Background: Ketosteroid dehydrogenases are enzymes of biotechnological relevance that introduce a double bond into steroids as a first step toward their degradation.
Results: First structures of a 3-ketosteroid-Δ4-(5α)-dehydrogenase combined with mutational analysis allowed the identification of residues essential for catalysis.
Conclusion: Tyr-319, Tyr-466, and Ser-468 have essential roles in catalysis.
Significance: These structures may facilitate the development of better catalysts for steroid conversion.
Keywords: Crystal Structure, Enzyme Kinetics, Enzyme Mechanisms, FAD, Steroid, Tuberculosis, Rhodococcus, Ketosteroid Dehydrogenase
Abstract
3-Ketosteroid Δ4-(5α)-dehydrogenases (Δ4-(5α)-KSTDs) are enzymes that introduce a double bond between the C4 and C5 atoms of 3-keto-(5α)-steroids. Here we show that the ro05698 gene from Rhodococcus jostii RHA1 codes for a flavoprotein with Δ4-(5α)-KSTD activity. The 1.6 Å resolution crystal structure of the enzyme revealed three conserved residues (Tyr-319, Tyr-466, and Ser-468) in a pocket near the isoalloxazine ring system of the FAD co-factor. Site-directed mutagenesis of these residues confirmed that they are absolutely essential for catalytic activity. A crystal structure with bound product 4-androstene-3,17-dione showed that Ser-468 is in a position in which it can serve as the base abstracting the 4β-proton from the C4 atom of the substrate. Ser-468 is assisted by Tyr-319, which possibly is involved in shuttling the proton to the solvent. Tyr-466 is at hydrogen bonding distance to the C3 oxygen atom of the substrate and can stabilize the keto-enol intermediate occurring during the reaction. Finally, the FAD N5 atom is in a position to be able to abstract the 5α-hydrogen of the substrate as a hydride ion. These features fully explain the reaction catalyzed by Δ4-(5α)-KSTDs.
Introduction
Rhodococcus is a genus of aerobic Gram-positive bacteria closely related to Mycobacterium and Corynebacterium (1). Its members show a broad catabolic diversity and a wide range of unique enzymatic capabilities, among which is the ability to degrade naturally occurring phytosterols. Intermediates of the phytosterol degradation pathway find application in the production of bioactive steroids, making Rhodococcus a particularly interesting organism for steroid bioconversion (2).
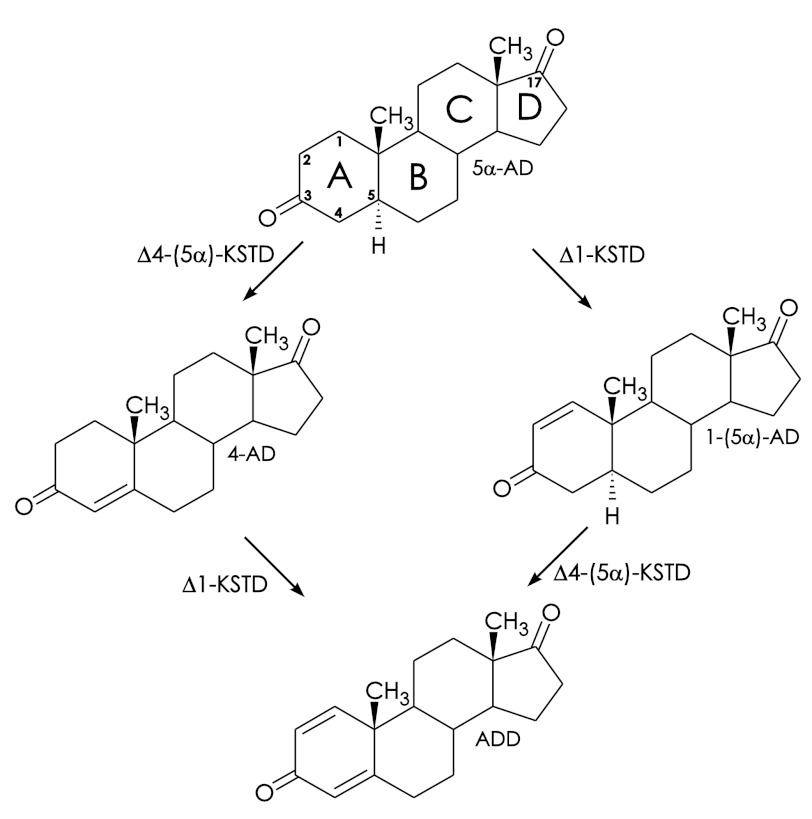
An important step in the microbial degradation of various naturally occurring sterols is the desaturation of the steroid A ring (Fig. 1). Several different 3-ketosteroid dehydrogenases (KSTDs)3 are known to promote this desaturation, such as 3-ketosteroid Δ1-dehydrogenase (Δ1-KSTD), 3-ketosteroid Δ4-(5α)-dehydrogenase (Δ4-(5α)-KSTD), and 3-ketosteroid Δ4-(5β)-dehydrogenase (Δ4-(5β)-KSTD), which are all FAD-containing enzymes. Although Δ1-KSTDs desaturate the C1-C2 position, Δ4-KSTDs introduce a double bond at the C4-C5 position (3, 4).
FIGURE 1.
Conversion of 5α-AD to ADD by Δ1-KSTD (EC 1.3.99.4) and Δ4-KSTD (EC 1.3.99.5). The two double bonds introduced by the enzymes are indicated. The intermediate products 1-(5α)-AD and 4-AD are 1-(5α)-androstene-3,17-dione and 4-androstene-3,17-dione, respectively.
Amino acid sequence analysis of (putative) KSTDs showed that Δ1-KSTDs and Δ4-KSTDs are clearly distinct, belonging to different branches of the phylogenetic tree. They display 20–24% sequence similarity to fumarate reductases (5). The highest homology is found in the FAD-binding domain; the substrate-binding domain shows lower sequence similarity. The residues involved in the dehydrogenation of the substrate by KSTDs are currently unknown, although chemical modification and mutagenesis studies on Δ1-KSTDs have implicated histidine and tyrosine residues in catalysis (6, 7). Mutational studies confirming the involvement of these residues in Δ4-(5α)-KSTDs have not been reported so far.
KSTDs also have an important function in Mycobacterium tuberculosis where they are part of the metabolic pathway for cholesterol degradation. In M. tuberculosis this pathway is central to the unusual ability of the bacterium to survive inside macrophages (8). Interestingly, transposon mutagenesis has suggested that the Δ4-KSTD homologue Rv1817 is involved in the pathogenicity of M. tuberculosis (9), and it has been proposed that KSTDs could be valuable targets for the development of antituberculosis drugs (8).
In the Rhodococcus jostii RHA1 genome (1), a single Δ4kstD gene (ro05698) is present. It codes for a protein of 490 amino acid residues, with an estimated molecular mass of 51.9 kDa. The gene has been cloned, and the enzyme has been overexpressed as a His6-tagged protein in Escherichia coli and purified (10). Spectroscopic analysis confirmed that it binds FAD, and the purified enzyme was shown to have 3-ketosteroid Δ4-(5α)-dehydrogenase activity. The enzyme has been crystallized, and its preliminary x-ray analysis has been published (10).
