Abstract
The molecular mechanisms governing self-renewal, differentiation, and lineage specification remain unknown. Transcriptional profiling is likely to provide insight into these processes but, as yet, has been confined to “static” molecular profiles of stem and progenitors cells. We now provide a comprehensive, statistically robust, and “dynamic” analysis of multipotent hemopoietic progenitor cells undergoing self-renewal in response to interleukin-3 (IL-3) and multilineage differentiation in response to lineage-affiliated cytokines. Cells undergoing IL-3-dependent proliferative self-renewal displayed striking complexity, including expression of genes associated with different lineage programs, suggesting a highly responsive compartment poised to rapidly execute intrinsically or extrinsically initiated cell fate decisions. A remarkable general feature of early differentiation was a resolution of complexity through the downregulation of gene expression. Although effector genes characteristic of mature cells were upregulated late, coincident with morphological changes, lineage-specific changes in gene expression were observed prior to this, identifying genes which may provide early harbingers of unilineage commitment. Of particular interest were genes that displayed differential behavior irrespective of the lineage elaborated, many of which were rapidly downregulated within 4 to 8 h after exposure to a differentiation cue. These are likely to include genes important in self-renewal, the maintenance of multipotentiality, or the negative regulation of differentiation per se.
Recent developments in both adult and embryonic stem cell biology have fueled excitement about the prospects for cell-based therapies in a number of clinical situations (33). Key to the long-term success of these strategies will be a molecular understanding of both the nature of “stemness” and the processes by which stem cells adopt specific cell fates. Stem and progenitor cells are characterized by their capacity to renew themselves and to give rise to more differentiated progeny. Although the extent of self-renewal and differentiation potentials varies between different classes of stem and progenitor cells and also between tissues, the fundamental problem of balancing these two processes is a common one (8).
The hemopoietic system is perhaps the most well characterized in terms of its stem and progenitor cell biology. Hemopoietic stem cells (HSC) and their more committed downstream progenitors sustain the production of at least eight different blood cell lineages throughout life. How the progeny of a stem cell selects a particular one of these multiple lineage fates remains unknown but is probably controlled by a set of regulatory rules that also coordinate proliferation, quiescence, and programmed cell death. Leukemia or aberrant hemopoeisis are major clinical consequences of subversion of the rules that govern stem and progenitor cell behavior (8).
Since the phenotype of any given cell is ultimately the product of the genes that it expresses or has expressed during the course of its lifetime, one approach to addressing how self-renewal and differentiation are regulated is to describe the complete gene expression programs of self-renewing and differentiating cells.
A number of molecular profiles of various classes of hemopoietic cells have been reported in the literature. A variety of different technical strategies have been used to characterize these cells including differential library construction, differential display, suppression PCR, as well as nylon and glass cDNA arrays and oligo-based Affymetrix GeneChip. For example, Phillips, et al. (37) generated HSC-enriched subtracted-cDNA libraries from fetal liver HSC by using PCR- and non-PCR-based methods. Terskikh at al. (47) generated a subtracted cDNA library from lineage-depleted adult bone marrow HSCs and mature blood cells and screened for HSC-specific genes. Park et al. (34), by using a similar approach, compared the expression profiles of HSC with multipotent progenitor cells by using cDNA macro (nylon)- and micro (glass)-arrays. Two groups (22, 40) have reported global expression profiling of purified HSCs on oligonucleotide arrays and compared these profiles to those of both neural and embryonic stem cells. Most recently, Li and coworkers (1) have compared different classes of prospectively isolated hemopoietic progenitors. Given limitations in cell numbers, target amplification has been prevalent, and sample replication has been minimal. In addition, most studies have provided only static snapshots of differently isolated cells. What has been lacking thus far is a dynamic analysis of hemopoietic cells undergoing self-renewal and differentiation down a number of different blood lineages.
Primitive hemopoietic cells cannot easily be maintained and expanded in vitro over a long period of time. In order to facilitate the investigation of the molecular mechanisms controlling hemopoeisis, in vitro hemopoietic cell line models have been developed. These included the multipotent FDCP-mix cells, which were derived from long-term cultures of mouse bone marrow (44). FDCP-mix cells are nonleukemic, have a normal diploid karyotype, and in early passage formed spleen colonies and displayed radioprotective ability when transplanted into irradiated animals. In vitro, FDCP-mix cells can self-renew and undergo multilineage differentiation in response to physiological cues such as stroma or growth factors. High concentrations of interleukin-3 (IL-3) stimulate self-renewal and the maintenance of the blast-like morphology. However, when cultured in the absence of high levels of IL-3 and in the presence of various other hemopoietic growth factor combinations, FDCP-mix cells can develop into many different myeloid cell lineages. These include erythroid, monocytic, neutrophilic, megakaryocytic, basophilic, and eosinophilic cells (18, 19, 38; C. Heyworth, unpublished observations). Thus, more than 90% of the colonies formed in soft agar are of a mixed composition; the cloning efficiency of FDCP-mix is 10 to 20% in soft-gel assays and 50% in liquid culture. Here we report the global gene expression profile of this multipotential progenitor cell line both under conditions of self-renewal and during multilineage differentiation.
The use of this cell model has allowed us to obtain sufficient cell numbers to analyze multilineage differentiation outputs at a number of time intervals from a common starting point. It has also allowed for triplication of the studies and obviated the need for probe amplification, which has facilitated the acquisition of a statistically robust data set. We first validate our experimental approach and cellular model system both by describing the behavior during differentiation of a number of genes whose expression characteristics within hemopoeisis are already known and by confirming for select genes the relative expression values, derived by microarray, by using real-time PCR analysis. Next, we analyze specific features of erythroid and neutrophil differentiation and the more common features of myeloid differentiation in general. Finally, we examine the nature of the multipotent state through analysis of FDCP-mix cells under conditions of self-renewal.
MATERIALS AND METHODS
Culture and analysis of FDCP-mix cells.
The cells were routinely cultured in Fischer's medium supplemented with 20% (vol/vol) horse serum and 2% (vol/vol) IL-3-conditioned medium (IL-3-CM) (23). Although the FDCP-mix cells were maintained routinely in IL-3-CM, all subsequent experiments were performed in the presence of recombinant murine IL-3 (R&D Systems, Abingdon, United Kingdom). Differentiation of the FDCP-mix cells was performed as previously described (17, 18). The growth factor concentrations used were as follows: condition 1, Epo (1 U/ml; Boehringer Mannheim), hemin (0.2 mM; Sigma), and IL-3 (0.05 ng/ml; R&D); condition 2, granulocyte colony-stimulating factor (G-CSF; 10,000 U/ml; Amgen) and stem cell factor (SCF; 100 ng/ml; Amgen); condition 3, IL-3 (0.05 ng/ml) and lung-conditioned medium (10% [vol/vol]); and condition 4, Tpo (50 ng/ml; R&D), Epo (1 U/ml), IL-6 (25 U/ml; R&D), and SCF (100 ng/ml). For colony assays, cells were washed twice and then resuspended in plating mixture containing recombinant IL-3 (10 ng/ml), fetal calf serum (20% [vol/vol]), bovine serum albumin (1 mg/ml), Noble agar (9% [wt/vol]), and Iscove medium. Routinely, 103 cells/ml were plated and grown for 7 days prior to scoring. At all of the time points, FDCP-mix cells, maintained in self-renewing growth factor conditions, were plated as a control for the variations in culture conditions. These cells had a plating efficiency of between 10 and 20%. Cell cytospins were stained with May-Grünwald-Giemsa and o-dianisidine (Koch-Light Laboratories) to determine cellular morphology. For analysis of cell surface markers, cells were washed and resuspended in fetal calf serum (2% [vol/vol]) in phosphate-buffered saline and incubated first with Rat monoclonal antibodies (Pharmingen) directed against Ter-119, Gr-1, or Mac-1, followed by incubation with fluorescein isothiocyanate-labeled anti-rat immunoglobulin G (Pharmingen). Stained samples were analyzed on a FACS-Vantage (Becton Dickinson), and cell cycle analysis was performed as described previously (16).
cRNA synthesis and hybridization to oligonucleotide probe arrays.
Total cellular RNA was isolated with TRIzol (Invitrogen, San Diego, Calif.) as recommended by the manufacturer. RNA concentration and integrity were determined by spectrophotometry and gel electrophoresis. Aliquots of the RNA samples were reserved for reverse transcription-PCR (RT-PCR) and real-time PCR analysis, whereas the bulk of the sample was labeled and hybridized to MG-U74Av2 Affymetrix GeneChip arrays (interrogating 12,488 genes and expressed sequence tags) as described previously (29). Raw fluorescence intensity data were acquired with Affymetrix microarray software (MAS 4.2). MAS 5.0 was used to calculate detection calls. All experiments were performed at least in triplicate.
