Abstract
BACKGROUND & AIMS
The pathogenesis of liver fibrosis involves activation of hepatic stellate cells, which is associated with depletion of intracellular lipid droplets. When hepatocytes undergo autophagy, intracellular lipids are degraded in lysosomes. We investigated whether autophagy also promotes loss of lipids in hepatic stellate cells to provide energy for their activation and extended these findings to other fibrogenic cells.
METHODS
We analyzed hepatic stellate cells from C57BL/6 wild-type, Atg7F/F, and Atg7F/F-GFAP-Cre mice, as well as the mouse stellate cell line JS1. Fibrosis was induced in mice using CCl4 or thioacetamide (TAA); liver tissues and stellate cells were analyzed. Autophagy was blocked in fibrogenic cells from liver and other tissues using small interfering RNAs against Atg5 or Atg7 and chemical antagonists. Human pulmonary fibroblasts were isolated from samples of lung tissue from patients with idiopathic pulmonary fibrosis or from healthy donors.
RESULTS
In mice, induction of liver injury with CCl4 or TAA increased levels of autophagy. We also observed features of autophagy in activated stellate cells within injured human liver tissue. Loss of autophagic function in cultured mouse stellate cells and in mice following injury reduced fibrogenesis and matrix accumulation; this effect was partially overcome by providing oleic acid as an energy substrate. Autophagy also regulated expression of fibrogenic genes in embryonic, lung, and renal fibroblasts.
CONCLUSIONS
Autophagy of activated stellate cells is required for hepatic fibrogenesis in mice. Selective reduction of autophagic activity in fibrogenic cells in liver and other tissues might be used to treat patients with fibrotic diseases.
Keywords: Myofibroblasts, Inflammation, Mouse Model, Energy Depletion
Fibrotic diseases account for up to 45% of worldwide mortality,1 yet the development of antifibrotic therapies remains elusive, underscoring the need for greater understanding of the pathogenesis of fibrosis. Myofibroblastic transformation of resident mesenchymal cells underlies the development of fibrosis in liver, lung, kidney, and other organs, suggesting that common pathways of wound repair are shared among different tissues.2 In liver, the activation of resident hepatic stellate cells into myofibroblasts through the acquisition of a contractile, proliferative, inflammatory, and fibrogenic phenotype is a critical event in fibrogenesis.3 Activated stellate cells/myofibroblasts are the primary source of extracellular matrix, whose accumulation leads to the progressive replacement of the liver parenchyma by scar.4
Among the most characteristic features of stellate cell activation is the loss of cytoplasmic lipid droplets (LDs), which are composed of retinyl esters and triglycerides,5 yet there has been little functional linkage between lipid loss and cellular activation. A recent study identifying the breakdown of intracellular lipids by macroautophagy (hereafter referred to as “autophagy”) to generate fatty acids for energy production in hepatocytes6 raised the possibility that the loss of LDs by autophagy drives the fibrogenic response in stellate cells. The aim of our study was to establish the role of autophagy in activation of hepatic stellate cells but also in other fibrogenic cells as embryonic myofibroblasts and fibrogenic cells from kidney and lung.
Materials and Methods
Cells and Cell Culture
Mouse hepatic stellate cells were isolated from C57BL/6 wild-type, Atg7F/F, and Atg7F/F-GFAP-Cre mice by enzymatic digestion and Percoll density gradient centrifugation with modifications.7 The mouse immortalized stellate cell line JS1, previously generated in our laboratory,8 was cultured with Dulbecco’s modified Eagle medium containing 10% fetal bovine serum. Mouse mesangial cells used were obtained as previously described.9 Human pulmonary fibroblasts were cultured from the lung tissues of patients with idiopathic pulmonary fibrosis undergoing lung transplant or from normal donor lungs that were not used for transplant.10 Some cells were treated with 10 mmol/L 3-methyladenine (3-MA; Sigma, St Louis, MO), 10 mmol/L chloroquine (Sigma), or 20 μmol/L etomoxir (Sigma). For lipid supplementation assays, either a 1:100 dilution of bovine serum albumin (BSA)– oleic acid (Sigma) or the equivalent concentration of BSA alone was added to cultured cells. Triglyceride measurements were performed using a triglyceride quantification kit (BioVision, Milpitas, CA).
Lentiviral Atg5 Small Interfering RNA Construction
Small hairpin (sense-loop-antisense) RNAs for Atg5 and Atg7 cloned into lentiviral vectors previously described6,11 were used. Knockdown and autophagy inhibition was optimum at 8 days of transduction, which was the time point used for all the experiments.
Animals
Atg7F/F mice previously described12 on the C57BL/6 background were crossed with a transgenic FVB line expressing cre recombinase under the control of the glial fibrillary acid protein (GFAP) promoter (GFAP-cre) to generate Atg7F/F-GFAP-cre mice with a stellate cell–specific knockout13 of Atg7.
All studies were approved by the Animal Care and Use Committee of the Mount Sinai School of Medicine and followed the National Institutes of Health guidelines for animal care.
Toxin-Induced Models of Acute and Chronic Liver Injury and Fibrosis
Carbon tetrachloride (Sigma) and thioacetamide (TAA; Acros Organics, Geel, Belgium) were used to induce hepatic fibrosis. For acute injury studies, mice received 3 intraperitoneal injections of 10% CCl4 (diluted in corn oil) at a dose of 0.5 μL/g body wt. In the chronic injury model, mice received intraperitoneal injections of 10% CCl4 (diluted in corn oil) at a dose of 0.5 μL/g body wt 3 times per week for 6 weeks. TAA (diluted in phosphate-buffered saline) treatment was administered by 3 intraperitoneal injections (100 mg/body wt) every other day. Under ketamine/xylazine anesthesia, animals were killed 48 hours after the last dose. All the experiments were performed with male mice at 4 months of age.