Here we present the crystal structure of Δ4-(5α)-KSTD (Ro05698) from R. jostii RHA1 in its steroid-free and product-bound form. These first structures of a KSTD enzyme allowed the identification of the active site residues involved in the dehydrogenation of 3-ketosteroid substrates and its reaction mechanism, which was confirmed through mutational analyses.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Rhodococcus strains were cultivated in LBP medium containing 1% (w/v) bacto-peptone (Difco, Detroit, MI), 0.5% (w/v) yeast extract (BD Biosciences) and 1% (w/v) NaCl at 30 °C and 200 rpm. E. coli strains were grown in Luria-Bertani broth (Sigma) at 37 °C unless stated otherwise.
KSTD Activity Staining
Initial Δ4-(5α)-KSTD activity was established by a native gel-based assay. Cell-free extracts of three Rhodococcus strains (Rhodococcus erythropolis SQ1, Rhodococcus rhodochrous, and R. jostii RHA1) were prepared as described (5). These were loaded on native PAGE gels and stained for KSTD activity following published procedures (5).
Protein Expression, Purification, and Characterization
Δ4-(5α)-KSTD was heterologously expressed in E. coli strain BL21(DE3) and subsequently purified and crystallized as described before (10). UV-visible spectra of purified enzyme measured in the range of 200 to 700 nm (Cary 100, Varian) with and without substrate were used to characterize the protein as a flavoprotein and to detect the reduction of the FAD upon addition of substrate.
Noncovalent binding of the flavin co-factor was determined as follows. Purified Δ4-(5α)-KSTD was applied to a SDS-PAGE gel. Following electrophoresis, the gel was treated with 5% acetic acid solution, and flavin was visualized by 254-nm UV irradiation. Lack of co-migration indicated noncovalent binding of the flavin co-factor to the Δ4-(5α)-KSTD protein.
Activity Assays and Product Identification
Because E. coli does not code for any KSTD enzymes, cell-free extracts were used for product formation assays as described before (5). Briefly, enzyme activities were determined spectrophotometrically at 30 °C in 50 mm Tris-HCl buffer, pH 7.4, using 80 μm dichlorophenolindophenol (DCPIP) as an artificial electron acceptor and 0–200 μm 1-(5α)-androstene-3,17-dione (1-(5α)-AD) as substrate (Steraloids). The initial reaction rates were determined from the absorbance change at 600 nm during the first 20 s of the reaction. The products were identified by HPLC following a reaction containing purified protein (50–200 μg), 200 μm 1-(5α)-AD dissolved in ethanol, and 200 μm DCPIP. No activity was detected in control reaction mixtures lacking steroids or purified enzyme or in extracts of E. coli with an empty pET15b expression vector.
Crystallization, Data Collection, and Processing
Δ4-(5α)-KSTD was crystallized at 293 K as described before, with a precipitant consisting of 200 mm NH4-acetate, 100 mm sodium citrate, pH 5.6, and 30% (w/v) PEG 4000 (10). Crystals were prepared for data collection by soaking them for ∼30 s in mother liquor, supplemented with 20% (w/v) glycerol, followed by cryo-cooling in liquid nitrogen. The 4-AD soaks were prepared by soaking crystals for ∼18 h in cryo-mother liquor, which had the 200 mm NH4-acetate replaced by 200 mm NH4Cl, and to which 10 μl of a saturated solution of 4-AD in ethanol had been added per 500 μl of cryo-mother liquor.
The diffraction data were collected at 100 K at the beamlines of the European Synchrotron Radiation Facility (Grenoble, France). The intensity data were processed using the programs MOSFLM (11) and SCALA (12) from the CCP4 package (13). A summary of the data collection statistics is shown in Table 1.
TABLE 1.
Data collection statistics
The values within parentheses are for the highest resolution shell, and the formulas for Rmerge and Rp.i.m. were taken from Weiss (52).
| Steroid-freea | 4-AD | |
|---|---|---|
| European Synchrotron Radiation Facility beamline | BM16 | ID14-1 |
| Wavelength (Å) | 1.00 | 0.93 |
| Space group | C2221 | C2221 |
| Unit cell parameters | ||
| a (Å) | 99.2 | 99.8 |
| b (Å) | 114.3 | 116.1 |
| c (Å) | 110.2 | 110.2 |
| α (°) | 90 | 90 |
| β (°) | 90 | 90 |
| γ (°) | 90 | 90 |
| Resolution (Å) | 1.6 (1.69–1.60) | 1.6 (1.69–1.60) |
| Rmergeb | 0.081 (0.319) | 0.110 (0.373) |
| Rp.i.mc | 0.023 (0.138) | 0.055 (0.201) |
| Total number of observations | 888,741 (80,282) | 390,566 (38,365) |
| Total number of unique reflections | 80,282 (9,999) | 80,072 (8,955) |
| Mean I/σ(I) | 22.6 (4.7) | 10.5 (3.2) |
| Completeness (%) | 97.0 (83.8) | 95.2 (74.1) |
| Multiplicity | 11.1 (6.1) | 4.9 (4.3) |
a Data collection statistics according to van Oosterwijk et al. (10).
b Rmerge = ΣhklΣi|Ii(hkl) − (I(hkl)|/ΣhklΣiIi(hkl).
c Rp.i.m. = Σhkl(1/(N − 1))½Σi|Ii(hkl) − (I(hkl)|/ΣhklΣiIi(hkl).
Phasing
The FFAS03 server (14) was used for identifying suitable models for molecular replacement. The structure with the highest sequence identity (24%) was that of flavocytochrome c3 fumarate reductase from Shewanella frigidimarina in the open conformation (Protein Data Bank entry 1qo8) (15). Because of the low sequence identity, the molecular replacement model was enhanced by incorporating all-serine structural information from various homologous proteins (17–24% identity; Protein Data Bank entries 1d4c (16), 1e39 (17), 1kf6 (18), 1nek (19), and 1zoy (20)). This ensemble of all-serine models was used to solve the structure of Δ4-(5α)-KSTD by molecular replacement at 2.5 Å resolution using the program Phaser (21). Several model building cycles, consisting of manual model building using COOT (22) and density modification with Resolve (23), were performed to improve the model obtained from Phaser until an R factor of 0.44%. Because of low sequence similarity and variable positions of residues 291–426 (substrate-binding domain) in the homologous structures, no interpretable density was observed for this region. However, manual model building and density modification improved the phases sufficiently to allow Arp/Warp (24) to build the complete model using data to 1.6 Å resolution.
Refinement
The model obtained from Arp/wArp was used as the starting model for the crystallographic refinement. First, manual model building using COOT (25) was performed to adjust minor discrepancies in the model. Then the model was refined with Refmac using TLS refinement (26) to a final R factor of 15.8% and an Rfree of 17.5% at 1.6 Å resolution. Most residues were visible in the electron density except for the first 25 residues (consisting of the His6 tag and 19 residues of the His6 tag linker) and the last residue. Two differences were found, A60T and T160A, compared with the published genome sequence. The final model contains FAD, one glycerol molecule, two acetate molecules, and 619 water molecules. Also 14 amino acids with double conformations were identified. In addition, the structure contains a chloride ion, of which the location was confirmed using a bromide MAD data set. The secondary structure was assigned using DSSP (27).
The product-bound structure was solved by rigid body fitting of the steroid-free structure followed by restrained refinement with REFMAC5. The geometric restraints for the FAD and 4-AD models were generated using the PRODRG2 server (28). These ligands were manually built into 2Fo − Fc and Fo − Fc electron density maps using COOT. The 4-AD model was refined to an R factor of 15.9% and an Rfree value of 18.1% at 1.6 Å resolution. A summary of the refinement statistics and the geometric quality of the models is given in Table 2. The quality of the models was analyzed using the program MolProbity (29). The figures were prepared using PyMOL (30). The atomic coordinates and experimental structure factor amplitudes have been deposited with the RCSB Protein Data Bank with Protein Data Bank codes 4AT0 (steroid-free) and 4AT2 (4-AD).
