Abstract
Peptoid molecules are biomimetic oligomers that can fold into unique three-dimensional structures. As part of an effort to advance computational design of folded oligomers, we present blind-structure predictions for three peptoid sequences using a combination of Replica Exchange Molecular Dynamics (REMD) simulation and Quantum Mechanical refinement. We correctly predicted the structure of a N-aryl peptoid trimer to within 0.2 Å rmsd-backbone and a cyclic peptoid nonamer to an accuracy of 1.0 Å rmsd-backbone. X-ray crystallographic structures are presented for a linear N-alkyl peptoid trimer and for the cyclic peptoid nonamer. The peptoid macrocycle structure features a combination of cis and trans backbone amides, significant nonplanarity of the amide bonds, and a unique “basket” arrangement of (S)-N(1-phenylethyl) side chains encompassing a bound ethanol molecule. REMD simulations of the peptoid trimers reveal that well folded peptoids can exhibit funnel-like conformational free energy landscapes similar to those for ordered polypeptides. These results indicate that physical modeling can successfully perform de novo structure prediction for small peptoid molecules.
Keywords: foldamer, molecular simulation
Foldamers are synthetic polymers that—like proteins—have the ability to self-assemble into unique folded structures (1). Examples of foldamer systems include β-peptides, γ-peptides, azapeptides, oligoureas, arylamides, oligohydrazides, polyphenylacetylenes, and peptoids, among others (2, 3). Of these, peptoids offer an attractive platform for designing functionalized, conformationally ordered molecular architectures (Fig. 1): they can be readily synthesized to incorporate chemically diverse side chains (4, 5), are resistant to proteolysis (6), and can retain structure and function in nonaqueous solvents. Peptoids have found capacity for diverse applications such as antimicrobials (7), drug delivery platforms (8), therapeutics (9), enantioselective catalysts (10), and nanostructured materials (11, 12).
Fig. 1.
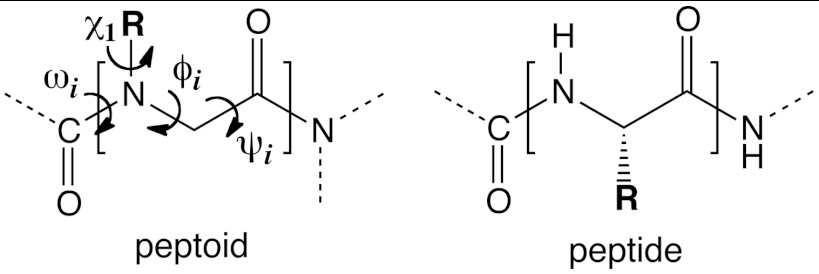
Peptoid vs. peptide architecture. Peptoids are oligomers of N-substituted glycine units. In contrast to peptides, peptoids feature an achiral backbone with side chain groups presented on the nitrogen atom. The relevant backbone dihedral angles are ω [Cα(i - 1); C(i - 1); N; Cα], φ [C(i - 1); N; Cα; C], ψ [N; Cα; C; N(i + 1)], and χ1 [Cα; N; NCα; Cβ].
Unlike peptides, peptoids lack the ability to form backbone hydrogen bonds and can readily populate both cis and trans backbone amide states. Thus, new strategies may be required to enable the rational design of ordered peptoid structures. For instance, even though the peptoid backbone is achiral, bulky chiral side chain groups such as 1-phenylethyl or 1-naphthylethyl can be used to induce stereocontrolled cis-amide helical structures resembling polyproline I (13–15). Such helices have been used to form tertiary assemblies (11, 16), and have been incorporated into enzymes with minimal loss of function (17). Alternatively, peptoid N-aryl side chains have been used to induce trans-amide helices that can mimic polyproline II structure (18, 19). It remains to be determined how these local rules can be used to control the global three-dimensional structure of peptoid macromolecules (20).
In order to design peptoids for applications, we need a way to predict their native structures from their sequences. Successes in designing protein molecules are largely attributable to the vast amount of information in the Protein Data Bank, of around 70,000 atomic-resolution structures. In contrast, very few peptoid structures are known (21). Hence, the current method of choice for predicting peptoid structures is physics-based computational modeling.
Are physical models up to the task of predicting peptoid structures? We performed here a blind test of a combined MD-quantum mechanical (QM) method, in the spirit of CASP [Critical Assessment of Structure Prediction (22)], used to test protein-structure prediction methods. The prediction targets included three peptoid sequences for which X-ray crystal structures were obtained, but not disclosed to the modelers. One of these structures has since been published, but the other two are reported here for the first time. Of these prediction targets, the largest was a cyclic peptoid nonamer. We used a hierarchical approach of replica-exchange molecular dynamics (REMD) (to identify low free-energy conformational basins) and QM calculations (for refinement of these conformations). This strategy correctly predicted the pattern of cis/transbackbone amides (cccctccct), and the structure of the peptoid nonamer to an accuracy of 1.0 Å rmsd-backbone. These results suggest that reliable ab initio peptoid structure prediction for complex three-dimensional folds is within reach.
Results
Blind Structure Prediction of Linear Peptoid—N-aryl Trimer.
The first two blind prediction targets were small N-alkyl and N-aryl peptoid trimers (compounds 1 and 2, Fig. 2). The latter structure has since been published (23), but the coordinates were unreleased at the time of the prediction. The trimers were modeled by both REMD and QM approaches; thus, for these trimers the QM and REMD components of our combined method could be directly compared.
