Abstract
Mainly regulated at the transcriptional level, the cellular cyclin-dependent kinase inhibitor, CDKN1A/p21WAF1 (p21), is a major cell cycle regulator of the response to DNA damage, senescence and tumor suppression. Here, we report that COUP-TF-interacting protein 2 (CTIP2), recruited to the p21 gene promoter, silenced p21 gene transcription through interactions with histone deacetylases and methyltransferases. Importantly, treatment with the specific SUV39H1 inhibitor, chaetocin, repressed histone H3 lysine 9 trimethylation at the p21 gene promoter, stimulated p21 gene expression and induced cell cycle arrest. In addition, CTIP2 and SUV39H1 were recruited to the silenced p21 gene promoter to cooperatively inhibit p21 gene transcription. Induction of p21WAF1 gene upon human immunodeficiency virus 1 (HIV-1) infection benefits viral expression in macrophages. Here, we report that CTIP2 further abolishes Vpr-mediated stimulation of p21, thereby indirectly contributing to HIV-1 latency. Altogether, our results suggest that CTIP2 is a constitutive p21 gene suppressor that cooperates with SUV39H1 and histone methylation to silence the p21 gene transcription.
Keywords: CTIP2, p21WAF1, SUV39H1, HIV-1
Introduction
The cyclin-dependent kinase inhibitor, CDKN1A/p21WAF1 (p21), is a key cell cycle controller that is mainly regulated at the transcriptional level (Gartel and Radhakrishnan, 2005). Induction of p21 results in G1-, G2- (Niculescu et al., 1998) or S-phase arrest (Ogryzko et al., 1997; Radhakrishnan et al., 2004). Both positive and negative regulations modulate p21 gene expression. A variety of cellular transcription factors, such as p53, Sp1 or C/EBP, stimulate p21 gene promoter activity (for review see Gartel and Tyner (1999)). However, little is known about the negative regulation of p21 (Gartel and Radhakrishnan, 2005). Previous studies have identified direct negative regulatory effects exerted by Myc and AP4 on the p21 gene promoter (Claassen and Hann, 2000; Gartel et al., 2001; Jung et al., 2008) and indirect effects through the inhibition of p53. Epigenetic modifications, such as DNA methylation and histone deacetylation, are crucial for p21 gene silencing (Suzuki et al., 2000; Gartel and Tyner, 2002; Lagger et al., 2003; Gartel and Radhakrishnan, 2005), but the underlying molecular mechanism through which the involved chromatin-modifying enzymes are recruited to the p21 gene promoter remain unclear. Although p21 has been known for some time to play an important part in cell cycle regulation and the pathogenesis of several disorders, including several cancers, p21 has been recently described as a pivotal facilitator of the human immunodeficiency virus 1 (HIV-1) life cycle in macrophages, a key target cell type in AIDS pathogenesis. HIV-1 infection activates p21 expression and forces a cell cycle arrest that is highly permissive for viral transcription in macrophages (Thierry et al., 2004; Vazquez et al., 2005). In hematopoietic cells, p21 expression is detrimental to HIV-1 replication (Zhang et al., 2007). In addition, HIV-1 has been shown to reduce p21 gene expression in T cells (Clark et al., 2000). In macrophages, HIV-1-induced p21 overexpression has been attributed to the HIV-1 Vpr protein. Vpr stimulates p21 gene transcription in cooperation with the cellular transcription factor Sp1, which binds to the p21 gene promoter (Amini et al., 2004). Vpr has many other cellular effects (for review see Moon and Yang (2006)), including indirect DNA damages, which also favor G2 arrest and apoptosis (Schrofelbauer et al., 2007). Moreover, p21 is required to sustain G2 arrest after DNA damage (Bunz et al., 1998).
We have previously reported that the cellular cofactor COUP-TF-Interacting Protein 2 (CTIP2) represses HIV-1 gene transcription in microglial cells, the central nervous system-resident macrophages. In microglial cells, CTIP2 binds the HIV-1 promoter through Sp1, and recruits histone deacetylase (HDAC) and histone methyltransferase (HMT) activities to promote heterochromatic environment and HIV-1 transcriptional silencing (Marban et al., 2005, 2007). The observation that CTIP2 can affect gene expression through HDAC and HMT recruitments to the viral promoter suggests that other genes might be targeted by similar mechanisms. CTIP2 has been observed to have many, sometimes apparently disparate, effects in a variety of cell types and biological systems, but little unifying mechanistic data have been reported. CTIP2 is expressed in the brain and the immune system (Leid et al., 2004), and plays a major role in the development of corticospinal motor neuron axonal projections to the spinal cord (Arlotta et al., 2005). In the SK-N-MC neuronal cell line, CTIP2 inhibits transcription of the cdk inhibitor p57KIP2 transcription (Topark-Ngarm et al., 2006). Mostly described as a corepressor, CTIP2 participates in the activation of IL2 gene expression in CD4 + T lymphocytes (Cismasiu et al., 2006). CTIP2 regulates both differentiation and survival of thymocytes, and is required for T-lymphocyte development. Thymocytes from CTIP2 KO mice are highly susceptible to apoptosis (Wakabayashi et al., 2003b), but CTIP2 gene deletions or point mutations induce thymic lymphomas in gamma-irradiated mice (Wakabayashi et al., 2003a). Recent studies confirmed that CTIP2 exerts anti-apoptotic activities in T cell lines (Grabarczyk et al., 2007; Kamimura et al., 2007). All these convergent data prompted us to investigate the impact of CTIP2 on the cell cycle, specifically in HIV-1-infected macrophages and microglial cells.
Here, we report that CTIP2 is a key transcriptional regulator of p21 gene expression. Recruited to the Sp1-binding sites of the p21 promoter, CTIP2 transcriptionally repressed constitutive p21 gene expression in cooperation with HDAC and HMT activities. Treatment with trichostatin A (TSA, a class-I and -II HDAC inhibitor) and with chaetocin (a specific inhibitor of the HMT SUV39H1) activated p21 gene promoter activity. Whereas SUV39H1 cooperated with CTIP2 to repress p21 gene transcription, chaetocin treatments induced cell cycle arrest. Interestingly, as p21 gene expression was enhanced upon HIV infection in macrophages and microglial cells, CTIP2 counteracted this Vpr-mediated effect and the related cell cycle arrest. CTIP2 and Vpr were shown to be recruited to the p21 promoter in vivo. However, CTIP2 overexpression displaced Vpr from the p21 gene promoter, thereby promoting p21 gene transcriptional silencing. Altogether, our results show histone methylation as an epigenetic modification involved in p21 transcriptional silencing, and suggest that CTIP2-mediated anti-HIV activity is a bimodal phenomenon. CTIP2 both recruits a repressive chromatin-modifying enzymatic complex to the HIV-1 promoter, as previously shown by our laboratory, and has an indirect effect on the cell cycle progression through p21 promoter repression.