TABLE 2.
Refinement statistics
| Steroid-free | 4-AD | |
|---|---|---|
| Resolution (Å) | 1.6 | 1.6 |
| Average B-factor | 16.9 | 14.1 |
| Ra | 15.8 | 15.9 |
| Rfreea | 17.5 | 18.1 |
| RMSD from target geometry | ||
| Bond lengths (Å) | 0.012 | 0.007 |
| Bond angles (°) | 1.44 | 1.25 |
| Total number of atoms | 4368 | 4353 |
| Number of amino acids | 482 | 482 |
| Number of Cl− ions | 1 | 1 |
| Number of acetate molecules | 2 | |
| Number of glycerol molecules | 1 | |
| Active site ligand | 4-AD | |
| Number of FAD molecules | 1 | 1 |
| Number of water molecules | 609 | 618 |
| Ramachandran angles (%)b | ||
| Favored | 95.4 | 95.5 |
| Allowed | 4.1 | 4.1 |
| Disallowed | 0.4 | 0.4 |
a R = Σhkl‖Fobs| − |Fcalc‖/Σhkl|Fobs|. Rfree is the R factor calculated with 5% of the reflections excluded from the refinement.
b Ramachandran statistics were obtained from COOT (22).
Site-directed Mutagenesis
Site-directed mutagenesis was performed using the QuikChangeTM site-directed mutagenesis kit (Stratagene) with plasmid pBluescript-ro05698 as template. The PCR product of ro05698 (10) was cloned into the EcoRV site of pBluescript (II) KS (Fermentas). Primers used for site-directed mutagenesis are listed in supplemental Table S1. Successful mutagenesis was confirmed by DNA sequencing. The resulting mutant genes were subcloned following NdeI/BamHI digestion into the NdeI/BamHI-digested pET15b (Novagen) for protein expression and protein purification. Mutation S468A in Ro05698 was obtained by ordering a synthetic DNA fragment (487 bp, AGOWA) of ro05698 containing the desired mutation. The synthetic fragment was digested with NotI/BamHI and subcloned into the NotI/BamHI digested pET15b-Ro05698 construct to obtain the full-length gene with the desired mutation.
RESULTS AND DISCUSSION
Ro05698 from R. jostii RHA1 Is a Δ4-(5α)-KSTD Enzyme
The genome of R. jostii RHA1 contains at least three probable Δ1-KSTD enzymes but only one single putative Δ4-(5α)-KSTD enzyme, encoded by ro05698, based on its 31% sequence identity to the Δ4-(5α)-KSTD of R. erythropolis (1, 5). Indeed, cell-free extracts of R. jostii RHA1 and two other Rhodococcus strains showed Δ4-(5α)-KSTD activity in a native PAGE assay with 1-(5α)-AD as substrate (supplemental Fig. S1). In R. jostii RHA1, this activity presumably originates from the product of the ro05698 gene.
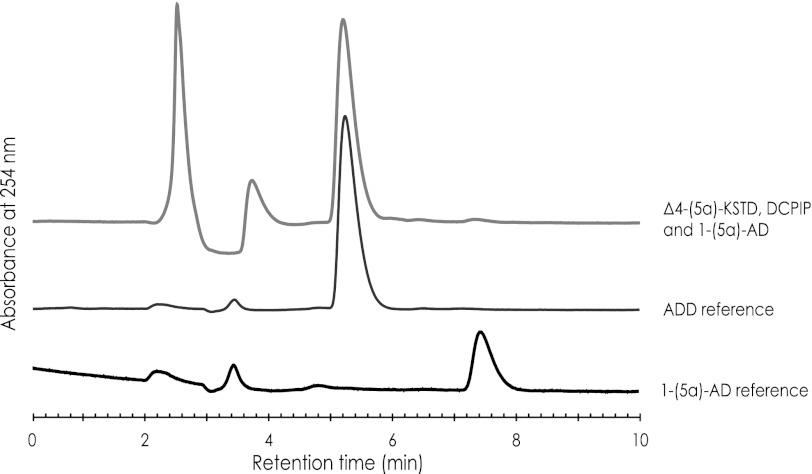
To firmly establish that ro05698 codes for an enzyme with Δ4-(5α)-KSTD activity, the gene was expressed in E. coli, and the Ro05698 protein was purified and incubated with the steroid substrate 1-(5α)-AD. HPLC analysis of the product confirmed the formation of 1,4-androstadiene-3,17-dione (ADD) (Fig. 2), indicating that ro05698 indeed codes for a Δ4-(5α)-KSTD. As expected, no activity was observed with the product 4-androstene-3,17-dione (4-AD) as substrate.
FIGURE 2.
HPLC analysis of substrates/products in a reaction mixture containing purified Δ4-(5α)-KSTD (50–200 μg), the electron acceptor DCPIP, and 1–5α-AD as substrates. The conversion of 1–5α-AD into ADD is ∼100%. Pure 1–5α-AD and ADD served as references for substrate/product identification.
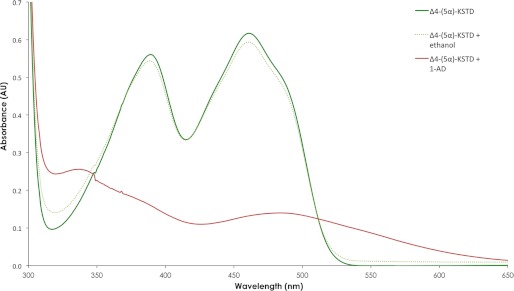
The absorption spectrum of purified Δ4-(5α)-KSTD protein is typical of that of a flavoprotein (31), with maxima at 461 and 389 nm (Fig. 3). The addition of 1-(5α)-AD led to the rapid disappearance of the maxima at ∼389 and 461 nm, indicating that the flavin co-factor is reduced along with the dehydrogenation of the steroid substrate. However, also a broad absorbance peak near 490 nm develops, which may indicate the formation of a charge-transfer complex of the electron-rich reduced FAD and the electron-poor oxidized substrate (32). This could suggest that the product does not dissociate from the reduced enzyme before the oxidizing substrate enters the active site.
FIGURE 3.
Absorption spectrum of purified Δ4-(5α)-KSTD from Rhodococcus jostii RHA1. The red line shows the absorption spectrum of Δ4-KSTD with substrate 1-(5α)-androstene-3,17-dione dissolved in ethanol (200 μm). The green line shows the absorption spectrum of Δ4-KSTD without steroid. The dotted green line shows the absorption spectrum of the Δ4-KSTD with ethanol as control. A Δ4-(5α)-KSTD concentration of 5.8 mg/ml in phosphate buffer with 10% glycerol, pH 7.2, was used.
A SDS-PAGE gel electrophoresis assay showed that the flavin co-factor did not co-migrate with the protein, suggesting that it is noncovalently bound to the Ro05698 protein. This was confirmed by the crystal structure of Δ4-(5α)-KSTD (see below).
