Abstract
Hemocytes are integral components of mosquito immune mechanisms such as phagocytosis, melanization, and production of antimicrobial peptides. However, our understanding of hemocyte-specific molecular processes and their contribution to shaping the host immune response remains limited. To better understand the immunophysiological features distinctive of hemocytes, we conducted genome-wide analysis of hemocyte-enriched transcripts, and examined how tissue-enriched expression patterns change with the immune status of the host. Our microarray data indicate that the hemocyte-enriched trascriptome is dynamic and context-dependent. Analysis of transcripts enriched after bacterial challenge in circulating hemocytes with respect to carcass added a dimension to evaluating infection-responsive genes and immune-related gene families. We resolved patterns of transcriptional change unique to hemocytes from those that are likely shared by other immune responsive tissues, and identified clusters of genes preferentially induced in hemocytes, likely reflecting their involvement in cell type specific functions. In addition, the study revealed conserved hemocyte-enriched molecular repertoires which might be implicated in core hemocyte function by cross-species meta-analysis of microarray expression data from Anopheles gambiae and Drosophila melanogaster.
Keywords: mosquito, hemocyte, phagocytosis, melanization, antimicrobial peptide, microarray
1. Introduction
Hemocytes are fundamental elements of the mosquito host defense. They mediate immune mechanisms such as phagocytosis, melanization, and production of antimicrobial peptides (AMPs) (Blandin and Levashina, 2007; Christensen et al., 2005; Strand, 2008). Phagocytosis and melanization responses can be initiated within minutes of non-self recognition, albeit to a varying degree depending on the pathogen class (Hillyer et al., 2003b, 2004). In comparison, AMP responses are characterized by transcriptional induction through Rel/NF-κB signaling pathways (Antonova et al., 2009; Shin et al., 2005). The emerging complexity of hemocyte biology and its multifaceted role in host defense lie not only in the temporally distinct and pathogen-specific manifestation of the effector mechanisms these cells mediate, but also because hemocyte responses occur in the context of an elaborate network of processes involving multiple physiological systems and immune-responsive tissues (Glenn et al., 2010; Hillyer, 2010; Schneider, 2009). The interactions between these components give rise to the function and behavior of the immune system as a whole. To begin to investigate the role of hemocytes in such interplay, a better understanding of the unique immunophysiological features of hemocytes becomes increasingly important. In this study, we utilized tissue-enriched expression profiles to critically evaluate hemocyte transcriptome responses to bacterial challenge by resolving patterns of transcriptional changes specific to circulating hemocytes from those that are likely shared by other immune responsive tissues. In addition, we identified conserved hemocyte-enriched molecular repertoires which might be implicated in core hemocyte function by cross-species meta-analysis of microarray expression data from Anopheles gambiae (Pinto et al., 2009) and Drosophila melanogaster (Irving et al., 2005).
Previous transcriptomic studies in dipteran species investigating the molecular physiology of circulating hemocytes (Baton et al., 2009; Irving et al., 2005; Pinto et al., 2009) have demonstrated that comparing hemocyte transcriptome profiles to carcass profiles can provide a useful metric for the screening of transcripts enriched in hemocytes. Tissue comparisons of this nature require careful analysis because (1) the numeric values of the resulting enrichment ratios do not have a readily interpretable biological meaning due to the undefined cellular composition of the carcass, and (2) high enrichment ratios themselves do not necessarily indicate tissue-specific gene expression. Nevertheless, relative rankings of the enrichment ratios in the context of a genome-wide screening can be highly informative because transcripts primarily or exclusively expressed in hemocytes will likely have higher enrichment ratios than most other transcripts. An added advantage of this approach is that it provides a potential means to guard against false-positive findings in studies comparing hemocyte samples, where results may be confounded by cell type heterogeneity. Provided that the proportion of contaminating cell type(s) in the hemocyte sample is lower than that in the carcass sample, enrichment ratios of the transcripts not expressed in hemocytes remain less than 1, and thus may be used as an additional criterion for critical evaluation of transcriptional profiles.
At any given time, the expression of many genes varies between different cell types and between different developmental and physiological states. Tissue-enriched genes, which are highly expressed in one particular tissue type and are either not expressed or are expressed at much lower levels in other tissues, have been hypothesized to be important in the specialized functions of the particular cell types in which they are expressed. Genes of the common host response may be induced in multiple tissue types during infection, whereas some clusters of genes are preferentially induced in specific cell types, likely reflecting their unique function in response to infection. Genes exhibiting tissue specificity during an immune response are particularly interesting in the context of hemocyte biology due to their possible involvement in cell type specific functions, such as intercellular signaling and communication that help coordinate the actions of different infection responsive tissues. Within this conceptual framework, the present study provides a detailed molecular perspective into the characteristic features of the hemocyte transcriptome in the mosquito Aedes aegypti by actively harnessing tissue-enriched expression profiles following bacterial challenge.
2. Materials and methods
2.1 Mosquito rearing and colony maintenance
The A. aegypti Liverpool strain was originally obtained from a colony from the University of London in 1977 and was reared as previously described (Christensen and Sutherland, 1984). Adult female mosquitoes were used for experimentation within 3 days of eclosion.
2.2 Experimental design and replication
Material for each experimental condition was generated from three (bacterial challenge) or four (naïve) separate generations of mosquitoes, and four separate microarray hybridizations using carcass and hemocyte material were performed. The first hybridization compared naïve and E.coli-challenged conditions, the second compared naïve and M. luteus-challenged conditions, and the third and fourth compared all three conditions.
2.3 Bacterial cultures and injection into mosquitoes
Bacteria used for intrathoracic injections were E. coli DH5α and M. luteus (University of Wisconsin-Madison). Cultures were grown to stationary phase in LB (Luria-Bertani) broth at 37 °C, with shaking at 300 rpm. A pulled glass capillary needle containing inoculum (0.5 µl undiluted bacterial culture) was inserted through the cervical membrane between the head and the thorax and the fluid injected as previously described (Hillyer et al., 2004). For each biological replicate, 50 individuals were injected per condition or remained naïve, and survivorship at 24 hours following injection was greater than 90%.
2.4 Mosquito tissue collection
At 24 hours post bacterial challenge, hemolymph was collected from 40 individuals by volume displacement (perfusion) as previously described (Beerntsen and Christensen, 1990). A tear was made above the penultimate abdominal segment of the mosquito, which was then placed on a vacuum saddle. A pulled glass capillary needle, attached to a syringe containing 1X HBSS (Invitrogen, Carlsbad, CA), was inserted through the cervical membrane between the head and the thorax. HBSS was slowly injected, and only the first drop of perfusate from each mosquito was collected into a microfuge tube containing cell lysis buffer (10% SDS, 1 M Tris pH 7.5, 5 mM EDTA), mixed well, and kept on ice. Each remaining carcass after perfusion was collected in parallel to the hemolymph sample by immediately freezing it in a tube on dry ice. Carcasses were stored at −80 °C.
2.5 RNA isolation and purification
Hemocyte total RNA was isolated from hemolymph immediately following collection. The isolation utilized a modification of a method originally developed for isolation of RNA from tissue culture cells (Peppel and Baglioni, 1990). Hemolymph was directly perfused into a 1.5 ml microfuge tube containing 150 µl 5X cell lysis buffer (10% SDS, 1 M Tris pH 7.5, 5 mM EDTA), mixed, and stored on ice. Once all the perfusate was collected, the solution was adjusted to 1X with HBSS and vortexed at medium speed for 5 seconds. Next, 0.3 volumes of ice-cold Solution 2 (42.9 g potassium acetate, 11.2 ml acetic acid, water to 100 ml) was added and the solution was vortexed at medium speed both upright and inverted for 10 seconds, centrifuged (14 k, 4 °C) for 15 min. and the supernatant transferred to a new tube. The solution was then centrifuged (14 k, 4 °C) for 15 min. and the supernatant transferred to a new tube. One half volume of buffered phenol and one half volume of chloroform/isoamyl 24:1 were added, mixed, and centrifuged (14 k, 4 °C) for 10 min. and the supernatant transferred to a new tube. One volume of chloroform/isoamyl 24:1 was added, mixed, centrifuged (14 k, 4 °C) for 10 min. and the supernatant transferred to a new tube. One volume of ice-cold isopropanol was added, mixed, and the RNA was precipitated overnight at −20 °C. RNA was pelleted (14 k, 4 °C) for 30 min. and the supernatant discarded. RNA was washed with 750 µl ice-cold 80% ethanol, centrifuged (14 k, 4 °C) for 10 min. and the supernatant discarded. The RNA was then air dried and resuspended in 10 µl nuclease-free water. Carcass total RNA was isolated using the single-step acid guanidinium thiocyanate-phenol-chloroform extraction method (Chomczynski and Sacchi, 1987) and resuspended in nuclease-free water. The quantity and purity of all RNA samples were measured on a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA). The measured RNA concentrations and RNA integrity were verified by denaturing gel electrophoresis using GelRed™ (Phenix Research Products, Candler, NC) staining.
