Abstract
Histone deacetylases (HDACs) are enzymes that catalyze the removal of acetyl groups from lysine residues of histone and nonhistone proteins. Recent studies suggest that they are key regulators of many cellular events, including cell proliferation and cancer development. Human class I HDACs possess homology to the yeast RPD3 protein and include HDAC1, HDAC2, HDAC3, and HDAC8. While HDAC1, HDAC2, and HDAC3 have been characterized extensively, almost nothing is known about HDAC8. Here we report that HDAC8 is phosphorylated by cyclic AMP-dependent protein kinase A (PKA) in vitro and in vivo. The PKA phosphoacceptor site of HDAC8 is Ser39, a nonconserved residue among class I HDACs. Mutation of Ser39 to Ala enhances the deacetylase activity of HDAC8. In contrast, mutation of Ser39 to Glu or induction of HDAC8 phosphorylation by forskolin, a potent activator of adenyl cyclase, decreases HDAC8's enzymatic activity. Remarkably, inhibition of HDAC8 activity by hyperphosphorylation leads to hyperacetylation of histones H3 and H4, suggesting that PKA-mediated phosphorylation of HDAC8 plays a central role in the overall acetylation status of histones.
In eukaryotes, genomic DNA is wrapped tightly around core histones to form nucleosomes, the fundamental building blocks of chromatin. Nucleosomes, once regarded as inert structural particles, are now considered integral and dynamic components of the machineries responsible for gene regulation. Many different enzymes and protein complexes are known to bring about changes in the state of chromatin by numerous mechanisms, with resultant effects on gene expression. One class of complexes alters DNA packaging (remodels chromatin) in an ATP-dependent manner (4, 29). Another class of chromatin-altering factors acts by covalently modifying histone proteins (5). These modifications include acetylation, phosphorylation, methylation, ubiquitination, and ADP-ribosylation. The best-characterized posttranslational histone modification is acetylation, which is catalyzed by histone acetyltransferase (HAT) enzymes. Histone acetylation is a reversible process that is regulated by the opposing activities of HATs and histone deacetylases (HDACs). Generally, hyperacetylation of histones results in transcriptional activation whereas deacetylation correlates with transcriptional silencing. Consistent with this generalization, transcriptional activators are often associated with HAT activity whereas HDACs frequently form complexes with transcriptional repressors (24). Therefore, these two regulatory processes work in harmony to achieve appropriate levels of gene expression. Several oncogenes and tumor suppressors (pRb, BRCA-1, BRCA-2, PML-RAR, and a zinc finger protein mutated in leukemia) have been shown to be associated with HATs or HDACs (41).
HDAC proteins are vital regulators of fundamental cellular events, including cell cycle progression, differentiation, and tumorigenesis (37, 45). A small-molecule inhibitor of HDAC, trichostatin A (TSA), arrests mammalian cells in both G1 and G2 (31, 44), while overexpression of HDAC1 in mouse cells reduces their growth rate by lengthening the duration of G2 and M (3). TSA induces terminal differentiation of mouse erythroleukemia cells and apoptosis of lymphoid and colorectal cancer cells. In addition, TSA treatment of cells expressing the PML zinc finger protein derepresses transcription and allows cells to differentiate normally (18). With this precedent, HDAC inhibitors are being actively explored as potential agents for the treatment of certain forms of cancer (22, 23, 27).
The human HDACs are organized into three different classes based on their similarity to yeast HDAC proteins (37, 45). Class I enzymes are ubiquitously expressed and include HDAC1, -2, -3, and -8, which are homologous to the yeast RPD3 protein. Class II includes HDAC4, -5, -6, -7, -9, and -10, which are similar to yeast HDA1 and are expressed in a tissue-specific manner. The Sir2-like class III HDACs, including SIRT1 to -7, require NAD+ for enzymatic activity. The most recent addition to the human HDAC family, HDAC11, uniquely shares sequence homology with the catalytic regions of both class I and II HDAC enzymes (15).
By far, the most frequently studied and best-characterized human HDACs are HDAC1 and HDAC2. Early studies elegantly demonstrated that HDAC1 and HDAC2 were associated with proteins that modulate their enzymatic activity and their recruitment to genomic regions. Three large multisubunit protein complexes, called Sin3, NuRD/Mi2, and CoREST, contain HDAC1 and HDAC2 (1, 17, 21, 25, 30, 38, 42, 46-48). In addition to complex formation, recent studies have revealed that the activity of class I HDACs is regulated by posttranslational modifications. For example, HDAC1 is a substrate for SUMO-1 (small ubiquitin-related modifier 1), and mutations of the target residues decrease transcriptional repression without affecting the ability of HDAC1 to associate with mSin3 (10). In addition, like those of many class II HDACs, the actions of HDAC1 and HDAC2 are regulated by phosphorylation. Phosphorylation of HDAC1 by protein kinase CK2 alters HDAC1's enzymatic activity and its capacity to form protein complexes (7, 13, 33). Similarly, phosphorylation of HDAC2 by protein kinase CK2 is essential for HDAC2's deacetylase activity and its association with mSin3, Mi2, Sp1, and Sp3 (36, 39). Our previous studies showed that, like HDAC1 and HDAC2, HDAC3 also is phosphorylated by protein kinase CK2 (39). Surprisingly, unlike other members of the class I HDAC family, HDAC8 is not phosphorylated by protein kinase CK2 (39). However, it is possible that kinases other than protein kinase CK2 phosphorylate HDAC8 and modulate its activity. A complete understanding of how phosphorylation regulates the actions of class I HDACs requires a thorough determination of whether HDAC8 is a phosphoprotein and, if so, what kinase is responsible and what the functional consequences are.
