Abstract
Plant life is strongly dependent on the environment, and plants regulate their growth and development in response to many different environmental stimuli. One of the regulatory mechanisms involved in these responses is phototropism, which allows plants to change their growth direction in response to the location of the light source. Since the study of phototropism by Darwin, many physiological studies of this phenomenon have been published. Recently, molecular genetic analyses of Arabidopsis have begun to shed light on the molecular mechanisms underlying this response system, including phototropin blue light photoreceptors, phototropin signaling components, auxin transporters, auxin action mechanisms and others. This review highlights some of the recent progress that has been made in further elucidating the phototropic response, with particular emphasis on mutant phenotypes.
Keywords: Arabidopsis thaliana, Auxin, Phototropin, Phototropism, Phytochrome, PIN
Introduction
Living plants show changes in their growth patterns in response to environmental stimuli such as light, gravity, water, temperature and touch. In 1880, Darwin outlined those various plant movements in detail in his book ‘The Power of Movement in Plants’. He also studied the phototropic response, which controls the direction of organ growth in accordance with the direction of the light source, and suggested the presence of some matter in the upper part which is acted on by light, and which transmits its effects to the lower part. This study contributed to the discovery of the first phytohormone, auxin.
The Cholodny–Went theory suggests that asymmetric distribution of the phytohormone auxin occurs in response to a tropic stimulus as the result of its lateral movement, and causes differential growth on the two sides of the plant organ and consequent organ bending (Fig. 1) (reviewed in Went and Thimann 1937, Tanaka et al. 2006, Whippo and Hangarter 2006). Many physiological studies have now suggested that phototropic responses are controlled by multiple photoreceptor systems, most notably the blue light photoreceptors, and many other studies have attempted to reveal how photoreceptors induce the asymmetric distribution of auxin in plant organs such as hypocotyls, coleoptiles and roots (reviewed in Briggs 1963, Poff et al. 1994, Iino 2001, Liscum 2002, Holland et al. 2009). Molecular genetic studies of these processes commenced in the 1990s using Arabidopsis thaliana mutants, and the complex physiological aspects of phototropism have begun to be systematically explained more recently in molecular terms. This review introduces the findings of these molecular genetic studies and assesses our current knowledge of phototropism in Arabidopsis.
Fig. 1.
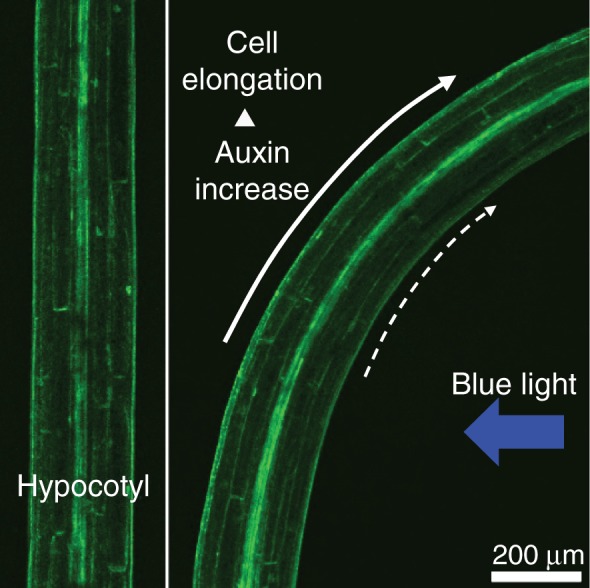
Distribution of auxin during hypocotyl phototropism in Arabidopsis. Two-day-old etiolated seedlings harboring the auxin reporter gene DR5rev:GFP were used. The hypocotyl was stimulated with unilateral irradiation of blue light for 3 h at 0.2 µmol m−2 s−1. GFP signals were detected with a confocal laser scanning microscope. The pictures show representative examples (K. Haga and T. Sakai, unpublished results).
Photoreceptors
Phototropins
Arabidopsis seedlings show positive hypocotyl phototropism and negative root phototropism under unilateral blue light irradiation. Previously, three research groups independently screened Arabidopsis mutants showing abnormalities in their phototropic responses. Hypocotyl phototropism mutants denoted as JK218, JK224, JK229 and JK345 were first isolated by Khurana and Poff (1989), and the mutants nonphototropic hypocotyl1 (nph1), nph2, nph3 and nph4 were described several years later by Liscum and Briggs (1995). Root phototropism mutants, root phototropism1 (rpt1), rpt2 and rpt3, were isolated by Okada and Shimura (1992, 1994). From the corresponding loci of these mutants, NPH1 was cloned and sequenced, and found to encode a 120 kDa UV-A/blue light photoreceptor protein, NPH1, that binds the blue light-absorbing chromophore, FMN, and that harbors two FMN-binding domains, light, oxygen, voltage1 (LOV1) and LOV2, at its N-terminus and a serine (Ser)/threonine (Thr) kinase domain at its C-terminus (Huala et al. 1997, Christie et al. 1998). Subsequent studies revealed that JK224 and rpt1 were allelic with nph1 (Liscum and Briggs 1995, Sakai et al. 2000). Although the nph1 mutants showed non-phototropic hypocotyl and root phenotypes under almost all experimental conditions (Liscum and Briggs 1995), these hypocotyls showed an obviously positive phototropic response at high fluence rates (10 and 100 µmol m−2 s−1) (Sakai et al. 2000). Thus, the evidence indicated that blue light photoreceptor(s) other than nph1 are involved in the signaling processes that trigger hypocotyl curvature upon illumination by a high fluence rate of blue light.
Arabidopsis also expresses an NPH1-homologous gene, NPH1-LIKE1 (NPL1) (Jarillo et al. 1998), which encodes a 110 kDa protein that also harbors two N-terminal LOV domains and a C-terminal Ser/Thr kinase domain (Kagawa et al. 2001). The npl1 mutant was isolated from a pool of T-DNA-tagged clones (Sakai et al. 2001). A single mutation in npl1 had no effect on the phototropic responses in hypocotyls and roots, but in a nph1 background caused a remarkable defect in the phototropic response of hypocotyls even under high fluence rates of blue light (Sakai et al. 2001). Therefore, it was concluded that nph1 and npl1 are major blue light photoreceptors in the phototropic response pathway in Arabidopsis and show partially overlapping functions in a fluence rate-dependent manner. These photoreceptors were subsequently renamed phototropin1 (phot1) and phototropin2 (phot2), with corresponding gene name changes (Briggs et al. 2001).
Blue light irradiation activates the kinase activity of the phototropins (reviewed by Christie 2007). Each FMN molecule associates with the LOV domains non-covalently in a dark state. Blue light absorption then causes the formation of a transient covalent adduct between the FMN and a critical cysteine residue in the LOV domain. This binding causes a conformational change in the phototropin proteins and activates their protein kinase activities. Activated phototropins are autophosphorylated at several serine residues (Inoue et al. 2008, Sullivan et al. 2008), among which the autophosphorylation of phot1 Ser851 in the activation loop between the LOV2 and kinase domain is critical to the signal transduction event that induces the phototropic responses. An alanine substitution at phot1 Ser851 causes a loss of function (Inoue et al. 2008). The Ser851 residue is also conserved in phot1 and phot2 from seed plants, ferns, mosses and green algae, suggesting that its autophosphorylation is a common event for phototropin-induced responses in plants (Inoue et al. 2008). In this context, Tseng and Briggs (2010) indicated that the roots curl in NPA1 (rcn1) mutation of the A1 subunit of Ser/Thr protein phosphatase 2A (PP2A) impairs the dephosphorylation of phot2 and enhances the phot2-mediating phototropic responses in hypocotyls, but does not impair either phot1 dephosphorylation or the phot1-mediating phototropic response. Thus, autophosphorylated phot2, which accumulates in the absence of RCN1, appears to be the active form of this protein and leads to enhanced phototropism. In Avena sativa (oat), unilateral blue light irradiation has been shown to induce a gradient of phot1 autophosphorylation across the coleoptiles, and its phosphorylation level is greater on the irradiated side than on the shaded side (Salomon et al. 1997a, Salomon et al. 1997b). Thus, varying light conditions cause differences in the phototropin activities between the irradiated and shaded sides of the plant, and this appears to result in asymmetric auxin distribution and the differential growth of the organ.
Expression analyses indicated that PHOT1 is expressed strongly in dividing and elongating cortical cells in the apical hook and in the root elongation zone in etiolated seedlings, although its expression is also observed in all except the root cap in the whole seedling (Sakamoto and Briggs 2002). Its expression pattern suggests that the perception of unilateral blue light irradiation leading to the induction of phototropic responses occurs mainly in the apex of the shoots and roots. PHOT2 expression is observed in the cotyledon and hook region, but not in the hypocotyls or roots (Kong et al. 2006). Northern blotting analysis also indicated that the expression of PHOT2 transcripts is quite low in roots (Kagawa et al. 2001), and this seems to underlie why phot1 mutants show phototropic responses in the hypocotyls, but not in the roots under high fluence rates of blue light. In plant cells kept in the dark, phot1 and phot2 are mainly localized at the inner surface of the plasma membrane (Fig. 2) (Sakamoto and Briggs 2002, Kong et al. 2006), even though these proteins contain no transmembrane domain. In response to blue light, a fraction of the phot1 photoreceptors is released into the cytoplasm (Sakamoto and Briggs 2002), and a fraction of the phot2 photoreceptors associates with the Golgi apparatus (Kong et al. 2006). Han et al. (2008) further indicated that pulse irradiation of red light at 2 h prior to phototropic induction by low fluence rates of blue light prevents blue light-inducible changes in the subcellular localization of phot1 in rapidly elongating regions of the hypocotyl in a phytochrome A (phyA)-dependent manner. Because this red light pre-irradiation enhances phototropic responses (Parks et al. 1996, Janoudi et al. 1997a, Stowe-Evans et al. 2001), these findings suggest that plasma membrane-localizing phot1 promotes the phototropic response and that the suppression of phot1 relocalization is a mechanism underlying the enhancement of the phototropic response by red light pre-irradiation.
Fig. 2.