2.6 Microarray hybridization
Agilent (Santa Clara, CA) 4×44k whole-genome Aedes aegypti microarrays (Nene et al., 2007) were used for hybridizations. To generate amino allyl-modified cDNA (aRNA) coupled with CyDye (GE Healthcare, Piscataway, NJ), Amino Allyl MessageAmp™ II aRNA Kit (Ambion AM1753, Austin, TX) was used according to the manufacturers’ instructions. The in vitro transcription was performed for 14 hours for all samples starting with 500 ng of total RNA. The quantity and purity of all resulting modified aRNA were measured on a NanoDrop spectrophotometer. The measured aRNA concentrations and aRNA size distributions were evaluated by denaturing gel electrophoresis using GelRed™ staining. In order to utilize dye-swap pairs for hybridization, each sample was coupled to both Cy3 and Cy5. The same amount of input aRNA (5–7 µg) was used for all samples within a biological replicate. The dye labeled aRNA concentration, pmol/µl dye and frequency of incorporation (FOI) were measured on a NanoDrop spectrophotometer. If the FOI of the labeled aRNA was outside of the desirable range the dye coupling reaction was repeated. Microarrays were hybridized using the Agilent Gene Expression Hybridization Kit according to manufacturers’ instructions. Two color hybridizations were performed using 125 pmol (700–1200 ng) of each labelled aRNA. Once assembled, arrays were secured in Agilent hybridization chambers and hybridized for 17 hours at 65 °C in an Agilent hybridization oven rotating at 10 rpm. Microarrays were washed according to the manufacturers’ instructions and scanned immediately.
2.7 Microarray data processing and Gene Ontology enrichment analysis
Fluorescence images were obtained from hybridized microarrays at 5 µm resolution using a GenePix 4000B array scanner (Molecular Devices, Foster City, CA) with the PMT gain settings automatically adjusted to a saturation tolerance level of 0.001%. A rray features were quantified in GenePix Pro 6.1 (Axon Instruments) and low quality fluorescence spots were flagged and excluded from subsequent analyses. Extracted results were imported into R environment for differential transcript abundance analysis using the Bioconductor LIMMA package (Smyth, 2004). “Normexp” method (Ritchie et al., 2007) was used for background correction with an offset of 50 for both channels to damp down the variability of the log ratios for low intensity spots while avoiding negative corrected intensities. The A (average intensities) and M (log-ratios) values were averaged among within-array replicate spots, followed by between-array normalization using the “Aquantile” method (Smyth and Speed, 2003) such that the A-values have the same empirical distribution across arrays but the M-values remain unchanged. Following the approach developed in LIMMA for separate channel analysis of two-color microarrays, a linear model was fit to log-intensity data rather than log-ratio data. Because the study utilized dye-swap technical replicates, each biological replicate was included as a coefficient in the linear model. After model fitting, relevant pairwise comparisons were made by specifying appropriate contrasts. Statistical significance of differential transcript abundance was assessed using moderated t-statistics, and p-values were adjusted for multiple testing to control false discovery rate using the Benjamini and Hochberg’s method. The microarray data from this study were deposited to GEO under accession number GSE38744. Gene Ontology (GO) analysis was performed in ErmineJ (Gillis et al., 2010; Lee et al., 2005) using GO annotations retrieved from the UniProtKB-GOA database (Camon et al., 2004) and VecterBase (Lawson et al., 2009). The gene score resampling (GSR) method was used to identify statistically overrepresented GO categories among genes showing high hemocyte-enriched expression pattern. P-values were adjusted for multiple testing to control false discovery rate using the Benjamini and Hochberg’s method. Hierarchical clustering analysis was performed in GeneSpring GX (Agilent Technologies) with average linkage using Pearson’s uncentered correlation coefficient as distance metric.
2.8 Microarray validation
qPCR (quantitative polymerase chain reaction) was used to validate the microarray expression ratios for a selected group of transcripts. PrimeTime™ qPCR Assays (Integrated DNA Technologies, Coralville, IA) were designed and resuspended in TE (10mM Tris, 0.1mM EDTA pH 8.0) according to the manufacturer’s recommendations. qPCR reactions (20 µl, 10 ng total RNA input) were executed using the TaqMan® RNA-to-Ct™ 1-Step Kit (Applied Biosystems, Carlsbad, CA) and an Applied Biosystems 7300 Real-Time PCR system according to the manufacturer’s recommendations. The Comparative Ct Method was utilized using one endogenous control (AAEL000987; 60S ribosomal protein L8) and four target genes (AAEL007967, AAEL000636, AAEL006355, AAEL006361). Validation experiments confirmed the endogenous control could be used for all four targets. All reactions were performed in triplicate. Primer sequences and results are summarized in Supplemental Dataset S1.
2.9 Orthologous group comparisons
A. gambiae hemocyte and carcass microarray data were obtained from (Pinto et al., 2009), and hemocyte enrichment ratios were computed using normalized intensity values reported in their S (stringent) list. D. melanogaster hemocyte and whole larvae data were obtained from (Irving et al., 2005), and processed similarly to generate tissue enrichment ratios, except that enrichment ratios were imputed for genes whose expression data exist only for hemocytes using the median (normalized intensity) value for the whole larvae microarray as the common denominator. Transcripts and their corresponding tissue enrichment ratios were mapped to genes and orthologous groups (OGs) to identify OGs that retained hemocyte-enriched expression patterns across A. aegypti, A. gambiae and D. melanogaster. An OG was considered as such, if there was at least one ortholog from each species that showed hemocyte enrichment. Insect non-supervised orthologous groups (inNOGs) from the eggNOG database (version 2.0) were used as a basis for these comparisons (Muller et al., 2010). For each species, an equal number of genes most highly enriched in hemocytes (as measured by enrichment ratios relative to the carcass or the whole animal within each dataset) were selected for Venn diagram analysis. To determine genes showing the most consistent pattern of high tissue enrichment across species, we compared orthologous groups using the geometric mean of the relative (i.e., within-study) enrichment ratio rankings of the constituent members.
3. Results and discussion
3.1 Hemocyte collection for transcriptome profiling
The methodologies previously employed to collect hemocytes for transcriptomic analyses undoubtedly contributed their own biases to both the transcripts that appear as significantly changed and the magnitude of their changes. In previous studies, hemocytes have been collected via displacement perfusion with cell lysis buffer (Bartholomay et al., 2007), collection from clipped probosces (Abraham et al., 2005; Chen and Laurence, 1987; Chun et al., 2000; Pinto et al., 2009), or collection in a capillary tube following the injection of buffer into the abdomen (Castillo et al., 2006). Not only do different collections contain varying proportions of contaminating cell types, but the hemocyte transcriptome itself can remain dynamic for differing amounts of time and be exposed to varying stimuli prior to either cell lysis or freezing in anticipation of RNA isolation. The displacement perfusion technique utilized in this study was designed to result in minimal contaminating cell types while rapidly initiating hemocyte lysis and capturing transcriptome RNA in the absence of influence from the collection conditions or factors outside of the mosquito hemocoel.
