Abstract
We used a genetic method, the yeast substrate-trapping system, to identify substrates for protein tyrosine phosphatases ζ (PTPζ/RPTPβ). This method is based on the yeast two-hybrid system, with two essential modifications: conditional expression of protein tyrosine kinase v-src (active src) to tyrosine-phosphorylate the prey proteins and screening by using a substrate-trap mutant of PTPζ (PTPζ-D1902A) as bait. By using this system, several substrate candidates for PTPζ were isolated. Among them, GIT1/Cat-1 (G protein-coupled receptor kinase-interactor 1/Cool-associated, tyrosine-phosphorylated 1) was examined further. GIT1/Cat-1 bound to PTPζ-D1902A dependent on the substrate tyrosine phosphorylation. Tyrosine-phosphorylated GIT1/Cat-1 was dephosphorylated by PTPζ in vitro. Immunoprecipitation experiments indicated that PTPζ-D1902A and GIT1/Cat-1 form a stable complex also in mammalian cells. Immunohistochemical analyses revealed that PTPζ and GIT1/Cat-1 were colocalized in the processes of pyramidal cells in the hippocampus and neocortex in rat brain. Subcellular colocalization was further verified in the growth cones of mossy fibers from pontine explants and in the ruffling membranes and processes of B103 neuroblastoma cells. Moreover, pleiotrophin, a ligand for PTPζ, increased tyrosine phosphorylation of GIT1/Cat-1 in B103 cells. All these results indicate that GIT1/Cat-1 is a substrate molecule of PTPζ.
Receptor-type protein tyrosine phosphatases (PTPs) are a structurally and functionally diverse family of enzymes (1, 2). To understand the functional roles of receptor-type PTPs, identification of their specific intracellular signaling cascade is essential. However, progress in this area has been limited to date because of the lack of efficient methods to isolate substrate molecules for receptor-type PTPs.
The two-hybrid system has been proven to be a powerful method for detecting molecular interactions between two proteins in yeast. The binding partners to SH2-containing cytosolic PTPs have been identified by a modified two-hybrid system using yeast cells expressing src tyrosine kinase (3, 4). However, direct screening of the substrate molecules for receptor-type PTPs by using such a modified two-hybrid system has not been reported. In this study, we attempted to identify the substrate(s) of PTPζ/RPTPβ by using a conditional yeast two-hybrid system, which we named the yeast substrate-trapping system. In this system, a substrate-trap mutant of PTPζ was adopted as bait that stably associates with its substrate (5), and active src (v-src) was conditionally expressed in yeast cells to tyrosine-phosphorylate the prey proteins because scarcely any tyrosine-phosphorylation occurs in normal yeast (6, 7).
PTPζ is a receptor-type PTP, expressed predominantly in the brain, that is synthesized as a chondroitin sulfate proteoglycan (8–12). There exist three splice variants of this molecule (13): a full-length transmembrane form (PTPζ-A); a short transmembrane form (PTPζ-B); and a soluble form (PTPζ-S), which is also known as 6B4 proteoglycan/phosphacan (8–12). It has been reported that the extracellular domain of PTPζ binds various cell adhesion molecules (Nr-CAM, L1/Ng-CAM, F3/contactin, N-CAM, and TAG1/axonin-1), growth factors [pleiotrophin (PTN)/heparin-binding growth-associated molecule (HB-GAM), midkine, and fibroblast growth factor-2 (FGF-2)], and extracellular matrix molecules (tenascin-C and tenascin-R) (14–18). However, little is known about the signal transduction mechanism of PTPζ, although sodium channel and β-catenin have been reported as candidates of the substrate for PTPζ very recently (19, 20). In addition, it is difficult to explain the variegated roles expected for PTPζ only with these two substrates.
We here identified GIT1/Cat-1 (G protein-coupled receptor kinase-interactor 1/Cool-associated, tyrosine-phosphorylated 1) as a substrate of PTPζ by using the yeast substrate-trapping system. The association between the substrate-trapping mutant of PTPζ and GIT1/Cat-1 was verified in NIH 3T3 and v-src-transformed 3Y1 cells by immunoprecipitation experiments. Subcellular colocalization of PTPζ and GIT1/Cat-1 was observed in the growth cones of mossy fibers from pontine explants and in the B103 neuroblastoma cells. Tyrosine phosphorylation of GIT1/Cat-1 in B103 cells was increased by stimulation with PTN. We also note that the yeast substrate-trapping system is widely applicable to isolation of substrate molecules for other PTPs.
Materials and Methods
The Yeast Substrate-Trapping System.
The cDNA fragment encoding the whole intracellular region (ICR) of rat PTPζ (amino acid residues 1748–2316; GenBank accession no. U09357), with a mutation by replacement of Asp at position 1902 to Ala (PTPζICR-D1902A), was prepared by PCR using specific primers (5′-GCTATGGGTGTTCCCGAGTACTCCCTAC-3′ and 5′-AGGCCACTGGGTGTAATGATACTGAGTG-3′) and the pBTM116 construct containing the sequence for the intracellular region of PTPζ (21) as a template. The fragments containing coding sequences for LexA and PTPζICR-D1902A were excised by SphI digestion and inserted into the SphI site of the pBridge vector (CLONTECH) to generate pBridgeLexA/ζICR-D1902A. The cDNA fragment encoding v-src was amplified by PCR using specific primers containing a NotI site (5′-GTGCAGGCGGCCGCCATGGGGAGTAGCAAG-3′ and 5′-ATTTTGCGGCCGCACTACTCAGCGACCTCC-3′) and the pBaBePurov-src plasmid (ref. 22; kindly provided by H. Sabe, Osaka Bioscience Institute, Osaka) as a template. This fragment was inserted into the NotI site of pBridgeLexA/ζICR-D1902A to generate pBridgeLexA/ζICR-D1902A/v-src. This construct thus expresses both the bait protein and v-src, the v-src expression being controlled by the Met25 promoter. This promoter responds to methionine levels in the medium; it is repressed in the presence of 1 mM methionine and expressed in the absence of methionine.