Immunoblot
Cell lysates were subjected to immunoblot analysis. Membranes were incubated with the following primary antibodies: rabbit anti-ATG7, rabbit anti–microtubule-associated light chain 3 (LC3), rabbit anti-ATG5 (Cell Signaling Technology, Boston, MA), chicken anti–adipocyte differentiation-related protein (ADRP) (Millipore, Billerica, MA), rabbit anti– glyceralde-hyde-3-phosphate dehydrogenase (GAPDH) (Sigma), rabbit anti–type I collagen (Rockland Inc, Gilbertsville, PA), rabbit anti—β–smooth muscle actin (SMA) (Millipore), rabbit anti—β–platelet-derived growth factor receptor (β-PDGFR; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti–matrix metalloproteinase 2 (MMP2) (Abcam, Cambridge, MA), mouse anti-tubulin (Sigma), rabbit anti-P62 (Enzo, New York, NY), and mouse anti-cre recombinase (Abcam). The reactions were detected with horseradish peroxidase– conjugated secondary antibodies. Blots were developed using the ECL Detection System (Amersham Pharmacia Biotech, Buckinghamshire, England).
Oil Red O Staining
Cells were stained with oil red O (ORO; 0.5% in propylene glycol) to visualize LDs. Nuclei were counterstained with hematoxylin for bright-field microscopic examination. ImageJ software was used for quantification.
Electron Microscopy
Cells and liver tissue were fixed with 3% glutaraldehyde in 0.2 mol/L sodium cacodylate, pH 7.4. Mouse livers were sufficiently perfused with the previously described solution before dissection. The specimens were then treated with 1% osmium tetroxide for 1 hour, followed by ethanol dehydration in graded steps through propylene oxide, and then embedded in Embed 812 (Electron Microscopy Sciences, Hatfield, PA). Ultra-thin sections were stained with uranyl acetate and lead citrate. Images were acquired on a Hitachi (Toronto, Ontario, Canada) H7650 transmission electron microscope.
Quantitative Real-Time Polymerase Chain Reaction
Cellular RNA was extracted using Qiagen mini columns (Qiagen, Germantown, MD) with an on-column deoxyribonuclease treatment. One microgram of RNA was reverse transcribed using RT Complete Double PrePrimed Kit (Clontech, Mountain View, CA). FastStart SYBR Green Master (Roche, Indianapolis, IN) was used for polymerase chain reaction. Samples were analyzed in triplicate in Microsoft Excel (Microsoft Corp, Redmond, WA) and normalized to β-actin expression.
Histologic and Immunohistochemical Studies
Liver samples were formalin fixed, paraffin embedded, sectioned at 4 μm, and processed routinely for H&E staining. Sirius Red (Sigma) was used to determine collagen deposition. A blinded pathologist scored 10 random areas per slide for fibrosis. Immunohistochemical staining of αSMA was performed with a rabbit polyclonal antibody (Abcam). Sections were stained with Sirius Red solution (saturated picric acid containing 0.1% Direct Red 80 and 0.1% Fast Green) to visualize collagen deposition.
Liver Tests
Quantitative determination of serum aspartate aminotransferase and alanine aminotransferase levels was performed using spectrophotometer analysis (Pointe Scientific, Inc, Canton, MI).
Statistics
Results are expressed as the mean and SEM. P values (Student 2-tailed, unpaired t test) of at least 3 independent determinations were calculated with Microsoft Excel software. Data were considered to be statistically significant at P < .05.
Results
Autophagic Function Is Increased With Hepatic Stellate Cell Activation In Vivo
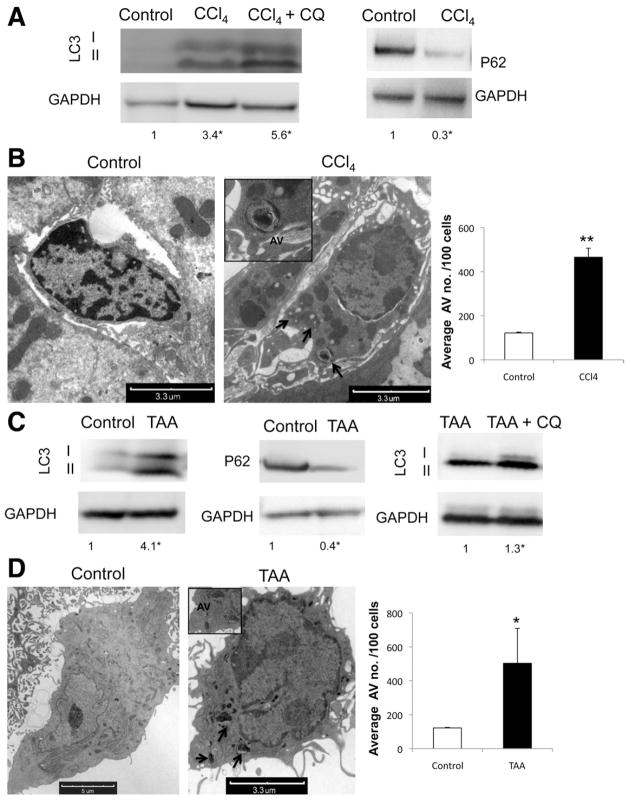
We first examined whether autophagic activity is altered during stellate cell activation in vivo. Cells isolated from wild-type mice following either 3 doses of CCl4 (0.5 μL/g body wt) or TAA (100 mg/g body wt), but not vehicle-treated animals, displayed increased accumulation of the conjugated form of LC3, LC3II, and decreased P62/SQSTM1 by immunoblot (Figure 1A and C); the latter is a selective autophagy adaptor molecule that brings ubiquitinated substrates to autophagosomes and is degraded by autophagy.14 There was a marked increase in the number of autophagic vacuoles (AVs) in these cells based on electron microscopy (EM) (Figure 1B and D). Autophagy flux assay was also performed using the lysosomal inhibitor chloroquine (CQ) (Figure 1A and C), confirming an up-regulation of autophagy after liver injury.
