SUMMARY
Heat-Shock Factor 1 (HSF1), master regulator of the heat-shock response, facilitates malignant transformation, cancer cell survival and proliferation in model systems. The common assumption is that these effects are mediated through regulation of heat-shock protein (HSP) expression. However, the transcriptional network that HSF1 coordinates directly in malignancy and its relationship to the heat-shock response have never been defined. By comparing cells with high and low malignant potential alongside their non-transformed counterparts, we identify an HSF1-regulated transcriptional program specific to highly malignant cells and distinct from heat shock. Cancer-specific genes in this program support oncogenic processes: cell-cycle regulation, signaling, metabolism, adhesion and translation. HSP genes are integral to this program, however, many are uniquely regulated in malignancy. This HSF1 cancer program is active in breast, colon and lung tumors isolated directly from human patients and is strongly associated with metastasis and death. Thus, HSF1 rewires the transcriptome in tumorigenesis, with prognostic and therapeutic implications.
Keywords: HSP90, HSP70, ChIP-Seq, genome-wide, outcome signature, Nurses’ Health Study, immunohistochemistry
INTRODUCTION
A wide variety of environmental stressors can damage proteins. These include elevated temperatures, oxidative agents, heavy metals and low pH. Organisms respond by inducing heat-shock proteins (HSPs), which act as molecular chaperones to restore protein homeostasis (Shamovsky and Nudler, 2008; Whitesell and Lindquist, 2005). This powerful adaptive mechanism, known as the heat-shock response, is unleashed by the heat-shock transcription factor HSF1. Upon heat shock, HSF1 is phosphorylated, trimerizes, and translocates to the nucleus. There, it induces chaperone gene expression by binding to DNA sequence motifs known as heat-shock elements (HSEs) (Pelham, 1982; Sakurai and Enoki, 2010). Major aspects of this classic response are conserved from yeast to humans and are vital in many stressful environments. HSF1 also functions as a critical regulator of longevity in some organisms (Chiang et al., 2012; Volovik et al., 2012). Consistent with this, recent work indicates that HSF1 helps cells accommodate the complex pathophysiological derangements in protein homeostasis that underlie many human diseases, especially those associated with aging (Morimoto, 2008).
We have previously shown in mice that HSF1 is co-opted by tumor cells to promote their survival, to the detriment of their hosts. The importance of HSF1 in supporting carcinogenesis, at least in model systems, is demonstrated by the dramatically reduced susceptibility of Hsf1-knockout mice to tumor formation. This has been established for cancers driven by oncogenic RAS, tumor suppressor p53 mutations, and chemical carcinogens (Dai et al., 2007; Jin et al., 2011; Min et al., 2007). In addition to its role in tumor formation in mice, HSF1 fosters the growth of human tumor cells in culture. Depleting HSF1 from established human cancer lines markedly reduces their proliferation and survival (Dai et al., 2007; Meng et al., 2010; Min et al., 2007; Santagata et al., 2012; Zhao et al., 2011).
In mouse models, HSF1 enables adaptive changes in a diverse array of cellular processes, including signal transduction, glucose metabolism and protein translation (Dai et al., 2007; Khaleque et al., 2008; Lee et al., 2008; Zhao et al., 2011; Zhao et al., 2009). The commonly held view is that HSF1 exerts this broad influence in cancer simply by allowing cells to manage the imbalances in protein homeostasis that arise in malignancy. According to this view, the main impact of HSF1 on tumor biology occurs indirectly, through the actions of molecular chaperones like HSP90 and HSP70 on their client proteins (Jin et al., 2011; Solimini et al., 2007). An alternate, and to date unexplored, possibility is that HSF1 plays a more direct role, rewiring the transcriptome and, thereby, the physiology of cancer cells.
To investigate the HSF1-regulated transcriptional program in cancer and how it relates to the classical heat-shock response, we first took advantage of human breast cancer cell lines with very different abilities to form tumors and metastasize (Ince et al., 2007). Two types of primary mammary epithelial cells (HMEC and BPEC) have been isolated from normal breast tissue derived from the same donor during reductive mammoplasty (Ince et al., 2007). These pairs of isogenic cells were established using different culture conditions that are believed to have supported the outgrowth of distinct cell types. The cells were immortalized (HME and BPE) and then transformed with an identical set of oncogenes (HMLER and BPLER). The resulting tumorigenic breast cell lines had very different malignant and metastatic potentials (low, HMLER and high, BPLER) supporting the concept that the cell type from which a cancer arises (“cell-of – origin”) can significantly influence its ultimate phenotype (Ince et al., 2007).
Here, using this well-controlled system, we identify changes in the HSF1 transcriptional program that occur during transformation and underlie the different malignant potentials of these cells. Chromatin immunoprecipitation coupled with massively parallel DNA sequencing (ChIP-Seq) revealed a surprisingly diverse transcriptional network coordinated by HSF1 in the highly malignant cells. We then extend analysis of this HSF1 cancer program to a wide range of well-established human cancer cell lines and to diverse types of tumors taken directly from patients. Finally, we establish the clinical relevance of our findings through in-depth analysis of HSF1 activation in cohorts of breast, colon, and lung cancer patients with known clinical outcomes. Thus, the breadth of HSF1 biology is far greater then previously appreciated.
RESULTS
HSF1 is activated in highly tumorigenic cells
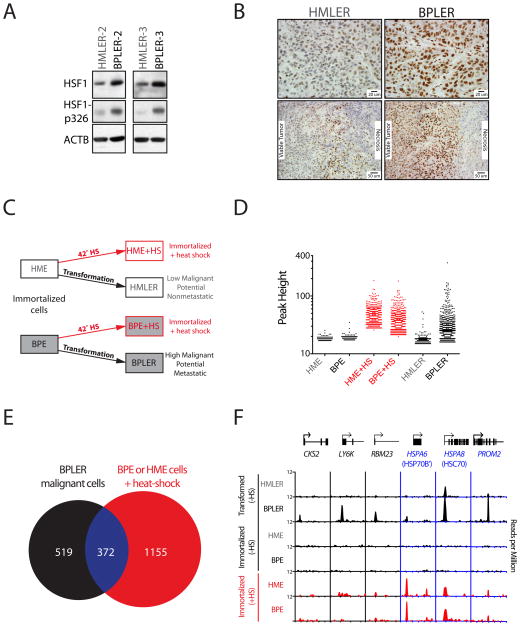
We first asked if HSF1 expression differed in the highly malignant BPLER and the much less malignant HMLER breast cancer cells (Ince et al., 2007). We used two sets of such cells, each pair derived independently from a different donor. In both, HSF1 protein expression was higher in the more malignant member of the pair, the BPLER cells (Figure 1A). The BPLER cells also had more phosphoserine-326-HSF1, a well established marker of HSF1 activation (Guettouche et al., 2005), than the HMLER cells (Figure 1A).
Figure 1. HSF1 is activated in metastatic and highly tumorigenic human mammary epithelial cell lines.
