Abstract
The effects of adiponectin on hepatic glucose and lipid metabolism at transcriptional level are largely unknown. We profiled hepatic gene expression in adiponectin knockout (KO) and wild-type (WT) mice by RNA sequencing. Compared with WT mice, adiponectin KO mice fed a chow diet exhibited decreased mRNA expression of rate-limiting enzymes in several important glucose and lipid metabolic pathways, including glycolysis, tricarboxylic acid cycle, fatty-acid activation and synthesis, triglyceride synthesis, and cholesterol synthesis. In addition, binding of the transcription factor Hnf4a to DNAs encoding several key metabolic enzymes was reduced in KO mice, suggesting that adiponectin might regulate hepatic gene expression via Hnf4a. Phenotypically, adiponectin KO mice possessed smaller epididymal fat pads and showed reduced body weight compared with WT mice. When fed a high-fat diet, adiponectin KO mice showed significantly reduced lipid accumulation in the liver. These lipogenic defects are consistent with the down-regulation of lipogenic genes in the KO mice.
Keywords: hepatic metabolism, lipogenesis, transcriptional regulation
Adipose tissue regulates energy homeostasis by secreting cytokines known as adipokines. Adiponectin, the most abundant adipokine in the serum, modulates lipid and glucose metabolism, including promotion of fatty acid oxidation and glucose utilization and repression of hepatic gluconeogenesis (1, 2). Several lines of adiponectin knockout (KO) or transgenic (Tg) mice have been generated in different laboratories (3, 4). Although the phenotypes reported are variable in some aspects, these mouse models all demonstrate that adiponectin deficiency predisposed mice to high-fat-diet (HFD)-induced insulin resistance (3–5). It was proposed that adiponectin is required to maintain a basal tone of insulin responsiveness (5). However, the way in which the basal tone of insulin responsiveness is established and how adiponectin maintains insulin sensitivity remain largely unknown.
Adiponectin is present in plasma at extremely high concentrations (2–10 μg/mL), in contrast to that of insulin, glucagon, and leptin, which are found at orders-of-magnitude smaller concentrations. Moreover, most metabolic hormones are released into the blood in a regulated pulsatile fashion (6); this process allows the insulin’s actions to respond to nutritional signals following meals or for glucagon to increase hepatic glucose output during hypoglycemia. In contrast, the blood levels of adiponectin are strikingly constant, making adiponectin a less-sensitive regulator in response to the acute changes in glucose and lipid levels following food intake or deprivation. We therefore hypothesize that adiponectin primarily regulates glucose and lipid metabolism at transcriptional levels besides its well described role that involves AMP-activated protein kinase (AMPK).
To test this hypothesis, we profiled hepatic gene expression in both wild-type (WT) and adiponectin KO mice fed a standard chow diet by high-throughput RNA sequencing (RNA-Seq). We found that adiponectin plays an important role in regulating the expression of rate-limiting enzymes in several important glucose and lipid metabolic pathways, including glycolysis, fatty acid activation, and synthesis. This role of adiponectin might be mediated by a master regulator of hepatic gene expression, the transcription factor (TF) Hnf4a.
Results
Rate-limiting enzymes catalyze the slowest or irreversible steps in metabolic pathways, which makes them the targets of transcriptional regulation and/or posttranslational modifications such as phosphorylation in the regulation of metabolism (7). Because adiponectin plays an important role in energy metabolism, our analysis on hepatic gene expression in adiponectin KO and WT mice was specifically focused on the expression levels of rate-limiting enzymes [retrieved from the Rate-Limiting Enzymes database (RLEdb); ref. 7] in glucose and lipid metabolic pathways. In addition, we examined the mRNA expression levels of important lipid binding proteins and transporters as well as components of the mitochondrial electron transport chain and oxidative phosphorylation.
Adiponectin KO Mice Exhibited a Significantly Decreased Expression of Key Lipogenic Genes in Liver.