Figure 1.
Autophagy is up-regulated in stellate cells after liver injury in vivo. (A and C) Immunoblots of stellate cells isolated from wild-type mice after acute liver injury induction with (A) CCl4 and (C) TAA with and without addition of CQ showing an increase of LC3II conversion and decrease in P62. (B and D) Electron micrographs of whole liver tissue, showing stellate cells after (B) CCl4 and (D) TAA treatment. Arrows indicate AV. (Right) Electron micrograph quantification of AV number per 100 cells (★P < .05, ★★P < .001; error bars indicate SEM). Protein ratios (normalized to GAPDH) were used to quantify fold change relative to control and are shown below each blot. Data represent the mean value of at least 3 experiments (★P < .05), and 3 animals per condition were used in this experiment.
We also evaluated autophagy levels in human cells (Supplementary Figure 1A–C). An increase in LC3II levels, as well as the number of AVs, together with a decrease in P62 was also evident in stellate cells isolated from hepatitis B virus–infected human liver compared with noninfected liver (Supplementary Figure 1A and B). We then analyzed fibroblasts from nonfibrotic lungs and those with idiopathic pulmonary fibrosis and found an increase in LC3II and decrease in P62 in idiopathic pulmonary fibrosis samples (Supplementary Figure 1C). These data indicate that increased autophagic function occurs during activation of fibrogenic cells in different tissues and from different species.
Blocking Autophagy in Stellate Cells Attenuates Fibrogenic Activity
To address the prospect that autophagy regulates stellate cell activation, we incubated an immortalized mouse stellate cell line, JS1,8 with 2 pharmacologic inhibitors of autophagy: 3-MA15 or CQ.16 Each compound decreased autophagy levels within 12 hours; 3-MA–treated cells showed a decreased number of autophagic vacuoles (Supplementary Figure 2B) and increased P62 (Supplementary Figure 2A). As expected, chloroquine blocked LC3II turnover, leading to its accumulation by immunoblot14 (Supplementary Figure 3A), concurrent with an increase in P62/SQSTM1 (Supplementary Figure 3B) and autophagosome number, which results from a block in autophagosome fusion with the lysosome (Supplementary Figure 3C). Cell viability was not affected by either 3-MA or CQ treatment (data not shown).
Because the effects of 3-MA and CQ (Supplementary Figure 5) are not entirely specific for autophagy, JS1 cells were also transduced with a lentiviral vector expressing small hairpin RNA to the essential autophagy genes Atg7 (siAtg7 cells) or Atg5 (siAtg5 cells) or with an empty lentivirus control (VEC) (Supplementary Figures 4 and 6). Knockdown of these 2 autophagy proteins was confirmed by immunoblot and led to significant decreases in LC3II (Supplementary Figures 4 and 6) and numbers of AVs (Supplementary Figure 4B).
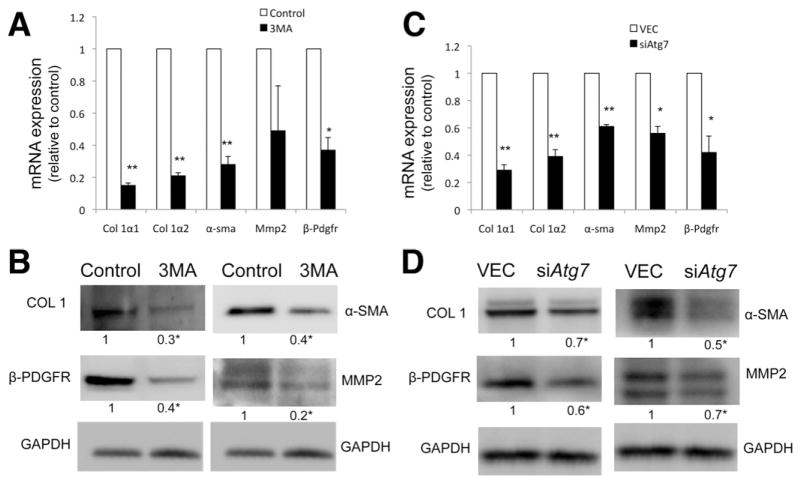
We next assessed whether specific inhibition of autophagy regulates a cardinal feature of stellate cell activation, the expression of fibrogenic genes. Inhibition of autophagy, either by pharmacologic means or by specific Atg5 or Atg7 knockdown, significantly reduced the expression of messenger RNAs encoding collagen α1(I) and α2(I) (Col 1α1, Col 1α2), β-Pdgfr, α-sma, and Mmp-2 (Figure 2A and C and Supplementary Figure 6B) and of their corresponding proteins (Figure 2B and D and Supplementary Figure 6C).