The Three-dimensional Structure of Δ4-(5α)-KSTD
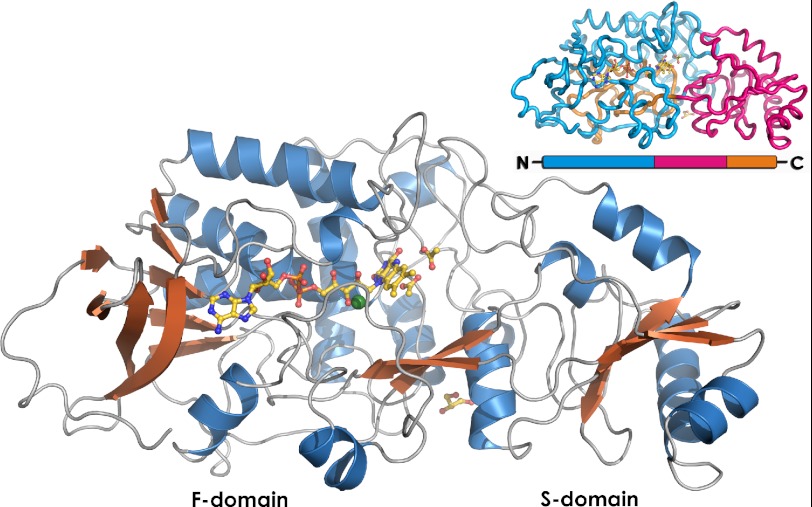
The crystal structure of Δ4-(5α)-KSTD was elucidated by molecular replacement using an ensemble of six structures with 17–24% sequence identity to Δ4-(5α)-KSTD as the starting model, and the resulting solution was refined to an R factor of 15.8% at 1.6 Å resolution (for details see Table 2). The crystals contain one Δ4-(5α)-KSTD molecule per asymmetric unit. Δ4-(5α)-KSTD has an ellipsoid shape with dimensions of 80 × 40 × 35 Å (Fig. 4). The overall fold resembles that of p-hydroxybenzoate hydroxylase (33), a fold often observed in flavoenzymes (34). The protein consists of two domains connected via a two-stranded antiparallel β-sheet (Fig. 4). One domain (F) contains a noncovalently bound FAD co-factor; it consists of a central five-stranded parallel β-sheet flanked on one side by four additional β-strands and on the other side by three α-helices. The other domain, or substrate-binding domain (S, residues 292–425) is an insert into domain F and divides it into two parts, F1 (residues 7–291) and F2 (residues 426–489). It consists of a four-stranded antiparallel β-sheet flanked on both sides by two α-helices. The active site is located at the interface between the F and S domains next to the isoalloxazine ring system of the FAD co-factor.
FIGURE 4.
Overview of the Δ4-(5α)-KSTD structure. FAD, glycerol, and the two acetate molecules bound in the active site are shown in yellow. The Cl− ion is shown in green. The inset shows the domain organization of Δ4-(5α)-KSTD. Domain F is shown in blue (F1) and orange (F2), and the S domain is in pink. Domain S is an insert into domain F, and the active site is located on the interface between the two domains.
The Δ4-(5α)-KSTD structure is well defined, with the exception of the His6 tag, the 19 residues of the linker, residues 1–6, and the C-terminal residue 490. In addition, two loops near the active site (residues 170–182 and 256–268; supplemental Fig. S3) are poorly visible in the electron density map, which may indicate flexibility. These loops may have a function in catalysis by shielding the FAD and substrate from the solvent (see below).
Comparison with Structurally Similar Proteins
A DALI (Distance-matrix ALIgnment) search (35) revealed that Δ4-(5α)-KSTD is structurally most closely related to cytochrome c3 fumarate reductase from S. frigidimarina (Z = 42.6, 27% sequence identity), cytochrome c fumarate reductase from Shewanella putrefaciens MR-1 (Z = 42.2, 28% identical residues), quinol-fumarate reductase from E. coli (Z = 32.6, 22% identical residues), and l-aspartate oxidase from E. coli (Z = 32.5, 21% sequence identity), which are all FAD-binding proteins with a p-hydroxybenzoate hydroxylase-like fold. Δ4-(5α)-KSTD has less, but still significant structural similarity to cholesterol oxidase from Brevibacterium sterolicum (1coy, Z = 16.2, 12% sequence identity), an enzyme that catalyzes a similar reaction on a similar steroid substrate as Δ4-(5α)-KSTD.
The highest structural similarity is found for the F domain with root mean square differences (RMSDs) on Cα atoms of 1.6–2.0 Å. The S-domain is less similar in structure, and only the fumarate reductases of S. frigidimarina and S. putrefaciens, two highly homologous proteins (59% sequence identity to each other), have a similar S domain with RMSDs of 1.9–2.0 Å (116 residues of 135). Nevertheless, although the overall fold of all these proteins is highly similar and all of them display a similar FAD binding mode, their catalytic centers show a huge variation, consistent with the large variation of their substrates, ranging from small compounds (e.g., fumarate) to sugars and steroids.
FAD Binding
The FAD molecule binds noncovalently in a long cleft in the F-domain (supplemental Fig. S2). Its ADP part is bound by a motif resembling the βαβαβ dinucleotide binding motif (Rossmann fold), frequently observed in flavoproteins (36). The adenine moiety is bound by two hydrogen bonds to the backbone amide and carbonyl group of Val-205. The two hydroxyl groups of the adenosine ribose are hydrogen-bonded to Glu-51. The two phosphate groups have hydrogen bonding interactions with the backbone amides of Ile-31, Ala-32, Ala-59, Thr-60, and Arg-456 and with the hydroxyl group of Thr-60. In addition there is a salt bridge between one of the phosphates and His-268. The d-ribitol binds in an extended conformation and is hydrogen-bonded to several water molecules and to the side chains of Ser-471 and Thr-60.
The isoalloxazine ring system is located close to the interface of the F and S domains. Its N5 nitrogen atom is hydrogen-bonded to the backbone amide of Gly-64, and the main chain NHs of Ser-471 and Leu-472 have hydrogen bonding interactions with the O2 oxygen of the pyrimidine ring. Ser-471 and Leu-472 are at the N-terminal end of the ∼25 Å long C-terminal α-helix formed by residues 470–489, and the dipole moment provided by this helix may stabilize the negative charge on the O2 oxygen atom in the anionic hydroquinone form of the FAD (37). Finally, the side chain hydroxyl group of Ser-471 points away from the N1 nitrogen of the flavin, but in a different rotamer conformation it would be at hydrogen bonding distance to the N1 nitrogen and could be involved in protonating it.
The isoalloxazine ring system is not planar; the butterfly angle between the pyrimidine and dimethylbenzene rings is ∼168°, similar to that observed in cholesterol oxidase (38). The color of the crystals and the spectrum of the protein sample used for crystallization indicate that the oxidized form of FAD is present in the structure.
The Active Site
In the steroid-free Δ4-(5α)-KSTD structure, the active site is open to the solvent and contains a well ordered network of water molecules and two acetate ions. A chloride ion is located ∼3.8 Å behind the dimethylbenzene ring of the isoalloxazine ring system (Fig. 5 and supplemental Fig. S2). It is bound in a partially hydrophobic environment (Phe-427 and the dimethylbenzene ring), and it is held in place by the backbone nitrogen of Ala-264. A negative charge close to the dimethylbenzene ring of the FAD may push electrons toward the pyrimidine ring and thereby lower the redox potential (39).
FIGURE 5.