A. Equal amounts of total protein were immunoblotted with the indicated antibody. B. IHC staining with anti-HSF1 antibody of the indicated tumors xenografted in mice. Upper panels show regions of viable tumor (high mag, scale bar 20 μm) and lower panels show the viable tumor/necrotic interface (low mag, scale bar 50 μm) C. Schematic of experimental groups analyzed by HSF1 ChIP-Seq. D. Graph of ChIP-Seq peak heights for each region of HSF1 occupancy, normalized by the number of reads in the dataset. E. Overlap of genes bound in malignant cells (BPLER, 37°C) and immortalized, non-tumorigenic cells after heat shock (BPE or HME cells, 42°C). F. Representative genes bound in BPLER cells (CKS2, LY6K, RBM23) and bound in both BPLER cells and heat-shocked HME and BPE cells (HSPA6, HSPA8, PROM2). X-axis: from −2kb from the transcription start site (TSS) to either +5, +6 or +10kb from the TSS for each gene; genes diagrams drawn to scale. See also Figure S1 and Tables S1 and S2.
To determine if these differences in HSF1 were simply an artifact of growth in cell culture, we implanted the cells into immunocompromised mice and allowed them to form tumors. HSF1 immunostaining was weak in the HMLER tumors. Moreover, it was largely restricted to nonmalignant, infiltrating stroma and to tumor areas bordering necrosis (Figure 1B), indicating that microenvironmental stress can influence the activation of HSF1. In BPLER tumors, however, HSF1 staining was strong, nuclear localized and very uniform thoughout (Figures 1B and S1A available online). Thus, the dramatic difference in HSF1 expression we observe between BPLER and HMLER cells is due to stable, cell-autonomous factors intrinsic to these distinct cell types (Ince et al., 2007).
Given this evidence for the activation of HSF1 in the BPLER cell type, we asked if such cells were more dependent on HSF1 than HMLER cells for growth and survival. Neither cell type was affected by negative control shRNA. With two independent shRNA that knockdown HSF1 expression, however, cell growth and viability were far more strongly reduced in the BPLER than the HMLER cells (Figure S1B).
HSF1 genome occupancy in cancer is distinct from heat-shock
To determine if the transcriptional program driven by HSF1 in highly malignant cells differs from that driven by a classical thermal stress, we used chromatin immunoprecipitation coupled with massively parallel DNA sequencing (ChIP-Seq), characterizing HSF1 binding sites genome-wide. We first assessed the immortalized non-transformed progenitor cells, HME and BPE, grown at 37°C or following a 42°C heat shock (Figure 1C). We then related the genome-wide distribution of HSF1 binding sites to those of the oncogenically transformed HMLER and BPLER cells grown at 37°C.
In the HME and BPE parental cell lines, a limited number of genes were bound by HSF1 in the absence of heat shock, and these were bound weakly (Figure 1D; Table S1). Heat shock drove robust binding of HSF1 to ~800 genes in HME cells and to ~1100 genes in BPE cells (Figure 1D; Table S1). These observations are consistent with a previous report that a large number of genes are bound by HSF1 in the mammalian heat-shock response (Page et al., 2006).
A small number of genes were bound by HSF1 under basal conditions in the transformed cells with low malignant potential, HMLER (37°C; Figure 1D). However, binding was more localized to promoter regions than in the parental cells (Figure S1C), suggesting some low level of HSF1 activation (MacIsaac et al., 2010). In sharp contrast, in the metastatic and highly tumorigenic BPLER cells, we identified ~900 genes bound by HSF1 at 37°C (Figure 1D; Table S1).
Surprisingly, a full 60% of the genes bound by HSF1 in BPLER cells were not bound in non-transformed parental lines, even after heat-shock (Figure 1E). Examples included (Figure 1F): cyclin-dependent kinase interacting protein, CKS2, which enables proliferation under conditions of replicative stress common to malignant cells (Liberal et al., 2011); LY6K which encodes a glycosylphosphatidyl-inositol (GPI)-anchored membrane protein implicated as a biomarker in lung and esophageal carcinomas (Ishikawa et al., 2007; Maruyama et al., 2010); and RBM23, which encodes an RNA-binding protein implicated in the regulation of estrogen-mediated transcription (Dowhan et al., 2005). Using the Molecular Signatures Database (MSigDB) (Subramanian et al., 2005) the genes bound by HSF1 in the BPLER cells, but not in either of the parental lines after heat shock, were most highly enriched in protein translation, RNA binding, metabolism, cell adhesion (Figure S1D; Table S2) and other processes vital in supporting the malignant state.
We analyzed the 100 bp genomic regions surrounding the peaks of HSF1 binding unique to BPLER cells using the ab initio motif discovery algorithm MEME (Machanick and Bailey, 2011). The canonical heat-shock element (HSE) was highly enriched in the HSF1-bound regions (p-value=1.4 × 10−97; Figure S1E) strongly suggesting the genes that are constitutively bound by HSF1 in malignant cells are bona fide HSF1-binding targets.
The remaining 40% of genes bound by HSF1 in BPLER cells under basal conditions were also bound in at least one of the two parental lines following heat-shock. As expected, these genes included many classical heat-shock genes, including HSPA8, the constitutively expressed HSC70 protein, and HSPD1/E1, which encodes HSP60 and HSP10 (Figure S1F). By the Molecular Signatures Database this large group of genes was enriched for protein folding categories (Figure S1D; Table S2).
Notably, however, for many of the genes bound in both cancer and heat shock, HSF1 binding differed. For example, the strongly heat-shock inducible HSPA6 gene (encoding HSP70B’) was highly bound in parental lines upon heat shock but only weakly bound in BPLER cells at 37°C (Figures 1F, S1G and S1H). Conversely, PROM2, which encodes a basal epithelial cell membrane glycoprotein, was weakly bound by HSF1 in parental lines following heat-shock, but highly bound in BPLER cells (Figure 1F). Thus, HSF1 engages a regulatory program in the highly malignant state that is distinct from the classic heat-shock response.
To assess the functional significance of the HSF1 cancer program, we asked if the genes comprising this program played a significant role in malignancy, using unbiased data from an independent investigation. The Elledge lab recently conducted a whole genome siRNA screen to identify genes that are required to maintain growth when cells are transformed with a malignantly activated Ras gene (Luo et al., 2009). Among the ~1600 genes identified in this screen our HSF1-bound gene set was very strongly enriched (73 gene overlap; p Value =7.95e-15, Table S2). The HSF1-bound genes we identified as unique to the malignant state were more strongly enriched (49 gene overlap; p Value = 1.1e-12) than those shared with heat-shocked cells (24 gene overlap; p Value = .0004), but both sets of genes were important in supporting the malignant state.