Figure 2.
Inhibition of autophagy decreases fibrogenesis. (A and C) Col 1α1, Col 1α2, α-sma, Mmp2, and β-Pdgfr messenger RNA (quantitative reverse-transcription polymerase chain reaction analysis) and (B and D) protein expression (immunoblot) in JS1 cells (A and B) after 24-hour treatment with 3-MA or (C and D) 8 days after transduction with siAtg7 lentivirus. All figures are representative of at least 3 independent experiments. Messenger RNA is expressed as fold change relative to control (★P < .05, ★★P < .001; error bars indicate SEM). Protein ratios (normalized to GAPDH) were used to quantify fold change relative to control and are shown below each blot. Data represent the mean value of at least 3 experiments (★P < .05).
To evaluate if autophagy is a generalized regulatory pathway in fibrosis, we analyzed the effect of autophagy inhibition with 3-MA in primary human stellate cells (Supplementary Figure 7A) and primary human lung fibroblasts (Supplementary Figure 7B). Embryonic fibroblasts from Atg5-deficient mice were also characterized (Supplementary Figure 8A), and autophagy was blocked with 3-MA or silencing Atg7 in mouse messengial cells (Supplementary Figure 8B and C), a fibrogenic cell type in kidney. The same protective effect of autophagy inhibition on fibrosis was corroborated in the different cell types, reinforcing the role of autophagy as a core pathway in fibrosis.
Blocking Autophagy in Stellate Cells Attenuates Liver Fibrosis In Vivo
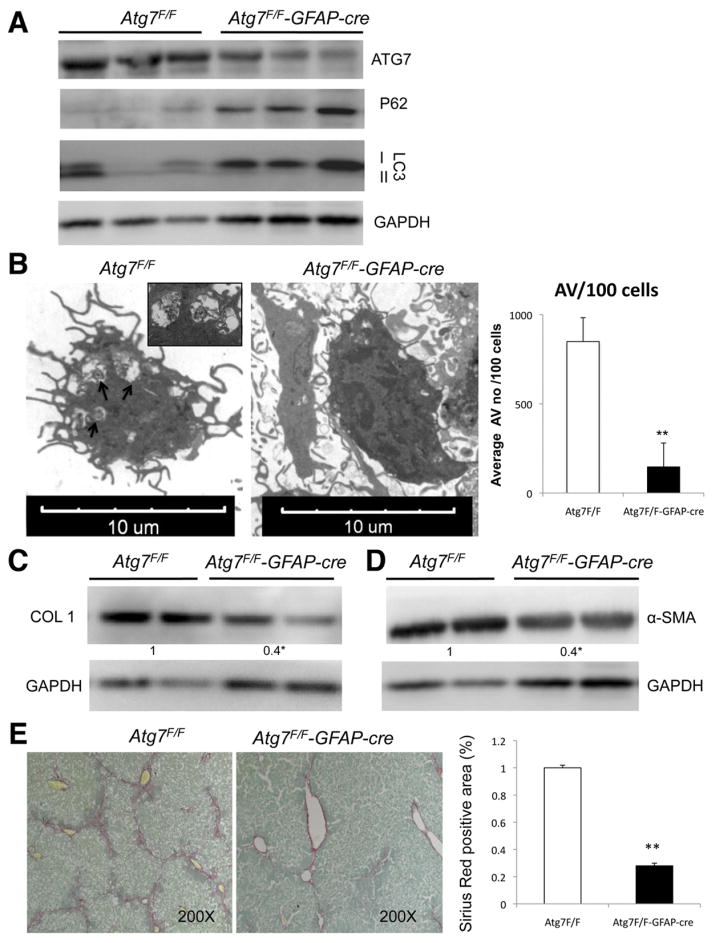
It remained critical to establish a role for autophagy in stellate cell activation in vivo. To do so, we generated a transgenic mouse line in which expression of Atg7 was specifically attenuated in stellate cells within liver by crossing Atg7F/F with animals expressing cre recombinase under the glial fibrillary acidic protein promoter (GFAP-cre).13 The cell type specificity of the Atg7 knockout was confirmed in primary isolated stellate cells from these animals by immunoblotting and quantitative real-time polymerase chain reaction (Supplementary Figure 9A and C) that was associated with increased cre recombinase protein expression (Supplementary Figure 9B). Stellate cells isolated from Atg7F/F-GFAP-cre contained reduced LC3-II levels, a compensatory increase in LC3-I levels, and P62 accumulation (Figure 3A). EM of these isolated cells displayed a significantly reduced number of AVs compared with their Atg7F/F littermates (Figure 3C). In Atg7F/F-GFAP-cre mice, the decrease in autophagy levels was specific to stellate cells, because there were no changes in hepatocyte AV number or Atg7 expression in other cell types as assessed by whole liver EM and Atg7 immunohistochemistry (Supplementary Figure 9D).
Figure 3.
Autophagy regulates stellate cell activation and fibrosis in vivo. (A) Immunoblots of stellate cells protein isolated from Atg7F/F and Atg7F/F-GFAP-cre mice showing decreased expression of ATG7, LC3-II, and increased P62. (B) Electron micrographs of stellate cells isolated from Atg7F/F and Atg7F/F-GFAP-cre mice and AV quantification (right) depict a significant decrease of AV in transgenic animals. Arrows indicate AV. (C and D) Immunoblots for (C) collagen type I and α-SMA in isolated stellate cells from Atg7F/F and Atg7F/F-GFAP-cre mice after chronic liver injury with CCl4. (E) Whole liver sections after chronic liver injury with CCl4 were stained for Sirius Red. (Right) Quantification of Sirius Red–positive area. ★P < .05, ★★P < .001. Error bars indicate SEM. Protein ratios (normalized to GAPDH) were used to quantify fold change relative to control and are shown below each blot. Data represent the mean value of at least 3 experiments (★P < .05) and a total of 18 animals: 9 Atg7F/F and 9 Atg7F/F-GFAP-cre.