Fig. 2.
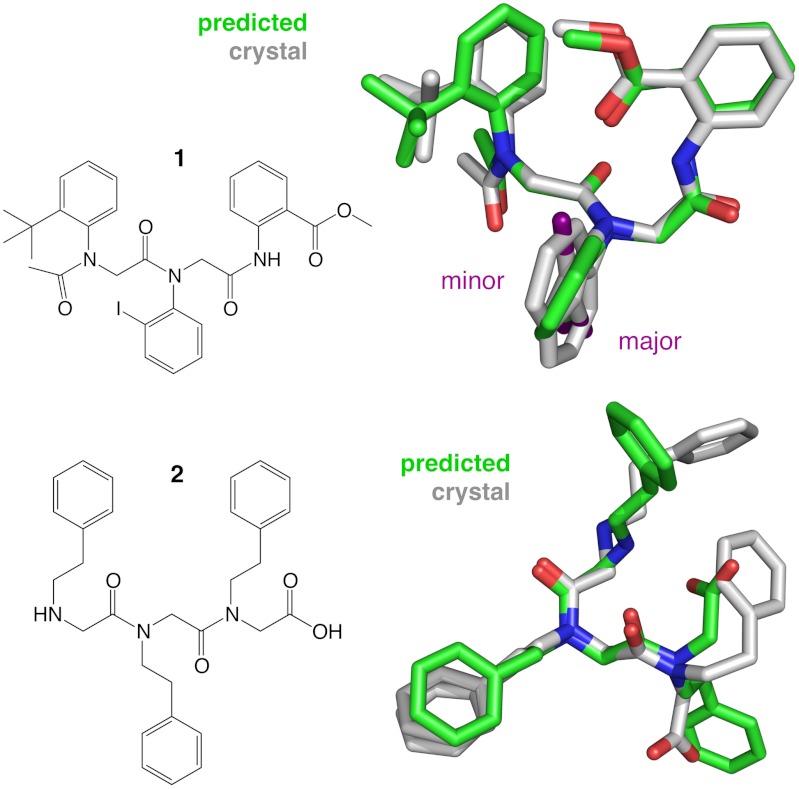
Chemical structures of N-aryl peptoid trimer 1 and N-alkyl peptoid trimer 2, shown with their respective solid-state conformations. The crystal structure of peptoid trimer 1 (gray) is represented by both the major conformer (approximately 98% occupancy, iodo group facing down) and the minor conformer (approximately 2% occupancy, iodo group facing up). Superimposed is the REMD prediction (green), which is in close concordance with the major conformer observed by X-ray crystallography (approximately 0.6 Å rmsd-backbone). The crystal structure of trimer 2 exhibits two alternative side chain conformers for residue 2 (69% and 31% occupancies). Superimposed is the lowest-energy conformation predicted by QM (approximately 0.8 Å rmsd-backbone).
The REMD simulations were performed as previously described (24). The Generalized AMBER Force Field (25) was used along with a popular Generalized Born implicit solvation model (26), with atomic partial charges generated by the AM1-BCC method (27); this potential has previously been shown to be accurate for known peptoid structures (24). Significant cis/trans amide isomerization barriers present a particular challenge when simulating peptoid conformational equilibria, which we are able to overcome by using REMD replicas up to 800 K. A ranked list of REMD predictions for the trimers was obtained by conformational free energy estimation of all possible backbone states. Discrete backbone states were defined by binary binning thresholds for each torsion angle: ω (cis vs. trans), φ (+ vs. -), and ψ, which was binned according to the closest (φ, ψ) dihedral minimum, αD = ( ± 90°,180°) or C7β = ( ± 120°,∓75°) (see SI Appendix for details).
The predictions for trimer 1, a heterotrimer of N(2-tert-butylphenyl) glycine, N(2-iodophenyl) glycine, and N(2-(carboxymethyl)phenyl) glycine, were then compared to the X-ray crystal structure (23), which includes both a major and a minor conformer (approximately 2% occupancy). We found that the top-ranked (i.e., lowest conformational free-energy) REMD prediction for the N-aryl trimer 1 was the best blind prediction, matching the major crystal conformer to a backbone rmsd of approximately 0.6 Å (Fig. 2).
QM studies of trimer 1 agree with the REMD results. We optimized 28 conformations, including conformers from the crystal structure, 20 alternative conformations, and six low energy REMD snapshots (see SI Appendix, Table S3). Similar to the REMD results, the lowest energy conformation identified by QM corresponds to the experimentally observed backbone geometry (approximately 0.2 Å rmsd-backbone). The major conformer is predicted to be approximately 0.9 kcal/mol more favorable than the minor conformer at the M052X/3-21G* level of theory. In previous work, we found ortho-iodo N-aryl peptoids favor by approximately 5 kcal/mol a geometry placing the iodo group distant from the following carbonyl group (23), but this effect is not predicted for trimer 1 due to the secondary (not tertiary) amide at residue 3. The optimized structure with the overall lowest energy differs slightly from the major crystal rotamer, in that the tert butyl group on residue 1 is rotated by 60°. This geometry is predicted to be only approximately 0.5 kcal/mol more favorable at levels of theory used here.
Experimental and Predicted Structures of a Linear Peptoid—N-alkyl Trimer.