HSF1 regulates the transcription of genes it binds in malignant cells
To investigate the consequences of HSF1 occupancy on gene expression, we compared RNA profiles in HMLER and BPLER cells transduced with control shRNA hairpins to those transduced with hairpins that knockdown HSF1. As we previously reported, the growth and survival of malignant cells is compromised by prolonged depletion of HSF1 (Dai et al., 2007). Therefore, we only analyzed mRNA expression in the early stages of shRNA inhibition, where HSF1 knockdown was still incomplete (Figure S2) but cell viability was unimpaired. This provides a conservative assessment of the effects of HSF1 on gene expression in malignant cells.
Control hairpins that did not reduce HSF1 levels (Scr and GFP; Figure S2), had minimal effects on the expression of HSF1-bound genes (Figure 2A; Table S3). Targeted hairpins that did reduce HSF1 had a minor impact in HMLER cells but markedly changed expression in BPLER cells. The expression of many genes decreased indicating that they were positively regulated by the HSF1 transcription factor. Many genes increased, indicating that a larger number of genes than previously appreciated are negatively regulated by HSF1. Genes unique to the malignant state and genes shareed by heat shocked cells were affected equivalently. For example, expression of the malignancy-associated genes CKS2 and RBM23 and the heat-shock protein genes HSPA8 (HSC70) and HSP90AA1 (HSP90) were all reduced (by ~50%) following HSF1 knockdown (Table S3).
Figure 2. The expression of HSF1-bound genes is altered by HSF1 depletion.
A. Relative gene expression levels following shRNA-mediated knockdown of HSF1 in indicated cells. Scr and GFP were negative control shRNA. B. Graph showing the number of genes positively regulated (reduced expression upon HSF1 depletion) or negatively regulated (increased expression upon HSF1 depletion) by HSF1 relative to site of gene occupancy by HSF1 (promoter versus distal). See also Figure S2 and Table S3.
Relating the effects of HSF1 knockdown on gene expression to our earlier ChIP-Seq analysis, ~70% of genes positively regulated by HSF1 were bound at the promoter while only ~30% of these genes were bound in distal regions (Figure 2B). Genes that were negatively regulated by HSF1, showed the opposite pattern (Figure 2B). This observation (p-value=0.00004) suggests that the direction of regulation (positive versus negative) in these cells is influenced by the location of the HSF1-binding site.
Next we examined the effects of HSF1 knockdown on gene expression in a cell line that had not been deliberately engineered. The MCF7 line was established from a human breast cancer metastasis. Moreover, as an estrogen receptor positive (ER+) line, its biology is fundamentally distinct from the hormone-receptor negative HMLER and BPLER cell lines. Despite these differences, the changes in gene expression caused by HSF1 knockdown was very similar in BPLER cells and MCF7 cells (Figure 2A).
HSF1 gene occupancy is conserved across a broad range of common human cancer cell lines
Next we used ChIP-qPCR to monitor HSF1 binding to a representative set of the HSF1-target genes in cell lines derived from patients with breast cancer. We used nine well-studied cancer lines (including MCF7 cells) representing all three major categories of breast cancer: ER+, HER2+and Triple Negative (TN). Under basal conditions (at 37°C) we detected HSF1 binding in each of the major breast cancer subtypes (Figure S3A). A range of binding intensities was observed. Most notably, however, the distinct pattern of HSF1 gene occupancy we had identified in the highly malignant BPLER cells was also present in these naturally-arising malignant cells. This included genes unique to malignant cells, such as CKS2 and RBM23, and genes shared by heat-shocked cells, such as HSPD1/E1. Again, the gene most strongly inducible by heat shock, HSPA6, was minimally bound across this entire panel of cancer lines under basal conditions (37°C; Figures S3A, S3B and S3C). We also analyzed HSF1 binding in the non-tumorigenic breast cell line MCF10A. Comparable to the low malignancy HMLER cells, MCF10A cells had low levels of HSF1 occupancy across all genes examined (Figures S3A and S3C).
These ChIP-PCR data spurred us to employ ChIP-Seq to generate additional genome-wide high-resolution maps of HSF1 occupancy. We performed ChIP-Seq analysis on the non-tumorigenic MCF10A cell line grown either at 37°C or following a 42°C heat-shock. We compared this data with our prior data from the non-tumorigenic cell lines HME and BPE and weakly tumorigenic HMLER cells. We then assessed HSF1 binding in a panel of human tumor lines that extended to other types of malignancy: duplicate samples of four breast, three lung and three colon cancer cell lines (Figures 3A and S3D), thus covering the human cancers with the highest total mortality in the developed world.
Figure 3. Genome-wide patterns of DNA occupancy by HSF1 across a broad range of common human cancer cell lines.
A. Heat map of ChIP-Seq read density for all HSF1 target regions (union of all HSF1-bound regions in all datasets). Genomic regions from −1kb to +1kb relative to the peak of HSF1 binding are shown. Regions are ordered the same in all datasets. Read density is depicted for non-tumorigenic cells at 37°C (green), cancer lines at 37°C (black) and non-tumorigenic (nt) lines following heat shock at 42°C (red). Asterisks indicate datasets also used for the analysis in Figure 1E. B. Principal component analysis (PCA) of HSF1 binding in heat-shocked parental lines (red) and cancer lines (black). C. ChIP-Seq density heat map of genomic regions differentially bound by HSF1 in cancer lines at 37°C, heat-shocked non-tumorigenic lines, and regions shared under both conditions. D. HSF1 binding of representative genes in cancer lines at 37°C (black) and heat-shocked non-tumorigenic lines (red). Examples of genes with distinct patterns of binding are presented: Enriched in cancer lines, heat-shocked non-tumorigenic lines, or both. E. Motif analysis of 100bp regions surrounding HSF1 binding peaks for genes enriched in cancer lines (BT20, NCIH838 and SKBR3), heat-shocked non-tumorigenic lines (HME, BPE, MCF10A) and both cancer and heat-shocked non-tumorigenic lines. See also Figure S3 and Table S1.
After heat shock, MCF10A cells exhibited an HSF1-binding profile that was comparable to that of heat-shocked HME and BPE cells. In the absence of heat shock the overall magnitude of HSF1 binding in all of the non-tumorigenic cell lines (nt) was uniformly very weak and the total number of bound genes was small (Figure 3A; Table S1). In contrast, in the cancer lines a range of HSF1 binding was observed at 37°C (Figure 3A). For example, robust binding was observed in the lung adenocarcinoma line NCI-H838 and in the TN breast carcinoma line BT20. Less pronounced overall binding was seen in others lines such as the weakly malignant HMLER. Binding in BPLER cells was intermediate.
Irrespective of the level of binding, the distribution of HSF1 occupancy on a genome-wide scale was remarkably similar among the cancer cell lines and distinct from the pattern of binding in the heat-shocked cells (Figure 3A). The differences between the heat-shocked and malignant states were further probed using principal component analysis (PCA; Figure 3B). This unsupervised method of clustering sets of data clearly distinguished one cluster containing all cell lines exposed to heat-shock and a second cluster containing all cancer cell lines. This analysis confirmed the global nature of the differences in the HSF1-binding profiles.