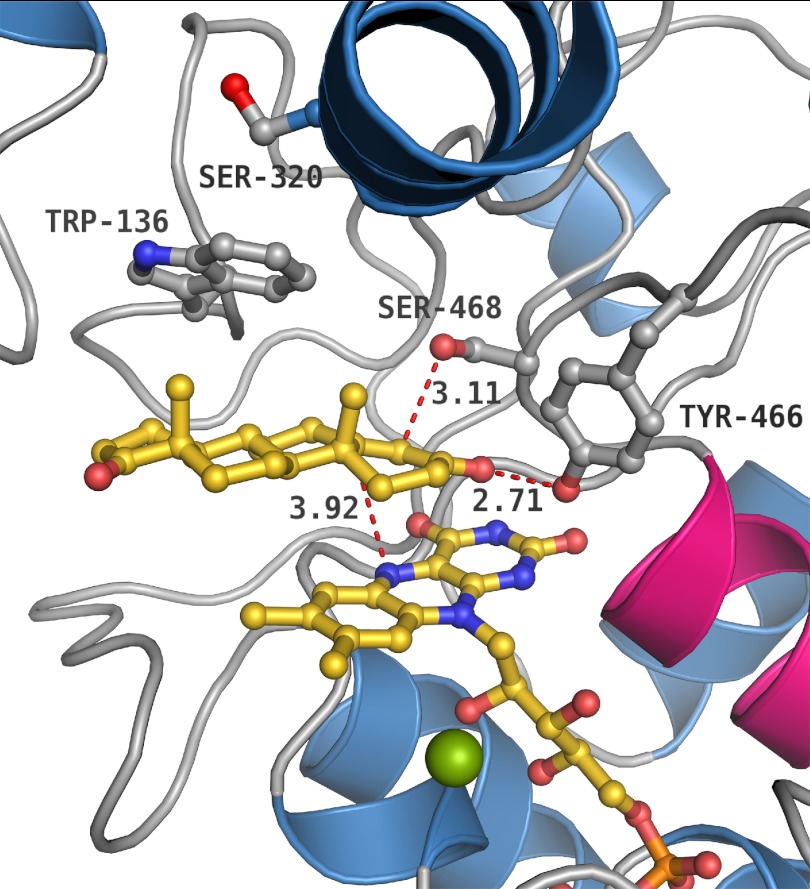
Product (4-AD) binding in Δ4-(5α)-KSTD. The most important active site residues are displayed. Trp-136 and Ser-320 display double conformations in the acetate bound structure but are fixed in the 4-AD bound structure. Trp-136 is stacking with the C and D ring of 4-AD.
Product Binding
The 1.6 Å resolution structure of Δ4-(5α)-KSTD with bound 4-androstene-3,17-dione (4-AD; Table 2) shows that 4-AD binds at the si face of the isoalloxazine ring system, displacing three water molecules and the two acetate molecules (Fig. 5). Binding of 4-AD does not cause significant changes in the overall protein structure; the all-atom RMSD between the structures is only 0.31 Å. The side chains of Trp-136 and Ser-320, which display a correlated double conformation in the steroid-free structure, become ordered upon binding of the steroid molecule. Trp-136 has a hydrophobic stacking interaction with the steroid, but it is conserved in only a few putative Δ4-KSTDs (5). A W136F substitution caused only a 3-fold reduction in apparent activity, but a W136A mutation resulted in inactive enzyme (Table 3), indicating that an aromatic residue capable of stacking is required at this position. Concomitantly with the rigidification of the Trp-136 side chain, also the Ser-320 side chain becomes ordered.
TABLE 3.
Apparent kinetic parameters of the R. jostii Ro05698 Δ4-(5α)-KSTD wild type and active site mutants using 1-(5α)-AD as substrate
| Vmax | Km | kcat | kcat/Km | |
|---|---|---|---|---|
| nmol·mg−1·min−1 | μm | s−1 | s−1·m−1 | |
| Wild typea | 27 × 102 ± 1.5 × 102 | 10 ± 2 | 2.4 | 2.4 × 106 |
| W136A | NDb | ND | ND | ND |
| W136F | 8 × 102 ± 3 × 102 | 66 ± 25 | 0.7 | 1 × 104 |
| Y319F | ND | ND | ND | ND |
| Y466A | ND | ND | ND | ND |
| Y466F | ND | ND | ND | ND |
| S468T | 19 × 102 ± 3.6 × 102 | 17 × 101± 3.3 × 101 | 1.6 | 9 × 103 |
| S468A | ND | ND | ND | ND |
a Contains mutations A60T (nucleotide G178A) and T160A (nucleotide A478G), but these mutations do not or hardly have an effect on activity.
b ND, not detectable.
4-AD binds with its C3 keto group at the same location as one of the acetate oxygen atoms in the steroid-free structure, forming a hydrogen bond to Tyr-466 (Fig. 5). Acetate is more often observed to mimic the binding of an oxygen atom of the substrate (40). The C4 carbon atom of 4-AD is near the Ser-468 hydroxyl group. Surprisingly, the C17 keto moiety of 4-AD has no interactions with the protein, but it is hydrogen-bonded to two water molecules. Apparently, the substituent at this position is not a very critical factor in substrate recognition, although preliminary results show that an aliphatic tail at C17, as in 5-α-cholestan-3-one, is not accepted at this position.4
Tyr-466 Is the Putative Catalytic Acid
The position of Tyr-466 and its hydrogen bonding interaction with the C3 keto group of 4-AD suggest that this residue may function as the catalytic acid that protonates the C3 keto group, promoting keto-enol tautomerization and labilization of the C4 hydrogen atoms. Formation of the enolate at the C3 keto group has previously been suggested as the initial catalytic step for these proteins (41, 42). Tyr-466 is conserved in all Δ4 and Δ1 KSTD enzymes. Site-directed mutagenesis (Y466A and Y466F) resulted in catalytically dead Δ4-(5α)-KSTD enzymes (Table 3), confirming the importance of Tyr-466 and showing that the hydroxyl group is critical for the reaction. Intriguingly, Tyr-466 is the only residue with a direct hydrogen bonding interaction with the product in the crystal structure and thus may also be a determinant for substrate recognition.
Ser-468 Is the Putative Catalytic Base
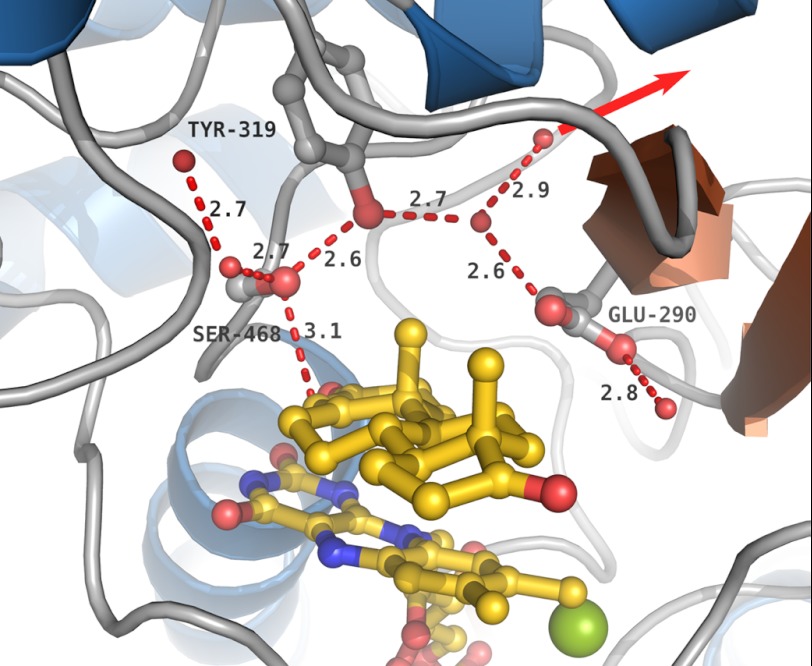
The Ser-468 hydroxyl group is at 3.1 Å from the C4 atom of 4-AD, a position compatible with a role of Ser-468 as the catalytic base, which abstracts a proton from C4. The C4 protons have been labilized because of keto-enol tautomerization of the keto group of the substrate upon interaction with the Tyr-466 hydroxyl group. The Ser-468 hydroxyl group is part of an extensive hydrogen bonding system, which can function as an effective system relaying protons in a concerted way to the solvent and which includes the Tyr-319 hydroxyl group, a water molecule, and the side chain of Glu-290 (Fig. 6). Because such concerted proton transfers do not involve high energy alkoxide intermediates (43), the proton can easily be transferred from the C4 atom to the solvent.