HDAC8 cDNA was identified initially by three independent groups using sequence homology database searches with class I HDAC proteins (6, 20, 40). The HDAC8 gene encodes a 377-amino-acid protein with a predicted molecular mass of 45 kDa and is located on the X chromosome at position q21.2-q21.3 or q13 (6, 40). Protein sequence comparisons of HDAC8 reveal a 37% similarity to HDAC1. In Northern blot analyses, the size of HDAC8 mRNA is between 1.7 and 2.4 kb, and HDAC8 mRNA is expressed in multiple human organs, including the liver, heart, brain, lung, pancreas, placenta, prostate, and kidney. Consistent with the presence of a stretch of basic residues that could serve as a nuclear localization signal, HDAC8 is predominantly located in the nucleus. A recent report suggests that the inv (16) fusion protein specifically associates with HDAC8 (11).
Although sequence analysis of HDAC8 revealed consensus phosphorylation sites for protein kinase A (PKA) and protein kinase CK2, our previous studies showed that HDAC8 was not phosphorylated by protein kinase CK2 in vitro (39). In the present study, we show that HDAC8 is phosphorylated instead by PKA both in vitro and in vivo. Most interestingly, phosphorylation of HDAC8 by PKA inhibits its deacetylase activity, which results in the hyperacetylation of histones H3 and H4. Thus, our findings uncover a novel mechanism of class I HDAC regulation.
MATERIALS AND METHODS
Viruses and plasmids.
A recombinant adenovirus that expresses Flag-tagged HDAC8 was generated by using the AdEasy system (19). pGEX4T-3-HDAC8, which expresses a glutathione S-transferase (GST)-HDAC8 fusion protein, and pCEP4F-HDAC8, which expresses Flag epitope-tagged HDAC8, have been described previously (20, 39). pGST-HDAC8(S39A) and pGST-HDAC8(S39E) were generated from pGEX4T-3-HDAC8 with the Quick-change site-directed mutagenesis kit (Stratagene) and confirmed by DNA sequencing. For the S39A mutant, oligodeoxynucleotides 5′-GATCCCCAAACGGGCCGCTATGGTGCATTCTTTG and 5′-CAAAGAATGCACCATAGCGGCCCGTTTGGGGATC were used as primers for PCR. For S39E, the primers were 5′-GATCCCCAAACGGGCCGAAATGGTGCATTCTTTG and 5′-CAAAGAATGCACCATTTCGGCCCGTTTGGGGATC. By identical strategies, pCEP4F-HDAC8(S39A) and pCEP4F-HDAC8(S39E) were generated from pCEP4F-HDAC8.
Cell culture and transfection.
HeLa cells were maintained in 75-cm2 tissue culture flasks in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 IU of penicillin per ml, and 100 μg of streptomycin per ml. Transfections were performed with Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer's instruction. Forty-eight hours after transfection, some cultures received forskolin, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), or the myristoylated PKA inhibitor 14-22 amide (PKI; Calbiochem) (9, 14, 35).
Immunoprecipitation and immunoblotting analysis.
Anti-Flag M2, anti-phosphoserine, anti-acetyl H3, and anti-acetyl H4 antibodies were purchased from Sigma, Zymed, Upstate, and Cell Signaling, respectively. Rabbit polyclonal anti-HDAC8 antibody was raised against a GST-tagged HDAC8 fusion protein containing the C-terminal region, residues 305 to 377, of HDAC8. For immunoprecipitations, cells were rinsed with ice-cold phosphate-buffered saline and lysed in 0.5 ml of modified radioimmunoprecipitation assay buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 1% NP-40, 5 mM NaF, 0.2 mM Na3VO4, and a cocktail of protease inhibitors. The lysates were immunoprecipitated with primary antibodies for 3 h to overnight at 4°C, and the immunocomplexes were collected, washed extensively, and resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). For immunoblot assays, samples were transferred onto nitrocellulose membranes. Membranes were then probed with the appropriate antibodies, and proteins recognized by the antibodies were detected with the Chemiluminescent Detection Kit (Pierce).
In vivo phosphate labeling.
HeLa cells infected with the Flag-HDAC8-expressing adenovirus for 48 h were preincubated with phosphate-free Dulbecco's modified Eagle's medium for 1 h before labeling. Cells were then treated with 32Pi at a concentration of 0.5 mCi/ml for 4 h at 37°C and lysed in a modified radioimmunoprecipitation assay buffer. In some experiments, cells were treated with H-89 for 45 min before harvest. Flag-HDAC8 protein was immunoprecipitated with M2 agarose and resolved by SDS-8% PAGE. Phosphorylated Flag-HDAC8 was detected by autoradiography.
Kinase assays.
In vitro PKA kinase assays were performed as previously described (39). Briefly, for each reaction, 1 μg of GST fusion protein was incubated with 2.5 U of the recombinant catalytic subunit of PKA (NEB) in the presence of 5 μCi of [γ-32P]ATP, 10 μM ATP, and manufacturer-supplied kinase buffers in a 20-μl total volume for 20 min at 30°C. Reactions were terminated by addition of SDS sample buffer, followed by heating at 100°C for 5 min. After electrophoresis, gels were stained with Coomassie blue, dried, and autoradiographed. Cellular PKA activation assays were performed as previously described with Kemptide (Upstate) as the substrate (28). For assays of HDAC8 phosphorylation by cellular PKA, GST-HDAC8 was conjugated to glutathione beads and incubated with 100 μg of HeLa cell extract overnight at 4°C. Beads were washed three times with lysis buffer and once with kinase buffer before kinase reactions were performed as described above.