Synthesis and crystallization of peptoid 2, a homotrimer of N(2-phenylethyl) glycine units, was performed as described in Materials and Methods. The high-resolution crystal structure has cis backbone amide bonds, with the side chain groups of residues 1 and 3 in close proximity. Two alternate side chain conformers (69% and 31% occupancies) are found for residue 2, each occupying similar molecular volumes. Backbone dihedral and side chain χ-angles are listed in Table 1. The 2-phenylethyl side chain groups participate in a layer of extensive aromatic π-π interactions between neighboring molecules in the crystal lattice (SI Appendix, Fig. S1). A layer of polar interactions is also formed across the amino- and carboxy-termini of neighboring peptoid units, the backbone amide groups, and bound water molecules. This hydrophobic/polar partitioning in the crystal is somewhat reminiscent of the nanosheet bilayers formed by charged periodic amphiphilic peptoid chains, which similarly incorporate N(2-phenylethyl) glycine residues (12).
Table 1.
Dihedral angles and side chain χ-angles for the crystal conformation of N-(2-phenylethyl) glycine peptoid trimer 2
| Residue | cis/trans | ω | φ | ψ | χ1 | χ2 | Occupancy |
| 1 | – | – | – | 177.5 | – | 166.3 | |
| 2 | cis | 9.8 | 77.6 | −175.8 | 111.9 | 174.9 | 0.69 |
| −91.0 | −172.4 | 0.31 | |||||
| 3 | cis | 1.9 | −95.1 | – | −125.8 | 51.5 |
Our predictions for the N-alkyl trimer 2 were less successful than those for trimer 1. REMD predicts a heterogeneous ensemble of states with similar conformational free energies, with the crystal conformation predicted to be approximately 2.5 kcal/mol above the top-ranked basin. Much of this discrepancy is likely due to the fact that crystal conformations of small molecules depend greatly on neighboring crystal lattice interactions, and are not necessarily well predicted by solution-state simulations. Quantum calculations further support conformational heterogeneity in trimer 2 (SI Appendix, Table S5). We find that the relative energies predicted by QM are dominated by the protonation state and orientation of the amine and carboxylic acid termini rather than by local backbone conformational constraints.
Conformational Landscapes of Foldable and Nonfoldable Peptoid Trimers.
Examination of the conformational free energy landscapes predicted for peptoid trimers 1 and 2 provides additional insight into their folding properties. The N-aryl trimer 1 has a funnel-like free energy landscape dominated by a small number of low-free energy states, a characteristic feature of foldable molecules (28, 29) (Fig. 3). About 10% of accessible states are found within approximately 3 kcal/mol above the lowest energy state. Indeed, NMR and HPLC studies (23) confirm the presence of a dominant pair of atropisomer solution conformations of N-aryl peptoid trimer 1. In contrast, the N-alkyl trimer 2 is predicted to exhibit a shallower free energy landscape, with 40% of all conformational states within approximately 3 kcal/mol of the ground state. These computational results suggest that the N-alkyl trimer 2 does not have a well defined solution structure.
Fig. 3.
Conformational free energy landscapes of N-aryl peptoid trimer 1 and N-alkyl peptoid trimer 2. Landscapes calculated from REMD simulations show a funnel-like landscape for 1, with a small number of low free energy torsional states. The observed crystal structure of 1 is found at the free energy minimum of this landscape (black arrow). The landscape of 2 is more degenerate, suggesting a more heterogeneous structure in solution. The crystal structure observed for 2 (red arrow) is estimated by REMD to be approximately 2.5 kcal/mol above the solution-phase ground state.
Several features of these small peptoid sequences contribute to their foldability. Conformational restriction of the main chain clearly helps narrow the number of low-energy conformations. For the N-aryl peptoid, local structure is controlled by incorporation of side chain groups that restrict the possible backbone conformational states through both steric and electronic effects. N-aryl side chains restrict the amide backbone nearly exclusively to the trans state (19) and the bulky iodo- and tert-butyl- substituents lead to additional biasing of the φ torsion (30). Unbranched N-alkyl side chains provide fewer local restrictions to the peptoid backbone (31), but can contribute to long-range interactions that can direct chain folding (12).
Blind Structure Prediction of a Cyclic Peptoid Nonamer.
For the more challenging problem of structure prediction for a cyclic nonamer, we used a hierarchical approach: first, we identified the lowest-free energy conformational basins using REMD, and then used QM to refine 273 representative structures taken from these basins, optimizing at the HF/3-21G* level and evaluating the energy in a number of DFT functionals. We found no great sensitivity to basis set, and present here the results of two different functionals, B3LYP/6-311G** and M052X/6-311G**, the latter of which has been reported to provide a more accurate description of dispersion interactions. Adhering to the conventions of CASP, we used conformational clustering to select and rank six diverse conformations for our final submitted prediction (see SI Appendix). Our top three predictions, ranked based on the DFT energies, exhibited the cccctccct pattern of cis/transamides, but differing φ-angle patterns. The remaining submitted predictions, chosen for diversity, had backbone structures ccccctttt, cccctcctt, and cccccccct.
Experimental Structure of a Cyclic Peptoid Nonamer.
Synthesis and purification of cyclic peptoid nonamer 3 incorporating (S)-N(1-phenylethyl) side chains were performed as previously described (32, 33). Needle-like crystals were obtained by slow evaporation in ethanol, and X-ray crystallographic direct methods were used to obtain a high-resolution structure (SI Appendix, Fig. S2). The unit cell includes two identical molecules in a monoclinic system with a P2(1) space group.