FIGURE 6.
The hydrogen bonding environment around Ser-468. The side chains of Ser-468, Tyr-319, and Glu-290 are shown. Dashed red lines indicate the hydrogen bonds and the contact between Ser-468 and the 4-AD C4 carbon. Distances are given in Å. The FAD and 4-AD are shown in yellow, and the chloride ion is in green. A red arrow marks the proposed proton relay pathway from the 4-AD C4 carbon to the solvent.
Ser-468 is conserved in Δ4-KSTD enzymes, including Rv1817 from M. tuberculosis H37Rv, but not in Δ1-KSTDs. Its hydroxyl group is essential for activity, because a S468A mutation abolished all catalytic activity, whereas a S468T mutant still retained activity. However, the apparent Km of this latter mutant protein was drastically increased, and the apparent Vmax was somewhat lower than that of wild-type enzyme (Table 3).
A Y319F mutant enzyme had also lost all activity on 1-(5α)-AD as substrate, showing the functional importance of the hydroxyl group of this tyrosine for activity (Table 3). In contrast, a E290Q mutation did not affect the catalytic activity of the enzyme, indicating that this residue is not essential for proton relay and that other relay routes may exist.
The Role of the FAD Co-factor in Catalysis
Flavoproteins involved in dehydrogenation reactions often display a few recurrent features, like the distance and angle between the FAD N5 nitrogen atom and the site of oxidative attack and the presence of a hydrogen bond donor close to the N5 nitrogen (44). In Δ4-(5α)-KSTD the N5 nitrogen is hydrogen-bonded to the backbone amide of Gly-64. It is at a distance of 3.9 Å from the C5 atom of the product, and the N10-N5-C5 angle is ∼108°. Both the distance and angle are in agreement with the values observed in other proteins (44). These values clearly suggest that the site of attack by the FAD is the C5 atom of the substrate. The position of the FAD N5 atom above and of Ser-468 below the 4-AD is in agreement with trans-dehydrogenation of the substrate (42).
In many flavoproteins, catalysis takes place at a location shielded from the solvent, which enhances the strength of polar interactions and is instrumental to substrate activation (34, 44). In some enzymes with the p-hydroxybenzoate hydroxylase fold loops move in upon substrate binding to cover the active site, and in others the complete S domain may relocate (15, 45). In Δ4-(5α)-KSTD two loops close to the active site (supplemental Fig. S3) show somewhat less defined electron density and may be flexible. However, in the 4AD-bound structure, no significant conformational differences are seen between the steroid-free and product-bound states. At present we cannot exclude that crystal contacts prevent large conformational changes or that the loops move to restrict solvent access to the FAD. Binding of the steroid substrate may also already provide sufficient shielding. Further research will be needed to resolve this question.
Comparison with Cholesterol Oxidase
Δ4-(5α)-KSTD is functionally and structurally related to cholesterol oxidase (45). The structures of the two proteins can be superimposed with an RMSD of 2.6 Å for 213 of 483 Cα atom positions with the highest similarity in the F-domain. Although Δ4-(5α)-KSTD catalyzes the dehydrogenation of the C4-C5 bond of steroids, cholesterol oxidase catalyzes the oxidation of steroids containing a 3β-hydroxyl group with concomitant isomerization of the Δ5 double bond to the Δ4 position. However, whereas in Δ4-(5α)-KSTD the substrate/product binds at the si face of the isoalloxazine ring system, in cholesterol oxidase the substrate binds in a pocket in front of the N5 nitrogen. This different substrate-binding mode positions the substrate such that in Δ4-(5α)-KSTD the C5 hydrogen atom is near the FAD N5 nitrogen, but in cholesterol oxidase it is the C3 hydroxyl group that is near the N5 atom of the FAD. This explains the different reaction specificity and regioselectivity of the two enzymes.
Cholesterol oxidase also contains an extra domain, which binds the apolar aliphatic tail connected to the C17 atom of the cholesterol. In the 4AD-bound structure of Δ4-(5α)-KSTD, the C17 keto group of the substrate/product is exposed to the solvent, in agreement with the wider range of substituents at this position that are accepted by Δ4-(5α)-KSTD.4
The Catalytic Mechanism
The oxidation of 5α-AD or 1-(5α)-AD requires the abstraction of two hydrogen atoms and the transfer of two electrons as a hydride ion to the FAD. It is not known whether these proton and hydride transfers occur in a concerted or stepwise manner. The two electrons will finally be transferred to a currently unknown electron acceptor, for example menaquinone (46).
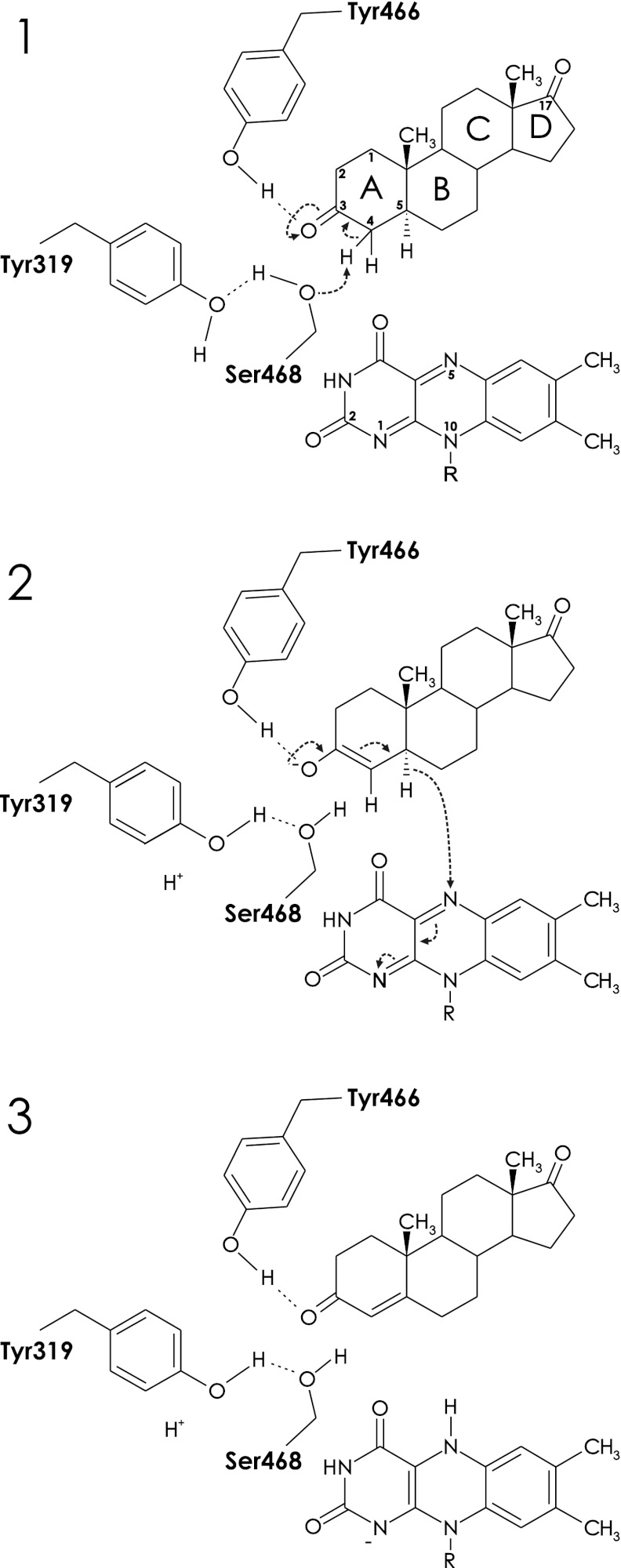
The crystal structure indicates Ser-468 as the putative base that abstracts the C4 α-hydrogen proton from the substrate and relays its own proton to the solvent via Tyr-139 (Fig. 7). The resulting deprotonated state of the substrate can be stabilized by the delocalization of the negative charge over the C3 keto group. In addition, Tyr-466, functioning as an acid, can stabilize the ensuing enolate by hydrogen bonding to the C3 oxygen atom. The FAD N5 atom is in a good position to abstract a hydride ion from the C5 atom of the enolate intermediate. In synchrony, the lone pair electrons of the negatively charged C3 oxygen atom move back toward C3 and the double bond between C3 and C4 shifts to the C4-C5 position, generating the product. The negative charge on the N5 of FAD can be delocalized over the N1-C = O2 region, but also the rest of the isoalloxazine ring may contribute (47). Two backbone amides stabilize the negative charge on O2 by hydrogen bonding, and the dipole moment of the C-terminal helix is also directed toward O2. The generated reducing equivalents may be donated to a respiratory chain, similar to what has been proposed for the Δ1-KSTD of Arthrobacter globiformis (48), but further studies are required to confirm this.