pBridgeLexA/ζICR-D1902A/v-src was transfected into Saccharomyces cerevisiae strain L40, which harbors reporter genes HIS3 and LacZ under the control of an upstream LexA-binding site, and transformants were selected by growth on tryptophan-minus plates in the presence of 1 mM methionine. To prepare competent cells, a transformant was cultured with shaking at 30°C for 48 h up to the stationary phase (OD600 > 1.5). The library screening was performed as described in ref. 23 and the manufacturer's manual (MATCHMAKER library user manual; CLONTECH). Approximately 2.6 × 106 and 1.1 × 106 clones were screened by using a young adult rat (10–12 weeks old) brain library and a 17-day mouse embryo MATCHMAKER cDNA library (CLONTECH), respectively. Positive clones were selected on plates lacking leucine, tryptophan, histidine, and methionine, and verified by a β-galactosidase filter-lift assay in the presence or absence of 1 mM methionine. The clones that showed an increase in blue color development induced by v-src were selected as candidate molecules for PTPζ substrates.
Antibodies.
The SalI fragment (nucleotide residues 751-1665) of GIT1/Cat-1 cDNA was fused with the glutathione S-transferase (GST) gene by using pGEX-4T-2 (Amersham Pharmacia), and the recombinant protein expressed in E. coli was purified by glutathione affinity chromatography according to the manufacturer's instructions. Rabbits were immunized with the purified GST-GIT1/Cat-1 protein, using Freund's complete adjuvant. The antibodies bound to the GST-GIT1/Cat-1-immobilized column were collected, and next applied to the GST-immobilized column. The flow-through fractions were used as anti-GIT1/Cat-1 antibodies. Biotinylated anti-GIT1/Cat-1 antibody was prepared by using the EZ-Link Sulfo-NHS-biotinylation Kit according to the manufacturer's instructions (Pierce).
Phosphatase Assay.
GST fusion proteins of the intracellular region of PTPζ (GST-PTPζICR) and GIT1/Cat-1 (amino acid residues 251–555; GST-GIT1/Cat-1) were expressed in E. coli by using pGEX-4T and pGEX-6P vectors (Amersham Pharmacia), respectively, and purified by glutathione affinity chromatography. GST-GIT1/Cat-1 fusion proteins were further treated with PreScission protease (Amersham Pharmacia), and GIT1/Cat-1 proteins were separated from the protease and GST by passing through a glutathione-Sepharose column. Purified GIT1/Cat-1 and reduced, carboxymethylated, and maleylated lysozyme (RCM lysozyme) were labeled at tyrosine by using [γ-32P]ATP and p60c-src (Upstate Biotechnology, Lake Placid, NY) as described (11). The phosphatase assay using 0.2 μM 32P-labeled substrates and 2 ng/10 μl of GST-PTPζICR was performed as described (11).
Expression in Cell Lines.
The full-length PTPζ-B cDNA (13) was inserted into the mammalian expression vector pZeoSV2 (Invitrogen) to yield pZeoPTPζ. Asp-1902 → Ala mutant expression plasmid, pZeoPTPζ-D1902A, was prepared by PCR with specific primers as described above, using pZeoPTPζ as a template. A DNA fragment covering the coding region of rat GIT1/Cat-1 was amplified by reverse transcription (RT)–PCR using specific primers (5′-CGGCCGAATTCCCGCTGAGGATGTCCCGGA-3′ and 5′-GGTAAGTCGACGGAAAGTGGTCACTGTTTC-3′) to make restriction sites for cloning. This fragment was inserted into the EcoRI/SalI sites of the expression vector pcDNA3.1 (Invitrogen) to yield pcDNAGIT1.
The plasmids were transfected into NIH 3T3 cells in 60-mm dishes by using Lipofectamine Plus Reagent (GIBCO/BRL). At 24 h after transfection, cells were serum-starved in DMEM (Nissui Seiyaku, Tokyo) containing 0.5% FCS for 24 h. Starved cells were trypsinized, washed with DMEM containing 0.5 mg/ml trypsin inhibitor (Wako Biochemicals, Osaka), and resuspended in DMEM. Cells were next plated and cultured for 1 h on the tissue culture plates pretreated with fibronectin (10 μg/ml, Wako Biochemicals). Then, cells were lysed with the lysis buffer [1% Nonidet P-40/20 mM Tris⋅HCl, pH 8.0/137 mM NaCl/1 mM phenylmethylsulfonyl fluoride (PMSF)/10 μg/ml leupeptin/10 μg/ml pepstatin A/0.1 mM sodium vanadate/1 mM NaF] for 20 min at 4°C. After centrifugation at 10,000 × g for 15 min, the supernatants were used for immunoprecipitation and Western blotting as described (13). Native 3Y1 and v-src-transformed 3Y1 cells were also transfected with the plasmids and cultured for 40 h. Then, cells were lysed and subjected to immunoprecipitation as above. Protein concentration was determined with a Micro BCA kit (Pierce), using BSA as a standard.
Immunohistochemistry.