Mice with autophagy-defective hepatic stellate cells displayed a normal liver architecture (Supplementary Figure 10A), and stellate cell ultrastructure was intact (data not shown). However, following liver injury, Atg7F/F-GFAP-cre mice had attenuated liver fibrosis. Chronic fibrosis was induced by 6 weeks of CCl4 in Atg7F/F-GFAP-cre and Atg7F/F mice. Collagen accumulation, as assessed by Sirius Red morphometry, was significantly reduced in Atg7F/F-GFAP-cre mice (Figure 3E). Moreover, whereas the number of α-SMA–positive stellate cells in transgenic animals was unchanged (data not shown), total α-SMA protein in isolated stellate cells was significantly reduced (Figure 3D), suggesting that loss of Atg7 in stellate cells attenuated α-SMA expression per cell but did not affect stellate cell number. In stellate cells isolated from the Atg7F/F-GFAP-cre animals, collagen I (COL 1) was markedly reduced compared with cells from Atg7F/F (Figure 3C). Atg7F/F-GFAP-cre and Atg7F/F mice displayed similar liver to body weight ratios (Supplementary Figure 10B) and comparable levels of liver injury, indicating that the effect of autophagy attenuation on hepatic fibrosis in stellate cells was not secondary to an alteration in the extent of liver injury (Supplementary Figure 10C). To establish that autophagy is a crucial regulator of stellate cell activation independent of the type of underlying injury, we also induced liver injury in Atg7F/F-GFAP-cre and Atg7F/F mice with TAA and confirmed the same protective effect of blocking autophagy on fibrosis development (Supplementary Figure 11).
Autophagy Stimulates Loss of LDs During Stellate Cell Activation to Provide Cellular Energy
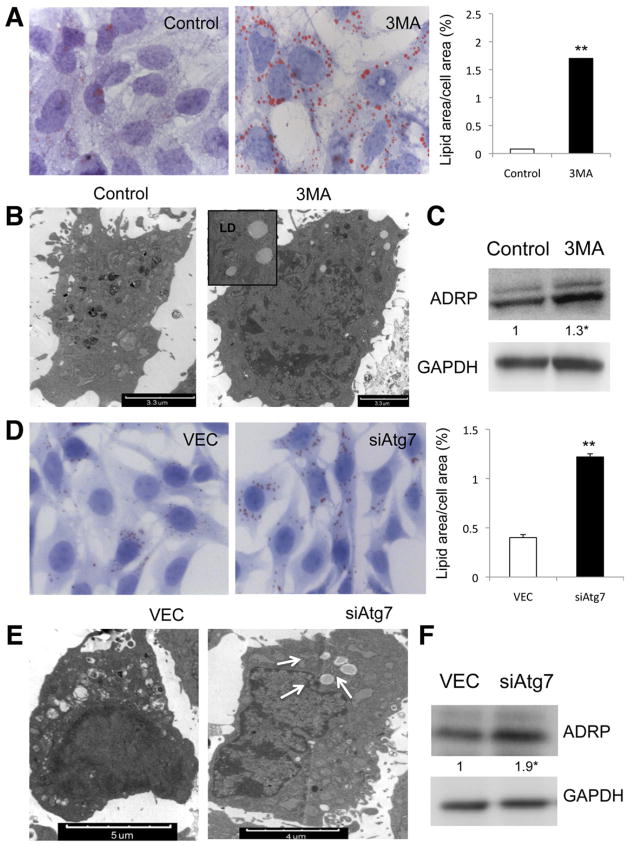
Although not previously examined, stellate cell activation is likely to be an intense energy-requiring process to fuel the pathways of cell proliferation, extracellular matrix secretion, and cellular contractility. Therefore, we reasoned that autophagy might provide a critical source of energy substrate in the form of triglyceride stored within cytoplasmic droplets. We therefore examined whether inhibition of autophagy attenuated the depletion of lipid content associated with cellular activation. ORO staining revealed an increased number of LDs in cells either treated with 3-MA or transduced with siAtg7 (Figure 4A and D) or siAtg5 (data not shown). These findings were associated with increased LDs on EM (Figure 4B and E), increased triglyceride content (Supplementary Figure 12A and B), and expression of ADRP in autophagy-deficient cells compared with control cells (Figure 4C and F). Stellate cells isolated from Atg7F/F-GFAP-cre contained increased LD content and ADRP (Supplementary Figure 13D and E) after activation induced by either 10 days in primary culture (data not shown) or in vivo following treatment with 3 doses of CCl4 and brief primary culture (Supplementary Figure 13B and C) compared with stellate cells from Atg7F/F mice, even though all cells had the same amount of LDs before activation (Supplementary Figure 13A).
Figure 4.
Autophagy deficiency in stellate cells leads to LD accumulation. Lipid content analysis in JS1 cells (A–C) treated with 3-MA or (D–F) transduced with siAtg7. (A and D) ORO staining and (right) quantification of ORO-stained area (★P < .05, ★★P < .001). Error bars indicate SEM. (B and E) Electron micrographs showing neutral LDs (arrows) and (C and F) immunoblots for ADRP (error bars indicate SEM). Protein ratios (normalized to GAPDH) were used to quantify fold change relative to control and are shown below each blot. Data represent the mean value of at least 3 experiments (★P < .05).