FIGURE 7.
Proposed reaction mechanism of Δ4-(5α)-KSTD. Panel 1, catalysis is initiated by the interaction of the O3 keto group of the substrate with the hydroxyl group of Tyr-466, which promotes keto-enol tautomerization and labilization of the C4 hydrogen atoms. Ser-468, acting as a base, abstracts a proton from the C4 atom of the substrate, with Tyr-319 serving as a proton relay system to the solvent. Panel 2, next, the FAD abstracts a hydride ion from the C5 carbon of the substrate, which, with a concomitant rearrangement, results in the formation of a double bond between C4 and C5. Panel 3, the product is formed and leaves the active site, perhaps only after the oxidizing substrate enters the active site. The negative charge on the reduced FAD is stabilized by the dipole moment of the C-terminal helix. Finally, the oxidized FAD is regenerated by an as yet unknown electron acceptor.
The Importance of Ketosteroid Dehydrogenases for Pathogenic Organisms
Δ4-(5α)-KSTD is the first ketosteroid dehydrogenase of which a crystal structure has been elucidated. The enzyme has an important role in the desaturation of the steroid A ring, which is a key step in the microbial degradation of saturated steroids. It has been proposed that saturated steroid intermediates are formed during cholesterol catabolism. Several pathogenic bacteria, including M. tuberculosis, Rhodococcus equi, and Mycobacterium bovis, contain a cholesterol catabolic pathway similar to that of R. jostii RHA1 (8, 49). In M. tuberculosis this pathway and in particular the Δ1-KSTD enzyme (Rv3537, which shows 28% sequence identity to R. jostii RHA1 Δ4-(5α)-KSTD) have been implicated to be important for growth of the intracellular pathogen in the hostile environment of macrophages (50, 51). This makes the cholesterol catabolic pathway a promising target for the development of therapeutic agents to combat M. tuberculosis (8). Our R. jostii RHA1 Δ4-(5α)-KSTD structure may facilitate the design of potent ketosteroid dehydrogenase inhibitors, as a first step toward the development of new antituberculosis drugs.
Supplementary Material
Acknowledgments
We are grateful to the scientists of Beamlines BM16 and ID14-1 (European Synchrotron Radiation Facility, Grenoble, France) for help during data collections.
This was project was supported in part by the Netherlands Ministry of Economic Affairs and the B-Basic partner organizations through B-Basic, a public-private Advanced Chemical Technologies for Sustainability program. This work was also supported by Schering-Plough.

This article contains supplemental Table S1 and Figs. S1–S3.
The atomic coordinates and structure factors (codes 4AT0 and 4AT2) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
J. Knol, D. van der Tuin, L. Dijkhuizen, and R. van der Geize, manuscript in preparation.
- KSTD
- ketosteroid dehydrogenase
- 5α-AD
- 5α-androstane-3,17-dione
- 1-(5α)-AD
- 1-(5α)-androstene-3,17-dione
- 4-AD
- 4-androstene-3,17-dione
- ADD
- 1,4-androstadiene-3,17-dione
- RMSD
- root mean square deviation
- DCPIP
- dichlorophenolindophenol.
REFERENCES
- 1. McLeod M. P., Warren R. L., Hsiao W. W., Araki N., Myhre M., Fernandes C., Miyazawa D., Wong W., Lillquist A. L., Wang D., Dosanjh M., Hara H., Petrescu A., Morin R. D., Yang G., Stott J. M., Schein J. E., Shin H., Smailus D., Siddiqui A. S., Marra M. A., Jones S. J., Holt R., Brinkman F. S., Miyauchi K., Fukuda M., Davies J. E., Mohn W. W., Eltis L. D. (2006) The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U.S.A. 103, 15582–15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van der Geize R., Dijkhuizen L. (2004) Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr. Opin. Microbiol. 7, 255–261 [DOI] [PubMed] [Google Scholar]
- 3. Horinouchi M., Hayashi T., Yamamoto T., Kudo T. (2003) A new bacterial steroid degradation gene cluster in Comamonas testosteroni TA441 which consists of aromatic-compound degradation genes for seco-steroids and 3-ketosteroid dehydrogenase genes. Appl. Environ. Microbiol. 69, 4421–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Levy H. R., Talalay P. (1959) Bacterial oxidation of steroids. II. Studies on the enzymatic mechanism of ring A dehydrogenation. J. Biol. Chem. 234, 2014–2021 [PubMed] [Google Scholar]
- 5. Knol J., Bodewits K., Hessels G. I., Dijkhuizen L., van der Geize R. (2008) 3-Keto-5α-steroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism. Biochem. J. 410, 339–346 [DOI] [PubMed] [Google Scholar]
- 6. Fujii C., Morii S., Kadode M., Sawamoto S., Iwami M., Itagaki E. (1999) Essential tyrosine residues in 3-ketosteroid-Δ1-dehydrogenase from Rhodococcus rhodochrous. J. Biochem. 126, 662–667 [DOI] [PubMed] [Google Scholar]
- 7. Matsushita H., Itagaki E. (1992) Essential histidine residue in 3-ketosteroid-Δ1-dehydrogenase. J. Biochem. 111, 594–599 [DOI] [PubMed] [Google Scholar]
- 8. Van der Geize R., Yam K., Heuser T., Wilbrink M. H., Hara H., Anderton M. C., Sim E., Dijkhuizen L., Davies J. E., Mohn W. W., Eltis L. D. (2007) A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 104, 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosas-Magallanes V., Stadthagen-Gomez G., Rauzier J., Barreiro L. B., Tailleux L., Boudou F., Griffin R., Nigou J., Jackson M., Gicquel B., Neyrolles O. (2007) Signature-tagged transposon mutagenesis identifies novel Mycobacterium tuberculosis genes involved in the parasitism of human macrophages. Infect. Immun. 75, 504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Oosterwijk N., Knol J., Dijkhuizen L., van der Geize R., Dijkstra B. W. (2011) Cloning, overexpression, purification, crystallization and preliminary x-ray analysis of 3-ketosteroid Δ4-(5α)-dehydrogenase from Rhodococcus jostii RHA1. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1269–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM. A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 13. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jaroszewski L., Rychlewski L., Li Z., Li W., Godzik A. (2005) FFAS03. A server for profile-profile sequence alignments. Nucleic Acids Res. 33, W284–W288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bamford V., Dobbin P. S., Richardson D. J., Hemmings A. M. (1999) Open conformation of a flavocytochrome c3 fumarate reductase. Nat. Struct. Biol. 6, 1104–1107 [DOI] [PubMed] [Google Scholar]