3.2 Microarray experimental design
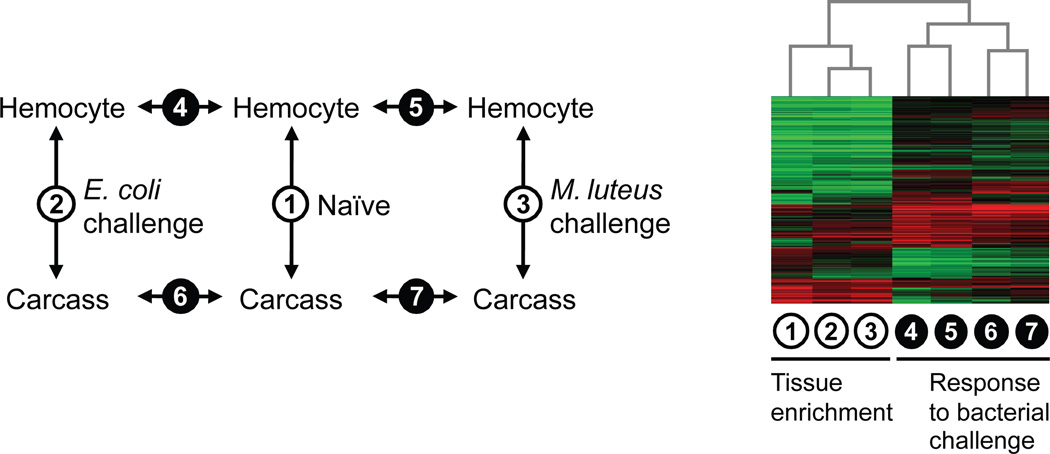
Using A. aegypti whole-genome microarrays (Nene et al., 2007), we profiled the transcriptome of circulating hemocytes and the remaining carcass in both naïve and bacteria-challenged adult female mosquitoes (Fig. 1A). For each biological replication, hemolymph perfusate and the remaining carcass were collected as paired samples at 24 hours post intra-hemocoelic injection of either Escherichia coli or Micrococcus luteus overnight culture (0.5 µl) in parallel with an age-matched naïve group from the same cohort. A paired design was employed so that, within each treatment group, variation in physiological or immunological state of the experimental animals would not be confounded with tissue comparison. Concomitantly-collected hemocyte and carcass samples were co-hybridized on microarrays in accordance with our primary emphasis on the accurate delineation of tissue enrichment ratios. However, ranking transcripts based on tissue enrichment was not the sole aim of the experimental design. We wanted to investigate how transcripts enriched in hemocytes change as a function of bacterial challenge, and conversely which infection-responsive transcripts show marked enrichment in hemocytes. To examine transcriptional changes associated with hemocoelic infection in distinct tissue samples, we adopted a linear model-based framework for analyzing dual-channel hybridizations (i.e., two-color microarrays) as separate single-channels (Smyth, 2004). This analytic approach benefited from consistent high quality of in situ synthesized Agilent microarrays and effective “within” and “between” array normalization methods in LIMMA (Bioconductor) (Ritchie et al., 2007; Smyth and Speed, 2003). The biological and technical dye-swap replicates of the 2×3 experimental conditions consisted of forty separate single-channel intensity profiles (Fig. S1). A hierarchical clustering of the complete dataset showed consistency among the biological replicates and, without exception, technical replicates clustered together suggesting that dye and array effects are smaller than the biological effects. Sample relations further indicated that tissue source is likely a more prominent factor influencing the overall transcriptome profile than infection status.
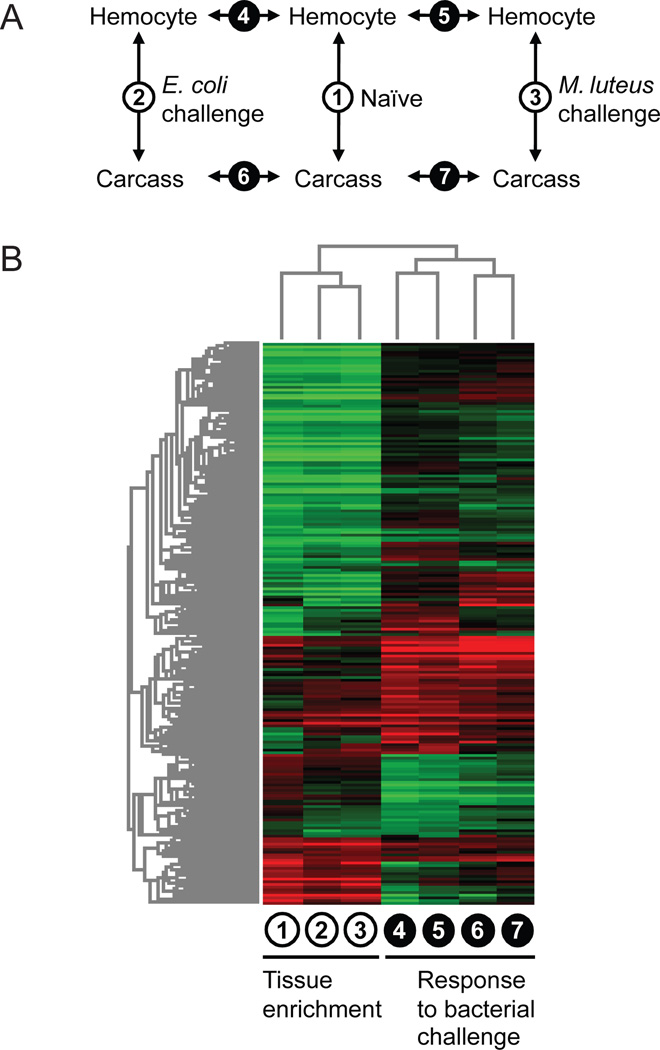
Fig. 1.
(A) Transcriptome profile comparisons among hemocyte and carcass samples from naïve and bacteria-challenged Aedes aegypti. (B) Hierarchical clustering of 3,324 transcripts displaying differential abundance in at least one of the seven pair-wise comparisons (P < 0.01 and fold change > 2).
3.3 Hemocyte-enriched transcriptome in relation to systemic bacterial challenge
We assessed differential transcript abundance between circulating hemocytes and the remaining carcass in naïve, E. coli-challenged and M. luteus-challenged mosquitoes. In addition, we compared E. coli- and M. luteus-challenged hemocyte and carcass transcriptomes to their corresponding naïves to derive infection response profiles for each challenge and tissue type (Fig. 1A). The expression ratios from these comparisons were tested for statistical significance using LIMMA’s moderated t-statistics (Smyth, 2004). In total, 3,324 transcripts displayed significant differences in their abundance (P < 0.01 and fold change > 2) in at least one of the seven pair-wise comparisons. To obtain a global view of the relationship among expression ratio profiles and partitioning of the transcripts in accordance with their tissue enrichment and infection-responsiveness, a two-way hierarchical clustering was performed on the differentially abundant transcripts (Fig. 1B). The resulting heat map indicates that the hemocyte-enriched transcriptome is dynamic and context-dependent. Responses to bacterial challenge in hemocytes and the carcass share common transcriptional patterns, but each also involves unique transcriptional changes, altering the tissue-enriched transcriptome in infected animals relative to the naïve. One implication of this finding is that transcripts specifically or preferentially induced in hemocytes during immune activation may not necessarily be tissue-enriched in naïve animals. Furthermore, the topology of the column dendrogram indicates that the hemocyte-enriched transcriptome profiles (i.e., expression ratios from tissue comparisons) in E. coli- and M. luteus-challenged mosquitoes are overall more similar to each other than either is to the naïve profile, suggesting common infection-associated transcriptional changes independent of the bacterial species used for immune challenge. However, because our hemocoelic infection model involved septic injury via injection, it is conceivable that some, if not most, of the shared transcriptional responses represent wound-healing processes.