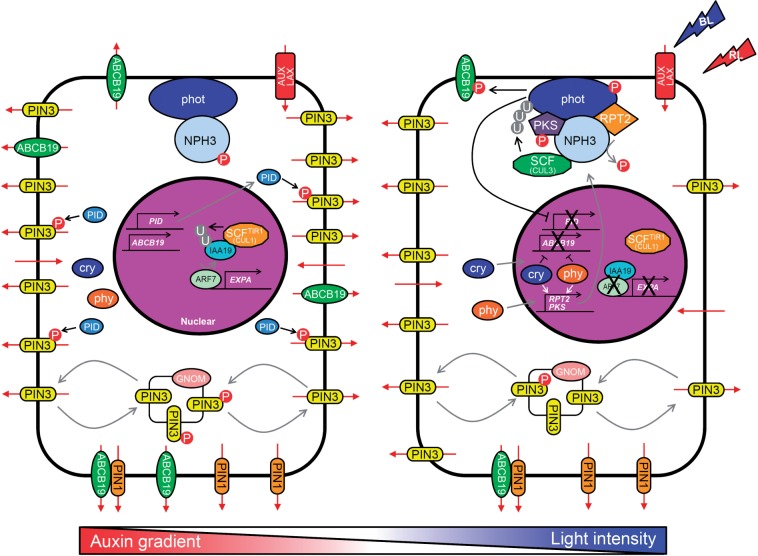
Schematic diagram of the cellular events related to signal transduction of phototropism following light stimulation. Light stimulation activates phototropins, cryptochromes and phytochromes, resulting in induction of many cellular events related to phototropism. The function of each molecule is described in the text. Red arrows indicate flow of auxin, black arrows show the action of molecules, and gray arrows illustrate the movements of molecules. BL, blue light; RL, red light; P, phosphorylation; U, ubiquitination.
There are some notable issues that remain to be resolved in relation to the photoreceptors. First, a green light photoreceptor that induces phototropic responses in the hypocotyls has not yet been identified. Arabidopsis hypocotyls show phototropic responses following illumination at wavelengths <560 nm (Steinitz et al. 1985), and phot1 has been found to be necessary for the green light (530 nm)-induced phototropic responses (Liscum and Briggs 1995). Thus, phot1 seems to function as a green light photoreceptor. However, the absorption of green light by phot1 and the subsequent activation of phot1 have not been observed. Secondly, phot1 phot2 double mutants show a slight phototropic curvature under blue light illumination at 10 µmol m−2 s−1 (Sakai et al. 2001). This observation suggests that some photoreceptors can induce the phototropic response independently of the phototropins in Arabidopsis. In this regard, double mutations of the blue light photoreceptors cryptochrome1 (cry1) and cry2 in the phot1 phot2 double mutant background cause a complete loss of phototropic responses under any light conditions (Ohgishi et al. 2004). Hence, crys may have partially overlapping functions with the phototropins. Millar et al. (2010) indicated that in the absence of a gravitropic response under microgravity conditions, red light induces a phy-dependent phototropic response in Arabidopsis hypocotyls. Kiss et al. (2003) also reported that red light irradiation induces root positive phototropism in a phyA- and phyB-dependent manner. Because phys also function as a blue light photoreceptor in many cases (reviewed by Wang 2005), these factors may partially induce a phototropic response in the absence of phot1 and phot2.
Phytochromes and cryptochromes
Although the phototropic response is primarily controlled by the phototropins, other photoreceptor families, namely phy and cry, also modulate these responses (reviewed in Hangarter 1997, Casal 2000, Iino 2006, Whippo and Hangarter 2006). In Arabidopsis, there are five PHY genes (PHYA–PHYE) encoding 120–130 kDa apoproteins (reviewed in Chen et al. 2004, Wang 2005). These apoproteins bind to the linear tetrapyrrole (phytochromobilin) of a chromophore and absorb primary red light, far-red light and secondary blue light. They harbor a C-terminal histidine kinase-related domain, and phyA shows phosphorylation activity toward several substrates in vitro including cry1 and cry2, the AUX/IAA proteins and phytochrome kinase substrate1 (PKS1) (Ahmad et al. 1998, Fankhauser et al. 1999, Colón-Carmona et al. 2000). The in vivo physiological relevance of these phosphorylation events in phototropism remains to be established. Phys localizing in the nucleus also appear to regulate gene expression (Fig. 2) (reviewed by Jiao et al. 2007), but the biological functions of cytosolic phys remain to be determined.
There are two CRY genes, CRY1 and CRY2, encoding 77 and 69 kDa apoproteins, respectively (reviewed in Chen et al. 2004, Wang 2005, Yu et al. 2010). They bind to two chromophores, a primary flavin (FADH−) and a second deazaflavin or pterin. Crys have two recognizable domains: an N-terminal photolyase-related domain that shares sequence homology with DNA photolyase, and a C-terminal domain that has little sequence similarity to any known proteins. The photolyase-related domain lacks photorepair activity, but shows blue/UV-A light-absorbing capacity through its binding to chromophores. By analogy to photolyase, the crys are expected to function by mediating light-dependent redox reactions. Crys also mainly localize in the nucleus (Fig. 2), and control the expression of a number of genes under blue light conditions (reviewed by Jiao et al. 2007).
Genetic studies have indicated that phys and crys function positively in phototropism. The first positive hypocotyl curvature (reviewed by Poff et al. 1994) in response to unilateral blue light pulse irradiation is reduced in both phyA phyB and cry1 cry2 double mutants (Janoudi et al. 1997b, Lascève et al. 1999). A pulse of red light given 2 h before unilateral blue light enhances the first positive curvature in a phyA- and phyB-dependent manner (Parks et al. 1996, Janoudi et al. 1997a, Stowe-Evans et al. 2001). The second positive hypocotyl curvature in response to unilateral blue light irradiation for ≥15 min (Janoudi et al. 1992) appears normal in phy and cry mutants, but is remarkably reduced in phyA cry1 cry2 triple mutants and phyA phyB cry1 cry2 quadruple mutants, particularly under high fluence rate conditions (Tsuchida-Mayama et al. 2010). These results suggest that phys and crys sometimes function redundantly in hypocotyl phototropism. Kang et al. (2008) indicated that the cry1 cry2 double mutation in the phot1 mutant background causes a remarkable decrease in hypocotyl curvatures during the phot2-mediated phototropic response under high-fluence blue light. Although Whippo and Hangarter (2003, 2004) reported that phyA and crys contribute to the attenuation of hypocotyl phototropism under high fluence rates of blue light (100 µmol m−2 s−1), this attenuation is probably caused by phyA- and cry-mediated hypocotyl growth inhibition under these light conditions.
The functional roles of phys and crys in phototropism appear to be exerted through the modulation of three distinct classes of target mechanism. The first of these is phototropin signaling. Activation of phys and crys promotes the transcription of the phototropin-binding protein RPT2, induction of which is required for phototropic responses, particularly at high fluence rates (Tsuchida-Mayama et al. 2010). Phys also control the subcellular localization of phot1 and the phosphorylation of PKS1 (Fankhauser et al. 1999, Han et al. 2008), although the significance of these events for the phototropic response is an unresolved issue. The second target mechanism is auxin signaling. Basipetal auxin transport in the hypocotyls of etiolated seedlings is suppressed by red light irradiation for 2 h in a phyA- and phyB-dependent manner (Nagashima et al. 2008a). The auxin transporter ATP-binding cassette subfamily B-type19 (ABCB19) protein plays an important role in this process and its expression in hypocotyls is suppressed by red light irradiation for ≥4 h in a phyA- and phyB-dependent manner (Blakeslee et al. 2007, Nagashima et al. 2008a). Activation of crys also causes suppression of ABCB19 expression (Nagashima et al. 2008a). In addition, because the abcb19 loss-of-function mutant shows significant phototropic curvature (Noh et al. 2003), the suppression of ABCB19 expression through the activation of phys and crys appears to contribute to the enhancement of phototropic responses. Light irradiation also influences other auxin transport-related genes, including PIN-FORMED1 (PIN1) (Blakeslee et al. 2007), PIN3 and PIN7 (Devlin et al. 2003), and PINOID (PID) (Ding et al. 2011), and these regulatory processes might also affect phototropic curvature. Continuous irradiations of red and blue light for 6 h cause about 25% decreases in the free IAA contents of the aerial portions of etiolated Arabidopsis seedlings in a phys- and cry-dependent manner, respectively (Nagashima et al. 2008a). In this context, red light irradiation induces the CYP83B1 gene encoding the enzyme that catalyzes the conversion of IAA precursors to indole glucosinolates and reduces IAA biosynthesis (Hoecker et al. 2004). Furthermore, phyA shows phosphorylation activities toward the AUX/IAA proteins in vitro (Colón-Carmona et al. 2000), and may modulate auxin-regulated gene expression. Thus, phys and crys may affect phototropic curvatures not only through auxin transport but also through auxin biosynthesis and/or metabolism and signaling.
The third target mechanism includes other pathways that indirectly affect the phototropic response. Because the gravitropic response counteracts the phototropic response, factors that control the gravitropic response indirectly regulate the phototropic response (reviewed in Hangarter 1997, Iino 2006). For example, the auxin transporter mutants aux1 and pin2/agravitropic1 (agr1), and the starchless mutants phosphoglucomutase1 (pgm1) and adp-glucose pyrophosphorylase1 (adg1) show a defective gravitropic response, resulting in the enhancement of the phototropic response in roots (Okada and Shimura 1992, Okada and Shimura 1994, Vitha et al. 2000). The findings of several studies have indicated that phys and crys suppress gravitropic responses in Arabidopsis hypocotyls (Robson and Smith 1996, Lariguet and Fankhauser 2004, Ohgishi et al. 2004, Nagashima et al. 2008a), and these effects appear to contribute to the enhancement of phototropic responses. Recently, Kim et al. (2011) have further clearly indicated that phyA and phyB inhibit hypocotyl negative gravitropism by converting starch-filled gravity-sensing endodermal amyloplasts to other plastids with chloroplastic or etioplastic features under exposure to red or far-red light.