Phosphopeptide mapping and phosphoamino acid analysis.
PKA-phosphorylated HDAC8 was resolved by SDS-PAGE and transferred to polyvinylidene difluoride membrane, and the membrane was exposed to X-ray film. The HDAC8-specific band was excised and digested with l-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK)-treated trypsin (Worthington) for 6 h at 37°C, followed by an additional 10 h incubation in the presence of fresh trypsin. Tryptic digests were separated by two-dimensional electrophoresis on thin-layer cellulose with the HTLE-700 system as described previously (26, 39). For phosphoamino acid analysis, 1/10 of the sample volume was subjected to partial HCl hydrolysis by incubation with 6 N HCl for 1 h at 110°C.
HDAC assay.
The enzymatic activity of HDAC8 was assayed with purified core histones as described previously (39).
RESULTS
Phosphorylation of HDAC8 by PKA.
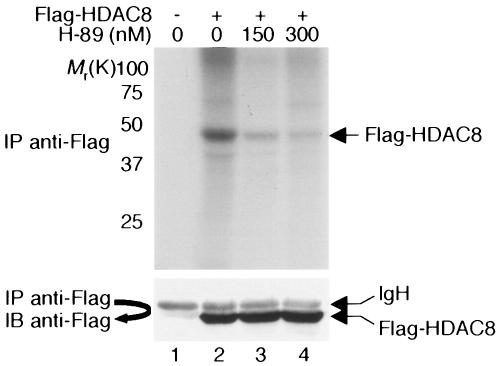
HDAC8 contains two potential phosphorylation sites for protein kinase CK2 and one potential phosphorylation site for PKA (illustrated in Fig. 1A and B) (20). The potential PKA phosphorylation site, located at Ser39 of HDAC8, is conserved between the human and mouse HDAC8s but not among different class I HDACs (Fig. 1B). Because previous studies did not detect HDAC8 phosphorylation by protein kinase CK2 (39), we asked whether PKA phosphorylated HDAC8. HeLa cells were infected with a recombinant adenovirus expressing Flag-HDAC8 and metabolically labeled with 32Pi. Cell extracts were immunoprecipitated with an anti-Flag antibody. As shown in Fig. 2 (lane 2, upper panel), HDAC8 incorporated 32P, indicating that it is phosphorylated in vivo. Treatment of cells with H-89, a potent inhibitor of PKA (9), reduced the phosphorylation of HDAC8 in a dose-dependent manner (lanes 3 and 4).
FIG. 1.
Amino acid sequences of HDAC8. (A) Amino acid sequence of human HDAC8 (AF230097). Residues within the putative catalytic domain are highlighted in bold. Potential phosphorylation sites, as determined by phosphobase detection (http://www.cbs.dtu.dk/databases/PhosphoBase/predict/predict.html), are underlined. The potential PKA phosphorylation site (Ser39) is marked with an asterisk. (B) Top, a potential PKA phosphorylation site in HDAC8 determined by database sequence analysis. Middle and bottom, comparison of the human HDAC8 sequence surrounding Ser39 with other human class I HDAC sequences and with the mouse HDAC8 sequence.
FIG. 2.
In vivo phosphorylation of HDAC8. HeLa cells were infected with a recombinant adenovirus that expresses Flag-HDAC8 and were labeled with 32Pi for 4 h. Cell extracts were immunoprecipitated with an anti-Flag antibody, and immunoprecipitates (IP) were resolved on an SDS-8% polyacrylamide gel. Phosphoproteins were visualized by autoradiography. As a negative control (lane 1), cells were infected with an adenovirus that expresses GFP alone. In lanes 3 and 4, cells were treated with the indicated dose of H-89 for 45 min before harvest. The positions of the molecular weight markers (weights are in thousands) are indicated on the left. Immunoprecipitates were immunoblotted with an anti-Flag antibody to ensure equal amounts of Flag-HDAC8 in lanes 2 to 4 (bottom panel). IgH, immunoglobulin H.
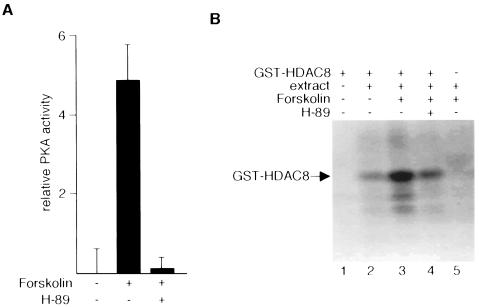
In a companion set of experiments, we examined the role of PKA in HDAC8 phosphorylation. We first treated HeLa cells with or without forskolin, a potent activator of adenyl cyclase, the enzyme that catalyzes the conversion of ATP to cyclic AMP (cAMP). PKA activity was monitored in HeLa cell lysates with Kemptide as the substrate. Treatment of cells with forskolin increased PKA activity, and this increase largely was abolished by cotreatment of cells with H-89 (Fig. 3A). Next, we incubated purified recombinant GST-HDAC8 with extracts of HeLa cells treated with or without forskolin for 45 min. Kinase assays were performed in the presence and absence of H-89. Lysates of forskolin-treated cells clearly phosphorylated HDAC8 to a greater extent than did lysates of untreated cells (Fig. 3B, compare lanes 2 and 3). Moreover, H-89 inhibited HDAC8 phosphorylation by lysates of forskolin-treated cells (lane 4). Together, these results strongly suggest that endogenous PKA phosphorylates HDAC8.