Data from these multiple cell lines allowed us to confidently identify regions of HSF1 binding that were strong in cancer cells but not in heat-shocked cells, weak in cancer but strong in heat-shock or similarly strong in both (Figure 3C). Examples of genes that were strongly bound in cancer but not in heat shock included CKS2, LY6K, RBM23, CCT6A, CKS1B, ST13, EIF4A2 (Figures S3E and 3D). Genes that were weakly bound in cancer lines but strongly bound in heat shock included HSPA6 and DNAJC7 (Figure 3D). Genes that were strongly bound in both cell types included HSPA4L and HSP90AB1 (Figure 3D).
We performed motif analysis to evaluate the 100 bp genomic regions surrounding the peaks of HSF1 binding in each of these groups. The HSE, comprised of adjacent inverted repeats of 5′-nGAAn-3′, was the most enriched motif in all three groups (Figure 3E). The regions strongly bound in cancer but not in heat-shock were enriched in HSEs that had three such repeats (p-value=8.8 × 10−106). They were also enriched in binding elements for YYI, the so called “ying-yang” transcription factor which is involved in activating and repressing a broad range of genes (p-value=3.7 × 10−7). The regions strongly bound in heat-shocked cells but not cancer were enriched for expanded HSEs, with a fourth 5′-nGAAn-3′ repeat (p-value=4.6 × 10−128). They also were enriched in an AP1/Fos/NRF2 (NFE2L2) binding site (p-value=1.4 × 10−24) as previously reported for mammalian heat-shock genes. This variation in binding motifs suggests the involvement of distinct co-regulators in establishing differential patterns of HSF1 occupancy. The regions strongly bound by HSF1 in both cancer and in heat shock, had features of both groups. They were enriched for HSEs with three inverted repeats (p-value=1.3 × 10−125). They were not enriched for the YY1 sites but were enriched for the AP1/Fos and NRF2 binding site (p-value=5.2 × 10−7).
HSF1-bound genes form distinct, coordinately-regulated modules
Integrating our diverse data sets, revealed a direct and pervasive role for HSF1 in cancer biology (Figure 4A). Extending far beyond protein folding and stress, HSF1-bound genes were involved in many facets of tumorigenesis, including the cell cycle, apoptosis, energy metabolism and other processes. To gain a more global view of the relationship between the genes most strongly bound by HSF1 in cancer cell lines, we generated an RNA expression correlation matrix through meta-analysis of pre-existing data sets (Figure 4B). We used the UCLA Gene Expression Tool (UGET) (Day et al., 2009) to query the extent to which the expression of each HSF1-bound gene correlated with every other HSF1-bound gene across all of the ~12,000 human expression profiles that have been generated with Affymetrix HG U133 Plus 2.0 arrays and made available through the Celsius database (Day et al., 2009) Hierarchical clustering of this gene-gene correlation matrix revealed five major transcription modules (Figure 4B).
Figure 4. Distinct, coordinately-regulated modules of HSF1-bound genes.
A. Graphical representation of the HSF1 cancer program integrating information on gene binding, regulation and function. The peak height is reflected in the diameter of the circle (log2 peak height: range ~3 to 9) and color intensity reflects gene regulation (average of log2 fold change in BPLER and MCF7 cells upon HSF1 knockdown; red - positively regulated; green – negatively regulated; gray – no data available). Well-bound, differentially regulated genes as well as several genes of biological interest are displayed. B. Gene-gene expression correlation matrix of HSF1-bound genes. Pair-wise correlation map is presented of the genes that were bound by HSF1 in at least two of the three cancer cell lines (BT20, NCIH38, and SKBR3). The Pearson correlation coefficient relating normalized mRNA expression data for each gene pair was assessed in nearly 12,000 expression profiles. Enriched GO (gene-ontology) categories for each module are shown. See also Figure S4.
The largest module was enriched for protein folding, translation and mitosis. Genes within this dominant module showed the strongest positive correlation with the expression of HSF1 mRNA itself. Many of these genes had indeed proven to be regulated by HSF1 in our HSF1 shRNA knockdown experiments (Figures 2, 4A and S4). A second, smaller module was positively correlated with the first and strongly enriched for RNA binding genes. Many of these genes, too, were positively regulated by HSF1 in our knockdown experiments (Figures 2 and 4A and S4). The remaining three modules (center to lower right of the matrix) were enriched for processes involved in immune functions, insulin secretion and apoptosis. All three of these modules were negatively correlated with the largest module, suggesting negative regulation by HSF1.
Activation of HSF1 in a broad range of cancer specimens taken directly from patients
Recently, we evaluated HSF1 expression and localization in a cohort of breast cancer patients culled from the Nurses’ Health Study (NHS) (Santagata et al., 2011). In that work, HSF1 was cytoplasmic and expressed at low levels in normal breast epithelial cells but it accumulated in the nucleus of the majority of tumor specimens. Here, we confirm that finding (Figures 5A, 5B and S5), combining samples from two independent breast cancer collections representing all three major clinical subtypes (see Supplemental Methods).
Figure 5. HSF1 is activated in a broad range of human tumors.
A. IHC shows strong nuclear HSF1 staining in human breast tumor cells (top) with adjacent normal breast epithelial cells (bottom) showing a lack of nuclear HSF1. B. Images of HSF1 IHC on breast cancer tissue microarray (TMA) cores. Heat map shows scoring of three TMAs. The top panel depicts data from two TMAs (BRC1501 and BRC1502), containing 138 breast tumors of all major breast cancer subtypes. Progesterone receptor (PR), ER, and HER2 were also evaluated. The middle panel shows data from 161 triple negative (TN) breast cancer cases. The bottom panel shows the lack of HSF1 nuclear expression in 16 normal mammary tissue sections. A summary is provided in the bar graph (right). C. HSF1 IHC showing high level nuclear staining in indicated tumors; T, Tumor; N, Normal adjacent tissue. A summary is provided in the bar graph. D. ChIP-Seq analysis of human breast and colon cancer surgical resection specimens (patient tumors). Heat map depicting ChIP-Seq read density for all HSF1 target regions defined in Figure 3A. For reference, the binding profiles for cancer cell lines in culture (black; average across BT20, NCIH838 and SKBR3) and parental heat-shocked cell lines (red) are included. HSF1 expression was evaluated by IHC in the same patient tumors used for ChIP-Seq (see Figure S5C) and scored as in Panel B. E. HSF1 binding in cell lines compared to patient tumors. Average binding across cancer cell lines in cell culture (black; average across BT20, NCIH838 and SKBR3), parental heat-shocked cell lines (red), and patient tumors (cyan) are depicted for the representative target genes indicated. F. PCA of HSF1 binding in heat-shocked parental cell lines (red), cancer cells lines (black) and patient tumors (cyan). See also Figure S5 and Table S1.