Statistical evaluation of replicate experiments.
Fluorescence intensity data were normalized by using dChip v1.0 (27, 28) and model-based expression indices (MBEI) calculated by using the PM/MM-difference model of Li and Wong (27, 28). Array outliers generated during these procedures were replaced by imputed values by using a k nearest-neighbor average. Differentially expressed genes were identified by significance analysis of microarrays (SAM), which is a nonparametric, permutation-based method (48). Lists of differentially expressed genes were derived at the minimum false discovery rate (FDR) (2). In addition, the lists were filtered for genes that displayed both a minimum difference of 100 between the highest and lowest expression values obtained and a minimum fold change of 2 between any two time points. The data for Affymetrix control probes sets were removed prior to analysis. Spiked control cRNAs were not used consistently. Cluster analysis of differentially expressed genes was performed and visualized by using Genesis (45), Cluster, and TreeView (7).
RT-PCR analysis.
Total RNA was extracted from cell pellets with TRIzol according to the manufacturer's recommendation and treated with RNase-free DNase I (Promega). Random hexamer-primed cDNA synthesis was performed with Superscript II reverse transcriptase (Invitrogen) according to manufacturer's specifications. RT-PCR was performed in a Perkin-Elmer GeneAmp PCR system 9700 with specific primers for 35 cycles. Real-time PCR was performed by the Quantitect SYBR Green PCR system (Qiagen) with an ABI Prism 7700 sequence detector. For each time point in each differentiation series, 500 ng of DNase-treated total RNA was reverse transcribed with SuperScript II reverse transcriptase by using random hexamer priming. Then, 50-pg equivalents of the resulting cDNAs were used in each subsequent real-time quantitative PCR (RQ-PCR). Oligonucleotide PCR primers were designed to have optimal annealing temperatures of ∼55°C and to generate PCR products of between 150 and 750 bp. For each individual PCR, the SYBR Green fluorescence values for all of the time points in each series were measured at the cycle when only one sample in the set had reached a fluorescence value of >10 standard deviations above baseline. The fluorescence of this sample (in which the PCR template would be most abundant) was arbitrarily given a “value” of 10, and those of the other samples in the series were assigned values relative to this by simple division. RT reactions were performed at least in duplicate, and the RQ-PCR scores provided for any given template-primer combination represents an arithmetic mean of these values. A similar “normalization” procedure was used for the corresponding microarray hybridization signal intensities, thereby allowing relative RQ-PCR fluorescence intensities and microarray hybridization signal intensities to be plotted on the same scale.
RESULTS
Experimental system.
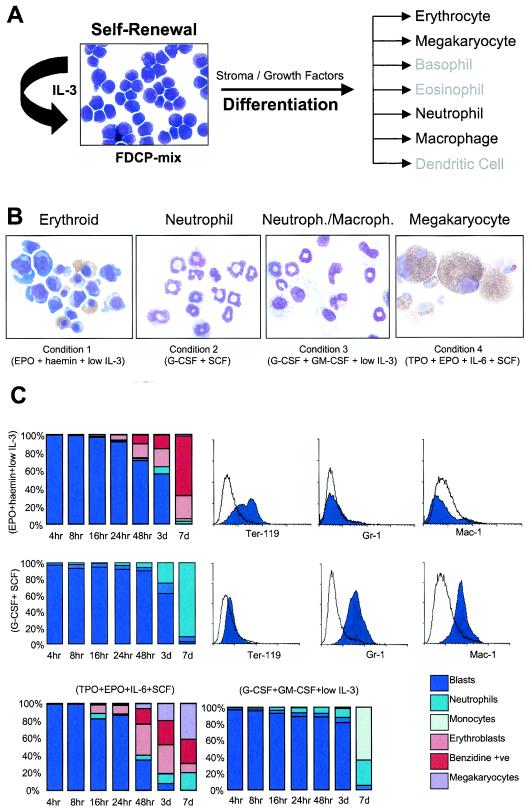
Self-renewing FDCP-mix cells were plated in triplicate under four different culture conditions that produced a variety of distinct developmental fates (Fig. 1). Over the first 16 h, cell numbers did not markedly increase and cellular viability was maintained at >92%. After this, the cells began proliferating and during, for example, erythroid development, a 14-fold increase in cell number was observed over the 7-day period. Conditions 1 and 2 produced >95% pure populations of erythroid and neutrophilic cells, respectively, whereas the other conditions produced mixtures of neutrophils and monocytes (condition 3) and erythroid cells and megakaryocytes (condition 4). Changes in morphology were reflected in the ability of the cell population to maintain its colony-forming potential (not shown). At 48 h the colony-forming potential had decreased by 20 to 50% of the initial value, this further decreased by day 3 and by the time cells exhibit a fully mature phenotype the colony-forming potential is negligible. Cell cycle analysis revealed that at time zero ca. 25 to 30% of cells were in S phase. During the course of the differentiation cultures there was a decrease in the proliferative activity rate that was similar in all of the different conditions (data not shown).
FIG. 1.
Experimental system. (A) Photomicrograph of self-renewing FDCP-mix cells and a scheme indicating their differentiation potentials in vitro. (B) Typical morphologies of cells produced under the cytokine conditions indicated. Day 7 cells were harvested, cytospun, and stained with May-Grünwald-Giemsa and o-dianisidine. Staining for acetyl cholinesterase was used to identify megakaryocytic cells. (C) After being stained the cells were morphologically assessed. The percentages of different cell types at various time points during differentiation are shown on the bar charts. The accompanying histograms present the results of fluorescence-activated cell sorting analyses of cell surface marker expression at T0 (unfilled) and day 7 (filled) during erythroid and neutrophilic differentiation, obtained with the antibodies indicated, as well as isotype controls (not shown).
Array analysis and validation.
In order to obtain a statistically robust data set, considerable attention was paid to experimental design with respect to sample replication, data acquisition, and subsequent mathematical treatment. Cultures sampled at 0, 4, 8, 16, 24, 48, 72, and 168 h of differentiation were analyzed by using Affymetrix GeneChip MG-U74Av2 arrays. Image files were inspected for artifacts, and chips with abnormal percentages of present calls or median intensities were excluded from further analysis. For three replicates per sample, CEL files were normalized and analyzed by using dChip (27, 28), and the resultant MBEIs were exported. M-versus-A plots (6) were then produced for all chips analyzed; those displaying abnormal or skewed distributions were discarded (see Appendix). The remaining chips were then subjected to reanalysis by dCHIP, and the resultant MBEIs were separately analyzed for each of the four differentiation pathways by SAM (version 1.13 [48]) to determine differentially expressed genes; gene lists corresponding to the minimum FDR observed were generated and filtered to exclude genes exhibiting <2-fold changes or signal differences between values of <100. Gene lists produced by using different versions of the dCHIP algorithm were compared, and the version yielding the best fit to the expected gene expression for genes whose behavior is known was selected for further analysis.
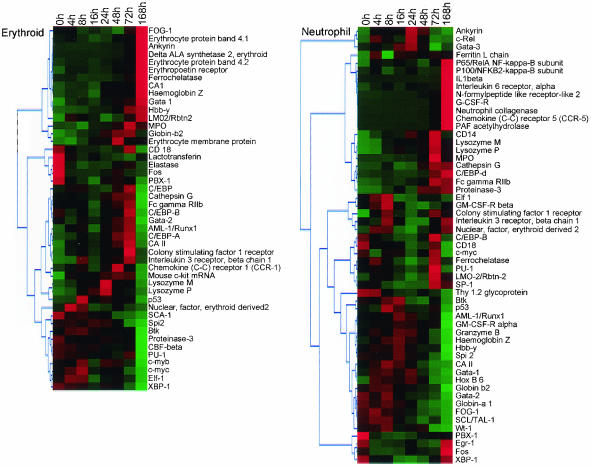
An analysis of the expression behavior, ascertained from the microarray, of genes with known or predictable expression within the hemopoietic differentiation hierarchy is presented in Fig. 2. Broadly, erythroid-affiliated genes (e.g., FOG-1, GATA-1, EPO-R, Globin, and CA1) were upregulated during erythroid differentiation, whereas neutrophil-affiliated genes were either not expressed, were unchanged, or were downregulated in the same differentiation series. An inverse pattern of gene expression was observed in the neutrophil differentiation series.
FIG. 2.
Validation of microarray results through analysis of the behavior of well characterized hemopoiesis-affiliated marker genes. Genes were selected for analysis based on (i) having a known profile of expression in hemopoietic cells, (ii) representation on the genechip, and (iii) exhibiting differential expression during differentiation of FDCP-mix. A diagrammatic interpretation of gene expression at different time points of erythroid (upper panel) and neutrophil (lower panel) differentiation is shown; green and red represent underexpression and overexpression, respectively, relative to the median, and genes exhibiting similar temporal behavior are clustered together. Details of the procedures by which differentially expressed genes were identified and subsequently clustered are explained in Materials and Methods and in the legend to Fig. 4.