Gibberellin signaling is also indirectly involved in the phototropic response. Five DELLA family proteins, GAI (gibberellin insensitive), RGA (repressor of ga1-3), RGA-like1 (RGL1), RGL2 and RGL3, are suppressors of gibberellin signaling (reviewed by Schwechheimer 2008), and gai rga rgl1 rgl2 rgl3 quintuple mutants show a slight decrease in phototropic and gravitropic curvatures (Tsuchida-Mayama et al. 2010). Treatment with the gibberellin biosynthesis inhibitor paclobutrazol (PAC) causes not only an enhancement of the gravitropic response in wild-type hypocotyls kept in the dark but also a partial recovery of the phototropic response in phyA cry1 cry2 triple mutants under high fluence rates of blue light. Gallego-Bartolomé et al. (2011) indicated that PAC treatment decreases the transcription of IAA19/MASSUGU2 (MSG2), a gene that encodes MSG2, a negative regulator of the auxin transcriptional factor NPH4/auxin-responsive factor7 (ARF7) (Tatematsu et al. 2004). ARF7 is a positive regulator of hypocotyl tropism (Harper et al. 2000). Thus, gibberellin signaling appears to have an inhibitory effect on hypocotyl tropism. The active gibberellin GA4 content in etiolated seedlings is dramatically decreased by blue light irradiation at high fluence rates in a cry-dependent manner (Zhao et al. 2007, Tsuchida-Mayama et al. 2010), suggesting that gibberellin suppresses hypocotyl bending under darkness and crys modulate the phototropic response through a reduction in gibberellin. On the other hand, gibberellin treatment does not suppress the phototropic response in wild-type seedlings (Tsuchida-Mayama et al. 2010). PhyA and/or crys might control not only the gibberellin content, but also gibberellin sensing and/or signaling processes that affect hypocotyl phototropism.
Phototropin Signaling Components
NPH3
nph3 mutants show a complete phototropic response defect under any light conditions, and NPH3 thus appears to be an essential factor for the signal transduction pathways of both phot1 and phot2 in phototropism (Liscum and Briggs 1996). The NPH3 gene was first identified by positional cloning in 1999 (Motchoulski and Liscum 1999). JK218 and rpt3 were found to be allelic with nph3 (Liscum and Briggs 1996, Sakai et al. 2000). NPH3 encodes a protein possessing two protein–protein interaction domains, a BTB/POZ (broad-complex, tramtrack and bric-à-brac/Pox virus and zinc finger) domain and a coiled-coil domain at the N- and C-termini, respectively, and appears to play a role as an adaptor protein (Motchoulski and Liscum 1999). The NPH3 proteins are localized to the plasma membrane and interact with phot1 and phot2 in vitro and in vivo (Fig. 2) (Motchoulski and Liscum 1999, Lariguet et al. 2006, de Carbonnel et al. 2010). Furthermore, the NPH3 ortholog in rice, coleoptile phototropism1 (CPT1), has been shown to be an essential mediator of auxin redistribution in coleoptiles during the phototropic response (Haga et al. 2005).
The NPH3 proteins are phosphorylated in etiolated seedlings under darkness and dephosphorylated by blue light irradiation in a phot1-dependent manner (Pedmale and Liscum 2007). Although these events seem to be involved in the regulation of the phototropic response, their biological significance remains to be determined. However, it is known that dephosphorylation of NPH3 is not essential for the phot2-induced phototropic response because phot1 mutants display phototropic responses without the dephosphorylation of NPH3 under high-intensity blue light conditions (Tsuchida-Mayama et al. 2008). These authors also identified three phosphorylated serine residues in a hinge region between the BTB domain and the NPH3 domains, and revealed that these phosphorylated residues are not required for the phot1-induced phototropic response under low-intensity blue light irradiation. A remaining question is whether the dephosphorylation of NPH3 is required for phot1 signaling in phototropic responses.
Roberts et al. (2011) recently indicated that NPH3 shows binding activity to cullin3a (CUL3a), a subunit of E3 ubiquitin ligase, in mammalian cells and Nicotiana benthamiana leaf epidermal cells, and they are necessary for both mono-/multi- and polyubiquitination of PHOT1 proteins in blue light-irradiated seedlings of Arabidopsis (Fig. 2). The Arabidopsis genome contains two CUL3 genes, CUL3a and CUL3b, and cul3a-1 cul3b-1 double mutants show an embryonic lethal phenotype (Figueroa et al. 2005, Thomann et al. 2005). Cul3a-3 cul3b-1 hypomorphic mutants (cul3a-3 is a weak allele) show a moderate phenotype in terms of hypocotyl phototropism, suggesting that CUL3–NPH3-containing E3 ubiquitin ligase ubiquitinates phot1 and controls hypocotyl phototropism (Roberts et al. 2011). The authors hypothesized that mono-/multiubiquitination of phot1 under low-intensity blue light causes phot1 internalization coupled to a blue light-induced relocalization of auxin transporters and polyubiquitination, and that subsequent degradation of phot1 under high-intensity blue light causes receptor desensitization. This model is not consistent with an earlier hypothesis that phyA enhances the phototropic response by suppressing phot1 internalization (Han et al. 2008). More detailed examinations of these possibilities are anticipated in the near future.
A recent study has identified an NPH3-binding protein, enhanced bending 1 (EHB1), as a small protein of 25 kDa with a C2/CaLB Ca2+-binding domain at its N-terminus and harboring a motif that is similar to the ZAC ARF-GAP (GTPase-activating protein for ADP-ribosylation factor) protein at its C-terminus (Knauer et al. 2011). Interestingly, ehb1 mutants show enhanced hypocotyl curvatures following phototropic and gravitropic stimulations, suggesting that this protein is a negative effector of blue light-induced and gravitropic bending (Knauer et al. 2011). EHB1 may also transduce signals from a phototropin- or auxin-driven calcium influx to vesicle trafficking mechanisms, although the function of Ca2+ influx in phototropism is not yet understood (Harada and Shimazaki 2007).
RPT2
rpt2 mutants are almost completely lacking in a root phototropic response, but show a moderate phenotype in terms of their hypocotyl phototropic response (Sakai et al. 2000). The nucleotide sequence of the RPT2 gene is similar to that of the NPH3 gene, and it encodes a protein harboring BTB/POZ, NPH3 and coiled-coil domains (Sakai et al. 2000). RPT2 proteins form complexes with phot1 and NPH3 in vivo, and localize at the plasma membrane (Fig. 2) (Inada et al. 2004). The phototropic curvature of rpt2 hypocotyls is induced by a low fluence rate of blue light, but the degree of this curvature decreases as the fluence rate increases. Thus, the rpt2 mutant phenotype appears to be an enhanced effector adaptation during the phototropic response. Genetic studies have now revealed that phot1 functions not only positively in the presence of RPT2 but also negatively in its absence during the phototropic response of hypocotyls to high fluence rates, suggesting that this negative effect causes the rpt2 phenotype and that RPT2 is involved in the modulation of the phot1 function, particularly under high-intensity blue light (Inada et al. 2004).
RPT2 transcripts are strongly induced by both blue and red light irradiation in etiolated seedlings (Sakai et al. 2000). Tsuchida-Mayama et al. (2010) indicated that the transcriptional induction of this gene under blue light conditions is dependent on phys and crys. A phyA cry1 cry2 mutant showed moderate curvatures under low fluence rates (0.01 and 0.1 µmol m−2 s−1) but barely any hypocotyl curvature under high fluence rates (10 and 100 µmol m−2 s−1). Thus, either phys or crys are required for hypocotyl phototropism under high fluence rates of blue light. This phenotype is similar to that of rpt2, and the constitutive expression of RPT2 driven by the Cauliflower mosaic virus 35S promoter (35S) partially complements the non-phototropic hypocotyl phenotype of phyA cry1 cry2 under high fluence rates of blue light (Tsuchida-Mayama et al. 2010). These results indicate that phys and crys control both the phot1 signaling pathway and phototropism through the induction of RPT2 expression.
The Arabidopsis genome encodes 32 genes belonging to the NPH3/RPT2-like (NRL) family (Sakai 2005), and the functions of other members of this family have been reported recently. Naked pins in yuc 1 (NPY1)/macchi-bou4 (MAB4)/At4g31820, NPY2/At2g14820, NPY3/At5g67440, NPY4/At2g23050 and NPY5/At4g37590 function redundantly in auxin-mediated organogenesis and root gravitropism with the AGC3 (protein kinase A, cGMP-dependent protein kinase and protein kinase C) kinase family (Cheng et al. 2007, Cheng et al. 2008, Li et al. 2011). SETH6/At2g47860 is required for pollen germination and pollen tube growth (Lalanne et al. 2004). A defect in the defectively organized tributaries 3 (dot3)/At5g10250 gene causes pleiotropic effects, and dot3 mutants develop a small body size, abnormal vein patterning, a stunted primary root, fused rosette leaves and low fertility (Petricka et al. 2008). NPH3-LIKE/At1g30440 was identified as a phot1-binding protein (Sullivan et al. 2009). These results suggest that NRL family members probably function in auxin-regulated cell elongation and differentiation. Further functional analyses of these factors would be very beneficial for our further understanding of the molecular functions of NPH3 and RPT2 in phototropism.
PKS family
PKS family members are also known as signal transducers of phototropism. PKS1, which was identified as a substrate for light-regulated phy kinase activity in vitro, is a phy signaling component belonging to a small protein family in Arabidopsis (PKS1–PKS4) (Fankhauser et al. 1999, Lariguet et al. 2006). PKS1 expression is transiently and precisely induced by light in the elongation zone of the root and hypocotyl, suggesting the possibility of its involvement in phototropism (Lariguet et al. 2003). Boccalandro et al. (2008) indicated that the PKS1 expression dependent on phyA is necessary for root negative phototropism, suggesting that the transcriptional regulation of PKS1, as well as that of RPT2, is crucial for the regulation of the phototropic response in roots by phy. Lariguet et al. (2006) observed that the hypocotyls of pks1, pks2 and pks4 mutants show phot1-induced phototropic response defects and that PKS1 interacts with phot1 and NPH3 in vivo (Fig. 2). Double pks mutants and pks1 pks2 pks4 triple mutants show enhanced phenotypes, suggesting that they function redundantly in hypocotyl phototropism. The molecular functions of the PKS proteins and the functional significance of their interactions with phys remain open questions in terms of further elucidating the phototropic response pathways.