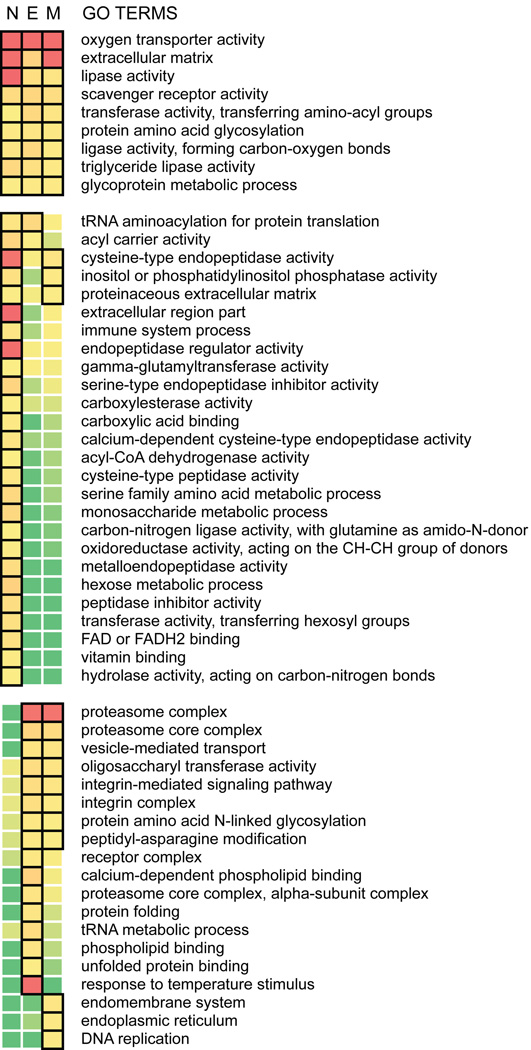
To examine the distinctive features of the hemocyte-enriched transcriptome, we evaluated the statistical association between specific gene ontology (GO) terms and high tissue enrichment ratios using the gene score resampling (GSR) method in ErmineJ (Lee et al., 2005). One advantage of this permutation-based approach, making it well suited for the study, is that it takes into account all measured enrichment ratios without arbitrary thresholding. In addition to complementing observations made at the level of individual transcripts by highlighting overrepresented gene groups and functional categories, this analysis captures the compositional changes in a hemocyte-enriched transcriptome following microbial challenge. Overall, our analyses identified 54 GO terms overrepresented among the hemocyte-enriched transcripts (P < 0.01; Fig. 2), which can be categorized into three broad groups: (1) GO terms that are significant in both naïve and the bacteria-challenged mosquitoes, (2) GO terms that are significant in the naïve mosquitoes but not in either of the bacteria-challenged mosquitoes, and (3) GO terms that are significant only in E. coli- or M. luteus-challenged mosquitoes.
Fig. 2.
GO categories overrepresented among hemocyte-enriched transcripts from Aedes aegypti. N: naïve, E: Escherichia coli-challenged, and M: Micrococcus luteus-challenged conditions. Heatmap represents the degree of statistical significance as determined by gene score resampling. P-value < 0.01 is boxed, and the color gradient of red to green indicates increasing P-values.
Considering the rapid initiation of phagocytosis and melanization reactions upon microbial recognition, it is likely that the molecular components involved in these processes are already in place in naïve mosquitoes. Constitutively-expressed, but highly hemocyte-enriched factors and structural components are particularly interesting in this regard. Members of the prophenoloxidase (PPO), fibrinogen and fibronectin related protein (FREP) and class C scavenger receptor (SCRC) families are among the most highly hemocyte-enriched transcripts in both naïve and bacteria-challenged mosquitoes (Dataset S2). Similar to PPOs and FREPs, carboxylesterase (AAEL004022-RA) and apolipoprotein D (AAEL002520-RA) also rank high when genes are sorted by enrichment ratios. Examination of GO categories reveals oxygen transporter, extracellular matrix, scavenger receptor and lipase activities displaying particularly significant association with tissue enrichment in hemocytes across infection status. These observations recapitulate the significance of hemocytes as the primary source of pattern recognition molecules and components of the PPO cascade. Genes associated with lipid metabolism are noteworthy in our ranked lists considering their potential involvement in phagocytosis, hemolymph clotting and coagulation (Theopold et al., 2004; Yeung and Grinstein, 2007). Cysteine-type endopeptidase and endopeptidase regulator activities, on the contrary, are significant in naïve mosquitoes but not in either of the bacteria-challenged mosquitoes. GO categories significant only in infected animals include integrin complex, integrin-mediated signaling pathway, vesicle-mediated transport, and proteasome complex. These are likely implicated in bacterial phagocytosis, cytoskeletal remodeling and degradation of ingested bacteria following phagocytosis (Hillyer et al., 2003b; Moita et al., 2006; Stuart et al., 2007).
An important limitation of ratio-based analyses using microarray data is the inherent inability to explicitly discern tissue-specific expression from tissue-enriched expression, which is further complicated by residual hemocytes in carcass samples. As a consequence, our data cannot be used to confirmatively establish tissue-specific expression of individual transcripts. Nonetheless, it remains reasonable to expect tissue-specific transcripts to display higher enrichment ratios than other transcripts in relative terms. Consistent with this idea is the observation that previously characterized hemocyte-specific transcripts, such as those encoding PPOs, rank particularly high in tissue enrichment as measured by our microarray approach. In addition, there is an apparent trend that, among the top-ranked transcripts, signal intensities in the carcass samples approach background levels, possibly reflecting their low abundance in most tissue types other than hemocytes (Fig. S2). Taken together, it is likely that our ranked lists capture biologically meaningful information concerning the characteristic features of the hemocyte transcriptome, and the top ranked transcripts represent viable candidates for future exploration of hemocyte-specific biomarkers.
3.4 Utilizing tissue-enriched expression profiles to elucidate hemocyte infection responses
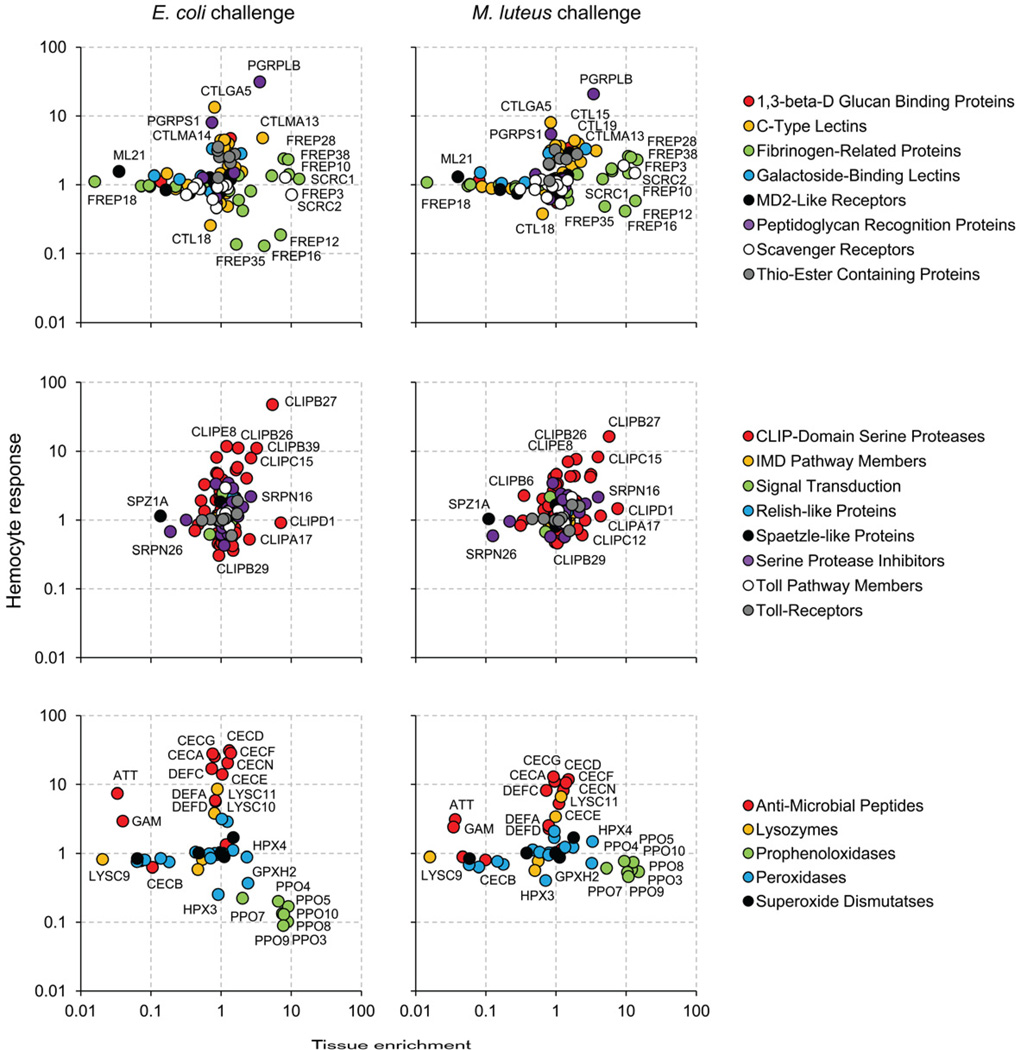
To date, transcriptomic analyses on hemocyte infection responses have mostly taken a one-dimensional view by focusing on the differences between hemocyte samples collected under various infection conditions. This approach, while effective in characterizing transcriptome changes within hemocytes, often failed to contextualize the findings in relation to changes occurring in other immune-responsive tissues. The lack of information regarding tissue specificity during infection response hinders assessment of whether an observed transcriptional change is likely to be distinctive of hemocytes or shared by multiple tissue types. In order to develop deeper insight into the transcriptome changes underlying hemocyte-specific processes, we took an integrative approach combining infection response profiles with tissue enrichment ratios. A two-dimensional representation of expression ratios, in which the infection response profiles are resolved along a second axis of post-infection tissue enrichment ratios, provides an insightful overview of hemocyte transcriptome dynamics (Fig. S3; Fig. 3). The distribution of genes on these Cartesian planes exposes the extent of heterogeneity in tissue enrichment among different infection responsive genes. For example, C-type lectins, CTLMA13 (AAEL011621-RA) and CTLMA14 (AAEL014382-RA) display similar levels of increase in hemocytes upon E. coli infection. However, their post-infection tissue enrichment ratios differ substantially. CTLMA13 is significantly enriched in hemocytes relative to the carcass, while CTLMA14 shows no enrichment, suggesting that transcriptional induction of CTLMA13 is likely more restricted to hemocytes.