FIG. 3.
Stimulation of HDAC8 phosphorylation by forskolin. (A) With filter-binding assays, PKA activities were determined in extracts prepared from HeLa cells treated with 10 μM forskolin for 45 min in the presence or absence of H-89 (10 μM). (B) Bacterially expressed, purified GST-HDAC8 was incubated with HeLa cell extracts prepared from cells that were untreated or treated with 10 μM forskolin for 45 min. H-89 (10 μM) was added to the in vitro phosphorylation reaction mixtures as indicated (lane 4). GST was used as a negative control (lane 5).
To show conclusively that PKA phosphorylates HDAC8, in vitro kinase assays were performed with purified GST-HDAC8 and catalytically active recombinant PKA. PKA phosphorylated HDAC8 in vitro (Fig. 4A, compare lanes 7 and 8), and phosphorylation was inhibited by H-89 (lanes 9 to 11). Maximal inhibition was achieved with 10 μM H-89. For comparison, we show that PKA phosphorylates HDAC8 as effectively as it phosphorylates the known PKA substrate histone H2B (lane 3). In addition, H-89 inhibited the phosphorylation of HDAC8 and H2B at similar doses (lanes 4 to 6).
FIG. 4.
In vitro phosphorylation of HDAC8 with purified PKA. (A) Purified GST-HDAC8 was incubated with PKA in the presence or absence of H-89 (0.15 μM in lanes 4 and 9, 2 μM in lanes 5 and 10, 10 μM in lanes 6 and 11), and in vitro kinase reactions were performed. Proteins were resolved by SDS-PAGE, and 32P-labeled proteins were visualized by autoradiography. GST (lane 1) and GST-H2B (lanes 2 to 6) were used as negative and positive controls, respectively. (B) GST-HDAC8 was phosphorylated by PKA in the presence of [γ-32P]ATP. Radiolabeled GST-HDAC8 was eluted from an SDS-polyacrylamide gel, digested with trypsin, and analyzed by TLC.
To determine the number of sites phosphorylated by PKA in vitro, purified GST-HDAC8 was incubated with PKA in the presence of [γ-32P]ATP and resolved by SDS-PAGE. Phosphorylated HDAC8 was eluted from the gel, digested with trypsin, and subjected to two-dimensional separation on a thin-layer-chromatography (TLC) plate. Consistent with the observation that HDAC8 contains one potential PKA site, tryptic digests contained one predominant phosphopeptide (Fig. 4B). Multiple minor spots were observed and may represent additional phosphorylation sites or, more likely, products of partial tryptic digestion.
PKA phosphorylates HDAC8 on serine.
GST-HDAC8 phosphorylated in vitro by PKA was excised from an SDS gel and subjected to phosphoamino acid analysis. Results of this experiment unambiguously show that PKA, a serine-threonine kinase, phosphorylates HDAC8 exclusively on serine (Fig. 5A). To confirm this, HeLa cells expressing Flag-HDAC8 were treated with forskolin and anti-Flag immunocomplexes were Western blotted with an anti-phosphoserine antibody. As shown in Fig. 5B (top), HDAC8 was indeed phosphorylated on serine in forskolin-treated cells (lane 3). In cells receiving both forskolin and H-89, little if any serine phosphorylation was seen (lane 4). Differences in the amounts of serine phosphorylation were not the result of differences in the amounts of Flag-HDAC8, which were comparable under all conditions (Fig. 5B, bottom).
FIG. 5.
Identification of serine as the phosphoacceptor residue in HDAC8. (A) HDAC8 was phosphorylated by PKA in the presence of [γ-32P]ATP and eluted from an SDS-polyacrylamide gel. After partial acid hydrolysis, the sample was analyzed by cellulose TLC. P-Ser, P-Thr, and P-Tyr indicate the positions of phosphoserine, phosphothreonine, and phosphotyrosine, respectively. (B) HeLa cells were transfected with a plasmid encoding Flag-HDAC8 and treated with forskolin (10 μM) or H-89 (10 μM) as indicated. Cell extracts were immunoprecipitated (IP) with an anti-Flag antibody and immunoblotted (IB) with an anti-phosphoserine antibody (top) or an anti-Flag antibody (bottom). (C) Extracts prepared from HeLa cells treated with forskolin (10 μM) or PKI (10 μM) were immunoprecipitated with an anti-HDAC8 antibody or preimmune serum and immunoblotted with an anti-phosphoserine antibody (top) or an anti-HDAC8 antibody (bottom).
Finally, we show that PKA phosphorylates endogenous HDAC8 on serine (Fig. 5C). HDAC8 was immunoprecipitated from extracts of untreated and forskolin-treated HeLa cells with an anti-HDAC8 antibody. Immune complexes were immunoblotted with an anti-phosphoserine antibody. Similar to our findings with overexpression of epitope-tagged HDAC8, endogenous HDAC8 was phosphorylated on serine in untreated cells (lane 3) and, to a greater extent, in cells receiving forskolin (lane 4). Like H-89, the highly specific PKA inhibitor PKI (14) blocked the serine phosphorylation of HDAC8 (lane 5).
Ser39 is the major phosphorylation site of HDAC8.