The crystal structure of peptoid 3 complements a corpus of previously solved peptoid X-ray structures [previous cyclic peptoid structures include tetramers, hexamers, and octamers (32, 33)]. The backbone atoms of the macrocycle form an irregular shape, with approximate dimensions 10.53 Å × 7.26 Å. The amide bonds exhibit a cis/transpattern of cccctccct, and all display significant distortions from planarity (Table 2). Most notable is residue 5, with an ω angle of 151.5° that deviates nearly 30° from planarity. QM calculations suggest this deviation may be enforced by a steric clash between the amide oxygen and (i-2) Hα atom (SI Appendix, Fig. S3). Increased propensities for ω angles near approximately 150° are also seen in the REMD simulations (SI Appendix, Fig. S4). Similarly, significant pyramidalization is found for backbone nitrogen centers in the crystal structure (SI Appendix, Table S1). The φ and ψ dihedral angles are consistent with previously reported peptoid structures; the φ angles do not deviate significantly from ± 70°, and ψ angles do not markedly deviate from ± 175°.
Table 2.
Dihedral angles for the crystal conformation of (S)-N(1-phenylethyl) glycine nonamer peptoid 3
| Residue | cis/trans | ω | φ | ψ | χ1 |
| 1 | cis | −2.3 | 73.4 | 175.2 | −98.9 |
| 2 | cis | 4.0 | −84.0 | −166.1 | −105.9 |
| 3 | cis | −8.1 | 75.4 | −172.0 | −152.1 |
| 4 | cis | −4.6 | 77.4 | 174.3 | −138.1 |
| 5 | trans | 151.5 | −64.3 | 173.7 | −114.6 |
| 6 | cis | −14.6 | 76.7 | 178.9 | −137.7 |
| 7 | cis | −5.4 | −75.3 | 174.9 | −118.2 |
| 8 | cis | −13.6 | −76.0 | −166.1 | −149.2 |
| 9 | trans | 170.7 | −61.4 | 142.2 | −132.3 |
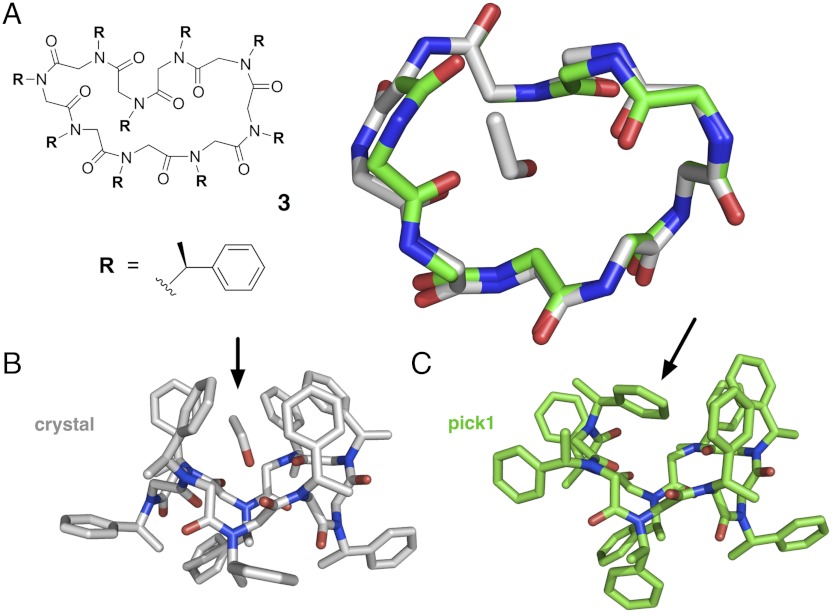
The χ1 angles for the N(1-phenylethyl) side chains range from -98.9° to -152.1° (Table 1). Interestingly, four side chain groups are oriented on one face of the macrocycle, while the other five side chains are directed towards the opposite face, resembling a molecular basket. Inside this “basket” is a single molecule of bound ethanol, with the ethyl group in contact with peptoid side chain groups, and the hydroxyl group favorably hydrogen bonded with a peptoid backbone carbonyl (Fig. 4B).
Fig. 4.
Experimental and predicted structures of cyclic peptoid nonamer 3, composed of (S)-N-(1-phenylethyl) glycine units. The amide bonds in the crystal structure were found to exhibit a cis/transpattern of cccctccct, with several significant deviations from amide planarity (see Table 1). (A) Shown superimposed are the backbone conformation of the compound 3 crystal structure (gray) and the top-ranked blind prediction (green), with a backbone-rmsd of approximately 1.0 Å. (B) The crystal structure shows the side chains oriented toward alternating faces of the macrocycle, with five side chain groups forming a “molecular basket” encompassing a single bound ethanol (arrow). (C) The predicted structure, which was modeled in the absence of ethanol, shows instead a peptoid side chain “filling” this basket (arrow), resulting in the rotation of two backbone ψ angles, compared to the crystal structure backbone.
NMR experiments were conducted to obtain structural information for nonamer 3 in solution. 1H-NMR spectra reveal excellent peak dispersion in CDCl3 (SI Appendix, Fig. S5). 1H-13C-HSQC spectra show 9 clearly resolved side chain methine and 18 backbone methylene resonances (SI Appendix, Fig. S6), and 2D-COSY spectra for the compound 3 also exhibit 9 off-diagonal correlations corresponding to the backbone methylene protons, all consistent with a single well defined structure in solution (SI Appendix, Fig. S7).
