Abstract
In contrast to a wealth of knowledge about the photoregulation of gibberellin metabolism in dicots, that in monocots remains largely unclear. In this study, we found that a blue light signal triggers reduction of active gibberellin content in rice seedlings with simultaneous repression of two gibberellin 20-oxidase genes (OsGA20ox2 and OsGA20ox4) and acute induction of four gibberellin 2-oxidase genes (OsGA2ox4–OsGA2ox7). For further examination of the regulation of these genes, we established a series of cryptochrome-deficient lines through reverse genetic screening from a Tos17 mutant population and construction of knockdown lines based on an RNA interference technique. By using these lines and phytochrome mutants, we elucidated that cryptochrome 1 (cry1), consisting of two species in rice plants (cry1a and cry1b), is indispensable for robust induction of the GA2ox genes. On the other hand, repression of the GA20ox genes is mediated by phytochromes. In addition, we found that the phytochromes also mediate the repression of a gibberellin 3-oxidase gene (OsGA3ox2) in the light. These results imply that, in rice seedlings, phytochromes mediate the repression of gibberellin biosynthesis capacity, while cry1 mediates the induction of gibberellin inactivation capacity. The cry1 action was demonstrated to be dominant in the reduction of active gibberellin content, but, in rice seedlings, the cumulative effects of these independent actions reduced active gibberellin content in the light. This pathway design in which different types of photoreceptors independently but cooperatively regulate active gibberellin content is unique from the viewpoint of dicot research. This redundancy should provide robustness to the response in rice plants.
Keywords: Cryptochrome, Gibberellin, Leaf sheath elongation, Photomorphogenesis, Phytochrome, Rice, Oryza sativa
Introduction
Rice is an important staple food that is eaten by billions of people worldwide (Hoshikawa 1989). To obtain an abundant harvest, formation of compact and sturdy seedlings is the first crucial task in the modern cultivation management of rice plants, which leads to robust growth in paddy fields at the vegetative stage. The shape of seedlings is influenced by various environment signals, including temperature, seeding density, soil moisture and light (Matsuo et al. 1995). The light signal has been demonstrated to be especially critical to the formation of compact seedlings through the drastic conversion of developmental programs from skotomorphogenesis to photomorphogenesis (Matsuo et al. 1995).
To monitor environmental light quality and quantity, plants are equipped with cryptochrome (cry), phytochrome (phy), phototropin and other photoreceptors (Lin 2002, Quail 2002, Demarsy and Fankhauser 2009). Cryptochrome is a blue/ultraviolet-A (B/UV-A) photoreceptor that is cognate with photolyase, a DNA repair enzyme (Cashmore et al. 1999). In angiosperms, cryptochrome genes form a small family. The Arabidopsis genome encodes at least two molecular species, cry1 and cry2, which display distinct properties. cry1 is a photostable species and is the major B light receptor for photomorphogenesis, while cry2 is photolabile (Lin et al. 1998). The cry2 species also mediates photomorphogenesis, but its action is limited to low-intensity B light (Lin et al. 1998). This species mainly mediates flowering time determination under natural conditions (Guo et al. 1998, El-Din El-Assal et al. 2001). We have identified three cryptochrome genes in the rice genome (Hirose et al. 2006). As phylogenetic investigation revealed that two of these genes encode a cry1-type receptor, they were named OsCRY1a (AB073546) and OsCRY1b (AB073547). The latter was categorized as cry2, named OsCRY2 (AB103094). Our previous study using transgenic rice plants overexpressing one of the three species suggested that all of the cryptochrome species in rice plants mediate B light perception to suppress elongation of leaf sheaths and blades (Hirose et al. 2006).
Phytochrome is a red/far-red (R/FR) photoreceptor that mediates various responses in plants, such as light-dependent germination, photomorphogenesis, shade avoidance and photoperiodic control of flowering (Franklin and Quail 2010). In contrast to the prevailing view, phytochrome can perceive a broad range of light wavelengths. In addition to effective perception of R/FR light, inefficient but certain perception of B light can be recognized in several plant species (Shinomura et al. 1996, Xie et al. 2007). Phylogenetic investigations indicated that phytochrome also forms a small gene family, and the most primitive family in angiosperms consists of three members: phyA, phyB and phyC (Clack et al. 1994, Mathews et al. 1995, Alba et al. 2000). Arabidopsis and several other dicots have additive species derived from phyB, resulting in an Arabidopsis genome encoding five phytochrome genes: PHYA, PHYB, PHYC, PHYD and PHYE (Sharrock and Quail 1989, Clack et al. 1994). In contrast, monocots, especially rice, have kept the most primitive set, with only three genes (PHYA, PHYB and PHYC) in the genome (Kay et al. 1989, Dehesh et al. 1991, Tahir et al. 1998, Basu et al. 2000). We have isolated a complete set of rice phytochrome single mutants and constructed all combinations of the double and triple mutants (Takano et al. 2001, Takano et al. 2005, Takano et al. 2009). These mutants have already contributed not only to the elucidation of basic features of the rice phytochromes, but also to the identification of their roles in various photoresponses in rice plants (Ishikawa et al. 2005, Xie et al. 2007, Shimizu et al. 2009).
Gibberellin is a tetracyclic diterpene phytohormone that promotes several responses in plants, such as germination, stem elongation and flower induction (Yamaguchi 2008). Stem elongation in etiolated seedlings is the major action of gibberellin, and gibberellin activity rapidly decreases following light irradiation, which stops stem elongation in the seedlings (Symons and Reid 2003, Zhao et al. 2007). This correlation contributes to the formation of compact seedlings in the light. In addition, semi-dwarf cultivars of wheat and rice bred through the introduction of dwarfing traits achieve a spectacular increase in yield. This increase, called the ‘Green Revolution’, has enlarged the agriculturally sustainable population of the world (Hargrove and Cabanilla 1979). Recently, it was elucidated that these dwarfing traits were attributable to a partial deficiency of gibberellin biosynthesis (Sasaki et al. 2002). This conclusion has led to the use of gibberellin biosynthesis inhibitors to form compact seedlings in farming technology. In this way, elucidation of the molecular mechanisms regulating gibberellin metabolism is an important target for agricultural researchers.
Gibberellin is a group of substances possessing the ent-gibberellane skeleton. To date, >100 gibberellin species have been identified from fungal and plant tissues, although the number of biologically active gibberellin species is quite limited (Hedden and Kamiya 1997). Only GA1, GA3 and GA4 have been confirmed to be endogenous active species in plants. Gibberellin is synthesized from trans-geranylgeranyl diphosphate through ent-kaurene (Hedden and Kamiya 1997, Olszewski et al. 2002, Yamaguchi 2008). A series of oxidations of ent-kaurene generates GA12, the first gibberellin product in the biosynthesis pathway. Further steps in the pathway are divided into two branches, the non-13-hydroxylation and early-13-hydroxylation pathways. In rice plants in the vegetative stage, active gibberellin is synthesized through the early-13-hydroxylation pathway (Kobayashi et al. 1988). In this pathway, GA53 is converted into the biologically active GA1 through a cascade of catalytic oxidations by two oxidases, gibberellin 20-oxidase (GA20ox) and gibberellin 3-oxidase (GA3ox). On the other hand, inactivation of gibberellin is also important for the regulation of gibberellin action. The most studied inactivation enzyme is gibberellin 2-oxidase (GA2ox), which catalyzes 2β-hydroxylation. This enzyme is recognized to be critical for the inactivation of gibberellin in various plants, especially during vegetative growth (Thomas et al. 1999).
Photoregulation of active gibberellin levels has been studied in several dicots, including lettuce (Toyomasu et al. 1992), pea (Reid et al. 2002) and Arabidopsis (Zhao et al. 2007). In pea seedlings during de-etiolation, a reduction of GA1 triggered by light exposure could be observed with simultaneous down-regulation of PsGA3ox1 and up-regulation of PsGA2ox2 (Reid et al. 2002). Because available photoreceptor mutants in pea are limited, only the minor contribution of phyA could be clarified in this regulation (Reid et al. 2002), but the major photoreceptor remains unclear. Zhao et al. (2007) analyzed the B light-induced repression of gibberellin biosynthetic-related genes (AtGA20ox1 and AtGA3ox1) and the expression of gibberellin inactivation-related genes (AtGA2ox1, AtGA2ox2, AtGA2ox6 and AtGA2ox8) in a series of photoreceptor mutants of Arabidopsis. They detected a weak contribution of phyA to the induction of the AtGA2ox1 gene, as in pea plants. In addition, they found that redundant actions of cry1 and cry2 mediate a large segment of the transcriptional regulation of these genes (Zhao et al. 2007). In contrast to the wealth of knowledge obtained from dicots, information about the photoregulation of gibberellin metabolism in monocots remains limited. In the present study, we focus on the photoregulation of gibberellin metabolism in rice seedlings. We found that a B light signal triggered a reduction of active gibberellin content with consistent changes in transcript levels of gibberellin biosynthesis- and inactivation-related genes. We utilized phytochrome-deficient mutants and newly established cryptochrome-deficient lines not only for identification of photoreceptors mediating the transcriptional regulations, but also for evaluation of each photoreceptor in the regulation of active gibberellin content. Our results indicate that cry1 and phyB independently regulate different sets of gibberellin-related genes, but their cumulative effects cooperatively mediate reduction of the active gibberellin content in rice seedlings in the light.