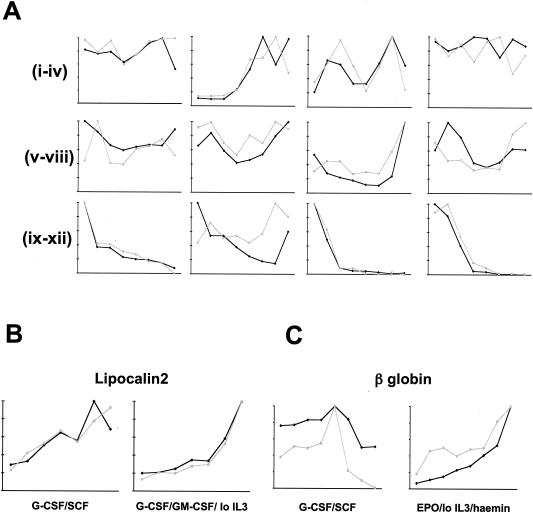
RQ-PCR was used to measure relative transcript abundances as an additional secondary screen. Transcripts were selected for this analysis with the aim of representing a broad cross-section of genes whose microarray signals varied in all possible permutations across each series. A comparison of the microarray and RQ-PCR data is presented in Fig. 3. Taken together, these studies encourage confidence in the array analysis and in FDCP-mix as a model of myeloid development.
FIG. 3.
Comparison of gene expression assayed by microarray and by RQ-PCR. For each series and for each gene analyzed, the values of both parameters at each time point were normalized relative to that with the highest value, which was arbitrarily assigned as 10. Microarray signal intensity is plotted in black, and RQ-PCR fluorescence in gray. (A) Analysis of arbitrarily selected genes during neutrophil differentiation of FDCP-mix cells supported by G-CSF plus SCF. Subpanels (from left to right): i, CD14; ii, MPO; iii, c-kit; vi, IL-3 receptor α chain; v, HPRT; vi, GAPDH; vii, schlafen 2; viii, β-catenin; ix, NDPP1; x, selenium-binding protein; xi, nephroblastoma overexpressed; xii, AEG-1 (acidic epididymal glycoprotein 1). (B) Analysis of lipocalin 2 (neutrophil marker) during neutrophil differentiation of FDCP-mix cells supported by G-CSF plus SCF (i) and during myelomonocytic differentiation of FDCP-mix cells supported by G-CSF plus GM-CSF plus low IL-3 (ii). (C) Analysis of β globin (erythroid marker) during neutrophil differentiation of FDCP-mix cells supported by G-CSF plus SCF (i) and during erythroid differentiation of FDCP-mix cells supported by EPO plus low IL-3 plus hemin (ii) as indicated.
Overview of gene expression changes.
Summing across all of the four differentiation series, the total number of genes exhibiting statistically significant differential expression was 3,560. In pure erythroid- and neutrophil-producing conditions, 1,721 and 2,108 genes, respectively, displayed differential expression (Table 1). Compared to conditions that do not produce erythroid cells (i.e., conditions 2 and 3 [see Materials and Methods]), 751 genes display differential behavior that is uniquely associated with erythroid commitment and differentiation; a similar analysis identified 1,238 genes as uniquely associated with neutrophil development. An investigation of the overall structure of the data revealed that, as expected, samples cluster according to time early in differentiation, regardless of lineage, whereas samples later segregate in a manner that reflects both lineage identity and the developmental relationships between lineages (see Appendix for additional details).
TABLE 1.
Summary of the number of genes identified as differentially expressed during FDCP-mix differentiationa
| Culture condition | Total no. of differentially expressed genes | Minimum FDR |
|---|---|---|
| Epo + hemin | 1,721 | 0.01894021 |
| G-CSF + SCF | 2,108 | 0.010401 |
| G-CSF + GM-CSF | 766 | 0.09393892 |
| Tpo + Epo + SCF | 2,070 | 0.01751727 |
| Total (unique counts) | 3,560 |
The number of differentially expressed genes in each differentiation pathway is shown, together with the minimum FDR used in each case. Also indicated is the total number of genes differentially expressed in at least one of the conditions.
Erythroid and neutrophil differentiation.
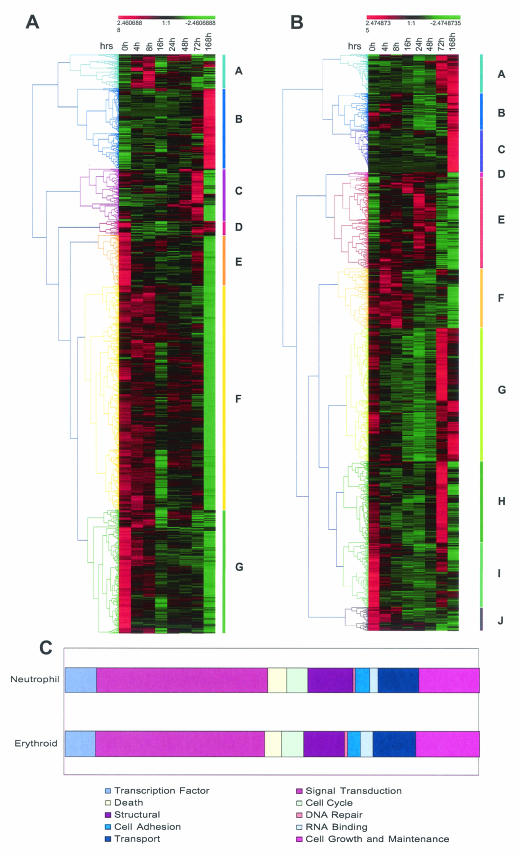
Genes that were differentially expressed in Epo-hemin-supported and G-CSF-SCF-supported differentiation series were hierarchically clustered (Fig. 4A) and classified according to function by using simplified ontologies (Fig. 4B). The gene lists include potentially novel lineage-specific genes and markers in the erythropoietic and neutrophilic pathways, as well as genes responsive to signaling induced by the cytokine combinations used. The most predominant feature of erythroid differentiation is the downregulation of gene expression. The limited clusters that show upregulation do so primarily at late time points in the differentiation series and comprise known markers of erythroid cells; examples include EpoR, Duffy, and rhesus blood group antigens; erythroid-specific forms of ankyrin and spectrin; globins; carbonic anhydrase; ALAS; and erythrocyte band protein 7.2. The erythroid-affiliated transcription factor GATA-1 also appears in the late erythroid “B” cluster. Other transcriptional regulators of note that cocluster with GATA-1 include Krüppel-like factor basic 3, the sine oculis- and Iroquois-related homeobox homologs Six3 and Irx3, the stem and erythroid cell-associated Lim-only protein Lmo2, and the polycomb group even-skipped homolog.
FIG. 4.
Analysis of differentially expressed genes in erythroid (A) and neutrophil (B) differentiation. The adjusted expression levels (mean = 0; variance = 1) at the indicated time points were hierarchically clustered by using uncentered Pearson correlation and average linkage clustering. The range of relative expression levels from lowest to highest is represented by the green and red shading, respectively. Colored bars along each graph highlight prominent gene clusters (A to G for erythrocytes and A to J for neutrophils) corresponding to the color-matched branches of the tree (45). (C) Bar charts showing the distribution of all differentially expressed genes according to function. Genes were annotated by using the Simplified Ontology tool in GeneSpring. It should be noted that this analysis is partially limited by the fact that annotations are available for only a subset of genes (>70%) represented on the genechip. Also, the criteria used to assign functional categories may result in nonredundant classification of genes (i.e., one gene may be assigned twice in different categories).
The two peaks of maximal gene expression seen (at 3 and 7 days) presumably relate to the early and late erythroid forms predominating at these time points in the culture. Interestingly, GATA-2, whose expression is thought to precede and possibly activate GATA-1, features in this earlier “C” cluster. Although analysis of the erythroid differentiation of FDCP-mix is limited by the varying and often incomplete extent of maturation obtained, these results indicate that the system can be used to examine the sequence of early molecular events associated with not only erythroid commitment but also the onset of maturation and terminal differentiation. Consistent with this, it is apparent that significant changes in gene expression occur prior to the appearance of phenotypically identifiable erythroid lineage cells. Although some of these changes in gene expression are also seen in other differentiation pathways, some are unique to erythropoiesis and may both constitute a “class predictor” for this pathway and may also be intimately associated with commitment per se. However, robust establishment of a “commitment signature” by class prediction approaches will require a substantially larger training and test set than is provided by the current analysis. The late bias in major gene expression changes is consistent with the view that whereas marking of erythroid territory may occur before erythroid commitment, major epigenetic and transcriptional changes must occur late in erythroid differentiation to produce the dramatic increase in expression of erythroid-associated luxury proteins that occurs in these stages.