The mRNA expression levels of rate-limiting enzymes in glucose and lipid metabolic pathways from both KO and WT mouse livers are presented in Table S1. The detailed pathways of glucose or lipid metabolism as well as the relative changes of genes between KO and WT mice are shown in Figs. S1, S2, and S3. Based on expression level, coverage depth, and fold change, we selected a smaller set of genes (shown in bold in Table S1) to validate the RNA-Seq data. To this end, real-time PCR assays were performed by using hepatic mRNAs from 9 or 10 individual WT and adiponectin KO mice. We confirmed that there is a good consistency between RNA-Seq and quantitative real-time PCR (RT-PCR) results (Figs. 1 and 2). Both RNA-Seq and RT-PCR showed that livers of adiponectin KO mice exhibited a remarkable down-regulation of key lipid metabolic enzymes, including ATP citrate lyase (Acly), fatty acid synthase (Fasn), and acetyl-CoA carboxylase alpha (Acc1) in fatty acid synthesis (Fig. 1A and Fig. S1A); stearoyl-CoA desaturase (Scd1) and fatty acid desaturase domain family member 6 (Fads6) in desaturation of fatty acids (Fig. 1A); glycerol kinase (Gyk) and glycerol-3-phosphate dehydrogenase (Gpd1) in triglyceride synthesis (Fig. 1A and Fig. S1B); acetyl-CoA acyltransferase (Acaa1b) and 3-hydroxy-3-methylglutaryl-CoA synthase 2 (Hmgcs2) in cholesterol synthesis (Fig. 1A and Fig. S1C); acetyl CoA synthetase long-chained (Acsl1 and Acsl5), acetyl CoA synthetase medium-chained (Acsm5), and fatty acid transporters (Fatp2 and Fatp5) in fatty acid activation (Fig. 1A); and acetyl-CoA carboxylase-β (Acc2) in fatty acid β oxidation (Fig. 1A and Fig. S1D).
Fig. 1.
RT-PCR validation of genes that are important in hepatic lipid metabolism in WT and adiponectin KO mice. The gene expression levels were examined by RT-PCR (n = 9 or 10 mice in each group) and presented in A Upper, B Upper. The corresponding expression levels by RNA-Seq are shown in A Lower, B Lower. Results are presented as mean ± SEM. *0.01 < P < 0.05; **0.001 < P < 0.01; ***P < 0.001; ****P < 0.0001. (A) Rate-limiting enzymes in fatty acid synthesis (Acly, Fasn, and Acc1), unsaturated fatty acid synthesis (Scd1 and Fads6), triglyceride synthesis (Gyk and Gpd1), cholesterol synthesis (Acaa1b and Hmgcs2), fatty acid activation (Acsl1, Acsl5, Acsm5, Fatp2, and Fatp5), and fatty acid β oxidation (Acc2). (B) Important lipid binding and transport proteins (Fabp, Apo, Scarb1, and Lcat).
Fig. 2.
RT-PCR validation of genes that are important in hepatic glucose metabolism, energy production, and detoxification functions in WT and KO mice. (Upper) The gene expression levels were examined by RT-PCR (n = 9 or 10). (Lower) The corresponding expression levels by RNA-Seq are shown. Results are presented as mean ± SEM. *0.01 < P < 0.05; **0.001 < P < 0.01; ***P < 0.001; ****P < 0.0001. (A) Rate-limiting enzymes in glycolysis (Gck, Pdhb, and Pfkfb1), gluconeogenesis (Pck1), and TCA cycle (Idh2). (B) Crucial gene components in electron transport chain and oxidative phosphorylation (ATP5a1, ATP5b, Ndufs6, Uqcr10, Uqcrq, Cox6a1, Cox6b1, and ATP5g3). (C) Rate-limiting genes involved in hepatic detoxification function. ATP, ATP synthase; NS, not significant.
Surprisingly, this list included not only key enzymes in fatty acid β oxidation but also rate-limiting enzymes in fatty acid synthesis and desaturation, fatty acid activation (a step required for both fatty acid oxidation and triglyceride synthesis), triglyceride synthesis, and cholesterol synthesis. Thus, adiponectin promotes fatty acid β oxidation not only through AMPK activation, but also through increasing the expression of rate-limiting enzymes in this pathway. On the other hand, down-regulation of important hepatic lipogenic genes in the absence of adiponectin suggests that there might be lipogenic defects in the livers of adiponectin KO mice, as we demonstrate later.
Apolipoprotein Genes Were Down-Regulated in KO Mice.
Apolipoproteins (Apos) are important lipid transporters. The dysregulation of Apos impairs the uptake of triglyceride and cholesterol in tissues and their subsequent metabolism. In adiponectin KO mice, we observed a general reduction in mRNAs encoding Apos, including Apoa2, a4, b, c1, c2, c3, and e. Apoa1 and c4 were the only two members unaffected among the genes we examined (Fig. 1B). Although Apoa1, the major component of high-density lipoprotein (HDL), was not affected in the KO mice, the expression levels of a receptor for HDL, the scavenger receptor class B member 1 (Scarb1), was significantly decreased in KO mice (Fig. 1B). Lecithin–cholesterol acyltransferase (Lcat), the enzyme converting free cholesterol into cholesteryl ester for packing HDL particle, was also down-regulated (Fig. 1B). However, the effects of down-regulated Scarb1 and Lcat in KO liver cholesterol clearance might be minimal, because ref. 5 showed that adiponectin deficiency did not affect the clearance of total cholesterol, free cholesterol, and cholesterol esters from the aorta.