Results
Gibberellin content in seedlings exposed to blue light
To evaluate the effect of B light irradiation on the regulation of gibberellin metabolism in rice seedlings, we measured the content of several molecular species of gibberellin in dark-grown seedlings before and after exposure to B light (10 µmol m−2 s−1) for 24 h (Fig. 1B). Only GA1 and GA20 showed significant reductions in their content after B light irradiation. GA1 is a major active gibberellin species in rice (Kobayashi et al. 1988). GA20 is an inactive gibberellin species, but is the precursor molecule immediately before GA1 in the biosynthesis pathway (Fig. 1A). On the other hand, the content of other gibberellin species was comparable between conditions. These results indicate that a B light signal triggers reductions of GA1 and GA20 content in rice etiolated seedlings.
Fig. 1.
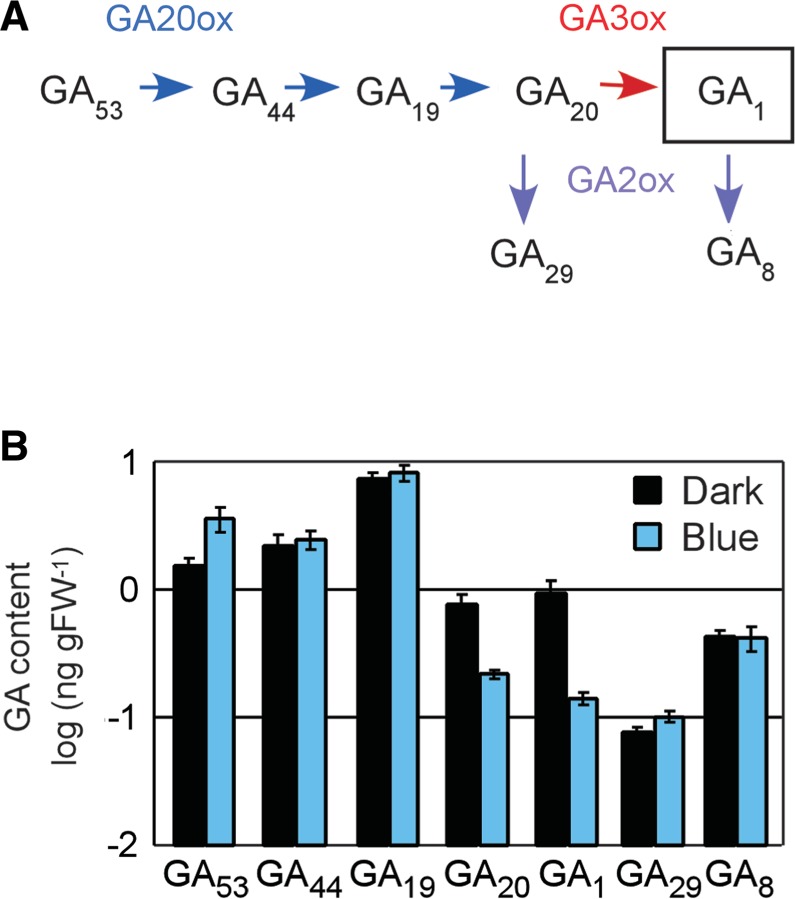
Endogenous gibberellin content in dark-grown WT seedlings and their transitions after B light irradiation. (A) Schematic drawing of the metabolic flow of gibberellin species in the early-13-hydroxylation pathway. Blue, red and purple arrows indicate steps catalyzed by GA20ox (blue), GA3ox (red) and GA2ox (purple). (B) Endogenous content of several gibberellin species in 3-day-old WT etiolated seedlings (black bars) and 3-day-old etiolated seedlings exposed to B light (10 µmol m−2 s−1) for another 24 h (blue bars). The contents of GA53, GA44, GA19, GA20, GA1, GA29 and GA8 are indicated by a logarithmic scale. All data are means of seven samples with standard errors.
Expression of genes involved in biosynthesis, inactivation and signaling of gibberellin under B light irradiation
To approach the molecular mechanism of the B light response observed by the change in gibberellin content, we investigated the expression profiles for the genes encoding gibberellin biosynthetic enzymes GA20ox and GA3ox under B light irradiation. These genes generally form a small family. In rice plants, the OsGA20ox family consists of four members and the OsGA3ox family consists of two members (Toyomasu et al. 1997, Itoh et al. 2001, Sasaki et al. 2002, Sakamoto et al. 2004). Fig. 2 and Supplementary Fig. S1 show their expression profiles in etiolated seedlings exposed to B light. Among the members of the OsGA20ox family, OsGA20ox2 and OsGA20ox4 were down-regulated by B light irradiation. The lowest levels of these transcripts were estimated to be <5% of their initial values. On the other hand, OsGA20ox1, OsGA20ox3, OsGA3ox1 and OsGA3ox2 are probably irrelevant to the B light-induced regulation of gibberellin content because OsGA20ox1 and OsGA3ox2 were constitutively expressed even under B light irradiation (Supplementary Fig. S1). In addition, the expression levels of OsGA20ox3 and OsGA3ox1 were quite low under our experimental conditions (data not shown).
Fig. 2.
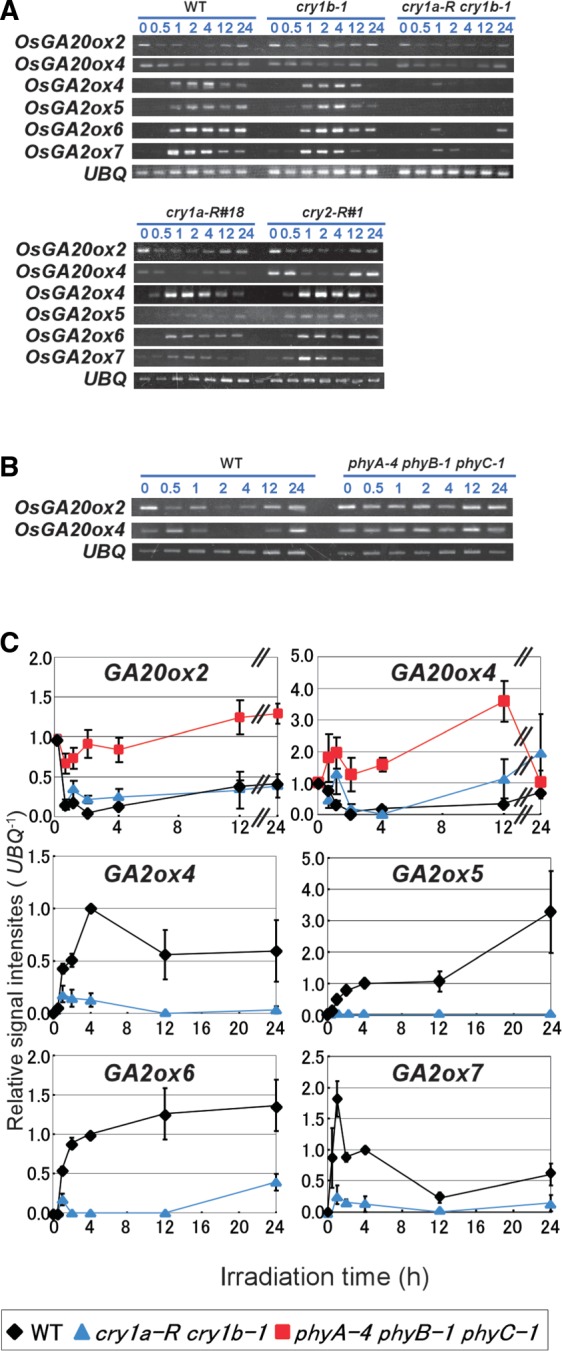
The effect of B light on the expression of gibberellin biosynthesis- and inactivation-related genes. Three-day-old etiolated seedlings of WT, cryptochrome-deficient and phytochrome-deficient lines were exposed to B light (10 µmol m−2 s−1) for the indicated periods. Transcripts of UBQ are shown as controls. (A) Expression of gibberellin biosynthesis- and inactivation-related genes in WT, cry1b-1, cry1a-R cry1b-1, cry1a-R and cry2-R lines. (B) Expression of OsGA20ox2 and OsGA20ox4 in WT seedlings and phyA-4 phyB-1 phyC-1 mutants. (C) Relative transcript levels and their changes of OsGA20ox2, OsGA20ox4 and OsGA2ox4-7 in WT, cry1a-R cry1b, and phyA-4 phyB-1 phyC-1 seedlings during B light irradiation.
We next examined the expression of gibberellin inactivation-related genes in the OsGA2ox family. Ten OsGA2ox genes (OsGA2ox1–OsGA2ox10) have been identified in the rice genome (Sakamoto et al. 2004, Lee and Zeevaart 2005, Lo et al. 2008; Supplementary Fig. S2). A series of semi-quantitative reverse transcription–PCR (RT–PCR) experiments revealed that the expression of OsGA2ox4–OsGA2ox7 genes was clearly induced by B light irradiation (Fig. 2A, C; Supplementary Fig. S1). The remaining members of the family are apparently not involved with the B light-induced changes in gibberellin content. Although the expression of OsGA2ox1, OsGA2ox2, OsGA2ox3, OsGA2ox8 and OsGA2ox9 could be observed, their levels were not affected by exposure to B light (Supplementary Fig. S1). Transcripts for OsGA2ox10 could not be detected under our conditions (data not shown), implying that its expression had little influence on gibberellin content.