HDAC8 contains one consensus PKA recognition motif, with Ser39 as the potential phosphoacceptor site (Fig. 1). Thus, it seemed likely that PKA phosphorylates HDAC8 at Ser39, and this was confirmed as follows. HeLa cells were transfected with plasmids encoding wild-type Flag-HDAC8 or Flag-HDAC8 containing a Ser-to-Ala substitution at residue 39. Cells were treated with or without forskolin, and anti-Flag immune complexes were Western blotted with anti-phosphoserine antibody. As shown in Fig. 6A (top), replacement of Ser39 with Ala greatly reduced the amount of serine-phosphorylated HDAC8 in forskolin-treated cells (compare lanes 2 and 3). In fact, the amounts of phosphorylated HDAC8(S39A) in forskolin-treated cells were less than the amounts of phosphorylated wild-type HDAC8 in untreated cells (compare lanes 3 and 4). We also found that recombinant PKA phosphorylated wild-type GST-HDAC8 to a much greater extent than it phosphorylated GST-HDAC8 containing an Ala-to-Ser substitution at residue 39 (Fig. 6B, top, compare lanes 4 and 6). Together, our data indicate that phosphorylation of HDAC8 by PKA occurs primarily at Ser39.
FIG. 6.
Identification of Ser39 as the phosphorylation site within HDAC8. (A) HeLa cells expressing Flag-HDAC8 or the Flag-HDAC8(S39A) mutant form were treated with forskolin (10 μM) as indicated. Cell extracts were immunoprecipitated (IP) with an anti-Flag antibody, and immune complexes were then immunoblotted (IB) with either an anti-phosphoserine antibody (top) or an anti-Flag antibody (bottom). (B) Purified GST-HDAC8 and GST-HDAC8(S39A) proteins were used as substrates for in vitro phosphorylation by PKA. The proteins were resolved by SDS-PAGE, and the 32P-radiolabeled proteins were visualized by autoradiography (top). Coomassie blue staining was performed before autoradiography to visualize the locations and amounts of the different proteins (bottom). The values on the left are molecular weights in thousands.
Phosphorylation of HDAC8 by PKA inhibits its enzymatic activity.
Unlike other class I HDACs, nothing is known about the mechanisms that regulate the activity of HDAC8. To determine whether phosphorylation of HDAC8 by PKA modulates its deacetylase activity, HeLa cells expressing Flag-HDAC8 were treated with or without forskolin and H-89, and anti-Flag immune complexes were assayed for deacetylase activity with core histones as the substrate. Treatment of cells with forskolin greatly reduced the deacetylase activity of HDAC8 (Fig. 7A, top, compare columns 2 and 3), and cotreatment of cells with H-89 negated the inhibitory effects of forskolin on HDAC8 activity (column 4). We also compared the deacetylase activities of wild-type HDAC8 and mutant HDACs in which Ser39 was replaced with Ala or Glu. Alanine substitution, which eliminates HDAC8 phosphorylation by PKA, had no effect on deacetylase activity (column 5), whereas Glu substitution, which mimics phosphorylation, reduced deacetylase activity as effectively as did forskolin treatment of cells expressing wild-type HDAC8 (column 6).
FIG. 7.
HDAC8 phosphorylation inhibits enzymatic activity. (A) HeLa cells were transfected with constructs expressing Flag-tagged wild-type or mutant HDAC8, as indicated. After treatment of cells with forskolin (10 μM) and H-89 (10 μM), Flag immunoprecipitates (IP) were assayed for HDAC activity. All experiments were normalized to equal amounts of DNA with parental expression vectors. Data shown are the average results ± the standard deviation from three separate transfections. Immunoprecipitates were immunoblotted (IB) with an anti-Flag antibody to ensure approximately equal amounts of Flag fusion proteins in reactions 2 to 6 (a representative blot is shown at the bottom). (B) Purified, GST, GST-HDAC8, GST-HDAC8(S39A), and GST-HDAC8(S39E) were assayed for deacetylase activity. All experiments were performed in triplicate, and the data shown are the average ± the standard deviation. Coomassie blue-stained gels were used to show approximately equal amounts of GST and GST fusion proteins used in each reaction mixture (a representative gel is shown at the bottom). The values on the left are molecular weights in thousands.
Unlike other class I HDACs, HDAC8 does not require protein cofactors for activity in vitro (20). Consistent with this earlier finding, purified wild-type HDAC8 deacetylated core histones in in vitro deacetylase assays (Fig. 7B, top, column 2). For unknown reasons, mutation of Ser39 to Ala slightly enhanced the deacetylase activity of HDAC8 (column 3). More importantly, and in agreement with the data in Fig. 7A, mutation of Ser39 to Glu significantly reduced the deacetylase activity of HDAC8 (column 4).
Phosphorylation of HDAC8 by PKA increases the acetylation of histones H3 and H4.