Fig. 3.
Transcriptional response of immune-related gene families in Aedes aegypti hemocytes after bacterial challenge. ImmunoDB gene families were grouped into recognition, signaling and effecter molecules (upper, middle and lower panels, respectively). X-axis: tissue enrichment relative to carcass after bacterial challenge. Y-axis: hemocyte response to bacterial challenge relative to naïve.
Using our integrative approach that combines infection response profiles with tissue enrichment ratios, we investigated further the transcriptional characteristics of immune-related gene families in A. aegypti (Waterhouse et al., 2007). The pattern of spatial segregation between distinct gene families and the level of variation among family members, bring to light the immunological characteristics of the hemocyte molecular repertoire within the currently understood framework of insect immunity (Fig. 3).
Members of the gene families implicated in pattern recognition form a distinct group displaying strong tissue enrichment in hemocytes. Both cell surface receptors and secreted proteins are represented, and many are constitutively expressed. Of particular interest is the FREP family, a phylogenetically conserved pattern recognition receptor that has undergone mosquito lineage-specific gene expansion with respect to D. melanogaster (Waterhouse et al., 2007). The present study indicates that at least 10 out of 37 FREPs in A. aegypti display a hemocyte-enriched expression to a varying degree (FREP3, 5, 10, 12, 16, 23, 26, 28, 35 and 38). Microbial binding of FREPs has been demonstrated in Armigeres subalbatus (Wang et al., 2005) and A. gambiae (Dong and Dimopoulos, 2009). FREP10, in particular, is a one-to-one ortholog of the A. gambiae FREP13 (FBN9) whose transcript knockdown alters Plasmodium infectivity as well as the host survival rate post bacterial challenge (Dong and Dimopoulos, 2009). Class C scavenger receptors, originally characterized in D. melanogaster hemocytes, recognize both gram-positive and -negative bacteria and a broad range of polyanionic ligands (Pearson et al., 1995; Ramet et al., 2001). SCRCs in A. aegypti contain two complement-control protein domains and one transmembrane domain like their homologs in D. melanogaster. These likely function as pattern recognition receptor for phagocytosis given the strong hemocyte-enriched expression. AAEL000636 and AAEL007967, homologs of the phagocytic receptors with EGF domains (eater and Nimrod C1) also display highly hemocyte-enriched expression patterns similar to that of the SCRCs (Kocks et al., 2005; Kurucz et al., 2007).
Of the 8 peptidoglycan recognition proteins (PGRPs) annotated in A. aegypti, our data indicate that PGRPLB and PGRPS1 (PGRP-SA in D. melanogaster) display substantial increases in transcript abundance in hemocytes, both after E. coli- and M. luteus-challenge. In Drosophila, the former degrades gram-negative bacteria peptidoglycan with its amidase activity in a negative feedback regulation of the Imd pathway (Zaidman-Remy et al., 2006), whereas the latter is involved in the recognition of Gram-positive bacteria and activation of the Toll pathway (Michel et al., 2001). Interestingly, our tissue enrichment data suggest that the induction of PGRPLB is more specific to hemocytes.
Members of intracellular immune signaling pathways, such as Toll and IMD, do not show particular enrichment in hemocytes, and are likely expressed in diverse (immune-responsive) tissues. Because signal transduction is mostly mediated by protein interactions, transcriptome data allow only indirect monitoring of the activity of these immune pathways via transcriptional changes of downstream target genes. In Drosophila larvae, hemocytes express the Toll ligand spätzle (SPZ) in a tissue-specific manner after bacterial challenge (Irving et al., 2005) and its knockdown in hemocytes blocks the expression of drosomycin gene in the fat body (Shia et al., 2009). Our data indicate a moderate induction of SPZ3A in both hemocytes and the carcass after bacteria challenge, and a carcass-enriched constitutive expression of SPZ1A. The latter has been shown previously to be induced in the fat body following fungal challenge (Shin et al., 2006). A large number of CLIP-domain serine proteases and serine protease inhibitors (serpins) are encoded in the genome of A. aegypti, many of which are induced upon infection. The present study indicates that only a few specific members of these families (e.g., CLIPB27 and CLIPD1) display distinct hemocyte-enriched expression patterns, suggesting their possible involvement in hemocyte-mediated immune mechanisms.
Tissue enrichment ratios of the AMP transcripts after bacterial challenge indicate that transcriptional induction of AMPs is not restricted to hemocytes. Moreover, the overall direction and magnitude of transcriptional change are highly similar between hemocytes and carcass samples. One notable exception to this pattern is a transcript encoding attacin that shows strong preferential increase in the carcass relative to hemocytes. A gambicin transcript also displays a similar pattern, but at a more moderate level. In our previous study, we reported dramatic increase in AMP transcripts in hemocytes upon bacterial challenge (Bartholomay et al., 2007). The current data suggest that, despite the comparatively large magnitude of induction, none of the AMP responses are unique to hemocytes. Although both studies highlight the contribution of hemocytes to AMP production during systemic infection, lack of tissue specificity suggests that AMP responses may not be central in directly mediating cellular function unique to hemocytes. This expression pattern is in sharp contrast to that of PPOs, the majority of which display strong hemocyte-enriched expression in both naïve and infected animals. Overall, these observations conform to earlier findings in mosquitoes and other insect species (Haine et al., 2008; Jiang et al., 2010; Lemaitre and Hoffmann, 2007; Yassine and Osta, 2010).
In addition to transcripts showing tissue enrichment following bacterial challenge (i.e., restricted expression in hemocytes), it is of interest to examine transcripts showing a disproportionate response in hemocytes relative to that observed in the carcass. Direct comparison of infection responses in hemocytes versus those in the carcass (Fig. S3) can identify transcriptional changes that are more pronounced in hemocytes. We compiled a list of genes showing infection-responsive transcriptional induction that is more restricted to or more pronounced in hemocytes; out of 625 transcripts that showed a greater than 2-fold increase (P < 0.01) in hemocytes at 24 hours post E. coli-challenge, 70 transcripts displayed a greater than 3-fold post-infection tissue enrichment (P < 0.01) or a magnitude of transcriptional response at least 3 times greater than that in the carcass (P < 0.01). M. luteus-challenge increased abundance levels of 534 hemocyte transcripts to more than 2-fold (P < 0.01) at 24 hours post immune challenge, and among these, 109 transcripts satisfied the above mentioned criteria of tissue enrichment or differential induction in hemocytes, and 47 transcripts were common between the two gene lists (E. coli and M. luteus challenge).