It is notable that the cyclic peptoid 3 characterized here is quite different than the previously described NMR solution structure of the analogous linear (S)-N(1-phenylethyl) peptoid oligomer in acetonitrile(34), shown to exhibit a “threaded loop” backbone arrangement in which the amide backbone pattern is cctttctc. The circular dichroism (CD) spectrum of cyclic peptoid 3 obtained in methanol reveals two relatively sharp minima at 210 and 220 nm (SI Appendix, Fig. S8), which also differs markedly from the linear analog, for which a single broad minima at 205 nm was observed (34).
Comparsion of Predicted and Experimental Structures of a Cyclic Peptoid Nonamer.
Our top-ranked prediction (pick1) agrees quite closely with the experimentally observed structure (Fig. 4), with the correct cccctccct pattern of cis/transamides. The predicted pattern of φ -angles (+−++−+++−) differs from experiment (+−++−+−−−) in only two successive residues, where + denotes φ ∼ +90° and - denotes φ approximately -90°. An overlay of the crystal structure and pick1 shows the two flipped φ angles result in one side chain residue rotating relative to the plane of the macrocycle ring. Other than this local inversion, the rest of the backbone atoms match closely, with a 1.0 Å backbone rmsd to the crystal structure, and 0.47 Å rmsd if the flipped backbone atoms are omitted from the comparison.
Free energies for backbone torsional states computed from the REMD simulations of the cyclic nonamer show a “foldable” landscape dominated by a handful of lowest free energy conformations. Only approximately 5% of accessible states are found within approximately 3 kcal/mol or less of the ground state. The submitted pick1 state (cccctccct, +−++−+++−) is predicted by REMD to be among the five lowest-free energy states (approximately 1.22 kcal/mol above the ground state), while the experimental crystal state (cccctccct, +−++−+−−−) is predicted to be among the 30 lowest-free energy conformations (approximately 3.08 kcal/mol) (SI Appendix, Fig. S9). Because the state corresponding to the experimental structure had a relatively high free energy, conformations from this basin were not selected for QM refinement. This finding indicates that our selection criteria, which were based on the cis/transstate alone, were too coarse.
We believe that improved agreement between prediction and experiment (beyond the intrinsic accuracy of DFT) may require a priori information about the crystal-packing environment and the solvent present during crystallization (in this case, ethanol). To support this idea, we performed computational studies comparing our top predicted structure to the experimental structure (full results described in the SI Appendix). Without the ethanol, the two DFT functionals considered here disagree as to whether the experimental or predicted structure is more favorable. Furthermore, when a bound ethanol is added to the predicted structure, DFT predictions unambiguously show a preference for the crystal conformation (see SI Appendix for details). These results suggest that it is important for high-resolution structure predictions to model crystal packing and potential solvent interactions (at least for small foldamers in which crystal and solvent contacts may outnumber intramolecular contacts). Computational studies of cyclo-(sarcosine)9 analogs show similar backbone conformational preferences, suggesting that the bulky N-1-phenylethyl side chains do not play a significant role in determining overall backbone geometries.
Discussion
Our blind prediction results suggest that a combined MD-QM computational approach can produce accurate ab initio peptoid structure predictions. This work is a tangible milestone on the path to reliable and efficient computational peptoid design.
We can provide some context regarding the significance of these results by comparing to similar advances in protein structure prediction. The cyclic peptoid nonamer contains 36 bonds that have at least two rotameric possibilities, while the N-aryl and N-alkyl peptoid trimers have 11 and 14 rotatable bonds, respectively. The nonamer search problem is similarly complex to chignolin (35) (10 residues, 29 bonds) or the C-terminal hairpin of protein G (36) (16 residues, 53 bonds), and the trimers similar to met-enkephalin (37) (5 residues, 15 bonds), all model peptide systems that were the subject of state-of the-art folding simulations a decade ago.
Continued progress in computational peptoid structure prediction and design will require further advances in forcefield development and sampling. One encouraging finding here is that currently available automated tools to provide transferrable forcefield parameters for arbitrary organic molecules can be used to predict folded peptoid structures accurately. This finding is a promising development for computational peptoid design, although further forcefield validation studies are required. We hope that the increasing number of high-resolution peptoid crystal structures (including those reported here) and solution-phase data can be used to develop and test even more accurate molecular models, as well as coarse-grained potentials that can model the thermodynamic driving forces responsible for the assembly of peptoid nanostructured materials (12, 38–40). As our computational work demonstrates, we can now accurately predict local backbone preferences of peptoids, but more work needs to be done to understand and predict nonlocal molecular packing interactions in the condensed phase, which may govern both self-assembly of longer peptoid chains (41) and binding interactions with proteins.
While here we have addressed the challenge of conformational sampling using REMD, a greater problem for computational design is efficient sampling of peptoid sequence space. Discrete rotamer and/or fragment-based search algorithms such as RosettaDesign have performed very efficiently for proteins (42). These same approaches can be applied to peptoids, although more information about specific sequence-structure relationships is needed. Backbone dihedral minima from previously published structures could aid in the prediction process, and molecular mechanics or QM models can be used to parameterize side chain scoring functions. Another promising strategy may be to construct kinetic network models (43) from simulated peptoid conformational ensembles so that the effects of sequence perturbations can be efficiently sampled. With simulation providing information about the entire conformational free energy landscape, both positive and negative design across many conformational states can be optimized simultaneously to design foldable molecules.