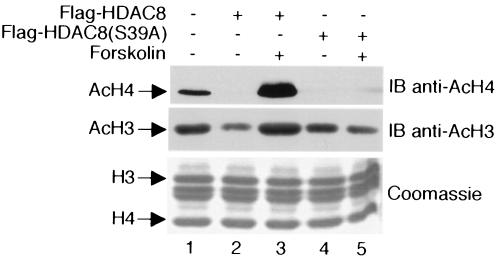
Earlier studies suggested that HDAC8 preferentially deacetylates histones H3 and H4 (6, 20, 40). To determine how decreases in HDAC8 activity resulting from PKA phosphorylation relate to histone acetylation in vivo, HeLa cells expressing wild-type HDAC8 or HDAC8(S39A) were treated with or without forskolin and core histones prepared from cell extracts were Western blotted with an antibody to acetylated histone H3 or H4. As expected, ectopic expression of wild-type HDAC8 in the absence of forskolin reduced the abundance of acetylated H3 and H4 (Fig. 8, compare lanes 1 and 2). Treatment of cells with forskolin, and consequent phosphorylation of HDAC8 by PKA, negated the capacity of HDAC8 to deacetylate H3 and H4 (lane 3). In fact, the amounts of acetylated H3 and H4 were greater in forskolin-treated cells expressing HDAC8 than in cells not expressing ectopic HDAC8 (compare lanes 1 and 3). HDAC8(S39A) also reduced the acetylation of H4 and, to a lesser extent, H3 (lane 4); however, the inhibitory effects of this mutant form were not reversed by forskolin (lane 5). These data show that phosphorylation of HDAC8 by PKA has a major effect on the acetylation status of histones H3 and H4.
FIG. 8.
HDAC8 phosphorylation increases histone H3 and H4 acetylation. HeLa cells were transfected with plasmids expressing Flag-HDAC8 or Flag-HDAC8(S39A). After treatment with or without forskolin (10 μM), core histones were prepared and analyzed by a Western blot (immunoblot [IB]) assay with an anti-acetylated-H4 antibody (top). Subsequently, the blot was stripped and reprobed with an anti-acetylated-H3 antibody (middle). A separate SDS-polyacrylamide gel was prepared in parallel to assess the qualities of core histones in each reaction (bottom).
DISCUSSION
HDAC proteins are key regulators of many cellular processes, and as anticipated, their activities are tightly controlled. Like many cellular enzymes, the activities of most, if not all, of the HDACs can be regulated by phosphorylation. For instance, phosphorylation of two class II enzymes, HDAC4 and HDAC5, results in recruitment of 14-3-3 proteins, which in turn elicits the nuclear export of HDACs and the derepression of HDAC target genes (16, 43). The actions of class I HDACs also are regulated by phosphorylation. Protein kinase CK2 phosphorylates human HDAC1 at Ser421 and Ser423 and human HDAC2 at Ser394, Ser411, and Ser424. Loss of phosphorylation of any of these residues by mutation or phosphatase treatment both reduces enzymatic activity and impedes protein complex formation (33, 36, 39). Conversely, hyperphosphorylation of HDAC1 and HDAC2 by phosphatase inhibition increases activity and changes complex formation (13).
In this study, we show that HDAC8 is a phosphoprotein and that PKA phosphorylates HDAC8 in vitro and in vivo. Unlike HDAC1 and HDAC2, which are phosphorylated on C-terminal residues, HDAC8 is phosphorylated in the N terminus at Ser39. The crystal structure of the Aquifex aeolicus HDAC homolog, HDLP, shows that Ser29 (corresponding to Ser39 of human HDAC8) lies within the second α helix, adjacent to the first loop, of HDLP (12). Although the overall homology between HDAC8 and other class I HDACs is high, the N-terminal portion of HDAC8 is not similar to the N-terminal region of other HDACs. This observation suggests that HDAC8 phosphorylation has consequences distinct from those resulting from the phosphorylation of other class I HDAC enzymes. In agreement with this idea, phosphorylation of HDAC1 and HDAC2 increases their deacetylation activity, whereas phosphorylation of HDAC8 by PKA reduces HDAC8's activity and results in the hyperacetylation of histones H3 and H4. Our data advocate that the cAMP signaling pathway mediated by PKA modulates the enzymatic activity of HDAC8, which in turn shifts the balance of histone acetylation and deacetylation. In this scenario, phosphorylation of HDAC8 by PKA promotes chromatin decondensation and transcriptional activation. Alternatively, phosphorylation of HDAC8 by PKA could conceivably enhance the acetylation of nonhistone proteins. An increase in acetylation of transcription factors, for example, may alter their abilities to initiate transcription. Further experiments to define nonhistone substrates for HDAC8 will help address this issue.
Besides deacetylation of core histones, nothing is known about the biological functions of HDAC8. However, the physiological responses mediated by PKA are known to include cellular proliferation, neuronal signaling, and cancer development (34). Major downstream targets of PKA include the cAMP-responsive element-binding protein (CREB), CREM, and NF-κB. CREB is phosphorylated by PKA at Ser133, and phosphorylation recruits coactivators such as CREB-binding protein and its homolog p300, both of which are HATs (2, 32). Interestingly, a recent study demonstrated that HDAC inhibitors augment CREB activity by prolonging CREB phosphorylation at Ser133 on chromosomes (28). Further, it was shown that HDAC1 associates with and blocks the Ser133 phosphorylation of CREB during the prestimulus and attenuation phases of the cAMP response. Moreover, HDAC1 promotes Ser133 dephosphorylation via a stable interaction with protein phosphatase 1 (PP1) (8). This finding suggests that the dephosphorylation of CREB and deacetylation of promoter-bound histone are coordinated by HDAC1-PP1 complexes that are important in silencing CREB activity in unstimulated cells. Surprisingly, the enzymatic activity of HDAC1 seems to be unrelated to its association with PP1. However, overexpression of HDAC1 does not completely inhibit CREB phosphorylation, signifying that HDAC1 cannot be solely responsible for the negative regulation in CREB activation. It is possible, therefore, that phosphorylation of HDAC8 by PKA may provide additional effects on the activation of CREB. In this case, by modulating its enzymatic activity, HDAC8 may play a distinct role as a signal-responsive suppressor that regulates CREB activity.