Many Key Hepatic Genes Involved in Glucose Metabolism Were Down-Regulated in Adiponectin KO Mice.
Compared with WT mice, adiponectin KO mice showed reduced expression of three major controlling enzymes in glycolysis, glucokinase (Gck), phosphofructokinase (Pfkfb1), and pyruvate dehydrogenase (Pdhb) (Fig. 2A and Fig. S2A); and down-regulated expression of isocitrate dehydrogenase (Idh2), a key enzyme in the tricarboxylic acid (TCA) cycle (Fig. 2A and Fig. S2B). Adiponectin has been reported to repress hepatic gluconeogenesis, an important mechanism to improve insulin sensitivity (2). In contrast to a general down-regulation of rate-limiting enzymes in many metabolic pathways, we found a slight, but statistically insignificant, increase of phosphoenolpyruvate carboxykinase (Pck1), the key rate-limiting enzyme for gluconeogenesis, in the KO mice (Fig. 2A and Fig. S2A).
Components of Hepatic Mitochondrial Electron Transport Chain and Oxidative Phosphorylation Were Insignificantly Affected in Adiponectin KO Mice.
Although one of the subunits of ubiquinol–cytochrome c reductase (Uqcr10) and several components of ATP synthase (Atp5a1 and Atp5b) were down-modulated in the KO mice, other genes that we examined remained unchanged. These included NADH dehydrogenase (ubiquinone) Fe-S protein (Ndufs6), ubiquinol–cytochrome c reductase (Uqcrq), cytochrome c oxidase (Cox6a1 and Cox6b1), and ATP synthase (Atp5g3) (Fig. 2B and Fig. S3). These data suggest that mitochondrial electron transport chain and oxidative phosphorylation pathways are not significantly affected in adiponectin KO mice.
Some Genes Involved in Liver Detoxification Functions Were Down-Regulated in KO Mice.
Metabolic detoxification represents another important metabolic function of liver: Harmful substances are constantly produced and removed by the liver. We observed a general reduction in expression of key enzymes in this process, including cytochrome P450 oxidases (Cyp2c50, Cyp2c54, Cyp2e1, Cyp2d11, Cyp2d12, Cyp3a44, Cyp3a59, and Cyp4a32), glutathione peroxidase (Gpx1 and Gpx4), aldo-keto reductases (Akr1c6 and Akr1c19), and UDP glycosyltransferases (Ugt1a1), indicating a reduced detoxification capacity of KO mouse liver (Fig. 2C).
Many Hepatic TFs Were Down-Regulated in the Absence of Adiponectin.
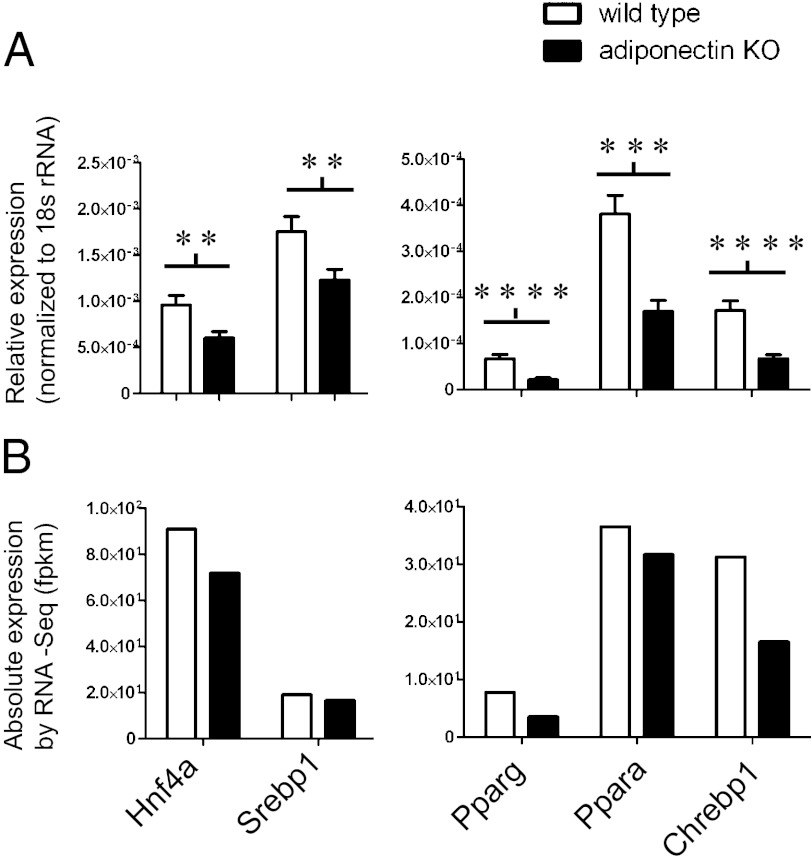
To understand how adiponectin deficiency leads to changes in the expression of such a wide range of genes, we examined whether adiponectin regulates expression of any TFs. To begin, all TFs that are expressed in mouse liver were retrieved from BioBase3 (9), and genes with <2 fragments per kilobase of exon per million fragments mapped (fpkm) in both WT and KO livers were removed from the analysis to reduce data noise. We found that many TFs were down-regulated in the KO mice (Table S2), among which are TFs important for hepatic glycolysis and