Next, because our ChIP-Seq analysis showed that the HSF1 cancer program is engaged not just in breast cancer cell lines but also in colon and lung cancer cell lines, we examined more than 300 formalin-fixed surgical specimens taken directly from patients. We included not only colon and lung cancer but also a wide variety of other tumor types. Normal cells adjacent to the tumor demonstrated low HSF1 levels and cytoplasmic localization of the protein. In contrast, high-level expression of HSF1 and nuclear localization was common (Figure 5C) across every cancer type we examined, including carcinomas of the cervix, colon, lung, pancreas and prostate as well as mesenchymal tumors such as meningioma. HSF staining was negative or weak in some tumors from each cancer type (Figure 5C). However, in those tumors, where expression was high, it was remarkably uniform across the sample, with nearly all tumor cells expressing similar levels of nuclear HSF1.
To determine if the high-level nuclear localization of HSF1 detected by immunostaining was truly indicative of its activation, we obtained human tumor samples from breast and colon adenocarcinomas that had been cryopreserved and were of a quality suitable for ChIP-Seq analysis (Figures 5D and S5). Obtaining and processing such human tumor specimens for a technique as demanding as genome-wide ChIP-Seq is highly challenging. In addition, many potentially confounding factors are unavoidable (e.g. cell-type heterogeneity due to the presence of blood and stromal elements, areas of necrosis and micro-environmental stress). Despite these difficulties, the distinct HSF1-binding profile we had established with cultured cancer cell lines was clearly conserved in those tumors that expressed high levels of HSF1. Genes (such as ST13 and EIF4A2) that were strongly bound by HSF1 in cancer lines but weakly bound after heat shock in non-transformed cells, were also strongly bound in these human tumor samples taken directly from patients (Figure 5E). Genes that were weakly bound by HSF1 in cancer lines but strongly bound after heat shock in non-transformed cells (such as HSPA6 and DNAJC7) were also weakly bound in patient tumor samples (Figure 5E). These global similarities in HSF1-binding profiles between cancer cell lines and tumor samples, as well as their divergence from heat shock profiles, were validated by principal component analysis (Figure 5F).
An HSF1-cancer signature identifies breast cancer patients with poor outcome
In our prior analysis of the Nurses’ Health cohort, HSF1 overexpression and nuclear localization was associated with reduced survival (Santagata et al., 2011). That work, however, was based entirely on HSF1 immunohistochemistry, an approach which is inherently only semi-quantitative. To acquire more precise and molecularly defined information about the effects of HSF1 activation in cancer, we asked if malignant potential and long-term outcomes correlate with the HSF1 transcriptional program identified above. We distilled an “HSF1-cancer signature” of 456 genes that were bound by HSF1 near their transcription start sites (Figure 2). Expression of these genes (Table S4) was interrogated in ten publicly available mRNA datasets derived from breast cancer patients that had been followed for an average of 7.58 years and had known clinical outcomes (referenced in Table S5). In total, these cohorts encompassed nearly 1,600 individuals of diverse national and ethnic origin. We divided each dataset into two groups, those with high (top 25%) and those with low (bottom 75%) expression of the HSF1-cancer signature. We performed Kaplan-Meier analysis independently on each dataset to assess potential associations between the HSF1-cancer signature and patient outcome: metastasis-free, relapse-free, or overall survival, depending on the reported outcome parameter for that dataset. One representative analysis is presented in Figure 6A, the remainder are shown in Figure S6A. High expression of our HSF1-cancer signature had a remarkable correlation with poor prognosis (HSF1-CaSig; Figures 6B and S6). In 9 of 10 independent datasets reported over the past 10 years, the P values ranged from 0.05 to <0.0001.
Figure 6. An HSF1-cancer signature is associated with reduced survival in patients with breast cancer.
A. Representative dataset (Pawitan et al., 2005) is shown from a meta-analysis of 10 publicly available mRNA expression datasets (Table S5) derived from human breast tumors with known clinical outcome and representing a total of 1594 patients. Each column corresponds to a tumor, and each row corresponds to a microarray probe for an HSF1-cancer signature (HSF1-CaSig) gene. Median levels of expression are depicted in black, increased expression in yellow, and decreased expression in blue. Tumors are ordered by average level of expression of the HSF1-cancer signature, from low (blue) to high (yellow). Red bars indicate deaths. Kaplan-Meier (KM) analysis of the tumors with high expression of the HSF1-cancer signature (top 25%, “High HSF1-CaSig”, yellow) versus low expressors (bottom 75%, “Low HSF1-CaSig”, blue) is shown. B. Log-rank p-values for each of the indicated classifiers were calculated for each dataset; results are displayed as a heat map. Corresponding KM curves are provided in Figure S6. C. Random gene signature analysis of a representative dataset (Pawitan et al., 2005). KM analysis on the dataset to evaluate associations between 10,000 individual randomly generated gene signatures and patient outcome. The random signatures are binned and ordered from least significant to most significant by the KM-generated test statistic. The Red arrow indicates the test statistic of the HSF1-CaSig. For reference, black arrows indicate the test statistic of the random signature with the median test statistic (5000th) and the random signature with the 95th percentile test statistic. D. KM analysis of individuals with ER+/Lymph node negative tumors (Wang et al., 2005) with Low HSF1-CaSig (blue) or High HSF1-CaSig (yellow). E. KM analysis of 947 individuals from the NHS with ER+, lymph-node negative tumors expressing no, low or high nuclear HSF1 as measured by IHC. Data are from the NHS (1976–1997). Log-rank p-values are shown. See also Figure S6 and Tables S4, S5 and S6.
Next, we considered a recent finding that many published cancer signatures are not significantly better outcome predictors than random signatures of identical size (Venet et al., 2011). We performed Kaplan-Meier analysis on independent datasets to evaluate associations between 10,000 individual randomly generated gene signatures and patient outcome (compiled data Table S4, example Fig 6C). A meta-analysis of the breast datasets showed that the HSF1-CaSig outperformed all 10,000 random gene signatures (Monte Carlo p Value across breast datasets < 0.0001, Table S4).
Our HSF1-cancer signature was more broadly associated with outcome than other well established prognostic indicators (Figures 6B and S6A) including the oncogene MYC, the proliferation marker Ki67 and even MammaPrint, an expression-based diagnostic tool used in routine clinical practice (Kim and Paik, 2010). Because various HSPs have been implicated as prognostic markers for a range of cancers including breast cancer (Ciocca and Calderwood, 2005), we also tested many individual HSP transcripts for possible association with outcome. None of these genes, or even a panel of HSP genes, was as strongly associated with poor outcome as our broader HSF1-cancer signature (Figures 6B and S6A).
HSF1 activation is an indicator of poor outcome in early breast cancer
At the time of diagnosis, the majority of breast cancer patients have ER+ tumors and early-stage disease (ER+/lymph-node negative tumors). A small fraction of these patients will experience a recurrence and might benefit from more aggressive treatment, but it is currently very difficult to identify them in advance. We found that our HSF1-cancer signature was significantly associated with metastatic recurrence in women initially diagnosed with ER+/lymph node negative tumors (p-value=0.0149) (Figure 6D).