How might phosphorylation of HDAC8 by PKA down-regulate its enzymatic activity? One possibility is that phosphorylation of HDAC8 by PKA causes a conformational change within HDAC8 that renders it less active. Consistent with this idea, we found that purified HDAC8(S39A) was slightly more active than wild-type HDAC8 in an in vitro deacetylase assay, whereas HDAC8(S39E) was less active. However, phosphorylation of HDAC8 by PKA or mutation of HDAC8 to mimic hyperphosphorylation (S39E) led to an almost complete loss of deacetylase activity in vivo. This intriguing finding suggests that, in addition to a possible conformational change, there must be alternative or additional mechanisms operating to down-modulate the activity of phospho-HDAC8 in vivo. Perhaps phosphorylation by PKA affects the cellular localization of HDAC8 or the ability of HDAC8 to interact with other proteins. Experiments designed to explore each of these possibilities are under way in our laboratory.
Our data showing that PKA phosphorylates HDAC8 provide yet another example of how the activities of human class I HDACs are regulated by protein kinases. A previous study shows that, in addition to kinases, protein phosphatases such as PP1 play a critical role in the phosphorylation status of HDAC1 and HDAC2 (13). What is not known is whether HDAC8 phosphorylation is also modulated by phosphatases. Also unclear is whether cross talk exists between the different posttranslational modifications of HDAC8. For example, a potential N-glycosylation site is present at Asn136 in HDAC8, and it is conceivable that phosphorylation of Ser39 may affect glycosylation of Asn136 and vice versa. Further experiments are necessary to address each of these important issues.
Acknowledgments
We thank Erding Hu and Stefan Kass for the HDAC8 cDNA, Wen Long Bai and Nancy Olashaw for discussion and critical reading of the manuscript, and the Moffitt Cancer Center Core Facility for technical support.
This work was supported by grants from the NIH (GM58486 and GM64850) and the Kaul Foundation to E.S. H.L. is a recipient of an American Heart Association postdoctoral fellowship.
REFERENCES
- 1.Ayer, D. E. 1999. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 9:193-198. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 3.Bartl, S., J. Taplick, G. Lagger, H. Khier, K. Kuchler, and C. Seiser. 1997. Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol. Cell. Biol. 17:5033-5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 5.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 6.Buggy, J. J., M. L. Sideris, P. Mak, D. D. Lorimer, B. McIntosh, and J. M. Clark. 2000. Cloning and characterization of a novel human histone deacetylase, HDAC8. Biochem. J. 350:199-205. [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, R., P. Kwon, Y. Yan-Neale, L. Sambuccetti, D. Fischer, and D. Cohen. 2001. Mammalian histone deacetylase 1 protein is posttranslationally modified by phosphorylation. Biochem. Biophys. Res. Commun. 283:445-453. [DOI] [PubMed] [Google Scholar]
- 8.Canettieri, G., I. Morantte, E. Guzman, H. Asahara, S. Herzig, S. D. Anderson, J. R. Yates III, and M. Montminy. 2003. Attenuation of a phosphorylation-dependent activator by an HDAC-PP1 complex. Nat. Struct. Biol. 10:175-181. [DOI] [PubMed] [Google Scholar]
- 9.Chijiwa, T., A. Mishima, M. Hagiwara, M. Sano, K. Hayashi, T. Inoue, K. Naito, T. Toshioka, and H. Hidaka. 1990. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J. Biol. Chem. 265:5267-5272. [PubMed] [Google Scholar]
- 10.David, G., M. A. Neptune, and R. A. DePinho. 2002. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem. 277:23658-23663. [DOI] [PubMed] [Google Scholar]
- 11.Durst, K. L., B. Lutterbach, T. Kummalue, A. D. Friedman, and S. W. Hiebert. 2003. The inv(16) fusion protein associates with corepressors via a smooth muscle myosin heavy-chain domain. Mol. Cell. Biol. 23:607-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finnin, M. S., J. R. Donigian, A. Cohen, V. M. Richon, R. A. Rifkind, P. A. Marks, R. Breslow, and N. P. Pavletich. 1999. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401:188-193. [DOI] [PubMed] [Google Scholar]
- 13.Galasinski, S. C., K. A. Resing, J. A. Goodrich, and N. G. Ahn. 2002. Phosphatase inhibition leads to histone deacetylases 1 and 2 phosphorylation and disruption of corepressor interactions. J. Biol. Chem. 277:19618-19626. [DOI] [PubMed] [Google Scholar]
- 14.Gangolli, E. A., M. Belyamani, S. Muchinsky, A. Narula, K. A. Burton, G. S. McKnight, M. D. Uhler, and R. L. Idzerda. 2000. Deficient gene expression in protein kinase inhibitor α null mutant mice. Mol. Cell. Biol. 20:3442-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, L., M. A. Cueto, F. Asselbergs, and P. Atadja. 2002. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J. Biol. Chem. 277:25748-25755. [DOI] [PubMed] [Google Scholar]
- 16.Grozinger, C. M., and S. L. Schreiber. 2000. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA 97:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 18.He, L. Z., F. Guidez, C. Tribioli, D. Peruzzi, M. Ruthardt, A. Zelent, and P. P. Pandolfi. 1998. Distinct interactions of PML-RARα and PLZF-RARα with co-repressors determine differential responses to RA in APL. Nat. Genet. 18:126-135. [DOI] [PubMed] [Google Scholar]