Auxin Transporters and Their Regulatory Factors
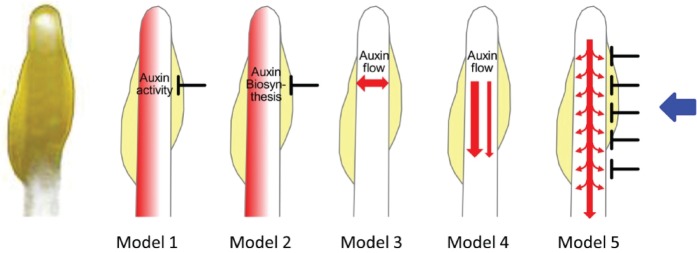
Asymmetric auxin distribution is caused by unilateral blue light irradiation and occurs in a phototropin signaling-dependent manner (Haga et al. 2005). Auxin induces the transcriptional activation of the expansin genes EXPA1 and EXPA8 (Esmon et al. 2006), which are involved in plant cell wall expansion (reviewed by Cosgrove 2000). Furthermore, many phototropism mutants show auxin-related signaling defects, as described in this review. These results support the Cholodny–Went theory, which suggests that asymmetric auxin distribution causes phototropic responses (reviewed in Went and Thimann 1937, Tanaka et al. 2006, Whippo and Hangarter 2006, Holland et al. 2009). Auxin biosynthesis appears to occur mainly in young leaves and shoots (reviewed by Benjamins et al. 2005). There are several hypotheses for the mechanisms causing asymmetric auxin distribution: a lateral accumulation of auxin on the shaded side of the plant occurs because of either light-induced inactivation (hypothetical model 1), light-induced inhibition of auxin biosynthesis (hypothetical model 2), light-induced lateral auxin transport from the irradiated side to the shaded side (hypothetical model 3), light-mediated inhibition of polar auxin transport from the apical to the basal parts (hypothetical model 4) or light-induced inhibition of lateral auxin transport from its main basipetal flow in the vasculature to the outer cell layers (hypothetical model 5) (Fig. 3). Among these possibilities, the functions of auxin metabolism and biosynthesis in phototropism have not yet been examined using molecular genetic approaches. Recent studies have revealed that the main auxin biosynthesis pathway is mediated by both tryptophan aminotransferase of Arabidopsis (TAA) and YUCCA (YUC) flavin monooxygenase-like proteins (Mashiguchi et al. 2011, Stepanova et al. 2011, Won et al. 2011). Consequently, the functions of these genes in phototropism will probably be elucidated in the near future. Physiological studies have suggested that the lateral and basipetal auxin transport systems are important for phototropism, and many studies have focused on elucidating the function of the auxin transporters in phototropism (reviewed in Briggs 1963, Poff et al. 1994, Iino 2001, Liscum 2002, Holland et al. 2009). Three kinds of auxin transporter families, the plant-specific PIN family, the ABCB transporters and the amino acid permease-like auxin-resistant1 (AUX1)/like-AUX (LAX) family, have been identified in plants (reviewed in Blakeslee et al. 2005, Robert and Friml 2009, Zažímalová et al. 2010). Reverse genetic approaches have also been employed to reveal the functions of these genes in phototropism.
Table 1.
Genes involved in phototropism in Arabidopsis
| Gene | Protein function | Mutant phenotypea | Reference(s) |
|---|---|---|---|
| Photoreceptors | |||
| CRY (family) | Blue/UV-A light photoreceptor | Reduced hypocotyl curvature (LOF) | Lascève et al. (1999), Whippo and Hangarter (2004), Kang et al. (2008), Tsuchida-Mayama et al. (2010) |
| PHOT1/NPH1/JK224 | Blue/UV-A light photoreceptor | No root curvature, no hypocotyl curvature under low fluence rate blue light (LOF) | Khurana and Poff (1989), Okada and Shimura (1992), Liscum and Briggs (1995), Sakai et al. (2000) |
| PHOT2/NPL1 | Blue/UV-A light photoreceptor | Reduced hypocotyl curvature in the phot1 mutant background | Sakai et al. (2001) |
| PHY (family) | Red/FR/blue light photoreceptor | Reduced hypocotyl curvature (LOF) | Parks et al. (1996), Janoudi et al. (1997a, 1997b), Stowe-Evans et al. (2001), Whippo and Hangarter (2003), Tsuchida-Mayama et al. (2010) |
| Phototropin signaling components | |||
| CUL3 (family) | CULLIN3 | Reduced hypocotyl curvature (LOF) | Roberts et al. (2011) |
| EHB1 | C2/CalB Ca2+-binding domain | Enhanced hypocotyl curvature (LOF) | Knauer et al. (2011) |
| NPH3/RPT3/JK218 | BTB | No hypocotyl/root curvature under any light conditions (LOF) | Khurana and Poff (1989), Okada and Shimura (1994), Liscum and Briggs (1996) |
| PKS (family) | Unknown | Reduced hypocotyl/root curvature (LOF) | Lariguet et al. (2006), Boccalandro et al. (2008) |
| RPT2 | BTB | Reduced hypocotyl/root curvature (LOF) | Okada and Shimura 1992, Sakai et al. (2000) |
| RCN1 | PP2A | Enhanced phot2-mediating phototropic responses in hypocotyls | Tseng and Briggs (2010) |
| Auxin transporters and their regulatory factors | |||
| ABCB19 | ABCB auxin transporter | Enhanced hypocotyl curvature (LOF) | Noh et al. (2003) |
| AGC3 (family) | AGC kinase | Reduced hypocotyl curvature (LOF and OE) | Ding et al. (2011) |
| AUX1/LAX (family) | Auxin influx carrier | Enhanced root curvature (LOF), reduced hypocotyl curvature in the nph4 mutant background (LOF), reduced hypocotyl curvature (LOF) | Okada and Shimura (1992), Watahiki et al. (1999), Stone et al. (2008), Christie et al. (2011) |
| GNOM | ARF-GEF | Reduced hypocotyl curvature (LOF) | Ding et al. (2011) |
| PIN2/AGR1 | Auxin efflux carrier | Enhanced root curvature (LOF) | Okada and Shimura (1994) |
| PIN3 | Auxin efflux carrier | Reduced (enhanced) hypocotyl curvature in etiolated (de-etiolated) seedlings (LOF) | Friml et al. (2002), Christie et al. (2011), Ding et al. (2011) |
| PIN7 | Auxin efflux carrier | Reduced hypocotyl curvature (LOF) | Christie et al. (2011), Ding et al. (2011) |
| Auxin action mechanisms | |||
| ABP1 | Unknown | Reduced hypocotyl curvature (LOF) | Effendi et al. (2011) |
| ARF7/NPH4 | Transcriptional factor | Reduced hypocotyl curvature (LOF) | Liscum and Briggs (1996) |
| ARF19 | Transcriptional factor | Recovered hypocotyl curvature in arf7 (OE) | Okushima et al. (2005), Stone et al. (2008) |
| AXR1 | Subunit of the RUB E1-activating enzyme | Reduced hypocotyl curvature (LOF) | Watahiki et al. (1999) |
| IAA19/MSG2 | Transcriptional repressor | Reduced hypocotyl curvature (GOF) | Tatematsu et al. (2004) |
| TIR1/AFB (family) | F-box protein | Reduced hypocotyl curvature (LOF) | Whippo and Hangarter (2005), Möller et al. (2010) |
| Others | |||
| ADG1 | ADP glucose pyrophosphorylase | Enhanced root curvature (LOF) | Vitha et al. (2000) |
| BAK | Leucine-rich repeat receptor-like protein kinase | Reduced hypocotyl curvature (OE) | Whippo and Hangarter (2005) |
| BIG/TIR3 | Calossin-like protein | Reduced hypocotyl curvature under high fluence rate blue light (LOF) | Whippo and Hangarter (2005) |
| DELLA (family) | Transcriptional regulators | Reduced hypocotyl curvature under high fluence rate blue light (LOF) | Tsuchida-Mayama et al. (2010) |
| HY5 | bZIP transcriptional factor | Reduced (enhanced) hypocotyl curvature under low (high) fluence rate blue light (LOF) | Whippo and Hangarter (2005) |
| PGM1 | Phosphoglucomutase | Enhanced root curvature (LOF) | Vitha et al. (2000) |
| SPY | O-GlcNac transferase | Reduced hypocotyl curvature under low fluence rate blue light(LOF) | Whippo and Hangarter (2005) |
a Mutant phenotype caused by: GOF, gain of function; LOF, loss of function; OE, overexpression.
Fig. 3.
Hypothetical models of auxin asymmetry during hypocotyl phototropism. The picture on the left shows the apical region of an etiolated Arabidopsis seedling taken from the opposite side of the cotyledon attachment side of the hook. Following phototropic stimulation, auxin asymmetry is established by either light-induced inactivation of auxin on the irradiated side (Model 1), light-induced inhibition of auxin biosynthesis on the irradiated side (Model 2), light-induced lateral transport of auxin from the irradiated side to the shaded side (Model 3), light-induced inhibition of basipetal transport of auxin on the irradiated side (Model 4) or light-induced inhibition of lateral transport of auxin from the main auxin flow on the irradiated side (Model 5). Red and blue arrows indicate flow of auxin and phototropic stimulation, respectively.
The PIN auxin efflux carrier family
The PIN family in Arabidopsis consists of eight members and is divided into two subclades that differ in terms of the length of a hydrophilic loop between the C-terminal and N-terminal transmembrane domains (reviewed by Zažímalová et al. 2010). PIN1, PIN2, PIN3, PIN4 and PIN7 have long hydrophilic loops, show a mostly polar plasma membrane localization, and function as efflux carriers that transport auxin from the cytosol to the extracellular matrix. In contrast to these long-loop PINs, the hydrophilic loop is short in PIN6, and almost absent in PIN5 and PIN8. These short-loop PINs do not localize to the plasma membrane, but to the endoplasmic reticulum (ER), and presumably mediate auxin flow from the cytosol to the lumen of the ER (Mravec et al. 2009). During phototropism, cell to cell auxin transport appears to be more important than the regulation of inner auxin homeostasis, and the functions of the long-loop PINs in the hypocotyls, particularly PIN1, PIN3, PIN4 and PIN7, have been well analyzed.