To further probe the potential prognostic value of HSF1 in this particularly challenging population, we returned to the Nurses’ Health Study cohort, because it provides one of the largest collections of patients with ER+/lymph node negative tumors for evaluation (n=947), and has the longest patient follow up. Because RNA samples are not available from this collection (initiated in 1976) we could assess only the levels and nuclear localization of HSF1. Survival decreased as HSF1 nuclear levels increased in a dose-dependent manner (p-value=0.0015; Figure 6E). This finding was validated by multivariate analysis which showed high level nuclear HSF1 to be associated with a nearly 100% increase in mortality (Table S6).
HSF1-cancer signature is associated with poor outcome in diverse human cancers
Next, we asked if the HSF1-cancer signature might have prognostic value beyond breast cancer. Analyzing multiple independent gene expression datasets that include outcomes data, increased expression of the HSF1 cancer program in colon and lung cancers was strongly associated with reduced survival (Figures 7A and 7B). The HSF1-CaSig outperformed all 10,000 random gene signatures in these datasets (Monte Carlo p Value across datasets < 0.0001, Table S4). Again, our HSF1-cancer signature was more significantly associated with outcome than any individual HSP transcript or even a panel of HSP genes (Figures 7B and S7). As expected, the MammaPrint expression signature, which was computationally derived using breast cancers, was a poor indicator of outcome in lung and colon cancers (significant in 1 of 4 datasets). Additional HSF1 signatures also comprising positively regulated genes (from Module 1 and 2 of our gene-gene correlation analysis; HSF1-CaSig2) or containing both positively and negatively regulated genes (HSF1-CaSig3) were also strongly associated with patient outcome across tumor types (Table S4). We conclude that the HSF1 cancer program that we have identified supports the malignant state in a diverse spectrum of cancers because it regulates core processes rooted in fundamental tumor biology that ultimately affect outcome.
Figure 7. An HSF1-cancer signature is associated with reduced survival in patients with colon or lung cancers.
A. KM analysis of survival in patients with colon or lung cancer based on Low HSF1-CaSig (blue) or High HSF1-CaSig (yellow). Log-rank p-values are shown. B. Heat map of log-rank p-values for each of the indicated classifiers in four datasets is shown. Corresponding KM curves are in Figure S7. See also Tables S4 and S5.
DISCUSSION
We have defined a distinct genome-wide transcriptional program that HSF1 coordinates in malignancy. This program includes some heat-shock proteins well known to be involved in oncogenic programs, such as Hsp90 (Whitesell and Lindquist, 2005). However, it differs fundamentally from the HSF1 program induced by thermal stress, in that it includes many genes that are not induced by heat shock and does not include many that typically are. This cancer program is commonly activated in a wide variety of human malignancies. It is strongly associated with metastasis and death in at least the three cancers responsible for ~30% of all cancer-related deaths worldwide: those of the breast, colon and lung. The very broad range of tumors in which we see immunohistochemical evidence of HSF1 activation suggests it will play a pervasive role throughout tumor biology.
What types of cellular processes does HSF1 regulate in cancer? They constitute an astonishingly diverse group that extends far beyond protein folding, and includes energy metabolism, cell cycle signalling, DNA repair, apoptosis, cell adhsion, extracellular matrix formation, and translation. Some of these processes were previously known to be affected by HSF1 (Dai et al., 2007; Jin et al., 2011; Zhao et al., 2009). However, the common assumption had been that HSF1s affects were mediated primarily by Hsp chaperone activities (Jin et al., 2011; Meng et al., 2010; Solimini et al., 2007). The remarkable breadth of the HSF1 cancer program in humans explains why HSF1 is such a powerful modifier of tumorigenesis in multiple animal models (Dai et al., 2007; Jin et al., 2011; Zhao et al., 2009) and why HSF1 was identified as one of only six potent metastasis-promoting genes in a genome-wide screen for enhancers of invasion by malignant melanoma cells (Scott et al., 2011).
Not only is the repertoire of HSF1-regulated genes in cancer much larger than just heat-shock genes, but even the manner in which some of the classical heat-shock genes are regulated differs. For example, HSPA6 (HSP70B’), a pillar of the heat-shock response, differs dramatically in these two states. Following heat stress, HSPA6 is the most highly induced of all mRNAs, yet, surprisingly in cancer, HSPA6 is only bound very weakly by HSF1. Its expression is not changed following HSF1 depletion and its transcript level does not correlate with that of HSF1 in our meta-analysis of 12,000 gene expression experiments.
What could account for activation of a distinct HSF1-regulated program in cancer? After many years of investigation, we do not yet fully understand how HSF1 activity is regulated during the classic heat-shock response. Multiple mechanisms have been described. These include the release of HSF1 from its normal sequestration by chaperones when unfolded substrates compete for chaperone binding. But in addition, HSF1 is subject to extensive array of post-translational modifications (at least 30) including acetylation, sumoylation and numerous phosphorylations (Anckar and Sistonen, 2011).
Some of these heat-shock regulatory mechanisms are likely shared by cancer cells. For instance, impaired protein homeostasis driven by the accumulation of mutant, misfolding-prone oncoproteins, aneuploidy and the increased rate of translation in cancer could chronically stimulate HSF1 activation by releasing it from sequestration from chaperones (Anckar and Sistonen, 2011). Dysregulation of signaling pathways in cancer could also drive post-translational modifications to HSF1. Some of these (such as those responsible for phosphorylation at serine 326) will likely be shared with heat-shocked cells. But others will likely be unique to cancer. Indeed, it seems extremely likely that different mechanisms of activiation will operate in different cancers. Several pathways activated in cancer such as EGFR/HER2 axis (Zhao et al., 2009), the RAS/MAPK (Stanhill et al., 2006) or the insulin/IGFI-like growth factor system (Chiang et al., 2012) have all been reported to alter HSF1 activity. Additional modes of cancer-specific regulation might include epigenetic states common to cancer and proliferating cells and transcriptional co-regulators.
How might the distinct transcriptional program regulated by HSF1 in malignancy have arisen? The association of this program with metastasis and death points to an evolutionary origin distinct from cancer itself. The broad range of cancer types in which we find HSF1 activated suggests that this program originated to support basic biological processes. Indeed, the sole heat-shock factor in yeast (yHSF), even at basal temperatures, binds many genes that are involved in a wide-range of core cellular functions (Hahn et al., 2004). These transcriptional targets allow yeast not only to adapt to environmental co ntingencies but also to modulate metabolism and maintain proliferation under normal growth conditions (Hahn et al., 2004; Hahn and Thiele, 2004). As a result, yHSF is essential for viability, paralleling the importance of HSF1 for the survival and proliferation of cancer cells (Dai et al., 2007). Activation of HSF1 may also be required in animals in states of high proliferation and altered metabolism such as immune activation and wound healing (Rokavec et al., 2012; Xiao et al., 1999; Zhou et al., 2008). Moreover, in diverse eukaryotes, HSF is a well-validated longevity factor; non-classical activation of this transcription factor could be highly relevant in this context (Chiang et al., 2012; Volovik et al., 2012)
Ironically, the evolutionarily ancient role played by HSF1 in helping cells to adapt, survive and proliferate is co-opted frequently to support highly malignant cancers. By enabling oncogenesis, the activation of this ancient pro-survival mechanism thereby actually impairs survival of the host. HSF1 activation in a particular tumor may reflect the degree to which accumulated oncogenic mutations have disrupted normal physiology even before overt invasion or metastasis occurs. This interpretation would explain the impressively broad prognostic value of our HSF1-cancer signature across disparate cancers and even at early stages of disease. Clinical implementation will require further refinement of the signature and validation in tissue and RNA samples from multiple clinical cohorts. Such studies are certainly warranted. As just one potential application, it might aid in the identification of indolent tumors that do not require intervention, reducing the burdens of unnecessary treatment (Kalager et al., 2012). In addition to its prognostic value, HSF1 and diverse regulators that activate it might prove useful targets for cancer therapeutics.