- 16. Leys D., Tsapin A. S., Nealson K. H., Meyer T. E., Cusanovich M. A., Van Beeumen J. J. (1999) Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR-1. Nat. Struct. Biol. 6, 1113–1117 [DOI] [PubMed] [Google Scholar]
- 17. Doherty M. K., Pealing S. L., Miles C. S., Moysey R., Taylor P., Walkinshaw M. D., Reid G. A., Chapman S. K. (2000) Identification of the active site acid/base catalyst in a bacterial fumarate reductase. A kinetic and crystallographic study. Biochemistry 39, 10695–10701 [DOI] [PubMed] [Google Scholar]
- 18. Iverson T. M., Luna-Chavez C., Croal L. R., Cecchini G., Rees D. C. (2002) Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site. J. Biol. Chem. 277, 16124–16130 [DOI] [PubMed] [Google Scholar]
- 19. Yankovskaya V., Horsefield R., Törnroth S., Luna-Chavez C., Miyoshi H., Léger C., Byrne B., Cecchini G., Iwata S. (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299, 700–704 [DOI] [PubMed] [Google Scholar]
- 20. Sun F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., Rao Z. (2005) Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 121, 1043–1057 [DOI] [PubMed] [Google Scholar]
- 21. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Terwilliger T. C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabsch W., Sander C. (1983) Dictionary of protein secondary structure. Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 28. Schüttelkopf A. W., van Aalten D. M. (2004) PRODRG. A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 29. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Delano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 31. Ghisla S., Massey V., Lhoste J. M., Mayhew S. G. (1974) Fluorescence and optical characteristics of reduced flavines and flavoproteins. Biochemistry 13, 589–597 [DOI] [PubMed] [Google Scholar]
- 32. Massey V., Ghisla S. (1974) Ann. N.Y. Acad. Sci. 227, 446–465 [Google Scholar]
- 33. Wierenga R. K., de Jong R. J., Kalk K. H., Hol W. G., Drenth J. (1979) Crystal structure of p-hydroxybenzoate hydroxylase. J. Mol. Biol. 131, 55–73 [DOI] [PubMed] [Google Scholar]
- 34. Mattevi A. (1998) The PHBH fold. Not only flavoenzymes. Biophys. Chem. 70, 217–222 [DOI] [PubMed] [Google Scholar]
- 35. Holm L., Sander C. (1996) Mapping the protein universe. Science 273, 595–603 [DOI] [PubMed] [Google Scholar]
- 36. Vallon O. (2000) New sequence motifs in flavoproteins. Evidence for common ancestry and tools to predict structure. Proteins Struct. Funct. Genet. 38, 95–114 [DOI] [PubMed] [Google Scholar]
- 37. Wierenga R. K., Drenth J., Schulz G. E. (1983) Comparison of the three-dimensional protein and nucleotide structure of the FAD-binding domain of p-hydroxybenzoate hydroxylase with the FAD- as well as NADPH-binding domains of glutathione reductase. J. Mol. Biol. 167, 725–739 [DOI] [PubMed] [Google Scholar]
- 38. Lyubimov A. Y., Heard K., Tang H., Sampson N. S., Vrielink A. (2007) Distortion of flavin geometry is linked to ligand binding in cholesterol oxidase. Protein Sci. 16, 2647–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mattevi A., Fraaije M. W., Mozzarelli A., Olivi L., Coda A., van Berkel W. J. (1997) Crystal structures and inhibitor binding in the octameric flavoenzyme vanillyl-alcohol oxidase. The shape of the active-site cavity controls substrate specificity. Structure 5, 907–920 [DOI] [PubMed] [Google Scholar]
- 40. Hallberg B. M., Leitner C., Haltrich D., Divne C. (2004) Crystal structure of the 270 kDa homotetrameric lignin-degrading enzyme pyranose 2-oxidase. J. Mol. Biol. 341, 781–796 [DOI] [PubMed] [Google Scholar]
- 41. Itagaki E., Matushita H., Hatta T. (1990) Steroid transhydrogenase activity of 3-ketosteroid-Δ1-dehydrogenase from Nocardia corallina. J. Biochem. 108, 122–127 [DOI] [PubMed] [Google Scholar]
- 42. Ringold H. J., Hayano M., Stefanovic V. (1963) Concerning the stereochemistry and mechanism of the bacterial C-1,2 dehydrogenation of steroids. J. Biol. Chem. 238, 1960–1965 [PubMed] [Google Scholar]
- 43. Fagan R. L., Nelson M. N., Pagano P. M., Palfey B. A. (2006) Mechanism of flavin reduction in class 2 dihydroorotate dehydrogenases. Biochemistry 45, 14926–14932 [DOI] [PubMed] [Google Scholar]
- 44. Fraaije M. W., Mattevi A. (2000) Flavoenzymes. Diverse catalysts with recurrent features. Trends Biochem. Sci. 25, 126–132 [DOI] [PubMed] [Google Scholar]
- 45. Li J., Vrielink A., Brick P., Blow D. M. (1993) Crystal structure of cholesterol oxidase complexed with a steroid substrate. Implications for flavin adenine dinucleotide dependent alcohol oxidases. Biochemistry 32, 11507–11515 [PubMed] [Google Scholar]
- 46. Abul-Hajj Y. J. (1978) Isolation of vitamin K2(35) from Nocardia restrictus and Corynebacterium simplex. A natural electron acceptor in microbial steroid ring A dehydrogenations. J. Biol. Chem. 253, 2356–2360 [PubMed] [Google Scholar]
- 47. Ghisla S., Massey V. (1989) Mechanisms of flavoprotein-catalyzed reactions. Eur. J. Biochem. 181, 1–17 [DOI] [PubMed] [Google Scholar]
- 48. Medentsev A. G., Arinbasarova A. Y., Koshcheyenko K. A., Akimenko V. K., Skryabin G. K. (1985) Regulation of 3-ketosteroid-1-en-dehydrogenase activity of Arthrobacter globiformis cells by a respiratory chain. J. Steroid Biochem. Mol. Biol. 23, 365–368 [DOI] [PubMed] [Google Scholar]
- 49. van der Geize R., Grommen A. W., Hessels G. I., Jacobs A. A., Dijkhuizen L. (2011) The steroid catabolic pathway of the intracellular pathogen Rhodococcus equi is important for pathogenesis and a target for vaccine development. PLoS Pathogens 7, e1002181-e1002181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rengarajan J., Bloom B. R., Rubin E. J. (2005) Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 102, 8327–8332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pandey A. K., Sassetti C. M. (2008) Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weiss M. S. (2001) J. Appl. Crystallogr. 34, 130–135 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.