We also investigated the expression of genes involved in gibberellin signaling: GID1 (Ueguchi-Tanaka et al. 2005), GID2 (Sasaki et al. 2003) and SLR1 (Ikeda et al. 2001). Their transcript levels were not influenced by B light irradiation (Supplementary Fig. S1), suggesting that gibberellin signaling is not controlled by B light, at least at their transcriptional level.
Construction of cryptochrome-deficient lines
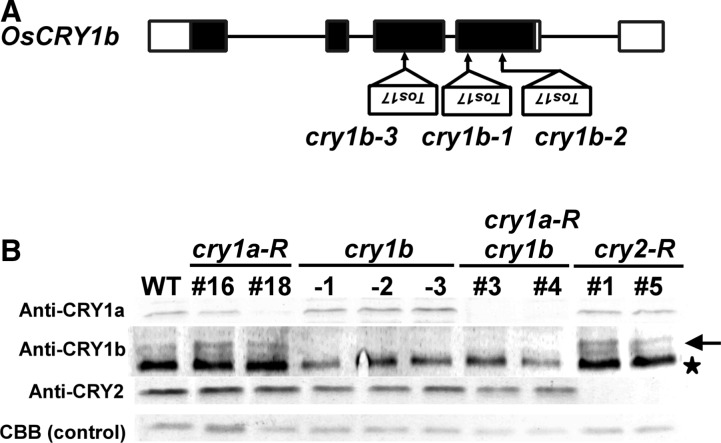
Mutants deficient in photoreceptors are important practically for identifying the roles of photoreceptors in various photoresponses. Because rice cryptochrome mutants had not been isolated previously, we attempted to isolate a series of them. In this work, we obtained three alleles of a cry1b mutant (cry1b-1, -2 and -3) from a mutant population generated by insertion of the endogenous retrotransposon Tos17 (Hirochika 1999). For each allele, a Tos17 insertion was detected in the portion of the sequence corresponding to the third or fourth exon of CRY1b (Fig. 3A). We estimated the protein levels of the cryptochromes in these mutants by immunoblot analysis using specific antibodies raised against CRY1a, CRY1b or CRY2 (Fig. 3B). CRY1b disappeared from the immunoblots for proteins of these mutants. Moreover, no nascent bands could be detected on the blots (data not shown), indicating that the insertions of Tos17 completely disrupt the CRY1b gene.
Fig. 3.
Basic features of the cryptochrome mutants and knockdown lines used in this work are shown. (A) Schematic drawing of the OsCRY1b gene structure with the Tos17 insertion sites of cry1b mutants isolated in this work. Introns (lines) and exons (boxes) are represented. Filled boxes indicate the coding regions, while open boxes correspond to 5′- and 3′-untranslated regions. (B) Western blotting analysis of OsCRY1a, OsCRY1b and OsCRY2 in cry1b mutants and cryptochrome-deficient transgenic lines. Proteins were extracted from 3-day-old etiolated seedlings of WT (Nipponbare), cry1a-R (#16 and #18), cry1b-1, cry1b-2, cry1b-3, cry1a-R cry1b-1 (#3 and #4) and cry2-R (#1 and #5) lines. A 50 µg aliquot of protein was used for each sample analyzed. Coomassie brilliant blue-stained bands served as a loading control. The anti-CRY1b antiserum used in this study detected two bands on the blots. The upper band (marked with an arrow) corresponds to CRY1b. The lower band (marked with a star) was confirmed to be a cross-reactant between the antiserum and a protein other than CRY1b.
Despite our efforts, cry1a and cry2 mutants could not be isolated from the mutant population; therefore, we constructed CRY1a and CRY2 knockdown transgenic lines (named cry1a-R and cry2-R, respectively) using an RNA interference (RNAi) technique (Hannon 2002, Miki and Shimamoto 2004). Fig. 3B shows that CRY2 was not observable in cry2-R lines. In the cry1a-R lines, CRY1a was slightly detected on blots. In this study, cry1a-R #18 was preferentially used because the repression of CRY1a in this line was the tightest obtained to date. We also made cry1a/b doubly deficient lines by introducing the cry1a-R construct into the cry1b-1 mutant (cry1a-R cry1b-1), because the lines are required for the definition of cry1 function under the mutually complementary relationship between cry1a and cry1b expected from their highly homologous amino acid sequences (Hirose et al. 2006, Zhang et al. 2006). Fig. 3B clearly shows that neither CRY1a nor CRY1b proteins could be detected in the cry1a-R cry1b-1 seedlings.
Identification of photoreceptors involved in transcriptional regulation of GA20ox and GA2ox genes under B light irradiation
We measured the transcript levels of GA20ox and GA2ox genes in response to B light treatment in cryptochrome-deficient lines and compared them with those in wild-type (WT) seedlings. The B light-induced expression of OsGA2ox4–OsGA2ox7 genes almost disappeared in cry1a-R cry1b-1, but remained unchanged in cry1a-R and cry1b-1 (Fig. 2A, C).
On the other hand, the repression of OsGA20ox2 and OsGA20ox4 by the B light treatment could be observed not only in the single crytochrome-deficient lines (cry1a-R, cry1b-1 and cry2-R), but also in the cry1-deficient line (cry1a-R cry1b-1), suggesting that photoreceptors other than cryptochromes may mediate the repression of these genes (Fig. 2A, C). We attempted to examine the involvement of phytochromes in this repression. As expected, the transient repression of the genes could not be detected in the phytochrome triple mutant (phyA-4 phyB-1 phyC-1; Fig. 2B).
Expression of genes involved in biosynthesis, inactivation and signaling of gibberellin under R light irradiation
To dissect the contribution of phytochrome to the regulation of gibberellin-related gene expression, we repeated semi-quantitative RT–PCR analyses for the estimation of a series of gibberellin-related transcripts under R light irradiation. We detected down-regulation of the OsGA20ox2 and OsGA20ox4 genes in WT seedlings during R light irradiation with almost identical kinetics to those induced by B light irradiation (Figs. 2, 4; Supplementary Fig. S1). This repression was significantly diminished in the phyB mutant and completely disappeared in the phyA phyB mutant (Fig. 4A, B). In contrast, nearly comparable down-regulation of these genes could be detected in the phyA, phyC and phyA phyC mutants (Fig. 4A). These results indicate that phyB is the major photoreceptor for the repression of OsGA20ox2 and OsGA20ox4, while phyA contributes to the regulation, but its action is supplemental and then appears only under the phyB-deficient background. The R light experiments revealed that the transcription of OsGA3ox2, which was not affected by B light irradiation, was repressed by R light irradiation. This repression was also mediated by phyB with a minor contribution from phyA (Fig. 4A, B; Supplementary Fig. S1). The most probable reason why this gene could not respond to the B light signal is that the PFR/PR ratio in the photoequilibrium state given under the B light signal might be too low to induce the response clearly.
Fig. 4.
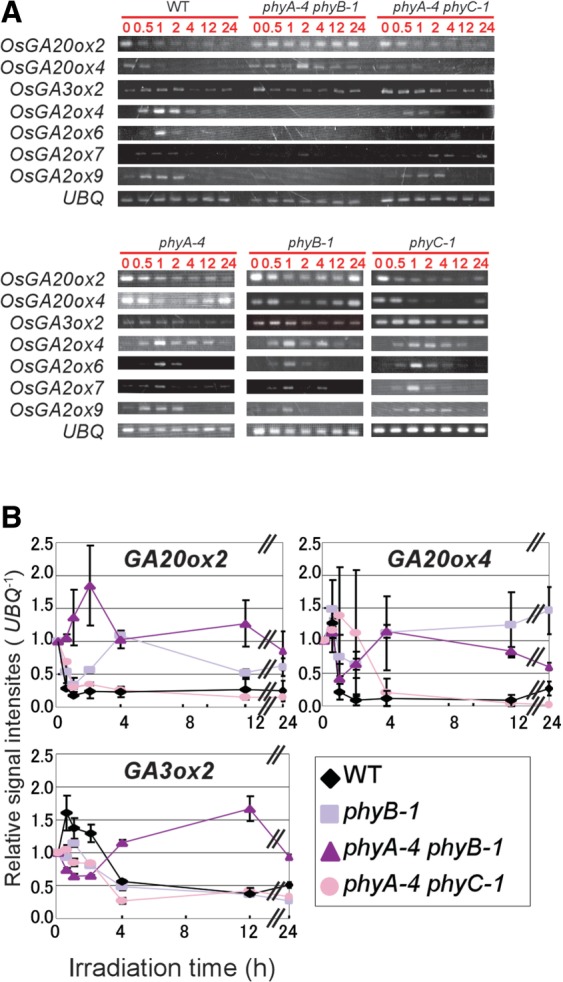
The R light-induced modulation of the expression of gibberellin biosynthesis- and inactivation-related genes in WT seedlings and various phytochrome mutants. Three-day-old etiolated seedlings of WT and phytochrome mutants were exposed to R light (10 µmol m−2 s−1) for the indicated period. Transcripts of UBQ are shown as controls. (A) The expression of gibberellin biosynthesis- and inactivation-related genes in WT, phyA-4 phyB-1, phyA-4 phyC-1, phyA-4, phyB-1 and phyC-1 lines during R light irradiation was examined. (B) Relative transcript levels of OsGA20ox2, OsGA20ox4 and OsGA3ox2 in WT, phyB-1, phyA-4 phyB-1 and phyA-4 phyC-1 were estimated from gel images of RT–PCR assays.