In contrast, neutrophil differentiation in this model is characterized by rather more different clusters of behavior and perhaps most notably by clusters that display a bimodal pattern of gene expression being expressed at T0 and then downregulated before being upregulated at the end of the differentiation series. This may reflect the phenomenon of “preview” described by Iscove and colleagues (personal communication), a default myelomonocytic as opposed to erythroid groundstate in self-renewing FDCP-mix cells, or both. In terms of clusters of genes that display upregulation during neutrophil differentiation, classic myeloid-associated genes include MPO, lysozyme, cathepsin G, neutrophil cytosolic factor 1, and the receptors for granulocyte-macrophage colony-stimulating factor (GM-CSF) and G-CSF, as well as neutrophil-affiliated transcriptional regulators such as C/EBPδ; RARα is also upregulated as previously documented for neutrophil differentiation of FDCP-mix and other cell types (50). Similarly, Mef2a, originally described as a myogenic-specific regulator, is upregulated during neutrophilic differentiation of FDCP-mix consistent with a role in myeloid differentiation demonstrated by studies of Mef2 family protein function in human HL-60 cells (43). A number of other regulatory molecules with as yet uncharacterized roles in neutrophil differentiation are present in these clusters, including Raf1, ETS2 (a homolog of EVI1), EVI5, LRF1 (also known as pokemon), and the microphthalmia-associated transcription factor Mitf1 to name but a few.
Signatures of self-renewal and differentiation.
The analysis of dynamic transcriptional profiles for multipotent progenitors undergoing differentiation to specific cell types such as neutrophils or erythroid cells is useful in identifying lineage-specific gene expression characteristics. In contrast, by comparing profiles associated with different differentiation outcomes and identifying genes whose expression is modulated in all cases, one may gain insight into general features of differentiation that are independent of the particular lineage specified. This analysis may also reveal genes whose expression is preferentially associated with self-renewal and the maintenance of multipotentiality.
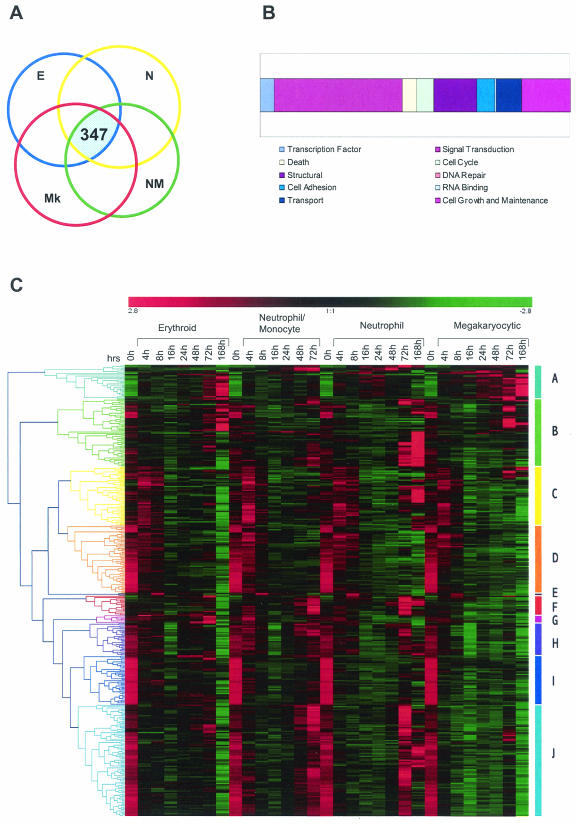
We compared the lists of differentially expressed genes from each of the differentiation conditions tested (see the experimental scheme in Fig. 1B) and identified 347 genes whose expression was modulated under all four differentiation regimes (Fig. 5A). These genes were categorized according to function as shown in Fig. 5B.
FIG.5.
Analysis of genes that are differentially expressed under all differentiation conditions. Gene expression levels for the four differentiation pathways analyzed (E, erythroid; N, neutrophil; NM, neutrophil/monocyte; Mk, megakaryocyte) were separately filtered by SAM as shown in Table 1. (A) The intersection of these four lists identifies 347 genes that are differentially expressed under all of the conditions tested. (B) Simplified ontologies were generated as in Fig. 4. (C) Hierarchical clustering of the 347 genes was performed as indicated in the legend for Fig. 4. Note that although all genes are differentially expressed in all four pathways they do not necessarily show similar expression profiles. For example, most of the genes in cluster D are similarly downregulated during differentiation in all pathways. However, genes belonging to cluster J show initial downregulation for all pathways and are upregulated only in neutrophil/monocyte and neutrophilic pathways at later time points.
Sets of genes exhibiting similar temporal expression characteristics during differentiation were identified by using hierarchical clustering (Fig. 5C). A number of different expression patterns are represented in this analysis that include genes that exhibit different patterns of behavior in different lineages. However, the most consistent pattern of behavior, accounting for ca. 25% of the genes, is seen in clusters D and I in Fig. 5C. Genes in these clusters are rapidly downregulated as multipotential cells undergo differentiation regardless of the cell lineage elaborated. Many functional classes of molecules are represented in this cluster, which may include genes important in self-renewal, the maintenance of multipotentiality, or the negative regulation of differentiation per se. Of these 83 genes, 61 are known, and these are listed in Table 2.
TABLE 2.
Annotated list of commonly downregulated genes in all four differentiation pathwaysa
| Gene | Accession no. | Definition |
|---|---|---|
| Ifi205 | M74123 | Interferon-activated gene 205 |
| Scya3 | NM_011337 | Small inducible cytokine A3 (MIP-1α) |
| Il4 | NM_021283 | IL-4 |
| Siat1 | NM_009175 | Sialyltransferase 1 (β-galactoside α-2,6-sialyltransferase) |
| Bnip3 | NM_009760 | BCL2/adenovirus E1B 19-kDa interacting protein 1 (NIP3) |
| Pfkp | NM_019703 | Phosphofructokinase, platelet |
| Gp49a | NM_008147 | Glycoprotein 49A |
| Map3k1 | AF117340 | Mitogen-activated protein kinase kinase kinase 1 |
| Net1 | NM_019671 | Neuroepithelial cell transforming gene 1 |
| Zfp106 | AWO48037 | Zinc-finger protein 106 |
| Ccr4 | AF199491 | Carbon catabolite repression 4 homolog (Saccharomyces cerevisiae) |
| Ctsc | NM_009982 | Cathepsin C |
| Ermelin (pending) | NM_139143 | Endoplasmic reticulum membrane protein |
| Dag1 | BC007150 | Dystroglycan 1 |
| Rnf19 | NM_013923 | Ring finger protein (C3HC4 type) 19 (XY body protein) |
| Rcn | NM_009037 | Reticulocalbin |
| Sc4mol | NM_025436 | Sterol-C4-methyl oxidase-like |
| Pbx1 | NM_008783 | Pre-B-cell leukemia transcription factor 1 |
| Hmgcr | M62766 | 3-Hydroxy-3-methylglutaryl-coenzyme A reductase |
| Lce (pending) | NM_130450 | Long-chain fatty acyl elongase |
| Hk2 | NM_013820 | Hexokinase 2 |
| Dhcr7 | NM_007856 | 7-Dehydrocholesterol reductase |
| Ak4 | NM_009647 | Adenylate kinase 4 |
| Ndr1 | NM_010884 | N-myc downstream regulated 1 |
| Ptgs2 | NM_011198 | Prostaglandin-endoperoxide synthase 2 (cyclooxygenase 2) |
| Bhlhb2 | NM_011498 | Basic helix-loop-helix domain containing, class B2 (Dec1) |
| Snap23 | NM_009222 | Synaptosomal-associated protein, 23 kDa |
| Cpe | NM_013494 | Carboxypeptidase E |
| Gzmc | NM_010371 | Granzyme C |
| Rbl2 | NM_011250 | Retinoblastoma-like 2 |
| Capn5 | NM_007602 | Calpain 5 |
| Pa26 (pending) | AI843106 | p53-regulated PA26 nuclear protein |
| Calca | AF330212 | Calcitonin/calcitonin-related polypeptide, alpha |
| Ak1 | NM_021515 | Adenylate kinase 1 |
| H2bfs | NM_023422 | H2B histone family, member S |
| Ccng2 | NM_007635 | Cyclin G2 |
| Tec | NM_013689 | Cytoplasmic tyrosine kinase, Dscr28C related (Drosophila) |
| S100a6 | NM_011313 | S100 calcium-binding protein A6 (calcyclin) |
| Serpinf1 | NM_011340 | Serine (or cysteine) proteinase inhibitor, clade F |
| Glb1 | NM_009752 | Galactosidase, beta 1 |
| Cd53 | NM_007651 | CD53 antigen |
| H1f0 | NM_008197 | H1 histone family, member 0 |
| Daf1 | NM_010016 | Decay-accelerating factor 1 |
| Daf2 | NM_007827 | Decay-accelerating factor 2 |
| Vamp5 | NM_016872 | Vesicle-associated membrane protein 5 |
| Prdx4 | NM_016764 | Peroxiredoxin 4 |
| Aeg1 | NM_009638 | Acidic epididymal glycoprotein 1 |
| Sparc | NM_009242 | Secreted acidic cysteine-rich glycoprotein |
| Hsd17b10 | NM_016763 | Hydroxysteroid (17-β) dehydrogenase 10 |
| Anxa2 | NM_007585 | Annexin A2 |
| Fkbp1a | NM_008019 | FK506-binding protein 1a (12 kDa) |
| Nov | NM_010930 | Nephroblastoma overexpressed gene |