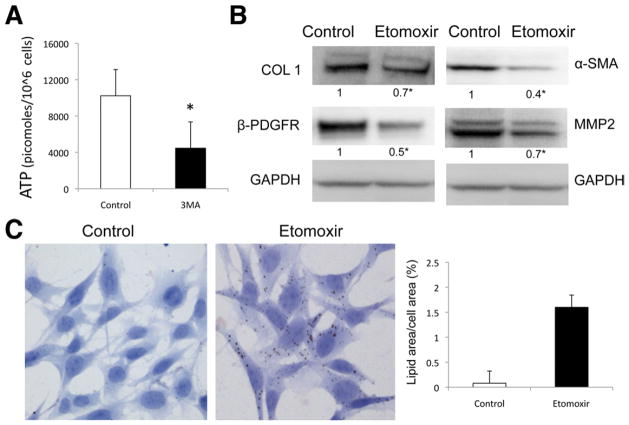
LDs are catabolized into free fatty acids (FFAs) by intracellular lipases to undergo mitochondrial β-oxidation, generating adenosine triphosphate (ATP). Because autophagy stimulates β-oxidation rates and energy production,6,17 we assessed the impact of inhibiting autophagy on ATP production in stellate cells. As predicted, inhibition of autophagy by 3-MA led to a decrease in ATP levels (Figure 5A). Moreover, treatment with etomoxir, an inhibitor of fatty acid oxidation, provoked accumulation of LDs in stellate cells (Figure 5C) and a decrease in COL1, α-SMA, MMP2, and β-PDGFR expression (Figure 5B) without affecting autophagic activity (data not shown).
Figure 5.
β-oxidation regulates stellate cell activation. (A) Autophagy was inhibited in JS1 cells with 3-MA, and ATP content was determined at 12 hours after treatment. (B and C) β-oxidation was blocked with etomoxir in JS1 cells after 12-hour expression of COL1, α-SMA, and β-PDGFR was determined by (B) immunoblot analysis and (C) LD content was examined by ORO staining (right, quantification of ORO-stained area). Levels of ATP are expressed in picomoles per 106 cells. Error bars indicate SEM. Protein ratios (normalized to GAPDH or tubulin) were used to quantify fold change relative to control and are shown below each blot. Data represent the mean value of at least 3 experiments (★P < .05).
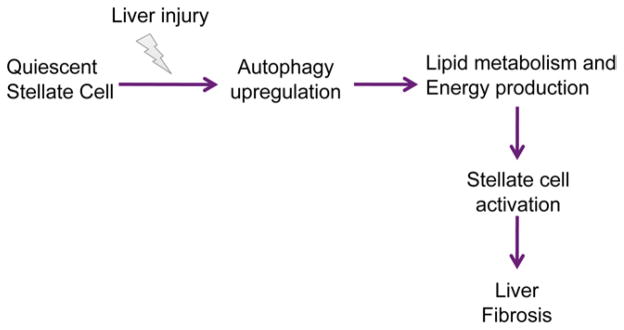
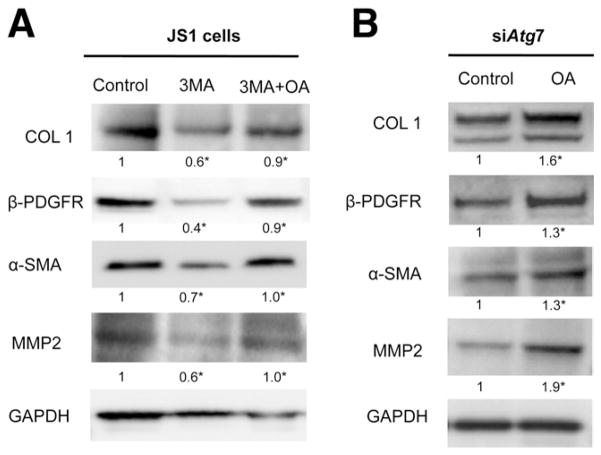
Together, these data suggest that autophagy normally provides a key source of FFAs that fuels stellate cell activation, and its inhibition impairs energy production (Figure 6). If this hypothesis is true, then providing an alternative source of FFAs in autophagy-deficient cells should at least partially rescue the attenuated activation. As predicted, addition of oleic acid (OA) after blocking autophagy in stellate cells with either 3-MA (Figure 7A) or siAtg7 (Figure 7B) was able to increase ATP production (data not shown) and significantly rescued the decrease in fibrogenesis mediated by a block in autophagy.
Figure 6.
Simplified model depicting the role of autophagy in stellate cell activation and fibrogenesis. The response of stellate cells to hepatic injury involves the up-regulation of autophagy, which fuels stellate cell activation, in part through its role in lipid breakdown and energy production, which drives the fibrogenic response of the liver.
Figure 7.
Decrease in fibrogenesis caused by autophagy inhibition can be rescued by exogenous FFAs. JS1 cells were treated with (A) 3-MA or transduced with (B) siAtg7 and supplemented with OA conjugated to BSA or with BSA alone. Expression of COL 1, α-SMA, MMP2, and β-PDGFR was determined by (A and B) immunoblot analysis. Protein ratios (normalized to GAPDH) were used to quantify fold change relative to control and are shown below each blot. Data represent the mean value of at least 3 experiments (★P < .05).