A series of R light experiments elucidated another minor effect of phytochromes on the light-induced expression of OsGA2ox genes. The B light induction of the OsGA2ox4, OsGA2ox6 and OsGA2ox7 genes was almost eliminated in the cry1a-R cry1b-1 line (Fig. 2A, C); however, weak transient induction remained. This residual induction might be attributable to a phytochrome action, because the weak induction could be reproduced by R light irradiation with similar kinetics to those induced by B light irradiation (Fig. 4A; Supplementary Fig. S1). In addition, the R light-mediated induction disappeared in the phyA phyB mutant (Fig. 4A).
Gibberellin content in cry1- or phytochrome-deficient seedlings under light conditions
In the previous sections, we elucidated the transcriptional regulation of genes encoding gibberellin biosynthesis and inactivation enzymes under light treatments. We also revealed the involvement of cryptochromes and phytochromes in this regulation. We next examined the direct consequences of photoreceptor deficiency on the reduction of active gibberellin content in the light. We measured the content of GA20 and GA1 in dark-grown WT, cry1a-R cry1b-1 and phyA-4 phyB-1 seedlings before and after light treatments (Fig. 5A, B). This figure has an unsolved problem in that GA1 content in the dark-grown cry1a-R cry1b-1 seedlings was slightly high compared with those of the WT and phyA-4 phyB-1 seedlings. We considered that the elevated GA1 level was likely to be a consequence of a somatic variation introduced through construction of the transgenic line. We supposed that values for the GA1 content in the cry1a-R cry1b-1 seedlings were elevated to a similar extent.
Fig. 5.
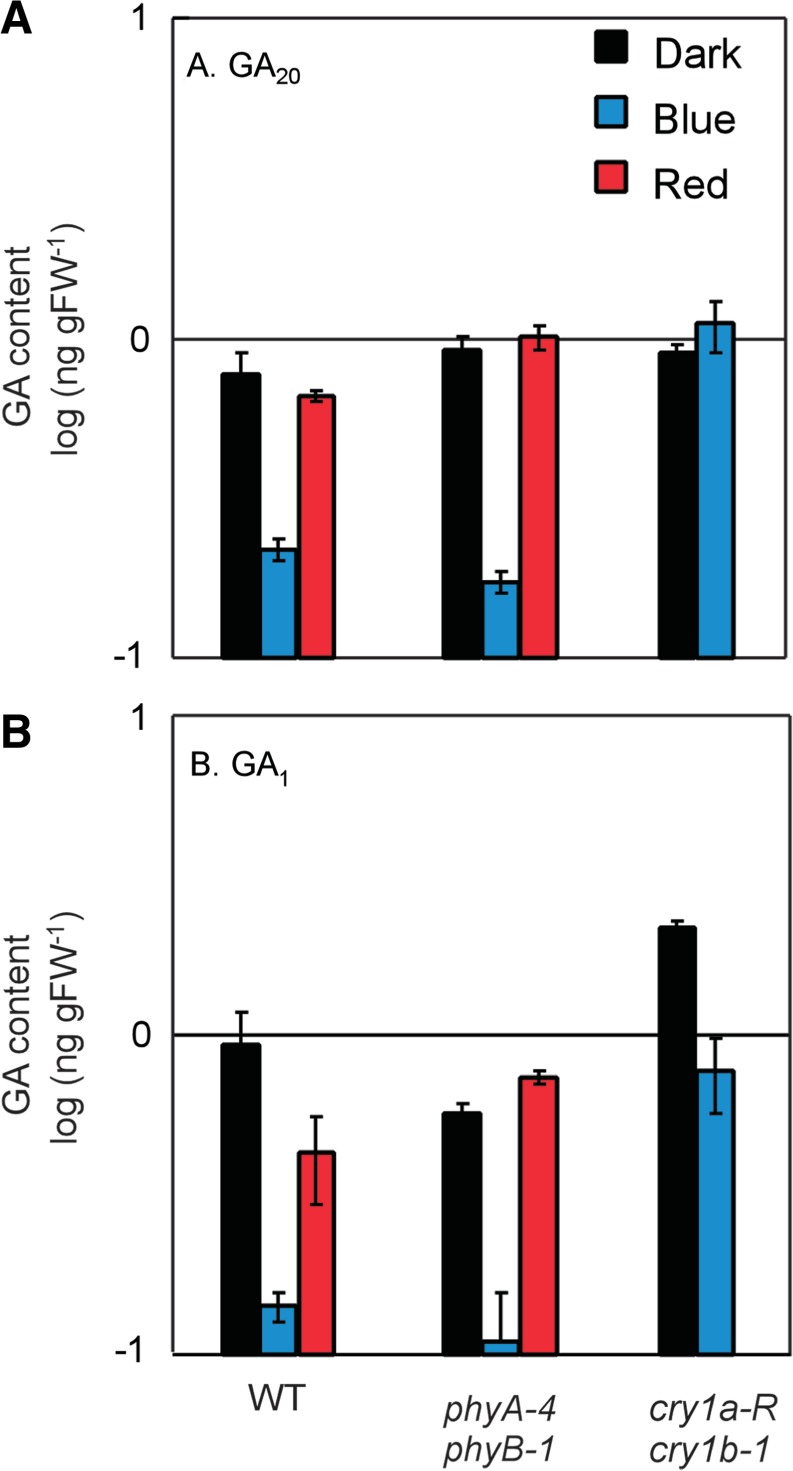
The endogenous content of GA20 (A) and GA1 (B) in 3-day-old dark-grown seedlings of cry1a-R cry1b-1, phyA-4 phyB-1 and WT lines and their transitions after exposure to B or R light for another 24 h are shown by a logarithmic scale. Black bars indicate their content in dark-grown seedlings, while red and blue bars represent their content after R or B light irradiation for another 24 h, respectively. All data are means estimated from at least three samples. Error bars indicate the SE of the data.
Fig. 5 displays the predominant implication of cry1 in the regulation of gibberellin metabolism. A drastic reduction of GA20 levels by B light treatment in WT seedlings completely disappeared in cry1a-R cry1b-1 seedlings. A similar reduction of GA1 content was significantly moderated in the cry1-deficient line. They were thought to be effects of cry1-mediated induction of GA2ox genes (Fig. 2A, C), which should increase GA2ox activity and then accelerate to reduce both GA1 and GA20 content through catalytic oxidation. If so, the drastic reduction of the GA1 and GA20 levels that was completely conserved even in the phyA-4 phyB-1 seedlings indicates the predominance of cry1 in the B light-induced reduction of the active gibberellin content in the seedlings. However, in parallel, we could recognize the substantial contribution of phytochromes to the regulation, which could be detected in observations that the GA1 content was reduced by R light irradiation in WT seedlings and the reduction completely disappeared in phyA-4 phyB-1 seedlings. In addition, moderate reduction of the GA1 content in cry1a-R cry1b-1 seedlings after B light irradiation might be attributed to the action of phytochrome. The phytochrome signal is likely to reduce only the GA1 content, which should result in the signal decreasing the biosynthesis of GA1 through the repression of GA20ox and GA3ox genes (Figs. 2, 4).
Lengths of leaf sheaths under B light irradiation
In this work, we revealed that cryptochromes and phytochromes mediate the reduction of active gibberellin content in rice seedlings, probably through transcriptional regulation of a limited number of gibberellin-related genes. These features might influence the morphological phenotypes of the seedlings, because gibberellin is a pivotal phytohormone for promoting stem elongation in rice plants. Therefore, we measured the lengths of the second leaf sheaths of the cryptochrome-deficient lines and WT seedlings grown under various fluence rates of B light and examined their fluence rate responsiveness (Supplementary Fig. S3B). In this figure, we do not show the dark control values, because we could not measure them from the dark-grown seedlings. A significant number of these seedlings displayed a dark phenotype in which elongation of the leaves and leaf sheaths was severely restricted and these parts could not grow from the coleoptiles.