| Slfn2 | NM_011408 | Schlafen 2 |
| Spint1 | NM_016907 | Serine protease inhibitor, Kunitz type 1 |
| Emr1 | U66888 | Epidermal growth factor-like module containing, mucin-like, hormone receptor-like sequence 1 |
| Enah | NM_010135 | Enabled homolog (Drosophila) |
| Scd1 | NM_009127 | Stearoyl-coenzyme A desaturase 1 |
| Glk | NM_016905 | Galactokinase |
| Reps1 | NM_009048 | RalBP1-associated Eps domain-containing protein |
| Bcl2 | NM_009741 | B-cell leukemia/lymphoma 2 |
| Tjp1 | NM_009386 | Tight junction protein 1 |
| Sqstm1* | NM_011018 | Sequestosome 1 |
| Fgf3* | NM_008007 | Fibroblast growth factor 3 |
| Elf1* | NM_007920 | E74-like factor 1 |
| Tie1* | NM_011587 | Tyrosine kinase receptor 1 |
| Ptgir* | D26157 | Prostaglandin I receptor (IP) |
| Phemx* | NM_020286 | Pan-hematopoietic expression |
| Cd9* | NM_007657 | CD9 antigen |
| Aqp9* | NM_022026 | Aquaporin 9 |
| Igfbp7* | NM_008048 | Insulin-like growth factor binding protein 7 |
| Wbscr5* | NM_022964 | Williams-Beuren syndrome chromosome region 5 homolog (human)/NTAL |
The first 61 genes belong to clusters D and I in Fig. 5C. Also included in this table are an additional 10 genes, indicated by an asterisk, sampled from the initial group of 347, which display downregulatory behavior when only the series that give rise to pure cell populations (neutrophil and erythroid) are examined. The names of genes, the GenBank accession numbers, and the definitions are given. Since the day 7 samples of the granulocyte-monocyte differentiation series failed at a technical level, only datum points up to 72 h were included. Inclusion of these data may therefore have resulted in an underestimate of this broad category of genes since cells at 72 h are not fully differentiated and retain some degree of self-renewal capacity as judged by colony-forming assays.
Of particular interest is the number of secreted molecules that feature in this list since these may provide insight into the extrinsic regulation of stem cell behavior. Investigation of fibroblast growth factor function on hemopoietic cell self-renewal and differentiation has been limited, but in other cell systems the use of basic fibroblast growth factors in serum-free conditions has produced extensive in vitro self-renewal in the absence of any marked signs of ageing (25). IL-4 enhances the survival of individually sorted FDCP-mix cells and also promotes the survival of the colony-forming cells over a 24- to 48-h time frame (unpublished observations). The expression profile of macrophage inflammatory protein 1α (MIP-1α) is consistent with its reported regulation of by IL-3. Moreover, the expression by FDCP-mix of the receptor for MIP-1α, CCR1, may provide for autocrine regulation of stem cell function.
The expression of NOV is also noteworthy, as is the rapidity of its downregulation of expression (within 4 h). Initially identified on the basis of its overexpression in nephroblastoma, it is part of the CCN family that also includes CTGF and the Wnt-induced secretory proteins (35). These molecules are now collectively termed insulin-like growth factor-binding proteins (IGFBPs) and may have both intracellular and extracellular functions. IGFBP7 (mac25) is also downregulated in the erythroid and neutrophil pathways and, although specific functions within hemopoietic cells for IGFBPs have not yet been described, these data suggest that such studies may be warranted.
AEG-1, normally produced by epididymal cells and involved in the fusion of egg and sperm, has been suggested to play a role in adaptive response to nerve injury (32). SPARC, a glycoprotein secreted by endothelial cells in response to injury (3), is also rapidly downregulated during FDCP-mix differentiation. The association between injury-responsive genes and stem cells may relate to a regenerative function. Alternatively, in the case of SPARC its expression in hemopoietic progenitors may relate to the common developmental propinquity of blood and endothelial lineages. The secreted protease inhibitors, SPINT1 and -2, inhibit hepatocyte growth factor (HGF) activity by inhibiting the action of HGF activator (31, 42). The HGF receptor c-Met is expressed on human hemopoietic progenitors, and SPINTs may modulate the activity of this or related signaling pathways.
Among the transcription factors that feature in this cluster, the ets family factor ELF-1 is hemopoiesis restricted and has been implicated as a regulator of SCL (9). The expression of the homeoprotein PBX1 and its subsequent downregulation may relate to its role in lymphoid malignancy where it is fused to the BHLH transcription factor E2A (26). Many BHLH proteins have been implicated in cell fate decisions. BHLHB2 (eip1/DEC1/stra13) is a widely expressed transcription factor that binds E-boxes but lacks the carboxy-terminal “WRPW domain” to which corepressors bind. BHLHB2 is involved in the control of the proliferation and differentiation of chondrocytes, nerve cells, fibroblasts, and T cells and is induced by hypoxia in several systems. Most notably, BHLHB2 is expressed in the suprachiasmic nucleus of the brain in a circadian fashion, and Dec1 and Dec2 are regulators of a mammalian molecular clock (20). CCR4/nocturnin exhibits circadian rhythmicity (11) and, interestingly, the long-term reconstitution capacity of stem cells is tightly regulated in a circadian manner (39).
Strikingly, a number of genes in this pan-downregulated cluster encode proteins such as SPARC, calcyclin, and Pbx1 that have been implicated as being either markers of, or pathogenetic in, various human malignancies. This reinforces the widely held view that many cancers can be regarded as diseases of stem cells (41). Finally, polymorphisms in the loci of a number of genes in this cluster have been linked with congenital hematological abnormalities in humans (such as chronic hemolytic anemia associated with adenylate kinase deficiency and Papillon Lefevre and Haim-Munk syndromes).
Groundstate analysis.
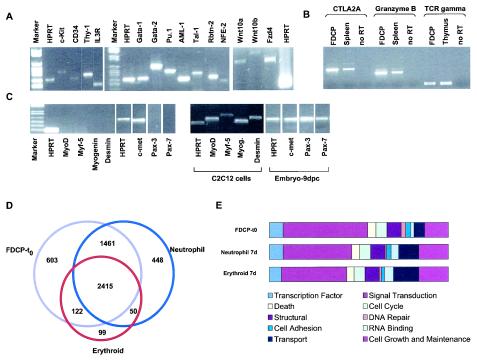
In contrast to most previous profiling experiments, which have compared static snapshots of different hemopoietic cell types, in this dynamic analysis of self-renewal and differentiation we have focused primarily on genes that exhibit differential expression over time. Nevertheless, a description of the static transcriptome of self-renewing FDCP-mix cells is of interest and would additionally afford comparison with the molecular signatures of other stem and progenitor cells. To gain an independent appreciation of the gene expression profile of self-renewing cells that does not rely on comparison to any other samples, we used a detection call algorithm (MAS 5.0) which simply evaluates the “presence” or “absence” of a transcript. The robustness of this method was assessed by RT-PCR, and sample data are shown in Fig. 6A. This analysis revealed considerable molecular complexity within stem cells, including the expression of a number of signaling systems, as well as components of different lineage-affiliated gene expression programs (multilineage priming). The complexity of gene expression afforded by this transcriptional priming may facilitate multipotent cells to execute cell fate decisions rapidly through integration of a broad range of extrinsic and intrinsic cues. We present below some evidence for multilineage priming and then briefly consider the range of signaling components expressed by self-renewing cells. Finally, we make some general comparisons with published results analyzing other stem cell systems.
FIG. 6.
