., Schmidt P. G. PMR or the self-complementary oligoribonucleotide CpCpGpG. Biochem Biophys Res Commun. 1974 Dec 23;61(4):1089–1094. doi: 10.1016/s0006-291x(74)80395-5. [DOI] [PubMed] [Google Scholar]
- Bolton P. H., Jones C. R., Bastedo-Lerner D., Wong K. L., Kearns D. R. Quantitative determination of the number of secondary and tertiary structure base pairs in transfer RNA in solution. Biochemistry. 1976 Oct 5;15(20):4370–4377. doi: 10.1021/bi00665a004. [DOI] [PubMed] [Google Scholar]
- Daniel W. E., Jr, Cohn M. Changes in tertiary structure accompanying a single base change in transfer RNA. Proton magnetic resonance and aminoacylation studies of Escherichia coli tRNAMet f1 and tRNAMet f3 and their spin-labeled (s4U8) derivatives. Biochemistry. 1976 Sep 7;15(18):3917–3924. doi: 10.1021/bi00663a003. [DOI] [PubMed] [Google Scholar]
- Daniel W. E., Jr, Cohn M. Proton nuclear magnetic resonance of spin-labeled Escherichia coli tRNAf1MET. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2582–2586. doi: 10.1073/pnas.72.7.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giessner-Prettre C., Pullman B., Borer P. N., Kan L. S., Ts'o P. O. Ring-current effects in the Nmr of nucleic acids: a graphical approach. Biopolymers. 1976 Nov;15(11):2277–2286. doi: 10.1002/bip.1976.360151114. [DOI] [PubMed] [Google Scholar]
- Giessner-Prettre C., Pullman B. Intermolecular nuclear shielding values for protons of purines and flavins. J Theor Biol. 1970 Apr;27(1):87–95. doi: 10.1016/0022-5193(70)90130-x. [DOI] [PubMed] [Google Scholar]
- Kallenbach N. R., Daniel W. E., Jr, Kaminker M. A. Nuclear magnetic resonance study of hydrogen-bonded ring protons in oligonucleotide helices involving classical and nonclassical base pairs. Biochemistry. 1976 Mar 23;15(6):1218–1224. doi: 10.1021/bi00651a007. [DOI] [PubMed] [Google Scholar]
- Kan L. S., Borer P. N., Ts'o P. O. Conformation and interaction of short nucleic acid double-stranded helices. II. Proton magnetic resonance studies on the hydrogen-bonded NH-N protons of ribosyl ApApGpCpUpU helix. Biochemistry. 1975 Nov 4;14(22):4864–4869. doi: 10.1021/bi00693a013. [DOI] [PubMed] [Google Scholar]
- Kan L. S., Ts'o P. O., von der Haar F., Sprinzl M., Cramer F. NMR study on the methyl and methylene proton resonances of tRNA Phe yeast. Biochem Biophys Res Commun. 1974 Jul 10;59(1):22–29. doi: 10.1016/s0006-291x(74)80168-3. [DOI] [PubMed] [Google Scholar]
- Kan L. S., Ts'o P. O., von der Haar F., Sprinzl M., Cramer F. Proton magnetic resonance studies on the conformation of the hexanucleotide, GmpApApYpApsiP, and Related fragments from the anticodong loop of baker's yeast phenylalanine transfer ribonucleic acid. Biochemistry. 1975 Jul 15;14(14):3278–3291. doi: 10.1021/bi00685a038. [DOI] [PubMed] [Google Scholar]
- Kearns D. R., Patel D. J., Shulman R. G. High resolution nuclear magnetic resonance studies of hydrogen bonded protons of tRNA in water. Nature. 1971 Jan 29;229(5283):338–339. doi: 10.1038/229338a0. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Atomic co-ordinates for yeast phenylalanine tRNA. Nucleic Acids Res. 1975 Sep;2(9):1629–1637. doi: 10.1093/nar/2.9.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J. E., Jack A., Robertus J. D., Brown R. S., Rhodes D., Clark B. F., Klug A. Structure of yeast phenylalanine transfer RNA at 2.5 A resolution. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4414–4418. doi: 10.1073/pnas.72.11.