The lengths of the second leaf sheaths of WT seedlings reflected obvious fluence rate-dependent shortening. The lengths of cry1a-R and cry1b-1 seedlings showed similar responses, but these values were slightly higher than those of WT seedlings under all fluence rates examined (Supplementary Fig. S3B). The lengths of cry2-R seedlings were longer than those of WT seedlings under weak B light; however, the tendency disappeared under intense B light. Under 30 µmol m−2 s−1 B light irradiation, the lengths of the second leaf sheaths of cry2-R were almost equivalent with those of WT seedlings (Supplementary Fig. S3B). In cry1a-R cry1b-1 seedlings, these lengths were significantly longer than those of WT seedlings, especially under a high fluence rate (≥10 µmol m−2 s−1) (Supplementary Fig. S3A, B). The result indicates that cry1 deficiency severely weakened the fluence rate-dependent shortening of leaf sheaths. However, it is noteworthy that the cry1-deficient line never lost its responsiveness, because the fluence rate response curve of the line was slightly downward in the graph (Supplementary Fig. S3B).
Effects of exogenous GA1 on elongation of the second leaf sheath under B light irradiation
In earlier sections, we demonstrated the predominance of cry1a/b actions in the B light-induced reduction of active gibberellin content in rice seedlings, which should be a consequence of rapid induction of the OsGA2ox4–OsGA2ox7 genes triggered by the B light irradiation. Robust induction of GA2ox enzymes can rapidly remove biologically active gibberellin from the cells, which implies that it is connected to quick suppression of leaf sheath elongation. We attempted to evaluate GA2ox activity in the leaf sheath cells through the effectiveness of exogenous GA1 on the elongation of second leaf sheaths. To remove any influences of endogenous active gibberellin from the assay, we added a gibberellin biosynthesis inhibitor, paclobutrazol (PAC), to the agar medium. PAC has been reported to inhibit ent-kaurene oxidase and consequently cause a drop in content of almost all molecular species of gibberellin (Hallahan et al. 1988). Even in the presence of PAC, leaf sheaths could elongate up to approximately 2.7 mm, which was probably due to gibberellin-independent elongation. The addition of PAC strongly inhibited further elongation of the second leaf sheaths of the cry1a-R cry1b-1 seedlings when grown under B light irradiation for 8 d (Fig. 6C), indicating that de novo synthesis of gibberellin is essential for the further elongation of leaf sheaths in this mutant. Next, we investigated the effect of exogenous GA1 on the elongation of the second leaf sheaths in WT seedlings and in cry1a-R cry1b-1 seedlings under B light irradiation in the presence of PAC. The second leaf sheaths of the GA1-treated cry1a-R cry1b-1 seedlings were significantly longer than those of the seedlings without GA1, while GA1 application had a weak effect on WT seedlings (Fig. 6B, C). The difference in effectiveness against the exogenous GA1 in WT seedlings and the cry1a-R cry1b-1 line implies two possibilities; one is that the cryptochrome mutations affect responsiveness to GA1, and the other is that the activity of a GA1-inactivating enzyme, probably GA2ox, is higher in WT seedlings than in cry1a-R cry1b-1 seedlings. The latter possibility is more probable because it correlates with our observations that WT seedlings under B light irradiation displayed robust induction of GA2ox genes, while the mutant lacked their induction (Fig. 2A, C).
Fig. 6.
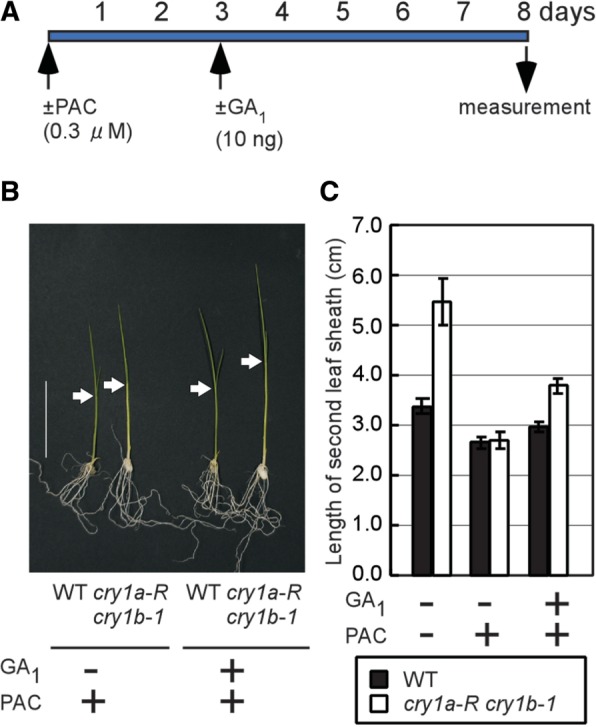
The effect of exogenous GA1 treatment on the suppression of leaf sheath elongation in the cry1-deficient line. (A) Schematic drawing of the experimental timetable for PAC and GA1 treatments. (B) This photograph shows typical seedlings grown under B light (10 µmol m−2 s−1) for 8 d at 28°C in the presence of PAC (0.3 µM) with and without GA1. Arrows indicate the lamina joints, a monocotyledonous structure between a leaf sheath and a leaf blade. Scale bar = 30 mm. (C) The lengths of second leaf sheaths in WT and cry1a-R cry1b-1 seedlings under B light with or without (+ or −) exogenous GA1 treatment in the presence or absence (+ or −) of PAC. Bars represent the lengths of second leaf sheaths of WT (black) and cry1a-R cry1b-1 (white) lines (n = 3–6).
Discussion
We started this work from measurements of endogenous gibberellin content in dark-grown seedlings before and after B light irradiation for 24 h (Fig. 1B). In our assays, the gibberellin species in the early-13-hydroxylation pathway could be detected at relatively high levels. On the other hand, the content of the gibberellin species in the non-13-hydroxylation pathway was relatively low. In particular, GA9, GA4 and GA34 could not be detected in our samples (data not shown). This is consistent with past reports that the early-13-hydroxylation pathway is predominant for gibberellin biosynthesis in rice seedlings during vegetative growth (Kobayashi et al. 1988). Fig. 1B displays the content of molecular species in the pathway, showing that the levels of GA1 and GA20 dropped significantly after B light irradiation. This result indicates that rice plants possess mechanisms to reduce the content of the biologically active gibberellin species GA1 along with that of its cognate precursor (GA20) after perception of B light signals.
To understand how B light stimuli decrease active gibberellin content in rice plants, we examined the transcript levels of GA20ox and GA3ox as gibberellin biosynthesis-related genes and those of GA2ox as gibberellin inactivation-related genes, because correlations between their transcript levels and active GA content have been reported in several plant species (Reid et al. 2002, Zhao et al. 2007). Fig. 2A and C and Supplementary Fig. S1 show that B light led to the repression of OsGA20ox2 and OsGA20ox4 and the induction of OsGA2ox4–OsGA2ox7, while the expression of other gibberellin biosynthesis- and inactivation-related genes was not affected. In addition, the gibberellin signaling-related genes GID1, GID2 and SLR1 did not respond to the B light. Thus, in rice plants, the transcript levels for limited members of the gibberellin biosynthesis- and inactivation-related enzymes were regulated by B light irradiation. These changes in their transcript levels are likely to be connected to the observed reductions of GA1 and GA20 after B light irradiation (Fig. 1B).
The application of cryptochrome-deficient lines and phytochrome mutants to the analyses of transcriptional profiles of the gibberellin-related genes opened up consideration of the functions of each photoreceptor in the regulation. For example, the induction of OsGA2ox4–OsGA2ox7 genes by B light irradiation disappeared in cry1a-R cry1b-1 seedlings, but remained unchanged in lines deficient in one of the cry1 species (cry1a-R and cry1b-1; Fig. 2A, C), indicating that cry1a and cry1b complementarily mediate the induction of OsGA2ox4–OsGA2ox7 genes. This feature is consistent with a wealth of past observations in dicots (Zhao et al. 2007). On the other hand, the repression of OsGA20ox2 and OsGA20ox4 under B light irradiation was not influenced by a deficiency of any cryptochromes (Fig. 2A, C). Surprisingly, the repression disappeared in phytochrome triple mutants (Fig. 2B, C), which indicates that the repression is mediated by phytochromes rather than cryptochromes. It is not unusual that rice phytochrome can conduct various B light responses, especially in juvenile seedlings (Takano et al. 2005, Xie et al. 2007). However, this phytochrome-dependent regulation of the GA20ox genes is unique from the viewpoint of past observations from Arabidopsis studies. In Arabidopsis, the GA20ox genes are also repressed by B light signals, but this repression is mediated by the cooperation of cry1 and cry2 (Zhao et al. 2007). A slight phyA contribution could be observed in the repression, but it was shown to be very weak. Furthermore, there has been no report suggesting a phyB contribution to the repression. Arabidopsis phyB was reported to control gibberellin biosynthesis-related genes (AtGA3ox1 and AtGA3ox2) and inactivation-related genes (AtGA2ox2), but the regulation was observed in seed germination (Yamaguchi et al. 1998, Oh et al. 2006). In addition, phyB reportedly mediated the induction of GA3ox1 and GA3ox2 expression and the reduction of GA2ox2 expression under light irradiation, which is in contrast to our observations.