Serial sections (5-μm-thick) of formalin-fixed and paraffin-embedded rat brains (Sprague-Dawley, female, 200 g) were deparaffinized in xylene and rehydrated in a graded ethanol series. The deparaffinized sections were washed with PBS, and microwaved in 10 mM citrate buffer, pH 6.0, for 5 min in a conventional microwave oven (500 W, MRO-N75, Hitachi, Tokyo). The sections were treated with 3.0% H2O2/PBS for 10 min to inactivate intrinsic peroxidase, blocked with 4% skim milk for 10 min, and then incubated overnight at 4°C with one of the following primary antibodies: 2.5 μg/ml monoclonal mouse anti-RPTPβ antibody (Transduction Laboratories, Lexington, KY) or 5.0 μg/ml purified rabbit anti-GIT1/Cat-1 antibody. The binding of specific antibodies was detected with an ABC-peroxidase kit (Vector Laboratories), using 3,3′-diaminobenzidine (DAB) and H2O2 as substrates.
PTN Beads Assay and Immunocytochemistry.
Latex beads (particle diameter, 3 μm; Sigma) were coated with PTN (20 μg/ml) or BSA (20 μg/ml) overnight at 4°C. The beads were washed three times with PBS before use. Wells of 48-well plates were coated with 40 μg/ml poly-L-lysine, and B103 cells (1 × 104 cells in 200 μl of DMEM containing 10% FCS) were added into each well and cultured for 48 h. The cells were then serum-starved by culturing in DMEM, containing 0.5% FCS, for 24 h and stimulated with the coated beads for various periods. Finally, the cells were lysed and subjected to the immunoprecipitation as described above.
Serum-starved B103 cells were incubated with the PTN-coated beads for 30 min. After fixation with 4% paraformaldehyde/PBS for 15 min, cells were permeabilized with 0.2% Triton X-100 (Wako Biochemicals)/PBS for 10 min. After blocking with 3% skim milk/PBS, cells were treated in the following way: incubation overnight with anti-6B4 proteoglycan antiserum (24) (anti-6B4 PG; 1/200) at 4°C, for 1 h with Alexa Fluor 594 anti-rabbit IgG conjugate (Molecular Probes) (1/100), and overnight with biotinylated anti-GIT1/Cat-1 antibody (1.0 μg/ml) at 4°C. Cells were then processed with a TSA Indirect kit according to the manufacturer's instructions (DuPont/NEN), and incubated with Alexa Fluor 488 streptavidin conjugate (Molecular Probes; 1/100). Finally, cells were mounted in PermaFluor and observed with a Zeiss fluorescence microscope. Pontine explant was cultured according to the method described by Hall et al. (25).
Results
The Yeast Substrate-Trapping System to Dissect the Signal Transduction Pathway of PTPζ.
The yeast substrate-trapping system is based on the yeast two-hybrid system with conditionally expressed tyrosine kinase. Because PTPζ is known to be enriched in growth cones of neurons, where src family kinases also play important regulatory functions (15), we used v-src to tyrosine phosphorylate the prey proteins. In addition, the whole intracellular domain of PTPζ with the Asp-1902 → Ala mutation (PTPζICR-D1902A) was used as bait. We anticipated that its affinity for a substrate(s) remains similar to that of the wild type, and that the PTPζ-substrate complex, once formed, would be stable enough to withstand screening because the catalytic activity is impaired (5). The trapping mechanism of the system is represented schematically in Fig. 1A. It was reported that expression of v-src in yeast S. cerevisiae resulted in rapid cell death (26, 27); however, the conditional expression of v-src under the control of the Met25 promoter yielded a stable yeast cell growth (data not shown) and a marked increase of tyrosine phosphorylation of yeast proteins (Fig. 2A).
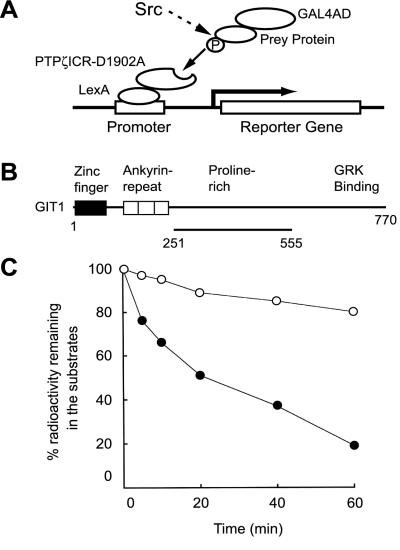
Figure 1.
Schematic representations of the yeast substrate-trapping system and GIT1/Cat-1 protein isolated by this system. (A) In the presence of 1 mM methionine when v-src is not induced, only the standard two-hybrid bindings occur (not shown). In the absence of methionine, prey proteins (substrates for PTPζ) could be phosphorylated by the induced v-src, and trapped by the bait consisting of the whole intracellular domain with an Asp-1902 → Ala mutation of PTPζ (PTPζICR-D1902A). This complex formation leads to activation of transcription of the reporter genes, HIS3 and LacZ. We performed screenings in two steps and selected the colonies that showed an increase in blue color development on expression of v-src (see Materials and Methods). (B) GIT1/Cat-1 contains a zinc-finger region, three ankyrin-repeat regions, a potential Src-homology 3 (SH3) binding site and a G protein-coupled receptor kinase (GRK) binding site. The region encoded by clone 10–1 is shown by the solid line underneath. (C) GIT1/Cat-1 (amino acid residues 251–555; ●) and RCM lysozyme (○) were tyrosine phosphorylated by p60c-src and then incubated with GST-PTPζICR for various periods. GST-PTPζICR efficiently dephosphorylated GIT1/Cat-1.
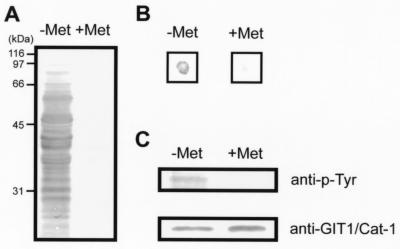
Figure 2.