Results
CTIP2 impairs constitutive p21 gene transcription
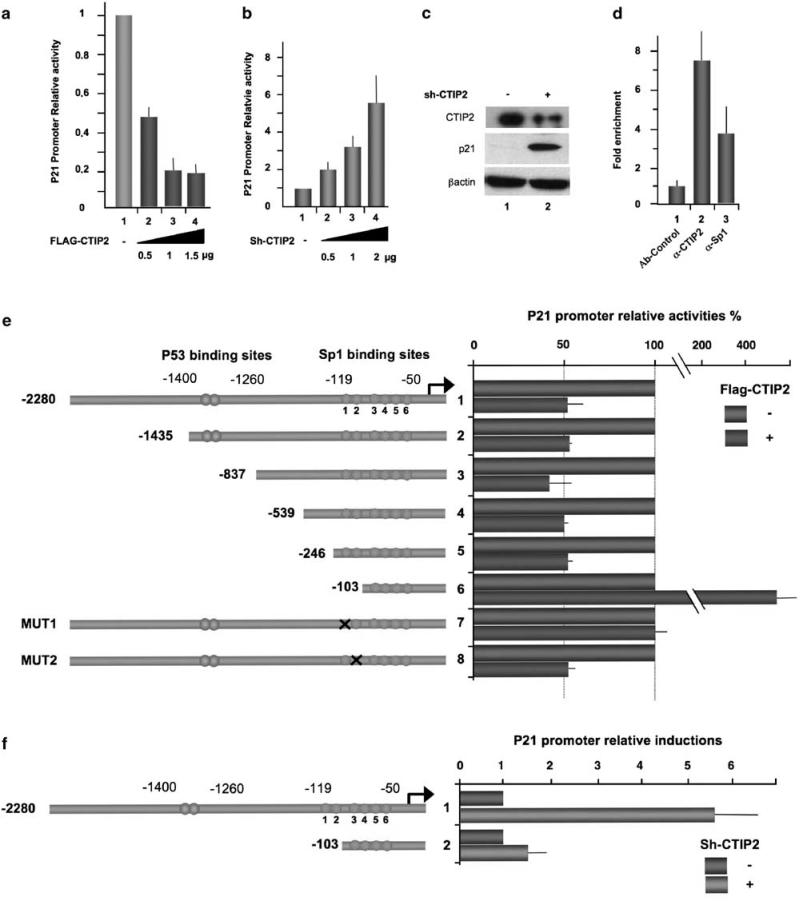
To assess the potential impact of CTIP2 on the transcriptional activity of the p21 gene promoter, we transfected microglial cells with a full-length p21 promoter–luciferase construct (p21-LUC) in the presence or not of CTIP2. Whereas overexpression of CTIP2 downregulated p21 gene transcription up to 80% (Figure 1a), knocking down CTIP2 strongly stimulated (up to sixfold) p21 promoter activity (Figure 1b and Supplementary Figures 1 and 6a). To evaluate p21 expression at the protein level, we performed immuno-detection experiments using extracts from CTIP2 knocked down cells (Figure 1c, column 2) and control cellular extracts (Figure 1c, column 1). The p21 protein was barely detectable in the control cell extracts, but was strongly increased in CTIP2 knocked down cells (Figure 1c, columns 1 and 2).
Figure 1.
Associated with the p21 proximal promoter region in vivo, COUP-TF-interacting protein 2 (CTIP2) impairs constitutive p21 gene transcription through the proximal –246/–103 Sp1-binding sites. Microglial cells (a and b) and 293T cells (c) were transfected with the indicated amounts of vectors in the presence of the p21-Luc plasmid. Luciferase activities (a and b) are presented relative to the activity of the p21-LUC cotransfected with the control empty vector. The protein levels (c) were accessed by western blot with the indicated antibodies 48-h post-transfection. (d) To detect association of CTIP2 and Sp1 with the p21 promoter, ChIP (chromatin immunoprecipitation) experiments were performed in microglial cells with the indicated antibodies. Immunoprecipitated DNA was subjected to real-time PCR quantification with primers targeting the endogenous p21 promoter proximal region. Specific enrichments of the p21 promoter in the immunoprecipitated material were normalized to enrichments in non-specific glyceraldehyde 3-phosphate dehydrogenase DNA and are presented relative to the control. (e and f) Microglial cells were transfected with the p21-LUC constructs in the presence or absence of the pFlag-CTIP2 or pShRNA-CTIP2 plasmids, as indicated. Luciferase assays were performed 48-h post-transfection. Results are presented relative to the basal level of each p21-LUC constructs taken as one.
These results showed that CTIP2 constitutively represses transcription of the p21 gene and its subsquent protein expression.
CTIP2 is recruited to the p21 gene proximal promoter in vivo to inhibit p21 gene transcription
To examine the presence of CTIP2 on the p21 gene promoter in vivo, we performed chromatin immunoprecipitation (ChIP) experiments using microglial cells and a set of primers flanking the proximal Sp1 binding sites of the endogenous p21 promoter. As shown in Figure 1d, CTIP2 as well as Sp1 associated with the proximal region of the p21 promoter. We recently reported that Sp1 protein acts as an anchor for CTIP2 on the HIV-1 promoter (Marban et al., 2005, 2007). As the p21 gene promoter harbors six Sp1-binding sites, we decided to map the Sp1 sites involved in CTIP2-mediated transcriptional repression by transfecting progressive 5′-truncated and mutated p21 reporter constructs. As shown in Figure 1e, 5′-truncations of the p21 promoter up to the Sp1-binding site region did not significantly impair CTIP2-mediated repression (Figure 1e, lanes 2–6). However, further deletion of the –246/–103 Sp1-binding sites (sites 1 and 2) not only abolished this repressive activity but also caused a 4.5-fold increase in luciferase activity (Figure 1e, compare lane 7 with lanes 2–6). Further mutations in the –103/+1 Sp1-binding sites (sites 3–6) did not change the CTIP2-mediated transcriptional effects observed with the –103 p21 promoter construct (data not shown). However, mutation of the Sp1 site 1 (Figure 1e, line 8), but not of the Sp1 site 2 (Figure 1e, line 9), impaired the CTIP2 repressive activity. Logically, whereas CTIP2 knockdown activated the full-length p21 promoter activity, 5′ deletion of the promoter region, including the Sp1 sites 1 and 2, abrogated this activation (Figure 1f, lane 3).