A series of B light experiments indicated the unique contribution of phytochromes in the repression of OsGA20ox genes in rice plants. If this is true, experiments using R light stimuli should emphasize the contribution; therefore, we examined and compared the expression of gibberellin biosynthesis- and inactivation-related genes under R light irradiation in WT seedlings and a set of phytochrome mutants (Fig. 4A, B; Supplementary Fig. S1). The R light irradiation of the WT seedlings led to repression of the gibberellin biosynthetic genes OsGA20ox2 and OsGA20ox4 with similar kinetics to those induced by B light irradiation. In addition, the R light irradiation also repressed OsGA3ox2 gene expression. Further examination showed another minor R light response as a transient induction of gibberellin inactivation-related genes (GA2ox4, Ga2ox6, GA2ox7 and GA2ox9). Analyses using a series of phytochrome mutants revealed that all of these R light responses were mediated by phyB with the synergistic support of phyA (Fig. 4A, B; Supplementary Fig. S1). These results can explain a phenotype with phyB-dependent suppression of leaf sheath elongation under R light (Takano et al. 2005). These results suggest that phyB practically mediates the regulation of gibberellin metabolism in rice plants. This feature is different from that in Arabidopsis, in which phyB tends to regulate gibberellin responsiveness rather than gibberellin metabolism during de-etiolation (Reed et al. 1996).
Our next efforts focused on evaluating photoreceptors in the regulation of active gibberellin content. We measured the content of GA1 and GA20 in WT, cry1-deficient (cry1a-R cry1b-1) and phytochrome double mutant (phyA-4 phyB-1) etiolated seedlings before and after light irradiation. In these measurements, the GA1 content in the dark-grown seedlings of cry1a-R cry1b-1, which is the control value for the line, was slightly high compared with those of WT and phyA-4 phyB-1 seedlings. The elevated GA1 level was likely to be a consequence of an abnormal mesocotyl elongation phenotype of the cry1a-R cry1b-1 seedlings grown in darkness, which was probably due to a somatic variation introduced through construction of the transgenic line. We could not determine the real reason for the results of the phenotype, but we thought that the basic photoresponse of GA1 content would not be influenced even with the abnormal phenotype.
These measurements revealed that B light effectively reduced both GA20 and GA1 content in the WT seedlings, which could be interpreted to reflect that the changes were attributed to the cumulative effects of the cry1-mediated induction of the GA2ox4-7 genes and the phytochrome-mediated repression of the GA20ox2 and GA20ox4 genes. On the other hand, R light irradiation moderately reduced only the GA1 content, possibly because R light irradiation repressed the expression of the biosynthesis-related genes GA20ox and GA3ox, which can reduce GA1 biosynthesis, but cannot consume previously synthesized GA1. Certainly, weak induction of GA2ox4, GA2ox6, GA20x7 and GA2ox9 by R light was detected, but it should not be enough for complete consumption of the previously synthesized GA1. Similar moderate reduction of the GA1 content could be observed in the cry1-deficient line under B light irradiation, which was probably due to the action of phytochromes.
The B light-irradiated phytochrome mutant showed drastic reductions in GA1 and GA20 content. This reduction could not be distinguished from that observed in WT seedlings, suggesting that cry1 action is predominant in the photoregulation of active gibberellin content. In summary, the cry1-dependent reduction of active gibberellin content, which is probably caused by the induction of GA2ox4–GA2ox7 genes, is more important for reducing the gibberellin content in this condition. However, our results simultaneously displayed the presence of a phytochrome-dependent mechanism for the regulation of GA1 content in rice plants.
In dicots, B light-induced suppression of stem elongation is considered in relation to the photoregulation of gibberellin metabolism (Reid et al. 2002, Zhao et al. 2007). Therefore, B light-induced reduction of active gibberellin content in rice seedlings should affect morphological phenotypes. In particular, the lengths of the leaf sheaths should be influenced by the changes in gibberellin content. We next examined the fluence rate dependence of the lengths of second leaf sheaths under B light irradiation using WT and various cryptochrome-deficient lines (Supplementary Fig. S3). The results suggest that cry2 mediates the suppression of the elongation of leaf sheaths under B light with fluence rates <10 µmol m−2 s−1, while cry1 can conduct the response over a wide range of fluence rates. In particular, cry1 preferentially transmits strong B light at fluence rates >10 µmol m−2 s−1. Similar fluence rate preferences have been reported for Arabidopsis cryptochromes (Lin et al. 1998), suggesting that each molecular species shares basic features with their orthologs. The fluence rate responses also indicated that weak responsiveness to B light was retained even in the cry1a-R cry1b-1 seedlings. The retained responsiveness might be attributed to the action of phytochromes.
In this study, we showed that five members of the OsGA2ox gene family (GA2ox4–GAox7 and GA2ox9) are photoregulated by cryptochromes, phytochromes or both (Figs. 4A, 5A), which probably contributes to the reduction of active gibberellin content under light irradiation. GA2ox members are subdivided into three classes (class I, II and III) based on phylogenetic relationships (Supplementary Fig. S2; Schomburg et al. 2003, Lee and Zeevaart 2005). OsGA2ox4 and OsGA2ox7 belong to class I, while OsGA2ox5, OsGA2ox6 and OsGA2ox9 are categorized into class III. We could not find any class II members that were under photoregulation. Recently, it has been reported that members of each class have individual substrate preferences in terms of the backbone structure of gibberellin (Schomburg et al. 2003, Lee and Zeevaart 2005). In the gibberellin biosynthesis pathway, almost all precursor species consist of 20 carbon atoms (C20-gibberellin). GA20ox converts a methyl group at the C-20 position of gibberellin to an aldehyde by sequential oxidations and finally removes this carbon atom, which produces a gibberellin species with 19 carbon atoms (C19-gibberellin). Therefore, products of GA20ox and their further metabolites, such as GA1, GA20, GA29 and GA8, are the C19-gibberellins (Olszewski et al. 2002, Yamaguchi 2008). Members of class I and II predominantly catalyze C19-gibberellins including active gibberellin species and close cognate precursors. In rice seedlings, the class I and II members seem to convert GA20 and GA1 into their respective inactive species, GA29 and GA8. In contrast, the class III members were able to catalyze only C20-gibberellin species, which are the upstream precursor species, namely GA53, GA19 and GA44, in the early-13-hydroxylation pathway. Therefore, the class III enzymes convert precursors into their metabolites to prevent their activation. In short, rice plants are equipped with two types of photoregulated GA2ox enzymes, one of which reduces active gibberellin content directly and the other eliminates precursor species under light irradiation.
We further investigated the importance of B light induction of class I GA2ox genes in the suppression of leaf sheath elongation through a comparison of the effectiveness of exogenous GA1 in WT and cry1a-R cry1b-1 seedlings (Fig. 6B, C). The effectiveness is probably correlated with the total activities of class I and II GA2ox. Exogenous GA1 induced the elongation of leaf sheaths in cry1a-R cry1b-1 seedlings, but not in WT seedlings (Fig. 6B, C), suggesting that the WT seedlings could eliminate exogenous active gibberellin species efficiently. This result is consistent with the fact that WT seedlings showed robust GA2ox expression (Fig. 2A). On the other hand, the cry1-deficient seedlings might not eliminate exogenous active gibberellin species, which could be attributable to a deficiency of induction of the OsGA2ox4 and OsGA2ox7 genes by B light irradiation (Fig. 2A). These results also suggest that the induction of class I OsGA2ox genes under B light irradiation is important for the suppression of leaf sheath elongation in the light.
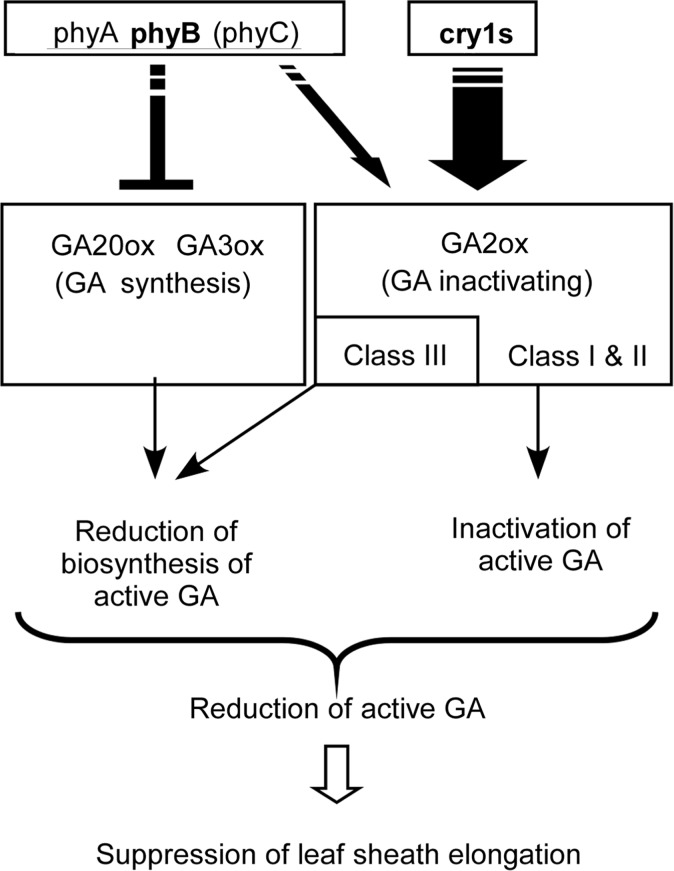
In summary, the results obtained from our experiments using monochromatic light sources and a series of photoreceptor mutants supply an opportunity to consider the mechanism that regulates gibberellin metabolism in natural light conditions (Fig. 7). Solar radiation, which consists of a broad range of wavelengths of visible light, can trigger two types of photoreceptors involved in the regulation, namely phytochromes and cryptochromes. Phytochromes mediate the repression of several members of the GA20ox and GA3ox gene families, which is connected to a reduction in the biosynthesis of GA1. In parallel, cryptochrome signals induce several GA2ox genes. GA2ox enzymes decrease active gibberellin levels through two processes: the reduction of precursor gibberellin species by class III enzymes and the reduction of active gibberellin species directly by class I enzymes. The cumulative effects of these independent actions are probably to decrease the GA1 content in rice seedlings, which should consequently suppress the elongation of leaf sheaths and blades and lead to the formation of compact seedlings. This pathway design, in which two different types of photoreceptors mediate the reduction of active gibberellin content independently, is significantly different from that in Arabidopsis. The redundant pathways provide robustness to the network in rice plants. Further research is required to clarify whether the individual pathway design reported here is specific to rice or is widely distributed in monocots.