Substrate identification with the yeast substrate-trapping system. (A) Yeast proteins were highly tyrosine-phosphorylated only when v-src was induced in the absence of methionine (−Met), whereas almost no tyrosine-phosphorylation was observed in the presence of methionine (+Met). Yeast lysates (70 μg) were analyzed by Western blotting with antiphosphotyrosine antibody. (B) β-Galactosidase filter-lift assay for clone 10–1. A filter placed on the plate lacking leucine, tryptophan, and methionine (Left) and lacking leucine and tryptophan (Right) was streaked directly with clone 10–1 and pBridgeLexA/ζICR-D1902A/v-src cotransformants, incubated at 30°C for 2 days, and developed for 3 h for color detection. (C) The prey consisting of the GAL4 activation domain fused to GIT1/Cat-1 was actually tyrosine-phosphorylated by v-src induction (−Met). Yeast cell lysates (750 μg) were immunoprecipitated with anti-GIT1/Cat-1 antiserum and analyzed by Western blotting with antiphosphotyrosine antibody (Upper) and anti-GIT1/Cat-1 antiserum (Lower).
Screening of candidate clones was performed in two steps. In the first step, yeast colonies were selected on the leucine-, tryptophan-, histidine-minus plate in the absence of methionine. As the second step, the positive clones thus identified were further examined by the β-galactosidase filter-lift assay in the presence or absence of methionine. In this assay, we selected the colonies that showed increased blue color development on the induction of v-src expression.
In the first-step screening, 5 and 23 positive clones were isolated from a rat brain library (2.6 × 106 independent clones) and a mouse embryo (1.1 × 106 independent clones) cDNA library, respectively. Many of them, including PSD-95/SAP90 family members, did not show any change in blue color development by v-src induction in the second-step screening (data not shown). Using the conventional yeast two-hybrid binding assay, we previously showed that the C terminus of PTPζ binds to the second PDZ domain of PSD-95/SAP90 family members (PSD-95/SAP90, SAP97/hdlg and SAP102) (21). The fact that the interaction between PTPζ and PSD-95/SAP90 family members was not affected by the v-src expression indicated that the basal two-hybrid binding system was not influenced by the tyrosine phosphorylation of many yeast proteins.
After the second-step screening, two and eight positive clones remained from the rat brain and mouse embryo cDNA libraries, respectively. Among them, clone 10–1 from the rat brain cDNA library showed the most remarkable increase in blue color development by v-src induction in the β-galactosidase filter-lift assay (Fig. 2B). Sequence analysis showed that clone 10–1 encodes a partial sequence of GIT1/Cat-1 (amino acid residues 251–555; Fig. 1B; refs. 28 and 29). GIT1/Cat-1 contains an ADP-ribosylation factor–GTPase-activating protein (ARF-GAP; ref. 1) domain at its N terminus that shares the zinc-finger region, and binds to G protein-coupled receptor kinases (GRKs) through a region located near the C terminus (28). The encoded region in clone 10–1 corresponded to the proline-rich region in-between these two domains, and contained six tyrosine residues. GIT1/Cat-1 is known to be tyrosine-phosphorylated by Src and Fak (29). The prey, which is a fusion protein consisting of GIT1/Cat-1 and GAL4 activation domain, was actually tyrosine-phosphorylated by induction of v-src (Fig. 2C). Incidentally, the GAL4 activation domain and LexA-coding sequence do not contain any tyrosine phosphorylation sites for v-src. To test whether PTPζ actually dephosphorylates GIT1/Cat-1, tyrosine-phosphorylated GIT1/Cat-1 protein (amino acid residues 251–555) was incubated with GST-PTPζICR. As shown in Fig. 1C, GST-PTPζICR efficiently dephosphorylated GIT1/Cat-1, whereas it did RCM lysozyme very poorly. Thus, we concluded that GIT1/Cat-1 is a promising physiological candidate for PTPζ substrate, and obtained the full-length clone by PCR amplification.
Interaction of PTPζ and GIT1/Cat-1 in Mammalian Cells.
It is known that tyrosine phosphorylation of GIT1/Cat-1 increases during cell spreading on fibronectin-coated substrates (29). We prepared NIH 3T3 cells cotransfected with GIT1/Cat-1 and PTPζ-D1902A expression plasmids, and cultured them on fibronectin-coated plates. Next, coimmunoprecipitation experiments were performed by using either antiserum against the extracellular region of PTPζ (anti-6B4 PG antiserum) or anti-GIT1/Cat-1 antiserum. As shown in Fig. 3 A and B, PTPζ and GIT1/Cat-1 were coprecipitated in the reciprocal immunoprecipitation experiments, whereas comparable amounts of preimmune serum failed to coprecipitate GIT1/Cat-1 or PTPζ. We could not detect src proteins in the immunoprecipitate with anti-6B4 PG antiserum, suggesting that the association between PTPζ and GIT1/Cat-1 is not mediated by src (data not shown).
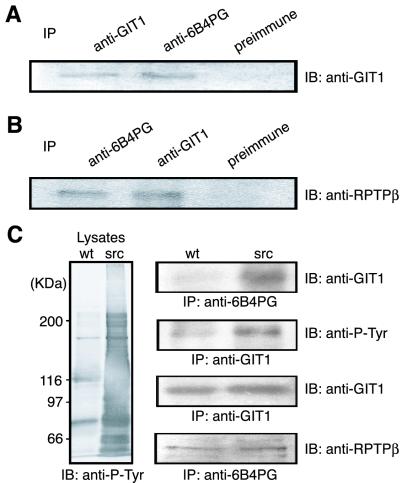
Figure 3.