Discussion
Our findings show that autophagic activity is required to establish an activated phenotype in hepatic stellate cells. In agreement with a recent study18 restricted to cultured cells, and using nonspecific inhibitors of autophagy, our data establish that inhibiting autophagy in hepatic stellate cells impairs their fibrogenic potential in vivo, leading to decreased expression of the fibrogenic genes α-sma, Collagen 1α1, Collagen 1α2, β-Pdgfr, and Mmp-2 without affecting cell viability and survival. These findings underscore the vital role of autophagy in generating energy to support stellate cell activation and drive extracellular matrix production. Genetic inhibition of autophagy in stellate cells attenuates fibrosis in 2 different injury models (CCl4 and TAA), because Atg7F/F-GFAP-cre mice displayed reduced stellate cell activation and fibrosis following liver injury. In addition, our data establish a role of autophagy not only in hepatic stellate cell activation and liver fibrosis but also the fibrogenic cells from other tissues (lung, kidney, embryonic fibroblasts) and in both mice and humans.
Basal autophagy is present in all cell types and is rapidly up-regulated as an adaptive response under conditions of cellular stress as a means to generate intracellular nutrients and energy.19,20 In normal liver, lysosomal activity is reported to be very low.21,22 Several insults to the liver (eg, alcohol consumption)23 have been identified as autophagy triggers; however, these studies are primarily focused on hepatocytes. We show here that the induction of autophagy in hepatic stellate cells occurs in response to injurious stimuli to the liver. Specifically, following CCl4 and TAA-induced liver injury, primary activated stellate cells up-regulate autophagy in parallel to the induction of fibrogenic markers.
Therefore, we hypothesized that autophagy may be essential to maintain hepatic stellate cell activation following metabolic stress during liver injury. In untreated mice with autophagy-defective hepatic stellate cells, liver architecture was preserved and stellate cell ultrastructure was intact, indicating that suppression of basal autophagy in stellate cells does not impact the maintenance of the quiescent state. However, following liver injury, when cells face an increased demand for intracellular energy, blocking autophagy impairs their activation and fibrogenic activity.
Quiescent hepatic stellate cells contain retinyl esters and triglycerides at similar concentrations,24 which together account for 75% of the total lipid content within cytoplasmic LDs.5 Loss of these droplets is a characteristic feature of stellate cell activation in liver injury,4,25 yet its functional link to this response has been obscure. Interestingly, autophagy contributes to the intracellular catabolism of lipids in hepatocytes, fibroblasts,6 and neurons,26 but a link to the broader tissue context has been lacking. Moreover, pharmacologic or genetic inhibition of autophagy in hepatocytes leads to reduced rates of β-oxidation and marked lipid accumulation in cytosolic LDs.6 Autophagy is an ATP-dependent process,27 and autophagy inhibition reduces mitochondrial β-oxidation rates6,17 and energy production. However, the role of LDs in providing energy to support stellate cell activation has not previously been examined. As shown in hepatocytes, here we show that autophagy-deficient stellate cells may be unable to process LDs by acid lipases, resulting in LD accumulation and decreased FFA availability, leading to decreased mitochondrial β-oxidation and ATP production. Moreover, autophagy-independent inhibition of β-oxidation with etomoxir mimics the effect of blocking autophagy on activation, fibrogenesis, and lipid accumulation, implicating β-oxidation and ATP production in stellate cell activation. Our findings, together with a prior study indicating that the absence of retinoid storage fails to affect hepatic stellate cell activation,28 suggest a critical function for lipids stored as triglycerides rather than retinoids in hepatic stellate cell activation.
Studies have linked FFAs to stimulation of au-tophagy29,30 and to complementing autophagy inhibition.17 Our previous study has shown that OA per se has no effect on hepatic stellate cell activation in autophagy-competent cells.31 However, the decrease in fibrogenesis resulting from autophagy inhibition can be partially rescued by exogenous OA complementation; conversely, in autophagy-competent cells, adding OA does not further augment fibrogenesis.
Our results suggest that autophagy provides energy that is essential to support stellate cell activation through LD mobilization, liberation of FFAs, and mitochondrial β-oxidation. These events allow the activated cell to maintain energy homeostasis in the face of increasing cellular energy demands conferred by fibrogenesis and proliferation. The findings provide a novel framework for understanding hepatic disease and the regulation of stellate cell activation. Moreover, because blocking autophagy in fibrogenic cells from other organs also attenuates fibrogenesis, autophagy appears to be an evolutionarily conserved core pathway1 that may contribute to the fibrotic response in a wide range of tissues.
Supplementary Material
Acknowledgments
The authors thank Beca Juan Rodés (Asociación Española para el Estudio del Hígado) and Beca de Formació al estranger (Societat Catalana de digestologia de l’Acadèmia de Ciéncies Médiques de Catalunya I de Balears) for support, Joseph Samet for technical assistance, Dr Masaaki Komatsu for the Atg7F/F mice, Dr Meena Bansal for the primary human stellate cells, Dr Carol Fenghali-Bostwick for the primary human pulmonary cells from normal and idiopathic pulmonary fibrosis lungs, Dr Erwin Bottinger for the mouse mesangial cells, and Dr Noboru Mizushima for the mouse embryonic fibroblasts.
Funding
Supported by US National Institutes of Health grants (RO1DK56621, R01 AA020709, KO5AA018408, P20AA017067, 5T32GM007280, R01DK044234, R01DK061498, and R01NS060123).