Etiolated seedlings of isolated pin3 mutants have been found to show a slight decrease in the phototropic curvature of their hypocotyls (Friml et al. 2002, Ding et al. 2011), suggesting that PIN3 has a role in hypocotyl phototropism. Ding et al. (2011) indicated that under conditions of darkness, PIN3 proteins are expressed in the hypocotyl endodermis, which constitutes a barrier between the vasculature and the outer cell layers, and show an apolar localization in endodermal cells. Under unilateral white light irradiation (2 µmol m−2 s−1), the PIN3 protein levels were found to be greatly decreased in the outer lateral side of the endodermal cells on the irradiated hypocotyl side in a phot1-dependent manner (Fig. 2). When the auxin distribution pattern was observed using the auxin reporter gene DR5rev (a synthetic auxin-inducible promoter):GFP (green fluorescent protein) (Friml et al. 2003), pin3 mutant seedlings showed less asymmetry in their DR5 activity between the epidermis of the irradiated hypocotyl side and the shaded hypocotyl side compared with wild-type seedlings. This suggests that the auxin concentration in the epidermis of the irradiated side of the seedling is lower than that in the shaded side. These observations suggest that PIN3 proteins in the endodermal cells of the irradiated hypocotyl side redirect auxin back into the vasculature and block its flow from the vasculature to the outer cell layers, most notably in the epidermis. This would promote the observed asymmetric auxin distribution and subsequent phototropic response in hypocotyls. Thus, the observed functions of PIN3 appear to be most consistent with hypothetical model 5 (Fig. 3).
Because the pin3 mutant phenotype is moderate, the redundant function of other PIN proteins in phototropism is under examination. Christie et al. (2011) evaluated the phototropic response in de-etiolated, dark-acclimated pin3 mutant Arabidopsis seedlings. Unexpectedly, pin3 mutants exhibited a higher curvature than the wild type, and analysis using DR5rev:GFP in this report indicated that the primary site of lateral auxin translocation is at and above the hypocotyl apex, as described in hypothetical model 3 (Fig. 3), rather than the hypocotyl elongation zone, at which PIN3 polar localization was reported in the study of Ding et al. (2011). These results also suggest that other key players in addition to PIN3 cause asymmetric auxin distribution during phototropic responses.
Ding et al. (2011) recently examined phototropic responses in etiolated seedlings of the pin3 pin7 and pin2 pin3 pin7 mutants, and their findings indicate that the phototropic curvature of pin3 is slightly decreased by the additional mutation in pin7, but not in pin2. Christie et al. (2011) also examined phototropic responses in other mutants including pin1, pin2, pin4 and pin7 using de-etiolated, dark-acclimated seedlings. The pin1, pin2 and pin4 mutants in this case showed normal phototropic responses, but the pin7 mutant exhibited a lower phototropic curvature than the wild type. These results suggest that PIN3 and PIN7 function in hypocotyl phototropism, but that this is altered in response to the status of seedling photomorphogenesis. Žádníková et al. (2010) indicated that the PIN7 promoter (pro):PIN7-GFP signal is strong in epidermal cells at the hypocotyl elongation zone in an apolar manner, and thus it is possible that PIN7 may contribute to auxin transport and accumulation in the epidermis of the shaded hypocotyl side.
Blakeslee et al. (2004) reported that PIN1 immunolocalization shows a gradient of delocalization, with a disruption of the basal localization of PIN1 in the cortical cells of the shaded side of the mid-hypocotyl region in a phot1-dependent manner, suggesting that PIN1 plays some role in the phototropic response. However, their later study indicated that PIN1pro:PIN1-GFP signals are restricted to the vascular tissue below the cotyledonary node and suggest that the PIN1 immunolocalization signal previously observed in cortical and bundle sheath cells (Noh et al. 2003, Blakeslee et al. 2004) was non-specific (Blakeslee et al. 2007). It is also noteworthy that none of the reported genetic studies to date has detected any contribution of PIN1 to phototropic responses.
Although a further examination of the phototropic responses in higher order pin mutants such as the pin1 pin3 pin4 pin7 quadruple mutant is strongly expected to reveal a redundancy of function among the PIN proteins, these multiple mutants show strong developmental defects (Friml et al. 2003), which will create technical challenges for such analyses. Hence, the significance of the PIN3 function in vivo has not been demonstrated through genetic approaches as yet, although one of the current models of phototropism involving PIN3 polar localization (hypothetical model 5 in Fig. 3) has potential and can elegantly explain some of the observed phenomena in relation to this process. Furthermore, how the phototropin signaling pathways promote PIN3 polar localization in the endodermis and what the functions of phototropin signaling are in other tissues including the epidermis and cortex remain to be determined.
AGC3 kinases: PID, WAG1 and WAG2
PINOID (PID), WAG1 and WAG2 protein kinases (AGC3 kinases) show strong homologies with the kinase domains of phototropins and are regulators of polar auxin transport (reviewed by Robert and Offringa 2008). A pid loss-of-function mutant was originally isolated as a clone showing apical organogenesis defects similar to those of the pin1 auxin efflux carrier mutant (Bennett et al. 1995). WAG1 and WAG2 genes were identified as PID homologs, and a reverse genetic approach revealed that wag1 wag2 double mutants exhibit a pronounced wavy root phenotype when grown vertically on agar plates (Santner and Watson 2006). Because the wavy root growth appeared to be involved in the tropic and auxin responses (Okada and Shimura 1994, Sakai et al. 2012), WAG1 and WAG2 therefore appear to control polar auxin transport. The overexpression of PID, WAG1 or WAG2 induces the movement of the auxin efflux carriers of PIN1, PIN2 and PIN4 from the basal to the apical side of the cells via phosphorylation of these PIN proteins (Friml et al. 2004, Dhonukshe et al. 2010). This phosphorylation-triggered apical PIN recycling competes with ARF-GEF (a guanine nucleotide exchange factor for ADP-ribosylation factor GTPase) GNOM-dependent basal recycling, and wag1 wag2 pid triple mutants show defects in apical PIN recycling (Dhonukshe et al. 2010). These kinases localize at the plasma membrane in an apolar manner and phosphorylate the middle serine in three conserved TPRXS(N/S) motifs within the PIN central hydrophilic loop (Dhonukshe et al. 2010). The TPRXS(N/S) motifs are well conserved in PIN1, PIN2, PIN3, PIN4, PIN6 and PIN7 (Huang et al. 2010), suggesting that these AGC3 kinases also phosphorylate PIN3, PIN6 and PIN7, and thereby also control the apical recycling of these proteins. These findings raise the possibility that AGC3 kinases function in the control of phototropic responses.
The role of PID function in tropic responses has recently been reported (Ding et al. 2011). The hypocotyls of 35S:PID seedlings, which strongly and constitutively express the PID gene, show significantly disrupted phototropic and gravitropic responses, but display normal hypocotyl growth (Ding et al. 2011, Rakusová et al. 2011). PID shows kinase activity toward the PIN3 proteins in vitro, and polarization of PIN3 in response to blue light and gravistimulation does not occur in the hypocotyls of 35S:PID seedlings (Ding et al. 2011, Rakusová et al. 2011). This suggests that the constitutive phosphorylation of PIN3 inhibits its polarization and subsequent asymmetric auxin distribution during tropic responses in 35S:PID seedlings. Furthermore, the transcription of PID was found to be repressed by white and blue light irradiation of seedlings, and PID expression appeared to be higher on the shaded side compared with the illuminated side. Therefore, Ding et al. (2011) hypothesized that the repression of PID transcription on the illuminated side of a seedling causes a decrease in PIN3 phosphorylation and subsequent enhancement of PIN3 relocalization, resulting in targeting of this protein to the inner cell side (Fig. 2). This in turn causes asymmetric auxin distribution and a differential phototropic response.
In contrast to gain-of-function mutants, a wag1 wag2 pid triple mutant shows opposite effects for the two different tropic responses. The triple mutant hypocotyls show a moderate phototropic curvature with suppression of blue light-induced PIN3 polarization, but a hypergravitropic curvature with gravity-induced PIN3 polarization. A simple explanation for this phenotype is that PID, WAG1 and WAG2 play a significant role in the phototropic response, but not in the gravitropic response, and that other regulators control the phosphorylation status and polarization of PIN3 proteins in response to gravistimulation. Dhounukshe et al. (2010) indicated that a basal to apical shift of PIN2 is not induced by AGC1 or AGC2 kinases (AGC kinases are described in later sections), suggesting that these kinases are not involved in this basal to apical shift regulation. The phosphorylation status of PIN1 is regulated antagonistically by PID and PP2A (Michniewicz et al. 2007). In addition to PIN1, the phosphorylation status of PIN3 may also be regulated by PP2A, and PIN3 dephosphorylation, rather than its phosphorylation, may be the crucial event in gravitropic responses. However, a loss-of-function mutant in one of the PP2A regulatory subunits, rcn1/pp2aa1, shows an enhanced hypocotyl gravitropic curvature similar to that of the wag1 wag2 pid triple mutant under darkness (Muday et al. 2006) and normal phot1-mediated phototropic responses (Tseng and Briggs 2010). Therefore, the role of PP2A in the regulation of the PIN3 phosphorylation status remains to be elucidated. There is also a possibility that the hypergravitropic response indirectly causes a decrease in the phototropic curvature in the hypocotyls of wag1 wag2 pid mutants and/or that PID kinases are required only to fine-tune the extent of hypocotyl bending in response to light and gravity. Thus, the functional role of AGC3 kinases in phototropic responses remains an open question.
If the phototropin signaling pathways promote phototropic responses through AGC3, an important mechanistic question is how phototropins control the activities of AGC3 kinases. Ding et al. (2011) reported that phot1 phot2 double mutants show lower light-mediated repression of PID transcription and that this repression causes PIN3 polarization on the illuminated side of the seedlings. However, their data appear to show that this repression in phot1phot2 seedlings occurs moderately. Moreover, phot1 single mutants do not show any abnormality in PID expression under blue light irradiation at 2 µmol m−2 s−1 (Ding et al. 2011), although phot1 mutants show an abnormal phototropic response under this condition (Sakai et al. 2001). Observations of PID protein distribution patterns may facilitate examination of this hypothesis.