Coimmunoprecipitation of PTPζ and GIT1/Cat-1. (A and B) NIH 3T3 cells were cotransfected with GIT1/Cat-1 and PTPζ-D1902A expression plasmids. Following the transfection, cells were plated on fibronectin. The cell lysates were immunoprecipitated with the appropriate antibodies, and the precipitated proteins were detected with anti-GIT1/Cat-1 (A) and anti-RPTPβ (B) antibodies. (C) Native 3Y1 (wt) and v-src-transformed 3Y1 (src) cells were cotransfected with GIT1/Cat-1 and PTPζ-D1902A expression plasmids. The cell lysates were analyzed by Western blotting using antiphosphotyrosine antibody (Left). The cell lysates were subjected to immunoprecipitation (Right) with anti-6B4 PG (first blot) or anti-GIT1/Cat-1 (second blot) antibodies. Western blotting indicated that GIT1/Cat-1 is highly tyrosine-phosphorylated (second blot) and is present in complex with PTPζ-D1902A (first blot) in the v-src-transformed 3Y1 cells. Both cells expressed comparable amounts of GIT1/Cat-1 and PTPζ-D1902A (third and fourth blots, respectively).
We further examined whether the interaction between PTPζ and GIT1/Cat-1 depends on the tyrosine-phosphorylation of GIT1/Cat-1. Native 3Y1 and v-src-transformed 3Y1 cells were cotransfected with GIT1/Cat-1 and PTPζ-D1902A expression plasmids. In the v-src-transformed cells, GIT1/Cat-1 was highly tyrosine-phosphorylated, whereas a very low level of tyrosine phosphorylation was observed in the normal cells (Fig. 3C). Immunoprecipitation experiments demonstrated that the complex formation between PTPζ-D1902A and GIT1/Cat-1 was detected only in the v-src-transformed cells (Fig. 3C), indicating that the association between these proteins depends on the tyrosine phosphorylation of GIT1/Cat-1.
Immunohistochemical Localization of PTPζ and GIT1/Cat-1.
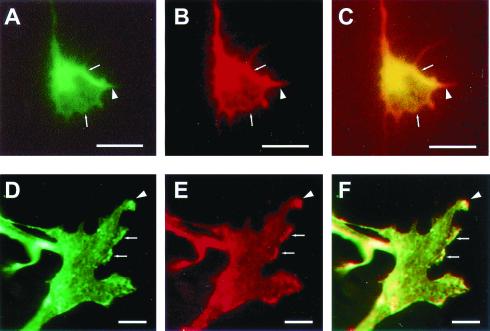
The distribution of GIT1/Cat-1 mRNA in rat tissues suggests that GIT1/Cat-1 is widely expressed in a variety of tissues, including brain (28). Immunohistochemical staining with a polyclonal antibody against GIT1/Cat-1 revealed that processes of the pyramidal cells in the hippocampus (Fig. 4A) and neocortex (Fig. 4C) were stained. Significant stainings with a monoclonal antibody against PTPζ were also observed in the processes of hippocampal and cortical pyramidal neurons (Fig. 4 B and D). Next, double-labeling experiments were performed on mossy fibers of pontine explants, which actively express PTPζ (10). Explants of pontine nuclei from postnatal-day-1 mice extended many axons on laminin-coated substrates. As shown in Fig. 5 A–C, the pontine axons were densely stained with both antibodies against PTPζ and GIT1/Cat-1. Colocalization of the two proteins was especially evident in the peripheral regions and filopodial processes of the growth cones. We also examined B103 neuroblastoma cells that endogenously express PTPζ and GIT1/Cat-1 by double labeling. The immunoreactivities by the antibodies against PTPζ and GIT1/Cat-1 were colocalized at the processes and the ruffling membranes of B103 cells (Fig. 5 D–F). These results suggest that there exists a recruitment system of GIT1/Cat-1 and PTPζ to specific membrane domains in neurons. Notwithstanding the colocalization, they were not coimmunoprecipitated from B103 cells (data not shown). This is probably because endogenous PTPζ in B103 cells is active and dephosphorylates GIT1/Cat-1, and accordingly does not make a stable complex with GIT1/Cat-1. Indeed, almost no tyrosine-phosphorylation of GIT1/Cat-1 was observed under the culture conditions used for immunocytochemistry and immunoprecipitation in our study (data not shown).
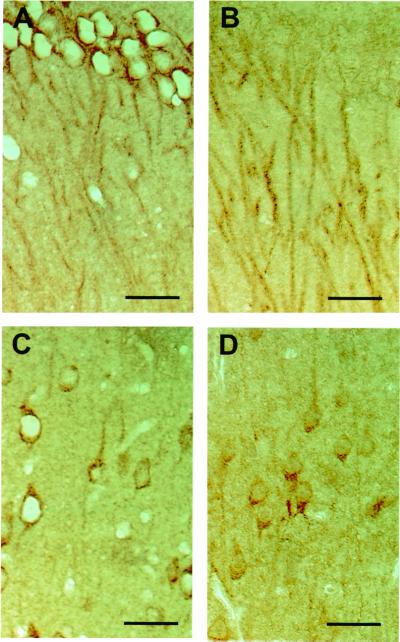
Figure 4.
Immunohistochemical localization of GIT1/Cat-1 and PTPζ in the rat brain. Rat brain serial sections were stained with anti-GIT1/Cat-1 (A and C) and anti-RPTPβ (B and D) antibodies, respectively. (A and B) Hippocampal CA1 region. The upper portion is the pyramidal cell layer. (C and D) Layer V of the frontal cortex. Layer IV is above. (Scale bars, 50 μm.)
Figure 5.