lipogenesis, such as Chrebp; the major regulator of hepatic lipid metabolism Ppara; the important lipid and glucose metabolic regulator Pparg; and the important cholesterol and fatty acid biosynthesis regulator Srebp1. Surprisingly, the expression of master regulators of liver-specific gene expression was also decreased in the KO mice. These include hepatocyte nuclear factor 4a (Hnf4a) and Ccaat-enhancer-binding proteins (C/EBPa and C/EBPb) (Table S2 and Fig. 3B). The highly significant decrease in the expression of these genes was confirmed by RT-PCR (Fig. 3A).
Fig. 3.
RT-PCR validation of TFs that are down-regulated in KO mice by RNA-Seq analysis. (A) The gene expression levels examined by RT-PCR (n = 9 or 10) are presented. (B) The corresponding expression levels by RNA-Seq (a pool of 9 or 10 mouse livers) are shown. Results are presented as mean ± SEM. *0.01 < P < 0.05; **0.001 < P < 0.01; ***P < 0.001; ****P < 0.0001.
Master Regulator of Hepatic Gene Expression Hnf4a Might Mediate the Role of Adiponectin in the Regulation of Key Metabolic Genes.
Hnf4a, one of the most important “master regulators” of hepatic gene expression, is one of the TFs down-regulated in adiponectin KO mice. Because many affected genes in the KO mice are known targets of Hnf4a—including Apoa2, b, and c3 (10); cytochrome P450 oxidases (11, 12); and fatty acid binding proteins (Fabps)—we determined whether the binding of Hnf4a to the promoter regions of many of its targets was affected in the absence of adiponectin. Using chromatin immunoprecipitation (ChIP) followed by PCR assays with specific primers designed for genes with Hnf4a binding elements in their promoter regions, we showed that the binding of Hnf4a to the promoters of Chrebp, Cyp2e1, Cyp8b1, and Scd1 was reduced significantly in adiponectin KO mice. The Hnf4a gene, which is autoregulated by HNF4a (14), was used as a positive control (Fig. 4). One sequence containing no Hnf4a binding elements—an insulin exon—was used as a negative control; as expected, this region showed no difference in Hnf4a binding between WT and KO mice. These data suggest that Hnf4a might be a mediator of adiponectin in regulating the expression of many important hepatic metabolic genes.
Fig. 4.
Hnf4a binding to the DNA regions of selected genes as measured by ChIP. Binding to promoters of Hnf4a, Chrebp, Cyp2e1, Cyp8b1, and Scd1 genes by Hnf4a was significantly lower in KO compared with WT livers. The genes examined were selected from genes down-regulated in the absence of adiponectin that had putative Hnf4a binding sites in their promoter regions. Insulin exon was used as negative controls. Data are presented as the mean ± SEM of four replicates from at least two independent ChIP experiments. *0.01 < P < 0.05; **0.001 < P < 0.01; ***P < 0.001.
Adiponectin KO Mice Exhibited Less Fat Accumulation and Body Weight Gain After 15–16 Wk of Age.