Gene expression in self-renewing FDCP-mix cells. The detection calls algorithm of MAS 5.0 was used to identify expressed, i.e., “present,” genes. Only genes that were designated as present in three of three replicates were considered for further analysis. (A) Some of the genes reproducibly called “present” and considered to be hallmarks of stem cells and/or of hemopoietic differentiation programs were verified by RT-PCR; confirmation of expression of Wnt signaling components is also shown. (B) Analysis of lymphoid priming in FDCP-mix cells. Spleen and thymus samples provide positive controls, and reactions performed on FDCP-mix cells in the absence of RT are indicated. (C) Analysis of muscle-affiliated gene expression programs in FDCP-mix cells. Muscle cell line (C2C12) and day 9 developing embryo samples served as positive controls. (D) Overlap in gene expression profiles of self-renewing (FDCP-t0) and erythroid or neutrophil differentiated FDCP-mix cells (day 7), identifies 603, 99, and 448 genes as uniquely expressed in the self-renewing, erythroid, and neutrophilic compartments, respectively. Functional annotation of these genes is shown in panel E. Simplified ontologies were generated as described for Fig. 4.
(i) Multilineage priming.
Evidence for this is apparent in the list of genes expressed in uncommitted cells at T0, which includes myeloid-affiliated effectors (e.g., MPO, lysozyme, and globin) and lymphoid-affiliated genes (e.g., TCRγ, CTLA2A, ly64, granzyme B, and serpin 2A) (13, 14). These latter two have been previously shown to be expressed in FDCP-mix and serpin 2A is expressed in freshly isolated murine HSC (47). RT-PCR analysis of TCRγ, CTLA2A, and granzyme B further confirmed the presence of lymphoid-affiliated programs in FDCP-mix cells (Fig. 6B). Control of access of transcriptional machinery to cis-regulatory elements is required for priming drawing attention to chromatin regulatory factors such as a SW1/SNF-related molecules, polycomb group chromobox homologs, Drosophila enhancer of zeste homologs, HMG box 2, and nucleosome-binding protein 1, as well as histone deacetylase, coactivator, and corepressor molecules. Recently, the corepressor Ncor has been shown to play a critical role in self-renewal of neural stem cells (15). The molecular mechanisms underlying multilineage priming are not clear but may involve some of the transcriptional components that specify HSC during ontogeny. GATA-2, TAL-1/SCL, and RUNX-1, which have been demonstrated to play critical roles in the specification or elaboration of HSC during ontogeny, score as present in FDCP-mix.
Cooperative and antagonistic interactions between lineage-affliliated transcription factors are thought to underlie a number of lineage specification decisions in hemopoeisis (10). The presence in self-renewing cells of GATA factors, PU.1, and the C/EBP family may provide starting points for these various potential lineage-determining regulatory loops of gene activity. Like Runx1, PU.1 and the C/EBPs have been implicated in the regulation of expression of a number of cytokine receptors, thereby facilitating the responsiveness of cells to extrinsic cues.
Interestingly, multilineage transcriptional priming does not appear to extend to muscle-related programs. Thus, key myogenic regulatory genes such as myf5 and Mef2 score as absent, and this is additionally borne out by the RT-PCR analysis presented in Fig. 6C.
(ii) Signaling components.
In terms of ligands, FDCP-mix cells appear to express the maintenance/expansion factor, IL-6, as well as the inhibitory cytokines transforming growth factor β, which has activity on FDCP-mix cells, and MIP-1α (see above). The presence of Wnt 10a was confirmed by RT-PCR (see Fig. 6A). This analysis also established the expression of Wnt 10b and Frizzled 4. These results are consistent with a role of Wnt signaling in multipotent stem or progenitor cells.
In terms of receptors, the presence of the IL-3 receptor is predictable, and expression of the leptin receptor is consistent with its stimulation of HSC proliferation in vitro (12). IGF1 and IGF2 receptors are expressed and, although IGFs are not classical hemopoietic growth factors, they are known to promote erythrocytes and lymphocytes, as well as the proliferation of leukemic cells (51). The ephrin receptor functions on both multipotent and erythroid cells (46, 49) and is also expressed in endothelial cells. The developmental relationship between hemopoietic and endothelial cells may explain the presense of the thrombin receptor, which recently has been identified as a key stem cell-defining gene (40). A role for purinergic receptor P2X in hemopoietic progenitors has not been reported, although these receptors feature strongly in recent reports of stem cell-enriched genes. Weissman and coworkers have argued for substantial overlaps in neural and hemopoietic cell programs (47); perhaps P2X expression in FDCP-mix is a remnant of such a process.
Other receptors present include those associated with stroma and adhesion (SDR1, SDR2, and CD18), as well as with chemotaxis and homing (C3ar1, S1P3, and CXCR4). The expression of CXCR4 is indicative of the primitive nature of FDCP-mix cells, as is the expression of receptors such as AA4.1 and endoglin, which have been used to identify and purify murine HSC (4, 36).
A number of downstream intracellular signaling components are present in FDCP-mix, perhaps most notably STAT4, STAT5A and -B, STAT6, STATIP1, and JAK2. STAT5/JAK2 mediate both IL-3 family-associated signaling and signaling through the Epo and TPO receptors. STAT4 is involved in leptin signaling, and STAT6 is involved in IL-4-dependent signaling (24). STATIP1 features as a pan-stem cell-enriched gene in recent studies reporting a generalized molecular signature of stemness.
(iii) General comparisons.
The comparison of self-renewing and differentiated cells identifies 603 genes as specific to the self-renewal compartment (SRC genes; Fig. 6D). The simplified ontologies of these are presented in Fig. 6E. Comparison of SRC genes with those previously reported as enriched in hemopoietic stem and progenitor cells by Ivanova et al. (22) identified an overlap of 94 genes. Similar comparisons to genes identified as being enriched in HSC, neural stem cells, or embryonic stem cells compared to their differentiated counterparts (40) revealed substantial overlaps of 188, 196, and 157 genes, respectively. Similar comparisons can now be made with other data sets, including the Stem Cell Database (http://stemcell.princeton.edu), but all of these various comparisons will inevitably be constrained by the difficulties associated with data compatibility between different studies.
DISCUSSION
In examining the gene lists generated it is important to consider some key aspects of the approaches used. Importantly, we have used a minimum of three independent samples per datum point. This, together with the lack of need for probe amplification afforded by the large cell numbers obtainable with FDCP-mix system, makes for a statistical robustness lacking in many earlier studies. It is also important to emphasize that our stringent filtering using minimum FDRs and a twofold change criterion is likely to have excluded a number of genes whose expression is genuinely differential. Also, some of the transcripts detected, or their differential in levels between samples, may not be translated into proteins or, more importantly, protein activity; relatively low levels of expression and also small changes in expression level may play important roles in early cell fate outcomes (16).
Multilineage priming.
The data provide additional evidence in support of the multilineage priming hypothesis (8, 21). This model argues that, under conditions of self-renewal, multipotent cells may simultaneously prime several different programs of lineage-affiliated gene activity. It is presumed that this hemopoietic noise is functional and provides the building blocks of future commitment decisions. In such a scheme, commitment and differentiation involves not only consolidation of appropriate programs but also repression of programs no longer required or compatible for the particular pathway selected. The presence of both myeloid and lymphoid features within these cells, together with their cycling characteristics, might indicate that FDCP-mix cells represent an in vitro equivalent of murine multipotent progenitor cells (1), which coexpress lymphoid and myeloid programs. In contrast, HSC appear to exclusively or predominantly exhibit only myeloid as opposed to lymphoid priming (1, 5, 30). Although FDCP-mix cells appear primed for lymphopoiesis both at the level of chromatin configuration and gene expression (8), they have not yet been shown to exhibit lymphoid differentiation potential, perhaps reflecting an abnormality in this program or the requirement for as-yet-unidentified extrinsic cues. Recently, it has been argued that the multilineage priming phenomenon in stem cells may extend to the priming of nonhemopoietic genes and thereby potentially provide a mechanistic explanation for plasticity or transdifferentiation (1). In single cell analyses of FCDP-mix, we failed to find expression of the myogenic regulator myogenin or the muscle-associated effector protein Desmin (21). Similarly, our current array analyses do not provide strong support for expression of muscle-associated gene expression programs, perhaps consistent with a multipotent progenitor cell assignment for FDCP-mix. However, limited evidence for the expression of endothelial cell- and neural cell-associated genes is available within these data, and the overlap in genes expressed in self-renewing FDCP-mix cells with those deemed as NSC and ES cells enriched by others warrants further analysis.
Self-renewal and differentiation.
The data provide a number of potential novel markers of differentiation for the pathways examined. Future analyses of the cis-acting regulatory elements that control the expression of these genes, particularly those that display similar kinetics of expression, may yield insight into coordinated regulatory mechanisms for these specific pathways. Similarly, a number of genes exhibit rapid expression changes (within 4 to 8 h) in response to the addition of the various cytokine cocktails used. Since cytokine combinations as opposed to individual treatments were applied, it is hard to relate specific changes to specific agonists. Nevertheless, these data argue that additional experiments with individual agents will be useful.