Altogether, both overexpression and knockdown experiments show that the CTIP2 cofactor negatively regulates p21 gene promoter activity through the Sp1-binding site 1 located 119 bp upstream of the initiation start site. Furthermore, these data and Supplementary Figure 3 suggest that Sp1 proteins bound to site 1 are involved in recruitment of CTIP2 to the p21 gene promoter.
CTIP2 cooperates with TSA-sensitive HDACs and with SUV39H1 to repress p21 expression
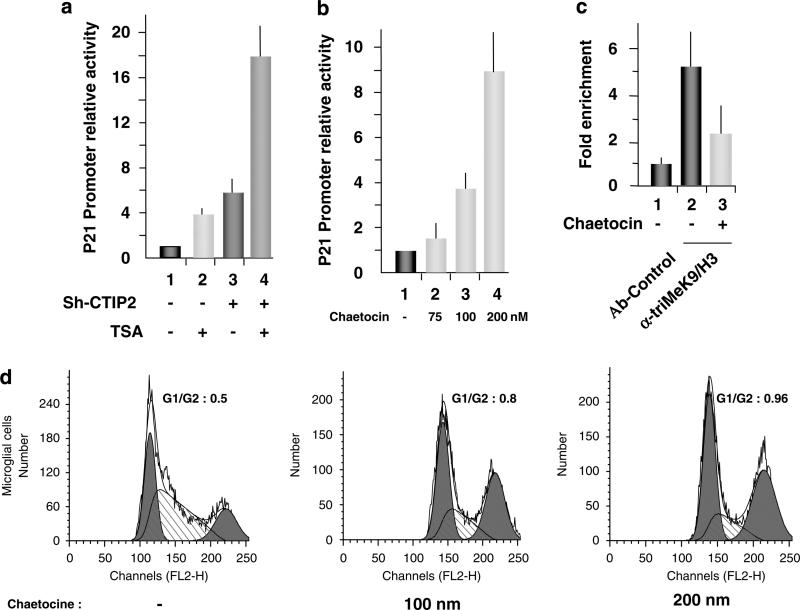
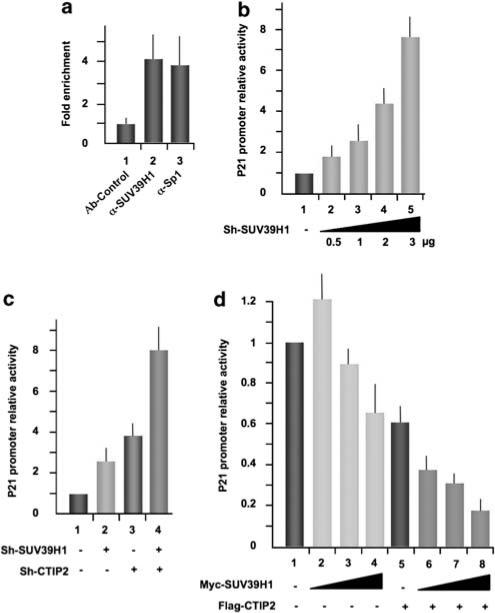
We have recently shown that CTIP2 is part of a chromatin-modifying multienzymatic complex that includes HDAC1, HDAC2 and SUV39H1 (Marban et al., 2007). To assess the potential role of these enzymes in p21 gene transcription, we transfected microglial cells with the full-length p21-LUC reporter plasmid. At 24-h post-transfection, cells were treated or not with TSA (Figure 2a). TSA treatment stimulated p21 gene promoter activity up to fourfold (Figure 2a, column 2), which is in agreement with the previously reported involvement of TSA-sensitive HDACs in the constitutive repression of p21 gene transcription (Murphy et al., 1999; Sowa et al., 1999). Furthermore, CTIP2 knockdown and TSA treatment synergistically activated p21 gene promoter activity up to 18-fold (Figure 2a, column 4). These data corroborate with the interactions among CTIP2, HDAC1 and HDAC2, which we have previously described (Marban et al., 2007), and with the previously described HDAC1 recruitment to the p21 promoter through Sp1 association (Lagger et al., 2003). Next, we focused on the potential role of SUV39H1 in p21 gene transcription. Treatments with increasing concentrations of chaetocin, a specific inhibitor of SUV39H1, activated p21 gene promoter activity in a dose-dependent manner (Figure 2b) and induced cell cycle arrest (Figure 2d and Supplementary Figure 5). Importantly, the level of trimethylation of histone H3 lysine 9 in the p21 promoter region decreased upon chaetocin treatment (Figure 2c), showing, in the context of the p21 promoter, the inhibitory effect of chaetocin on SUV39H1-mediated trimethylation. To further study the role of SUV39H1 in p21 gene repression, we investigated the recruitment of this HMT to the p21 gene promoter using ChIP assays. As shown in Figure 3a, SUV39H1 was constitutively associated with the p21 promoter in vivo. Functionally, SUV39H1 knockdown strongly activated the p21 promoter activity in a dose-dependent manner (Figure 3b and Supplementary Figure 6b). We have recently reported that CTIP2 interacts with SUV39H1 and favors its recruitment to the HIV-1 promoter (Marban et al., 2007). To examine whether SUV39H1 cooperates with CTIP2 in p21 promoter repression, we performed CTIP2 and SUV39H1 knockdown and overexpression experiments in microglial cells expressing p21-promoter–reporter constructs (Figure 3c). Knocking down SUV39H1 expression caused a threefold stimulation of p21 gene promoter activity (Figure 3c, column 2), and the combined knockdown of both SUV39H1 and CTIP2 cooperated to stimulate p21 gene promoter activity up to ninefold (Figure 3c, column 4). Moreover, although overexpression of SUV39H1 alone only slightly repressed p21 gene promoter activity in a dose-dependent manner (Figure 3d, columns 2–4), the combined over-expression of both CTIP2 and SUV39H1 synergistically repressed p21 promoter activity up to 80% (Figure 3d, columns 6–8). This functional repressive cooperation supports the model that CTIP2 mediates recruitment of SUV39H1 to the p21 promoter, as we have observed in the context of the HIV-1 promoter (Marban et al., 2007). Altogether, these results show the involvement of SUV39H1 and the cooperation of CTIP2 and SUV39H1 in p21 gene transcriptional silencing and in cell cycle control.
Figure 2.
Trichostatin A (TSA) and chaetocin stimulate p21 gene transcription. (a, b) Microglial cells were transfected with the p21-LUC reporter plasmid in the presence or absence of pSuper-shRNA-CTIP2, as indicated. At 24-h post-transfection, cells were treated with 450 μm TSA or the indicated amount of chaetocin for 24 h and assessed for LUC activity. (c) Microglial cells were treated or not with 200 nm of chaetocin and subjected to ChIP (chromatin immunoprecipitation) experiments with anti-H3 trimethylated lysine 9. Immunoprecipitated DNA was subjected to real-time PCR quantification with primers targeting the endogenous p21 promoter proximal region. Specific enrichment of the p21 promoter in the immunoprecipitated material were normalized to enrichments in nonspecific glyceraldehyde 3-phosphate dehydrogenase DNA and presented relative to the control. (d) Microglial cells were treated with the indicated amount of chaetocin for 24 h before being assessed for cell cycle status by flow cytometry.