Our understanding of the extensive role played by HSF1 in supporting cancers continues to mature. The protein has been defined for decades by its ability to coordinate chaperone protein expression and enhance survival in the face of heat stress (Christians et al., 2002; Ritossa, 1962). While appreciating the importance of these classical mechanisms, the role of HSF1 is clearly much broader and deeper.
MATERIALS AND METHODS
Cell culture methods
Cell lines were cultured as detailed in Supplemental Methods.
ChIP-Seq and ChIP-qPCR
ChIP-qPCR and ChIP-Seq experiments were performed as described previously (Lee et al., 2006), with modifications detailed in Supplemental Methods.
Gene expression Analysis
Lentiviral shRNA methods were described previously (Dai et al., 2007). Gene expression analysis was performed as described in Supplemental Methods. Microarray data was deposited in NCBI Gene Expression Omnibus. RT-PCR analysis, gene-gene correlation analysis of HSF1-bound genes and correlation of HSF1-bound gene expression with outcome is detailed in Supplemental Methods.
Immunohistochemistry and The Nurses’ Health Study (NHS) analysis
Paraffin sections were stained using HSF1 antibody (Thermo Scientific, RT-629-PABX) as detailed in Supplemental Methods. The NHS is a prospective cohort study initiated in 1976 (Hu et al., 2011; Tamimi et al., 2008). For design and study population, and analysis, see Supplemental Methods.
Statistical Analysis
Correlation of gene expression with location of HSF1 occupancy was performed using a two-tailed Fisher’s Exact Test. P-values for significance of overlap between pairs of gene sets were generated using the hypergeometric distribution. Statistical methods for ChIP-Seq analysis and the Nurses’ Health Study outcome data analysis are detailed in Supplemental Methods. Kaplan-Meier analysis was used to compare outcome events. P-values were generated using the logrank test. For all other data, mean +/− standard deviation is reported and statistical significance between means was determined using a two-tailed t test.
Supplementary Material
HIGHLIGHTS.
Comprehensive study of the direct transactivating effects of HSF1 in cancer
HSF1 regulates diverse cellular processes that extend far beyond heat-shock genes
Fundamental differences in HSF1 program in cancer versus heat shock
HSF1 activation in multiple cancers is strongly associated with metastasis and death
Acknowledgments
We thank G. Frampton, I. Barrasa and S.Gupta for bioinformatic assistance. We thank T. Mazor, T. Volkert and the WIBR-GTC for sequencing support. We thank the Lindquist lab and T. Lee for discussion and K. Allendoerfer, B. Bevis, G. Karras, R. Shouval and K. Matlock for comments. The work was supported by the J&J COSAT focused funding program (L.W.) and the Marble Fund (S.L.). S.L. is an Investigator of the Howard Hughes Medical Institute. M.L.M. is supported by American Cancer Society New England Division-SpinOdyssey (#PF-09-253-01-DMC). S.S. is supported by NIH (K08NS064168), the Brain Science Foundation and the V Foundation. Additional support was provided by GSK (WE234 EPI40307); Public Health Service Grants CA087969, and SPORE in Breast Cancer CA089393, from the NCI, NIH, Dept. of Health and Human Services. T.A.I is supported by the Breast Cancer Research Foundation, NCI (R01-CA146445-01) and DoD-CDMRP Breast Cancer Research Program (W81XWH-08-1-0282 BC-07456). We thank the participants and staff of the NHS, and state cancer registries: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anckar J, Sistonen L. Regulation of HSF1 Function in the Heat Stress Response: Implications in Aging and Disease. Annu Rev Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- Chiang WC, Ching TT, Lee HC, Mousigian C, Hsu AL. HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell. 2012;148:322–334. doi: 10.1016/j.cell.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians ES, Yan LJ, Benjamin IJ. Heat shock factor 1 and heat shock proteins: Critical partners in protection against acute cell injury. Crit Care Med. 2002;30:S43–S50. [PubMed] [Google Scholar]
- Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10:86–103. doi: 10.1379/CSC-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A, Dong J, Funari VA, Harry B, Strom SP, Cohn DH, Nelson SF. Disease gene characterization through large-scale co-expression analysis. PLoS One. 2009;4:e8491. doi: 10.1371/journal.pone.0008491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O’Malley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Guettouche T, Boellmann F, Lane WS, Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ, Iyer VR. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol. 2004;24:5249–5256. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JS, Thiele DJ. Activation of the Saccharomyces cerevisiae heat shock transcription factor under glucose starvation conditions by Snf1 protein kinase. J Biol Chem. 2004;279:5169–5176. doi: 10.1074/jbc.M311005200. [DOI] [PubMed] [Google Scholar]
- Hu R, Dawood S, Holmes MD, Collins LC, Schnitt SJ, Cole K, Marotti JD, Hankinson SE, Colditz GA, Tamimi RM. Androgen receptor expression and breast cancer survival in postmenopausal women. Clin Cancer Res. 2011;17:1867–1874. doi: 10.1158/1078-0432.CCR-10-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE, Iglehart JD, Weinberg RA. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- Ishikawa N, Takano A, Yasui W, Inai K, Nishimura H, Ito H, Miyagi Y, Nakayama H, Fujita M, Hosokawa M, et al. Cancer-testis antigen lymphocyte antigen 6 complex locus K is a serologic biomarker and a therapeutic target for lung and esophageal carcinomas. Cancer Res. 2007;67:11601–11611. doi: 10.1158/0008-5472.CAN-07-3243. [DOI] [PubMed] [Google Scholar]
- Jin X, Moskophidis D, Mivechi NF. Heat Shock Transcription Factor 1 Is a Key Determinant of HCC Development by Regulating Hepatic Steatosis and Metabolic Syndrome. Cell Metab. 2011;14:91–103. doi: 10.1016/j.cmet.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalager M, Adami HO, Bretthauer M, Tamimi RM. Overdiagnosis of Invasive Breast Cancer Due to Mammography Screening: Results From the Norwegian Screening Program. Ann Intern Med. 2012;156:491–499. doi: 10.7326/0003-4819-156-7-201204030-00005. [DOI] [PubMed] [Google Scholar]
- Khaleque MA, Bharti A, Gong J, Gray PJ, Sachdev V, Ciocca DR, Stati A, Fanelli M, Calderwood SK. Heat shock factor 1 represses estrogen-dependent transcription through association with MTA1. Oncogene. 2008;27:1886–1893. doi: 10.1038/sj.onc.1210834. [DOI] [PubMed] [Google Scholar]