Immunohistochemical localization of GIT1/Cat-1 and PTPζ in the growth cones and B103 neuroblastoma cells. The mossy fibers extending from pontine explants were doubly immunostained with anti-GIT1/Cat-1 (A) and anti-6B4 PG (B) antibodies. A combined image is shown in C. The colocalization of both proteins was evident at the peripheral regions (arrows) and filopodial processes (arrowhead) of the growth cones. B103 cells were doubly immunostained with anti-GIT1/Cat-1 (D) and anti-6B4 PG (E) antibodies. A combined image is shown in F. GIT1/Cat-1 and PTPζ were colocalized in the processes (arrowhead) and the ruffling membranes (arrows) of the cells. [Scale bars, 5 μm (A–C) and 10 μm (D–F).]
PTN Stimulates Tyrosine Phosphorylation of GIT1/Cat-1.
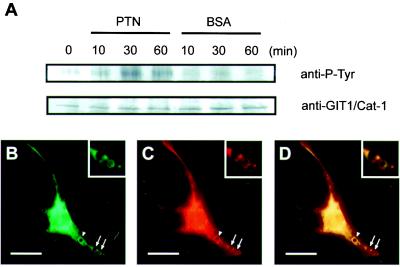
We have shown that PTN interacts with PTPζ as a ligand (14, 15, 19). To examine whether GIT1/Cat-1 is involved in the signal transduction of PTN through PTPζ, a beads assay was performed by using B103 cells. Ligand-coated beads attached to the cell surface trigger the signal transduction and also lead to visualization of the local signal transduction responses around the beads (30). When B103 cells were stimulated with PTN-coated beads, a transient increase in GIT1/Cat-1 tyrosine phosphorylation was detected with a maximum at around 30 min after the stimulation (Fig. 6A). The tyrosine-phosphorylation level of GIT1/Cat-1 was still higher than the basal level at 60 min after stimulation (Fig. 6A). In contrast, BSA-coated beads used as a control scarcely influenced the phosphorylation level of GIT1/Cat-1 (Fig. 6A). Double labeling of B103 cells incubated with PTN-coated beads indicated that both PTPζ and GIT1/Cat-1 were recruited to the surrounding regions of the beads (Fig. 6 B–D), suggesting that GIT1/Cat-1 is associated with PTPζ in the signal transduction pathway of PTN.
Figure 6.
PTN stimulates tyrosine phosphorylation of GIT1/Cat-1 in the B103 cells. (A) Time course of the tyrosine phosphorylation of GIT1/Cat-1 in response to polystyrene beads coated with PTN (20 μg/ml) or BSA (20 μg/ml). B103 cell lysates (800 μg) were immunoprecipitated with anti-GIT1/Cat-1 antiserum and analyzed by Western blotting with antiphosphotyrosine antibody (Upper) and anti-GIT1/Cat-1 antibody (Lower). PTN-coated beads induced accumulations of GIT1/Cat-1 (B) and PTPζ (C) around the beads after 30 min of incubation (arrows and arrowheads). The combined image (D) shows that the two proteins were coaggregated around the beads. (Insets) A magnified view of the beads marked by arrows. A similar accumulation was also observed around BSA-coated beads (data not shown). [Scale bars, 20 μm (B–D).]
Discussion
In the present study, we developed the yeast substrate-trapping system to dissect the signal transduction pathway of PTPζ. In this system, we used the whole intracellular domain of PTPζ with the Asp-1902 → Ala mutation as bait. This Asp residue is known to act as a general acid to facilitate protonation of the phenolic oxygen atom of the tyrosyl leaving group (31, 32). In addition to this type of mutation, alteration of the catalytically essential Cys to Ser or Ala was also used for substrate-trapping of some PTPs by pulldown assay (31, 33). We also tried the Cys-1934 → Ser mutant of PTPζ as bait, but could not isolate any positive clones from the screening of ≈2.5 × 106 and 2.3 × 106 clones of the rat brain and mouse embryo cDNA libraries, respectively.
Using this system, we identified GIT1/Cat-1 as a molecule that associates with PTPζ dependent on the v-src induction. Our experiments showed that (i) PTPζ dephosphorylated tyrosine-phosphorylated GIT1/Cat-1 in vitro; (ii) GIT1/Cat-1 and PTPζ-D1902A were coimmunoprecipitated from NIH 3T3 cell lysates; (iii) the association between PTPζ-D1902A and GIT1/Cat-1 depended on the tyrosine-phosphorylation of GIT1/Cat-1 in 3Y1 cells; (iv) GIT1/Cat-1 and PTPζ were colocalized in the growth cones of pontine axons and in B103 cells; and (v) PTN, a ligand for PTPζ, increased tyrosine phosphorylation of GIT1/Cat-1 in B103 cells. These results indicate that PTPζ is functionally coupled with GIT1/Cat-1 in the signal transduction pathway of PTN. In B103 cells, colocalization of endogenous PTPζ and GIT1/Cat-1 was observed immunocytochemically independent of the tyrosine phosphorylation of GIT1/Cat-1, where the association of these proteins was not detected by immunoprecipitation. This suggests that these proteins are constitutively associated weakly in the signaling complex, and that the tyrosine phosphorylation level of GIT1/Cat-1 is regulated by the ligand binding to PTPζ. Recently, Meng et al. (19) reported that PTN inactivates the tyrosine phosphatase activity of PTPζ and increases the tyrosine phosphorylation level of β-catenin. This is also the case for GIT1/Cat-1: PTN increases the tyrosine phosphorylation of GIT1/Cat-1, presumably through reduction of the catalytic activities of PTPζ. Thus, PTPζ is likely to be constitutively active in the basal condition.