Figure 3.
COUP-TF-interacting protein 2 (CTIP2) cooperates with SUV39H1 to repress p21 gene transcription. (a) ChIP (chromatin immunoprecipitation) experiments were performed in microglial cells with the indicated antibodies. Immunoprecipitated DNA was subjected to real-time PCR quantification with primers targeting the endogenous p21 promoter proximal region. Specific enrichment of the p21 promoter in the immunoprecipitated material were normalized to enrichments in non-specific glyceraldehyde 3-phosphate dehydrogenase DNA and presented relative to the control. (b–d) Microglial cells were transfected with the p21-LUC reporter plasmid in the presence or absence of the indicated vectors. LUC assays were performed 48-h post-transfection and results presented relative to the basal level.
p21 gene transcription is upregulated upon HIV-1 infection in primary ‘microglial-like’ cells
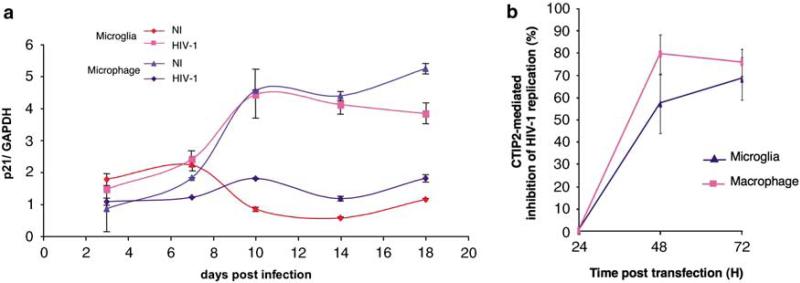
Infection of macrophages by HIV-1 activates p21 mRNA production and promotes G2/M cell cycle arrest. Both events seem to favor HIV-1 replication (Thierry et al., 2004; Vazquez et al., 2005). To examine whether p21 expression is modified in microglial cells, we infected human primary macrophages and micro-glial-like cells with the HIV-1 Bal isolate and quantified p21 mRNA by quantitative reverse transcriptase PCR (Figure 4a). Given that the fluctuations of the p21 mRNA level in non-infected cells may result from the culture conditions, p21 mRNA production was stimulated by HIV-1 infection in primary macrophages, as previously described (Vazquez et al., 2005). Interestingly, the same range of activation was observed in infected primary microglial-like cells, suggesting the need for p21-induced events for efficient HIV-1 expression in microglial cells too. More importantly, these results suggest that p21 induction may be a good target for transcriptional silencing factors involved in HIV-1 latency. Thus, we logically looked at CTIP2-mediated activity in both HIV-1-infected primary monocyte-derived cell types.
Figure 4.
As P21 gene expression is upregulated by human immunodeficiency virus 1 (HIV-1) infection, COUP-TF-interacting protein 2 (CTIP2) inhibits HIV-1 replication in human primary macrophages and microglial-like cells. (a) RNAs from HIV-1/Bal-infected cells were extracted and assessed for p21 mRNA expression. Inductions at each time points are presented relative to the control glyceraldehyde 3-phosphate dehydrogenase level. (b) Cells were transfected with HIV-1 pNL4-3 in the presence or absence (control cells) of the Flag-CTIP2 expression vector. HIV-1 reverse transcriptase activity was assessed 24-, 48- and 72-h post-transfection. Inhibition of HIV-1 replication is presented relative to the reverse transcriptase (RT) activity detected with the HIV-1-infected control cells. RT activity in control cells (without Flag-CTIP2) varied between 0 and 197 pg/ml.
CTIP2 represses HIV-1 replication in human primary macrophages and microglial-like cells
To investigate CTIP2 function in HIV-1-infected primary cells, we transfected primary macrophages and primary macrophage-derived microglial-like cells with a complete HIV-1 infectious provirus (pNL4-3) in the presence or not of CTIP2 overexpression. As shown in Figure 4b, CTIP2 overexpression repressed HIV-1 replication up to 70% in both human primary cell types tested. These results confirmed, in cultured primary cells, the repressive activity of CTIP2 previously observed in a microglial cell line. Moreover, these results prompted us to investigate, in the context of HIV-1 infection, the impact of CTIP2 on p21 gene expression and the resulted cell cycle status.
CTIP2 impairs Vpr-mediated stimulation of p21 gene transcription and the resulting cell cycle arrest
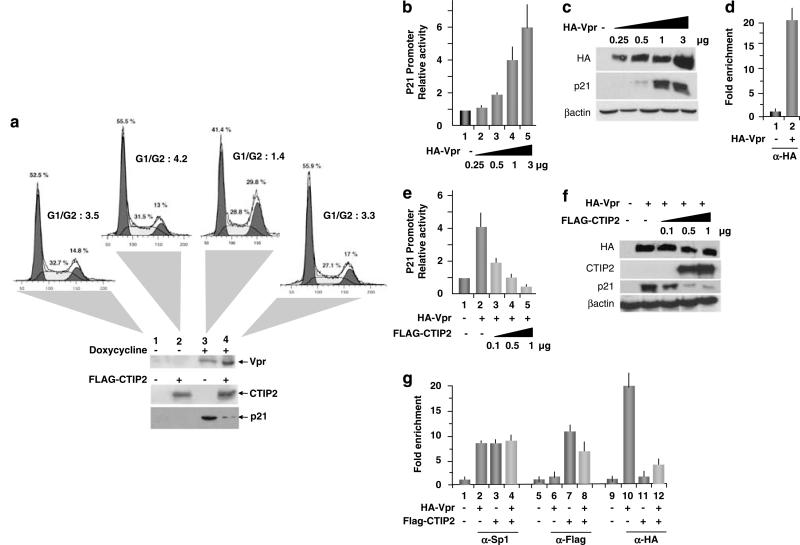
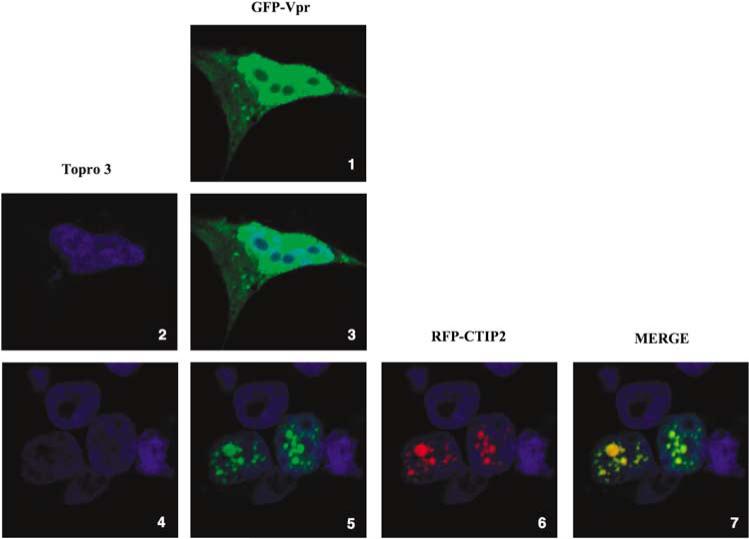