Foldable peptoid architectures, such as the cyclic nonamer we characterize here, may be good starting points for the design of binding and catalytic functions, as they offer a way to position diverse chemical moieties in well defined spatial arrangements. Because cyclic structures have smaller conformational entropies in the unfolded state compared to linear structures, they may be more suited for these purposes. The presence of bound ethanol in the cyclic nonamer crystal structure additionally suggests that the basket of (S)-N(1-phenylethyl) side chains on the top face of the macrocycle constitutes a defined binding pocket that could be elaborated in future design efforts. Indeed, our computational modeling predicts a significant energetic reward for filling this pocket, at the expense of a subtle backbone rearrangement (see Fig. 4C).
Conclusion
In this work, we have presented blind structure predictions for three peptoid sequences using physical computational approaches. We subsequently obtained high-resolution crystal structures for these targets, two of which we publish here. One of the targets is a macrocyclic nonamer, the largest cyclic peptoid structure published to date, with a cccctccct pattern of cis/transbackbone amides. Distinctive features of this new structure include significant nonplanarity of amide bonds and a basket arrangement of (S)-N(1-phenylethyl) side chains containing a bound ethanol molecule.
A principal aspect of this work was our blind predictions of structure using REMD simulations and QM, prior to determining the crystal structures. These predictions validate current modeling strategies. We predicted the structure of an N-aryl glycine peptoid trimer to 0.6 Å rmsd-backbone, and approximately 0.2 Å rmsd-backbone using QM. Using a hierarchical combination of REMD and QM refinement, we correctly predicted the backbone amide pattern of the (S)-N(1-phenylethyl) glycine nonamer, and the three-dimensional structure to within an accuracy of 1.0 Å rmsd-backbone. Our REMD simulations reveal that foldable peptoids have funnel-like conformational free energy landscapes. These results suggest that physical modeling approaches can successfully make de novo predictions of the structures of small peptoids.
Materials and Methods
Synthesis and Characterization of N-alkyl Peptoid Trimer 2.
The N-(2-phenylethyl) glycine tripeptoid C-terminal acid was synthesized on 2-chlorotrityl resin (Chem-Impex International) using the submonomer solid-phase peptoid synthesis method (4). The crude product was purified by reverse-phase HPLC to afford a white powder of 95% purity [calculated mass: 501.3; observed mass (MH+): 501.8].
Single Crystal X-Ray Diffraction of Peptoid 2.
Diffraction quality crystals were obtained by vapor diffusion. 1.0 mg of purified 2 was added to 0.5 mL of DMF in a small vial which was then placed within a 20 mL scintillation vial containing 3 mL of water and then capped. The sample was then left undisturbed for a week and long thin needles of X-ray quality were obtained. Data were collected on a colorless needle 0.16 × 0.10 × 0.06 mm3 in size. Data reduction was performed using SAXS Area INTegration and SAXS Area Detector ABSorption, and the structure was solved by SIR-2004 and refined with SHELXL-97. Crystallography data: orthorhombic, Pbca, a = 19.0847(14) Å, b = 9.9380(8) Å, c = 29.717(3) Å, α = 90°, β = 90°, γ = 90°, Z = 8, V = 5636.3(8) Å3, ρcalcd = 1.225 g/cm3.
Synthesis of Cyclic Peptoid Nonamer 3.
The linear (S)-N(1-phenylethyl) glycine nonamer was prepared on an automated synthesizer (Charybdis Technologies Inc.) using a slightly modified form of the submonomer approach. The oligomer was synthesized on 2-chlorotrityl chloride resin (NovaBiochem; 100–200 mesh; 1.3 mmol/g). (S)-(-)-1-Phenylethylamine (Alfa Aesar) was used as the submonomer reagent to provide the N-(S)-phenylethylamine monomer units. The linear oligomer was subjected to cyclization using previously published protocols(33). Full details are described in the SI Appendix.
Single Crystal X-Ray Diffraction of Cyclic Peptoid Nonamer 3.
A solution of (S)-N(1-phenylethyl) glycine was prepared (12 mg peptoid dissolved in 800 μL ethanol) in an NMR tube and subject to slow evaporation at room temperature. Colorless needle-like crystals were observed over the course of several days. Crystallography data: 0.49 × 0.30 × 0.12 mm, monoclinic, P2(1), a = 12.8164(3) Å, b = 26.1272(7) Å, c = 13.4896(3) Å, α = 90°, β = 99.5160(10)°, γ = 90°, Z = 2, V = 4454.92(19) Å3, ρcalcd = 1.219 g/cm3.
Circular Dichroism.
CD measurements were performed with an Aviv 202 SF Circular Dichroism Spectrophotometer using a 1 mm pathlength fused quartz cell. Peptoid concentrations were approximately 50 μM.
NMR.
NMR data was collected with an Avance-400 NMR Spectrometer (Bruker). 12 mg of pure cyclo-(Nspe)9 was dissolved in 700 uL of CDCl3. Proton, HQSC and COSY data are described in the SI Appendix.
Molecular Dynamics Simulation.
REMD calculations were performed using AMBER9. Constant-energy dynamics runs were swapped every 1 ps, each time initialized with Boltzmann-distributed velocities for replica temperatures 300–800 K. A 2 fs time step was used with the SHAKE algorithm to constrain hydrogen bond lengths. For the peptoid trimers, 32 replicas were used, each seeded with a different initial conformation. Trimer simulation lengths were 500 ns × 32 replicas = 16 μs of trajectory data, with an acceptance ratio of approximately 80%. The peptoid nonamer 3 was simulated for 1,000 ns × 15 replicas = 15 μs of trajectory data, with an acceptance ratio of approximately 45%, seeded with conformations similar to the threaded-loop structure of linear (S)-N(1-phenylethyl) glycine (34). Full details are in the SI Appendix.