GIT1 was originally identified as a binding partner for the β-adrenergic receptor kinase, GRK2 (28). GIT1 has ADP-ribosylation factor–GTPase-activating protein (ARF-GAP) activity, and overexpression of GIT1 in HEK293 cells inhibits G protein-coupled receptor internalization (28, 34). This inhibition requires the ARF-GAP domain of its N terminus, suggesting that GTP-GDP cycling of ARF proteins is involved in this process (28). ARF proteins are now recognized as key players in numerous vesicular trafficking events, ranging from the formation and fusion of vesicles in the Golgi apparatus to exocytosis and endocytosis (35). Because GIT1 appears to alter receptor internalization by a general mechanism and is widely expressed in tissues, it may be a common component of G protein-coupled receptor signaling cascades and may play a role in coordinating internalization of a variety of cell-surface proteins.
On the other hand, it was also reported that tyrosine phosphorylation of Cat-1 is increased following cell spreading on fibronectin, and is also dynamically regulated during the cell cycle (29). Moreover, Cat-1 plays a role in the p21-activated serine threonine kinase (Pak) regulation. Pak serves as a target for the small GTP-binding proteins, Cdc42 and Rac, and participates in a wide range of biological activities, including cytoskeletal and cell morphological changes, which are essential for neurite outgrowth and neuronal migration (36). We recently demonstrated that the ligand-receptor relationship between PTN and PTPζ is involved in the neuronal migration and neurite extension of cortical neurons (14, 15), and that PSD-95 family members bind to the C terminus of PTPζ (21).
In the present study, we showed that GIT1/Cat-1 is a downstream mediator of PTN signaling. Therefore, PTN, PTPζ, GIT1/Cat-1, and the Pak signaling pathway might participate in the signal transduction mechanism of neurons underlying neuronal migration, neurite extension, and regulation of synaptic transmission. Consistently, we recently found that PTPζ, c-src, phosphatidylinositol 3-kinase, and mitogen-activated protein (MAP) kinases are involved in the haptotactic migration of UMR106 cells induced by midkine (37), which is a heparin-binding growth factor that is highly related to PTN and binds to PTPζ with high affinity (16). It is also reported that GAP activity of GIT/Cat proteins is stimulated by phosphatidylinositol 3,4,5-triphosphate (38) and that c-src, phosphatidylinositol 3-kinase, and MAP kinases are important constituents of the Pak signaling mechanism (39, 40). Thus, it is suggested that GIT/Cat proteins serve as convergence points between G protein-coupled receptors, integrins, vesicular trafficking regulators, cell cycle regulators, and Cdc42/Rac/Pak signaling pathways (29, 35). The diverse roles of PTPζ might be understood as a regulator of its substrate molecule, GIT1/Cat-1.
Acknowledgments
We thank Dr. H. Sabe for providing us with pBaBePurov-src vector, 3Y1, and v-src-transformed 3Y1 cells, and Drs. Paul Bartel and Stan Fields for pBTM116 vector. We also thank Ms. Megumi Gotoh, Masae Mizoguchi, and Kaoru Yamada for technical assistance and Ms. Akiko Kodama for secretarial assistance. This work was supported by grants-in-aid from the Ministry of Education, Science, Sports and Culture of Japan, and Japan Science and Technology Corporation (CREST).
Abbreviation
- PTP
protein tyrosine phosphatase
- GIT1
G protein-coupled receptor kinase-interactor 1
- Cat-1
Cool-associated, tyrosine-phosphorylated 1
- PTN
pleiotrophin
- ICR
intracellular region
- RPTP
receptor-type protein tyrosine phosphatase
- GST
glutathione S-transferase
- RCM
reduced, carboxymethylated, and maleylated
- v-src
active src
- Pak
p21-activated serine threonine kinase
References
- 1.Kavin M, Walton K M, Dixon J E. Annu Rev Biochem. 1993;62:101–120. doi: 10.1146/annurev.bi.62.070193.000533. [DOI] [PubMed] [Google Scholar]
- 2.Sun H, Tonks N K. Trends Biol Sci. 1994;19:480–485. doi: 10.1016/0968-0004(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 3.Keegan K, Cooper J A. Oncogene. 1996;12:1537–1544. [PubMed] [Google Scholar]
- 4.Mousseau D D, Banville D, L'Abbe D, Bouchard P, Shen S-H. J Biol Chem. 2000;275:4467–4474. doi: 10.1074/jbc.275.6.4467. [DOI] [PubMed] [Google Scholar]
- 5.Flint A J, Tiganis T, Barford D, Tonks N K. Proc Natl Acad Sci USA. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castellanos R M P, Mazon M J. J Biol Chem. 1985;260:8240–8242. [PubMed] [Google Scholar]
- 7.Navon A, Schwarz Y, Hazan B, Kassir Y, Nir U. Mol Gen Genet. 1994;244:160–167. doi: 10.1007/BF00283517. [DOI] [PubMed] [Google Scholar]
- 8.Krueger N X, Saito H. Proc Natl Acad Sci USA. 1992;89:7417–7421. doi: 10.1073/pnas.89.16.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy J B, Canoll P D, Silvennoinen O, Barnea G, Morse B, Honegger A M, Huang J-T, Cannizzaro L A, Park S-H, Druck T, et al. J Biol Chem. 1993;268:10573–10581. [PubMed] [Google Scholar]