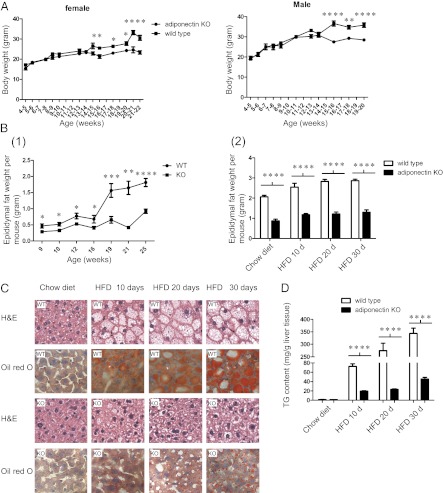
Down-regulation of key lipogenic genes suggests a lipogenic defect in adiponectin KO mice, and we therefore determined whether down-regulation of these genes leads to any phenotypic changes. Although at 6 wk the adiponectin KO mice did not exhibit any significant difference in body weight gain from that of the WT mice (4), our study of KO mice showed that, when fed a chow diet, both female and male KO mice displayed modest, but significant, decreased body weight starting from 15–16 wk of age. An average of 15–20% reduction in weight gain was observed in both male and female KO mice (Fig. 5A). Moreover, male KO mice maintained on a normal chow diet (age range: 9–25 wk) possessed smaller epididymal fat pads compared with WT mice; the average epididymal fat pad weights of KO mice in this age range were 30–70% lower than those of WT mice (Fig. 5B Left). In addition, after mice (18–20 wk old) were fed a HFD for 10, 20, or 30 d, there were reduced epididymal fat pad weights in the KO mice (Fig. 5B Right).
Fig. 5.
Adiponectin KO mice showed reduced body weight after 15–16 wk of age and possessed smaller epididymal fat pads compared with WT mice. When fed a HFD for 10, 20, and 30 d, KO mice exhibited a dramatically reduced fat accumulation in their livers. Results were presented as mean ± SEM. *0.01 < P < 0.05; **0.001 < P < 0.01; ***P < 0.001; ****P < 0.0001. (A) Comparison of body weight (grams) between female and male WT (■) and adiponectin KO (●) mice fed a chow diet at the indicated age (n = 6∼15). (B Left) Epididymal fat pad weights of WT and adiponectin KO male mice (age 9∼25 wk) maintained on a normal chow diet (n = 3∼9). (Right) Epididymal fat pad weight of WT and adiponectin KO male mice (age 18∼20 wk) following a control (chow diet) and a HFD for 10, 20, or 30 d (n = 5∼7). (C) H&E and Oil Red O staining of livers of male WT and adiponectin KO mice (age 18–20 wk), after being fed a control diet or a HFD for 10, 20, and 30 d. (D) Effects of a control and a HFD for 10, 20, and 30 d on liver triglyceride levels (TG) in male WT and KO mice (n = 5∼7).
To investigate lipogenesis in adiponectin KO mice, we fed 18- to 20-wk-old male mice with either a chow diet for 30 d or a HFD for 10, 20, or 30 d. The liver, white adipose, and skeletal muscle tissues of these mice were collected and subjected to histological analysis by H&E and Oil Red O staining. Although we did not detect any morphological difference in white adipose and skeletal muscle tissues between adiponectin KO and WT mice (Fig. S4), histological examination of the livers clearly identified prominent lipid droplets in hepatocytes in all three groups of WT mice fed a HFD (Fig. 5C). Adiponectin KO mouse livers, conversely, only exhibited a minor degree of intracellular lipid accumulation (Fig. 5C). Hepatic triglyceride quantification indicated a remarkably reduced fat accumulation in the KO mouse liver (Fig. 5D). Together our data suggest that major lipogenic defects are associated with adiponectin deficiency due to the decreased expression of lipogenic genes.
Discussion
In this study we compared the hepatic gene expression profiles in adiponectin KO and WT mice using RNA-Seq technology followed by RT-PCR analyses. Genes with altered expression levels in adiponectin KO mouse liver include those encoding rate-limiting enzymes in glycolysis, TCA cycle, fatty acid activation, fatty acid synthesis and desaturation, triglyceride synthesis, and cholesterol synthesis. Our data clearly demonstrate that adiponectin regulates hepatic glucose and lipid metabolism at the transcriptional level.