As regards self renewal, The JAK/STAT signaling pathway is prevalent in FDCP-mix cells, and JAK2 is phosphorylated in response to IL-3 signaling in these cells (unpublished observations). The JAK/STAT pathway is involved in ES cell self-renewal and may be an important hallmark of “stemness” in general (40). Genes whose expression is downregulated in all FDCP-mix differentiation series are predicted to include self-renewal genes and secreted molecules such as NOV, with growth-altering properties, are particularly worthy of consideration in this regard. Genes involved in IL-3 signaling are also likely to be included here, as are genes that have been primed in the multipotent state and are affiliated with differentiation programs that were not elaborated under the conditions used. It is perhaps important to emphasize that we have examined “self-renewal” specifically in the context of the replicative or proliferative maintenance of multipotentiality of factor-dependent cells in vitro in response to IL-3. Whether self-renewal is orchestrated at a mechanistic level in the same way at different levels of the hemopoietic hierarchy or indeed in different classes of tissue-restricted stem cells and, additionally, in totipotent cells remains an open question.
The extent to which these studies will immediately reveal molecules involved in lineage commitment depends on the extent to which these decisions are normally orchestrated by stochastic or instructive processes. Nevertheless, the identification of gene expression changes that are (i) unique to the specific lineage outputs obtained and (ii) precede phenotypically recognizable lineage features should provide a good starting point for examining early events associated with commitment.
Acknowledgments
We thank Gillian May and Mel Greaves for critical reading of the manuscript and Tim Dexter for technical assistance with the M-versus-A plots.
Appendix
Quality control. M-versus-A plots provided a key measure of the quality of the data used in the present study. The plots for these data are shown in Fig. A1. The plots show the log ratios M = log2Eij − log2Emedi versus the abundance A = −(log2Eij + log2Emed i), where Eij is the expression level of the ith gene in the array j and Emed i is the median expression level of the ith gene across all 84 arrays in the experiment. Vertical lines indicate the positions of the 10th, 25th, 50th, 75th, and 90th percentiles, respectively. The plots were obtained by using dChip v1.0 (27, 28) derived MBEIs after 84 arrays were normalized on the feature level. Array outliers were replaced by SAM (48) k nearest-neighbor imputer, and low expression values were truncated to a floor value of 20. Dudoit et al. (6) applied similar plots to detect intensity-dependent biases in the log ratios. We used the M-versus-A plot to discern intensity-dependent differences between individual arrays and a control array, containing median expression levels for each probe set, and to assay the quality of the data and the success of the applied normalization.
FIG. A1.
M-versus-A plots of all samples used in the present study.
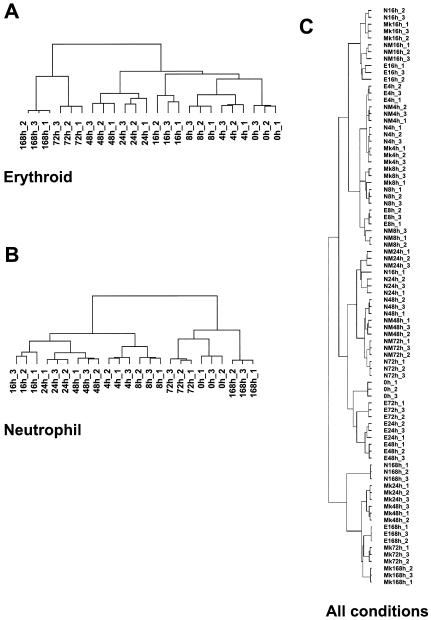
Clustering of replicates. Cluster analysis of individual sample replicates gives an appreciation of the overall “shape” for these data. The data for the erythroid (Fig. A2A) and neutrophil (Fig. A2B) pathways were filtered for differential gene expression with SAM, resulting in 1,721 and 2,108 probe sets, respectively. Filtered arrays (three replicates for all time points) were hierarchically clustered by using uncentered Pearson correlation and average linkage clustering as implemented in the Cluster and TreeView programs (7). Note that the individual replicates cluster tightly according to time points in both erythroid and neutrophil differentiation series. In the erythroid series, the relatedness of individual time points precisely reflects the temporal progression of differentiation, whereas this relationship is more discontinuous in the case of neutrophils. Note that when these data are compared to those presented in Fig. 2 that the latter analysis was restricted to only a few selected genes; thus, the overall patterns observed may be somewhat different. In Fig. A2C, data from all 84 arrays were filtered with SAM. Filtered arrays containing the differentially expressed genes obtained (three replicates for all time points in the four pathways) were clustered as described above. The principal feature revealed by this comparative analysis of differentiation is that, early in their differentiation, samples cluster according to time point rather than according to the differentiation pathway selected. As might be expected, later in differentiation the samples cluster according to the lineage specified.
FIG. A2.
Clustering of sample replicates. Abbreviations: E, erythroid; N, neutrophil; NM, neutrophil/monocyte; Mk, megakaryocyte.
REFERENCES
- 1.Akashi, K., X. He, J. Chen, H. Iwasaki, C. Niu, B. Steenhard, J. Zhang, J. Haug, and L. Li. 2003. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood 101:383-389. [DOI] [PubMed] [Google Scholar]
- 2.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289-300. [Google Scholar]
- 3.Bradshaw, A. D., and E. H. Sage. 2001. SPARC, a matricellular protein that functions in cellular differentiation and tissue response to injury. J. Clin. Investig. 107:1049-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, C. Z., M. Li, D. de Graaf, S. Monti, B. Gottgens, M. J. Sanchez, E. S. Lander, T. R. Golub, A. R. Green, and H. F. Lodish. 2002. Identification of endoglin as a functional marker that defines long-term repopulating hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 99:15468-15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delassus, S., I. Titley, and T. Enver. 1999. Functional and molecular analysis of hematopoietic progenitors derived from the aorta-gonad-mesonephros region of the mouse embryo. Blood 5:1495-1503. [PubMed] [Google Scholar]
- 6.Dudoit, S., Y. H. Yang, M. J. Callow, and T. P. Speed. 2002. Statistical methods for identifying genes with differential expression in replicated cDNA microarray experiments. Stat. Sin. 12:111-139. [Google Scholar]
- 7.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enver, T., and M. Greaves. 1998. Loops, lineage, and leukemia. Cell 94:9-12. [DOI] [PubMed] [Google Scholar]
- 9.Gottgens, B., A. Nastos, S. Kinston, S. Piltz, E. C. Delabesse, M. Stanley, M. J. Sanchez, A. Ciau-Uitz, R. Patient, and A. R. Green. 2002. Establishing the transcriptional program for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 21:3039-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graf, T. 2002. Differentiation plasticity of hematopoietic cells. Blood 99:3089-3101. [DOI] [PubMed] [Google Scholar]
- 11.Green, C. B., and J. C. Besharse. 1996. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proc. Natl. Acad. Sci. USA 93:14884-14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haluzik, M., M. Markova, J. J. Slaby, J. Jiskra, J. Krizova, and T. Hass. 2002. The changes of serum leptin and soluble leptin receptor levels in patients undergoing mobilization of peripheral blood stem cells before autologous stem cells transplantation. Endocrinol. Res. 28:189-197. [DOI] [PubMed] [Google Scholar]
- 13.Hampson, I. N., M. A. Cross, C. M. Heyworth, L. Fairbairn, E. Spooncer, G. J. Cowling, and T. M. Dexter. 1992. Expression and downregulation of cytotoxic cell protease 1 or Granzyme “B” transcripts during myeloid differentiation of interleukin-3-dependent murine stem cell lines. Blood 80:3097-3105. [PubMed] [Google Scholar]
- 14.Hampson, I. N., L. Hampson, M. Pinkoski, M. Cross, C. M. Heyworth, R. C. Bleackley, E. Atkinson, and T. M. Dexter. 1997. Identification of a serpin specifically expressed in multipotent and bipotent hematopoietic progenitor cells and in activated T cells. Blood 89:108-118. [PubMed] [Google Scholar]