Fig. 7.
A model for the photoregulation of gibberellin content in rice seedlings. The cry1 (cry1a/b)-mediated rapid induction of a limited number of the OsGA2ox gene family members (OsGA2ox4–OsGA2ox7). Phytochromes mediated the repression of gibberellin biosynthesis-related genes (OsGA20ox2, OsGA20ox4 and OsGA3ox2) and weak induction of gibberellin inactivation-related genes (OsGA2ox4, OsGA2ox6, OsGA2ox7 and OsGA2ox9). These independent actions cumulatively mediate a reduction in active gibberellin content in the cells, which is probably connected to the suppression of leaf sheath elongation in rice seedlings under the light.
Materials and Methods
Plant materials and growth conditions
We used the cultivar Nipponbare as the WT strain in this study. In addition, the rice cryptochrome-deficient lines newly established in this study and a series of phytochrome mutants (phyA-4, phyB-1, phyC-1, phyA-4 phyB-1, phyA-4 phyC-1 and phyA-4 phyB-1 phyC-1; Takano et al. 2001, Takano et al. 2005, Takano et al. 2009) were used. Dehusked seeds were surface-sterilized in liquid sodium hypochlorite, sown onto 0.5% (w/v) agar and grown at 28°C under the various light conditions mentioned in the text. An R light-emitting diode panel (Model LED-R, EYELA) and a B light-emitting diode panel (Model LED-B, EYELA) were used as monochromatic light sources.
Measurement of gibberellin content
Rice seedlings grown in the dark for 3 d or in the dark for 3 d with 24 h B or R light irradiation were harvested and the samples were kept at −80°C until gibberellin extraction. Gibberellin levels were determined by liquid chromatography-selected reaction monitoring on a quadrupole/time-of-flight tandem mass spectrometer (Q-Tof Premier, Waters) connected to an Acquity Ultra Performance liquid chromatograph equipped with a reverse phase column (Acquity UPLC BEH-C18, Waters) as described previously (Varbanova et al. 2007). We used 2H-labeled gibberellin species as internal standards. The labeled gibberellin species were purchased from Professor Lewis Mander (Australian National University, Canberra, Australia).
RT–PCR
Total RNA was isolated from aerial parts of the seedlings using an RNeasy Plant Mini kit (QIAGEN). To remove any genomic DNA contamination, the RNA samples were treated with RNase-free DNase I (QIAGEN) according to the manufacturer’s instructions. A 1 µg aliquot of total RNA was used as a template to synthesize cDNA using a ReverTra Ace kit (TOYOBO). Gibberellin-related genes were amplified from the cDNA using gene-specific primers (see Supplementary Table S2). In these reactions, 1 µl of cDNA products was added to 20 µl of PCR mixture and the total number of amplification cycles was adjusted to keep the results semi-quantitative. After PCR, the products were examined by agarose gel electrophoresis with ethidium bromide staining. At least three biological repetitions of RT–PCR assays were carried out to maintain statistical reliability. Gel images were obtained with the AlphaImager Mini System (ProteinSimple) and the relative amounts of transcripts were estimated from the gel images using ImageJ software (http://rsb.info.nih.gov/ij/). Levels of OsUBQ (D12629) transcript were stable during our experimental conditions (data not shown); therefore, we used this transcript as the quantitative standard. Representative gel images of the RT–PCR analyses are shown in each figure.
Screening of cryptochrome mutants
The cry1b mutants were isolated from a mutant population of rice (Oryza sativa cv. Nipponbare) generated by random insertion of an endogenous retrotransposon, Tos17 (Hirochika 1999), which is available from the Rice Genome Resource Center (RGRC) of the National Institute of Agrobiological Sciences (Tsukuba, Japan, http://www.rgrc.dna.affrc.go.jp/index.html). The first screening was carried out by PCR against three-dimensional panels of mutant DNA pools supplied from the RGRC using primer sets designed for the amplification of a Tos17-tagged CRY1b gene. To eliminate false amplifications, nested PCR was conducted against PCR products obtained in the first screening. Primer sequences used in the screening are listed in Supplementary Table S1. The amplified DNA fragments were cloned and sequenced to confirm the insertion of Tos17 into the CRY1b gene. The matrix of appearance of PCR products on three-dimensional panels enabled us to identify candidate lines in the mutant population. Consequently, three alleles having a cry1b mutation were screened from the population. M2 seeds of the lines supplied from the RGRC were cultivated for the establishment of cry1b homozygous lines (cry1b-1, -2 and -3). Isolation of cry1a and cry2 mutants was attempted in the same manner, but they could not be obtained from this population.
Construction of transgenic plants
To construct CRY1a and CRY2 knockdown transgenic lines using an RNAi technique (Miki and Shimamoto 2004), each unique sequence of these genes was amplified by PCR from each cDNA clone isolated in our previous work (Hirose et al. 2006). The sequences of primer pairs for amplification of CRY1a were 5′-CACCTTTCAGACTACAATTCACCGGG-3′ and 5′-CACTTGATTCATGCACACCAAG-3′, and those for CRY2 were 5′-CACCAGATGGTGAGGTTGTGGAGGA-3′ and 5′-CCGTGTCAGCTCAGTTCTCAG-3′. The PCR products were subcloned into a Gateway entry vector, pENTR/D-TOPO (Invitrogen). The inserts in the vector were transferred into an RNAi vector for plant research, pANDA (Miki and Shimamoto 2004), by an LR clonase reaction. The RNAi constructs were then introduced into rice (O. sativa cv. Nipponbare or the cry1b-1 mutant) through an Agrobacterium-mediated procedure (Toki 1997). Agrobacterium tumefaciens strain EHA101 (Hood et al. 1986) was used in this transformation. The transformants obtained were cultivated for at least two generations to establish homozygous transgenic lines (cry1a-R, cry2-R and cry1a-R cry1b-1).
Western blotting analysis
Rice seedlings grown in the dark for 3 d were harvested using a night vision device. Aerial parts of the seedlings were ground using a mortar and pestle in protein elution buffer (100 mM Tris–HCl, pH 8.3, 5 mM EDTA, 0.2% 2-mercaptoethanol; Nagatani et al. 1993) containing 1× Complete Protease Inhibitor Cocktail (Roche). Proteins in the extracts were precipitated with 40% saturated ammonium sulfate. After centrifugation, proteins in the precipitate were resuspended in the same buffer. The protein concentration of the extracts was determined using Coomassie PLUS Protein Assay Reagent (Pierce). A 50 µg aliquot of protein was subjected to SDS–PAGE using gels containing 12% (w/v) acrylamide. Separated proteins in the gels were blotted onto PVDF membranes (Millipore) electrophoretically. The antibodies used and the procedures for immunochemical detection followed an earlier study (Hirose et al. 2006) using BCIP/NBT Color Development Substrate (Promega).
Growth measurement of seedlings grown under blue light
Rice seedlings grown under various fluence rates (0.1, 1, 10 or 30 µmol m−2 s−1) of B light for 8 d at 28°C were picked up and scanned with a flatbed scanner to obtain images for computer analyses. The lengths of their second leaf sheaths were measured from the images using ImageJ software.
The effect of exogenous GA1 on leaf elongation
A microdrop assay (Murakami 1972) was conducted to examine the effects of exogenous GA1 on the seedlings. For our purposes, the assay was slightly modified in terms of conditions of light treatments. A 1 µl drop of ethanol containing 10 ng of GA1 (OlChemIm) was put onto shoots of WT and cry1a-R cry1b-1 seedlings that were grown under B light on agar medium in the presence of 0.3 µM PAC (Wako Pure Chemical Industries) for 3 d. On the fifth day after the application of GA1, images of the seedlings were acquired with a flatbed scanner and the lengths of the second leaf sheaths were measured using ImageJ image processing software.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Ministry of Agriculture, Forestry and Fisheries of Japan [Genomics for Agricultural Innovation, grant No. GPN-0003].
Supplementary Material
Acknowledgments
We thank Drs. Ko Shimamoto and Daisuke Miki for providing the pANDA vector. We also thank Koji Harada, Haruhiro Sato, Yumiko Iguchi and Kazuko Yagi for their technical assistance.