- 19.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, E., Z. Chen, T. Fredrickson, Y. Zhu, R. Kirkpatrick, G. F. Zhang, K. Johanson, C. M. Sung, R. Liu, and J. Winkler. 2000. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J. Biol. Chem. 275:15254-15264. [DOI] [PubMed] [Google Scholar]
- 21.Humphrey, G. W., Y. Wang, V. R. Russanova, T. Hirai, J. Qin, Y. Nakatani, and B. H. Howard. 2001. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 276:6817-6824. [DOI] [PubMed] [Google Scholar]
- 22.Johnstone, R. W. 2002. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat. Rev. Drug Discov. 1:287-299. [DOI] [PubMed] [Google Scholar]
- 23.Kelly, W. K., O. A. O'Connor, and P. A. Marks. 2002. Histone deacetylase inhibitors: from target to clinical trials. Expert Opin. Investig. Drugs 11:1695-1713. [DOI] [PubMed] [Google Scholar]
- 24.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 25.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 26.Lee, H., and W. Bai. 2002. Regulation of estrogen receptor nuclear export by ligand-induced and p38-mediated receptor phosphorylation. Mol. Cell. Biol. 22:5835-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melnick, A., and J. D. Licht. 2002. Histone deacetylases as therapeutic targets in hematologic malignancies. Curr. Opin. Hematol. 9:322-332. [DOI] [PubMed] [Google Scholar]
- 28.Michael, L. F., H. Asahara, A. I. Shulman, W. L. Kraus, and M. Montminy. 2000. The phosphorylation status of a cyclic AMP-responsive activator is modulated via a chromatin-dependent mechanism. Mol. Cell. Biol. 20:1596-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neely, K. E., and J. L. Workman. 2002. The complexity of chromatin remodeling and its links to cancer. Biochim. Biophys. Acta 1603:19-29. [DOI] [PubMed] [Google Scholar]
- 30.Ng, H. H., and A. Bird. 2000. Histone deacetylases: silencers for hire. Trends Biochem. Sci. 25:121-126. [DOI] [PubMed] [Google Scholar]
- 31.Ogryzko, V. V., T. H. Hirai, V. R. Russanova, D. A. Barbie, and B. H. Howard. 1996. Human fibroblast commitment to a senescence-like state in response to histone deacetylase inhibitors is cell cycle dependent. Mol. Cell. Biol. 16:5210-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 33.Pflum, M. K., J. K. Tong, W. S. Lane, and S. L. Schreiber. 2001. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J. Biol. Chem. 276:47733-47741. [DOI] [PubMed] [Google Scholar]
- 34.Robinson-White, A., and C. A. Stratakis. 2002. Protein kinase A signaling: “cross-talk” with other pathways in endocrine cells. Ann. N. Y. Acad. Sci. 968:256-270. [DOI] [PubMed] [Google Scholar]
- 35.Schmitt, J. M., and P. J. Stork. 2001. Cyclic AMP-mediated inhibition of cell growth requires the small G protein Rap1. Mol. Cell. Biol. 21:3671-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun, J. M., H. Y. Chen, M. Moniwa, D. W. Litchfield, E. Seto, and J. R. Davie. 2002. The transcriptional repressor Sp3 is associated with CK2-phosphorylated histone deacetylase 2. J. Biol. Chem. 277:35783-35786. [DOI] [PubMed] [Google Scholar]
- 37.Thiagalingam, S., K. H. Cheng, H. J. Lee, N. Mineva, A. Thiagalingam, and J. F. Ponte. 2003. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann. N. Y. Acad. Sci. 983:84-100. [DOI] [PubMed] [Google Scholar]
- 38.Tong, J. K., C. A. Hassig, G. R. Schnitzler, R. E. Kingston, and S. L. Schreiber. 1998. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395:917-921. [DOI] [PubMed] [Google Scholar]
- 39.Tsai, S. C., and E. Seto. 2002. Regulation of histone deacetylase 2 by protein kinase CK2. J. Biol. Chem. 277:31826-31833. [DOI] [PubMed] [Google Scholar]
- 40.Van den Wyngaert, I., W. de Vries, A. Kremer, J. Neefs, P. Verhasselt, W. H. Luyten, and S. U. Kass. 2000. Cloning and characterization of human histone deacetylase 8. FEBS Lett. 478:77-83. [DOI] [PubMed] [Google Scholar]
- 41.Vigushin, D. M., and R. C. Coombes. 2002. Histone deacetylase inhibitors in cancer treatment. Anticancer Drugs 13:1-13. [DOI] [PubMed] [Google Scholar]
- 42.Wade, P. A., A. Gegonne, P. L. Jones, E. Ballestar, F. Aubry, and A. P. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 23:62-66. [DOI] [PubMed] [Google Scholar]
- 43.Wang, A. H., M. J. Kruhlak, J. Wu, N. R. Bertos, M. Vezmar, B. I. Posner, D. P. Bazett-Jones, and X. J. Yang. 2000. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 20:6904-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wharton, W., J. Savell, W. D. Cress, E. Seto, and W. J. Pledger. 2000. Inhibition of mitogenesis in Balb/c-3T3 cells by trichostatin A. Multiple alterations in the induction and activation of cyclin-cyclin-dependent kinase complexes. J. Biol. Chem. 275:33981-33987. [DOI] [PubMed] [Google Scholar]
- 45.Yang, X. J., and E. Seto. 2003. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr. Opin. Genet. Dev. 13:143-153. [DOI] [PubMed] [Google Scholar]
- 46.You, A., J. K. Tong, C. M. Grozinger, and S. L. Schreiber. 2001. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 98:1454-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]