Ab Initio QM Calculations.
QM calculations used Gaussian03 and model chemistries and basis sets are indicated in figures and tables in the SI Appendix.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Pascal Wassam, who provided crucial help in modifying AMBER9 to incorporate GB radii for iodine. We thank Dr. Antonio DiPasquale of the UC Berkeley College of Chemistry X-ray Crystallography Facility for helpful discussions. We thank the Institute of Computational Molecular Science and the Temple Owlsnest high performing computing cluster for computational resources used for validation simulations and data analysis. Portions of this work were performed at the Molecular Foundry, Lawrence Berkeley National Laboratory, which is supported by the Office of Science, Office of Basic Energy Sciences, of the Department of Energy under Contract No. DE-AC02—05CH11231. Portions of this work were funded by the Defense Threat Reduction Agency under IACRO B1144571. This work was supported by the National Science Foundation (NSF) through award CHE-1152317 (to K.K.). G.L.B. and R.N.B. were funded by National Institutes of Health (NIH) PN2 EY016586-06, NIH U54CA143907-01, and NSF IOS-1126971. This research was supported in part by the National Science Foundation through major research instrumentation Grant number CNS-09-58854. This paper is dedicated to the memory of our friend and colleague Dr. Samuel Hawxwell, who illuminated our work with his enthusiasm and insight.
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystal structure of cyclic nonamer peptoid 3 and N-aryl peptoid 2 have been deposited at the Crystallographic Data Centre under deposition numbers CCDC 890788 and 890789, respectively.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209945109/-/DCSupplemental.
References
- 1.Gellman SH. Foldamers: A manifesto. Acc Chem Res. 1998;31:173–180. [Google Scholar]
- 2.Goodman CM, Choi S, Shandler S, DeGrado WF. Foldamers as versatile frameworks for the design and evolution of function. Nat Chem Biol. 2007;3:252–262. doi: 10.1038/nchembio876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill DJ, Mio MJ, Prince RB, Hughes TS, Moore JS. A field guide to foldamers. Chem Rev. 2001;101:3893–4012. doi: 10.1021/cr990120t. [DOI] [PubMed] [Google Scholar]
- 4.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. Efficient method for the preparation of peptoids [oligo (N-substituted glycines)] by submonomer solid-phase synthesis. J Am Chem Soc. 1992;114:10646–10647. [Google Scholar]
- 5.Culf AS, Ouellette RJ. Solid-phase synthesis of N-substituted glycine oligomers (α-peptoids) and derivatives. Molecules. 2010;15:5282–5335. doi: 10.3390/molecules15085282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller SM, et al. Comparison of the proteolytic susceptibilities of homologous L-amino acid, D-amino acid, and N-substituted glycine peptide and peptoid oligomers. Drug Dev Res. 1995;35:20–32. [Google Scholar]
- 7.Chongsiriwatana NP, et al. Peptoids that mimic the structure, function, and mechanism of helical antimicrobial peptides. Proc Natl Acad Sci USA. 2008;105:2794–2799. doi: 10.1073/pnas.0708254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schröder T, Quintilla A, Setzler J. Joint experimental and theoretical investigation of the propensity of peptoids as drug carriers. WSEAS Trans Biol Biomed. 2007;4:145–148. [Google Scholar]
- 9.Chen X, et al. Expanded polyglutamine-binding peptoid as a novel therapeutic agent for treatment of Huntington’s disease. Chem Biol. 2011;18:1113–1125. doi: 10.1016/j.chembiol.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maayan G, Ward MD, Kirshenbaum K. Folded biomimetic oligomers for enantioselective catalysis. Proc Natl Acad Sci USA. 2009;106:13679–13684. doi: 10.1073/pnas.0903187106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee B-C, Chu TK, Dill KA, Zuckermann RN. Biomimetic nanostructures: Creating a high-affinity zinc-binding site in a folded nonbiological polymer. J Am Chem Soc. 2008;130:8847–8855. doi: 10.1021/ja802125x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nam KT, et al. Free-floating ultrathin two-dimensional crystals from sequence-specific peptoid polymers. Nat Mater. 2010;9:454–460. doi: 10.1038/nmat2742. [DOI] [PubMed] [Google Scholar]
- 13.Stringer JR, Crapster JA, Guzei IA, Blackwell HE. Extraordinarily robust polyproline type I peptoid helices generated via the incorporation of α-chiral aromatic N-1-naphthylethyl side chains. J Am Chem Soc. 2011;133:15559–15567. doi: 10.1021/ja204755p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu CW, Sanborn TJ, Huang K, Zuckermann RN, Barron AE. Peptoid oligomers with α-chiral, aromatic side chains: Sequence requirements for the formation of stable peptoid helices. J Am Chem Soc. 2001;123:6778–6784. doi: 10.1021/ja003154n. [DOI] [PubMed] [Google Scholar]