Abbreviations used in this paper
- ADRP
adipocyte differentiation-related protein
- AV
autophagic vacuole
- BSA
bovine serum albumin
- CQ
chloroquine
- EM
electron microscopy
- FFA
free fatty acid
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFAP
glial fibrillary acid protein
- LC3
microtubule-associated light chain 3
- LD
lipid droplet
- 3-MA
3-methyladenine
- MMP2
matrix metalloproteinase 2
- OA
oleic acid
- ORO
oil red O
- PDGFR
platelet-derived growth factor receptor
- SMA
smooth muscle actin
- TAA
thioacetamide
Footnotes
Conflicts of interest
The authors disclose no conflicts.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at doi: 10.1053/j.gastro.2011.12.044.
References
- 1.Mehal WZ, Iredale J, Friedman SL. Scraping fibrosis: expressway to the core of fibrosis. Nat Med. 2011;17:552–553. doi: 10.1038/nm0511-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaner WS, O’Byrne SM, Wongsiriroj N, et al. Hepatic stellate cell lipid droplets: a specialized lipid droplet for retinoid storage. Biochim Biophys Acta. 2009;1791:467–473. doi: 10.1016/j.bbalip.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blomhoff R, Berg T. Isolation and cultivation of rat liver stellate cells. Methods Enzymol. 1990;190:58–71. doi: 10.1016/0076-6879(90)90009-p. [DOI] [PubMed] [Google Scholar]
- 8.Guo J, Loke J, Zheng F, et al. Functional linkage of cirrhosis-predictive single nucleotide polymorphisms of Toll-like receptor 4 to hepatic stellate cell responses. Hepatology. 2009;49:960–968. doi: 10.1002/hep.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satriano JA, Banas B, Luckow B, et al. Regulation of RANTES and ICAM-1 expression in murine mesangial cells. J Am Soc Nephrol. 1997;8:596– 603. doi: 10.1681/ASN.V84596. [DOI] [PubMed] [Google Scholar]
- 10.Pilewski JM, Liu L, Henry AC, et al. Insulin-like growth factor binding proteins 3 and 5 are overexpressed in idiopathic pulmonary fibrosis and contribute to extracellular matrix deposition. Am J Pathol. 2005;166:399– 407. doi: 10.1016/S0002-9440(10)62263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh R, Xiang Y, Wang Y, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komatsu M, Wang QJ, Holstein GR, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Jung Y, Omenetti A, et al. Fate-mapping evidence that hepatic stellate cells are epithelial progenitors in adult mouse livers. Stem Cells. 2008;26:2104–2113. doi: 10.1634/stemcells.2008-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blommaart EF, Krause U, Schellens JP, et al. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 16.Blommaart EF, Luiken JJ, Meijer AJ. Regulation of hepatic protein degradation. Contrib Nephrol. 1997;121:101–108. doi: 10.1159/000059859. [DOI] [PubMed] [Google Scholar]
- 17.Heaton NS, Perera R, Berger KL, et al. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thoen LF, Guimaraes EL, Dolle L, et al. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55:1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehrpour M, Esclatine A, Beau I, et al. Autophagy in health and disease. 1. Regulation and significance of autophagy: an overview. Am J Physiol Cell Physiol. 2010;298:C776–C785. doi: 10.1152/ajpcell.00507.2009. [DOI] [PubMed] [Google Scholar]
- 21.Cuervo AM, Knecht E, Terlecky SR, et al. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol. 1995;269:C1200–C1208. doi: 10.1152/ajpcell.1995.269.5.C1200. [DOI] [PubMed] [Google Scholar]
- 22.Jones RG, Plas DR, Kubek S, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Ding WX, Li M, Chen X, et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139:1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senoo H, Yoshikawa K, Morii M, et al. Hepatic stellate cell (vitamin A-storing cell) and its relative—past, present and future. Cell Biol Int. 2010;34:1247–1272. doi: 10.1042/CBI20100321. [DOI] [PubMed] [Google Scholar]
- 25.Friedman SL, Wei S, Blaner WS. Retinol release by activated rat hepatic lipocytes: regulation by Kupffer cell-conditioned medium and PDGF. Am J Physiol. 1993;264:G947–G952. doi: 10.1152/ajpgi.1993.264.5.G947. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Vicente M, Talloczy Z, Wong E, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol. 2004;36:2445–2462. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Kluwe J, Wongsiriroj N, Troeger JS, et al. Absence of hepatic stellate cell retinoid lipid droplets does not enhance hepatic fibrosis but decreases hepatic carcinogenesis. Gut. 2011;60:1260–1268. doi: 10.1136/gut.2010.209551. [DOI] [PubMed] [Google Scholar]
- 29.Komiya K, Uchida T, Ueno T, et al. Free fatty acids stimulate autophagy in pancreatic beta-cells via JNK pathway. Biochem Biophys Res Commun. 2010;401:561–567. doi: 10.1016/j.bbrc.2010.09.101. [DOI] [PubMed] [Google Scholar]
- 30.Choi SE, Lee SM, Lee YJ, et al. Protective role of autophagy in palmitate-induced INS-1 beta-cell death. Endocrinology. 2009;150:126–134. doi: 10.1210/en.2008-0483. [DOI] [PubMed] [Google Scholar]
- 31.Lee TF, Mak KM, Rackovsky O, et al. Downregulation of hepatic stellate cell activation by retinol and palmitate mediated by adipose differentiation-related protein (ADRP) J Cell Physiol. 2010;223:648– 657. doi: 10.1002/jcp.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.