PID kinase activity is negatively regulated by calcium in vivo (Benjamins et al. 2003). PID and other AGC3 kinase activities are repressed by binding of TOUCH3 (TCH3) and enhanced by binding of PINOID BINDING PROTEIN1 (PBP1), in a calcium-dependent manner (Benjamins et al. 2003, Robert and Offringa 2008). The activation of phototropins induces an increase in the cytosolic Ca2+ concentration (Baum et al. 1999, Babourina et al. 2002, Harada et al. 2003, Stoelzle et al. 2003). Although the function of calcium signaling in phototropism is not yet clear (Harada and Shimazaki 2007), it has been proposed that phototropin signaling may initiate the relocalization of PINs by modulating the activity of AGC3 kinases through their calcium-induced interaction with TCH3 and PBP1 (Robert and Offringa 2008).
PID, WAG1 and WAG2 protein kinases belong to the AGCVIII plant-specific subfamily of AGC kinases (Bögre et al. 2003, Galván-Ampudia and Offringa 2007). The Arabidopsis genome encodes 37 AGC kinases, of which 23 are classified as AGCVIII group members (Bögre et al. 2003, Galván-Ampudia and Offringa 2007). Galván-Ampudia and Offringa (2007) further classified the AGCVIII kinases into four distinct groups, AGC1–AGC4. PID, WAG1 and WAG2 belong to the AGC3 group, with an additional member, AGC3-4. Importantly, PHOT1 and PHOT2 also belong to the AGCVIII kinase family and form the AGC4 group. Both the AGC3 and AGC4 kinases appear to control polar auxin transport along with NRL family members, including NPH3, RPT2 and NPYs, and a symmetry between these two signaling pathways has been proposed (Galván-Ampudia and Offringa 2007, Robert and Offringa 2008). Although phot1 does not show PIN3 phosphorylation activity in vitro (Ding et al. 2011), it is possible that phototropin AGC4 kinases may phosphorylate PIN proteins and control their polarization in vivo.
ARF-GDP/GTP exchange factor GNOM
GNOM is an ARF-GEF factor that is important for coat recruitment and cargo-selective vesicle trafficking, and is necessary for embryonic axis formation and polar auxin transport mediated by PIN auxin efflux carriers (Fig. 2) (Mayer et al. 1993, Steinmann et al. 1999, Donaldson and Jackson 2000, Geldner et al. 2003). Its loss-of-function mutant lacks a primary root and shows an invariable phenotype (Mayer et al. 1993, Steinmann et al. 1999). The partial loss-of-function gnomR5 allele shows strong defects in hypocotyl phototropism and gravitropism (Ding et al. 2011, Rakusová et al. 2011), in addition to defects in root gravitropism and hydrotropism (Geldner et al. 2004, Miyazawa et al. 2009). This evidence suggests that GNOM-dependent PIN polarization is required for differential growth in response to light, gravity and water. However, the plasma membrane localization of phot1 and phot2 is sensitive to the vesicle trafficking inhibitor brefeldin A (BFA), which inhibits GNOM-mediated vesicle trafficking (Kong et al. 2006, Kaiserli et al. 2009). Hence, the phototropic response appears to require GNOM-mediated vesicle trafficking, which transports not only the PIN auxin efflux carriers, but also many other plasma membrane-associated proteins including the phototropins.
The ABCB19 auxin transporter
The ABCB/PGP (P-glycoprotein)/MDR (multidrug resistance) transporters harbor two ABC domains and are well conserved among prokaryotes and eukaryotes (reviewed by Martinoia et al. 2002). The Arabidopsis genome encodes 22 ABCB genes and, although most ABCB transporters have yet to be functionally characterized, three of these proteins, ABCB1, ABCB4 and ABCB19, have been identified as auxin transporters (reviewed by Titapiwatanakun and Murphy 2009). ABCB1 and ABCB19 show auxin efflux activity, and ABCB4 shows both efflux and influx characteristics. abcb19 mutants show strong hypocotyl curvatures in response to light and gravity, indicating that ABCB19 is a negative regulator of hypocotyl bending (Noh et al. 2003).
Basipetal auxin transport is strongly disrupted in hypocotyls of etiolated seedlings of abcb19 mutants (Noh et al. 2001), suggesting that ABCB19 is required for this shoot apex to root transport mechanism. Christie et al. (2011) reported that phot1 activation reduces basipetal auxin transport in hypocotyls and that phot1 directly phosphorylates ABCB19 proteins in vitro and suppresses its auxin transport activity in HeLa cells. Furthermore, in de-etiolated, dark-acclimated seedlings of abcb19 mutants, DR5rev:GFP expression has been shown to be markedly decreased in the vasculature of hypocotyls and increased in the epidermis of the mid-hypocotyl shaded side. These findings suggest that ABCB19 functions as a target for phot1 action in shoot apical tissues to halt vertical growth initially and concentrate auxin within this region. The suppression of ABCB19 activity by phot1-dependent phosphorylation may cause a redistribution of auxin flow from the vasculature to the epidermis of the shaded hypocotyl side, which causes enhancement of phototropic curvatures. These observations therefore appear to modify hypothetical model 4 (Fig. 3), i.e. that light irradiation suppresses basipetal auxin transport in the vasculature and enhances it in the epidermis of the shaded side of hypocotyls.
Nagashima et al. (2008a) indicated that the ABCB19 proteins are decreased in the upper half of hypocotyls by red or blue light irradiation for ≥4 h. The red light effects are dependent on the phys (phyA and phyB), and the blue light effects require either phys or crys. Consistent with these results, red light irradiation reduces basipetal auxin transport in a phyA- and phyB-dependent manner, suggesting that the suppression of this transport is caused by a reduction in ABCB19 protein levels under red light conditions. Thus, ABCB19 appears to be functionally suppressed under blue light conditions in a rapid manner via protein phosphorylation mediated through phot1 activity, and then by a reduction in its protein levels through the activities of phys or crys, or both (Fig. 2). These molecular mechanisms probably also enhance the phototropic responses in hypocotyls.
Although ABCB19 can function as an auxin efflux transporter by itself, it can also control the auxin transport activity of PIN1 (reviewed by Titapiwatanakun and Murphy 2009). ABCB19 proteins bind to PIN1 proteins in vivo and in yeast, and stabilize their localization at the plasma membrane (Blakeslee et al. 2007, Titapiwatanakun et al. 2009). The co-expression of ABCB19 and PIN1 enhances auxin efflux activity, inhibitor sensitivity and substrate specificity in HeLa cells (Blakeslee et al. 2007). ABCB19 and PIN1 expression patterns overlap at the shoot apex and vasculature in hypocotyls, but polar ABCB19 signals in the bundle sheath and cortical cells do not overlap with PIN1pro:PIN1-GFP signals that are restricted to cells of the vascular parenchyma, suggesting that the direct interaction of ABCB19 and PIN1 is tissue dependent (Blakeslee et al. 2007). Moreover, because ABCB19 also shows binding activity to PIN2 in yeast and in vitro, and its tissue expression overlaps with that of PIN3 and PIN4 in the cortex, this protein may control the activities of other PIN transport pathways including those of PIN3 and PIN4.
ABCB19 proteins bind to a well-characterized inhibitor of auxin efflux, N-1-naphthylphthalamic acid (NPA) (Noh et al. 2001). The inhibitory effects of NPA on auxin transport systems lead to the suppression of both elongation and differential growth in Arabidopsis hypocotyls (Jensen et al. 1998, Friml et al. 2002). Recently, Nagashima et al. (2008b) reported that ABCB19 is required for NPA to exert its inhibitory effects on phototropic and gravitropic responses, but not on elongation, in hypocotyls. Expression analysis of the DR5:GUS (β-glucuronidase) auxin reporter gene has suggested that NPA inhibits hypocotyl tropisms by suppressing the asymmetric distribution of auxin in an ABCB19-dependent manner. NPA may thus exert suppressive effects upon the auxin efflux activities of PINs, which are involved in asymmetric auxin distribution through ABCB19. Further analyses are required to elucidate the relationship between ABCB19 and PINs in the phototropic response.
The AUX1/LAX auxin influx carriers
The AUX1/LAX genes, which encode auxin influx carrier proteins, are related to a family of amino acid/auxin:proton symport permeases, and four of these genes, AUX1, LAX1, LAX2 and LAX3, are present in the Arabidopsis genome (reviewed in Titapiwatanakun and Murphy 2009, Zažímalová et al. 2010). aux1 mutants show normal phototropic responses in their hypocotyls and enhanced phototropic responses in their roots (Okada and Shimura 1992, Watahiki et al. 1999). Genetic studies also indicated that the hypocotyls of de-etiolated seedlings of aux1 lax2 lax3 triple mutants show a slight phototropic curvature decrease in de-etiolated, dark-acclimated seedlings (Christie et al. 2011) and that the aux1 mutation enhances the hypocotyl phototropism phenotype of nph4/msg1 mutants (Watashiki et al. 1999, Stone et al. 2008). However, whether the phototropins control the function of AUX1/LAX auxin influx carriers remains unknown. The evidence obtained thus far indicates that this auxin transporter family makes some contribution to the phototropic response in Arabidopsis hypocotyls.
Auxin Action Mechanisms
The auxin receptors TIR1/AFB and ABP1
The transport inhibitor-resistant1 (TIR1) F-box protein is the first identified auxin receptor (reviewed by Mockaitis and Estelle 2008). The Arabidopsis TIR1/auxin-binding F-box (AFB) family contains six members (TIR1 and AFB1–AFB5) that localize in the nucleus (Dharmasiri et al. 2005, Parry et al. 2009). Recent genetic studies have indicated that TIR1, AFB1, AFB2 and AFB3 act as positive regulators, and that AFB4 is a negative regulator, of auxin signaling (Dharmasiri et al. 2005, Parry et al. 2009, Greenham et al. 2011). Hypocotyls of tir1 afb1 afb2 afb3 quadruple mutants show a severe phototropism defect and a moderate gravitropism phenotype (Möller et al. 2010), suggesting that TIR1/AFB auxin receptor signaling causes differential growth through the regulation of ARF7 and/or other transcriptional factors in response to an asymmetric auxin distribution (see subsequent sections). On the other hand, Schneck et al (2010) indicated that hypocotyl segments of a tir1 afb1 afb2 afb3 quadruple mutant show normal auxin-induced elongation >4 h after IAA treatment, and propose that the TIR1/AFB family is not involved in the rapid cell expansion that occurs in response to auxin. The relationship between the roles of TIR1/AFB in auxin-induced cell expansion in hypocotyl segments and the phototropic responses of seedling hypocotyls is unclear. Moreover, Möller et al. (2010) highlighted differences in TIR1/AFB function between phototropic and gravitropic responses. Further analyses will be necessary to elucidate properly the functional role of TIR1/AFB in tropic responses and in auxin-induced cell elongation.