Glossary
Abbreviations
- B
blue
- cry
cryptochrome
- FR
far-red
- GA2ox
gibberellin 2-oxidase
- GA20ox
gibberellin 20-oxidase
- GA3ox
gibberellin 3-oxidase
- PAC
paclobutrazol
- phy
phytochrome
- R
red
- RNAi
RNA interference
- RT–PCR
reverse transcription–PCR
- WT
wild type.
References
- Alba R, Kelmenson PM, Cordonnier-Pratt MM, Pratt LH. The phytochrome gene family in tomato and the rapid differential evolution of this family in angiosperms. Mol. Biol. Evol. 2000;17:362–373. doi: 10.1093/oxfordjournals.molbev.a026316. [DOI] [PubMed] [Google Scholar]
- Basu D, Dehesh K, Schneider-Poetsch HJ, Harrington SE, McCouch SR, Quail PH. Rice PHYC gene: structure, expression, map position and evolution. Plant Mol. Biol. 2000;44:27–42. doi: 10.1023/a:1006488119301. [DOI] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: blue light receptors for plants and animals. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol. Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- Dehesh K, Tepperman J, Christensen AH, Quail PH. phyB is evolutionarily conserved and constitutively expressed in rice seedling shoots. Mol. Gen. Genet. 1991;225:305–313. doi: 10.1007/BF00269863. [DOI] [PubMed] [Google Scholar]
- Demarsy E, Fankhauser C. Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol. 2009;12:69–74. doi: 10.1016/j.pbi.2008.09.002. [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S, Alonso-Blanco C, Peeters AJ, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- Hallahan DL, Heasman AP, Grossel MC, Quigley R, Hedden P, Bowyer JR. Synthesis and biological activity of an azido derivative of paclobutrazol, an inhibitor of gibberellin biosynthesis. Plant Physiol. 1988;88:1425–1429. doi: 10.1104/pp.88.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Hargrove TR, Cabanilla VL. The impact of semidwarf varieties on Asian rice-breeding programs. Biosicence. 1979;29:731–735. [Google Scholar]
- Hedden P, Kamiya Y. Gibberellin biosynthesis: enzymes, genes and their regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:431–460. doi: 10.1146/annurev.arplant.48.1.431. [DOI] [PubMed] [Google Scholar]
- Hirochika H. Retrotransposons of rice as a tool for forward and reverse genetics. In: Shimamoto K, editor. Molecular Biology of Rice. Tokyo: Springer-Verlag; 1999. pp. 43–58. [Google Scholar]
- Hirose F, Shinomura T, Tanabata T, Shimada H, Takano M. Involvement of rice cryptochromes in de-etiolation responses and flowering. Plant Cell Physiol. 2006;47:915–925. doi: 10.1093/pcp/pcj064. [DOI] [PubMed] [Google Scholar]
- Hood EE, Helmer GL, Fraley RT, Chilton MD. The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J. Bacteriol. 1986;168:1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshikawa K, editor. The Growing Rice plant. Tokyo: Rural Culture Association; 1989. [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, et al. slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Tamaki S, Yokoi S, Inagaki N, Shinomura T, Takano M, et al. Suppression of the floral activator Hd3a is the principal cause of the night break effect in rice. Plant Cell. 2005;17:3326–3336. doi: 10.1105/tpc.105.037028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sentoku N, Kitano H, Matsuoka M, Kobayashi M. Cloning and functional analysis of two gibberellin 3β-hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl Acad. Sci. USA. 2001;98:8909–8914. doi: 10.1073/pnas.141239398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay SA, Keith B, Shinozaki K, Chua NH. The sequence of the rice phytochrome gene. Nucleic Acids Res. 1989;17:2865–2866. doi: 10.1093/nar/17.7.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Yamaguchi I, Murofushi N, Ota Y, Takahashi N. Fluctuation and localization of endogenous gibberellins in rice. Agric. Biol. Chem. 1988;52:1189–1194. [Google Scholar]
- Lee DJ, Zeevaart JA. Molecular cloning of GA 2-oxidase3 from spinach and its ectopic expression in Nicotiana sylvestris. Plant Physiol. 2005;138:243–254. doi: 10.1104/pp.104.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl Acad. Sci. USA. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. Blue light receptors and signal transduction. Plant Cell. 2002;14(Suppl):S207–S225. doi: 10.1105/tpc.000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JA, Chen LJ, et al. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 2008;20:2603–2618. doi: 10.1105/tpc.108.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S, Lavin M, Sharrock R. Evolution of the phytochrome gene family and its utility for phylogenetic analyses of angiosperms. Ann. MO Bot. Gard. 1995;82:296–321. [Google Scholar]
- Matsuo T, Kumazawa K, Ishii R, Hirata H, editors. Science of the Rice Plant. Tokyo: Rural Culture Association; 1995. [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Murakami Y. Dwarfing genes in rice and their relation to gibberellin biosynthesis. In: Carr DJ, editor. Plant Growth Substances. Berlin: Springer-Verlag; 1972. pp. 166–174. [Google Scholar]
- Nagatani A, Reed JW, Chory J. Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 1993;102:269–277. doi: 10.1104/pp.102.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung WI, Choi G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006;47:124–139. doi: 10.1111/j.1365-313X.2006.02773.x. [DOI] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F. Gibberellin signaling: biosynthesis, inactivation, and response pathways. Plant Cell. 2002;14(Suppl):S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- Reed JW, Foster KR, Morgan PW, Chory J. Phytochrome B affects responsiveness to gibberellins in Arabidopsis. Plant Physiol. 1996;112:337–342. doi: 10.1104/pp.112.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB, Botwright NA, Smith JJ, O’Neill DP, Kerckhoffs LH. Control of gibberellin levels and gene expression during de-etiolation in pea. Plant Physiol. 2002;128:734–741. doi: 10.1104/pp.010607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134:1642–1653. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, et al. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, et al. Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science. 2003;299:1896–1898. doi: 10.1126/science.1081077. [DOI] [PubMed] [Google Scholar]
- Schomburg FM, Bizzell CM, Lee DJ, Zeevaart JA, Amasino RM. Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. Plant Cell. 2003;15:151–163. doi: 10.1105/tpc.005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Tanabata T, Xie X, Inagaki N, Takano M, Shinomura T, et al. Phytochrome-mediated growth inhibition of seminal roots in rice seedlings. Physiol. Plant. 2009;137:289–297. doi: 10.1111/j.1399-3054.2009.01277.x. [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons GM, Reid JB. Hormone levels and response during de-etiolation in pea. Planta. 2003;216:422–431. doi: 10.1007/s00425-002-0860-z. [DOI] [PubMed] [Google Scholar]
- Tahir M, Kanegae H, Takano M. Phytochrome C (PHYC) gene in rice: isolation and characterization of a complete coding sequence. Plant Physiol. 1998;118:1535. [Google Scholar]
- Takano M, Inagaki N, Xie X, Kiyota S, Baba-Kasai A, Tanabata T, et al. Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice. Proc. Natl Acad. Sci. USA. 2009;106:14705–14710. doi: 10.1073/pnas.0907378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, Ishizuka T, et al. Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell. 2005;17:3311–3325. doi: 10.1105/tpc.105.035899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M, Kanegae H, Shinomura T, Miyao A, Hirochika H, Furuya M. Isolation and characterization of rice phytochrome A mutants. Plant Cell. 2001;13:521–534. doi: 10.1105/tpc.13.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P. Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl Acad. Sci. USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki S. Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol. Biol. Rep. 1997;15:16–21. [Google Scholar]
- Toyomasu T, Kawaide H, Sekimoto H, vonNumers C, Phillips AL, Hedden P, et al. Cloning and characterization of a cDNA encoding gibberellin 20-oxidase from rice (Oryza sativa) seedlings. Physiol. Plant. 1997;99:111–118. [Google Scholar]
- Toyomasu T, Yamane H, Yamaguchi I, Murofushi N, Takahashi N, Inoue Y. Control by light of hypocotyl elongation and levels of endogenous gibberellins in seedlings of Lactuca sativa L. Plant Cell Physiol. 1992;33:695–701. [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- Varbanova M, Yamaguchi S, Yang Y, McKelvey K, Hanada A, Borochov R, et al. Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell. 2007;19:32–45. doi: 10.1105/tpc.106.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Shinomura T, Inagaki N, Kiyota S, Takano M. Phytochrome-mediated inhibition of coleoptile growth in rice: age-dependency and action spectra. Photochem. Photobiol. 2007;83:131–138. doi: 10.1562/2006-03-17-RA-850. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T. Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell. 1998;10:2115–2126. doi: 10.1105/tpc.10.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YC, Gong SF, Li QH, Sang Y, Yang HQ. Functional and signaling mechanism analysis of rice CRYPTOCHROME 1. Plant J. 2006;46:971–983. doi: 10.1111/j.1365-313X.2006.02753.x. [DOI] [PubMed] [Google Scholar]
- Zhao X, Yu X, Foo E, Symons GM, Lopez J, Bendehakkalu KT, et al. A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiol. 2007;145:106–118. doi: 10.1104/pp.107.099838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.