- 15.Kirshenbaum K, et al. Sequence-specific polypeptoids: A diverse family of heteropolymers with stable secondary structure. Proc Natl Acad Sci USA. 1998;95:4303–4308. doi: 10.1073/pnas.95.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee B-C, Zuckermann RN, Dill KA. Folding a nonbiological polymer into a compact multihelical structure. J Am Chem Soc. 2005;127:10999–11009. doi: 10.1021/ja0514904. [DOI] [PubMed] [Google Scholar]
- 17.Lee BC, Zuckermann RN. Protein side-chain translocation mutagenesis via incorporation of peptoid residues. ACS Chem Biol. 2011;6:1367–1374. doi: 10.1021/cb200300w. [DOI] [PubMed] [Google Scholar]
- 18.Stringer JR, Crapster JA, Guzei IA, Blackwell HE. Construction of peptoids with all trans-amide backbones and peptoid reverse turns via the tactical incorporation of N-aryl side chains capable of hydrogen bonding. J Org Chem. 2010;75:6068–6078. doi: 10.1021/jo101075a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah NH, et al. Oligo( N-aryl glycines): A new twist on structured peptoids. J Am Chem Soc. 2008;130:16622–16632. doi: 10.1021/ja804580n. [DOI] [PubMed] [Google Scholar]
- 20.Wetzler M, Barron AE. Commentary: Progress in the de novo design of structured peptoid protein mimics. Biopolymers. 2011;96:556–560. doi: 10.1002/bip.21621. [DOI] [PubMed] [Google Scholar]
- 21.Butterfoss GL, Renfrew PD, Kuhlman B, Kirshenbaum K, Bonneau R. A preliminary survey of the peptoid folding landscape. J Am Chem Soc. 2009;131:16798–16807. doi: 10.1021/ja905267k. [DOI] [PubMed] [Google Scholar]
- 22.Moult J, Fidelis K, Kryshtafovych A, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)-round IX. Proteins. 2011;79(Suppl 10):1–5. doi: 10.1002/prot.23200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul B, et al. Peptoid atropisomers. J Am Chem Soc. 2011;133:10910–10919. doi: 10.1021/ja2028684. [DOI] [PubMed] [Google Scholar]
- 24.Voelz VA, Dill KA, Chorny I. Peptoid conformational free energy landscapes from implicit-solvent molecular simulations in AMBER. Biopolymers. 2010;96:639–650. doi: 10.1002/bip.21575. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 26.Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 27.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 28.Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 29.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 30.Paul B, et al. N-naphthyl peptoid foldamers exhibiting atropisomerism. Org Lett. 2012;14:926–929. doi: 10.1021/ol203452f. [DOI] [PubMed] [Google Scholar]
- 31.Fafarman AT, Borbat PP, Freed JH, Kirshenbaum K. Characterizing the structure and dynamics of folded oligomers: Pulsed ESR studies of peptoid helices. Chem Commun. 2007:377–379. doi: 10.1039/b612198e. [DOI] [PubMed] [Google Scholar]
- 32.Yoo B, Shin SBY, Huang ML, Kirshenbaum K. Peptoid macrocycles: Making the rounds with peptidomimetic oligomers. Chemistry. 2010;16:5528–5537. doi: 10.1002/chem.200903549. [DOI] [PubMed] [Google Scholar]
- 33.Shin SBY, Yoo B, Todaro LJ, Kirshenbaum K. Cyclic peptoids. J Am Chem Soc. 2007;129:3218–3225. doi: 10.1021/ja066960o. [DOI] [PubMed] [Google Scholar]
- 34.Huang K, et al. A threaded loop conformation adopted by a family of peptoid nonamers. J Am Chem Soc. 2006;128:1733–1738. doi: 10.1021/ja0574318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honda S, Yamasaki K, Sawada Y, Morii H. 10 residue folded peptide designed by segment statistics. Structure. 2004;12:1507–1518. doi: 10.1016/j.str.2004.05.022. (Cambridge, MA) [DOI] [PubMed] [Google Scholar]
- 36.Blanco FJ, Rivas G, Serrano L. A short linear peptide that folds into a native stable β-hairpin in aqueous solution. Nat Struct Biol. 1994;1:584–590. doi: 10.1038/nsb0994-584. [DOI] [PubMed] [Google Scholar]
- 37.Isogai Y, Némethy G, Scheraga HA. Enkephalin: Conformational analysis by means of empirical energy calculations. Proc Natl Acad Sci USA. 1977;74:414–418. doi: 10.1073/pnas.74.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lau KHA, Ren C, Park SH, Szleifer I, Messersmith PB. An experimental—Theoretical analysis of protein adsorption on peptidomimetic polymer brushes. Langmuir. 2012;28:2288–2298. doi: 10.1021/la203905g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park SH, Szleifer I. Structural and dynamical characteristics of peptoid oligomers with achiral aliphatic side chains studied by molecular dynamics simulation. J Phys Chem B. 2011;115:10967–10975. doi: 10.1021/jp2025957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X, Ding K, Ayres N. Investigation into fiber formation in N-alkyl urea peptoid oligomers and the synthesis of a water-soluble PEG/N-alkyl urea peptoid oligomer conjugate. Polym Chem. 2011;2:2635–2642. [Google Scholar]
- 41.Rosales AM, Murnen HK, Zuckermann RN, Segalman RA. Control of crystallization and melting behavior in sequence specific polypeptoids. Macromolecules. 2010;43:5627–5636. [Google Scholar]
- 42.Leaver-Fay A, et al. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voelz VA, Bowman GR, Beauchamp K, Pande VS. Molecular simulation of ab initio protein folding for a millisecond folder NTL9(1–39) J Am Chem Soc. 2010;132:1526–1528. doi: 10.1021/ja9090353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.