Another type of auxin receptor, auxin-binding protein1 (ABP1), is a unique protein that localizes in the ER and on the outer surface of the plasma membrane in Arabidopsis (reviewed by Tromas et al. 2010). The optimal pH for the binding of the synthetic auxin NAA is 5.5, and essentially no binding is obtained at pH values of around 7.0. Because the pH in the ER lumen is close to 7.0, and that in the extracellular matrix is near 5.5, it is likely that ABP1 works as an auxin receptor at the outer surface of the plasma membrane. Although genetic analysis of the ABP1 gene is made difficult by the fact that its null mutant is embryonic lethal (Chen et al. 2001), Effendi et al. (2011) have recently reported that a heterozygous abp1/ABP1 mutant shows moderate phototropic and gravitropic response phenotypes. This finding suggests that the ABP1 signaling pathway is crucial for the auxin action mechanisms that control differential growth in response to light and gravity.
ABP1 shows a wide range of regulatory targets including proton pumps and ion channels at the plasma membrane, endocytosis and gene expression (reviewed in Sauer and Kleine-Vehn 2011, Scherer 2011). It has been observed that auxin binding by ABP1 rapidly stimulates the activity of the plasma membrane H+-ATPase and K+-inward channels via unknown mechanisms. Acidification of the extracellular matrix by activation of plasma membrane H+-ATPase can activate cell wall expansins, which are the major agents for cell wall loosening (reviewed by Cosgrove 2000), thus facilitating cell expansion. Cellular water uptake with K+ influx also allows for turgor-induced cell expansion. These activities appear to be crucial for rapid cell expansion in response to auxin and the differential growth of organs. A recent study has also indicated that ABP1 promotes clathrin-dependent endocytosis and that the auxin binding of ABP1 negatively regulates this activity, and therefore negatively regulates the internalization of auxin efflux carrier PIN proteins (Robert et al. 2010). The regulation of endocytosis appears to be one of the underlying mechanisms that control the function of PINs, so this particular activity of ABP1 may contribute to the generation of asymmetric auxin distribution during the phototropic response. Furthermore, and also as a consequence of this process, the regulation of the internalization of auxin transporters appears to control the cytosolic auxin concentration and the transcription of the auxin early responsive genes, including IAA, SAUR, GH3.5 and ABP1 itself (Effendi et al. 2011). These functions of ABP1 probably contribute overall to phototropic responses.
The AXR1 subunit of the RUB E1-activating enzyme
The auxin-resistant mutant axr1 shows small hypocotyl curvatures as a result of both phototropic and gravitropic responses (Lincoln et al. 1990, Watahiki et al. 1999). The AXR1 gene encodes a subunit of the RELATED TO UBIQUITIN (RUB) E1-activating enzyme, and heterodimers of AXR1 and ECR1 function as RUB E1 (del Pozo et al. 2002). The Arabidopsis genome contains a single gene AXR1-like (AXL) with strong similarity to AXR1 (Dharmasiri et al. 2007). AXR1 and AXL have redundant functions in RUB conjugation, and the majority of axr1 axl double mutants are embryonic or seedling lethal (Dharmasiri et al. 2007). In Arabidopsis, CUL1, CUL3a/b and CUL4 are known to be the main targets of RUB modification (reviewed by Hotton and Callis 2008). A phototropism defect in the axr1 mutants may be caused by an abnormality in the RUB modification of CUL1, which is a subunit of the SCFTIR1/AFB (Skp1–Cullin1–TIR1/AFB F-box) E3 ubiquitin ligase that controls the activities of the ARF transcriptional factors including ARF7/NPH4. However, Watahiki et al. (1999) indicated that the axr1 mutation in an msg1/nph4 mutant background enhances its phenotype and causes a severe hypocotyl phototropism defect. This suggests that the AXR1 and ARF7 genes may act independently during phototropism. Another recent study has indicated that CUL3 is involved in phototropism through the function of NPH3 (Roberts et al. 2011). RUB modifications of CUL3, in addition to CUL1, may also be involved in the phototropic responses.
The NPH4/ARF7 auxin-responsive transcription factor
nph4 mutants show moderate hypocotyl phototropism and gravitropism phenotypes (Liscum and Briggs 1996), and the NPH4 gene appears to function directly in differential growth responses. Harper et al. (2000) cloned and sequenced NPH4, which encodes the auxin-regulated transcriptional activator ARF7, one of 23 members of the ARF family of transcriptional regulators (reviewed by Guilfoyle and Hagen 2007). ARF7 function is suppressed by the binding of the transcriptional repressor IAA19/MSG2 (Tatematsu et al. 2004). Auxin binds to the TIR1/AFB subunit of the SCFTIR1/AFB ubiquitin ligase, and promotes degradation of IAA19 proteins through a ubiquitin–proteasome pathway (Fig. 2) (reviewed in Guilfoyle and Hagen 2007, Mockaitis and Estelle 2008). Free ARF7 proteins appear to form homodimers and/or heterodimers with the other transcriptional activators (Ulmasov et al. 1999, Guilfoyle and Hagen 2007), such as ARF19 or MYB77 (Wilmoth et al. 2005, Shin et al. 2007). ARF7 thereby promotes the transcription of auxin-responsive genes including the expansin genes EXPA1 and EXPA8 (Fig. 2) (Esmon et al. 2006), which are involved in cell wall expansion (reviewed by Cosgrove et al. 2000).
nph4/arf7 Arabidopsis mutants show a moderate phototropic phenotype (Liscum and Briggs 1996) that is suppressed by ethylene or by red light irradiation (Harper et al. 2000, Stowe-Evans et al. 2001). Furthermore, phototropic curvatures observed in msg2-1/iaa19 dominant mutants are smaller than those of nph4 null mutants (Tatematsu et al. 2004). These results suggest that one or more additional ARFs function in phototropism. ARF19 is involved in phototropism in nph4 mutants under ethylene exposure, which induces its expression and recovers the phototropic responses (Li et al. 2006, Stone et al. 2008). However, the arf19 single mutant shows normal phototropic responses, and the arf7 arf19 double mutants also show moderate phototropic curvatures similar to those of the arf7 single mutants (Okushima et al. 2005, Stone et al. 2008). Stone et al. (2008) attempted to identify the locus at which a mutation produces an enhancing effect on the nph4 phenotype on the hypocotyl phototropic response, but the involvement of additional ARFs has not been identified as yet. Thus, it has not yet been determined whether phototropic bending is promoted by the ARF transcription factors in a functionally redundant manner or by other transcription factors and/or mechanisms.
Perspectives
Since the identification of NPH1/phot1 in 1997, significant progress has been made in our understanding of the molecular mechanisms governing the phototropic responses in plants, including the functional redundancy of phot1 and phot2, various effects of phys and crys, the components of phototropin signaling, auxin transporters and auxin signaling factors. However, many questions regarding the molecular basis of phototropism remain to be answered; for example, the nature of how phototropin signaling induces the asymmetric distribution of auxin, a lack of any clear phototropism phenotypes in pin and pid multiple mutants and whether the ubiquitination of phot1 by NPH3 is important for the phototropic response. Further studies of these and other issues will ultimately elucidate the phototropin signaling pathways and the complex mechanisms that underlie the responses of plants to light stimuli.
Funding
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology, Japan [a Grant-in-Aid for Scientific Research on Innovative Areas ‘Plant Environmental Sensing’ (No. 23120510) to T.S.].
Glossary
Abbreviations
- 35S
Cauliflower mosaic virus 35S promoter
- ABCB
ATP-binding cassette subfamily B-type
- ABP
auxin-binding protein
- ADG
ADP glucose pyrophosphorylase
- AFB
auxin-binding F-box
- AGC
protein kinase A, cGMP-dependent protein kinase and protein kinase C
- AGR
agravitropic
- ARF
auxin-responsive factor
- AXL
AXR1-like
- AXR
auxin resistant
- ARF-GAP
ADP-ribosylation factor–GTPase-activating protein
- ARF-GEF
ADP-ribosylation factor–guanine nucleotide exchange factor
- AUX
auxin resistant
- BFA
brefeldin A
- BTB/POZ
broad-complex, tramtrack and bric-à-brac/Pox virus and zinc finger
- CPT
coleoptile phototropism
- cry
cryptochrome
- CUL
cullin
- CYP
cytochrome P450
- DOT
defectively organized tributaries
- EHB
enhanced bending
- ER
endoplasmic reticulum
- EXPA
expansin
- GAI
gibberellin insensitive
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- LAX
like-AUX
- LOV
light, oxygen, voltage
- MAB
macchi-bou
- MDR
multidrug resistance
- MSG
massugu
- NPH
nonphototropic hypocotyl
- NPL
NPH1-like
- NPY
naked pins in yuc
- NRL
NPH3/RPT2-like
- PAC
paclobutrazol
- PBP1
pinoid-binding protein 1
- PGM
phosphoglucomutase
- PGP
P-glycoprotein
- phot
phototropin
- phy
phytochrome
- PID
pinoid
- PIN
pin-formed
- PKS
phytochrome kinase substrate
- PP2A
protein phosphatase 2A
- pro
promoter
- RCN
roots curl in NPA
- RGA
repressor of ga1-3
- RGL
RGA-like
- RPT
root phototropism
- RUB
related to ubiquitin
- SCF
Skp1–Cullin1–F-box
- TAA
tryptophan aminotransferase of Arabidopsis
- TCH3
TOUCH3
- TIR
transport inhibitor-resistant
- YUC
yucca.