To determine whether CTIP2 impacts Vpr-induced cell cycle arrest, we examined the cell cycle status of doxycycline-inducible Vpr-expressing cells (Yoshizuka et al., 2005) with or without CTIP2 overexpression. Quantification of DNA content by flow cytometry confirmed the ability of Vpr to induce G2/M cell cycle arrest (Figure 5a, panel 3). Whereas the cell cycle status was slightly impacted by CTIP2 overexpression in the absence of Vpr (Figure 5a, panel 2), Vpr-induced G2/M block was completely abolished (Figure 5a, panel 4). Vpr, CTIP2 and p21 expression were verified by western blot experiments (Figure 5a, lower panels). Vpr-induced cell cycle arrest could be a consequence of p21 gene transcriptional activation. As shown in Figures 5b and c, Vpr expression stimulated p21 gene promoter activity and subsequent p21 protein expression. To examine whether Vpr directly affected p21 gene promoter activity in vivo, we performed ChIP experiments targeting the proximal region of the promoter. As shown in Figure 5d, Vpr was found associated with the p21 gene proximal promoter in vivo. Interestingly, overexpression of CTIP2 repressed Vpr-induced p21 gene transcription (Figure 5e, columns 3–5) and p21 protein expression (Figure 5f), as suggested by the ability of CTIP2 to impair Vpr-mediated cell cycle arrest. To examine the recruitment of CTIP2 and Vpr to the p21 promoter, we performed additional ChIP experiments. As shown in Figure 5g, as Vpr expression slightly affected CTIP2 recruitment to the p21 promoter proximal region (Figure 5g, column 8), CTIP2 over-expression impaired Vpr association with the same region (Figure 5g, column 12). As a control, expression of Vpr and CTIP2 did not significantly modify Sp1 binding to the GC-rich regions of the p21 promoter (Figure 5g, columns 2–4). To assess CTIP2-mediated relocation of Vpr in microglial cells, we observed CTIP2 and Vpr cellular localizations by confocal microscopy. As Vpr–GFP (green fluorescent protein) alone localized in the nucleus and the cytoplasm of the cells (Figure 6, panels 1 and 2), coexpression of red fluorescent protein (RFP)–CTIP2 with Vpr–GFP relocated Vpr in characteristic CTIP2-induced ball-like structures (Figure 6, panels 5–7). To visualize microglial cell nuclei, genomic DNA was stained (Figure 6, panels 2–7). As shown, CTIP2 and Vpr colocalized in the same structures (Figure 6, panel 7), suggesting that CTIP2 may displace Vpr from the p21 promoter to subnuclear structures. As a control, GFP–SUV39H1 expression alone did not affect Vpr localization (Supplementary Figure 7). Altogether, these results revealed CTIP2 as a major p21 gene transcriptional silencer and a potent anti-Vpr cellular factor.
Figure 5.
COUP-TF-interacting protein 2 (CTIP2) impairs Vpr-mediated stimulation of p21 gene transcription and the resulting cell cycle arrest. (a) 293T cells inducible for HIV-1 Vpr expression, were transfected with the Flag-CTIP2 expression vector or the control vector 24 h before being treated with doxycyclin. Cell cycle status was assessed by flow cytometry. Controls of Vpr, CTIP2 and p21 expressions are presented. Microglial cells (b and e) and 293 T cells (c and f) were transfected with the indicated amounts of expression vectors in the presence of the p21-Luc plasmid. Luciferase activities (b and e) are presented relative to the activity of the p21-LUC complemented by the control empty vector. Protein levels (c and f) were assessed by western blot using the indicated antibodies 48-h post-transfection. (d and g) 293T cells were transfected or not with the HA-Vpr and the Flag-CTIP2 expression vectors 48 h before being processed for ChIP (chromatin immunoprecipitation) assays. Immunoprecipitated DNA was subjected to real-time PCR quantification with primers targeting the endogenous p21 promoter proximal region. Specific enrichments of the p21 promoter in the immunoprecipitated material were normalized to enrichments in non-specific glyceraldehyde 3-phosphate dehydrogenase DNA and presented relative to the control.
Figure 6.
COUP-TF-interacting protein 2 (CTIP2) relocates Vpr to subnuclear structures. Microglial cells were transfected with pVpr-GFP alone (panels 1, 2 and 3) or together with pRFP-CTIP2 plasmid (panels 4–10). Cells were fixed 24-h post-transfection and confocal analysis was performed.
Discussion
In addition to its role in DNA damage response, p21 is involved in differentiation, senescence, tumor suppression and HIV-1 expression (Gartel and Tyner, 2002; Thierry et al., 2004; Vazquez et al., 2005).