- 10.Maeda N, Matsui F, Oohira A. Dev Biol. 1992;151:564–574. doi: 10.1016/0012-1606(92)90194-l. [DOI] [PubMed] [Google Scholar]
- 11.Maeda N, Hamanaka H, Shintani T, Nishiwaki T, Noda M. FEBS Lett. 1994;354:67–70. doi: 10.1016/0014-5793(94)01093-5. [DOI] [PubMed] [Google Scholar]
- 12.Maurel P, Rauch U, Flad M, Margolis R K, Margolis R U. Proc Natl Acad Sci USA. 1994;91:2512–2516. doi: 10.1073/pnas.91.7.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishiwaki T, Maeda N, Noda M. J Biochem. 1998;123:458–467. doi: 10.1093/oxfordjournals.jbchem.a021959. [DOI] [PubMed] [Google Scholar]
- 14.Maeda N, Nishiwaki T, Shintani T, Hamanaka H, Noda M. J Biol Chem. 1996;271:21446–21452. doi: 10.1074/jbc.271.35.21446. [DOI] [PubMed] [Google Scholar]
- 15.Maeda N, Noda M. J Cell Biol. 1998;142:203–216. doi: 10.1083/jcb.142.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda N, Ichihara-Tanaka K, Kimura T, Kadomatsu K, Muramatsu T, Noda M. J Biol Chem. 1999;274:12474–12479. doi: 10.1074/jbc.274.18.12474. [DOI] [PubMed] [Google Scholar]
- 17.Milev P, Monnerie H, Popp S, Margolis R K, Margolis R U. J Biol Chem. 1998;273:21439–21442. doi: 10.1074/jbc.273.34.21439. [DOI] [PubMed] [Google Scholar]
- 18.Peles E, Schlessinger J, Grumet M. Trends Biochem Sci. 1998;23:121–124. doi: 10.1016/s0968-0004(98)01195-5. [DOI] [PubMed] [Google Scholar]
- 19.Meng K, Rodriguez-Pena A, Dimitrov T, Chen W, Yamin M, Noda M, Deuel T F. Proc Natl Acad Sci USA. 2000;97:2603–2608. doi: 10.1073/pnas.020487997. . (First Published March 7, 2000; 10.1073/pnas.020487997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratcliffe C F, Qu Y, McCormick K A, Tibbs V C, Dixon J E, Scheuer T, Catterall W A. Nat Neurosci. 2000;3:437–444. doi: 10.1038/74805. [DOI] [PubMed] [Google Scholar]
- 21.Kawachi H, Tamura H, Watakabe I, Shintani T, Maeda N, Noda M. Mol Brain Res. 1999;72:47–54. doi: 10.1016/s0169-328x(99)00204-1. [DOI] [PubMed] [Google Scholar]
- 22.Sabe H, Okada M, Nakagawa H, Hanafusa H. Mol Cell Biol. 1992;12:4706–4713. doi: 10.1128/mcb.12.10.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields S, Song O-K. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 24.Maeda N, Noda M. Development. 1996;122:647–658. doi: 10.1242/dev.122.2.647. [DOI] [PubMed] [Google Scholar]
- 25.Hall A C, Lucas F R, Salinas P C. Cell. 2000;100:525–535. doi: 10.1016/s0092-8674(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 26.Brugge J S, Jarosik G, Andersen J, Queral-Lustig A, Fedor-Chaiken M, Broach J R. Moll Cell Biol. 1987;7:2180–2187. doi: 10.1128/mcb.7.6.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kornbluth S, Jove R, Hanafusa H. Proc Natl Acad Sci USA. 1987;84:4455–4459. doi: 10.1073/pnas.84.13.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Premont R T, Claing A, Vitale N, Freeman J L R, Pitcher J A, Patton W A, Moss J, Vaughan M, Lefkowitz R J. Proc Natl Acad Sci USA. 1998;95:14082–14087. doi: 10.1073/pnas.95.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagrodia S, Bailey D, Lenard Z, Hart M, Guan J L, Premont R T, Taylor S J, Cerione R A. J Biol Chem. 1999;274:22393–22400. doi: 10.1074/jbc.274.32.22393. [DOI] [PubMed] [Google Scholar]
- 30.Miyamoto S, Teramoto H, Coso O A, Gutkind J S, Burbelo P D, Akiyama S K, Yamada K M. J Cell Biol. 1995;131:791–805. doi: 10.1083/jcb.131.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia Z, Barford D, Flint A J, Tonks N K. Science. 1995;268:1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z-Y, Wang Y, Dixon J E. Proc Natl Acad Sci USA. 1994;91:1624–1627. doi: 10.1073/pnas.91.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milarski K L, Zhu G, Pearl C G, McNamara D J, Dobrusin E M, MacLean D, Thieme-Sefler A, Zhang Z-Y, Sawyer T, Decker S J, et al. J Biol Chem. 1993;268:23634–23639. [PubMed] [Google Scholar]
- 34.Claing A, Perry S J, Achiriloaie M, Walker J K L, Albanesi J P, Lefkowitz R J, Premont R T. Proc Natl Acad Sci USA. 2000;97:1119–1124. doi: 10.1073/pnas.97.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moss J, Vaughan M. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 36.Bagrodia S, Cerione R A. Trends Cell Biol. 1999;9:350–355. doi: 10.1016/s0962-8924(99)01618-9. [DOI] [PubMed] [Google Scholar]
- 37.Qi M, Ikematsu S, Maeda N, Ichihara-Tanaka K, Sakuma S, Noda M, Muramatsu T, Kadomatsu T. J Biol Chem. 2001;276:15868–15875. doi: 10.1074/jbc.m005911200. [DOI] [PubMed] [Google Scholar]
- 38.Vitale N, Patton W A, Moss J, Vaughan M, Lefkowitz R J, Premont R T. J Biol Chem. 2000;275:13901–13906. doi: 10.1074/jbc.275.18.13901. [DOI] [PubMed] [Google Scholar]
- 39.Yoshii S, Tanaka M, Otsuki Y, Wang D-Y, Guo R-J, Zhu Y, Takeda R, Hanai H, Kaneko E, Sugimura H. Oncogene. 1999;18:5680–5690. doi: 10.1038/sj.onc.1202936. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Z-S, Manser E, Loo T-H, Lim L. Mol Cell Biol. 2000;20:6354–6363. doi: 10.1128/mcb.20.17.6354-6363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]