Our compiled list contains a number of cytoskeletal components, adhesion molecules, cell-cycle associated proteins, calmodulins and a nitric oxide synthase, in addition to the members of the canonical immune gene families (Table 1; Dataset S3). The most noticeable group, however, is the small heat shock proteins (HSPs) with alpha-crystallin domain. These likely function as molecular chaperones alleviating malfolded protein aggregation during cellular stress. In vertebrate systems, increasing evidence suggests that extracellular and membrane-bound HSPs display immunomodulatory properties (Henderson and Pockley, 2010), and it remains to be investigated whether any of the hemocyte-derived HSPs contribute to systemic immune responses beyond maintaining the cellular homeostasis of phagocytes. It may be possible that these HSPs constitute “danger” signals for the host to modulate the level of immune activation based on the balance between nonself elicitors and endogenous danger signals (Lazzaro and Rolff, 2011).
Table 1.
Genes displaying transcriptional induction (logFC) more restricted to or more pronounced in Aedes aegypti hemocytes, with respect to carcass following Escherichia coli or Micrococcus luteus challengea.
| VectorBase ID | Description | Hemocyte responseb | Tissue enrichmentc | Differential responsed | |||
|---|---|---|---|---|---|---|---|
| E. coli | M. luteus | E. coli | M. luteus | E. coli | M. luteus | ||
| AAEL013350-RA | heat shock protein 26kD | 6.49 | 3.10 | 3.04 | 1.98 | 2.95 | 1.91 |
| AAEL013281-RA | serine-type enodpeptidase | 3.16 | 1.71 | 2.73 | 1.82 | 2.62 | 1.71 |
| AAEL010171-RA | peptidoglycan recognition protein sb2 | 4.97 | 4.39 | 1.83 | 1.77 | 2.56 | 2.46 |
| AAEL006479-RA | conserved hypothetical protein | 3.59 | 2.65 | 2.28 | 1.93 | 2.18 | 1.87 |
| AAEL009537-RA | hypothetical protein | 5.79 | 4.35 | 3.98 | 3.81 | 1.65 | |
| AAEL001241-RA | conserved hypothetical protein | 3.65 | 2.50 | 3.67 | 2.93 | 2.31 | |
| AAEL006921-RA | Calmodulin | 4.51 | 3.47 | 2.56 | 2.52 | 1.63 | |
| AAEL006533-RA | Ets domain-containing protein | 3.61 | 2.07 | 2.45 | 1.90 | 1.98 | |
| AAEL010802-RB | conserved hypothetical protein | 3.02 | 2.01 | 2.41 | 1.82 | 2.03 | |
| AAEL013349-RA | lethal(2)essential for life protein | 5.46 | 2.12 | 3.11 | 3.17 | 1.61 | |
| AAEL013344-RA | lethal(2)essential for life protein | 5.18 | 1.65 | 2.16 | 3.93 | 1.68 | |
| AAEL008489-RA | calcyphosine/tpp | 4.20 | 3.04 | 1.86 | 2.76 | 2.20 | |
see Dataset S3 for a complete list.
Hemocyte response to bacterial challenge relative to naïve.
Tissue enrichment relative to carcass after bacterial challenge.
Differential response in hemocytes relative to carcass.
3.5 Hemocyte transcriptome responses to E. coil vs. M. luteus
We examined transcripts showing differential response to E. coli as compared to M. luteus challenge. Because we based our analysis of differential transcript abundance on linear models (Smyth, 2004), it was possible to directly contrast a pair of expression ratio profiles (i.e., “ratio of ratios”), rather than making a comparison at the level of gene lists. Interestingly, the number of transcripts with a differential response (P < 0.01 and fold change > 3) was greater in hemocytes than in carcass (51 and 16, respectively; Fig. S4; Dataset S2). A closer look at these transcripts suggests that hemocyte response differs between the two immune elicitors in three distinct ways: (1) elevation of HSP transcript abundance is more pronounced following E. coli challenge, and (2) PPO and (3) cell-cycle related transcripts are present at higher levels after M. luteus challenge. These observations are consistent with our GO analysis indicating significant overrepresentation of the GO terms, response to temperature stimulus (GO:0009266) and DNA replication (GO:0006260), among hemocyte-enriched transcripts after E. coli and M. luteus challenge, respectively (Fig. 2). The transcriptomic differences are intriguing, especially the differences in PPO levels, because E. coli predominantly elicits phagocytosis, whereas M. luteus induces a melanization response (but not in a mutually exclusive manner) (Hillyer et al., 2003a, b). In addition, the variation in HSP levels could be a reflection of the different phagocytic loads in hemocytes. However, the inferences we can make on the molecular basis of the differential hemocyte response remain limited, not only because of the shortcomings of our experimental design (e.g., temporal resolution, dosage coverage, etc.) but also because the initiation of phatocytosis and melanization responses likely depend primarily on protein level interactions that cannot be examined using a transcriptomic approach.
3.6 Hemocyte-enriched transcripts conserved among A. aegypti, A. gambiae and D. melanogaster
A meta-analysis was performed on published data to investigate the hemocyte-enriched transcripts conserved among A. aegypti, A. gambiae and D. melanogaster. Transcripts enriched in hemocytes in adult female A. gambiae (relative to the carcass) and in larval D. melanogaster (relative to the whole animal) were obtained from previous microarray studies (Irving et al., 2005; Pinto et al., 2009). A three-way comparison was made at the level of orthologous groups delineated by eggNOG (evolutionary genealogy of genes: Non-supervised Orthologous Groups) database (Muller et al., 2010). An orthologous group was considered to retain hemocyte-enriched expression pattern if there was at least one ortholog from each species that showed tissue enrichment. Transcripts with the highest enrichment ratio were mapped to genes and orthologous groups when alternatively spliced transcripts or in-paralogs were present. Relative ranking of tissue enrichment ratios within each study then was used as a common basis for comparison to alleviate the effects of study specific differences in transcriptome quantification. The most highly hemocyte enriched transcripts (with an orthologous group assignment) from each species were compared to identify orthologous groups of genes that retained hemocyte enriched expression patterns among these dipterans. A Venn diagram analysis including the top 500 transcripts from each species indicated that 117 and 87 A.aegypti transcripts had orthologs with conserved hemocyte-enriched expression in A. gambiae and D. melanogaster, respectively, suggesting a rather low level of overall conservation (Fig. 4). This observation is in agreement with previous comparisons in which the A. aegypti hemocyte transcriptome was represented by EST datasets (Baton et al., 2009; Pinto et al., 2009). However, considering biological and methodological factors that confounded our analysis (i.e., variation in developmental stages, hemocyte collection methods, mRNA amplification methods, microarray platforms, normalization strategies, etc.) the observed level of differences in hemocyte molecular repertoires among these dipterans may not be due solely to their phylogenic divergence, making it difficult to analyze the distinguishing transcriptional characteristics of each species with high confidence. In particular, stringent data filtering performed in each study could have resulted in an increase in false-negatives that might have inflated the number of hemocyte-enriched transcripts that appear unique to each species. On the other hand, the current meta-analysis revealed 38 orthologous groups with hemocyte-enriched expression conserved in all three species. We further determined genes showing the most consistent pattern of high tissue enrichment across species using the geometric mean of the relative ranks (Table 2; Dataset S3). Given the nature of our meta-analytic approach, these likely represent true positives, albeit a partial list. Prophenoloxidase (AAEL011763; AGAP006258; CG5779), extracellular ligand-gated ion channel (AAEL004958; AGAP010580; CG6698), Wnt-protein binding (AAEL007967; AGAP009762; CG6124: eater/Nimrod homologs) and C-type scavenger receptor (AAEL006361; AGAP011974; CG4099) were at the top of this ranked list. Several serine proteases, structural proteins such as laminin (implicated in cell migration and adhesion), and calcium ion binding proteins also were represented. Phylogenetically conserved tissue-enriched expression could imply functional constraints (Piro et al., 2011), possibly suggesting that these genes are indispensable for hemocyte function. Because our approach effectively captured genes previously described in association with hemocyte-specific processes (e.g., PPOs and phagocytic receptors), top ranked orthologous groups with uncharacterized function strongly merit further experimental investigation in relation to hemocyte biology.