Here, we show that CTIP2 is a constitutive transcriptional repressor of p21 gene transcription. The observed presence of endogenous CTIP2 on the p21 gene promoter in vivo was correlated with the undetectable level of the p21 protein under basal conditions. Moreover, the activation of p21 expression in CTIP2 knocked down cells provided further evidence for the constitutive repressive activity of CTIP2. As we have previously shown that CTIP2 is recruited to the HIV-1 promoter through Sp1 (Marban et al., 2005), we decided to examine the Sp1-binding sites of the p21 promoter involved in CTIP2-mediated transcriptional repression. The Sp1-binding site 1 (–119 bp region) seemed necessary for CTIP2-mediated repression and for its anchorage at the p21 promoter by Sp1 proteins (Supplementary Figure 3). The association of CTIP2 with HDAC1 and HDAC2 has been well documented (Marban et al., 2007) (Cismasiu et al., 2005; Topark-Ngarm et al., 2006). Thus, the surprising stimulation of the –103/+ 1 p21 promoter construct mediated by CTIP2 overexpression may be explained by an artificial titration of HDAC1 that would favor p53-mediated transactivation of p21 expression, as suggested by Lagger et al., 2003. The fact that CTIP2 knockdown did not affect the –103/+1 transcriptional activity further supports this hypothesis. We did not observe significant CTIP2-mediated modification of p53 expression, localization, phosphorylation or acetylation (Supplementary Figure 2b and data not shown). However, CTIP2 repressed p53-dependent activation of the full-length p21 gene promoter (Supplementary Figure 2a), suggesting that CTIP2 would need to be associated with the p21 promoter to inhibit p53. To further explore in molecular detail the role of CTIP2 in p21 repression, we focused on CTIP2-associated enzymatic activities. As HDAC inhibitors have been previously shown to favor p21 expression (for review see Ocker and Schneider-Stock (2007)), we limited our investigations in this field. As previously shown, we show that the inhibitor TSA induced p21 expression. Moreover, we observed that CTIP2 knockdown cooperated with TSA treatment to stimulate the p21 promoter. These results revealed that the functional cooperation of CTIP2 with TSA-sensitive HDAC (here HDAC1 and HDAC2) is not limited to the HIV-1 promoter (Marban et al., 2007) and confirmed the CTIP2-mediated recruitment of the enzymes to the targeted promoters. More interesting was the stimulation observed after Chaetocin treatment. The SUV39H1 inhibitor (Greiner et al., 2005) strongly stimulated p21 gene transcription and induced cell cycle arrest. These observations were consistent with an observed decrease in methylation of the histone H3 lysine 9 at the p21 promoter confirming an effective inhibition of endogenous SUV39H1 activity. In line with these observations, we found endogenous SUV39H1 associated with the p21 promoter in vivo and we showed its functional cooperation with CTIP2 in p21 gene transcriptional silencing. These results provide the first evidence of SUV39H1 involvement in p21 gene transcriptional silencing. In addition, they also suggest that chaetocin could be used as an anti-proliferative pharmacological tool. Recently, chaetocin has been proposed as a promising anti-myeloma agent (Isham et al., 2007). As previously suggested for HDAC inhibitors, chaetocin anti-myeloma activity has been linked to oxidative stress (Pei et al., 2004). Our results suggest that pro-apoptotic activity of chaetocin may be mediated, at least in part, by inhibition of SUV39H1 HMT activity at the p21 gene promoter. We report that SUV39H1 and CTIP2 cooperate to silence p21 gene transcription and thereby favor cell cycle progression. The previously reported anti-apoptotic activities of CTIP2 have been attributed to its involvement in the maintenance of genomic integrity (Kamimura et al., 2007) and in the modulation of intrinsic apoptotic pathways (Grabarczyk et al., 2007). Althought our results show that CTIP2 impairs p21 gene expression, the regulatory link between the potential anti-apoptotic function of CTIP2 and p21 needs further investigations.
As HIV-1 infection reduces p21 gene expression in T lymphocytes (Clark et al., 2000), infection of macrophages stimulates p21 expression and induces cell cycle arrest (Vazquez et al., 2005). Interestingly, G2 arrest favors HIV-1 expression as a result of nuclear factor κB, C-Jun and CREB-binding protein recruitment to the viral promoter (Thierry et al., 2004). However, the physiological mechanism underlying p21-mediated facilitation of HIV-1 expression needs to be further clarified in non-dividing cells. Mechanistically, HIV-1-induced p21 expression and cell cycle arrest have been attributed to HIV-1 Vpr functions. Apart from CTIP2 repression of HIV-1 replication in human primary macrophage and microglial cells, our results showed that CTIP2 abrogates Vpr-induced cell cycle arrest, suggesting that CTIP2 may also impact Vpr-mediated transcriptional activation of the p21 gene. Vpr has been described to collaborate with the cellular Sp1 transcription factor to stimulate p21 promoter in human astrocyte (Amini et al., 2004). In microglial cells, we observed a Vpr-mediated transcriptional stimulation of p21 gene expression as a result of Vpr recruitment at the endogenous p21 gene promoter in vivo. Interestingly, CTIP2 repressed Vpr-mediated transcriptional activation of p21 expression. Our ChIP assays suggest that CTIP2 antagonises Vpr binding to the p21 gene promoter and thereby counteracts Vpr function. However, as Vpr-induced G2 arrest may be explained by multiple molecular mechanisms, including association with the Cul4–DDB1 E3 ligase complex (Le Rouzic et al., 2007; Andersen et al., 2008), more investigations need to be engaged to determine all CTIP2-impacted functions of Vpr (Supplementary Figure 4).
Altogether, CTIP2 cooperates with SUV39H1 and HDACs to suppress endogenous and Vpr-induced p21 gene expression.
Materials and methods
Plasmids
Constructs used in this study have been described previously: pcDNA3, pFLAG-CTIP2, pNL4. 3, pVSV.G and pRFPCTIP2 (Marban et al., 2005), pMyc-SUV39H1, pshRNASUV39H1 (Schotta et al., 2004), pshRNA-CTIP2 (Marban et al., 2007), pHA-Vpr and the different deletion mutants of p21-LUC (Koutsodontis et al., 2002; Amini et al., 2004).
Cell culture
The human microglial cell line (Janabi et al., 1995) and HEK 293T cell line were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum and 100 U/ml penicillin–streptomycin. Primary macrophages and monocyte-derived microglia-like cells were isolated, prepared and cultured as described (Leone et al., 2006). Inducible Vpr-expressing HEK cells were described (Yoshizuka et al., 2005).
Cell cycle analysis
Cell cycle status has been determined by propidium iodide DNA staining and flow cytometry analyses. Briefly, trypsinized cells were washed and resuspended in propidium iodide staining solution for 30 min. Flow cytometric analysis was performed in a FACScalibur instrument (Becton Dickinson, San Jose, CA, USA). Cell cycle analysis was performed using CellQuest software (Becton Dickinson).
SDS–polyacrylamide gel electrophoresis and western blot analysis
Sodium dodecyl sulfate polyacrylamide gel electrophoresis was performed using standard techniques. Proteins were detected using antibodies directed against the FLAG epitope (M2 mouse monoclonal from Sigma, St Louis, MO, USA), CTIP2 (Santa Cruz, Santa Cruz, CA, USA), p21 (Santa Cruz), the HA epitope (Covance, Princeton, NJ, USA) and β-actin (Sigma). Proteins were visualized by chemiluminescence using the Super Signal Chemiluminescence Detection System (Pierce, Rockford, IL, USA).
Luciferase assays
Microglial cells cultured in 48-well plates were transfected with the indicated vectors and the Renilla control vector using the calcium phosphate co-precipitation method. Two days later, cells were collected and firefly luciferase activity was determined using the Dual-Glo Luciferase Assay System (Promega, Madison, WI, USA) and normalized to the Renilla luciferase activity. Values correspond to an average of at least three independent experiments performed in duplicates.