Adiponectin KO mice possessed smaller epididymal fat pads at ages ranging from 9 to 25 wk when maintained on a normal chow diet. When exposed to a HFD, 18- to 20-wk-old adiponectin KO mice not only possessed smaller epididymal fat pads but also exhibited a significant reduction in hepatic lipid accumulation. AMPK activation by adiponectin cannot explain these findings because any reduction of AMPK activity in the KO mice will promote lipid synthesis. Consistent with our results, Combs et al. (15) reported that with the chronic elevation of adiponectin, female Tg mice exhibit an increase in adiposity without changes in food intake, average 24-h energy expenditure, or the respiratory quotient. The finding also excluded the involvement of AMPK in these phenotypes. The defective lipogenesis in KO mice, however, is explained very well by our gene profiling data. First, many critical genes involved in fatty acid synthesis were down-regulated in the livers of KO mice, including genes controlling fatty acid biosynthesis (Acly, Acc1, and Fasn), and the subsequent formation of unsaturated fatty acids (Scd1 and Fads6). Second, triglyceride formation, as well as cholesterol synthesis, was also affected in the KO mice, reflected by the decreased levels of rate-limiting enzymes in these pathways (Gpd, Gyk, Acaa, and Hmgcs). Third, important regulatory TFs in the hepatic lipogenic program, including Chrebp and Srebp1, were also down-regulated. The changes in these key genes and gene regulators provide a genetic basis for the observed defects in lipogenesis observed in the KO mice fed a HFD. Our RNA-Seq analysis was based on mice at age 12 wk when KO mice exhibited only weak lipogenic defects (Fig. S5). This defect became more evident with KO mice at 18–20 wk of age. We believe that the lipogenic defects in adiponectin KO mice exist at all ages, although they are more overt in conditions when fat formation dominates expenditure, such as in aging or when exposed to a HFD.
The adiponectin KO mice used in this study exhibited delayed clearance of free fatty acids (FFAs) in plasma (16). This delayed clearance is explained by our findings of significantly lower levels of mRNAs encoding lipid transport proteins (Apos and Fabps) and proteins important for fatty acid absorption, activation, and oxidation [acyl-CoA synthetases (Acsl and Fatp) and Acc2]. More importantly, the mRNA encoding Ppara, the major transcriptional regulator of lipid metabolism in the liver—whose functions include promotion of fatty acid uptake and oxidation by up-regulation of genes involved in fatty acid transport and fatty acid β oxidation—is also down-regulated in the KO mice. The down-modulation of Ppara could be responsible for the changes in these genes because many of the genes described above are transcriptional targets of Ppara.
The complicated functions of the liver are regulated primarily by a limited number of liver-enriched TFs. These include hepatocyte nuclear factor 1 (Hnf1), Hnf3, Hnf4a, and Hnf6, as well as members of the C/EBP family (17, 18). TFs in these families cooperatively function in the liver as master regulators, with Hnf1a, Hnf4a, and Hnf6 being the center of the regulatory network (19, 20). Here we showed that adiponectin normally up-regulates the transcript levels of TFs, including Hnf4a. Odom et al. (20) showed that Hnf4a binds directly to almost 50% of the actively transcribed hepatic genes, whereas no other TF binds to more than one-fifth of that number. The actual genes regulated by Hnf4a may be much larger because many of the Hnf4a-targeted genes themselves are TFs and transcription cofactors. For example, Hnf4a controls the expression of another master regulator, Hnf1, in hepatocytes (21). Moreover, the two crucial regulatory TFs regulating hepatic lipogenesis, Srebp1 and Chrebp1, are also targets of Hnf4a, demonstrated by other groups and this study, respectively (22). Reduced levels of Hnf4a have been implicated in the pathogenesis of type 2 diabetes mellitus and type 1 form of maturity-onset diabetes of the young (13, 23–26). The large number of hepatic genes affected by the misregulation of Hnf4a is thought to cause the impaired insulin signaling in these metabolic syndromes (20).
Proper energy deposition and expenditure are keys for a balanced metabolism, which underlies the sensitive responsiveness of peripheral metabolic organs to the actions of insulin. Adiponectin KO mice exhibited a HFD-induced insulin resistance (3–5), suggesting an important role of adiponectin in maintaining insulin sensitivity. Here we showed that adiponectin serves as a regulator of many important hepatic genes in glucose and lipid biosynthesis and catabolism; the cumulative effects of all these genes expressed at the optimal levels create the basal tone essential for maintaining sensitive insulin responsiveness. For example, increased FFAs in plasma induce insulin resistance. Adiponectin promotes the expression of key hepatic genes in triglyceride formation and lipid clearance, which promotes FFA clearance from plasma and to decreased plasma FFAs. When the serum adiponectin level is reduced in obesity or other metabolic syndromes, maintenance of this basal tone of insulin sensitivity by adiponectin is impaired, causing insulin resistance.
Experimental Procedures
Mice and Animal Studies.
Adiponectin KO mice were a gift from Yuji Matsuzawa (Osaka University, Osaka) (3). WT and adiponectin KO mice were maintained in a C57BL/6J genetic background. Body weight of each mouse was measured at the indicated age. Epididymal fat pads were harvested from mice at 18–20 wk of age.
RNA-Seq.