Fig. 4.
Orthologous groups displaying hemocyte-enriched expression identified through meta-analysis of microarray studies.
Table 2.
Orthologous groups among Aedes aegypti, Anopheles gambiae (Pinto et al., 2009) and Drosophila melanogaster (Irving et al., 2005) with hemocyte-enriched expressiona.
| Orthologous groupb | Description | A. aegypti | A. gambiae | D. melanogaster | Geometric mean rank | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IDc | logFC | Rank | IDc | logFC | Rank | IDc | logFC | Rank | |||
| inNOG07770 | Prophenoloxidase | 11763 | 3.96 | 1 | 006258 | 6.81 | 1 | 5779 | 3.75 | 32 | 3.2 |
| inNOG10178 | Extracellular ligand-gated ion channel | 04958 | 3.25 | 7 | 010580 | 5.37 | 2 | 6698 | 5.44 | 3 | 3.5 |
| inNOG06548 | Wnt-Protein binding (Eater/Nimrod) | 07967 | 2.65 | 13 | 009762 | 4.73 | 6 | 6124 | 5.45 | 2 | 5.4 |
| inNOG09691 | Scavenger receptor class C, type | 06361 | 3.70 | 4 | 011974 | 3.85 | 18 | 4099 | 4.67 | 13 | 9.8 |
| inNOG07282 | Serine protease | 07796 | 2.73 | 12 | 002422 | 4.26 | 9 | 9372 | 4.42 | 17 | 12.2 |
| inNOG10070 | Unknown function | 02520 | 3.38 | 6 | 000790 | 5.17 | 4 | 4139 | 2.92 | 87 | 12.8 |
| inNOG10383 | CG10824-PA protein | 08658 | 2.63 | 15 | 007045 | 2.87 | 41 | 4950 | 5.05 | 5 | 14.5 |
| inNOG09987 | Unknown function | 09086 | 2.59 | 17 | 005820 | 4.24 | 11 | 6310 | 3.23 | 61 | 22.5 |
| inNOG08715 | Beta chain protein | 02848 | 2.19 | 28 | 010510 | 2.88 | 40 | 3401 | 4.22 | 20 | 28.2 |
| inNOG07005 | Protein involved in regulation of embryonic development | 08773 | 2.08 | 32 | 004993 | 3.15 | 31 | 10236 | 3.60 | 40 | 34.1 |
| inNOG07544 | Calcium ion binding protein | 13656 | 2.16 | 30 | 000305 | 2.73 | 46 | 6378 | 3.78 | 29 | 34.2 |
| inNOG06226 | Serine-Type peptidase | 05521 | 1.04 | 145 | 000994 | 4.26 | 10 | 2493 | 3.70 | 34 | 36.7 |
| inNOG08907 | Beta-1,3-Glucuronyltransferase | 06254 | 1.97 | 39 | 001367 | 2.92 | 39 | 6207 | 3.13 | 67 | 46.7 |
| inNOG05564 | Laminin beta protein | 03658 | 1.03 | 152 | 001381 | 2.98 | 37 | 7123 | 4.13 | 22 | 49.8 |
| inNOG08874 | Procollagen-Lysine protein | 08099 | 1.41 | 77 | 001507 | 2.67 | 51 | 6199 | 3.51 | 43 | 55.3 |
see Dataset S3 for a complete list.
eggNOG (evolutionary genealogy of genes: Non-supervised Orthologous Groups) database
AAEL00, AGAP0, and CG were omitted from A. aegypti, A. gambiae, and D. melanogaster gene IDs, respectively.
Supplementary Material
Dataset S1. qPCR validation results and primer sequences.
Dataset S2. Log expression ratios (logFC) and p-values from differential transcript abundance analysis using LIMMA.
Dataset S3. Complete Table 1 and 2.
Highlights.
Hemocyte-enriched transcripts were identified using microarrays.
Host immune status alters tissue-enriched expression.
Transcriptional patterns unique to hemocytes were resolved from those likely shared by other immune responsive tissues.
Meta-analysis revealed orthologous groups with conserved hemocyte-enriched expression.
Acknowledgements
The authors gratefully acknowledge support from NIH grants AI 067698 and AI 019769.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham EG, Pinto SB, Ghosh A, Vanlandingham DL, Budd A, Higgs S, Kafatos FC, Jacobs-Lorena M, Michel K. An immune-responsive serpin, SRPN6, mediates mosquito defense against malaria parasites. Proc Natl Acad Sci U S A. 2005;102:16327–16332. doi: 10.1073/pnas.0508335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonova Y, Alvarez KS, Kim YJ, Kokoza V, Raikhel AS. The role of NF-kappaB factor REL2 in the Aedes aegypti immune response. Insect Biochem Mol Biol. 2009;39:303–314. doi: 10.1016/j.ibmb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomay LC, Mayhew GF, Fuchs JF, Rocheleau TA, Erickson SM, Aliota MT, Christensen BM. Profiling infection responses in the haemocytes of the mosquito, Aedes aegypti. Insect Mol Biol. 2007;16:761–776. doi: 10.1111/j.1365-2583.2007.00773.x. [DOI] [PubMed] [Google Scholar]
- Baton LA, Robertson A, Warr E, Strand MR, Dimopoulos G. Genome-wide transcriptomic profiling of Anopheles gambiae hemocytes reveals pathogen-specific signatures upon bacterial challenge and Plasmodium berghei infection. BMC Genomics. 2009;10:257. doi: 10.1186/1471-2164-10-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerntsen BT, Christensen BM. Dirofilaria immitis: effect on hemolymph polypeptide synthesis in Aedes aegypti during melanotic encapsulation reactions against microfilariae. Exp Parasitol. 1990;71:406–414. doi: 10.1016/0014-4894(90)90066-l. [DOI] [PubMed] [Google Scholar]
- Blandin SA, Levashina EA. Phagocytosis in mosquito immune responses. Immunol Rev. 2007;219:8–16. doi: 10.1111/j.1600-065X.2007.00553.x. [DOI] [PubMed] [Google Scholar]
- Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J, Binns D, Harte N, Lopez R, Apweiler R. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004;32:D262–D266. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo JC, Robertson AE, Strand MR. Characterization of hemocytes from the mosquitoes Anopheles gambiae and Aedes aegypti. Insect Biochem Mol Biol. 2006;36:891–903. doi: 10.1016/j.ibmb.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Laurence BR. In vitro study on humoral encapsulation of microfilariae: establishment of technique and description of reactions. Int J Parasitol. 1987;17:781–787. doi: 10.1016/0020-7519(87)90059-2. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Christensen B, Li J, Chen C, Nappi A. Melanization immune responses in mosquito vectors. Trends Parasitol. 2005;21:192–199. doi: 10.1016/j.pt.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Christensen BM, Sutherland DR. Brugia pahangi: exsheathment and midgut penetration in Aedes aegypti. Trans Am Microsc Soc. 1984;103:423–433. [Google Scholar]
- Chun J, McMaster J, Han Y, Schwartz A, Paskewitz SM. Two-dimensional gel analysis of haemolymph proteins from Plasmodium-melanizing and -non-melanizing strains of Anopheles gambiae. Insect Mol Biol. 2000;9:39–45. doi: 10.1111/j.1365-2583.2000.00157.x. [DOI] [PubMed] [Google Scholar]
- Dong Y, Dimopoulos G. Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J Biol Chem. 2009;284:9835–9844. doi: 10.1074/jbc.M807084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis J, Mistry M, Pavlidis P. Gene function analysis in complex data sets using ErmineJ. Nat Protoc. 2010;5:1148–1159. doi: 10.1038/nprot.2010.78. [DOI] [PubMed] [Google Scholar]
- Glenn JD, King JG, Hillyer JF. Structural mechanics of the mosquito heart and its function in bidirectional hemolymph transport. J Exp Biol. 2010;213:541–550. doi: 10.1242/jeb.035014. [DOI] [PubMed] [Google Scholar]
- Haine ER, Moret Y, Siva-Jothy MT, Rolff J. Antimicrobial defense and persistent infection in insects. Science. 2008;322:1257–1259. doi: 10.1126/science.1165265. [DOI] [PubMed] [Google Scholar]
- Henderson B, Pockley AG. Molecular chaperones and protein-folding catalysts as intercellular signaling regulators in immunity and inflammation. J Leukoc Biol. 2010;88:445–462. doi: 10.1189/jlb.1209779. [DOI] [PubMed] [Google Scholar]
- Hillyer JF. Mosquito immunity. Adv Exp Med Biol. 2010;708:218–238. doi: 10.1007/978-1-4419-8059-5_12. [DOI] [PubMed] [Google Scholar]
- Hillyer JF, Schmidt SL, Christensen BM. Hemocyte-mediated phagocytosis and melanization in the mosquito Armigeres subalbatus following immune challenge by bacteria. Cell Tissue Res. 2003a;313:117–127. doi: 10.1007/s00441-003-0744-y. [DOI] [PubMed] [Google Scholar]