- Kim C, Paik S. Gene-expression-based prognostic assays for breast cancer. Nat Rev Clin Oncol. 2010;7:340–347. doi: 10.1038/nrclinonc.2010.61. [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Lee HJ, Lee JS, Jeoung D, Kang CM, Bae S, Lee SJ, Kwon SH, Kang D, Lee YS. A novel function for HSF1-induced mitotic exit failure and genomic instability through direct interaction between HSF1 and Cdc20. Oncogene. 2008;27:2999–3009. doi: 10.1038/sj.onc.1210966. [DOI] [PubMed] [Google Scholar]
- Liberal V, Martinsson-Ahlzen HS, Liberal J, Spruck CH, Widschwendter M, McGowan CH, Reed SI. Breast Cancer Special Feature: Cyclin-dependent kinase subunit (Cks) 1 or Cks2 overexpression overrides the DNA damage response barrier triggered by activated oncoproteins. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1102434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27:1696–1697. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac KD, Lo KA, Gordon W, Motola S, Mazor T, Fraenkel E. A quantitative model of transcriptional regulation reveals the influence of binding location on expression. PLoS Comput Biol. 2010;6:e1000773. doi: 10.1371/journal.pcbi.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama M, Yoshitake H, Tsukamoto H, Takamori K, Araki Y. Molecular expression of Ly6k, a putative glycosylphosphatidyl-inositol-anchored membrane protein on the mouse testicular germ cells. Biochem Biophys Res Commun. 2010;402:75–81. doi: 10.1016/j.bbrc.2010.09.117. [DOI] [PubMed] [Google Scholar]
- Meng L, Gabai VL, Sherman MY. Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene. 2010;29:5204–5213. doi: 10.1038/onc.2010.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JN, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene. 2007;26:5086–5097. doi: 10.1038/sj.onc.1210317. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page TJ, Sikder D, Yang L, Pluta L, Wolfinger RD, Kodadek T, Thomas RS. Genome-wide analysis of human HSF1 signaling reveals a transcriptional program linked to cellular adaptation and survival. Mol Biosyst. 2006;2:627–639. doi: 10.1039/b606129j. [DOI] [PubMed] [Google Scholar]
- Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, Han X, Holmberg L, Huang F, Klaar S, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–964. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR. A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell. 1982;30:517–528. doi: 10.1016/0092-8674(82)90249-5. [DOI] [PubMed] [Google Scholar]
- Ritossa F. A new puffing pattern induced by temperature shock and DNP in Drosophila. Experimentia. 1962;18:571–573. [Google Scholar]
- Rokavec M, Wu W, Luo JL. IL6-Mediated Suppression of miR-200c Directs Constitutive Activation of Inflammatory Signaling Circuit Driving Transformation and Tumorigenesis. Mol Cell. 2012;45:777–789. doi: 10.1016/j.molcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H, Enoki Y. Novel aspects of heat shock factors: DNA recognition, chromatin modulation and gene expression. FEBS J. 2010;277:4140–4149. doi: 10.1111/j.1742-4658.2010.07829.x. [DOI] [PubMed] [Google Scholar]
- Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, Schnitt SJ, Whitesell L, Tamimi RM, Lindquist S, et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18378–18383. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Xu YM, Wijeratne EM, Kontnik R, Rooney C, Perley CC, Kwon H, Clardy J, Kesari S, Whitesell L, et al. Using the heat-shock response to discover anticancer compounds that target protein homeostasis. ACS Chem Biol. 2012;7:340–349. doi: 10.1021/cb200353m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KL, Nogueira C, Heffernan TP, van Doorn R, Dhakal S, Hanna JA, Min C, Jaskelioff M, Xiao Y, Wu CJ, et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer Cell. 2011;20:92–103. doi: 10.1016/j.ccr.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamovsky I, Nudler E. New insights into the mechanism of heat shock response activation. Cell Mol Life Sci. 2008;65:855–861. doi: 10.1007/s00018-008-7458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solimini NL, Luo J, Elledge SJ. Non-oncogene addiction and the stress phenotype of cancer cells. Cell. 2007;130:986–988. doi: 10.1016/j.cell.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Stanhill A, Levin V, Hendel A, Shachar I, Kazanov D, Arber N, Kaminski N, Engelberg D. Ha-ras(val12) induces HSP70b transcription via the HSE/HSF1 system, but HSP70b expression is suppressed in Ha-ras(val12)-transformed cells. Oncogene. 2006;25:1485–1495. doi: 10.1038/sj.onc.1209193. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamimi RM, Baer HJ, Marotti J, Galan M, Galaburda L, Fu Y, Deitz AC, Connolly JL, Schnitt SJ, Colditz GA, et al. Comparison of molecular phenotypes of ductal carcinoma in situ and invasive breast cancer. Breast Cancer Res. 2008;10:R67. doi: 10.1186/bcr2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venet D, Dumont JE, Detours V. Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput Biol. 2011;7:e1002240. doi: 10.1371/journal.pcbi.1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volovik Y, Maman M, Dubnikov T, Bejerano-Sagie M, Joyce D, Kapernick EA, Cohen E, Dillin A. Temporal requirements of heat shock factor-1 for longevity assurance. Aging Cell. 2012;11:491–499. doi: 10.1111/j.1474-9726.2012.00811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, Benjamin IJ. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 1999;18:5943–5952. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Liu H, Liu Z, Ding Y, Ledoux SP, Wilson GL, Voellmy R, Lin Y, Lin W, Nahta R, et al. Overcoming Trastuzumab Resistance in Breast Cancer by Targeting Dysregulated Glucose Metabolism. Cancer Res. 2011;71:4585–4597. doi: 10.1158/0008-5472.CAN-11-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YH, Zhou M, Liu H, Ding Y, Khong HT, Yu D, Fodstad O, Tan M. Upregulation of lactate dehydrogenase A by ErbB2 through heat shock factor 1 promotes breast cancer cell glycolysis and growth. Oncogene. 2009;28:3689–3701. doi: 10.1038/onc.2009.229. [DOI] [PubMed] [Google Scholar]
- Zhou JD, Luo CQ, Xie HQ, Nie XM, Zhao YZ, Wang SH, Xu Y, Pokharel PB, Xu D. Increased expression of heat shock protein 70 and heat shock factor 1 in chronic dermal ulcer tissues treated with laser-aided therapy. Chin Med J (Engl) 2008;121:1269–1273. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.