Liver tissues from WT or adiponectin KO male mice (n = 9 or 10) at 12 wk of age were harvested and subjected to total RNA extraction. Total RNA from 9 or 10 mice of the same strain was pooled together as one biological sample. The mRNA sequencing samples were prepared by using the Illumina sample preparation protocol (RS-930-1001; Illumina). Briefly, mRNA was isolated from total RNA (10 μg) and then was used to synthesize cDNA. The resulting cDNA (fragments of 200–250 bp) was purified and enriched by PCR. The cDNA libraries were sequenced by an Illumina Genome analyzer II at the Whitehead Genome Technology Core.
Read Mapping and Expression-Level Normalization.
A total of 21.8 million and 22.7 million 36-bp single reads were obtained from the RNA of adiponectin KO and WT mice, respectively. TopHat (27) was used for mapping the reads to mouse mm9 genome, guided with Ensembl (NCBIM37 62) gtf file. The segment size for TopHat was reduced to 18, and the maximum mismatch within each segment was set to 1. More than 82% of the reads from both samples were mapped. Gene fpkms were calculated and compared with Cuffdiff (8) with –b and –u options, and rRNAs downloaded from Ensembl were excluded with –M option. Only genes with “Test Status” as OK and with at least 2 fpkm were kept for further analysis.
RT-PCR.
Total RNA from liver tissues of WT or adiponectin KO mice (n = 9 or 10) was isolated, and qRT-PCR was performed on an ABI Prism 7900HT Sequence Detector (Applied Biosystems) from 2 ng of total RNA after reverse transcription. 18S was used as the internal control. Data were analyzed by the relative quantification (ΔΔCt) method. Primer sequences are available upon request.
HFD Study.
Eighteen- to 20-wk-old male WT or adiponectin KO mice were divided randomly into four groups. Three groups were fed a HFD D12492 (60% kcal% fat) (Research Diets) for 10, 20, and 30 d, and one group was fed a chow diet for 30 d (11.8% kcal% fat) (LabDiet; 5P00 ProLab RMH 3000).
Hepatic Triglyceride Contents.
Liver tissues from mice under HFD studies were homogenized and extracted with chloroform/methanol (2:1; vol/vol) three times. The organic layer was collected after centrifugation and dried. The resulting samples were resuspended in 3 M KOH at 70 °C. Triglyceride level was measured by using a serum triglyceride determination kit (TR0100; Sigma).
Histology.
Liver, white adipose, and skeletal muscle tissues harvested from WT or adiponectin KO mice were stained with H&E or Oil Red O staining. All histology procedures were performed by the Histology Core Facility at the Koch Institute, Massachusetts Institute of Technology.
ChIP Experiment.
ChIP was performed as described (28, 29) with modifications. Five micrograms of specific or control antibody (HNF4-α, sc-8971; rabbit IgG, sc-2027) was incubated with preblocked Dyanbead Protein A (100.02D; Invitrogen) overnight. The specificity of the antibodies was described (30). Livers from WT or adiponectin KO mice were cross-linked, lysed, and then sonicated (28). The resulting chromatin was incubated with the antibody-coupled beads overnight. The beads were then washed, and the chromatin was eluted in ChIP elution buffer, reverse-cross-linked at 65 °C overnight, and treated with RNase and Proteinase K. The DNA was extracted, and 2 μL of DNA was used for each ChIP-PCR experiment. Primer sequences are available upon request.
Statistical Analysis.
Results were presented as mean ± SEM. Differences between groups were examined for statistical significance by unpaired one-tailed Student's t test or two-way ANOVA. P < 0.05 was regarded as statistically significant.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE38144).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211611109/-/DCSupplemental.