HIV-1 infection and viral replication
Macrophages and microglial-like cells were cultured and prepared as previously described (Leone et al., 2006). Cultured in 24-well plates, cells were transfected using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) with the complete HIV-1 infectious molecular clone pNL4-3 and the expression plasmids, as indicated. Total amounts of DNA were normalized with the corresponding empty vector. For infection experiments, cells were infected with HIV-1 Bal. HIV-1 replication was monitored as described previously (Leone et al., 2006). Values correspond to an average of at least three independent experiments carried out in duplicates.
Immunofluorescence staining and confocal microscopy observation
Microglial cells cultured in 24-well plates were transfected using Lipofectamine 2000 reagent with pRFP-CTIP2 and pVpr-GFP expression vectors. Cells were fixed and permeabilized as previously described (Rohr et al., 2003). The coverslips were then incubated for 10 min at room temperature with TOPRO3 (Molecular Probes, Eugene, OR, USA). The stained cells were analysed by confocal microscopy using a Zeiss laser scanning microscope (Zeiss, Jena, Germany; model 510 inverted) equipped with a Planapo oil (×63) immersion lens (numerical aperture = 1,4).
Chromatin immunoprecipitation assays
The HEK 293T and microglial cells cultured in 100-mm diameter dishes were transfected (or not as indicated) using the calcium phosphate co-precipitation method with the indicated vectors. ChIP assays were performed using the ChIP assay kit (Milipore Inc., Billerica, MA, USA) 48-h post-transfection. The primary antibodies used for ChIP were as follows: anti-CTIP2 (Abcam), anti-Sp1 (Milipore Inc.), anti-HA (Covance), anti-triMeK9/H3, anti-SUV39H1 (Abcam, Cambridge, UK). Treatments with chaetocin were performed at a final concentration of 200 nm for 24 h. Immunoprecipitated DNA was subjected to real-time PCR quantification and normalized to the non-specific glyceraldehyde 3-phosphate dehydrogenase promoter enrichment.
mRNA Quantification
The RNAs from HIV-1 Bal-infected cells were extracted with RNeasy Plus Mini Kit (Qiagen, Germantown, MD, USA) and retrotranscription was performed with Superscript III (Invitrogen). cDNA were quantified and normalized to the β-actin mRNA level.
Acknowledgements
We thank Dr D Kardassis for providing us with p21-LUC plasmids and Dr Yannick Goumon for his technical support. This work was supported by the Institut National de la Santéet de la Recherche Médicale (INSERM), by grants from the Agence Nationale de Recherches sur le SIDA (ANRS) to OR and CVL, from Sidaction to OR and TC and from the French Ministry of Research (‘ACI JC 5364’ to OR and doctoral grant to TC and SS) The work in CVL's laboratory was supported by grants from the ‘Fonds National de la Recherche Scientifique’ (FNRS, Belgium), the Télévie-Program of the FNRS, from the ‘Action de Recherche concertée du Ministère de la Communauté Française’ (ULB, ARC program no. 04/09-309), from the Internationale Brachet Stiftung (IBS), from the Région Wallonne-Commission Européenne FEDER (Intergenes Project, Interreg III program) and from the Theyskens-Mineur Foundation. CVL is ‘Directeur de Recherches’ of the FNRS. This work was supported by NIH to BES.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Amini S, Saunders M, Kelley K, Khalili K, Sawaya BE. Interplay between HIV-1 Vpr and Sp1 modulates p21(WAF1) gene expression in human astrocytes. J Biol Chem. 2004;279:46046–46056. doi: 10.1074/jbc.M403792200. [DOI] [PubMed] [Google Scholar]
- Andersen JL, Le Rouzic E, Planelles V. HIV-1 Vpr: mechanisms of G2 arrest and apoptosis. Exp Mol Pathol. 2008;85:2–10. doi: 10.1016/j.yexmp.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Cismasiu VB, Adamo K, Gecewicz J, Duque J, Lin Q, Avram D. BCL11B functionally associates with the NuRD complex in T lymphocytes to repress targeted promoter. Oncogene. 2005;24:6753–6764. doi: 10.1038/sj.onc.1208904. [DOI] [PubMed] [Google Scholar]
- Cismasiu VB, Ghanta S, Duque J, Albu DI, Chen HM, Kasturi R, et al. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood. 2006;108:2695–2702. doi: 10.1182/blood-2006-05-021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassen GF, Hann SR. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta -induced cell-cycle arrest. Proc Natl Acad Sci USA. 2000;97:9498–9503. doi: 10.1073/pnas.150006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E, Santiago F, Deng L, Chong S, de La Fuente C, Wang L, et al. Loss of G(1)/S checkpoint in human immunodeficiency virus type 1-infected cells is associated with a lack of cyclin-dependent kinase inhibitor p21/Waf1. J Virol. 2000;74:5040–5052. doi: 10.1128/jvi.74.11.5040-5052.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartel AL, Radhakrishnan SK. A novel transcriptional inhibitor induces apoptosis in tumor cells and exhibits antiangiogenic activity. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-05-3940. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL. The growth-regulatory role of p21 (WAF1/CIP1). Exp Cell Res. 1999;246:280–289. doi: 10.1006/excr.1998.4319. [DOI] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther. 2002;1:639–649. [PubMed] [Google Scholar]
- Gartel AL, Ye X, Goufman E, Shianov P, Hay N, Najmabadi F, et al. Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc Natl Acad Sci USA. 2001;98:4510–4515. doi: 10.1073/pnas.081074898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarczyk P, Przybylski GK, Depke M, Volker U, Bahr J, Assmus K, et al. Inhibition of BCL11B expression leads to apoptosis of malignant but not normal mature T cells. Oncogene. 2007;26:3797–3810. doi: 10.1038/sj.onc.1210152. [DOI] [PubMed] [Google Scholar]
- Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- Isham CR, Tibodeau JD, Jin W, Xu R, Timm MM, Bible KC. Chaetocin: a promising new antimyeloma agent with in vitro and in vivo activity mediated via imposition of oxidative stress. Blood. 2007;109:2579–2588. doi: 10.1182/blood-2006-07-027326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janabi N, Peudenier S, Heron B, Ng KH, Tardieu M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci Lett. 1995;195:105–108. doi: 10.1016/0304-3940(94)11792-h. [DOI] [PubMed] [Google Scholar]