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot D. R., Wong K. L., Kearns D. R., Reid B. R., Shulman R. G. Assignment of the low field proton nuclear magnetic resonance spectrum of yeast phenylalanine transfer RNA to specific base pairs. J Mol Biol. 1973 Jun 25;78(1):71–89. doi: 10.1016/0022-2836(73)90429-4. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Canuel L. Nuclear magnetic resonance studies of the helix-coil transition of poly (dA-dT) in aqueous solution. Proc Natl Acad Sci U S A. 1976 Mar;73(3):674–678. doi: 10.1073/pnas.73.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. J. Proton and phosphorus NMR studies of d-CpG(pCpG)n duplexes in solution. Helix-coil transition and complex formation with actinomycin-D. Biopolymers. 1976 Mar;15(3):533–558. doi: 10.1002/bip.1976.360150310. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Tonelli A. E. Assignment of the proton Nmr chemical shifts of the T-N3H and G-N1H proton resonances in isolated AT and GC Watson-Crick base pairs in double-stranded deoxy oligonucleotides in aqueous solution. Biopolymers. 1974;13(10):1943–1964. doi: 10.1002/bip.1974.360131003. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Seeman N. C., Wang A. H., Suddath F. L., Rich A. Yeast phenylalanine transfer RNA: atomic coordinates and torsion angles. Nucleic Acids Res. 1975 Dec;2(12):2329–2341. doi: 10.1093/nar/2.12.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Wang A. H., Seeman N. C., Suddath F. L., Rich A., Sussman J. L., Kim S. H. Hydrogen bonding in yeast phenylalanine transfer RNA. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4866–4870. doi: 10.1073/pnas.72.12.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid B. R., Ribeiro N. S., Gould G., Robillard G., Hilbers C. W., Shulman R. G. Tertiary hydrogen bonds in the solution structure of transfer RNA. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2049–2053. doi: 10.1073/pnas.72.6.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid B. R., Robillard G. T. Demonstration and origin of six tertiary base pair resonances in the NMR spectrum of E. coli tRNA1Val. Nature. 1975 Sep 25;257(5524):287–291. doi: 10.1038/257287a0. [DOI] [PubMed] [Google Scholar]
- Robillard G. T., Tarr C. E., Vosman F., Berendsen H. J. Similarity of the crystal and solution structure of yeast tRNAPhe. Nature. 1976 Jul 29;262(5567):363–369. doi: 10.1038/262363a0. [DOI] [PubMed] [Google Scholar]
- Shulman R. G., Hilbers C. W. Ring-current shifts in the 300 MHz nuclear magnetic resonance spectra of six purified transfer RNA molecules. J Mol Biol. 1973 Jun 25;78(1):57–69. doi: 10.1016/0022-2836(73)90428-2. [DOI] [PubMed] [Google Scholar]
- Stout C. D., Mizuno H., Rubin J., Brennan T., Rao S. T., Sundaralingam M. Atomic coordinates and molecular conformation of yeast phenylalanyl tRNA. An independent investigation. Nucleic Acids Res. 1976 Apr;3(4):1111–1123. doi: 10.1093/nar/3.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman J. L., Kim S. H. Idealized atomic coordinates of yeast phenylalanine transfer RNA. Biochem Biophys Res Commun. 1976 Jan 12;68(1):89–96. doi: 10.1016/0006-291x(76)90014-0. [DOI] [PubMed] [Google Scholar]
- Wong K. L., Bolton P. H., Kearns D. R. Tertiary structure in E. coli tRNA Arg and tRNA Val. Biochim Biophys Acta. 1975 Apr 2;383(4):446–451. [PubMed] [Google Scholar]