References
- 1.Zhou H, et al. Adiponectin represses gluconeogenesis independent of insulin in hepatocytes. Biochem Biophys Res Commun. 2005;338:793–799. doi: 10.1016/j.bbrc.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Combs TP, Berg AH, Obici S, Scherer PE, Rossetti L. Endogenous glucose production is inhibited by the adipose-derived protein Acrp30. J Clin Invest. 2001;108:1875–1881. doi: 10.1172/JCI14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maeda N, et al. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat Med. 2002;8:731–737. doi: 10.1038/nm724. [DOI] [PubMed] [Google Scholar]
- 4.Kubota N, et al. Disruption of adiponectin causes insulin resistance and neointimal formation. J Biol Chem. 2002;277:25863–25866. doi: 10.1074/jbc.C200251200. [DOI] [PubMed] [Google Scholar]
- 5.Nawrocki AR, et al. Mice lacking adiponectin show decreased hepatic insulin sensitivity and reduced responsiveness to peroxisome proliferator-activated receptor gamma agonists. J Biol Chem. 2006;281:2654–2660. doi: 10.1074/jbc.M505311200. [DOI] [PubMed] [Google Scholar]
- 6.Jiang G, Zhang BB. Glucagon and regulation of glucose metabolism. Am J Physiol Endocrinol Metab. 2003;284:E671–E678. doi: 10.1152/ajpendo.00492.2002. [DOI] [PubMed] [Google Scholar]
- 7.Zhao M, Chen X, Gao G, Tao L, Wei L. RLEdb: A database of rate-limiting enzymes and their regulation in human, rat, mouse, yeast and E. coli. Cell Res. 2009;19:793–795. doi: 10.1038/cr.2009.61. [DOI] [PubMed] [Google Scholar]
- 8.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matys V, et al. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34(Database issue):D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vergnes L, Taniguchi T, Omori K, Zakin MM, Ochoa A. The apolipoprotein A-I/C-III/A-IV gene cluster: ApoC-III and ApoA-IV expression is regulated by two common enhancers. Biochim Biophys Acta. 1997;1348:299–310. doi: 10.1016/s0005-2760(97)00071-4. [DOI] [PubMed] [Google Scholar]
- 11.Chen D, Lepar G, Kemper B. A transcriptional regulatory element common to a large family of hepatic cytochrome P450 genes is a functional binding site of the orphan receptor HNF-4. J Biol Chem. 1994;269:5420–5427. [PubMed] [Google Scholar]
- 12.Chen D, Park Y, Kemper B. Differential protein binding and transcriptional activities of HNF-4 elements in three closely related CYP2C genes. DNA Cell Biol. 1994;13:771–779. doi: 10.1089/dna.1994.13.771. [DOI] [PubMed] [Google Scholar]
- 13.Stoffel M, Duncan SA. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4alpha regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci USA. 1997;94:13209–13214. doi: 10.1073/pnas.94.24.13209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Späth GF, Weiss MC. Hepatocyte nuclear factor 4 expression overcomes repression of the hepatic phenotype in dedifferentiated hepatoma cells. Mol Cell Biol. 1997;17:1913–1922. doi: 10.1128/mcb.17.4.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Combs TP, et al. A transgenic mouse with a deletion in the collagenous domain of adiponectin displays elevated circulating adiponectin and improved insulin sensitivity. Endocrinology. 2004;145:367–383. doi: 10.1210/en.2003-1068. [DOI] [PubMed] [Google Scholar]
- 16.Matsuda M, et al. Role of adiponectin in preventing vascular stenosis. The missing link of adipo-vascular axis. J Biol Chem. 2002;277:37487–37491. doi: 10.1074/jbc.M206083200. [DOI] [PubMed] [Google Scholar]
- 17.Cereghini S. Liver-enriched transcription factors and hepatocyte differentiation. FASEB J. 1996;10:267–282. [PubMed] [Google Scholar]
- 18.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo CJ, et al. A transcriptional hierarchy involved in mammalian cell-type specification. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- 20.Odom DT, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffo G, et al. HNF4 and HNF1 as well as a panel of hepatic functions are extinguished and reexpressed in parallel in chromosomally reduced rat hepatoma-human fibroblast hybrids. J Cell Biol. 1993;121:887–898. doi: 10.1083/jcb.121.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto T, et al. SREBP-1 interacts with hepatocyte nuclear factor-4 alpha and interferes with PGC-1 recruitment to suppress hepatic gluconeogenic genes. J Biol Chem. 2004;279:12027–12035. doi: 10.1074/jbc.M310333200. [DOI] [PubMed] [Google Scholar]
- 23.Winckler W, et al. Association testing of variants in the hepatocyte nuclear factor 4alpha gene with risk of type 2 diabetes in 7,883 people. Diabetes. 2005;54:886–892. doi: 10.2337/diabetes.54.3.886. [DOI] [PubMed] [Google Scholar]
- 24.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 25.Saif-Ali R, Harun R, Al-Jassabi S, Wan Ngah WZ. Hepatocyte nuclear factor 4 alpha P2 promoter variants associate with insulin resistance. Acta Biochim Pol. 2011;58:179–186. [PubMed] [Google Scholar]
- 26.Hani EH, et al. A missense mutation in hepatocyte nuclear factor-4 alpha, resulting in a reduced transactivation activity, in human late-onset non-insulin-dependent diabetes mellitus. J Clin Invest. 1998;101:521–526. doi: 10.1172/JCI1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lo KA, et al. Genome-wide profiling of H3K56 acetylation and transcription factor binding sites in human adipocytes. PLoS ONE. 2011;6:e19778. doi: 10.1371/journal.pone.0019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt D, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.