- Jung P, Menssen A, Mayr D, Hermeking H. AP4 encodes a c-MYC-inducible repressor of p21. Proc Natl Acad Sci USA. 2008;105:15046–15051. doi: 10.1073/pnas.0801773105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura K, Mishima Y, Obata M, Endo T, Aoyagi Y, Kominami R. Lack of Bcl11b tumor suppressor results in vulnerability to DNA replication stress and damages. Oncogene. 2007;26:5840–5850. doi: 10.1038/sj.onc.1210388. [DOI] [PubMed] [Google Scholar]
- Koutsodontis G, Moustakas A, Kardassis D. The role of Sp1 family members, the proximal GC-rich motifs, and the upstream enhancer region in the regulation of the human cell cycle inhibitor p21WAF-1/Cip1 gene promoter. Biochemistry. 2002;41:12771–12784. doi: 10.1021/bi026141q. [DOI] [PubMed] [Google Scholar]
- Lagger G, Doetzlhofer A, Schuettengruber B, Haidweger E, Simboeck E, Tischler J, et al. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol Cell Biol. 2003;23:2669–2679. doi: 10.1128/MCB.23.8.2669-2679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic E, Belaidouni N, Estrabaud E, Morel M, Rain JC, Transy C, et al. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle. 2007;6:182–188. doi: 10.4161/cc.6.2.3732. [DOI] [PubMed] [Google Scholar]
- Leid M, Ishmael JE, Avram D, Shepherd D, Fraulob V, Dolle P. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr Patterns. 2004;4:733–739. doi: 10.1016/j.modgep.2004.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone C, Le Pavec G, Meme W, Porcheray F, Samah B, Dormont D, et al. Characterization of human monocyte-derived micro-glia-like cells. Glia. 2006;54:183–192. doi: 10.1002/glia.20372. [DOI] [PubMed] [Google Scholar]
- Marban C, Redel L, Suzanne S, Van Lint C, Lecestre D, Chasserot-Golaz S, et al. COUP-TF interacting protein 2 represses the initial phase of HIV-1 gene transcription in human microglial cells. Nucleic Acids Res. 2005;33:2318–2331. doi: 10.1093/nar/gki529. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C, et al. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 2007;26:412–423. doi: 10.1038/sj.emboj.7601516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon HS, Yang JS. Role of HIV Vpr as a regulator of apoptosis and an effector on bystander cells. Mol Cells. 2006;21:7–20. [PubMed] [Google Scholar]
- Murphy M, Ahn J, Walker KK, Hoffman WH, Evans RM, Levine AJ, et al. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu AB, 3rd, Chen X, Smeets M, Hengst L, Prives C, Reed SI. Effects of p21(Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol Cell Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocker M, Schneider-Stock R. Histone deacetylase inhibitors: signalling towards p21cip1/waf1. Int J Biochem Cell Biol. 2007;39:1367–1374. doi: 10.1016/j.biocel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Wong P, Howard BH. WAF1 retards S-phase progression primarily by inhibition of cyclin-dependent kinases. Mol Cell Biol. 1997;17:4877–4882. doi: 10.1128/mcb.17.8.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei XY, Dai Y, Grant S. Synergistic induction of oxidative injury and apoptosis in human multiple myeloma cells by the proteasome inhibitor bortezomib and histone deacetylase inhibitors. Clin Cancer Res. 2004;10:3839–3852. doi: 10.1158/1078-0432.CCR-03-0561. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan SK, Feliciano CS, Najmabadi F, Haegebarth A, Kandel ES, Tyner AL, et al. Constitutive expression of E2F-1 leads to p21-dependent cell cycle arrest in S phase of the cell cycle. Oncogene. 2004;23:4173–4176. doi: 10.1038/sj.onc.1207571. [DOI] [PubMed] [Google Scholar]
- Rohr O, Lecestre D, Chasserot-Golaz S, Marban C, Avram D, Aunis D, et al. Recruitment of Tat to heterochromatin protein HP1 via interaction with CTIP2 inhibits human immunodeficiency virus type 1 replication in microglial cells. J Virol. 2003;77:5415–5427. doi: 10.1128/JVI.77.9.5415-5427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B, Hakata Y, Landau NR. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Proc Natl Acad Sci USA. 2007;104:4130–4135. [Google Scholar]
- Sowa Y, Orita T, Minamikawa-Hiranabe S, Mizuno T, Nomura H, Sakai T. Sp3, but not Sp1, mediates the transcriptional activation of the p21/WAF1/Cip1 gene promoter by histone deacetylase inhibitor. Cancer Res. 1999;59:4266–4270. [PubMed] [Google Scholar]
- Suzuki T, Yokozaki H, Kuniyasu H, Hayashi K, Naka K, Ono S, et al. Effect of trichostatin A on cell growth and expression of cell cycle- and apoptosis-related molecules in human gastric and oral carcinoma cell lines. Int J Cancer. 2000;88:992–997. doi: 10.1002/1097-0215(20001215)88:6<992::aid-ijc24>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Thierry S, Marechal V, Rosenzwajg M, Sabbah M, Redeuilh G, Nicolas JC, et al. Cell cycle arrest in G2 induces human immunodeficiency virus type 1 transcriptional activation through histone acetylation and recruitment of CBP, NF-kappaB, and c-Jun to the long terminal repeat promoter. J Virol. 2004;78:12198–12206. doi: 10.1128/JVI.78.22.12198-12206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topark-Ngarm A, Golonzhka O, Peterson VJ, Barrett B, Jr, Martinez B, Crofoot K, et al. CTIP2 associates with the NuRD complex on the promoter of p57KIP2, a newly identified CTIP2 target gene. J Biol Chem. 2006;281:32272–32283. doi: 10.1074/jbc.M602776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez N, Greenwell-Wild T, Marinos NJ, Swaim WD, Nares S, Ott DE, et al. Human immunodeficiency virus type 1-induced macrophage gene expression includes the p21 gene, a target for viral regulation. J Virol. 2005;79:4479–4491. doi: 10.1128/JVI.79.7.4479-4491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi Y, Inoue J, Takahashi Y, Matsuki A, Kosugi-Okano H, Shinbo T, et al. Homozygous deletions and point mutations of the Rit1/Bcl11b gene in gamma-ray induced mouse thymic lymphomas. Biochem Biophys Res Commun. 2003a;301:598–603. doi: 10.1016/s0006-291x(02)03069-3. [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, et al. Bcl11b is required for differentiation and survival of alpha beta T lymphocytes. Nat Immunol. 2003b;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- Yoshizuka N, Yoshizuka-Chadani Y, Krishnan V, Zeichner SL. Human immunodeficiency virus type 1 Vpr-dependent cell cycle arrest through a mitogen-activated protein kinase signal transduction pathway. J Virol. 2005;79:11366–11381. doi: 10.1128/JVI.79.17.11366-11381.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Scadden DT, Crumpacker CS. Primitive hematopoietic cells resist HIV-1 infection via p21. J Clin Invest. 2007;117:473–481. doi: 10.1172/JCI28971. [DOI] [PMC free article] [PubMed] [Google Scholar]