- 15.Hermanson, O., K. Jepsen, and M. G. Rosenfeld. 2002. N-CoR controls differentiation of neural stem cells into astrocytes. Nature 419:934-939. [DOI] [PubMed] [Google Scholar]
- 16.Heyworth, C., K. Gale, M. Dexter, G. May, and T. Enver. 1999. A GATA-2/estrogen receptor chimera functions as a ligand-dependent negative regulator of self-renewal. Genes Dev. 13:1847-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heyworth, C. M., M. Alauldin, M. A. Cross, L. J. Fairbairn, T. M. Dexter, and A. D. Whetton. 1995. Erythroid development of the FDCP-Mix A4 multipotent cell line is governed by the relative concentrations of erythropoietin and interleukin 3. Br. J. Haematol. 91:15-22. [DOI] [PubMed] [Google Scholar]
- 18.Heyworth, C. M., T. M. Dexter, O. Kan, and A. D. Whetton. 1990. The role of hemopoietic growth factors in self-renewal and differentiation of IL-3-dependent multipotential stem cells. Growth Factors 2:197-211. [DOI] [PubMed] [Google Scholar]
- 19.Heyworth, C. M., M. A. Pearson, T. M. Dexter, G. Wark, P. J. Owen-Lynch, and A. D. Whetton. 1995. Macrophage inflammatory protein-1α mediated growth inhibition in a hemopoietic stem cell line is associated with inositol 1,4,5 triphosphate generation. Growth Factors 12:165-172. [DOI] [PubMed] [Google Scholar]
- 20.Honma, S., T. Kawamoto, Y. Takagi, K. Fujimoto, F. Sato, M. Noshiro, Y. Kato, and K. Honma. 2002. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 419:841-844. [DOI] [PubMed] [Google Scholar]
- 21.Hu, M., D. Krause, M. Greaves, S. Sharkis, M. Dexter, C. Heyworth, and T. Enver. 1997. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11:774-785. [DOI] [PubMed] [Google Scholar]
- 22.Ivanova, N. B., J. T. Dimos, C. Schaniel, J. A. Hackney, K. A. Moore, and I. R. Lemischka. 2002. A stem cell molecular signature. Science 298:601-604. [DOI] [PubMed] [Google Scholar]
- 23.Karasuyama, H., and F. Melchers. 1988. Establishment of mouse cell lines which constitutively secrete large quantities of interleukin 2, 3, 4 or 5, using modified cDNA expression vectors. Eur. J. Immunol. 18:97-104. [DOI] [PubMed] [Google Scholar]
- 24.Kisseleva, T., S. Bhattacharya, J. Braunstein, and C. W. Schindler. 2002. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285:1-24. [DOI] [PubMed] [Google Scholar]
- 25.Kondo, T., and M. Raff. 2000. Oligodendrocyte precursor cells reprogramd to become multipotential CNS stem cells. Science 289:1754-1757. [DOI] [PubMed] [Google Scholar]
- 26.LeBrun, D. P., and M. L. Cleary. 1994. Fusion with E2A alters the transcriptional properties of the homeodomain protein PBX1 in t(1;19) leukemias. Oncogene 9:1641-1647. [PubMed] [Google Scholar]
- 27.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:0032.1-0032.11. [DOI] [PMC free article] [PubMed]
- 29.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 30.Miyamoto, T., H. Iwasaki, B. Reizis, M. Ye, T. Graf, I. L. Weissman, and K. Akashi. 2002. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell 1:137-147. [DOI] [PubMed] [Google Scholar]
- 31.Muller-Pillasch, F., C. Wallrapp, K. Bartels, G. Varga, H. Friess, M. Buchler, G. Adler, and T. M. Gress. 1998. Cloning of a new Kunitz-type protease inhibitor with a putative transmembrane domain overexpressed in pancreatic cancer. Biochim. Biophys. Acta 1395:88-95. [DOI] [PubMed] [Google Scholar]
- 32.Newton, R. A., S. Bingham, P. D. Davey, A. D. Medhurst, V. Piercy, P. Raval, A. A. Parsons, G. J. Sanger, C. P. Case, and S. N. Lawson. 2000. Identification of differentially expressed genes in dorsal root ganglia following partial sciatic nerve injury. Neuroscience 95:1111-1120. [DOI] [PubMed] [Google Scholar]
- 33.Orkin, S. H., and S. J. Morrison. 2002. Stem-cell competition. Nature 418:25-27. [DOI] [PubMed] [Google Scholar]
- 34.Park, I. K., Y. He, F. Lin, O. D. Laerum, Q. Tian, R. Bumgarner, C. A. Klug, K. Li, C. Kuhr, M. J. Doyle, T. Xie, M. Schummer, Y. Sun, A. Goldsmith, M. F. Clarke, I. L. Weissman, L. Hood, and L. Li. 2002. Differential gene expression profiling of adult murine hematopoietic stem cells. Blood 99:488-498. [DOI] [PubMed] [Google Scholar]
- 35.Perbal, B. 2001. NOV (nephroblastoma overexpressed) and the CCN family of genes: structural and functional issues. Mol. Pathol. 54:57-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrenko, O., A. Beavis, M. Klaine, R. Kittappa, I. Godin, and I. R. Lemischka. 1999. The molecular characterization of the fetal stem cell marker AA4. Immunity 10:691-700. [DOI] [PubMed] [Google Scholar]
- 37.Phillips, R. L., R. E. Ernst, B. Brunk, N. Ivanova, M. A. Mahan, J. K. Deanehan, K. A. Moore, G. C. Overton, and I. R. Lemischka. 2000. The genetic program of hematopoietic stem cells. Science 288:1635-1640. [DOI] [PubMed] [Google Scholar]
- 38.Pierce, A., A. D. Whetton, P. J. Owen-Lynch, J. Tavernier, E. Spooncer, T. M. Dexter, and C. M. Heyworth. 1998. Ectopic interleukin-5 receptor expression promotes proliferation without development in a multipotent hematopoietic cell line. J. Cell Sci. 111(Pt. 6):815-823. [DOI] [PubMed] [Google Scholar]
- 39.Quesenberry, P., H. Habibian, M. Dooner, C. McAuliffe, J. F. Lambert, G. Colvin, C. Miller, A. Frimberger, and P. Becker. 2001. Physical and physiological plasticity of hematopoietic stem cells. Blood Cells Mol. Dis. 27:934-937. [DOI] [PubMed] [Google Scholar]
- 40.Ramalho-Santos, M., S. Yoon, Y. Matsuzaki, R. C. Mulligan, and D. A. Melton. 2002. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298:597-600. [DOI] [PubMed] [Google Scholar]
- 41.Reya, T., S. J. Morrison, M. F. Clarke, and I. L. Weissman. 2001. Stem cells, cancer, and cancer stem cells. Nature 414:105-111. [DOI] [PubMed] [Google Scholar]
- 42.Shimomura, T., K. Denda, A. Kitamura, T. Kawaguchi, M. Kito, J. Kondo, S. Kagaya, L. Qin, H. Takata, K. Miyazawa, and N. Kitamura. 1997. Hepatocyte growth factor activator inhibitor, a novel Kunitz-type serine protease inhibitor. J. Biol. Chem. 272:6370-6376. [DOI] [PubMed] [Google Scholar]
- 43.Shin, H. H., J. Y. Seoh, H. Y. Chung, S. J. Choi, M. J. Hahn, J. S. Kang, M. S. Choi, and T. H. Han. 1999. Requirement of MEF2D in the induced differentiation of HL60 promyeloid cells. Mol. Immunol. 36:1209-1214. [DOI] [PubMed] [Google Scholar]
- 44.Spooncer, E., C. M. Heyworth, A. Dunn, and T. M. Dexter. 1986. Self-renewal and differentiation of interleukin-3-dependent multipotent stem cells are modulated by stromal cells and serum factors. Differentiation 31:111-118. [DOI] [PubMed] [Google Scholar]
- 45.Sturn, A., J. Quackenbush, and Z. Trajanoski. 2002. Genesis: cluster analysis of microarray data. Bioinformatics 18:207-208. [DOI] [PubMed] [Google Scholar]
- 46.Suenobu, S., N. Takakura, T. Inada, Y. Yamada, H. Yuasa, X. Q. Zhang, S. Sakano, Y. Oike, and T. Suda. 2002. A role of EphB4 receptor and its ligand, ephrin-B2, in erythropoiesis. Biochem. Biophys. Res. Commun. 293:1124-1131. [DOI] [PubMed] [Google Scholar]
- 47.Terskikh, A. V., M. C. Easterday, L. Li, L. Hood, H. I. Kornblum, D. H. Geschwind, and I. L. Weissman. 2001. From hematopoiesis to neuropoiesis: evidence of overlapping genetic programs. Proc. Natl. Acad. Sci. USA 98:7934-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Y., S. Ota, H. Kataoka, M. Kanamori, Z. Li, H. Band, M. Tanaka, and H. Sugimura. 2002. Negative regulation of EphA2 receptor by Cbl. Biochem. Biophys. Res. Commun. 296:214-220. [DOI] [PubMed] [Google Scholar]
- 50.Zhu, J., C. M. Heyworth, A. Glasow, Q. H. Huang, K. Petrie, M. Lanotte, G. Benoit, R. Gallagher, S. Waxman, T. Enver, and A. Zelent. 2001. Lineage restriction of the RARalpha gene expression in myeloid differentiation. Blood 98:2563-2567. [DOI] [PubMed] [Google Scholar]
- 51.Zumkeller, W. 2002. The insulin-like growth factor system in hematopoietic cells. Leuk. Lymphoma 43:487-491. [DOI] [PubMed] [Google Scholar]