- Hillyer JF, Schmidt SL, Christensen BM. Rapid phagocytosis and melanization of bacteria and Plasmodium sporozoites by hemocytes of the mosquito Aedes aegypti. J Parasitol. 2003b;89:62–69. doi: 10.1645/0022-3395(2003)089[0062:RPAMOB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Hillyer JF, Schmidt SL, Christensen BM. The antibacterial innate immune response by the mosquito Aedes aegypti is mediated by hemocytes and independent of Gram type and pathogenicity. Microbes Infec. 2004;6:448–459. doi: 10.1016/j.micinf.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Irving P, Ubeda JM, Doucet D, Troxler L, Lagueux M, Zachary D, Hoffmann JA, Hetru C, Meister M. New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell Microbiol. 2005;7:335–350. doi: 10.1111/j.1462-5822.2004.00462.x. [DOI] [PubMed] [Google Scholar]
- Jiang H, Vilcinskas A, Kanost MR. Immunity in lepidopteran insects. Adv Exp Med Biol. 2010;708:181–204. doi: 10.1007/978-1-4419-8059-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, Strom C, Conto SL, Hetru C, Stuart LM, Stehle T, Hoffmann JA, Reichhart JM, Ferrandon D, Ramet M, Ezekowitz RA. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123:335–346. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Kurucz E, Markus R, Zsamboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, Udvardy A, Krausz I, Lukacsovich T, Gateff E, Zettervall CJ, Hultmark D, Ando I. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. 2007;17:649–654. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, Hammond M, Hill CA, Konopinski N, Lobo NF, MacCallum RM, Madey G, Megy K, Meyer J, Redmond S, Severson DW, Stinson EO, Topalis P, Birney E, Gelbart WM, Kafatos FC, Louis C, Collins FH. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37:D583–D587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro BP, Rolff J. Danger, Microbes, and Homeostasis. Science. 2011;332:43–44. doi: 10.1126/science.1200486. [DOI] [PubMed] [Google Scholar]
- Lee HK, Braynen W, Keshav K, Pavlidis P. ErmineJ: tool for functional analysis of gene expression data sets. BMC Bioinformatics. 2005;6:269. doi: 10.1186/1471-2105-6-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Michel T, Reichhart J-M, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- Moita LF, Vriend G, Mahairaki V, Louis C, Kafatos FC. Integrins of Anopheles gambiae and a putative role of a new beta integrin, BINT2, in phagocytosis of E. coli. Insect Biochem Mol Biol. 2006;36:282–290. doi: 10.1016/j.ibmb.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Muller J, Szklarczyk D, Julien P, Letunic I, Roth A, Kuhn M, Powell S, von Mering C, Doerks T, Jensen LJ, Bork P. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 2010;38:D190–D195. doi: 10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O'Leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson A, Lux A, Krieger M. Expression cloning of dSR-CI, a class C macrophage-specific scavenger receptor from Drosophila melanogaster. Proc Natl Acad Sci U S A. 1995;92:4056–4060. doi: 10.1073/pnas.92.9.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppel K, Baglioni C. A simple and fast method to extract RNA from tissue culture cells. Biotechniques. 1990;9:711–713. [PubMed] [Google Scholar]
- Pinto SB, Lombardo F, Koutsos AC, Waterhouse RM, McKay K, An C, Ramakrishnan C, Kafatos FC, Michel K. Discovery of Plasmodium modulators by genome-wide analysis of circulating hemocytes in Anopheles gambiae. Proc Natl Acad Sci U S A. 2009;106:21270–21275. doi: 10.1073/pnas.0909463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro RM, Ala U, Molineris I, Grassi E, Bracco C, Perego GP, Provero P, Di Cunto F. An atlas of tissue-specific conserved coexpression for functional annotation and disease gene prediction. Eur J Hum Genet. 2011;19:1173–1180. doi: 10.1038/ejhg.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramet M, Pearson A, Manfruelli P, Li X, Koziel H, Gobel V, Chung E, Krieger M, Ezekowitz RA. Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity. 2001;15:1027–1038. doi: 10.1016/s1074-7613(01)00249-7. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- Schneider D. Physiological integration of innate immunity. In: Rolff J, Reynolds SE, editors. Insect Infection and Immunity: Evolution, Ecology, and Mechanisms. Oxford University Press; 2009. pp. 106–116. [Google Scholar]
- Shia AK, Glittenberg M, Thompson G, Weber AN, Reichhart JM, Ligoxygakis P. Toll-dependent antimicrobial responses in Drosophila larval fat body require Spatzle secreted by haemocytes. J Cell Sci. 2009;122:4505–4515. doi: 10.1242/jcs.049155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SW, Bian G, Raikhel AS. A toll receptor and a cytokine, Toll5A and Spz1C, are involved in toll antifungal immune signaling in the mosquito Aedes aegypti. J Biol Chem. 2006;281:39388–39395. doi: 10.1074/jbc.M608912200. [DOI] [PubMed] [Google Scholar]
- Shin SW, Kokoza V, Bian G, Cheon HM, Kim YJ, Raikhel AS. REL1, a homologue of Drosophila dorsal, regulates toll antifungal immune pathway in the female mosquito Aedes aegypti. J Biol Chem. 2005;280:16499–16507. doi: 10.1074/jbc.M500711200. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Strand MR. Insect hemocytes and their role in immunity. In: Beckage NE, editor. Insect Immunology. Academic Press; 2008. pp. 25–47. [Google Scholar]
- Stuart LM, Boulais J, Charriere GM, Hennessy EJ, Brunet S, Jutras I, Goyette G, Rondeau C, Letarte S, Huang H, Ye P, Morales F, Kocks C, Bader JS, Desjardins M, Ezekowitz RAB. A systems biology analysis of the Drosophila phagosome. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- Theopold U, Schmidt O, Soderhall K, Dushay MS. Coagulation in arthropods: defence, wound closure and healing. Trends Immunol. 2004;25:289–294. doi: 10.1016/j.it.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhao Q, Christensen BM. Identification and characterization of the fibrinogen-like domain of fibrinogen-related proteins in the mosquito, Anopheles gambiae, and the fruitfly, Drosophila melanogaster, genomes. BMC Genomics. 2005;6:114. doi: 10.1186/1471-2164-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, Dong Y, Jiang H, Kanost MR, Koutsos AC, Levashina EA, Li J, Ligoxygakis P, Maccallum RM, Mayhew GF, Mendes A, Michel K, Osta MA, Paskewitz S, Shin SW, Vlachou D, Wang L, Wei W, Zheng L, Zou Z, Severson DW, Raikhel AS, Kafatos FC, Dimopoulos G, Zdobnov EM, Christophides GK. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine H, Osta MA. Anopheles gambiae innate immunity. Cell Microbiol. 2010;12:1–9. doi: 10.1111/j.1462-5822.2009.01388.x. [DOI] [PubMed] [Google Scholar]
- Yeung T, Grinstein S. Lipid signaling and the modulation of surface charge during phagocytosis. Immunological Reviews. 2007;219:17–36. doi: 10.1111/j.1600-065X.2007.00546.x. [DOI] [PubMed] [Google Scholar]
- Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, Oh BH, Ueda R, Mengin-Lecreulx D, Lemaitre B. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset S1. qPCR validation results and primer sequences.
Dataset S2. Log expression ratios (logFC) and p-values from differential transcript abundance analysis using LIMMA.
Dataset S3. Complete Table 1 and 2.