References
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol. Cell. 1998;1:939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- Babourina O, Newman I, Shabala S. Blue light-induced kinetics of H+ and Ca2+ fluxes in etiolated wild-type and phototropin-mutant Arabidopsis seedlings. Proc. Natl Acad. Sci. USA. 2002;99:2433–2438. doi: 10.1073/pnas.042294599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum G, Long JC, Jenkins GI, Trewavas AJ. Stimulation of the blue light phototropic receptor NPH1 causes a transient increase in cytosolic Ca2+ Proc. Natl Acad. Sci. USA. 1999;96:13554–13559. doi: 10.1073/pnas.96.23.13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Ampudia CS, Hooykaas PJ, Offringa R. PINOID-mediated signaling involves calcium-binding proteins. Plant Physiol. 2003;132:1623–1630. doi: 10.1104/pp.103.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R, Malenica N, Luschnig C. Regulating the regulator: the control of auxin transport. BioEssays. 2005;27:1246–1255. doi: 10.1002/bies.20322. [DOI] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR. Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 1995;8:505–520. [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell. 2007;19:131–147. doi: 10.1105/tpc.106.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Peer WA, Makam SN, Murphy AS. Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol. 2004;134:28–31. doi: 10.1104/pp.103.031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee JJ, Peer WA, Murphy AS. Auxin transport. Curr. Opin. Plant Biol. 2005;8:494–500. doi: 10.1016/j.pbi.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Boccalandro HE, De Simone SN, Bergmann-Honsberger A, Schepens I, Fankhauser C, Casal JJ. PHYTOCHROME KINASE SUBSTRATE1 regulates root phototropism and gravitropism. Plant Physiol. 2008;146:108–115. doi: 10.1104/pp.107.106468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L, Ökrész L, Henriques R, Anthony RG. Growth signalling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci. 2003;8:424–431. doi: 10.1016/S1360-1385(03)00188-2. [DOI] [PubMed] [Google Scholar]
- Briggs WR. The phototropic responses of higher plants. Annu. Rev. Plant Physiol. 1963;14:311–352. [Google Scholar]
- Briggs WR, Beck CF, Cashmore AR, Christie JM, Hughes J, Jarillo JA, et al. The phototropin family of photoreceptors. Plant Cell. 2001;13:993–997. doi: 10.1105/tpc.13.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. Phytochromes, cryptochromes, phototropin: photoreceptor interactions in plants. Photochem. Photobiol. 2000;71:1–11. doi: 10.1562/0031-8655(2000)071<0001:pcppii>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Young JC, Sussman MR, Jones AM. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 2001;15:902–911. doi: 10.1101/gad.866201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu. Rev. Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl Acad. Sci. USA. 2007;104:18825–18829. doi: 10.1073/pnas.0708506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y. NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc. Natl Acad. Sci. USA. 2008;105:21017–21022. doi: 10.1073/pnas.0809761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM. Phototropin blue-light receptors. Annu. Rev. Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, et al. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- Christie JM, Yang H, Richter GL, Sullivan S, Thomson CE, Lin J, et al. phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol. 2011;9:e1001076. doi: 10.1371/journal.pbio.1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona A, Chen DL, Yeh KC, Abel S. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol. 2000;124:1728–1738. doi: 10.1104/pp.124.4.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Loosening of plant cell walls by expansins. Nature. 2000;407:321–326. doi: 10.1038/35030000. [DOI] [PubMed] [Google Scholar]
- Darwin C. London: John Murray; 1880. The Power of Movement in Plants. [Google Scholar]
- de Carbonnel M, Davis P, Roelfsema MR, Inoue S, Schepens I, Lariguet P, et al. The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol. 2010;152:1391–1405. doi: 10.1104/pp.109.150441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M. AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis Cullin AtCUL1 is required for auxin response. Plant Cell. 2002;14:421–433. doi: 10.1105/tpc.010282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 2003;133:1617–1629. doi: 10.1104/pp.103.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Karunarathna N, Jurgens G, Estelle M. AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J. 2007;52:114–123. doi: 10.1111/j.1365-313X.2007.03211.x. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Huang F, Galván-Ampudia CS, Mähönen AP, Kleine-Vehn J, Xu J, et al. Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development. 2010;137:3245–3255. doi: 10.1242/dev.052456. [DOI] [PubMed] [Google Scholar]
- Ding Z, Galván-Ampudia CS, Demarsy E, Łangowski L, Kleine-Vehn J, Fan Y, et al. Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 2011;13:447–452. doi: 10.1038/ncb2208. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 2000;12:475–482. doi: 10.1016/s0955-0674(00)00119-8. [DOI] [PubMed] [Google Scholar]
- Effendi Y, Rietz S, Fischer U, Scherer GF. The heterozygous abp1/ABP1 insertional mutant has defects in functions requiring polar auxin transport and in regulation of early auxin-regulated genes. Plant J. 2011;65:282–294. doi: 10.1111/j.1365-313X.2010.04420.x. [DOI] [PubMed] [Google Scholar]
- Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E. A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc. Natl Acad. Sci. USA. 2006;103:236–241. doi: 10.1073/pnas.0507127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Yeh K-C, Lagarias JC, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Figueroa P, Gusmaroli G, Serino G, Habashi J, Ma L, Shen Y, et al. Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell. 2005;17:1180–1195. doi: 10.1105/tpc.105.031989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Schwarz H, Hamann T, Offringa R, et al. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature. 2002;415:806–809. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, et al. A PINOID-dependent binary switch in apical–basal PIN polar targeting directs auxin efflux. Science. 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Kami C, Fankhauser C, Alabadí D, Blázquez MA. A hormonal regulatory module that provides flexibility to tropic responses. Plant Physiol. 2011;156:1819–1825. doi: 10.1104/pp.111.173971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván-Ampudia CS, Offringa R. Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci. 2007;12:541–547. doi: 10.1016/j.tplants.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, et al. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112:219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Geldner N, Richter S, Vieten A, Marquardt S, Torres-Ruiz RA, Mayer U, et al. Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development. 2004;131:389–400. doi: 10.1242/dev.00926. [DOI] [PubMed] [Google Scholar]
- Greenham K, Santner A, Castillejo C, Mooney S, Sairanen I, Ljung K, et al. The AFB4 auxin receptor is a negative regulator of auxin signaling in seedlings. Curr. Biol. 2011;21:520–525. doi: 10.1016/j.cub.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guilfoyle TJ, Hagen G. Auxin response factors. Curr. Opin. Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Haga K, Takano M, Neumann R, Iino M. The rice COLEOPTILE PHOTOTROPISM1 gene encoding an ortholog of Arabidopsis NPH3 is required for phototropism of coleoptiles and lateral translocation of auxin. Plant Cell. 2005;17:103–115. doi: 10.1105/tpc.104.028357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han IS, Tseng TS, Eisinger W, Briggs WR. Phytochrome A regulates the intracellular distribution of phototropin 1–green fluorescent protein in Arabidopsis thaliana. Plant Cell. 2008;20:2835–2847. doi: 10.1105/tpc.108.059915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangarter RP. Gravity, light and plant form. Plant Cell Environ. 1997;20:796–800. doi: 10.1046/j.1365-3040.1997.d01-124.x. [DOI] [PubMed] [Google Scholar]
- Harada A, Sakai T, Okada K. Phot1 and phot2 mediate blue light-induced transient increases in cytosolic Ca2+ differently in Arabidopsis leaves. Proc. Natl Acad. Sci. USA. 2003;100:8583–8588. doi: 10.1073/pnas.1336802100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Shimazaki K. Phototropins and blue light-dependent calcium signaling in higher plants. Photochem. Photobiol. 2007;83:102–111. doi: 10.1562/2006-03-08-IR-837. [DOI] [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, et al. The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell. 2000;12:757–770. doi: 10.1105/tpc.12.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Toledo-Ortiz G, Bender J, Quail PH. The photomorphogenesis-related mutant red1 is defective in CYP83B1, a red light-induced gene encoding a cytochrome P450 required for normal auxin homeostasis. Planta. 2004;219:195–200. doi: 10.1007/s00425-004-1211-z. [DOI] [PubMed] [Google Scholar]
- Holland JJ, Roberts D, Liscum E. Understanding phototropism: from Darwin to today. J. Exp. Bot. 2009;60:1969–1978. doi: 10.1093/jxb/erp113. [DOI] [PubMed] [Google Scholar]
- Hotton SK, Callis J. Regulation of cullin RING ligases. Annu. Rev. Plant Biol. 2008;59:467–489. doi: 10.1146/annurev.arplant.58.032806.104011. [DOI] [PubMed] [Google Scholar]
- Huala E, Oeller PW, Liscum E, Han IS, Larsen E, Briggs WR. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- Huang F, Zago MK, Abas L, van Marion A, Galván-Ampudia CS, Offringa R. Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell. 2010;22:1129–1142. doi: 10.1105/tpc.109.072678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino M. Phototropism in higher plants. In: Häder D-P, Lebert M, editors. Photomovement. Amsterdam: Elsevier Science B.V.; 2001. pp. 659–811. [Google Scholar]
- Iino M. Toward understanding the ecological functions of tropisms: interactions among and effects of light on tropisms. Curr. Opin. Plant Biol. 2006;9:89–93. doi: 10.1016/j.pbi.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Inada S, Ohgishi M, Mayama T, Okada K, Sakai T. RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. Plant Cell. 2004;16:887–896. doi: 10.1105/tpc.019901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Kinoshita T, Matsumoto M, Nakayama KI, Doi M, Shimazaki K. Blue light-induced autophosphorylation of phototropin is a primary step for signaling. Proc. Natl Acad. Sci. USA. 2008;105:5626–5631. doi: 10.1073/pnas.0709189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoudi AK, Gordon WR, Wagner D, Quail P, Poff KL. Multiple phytochromes are involved in red-light-induced enhancement of first-positive phototropism in Arabidopsis thaliana. Plant Physiol. 1997a;113:975–979. doi: 10.1104/pp.113.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoudi AK, Konjević R, Apel P, Poff KL. Time threshold for second positive phototropism is decreased by a preirradiation with red light. Plant Physiol. 1992;99:1422–1425. doi: 10.1104/pp.99.4.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoudi AK, Konjević R, Whitelam G, Gordon W, Poff KL. Both phytochrome A and phytochrome B are required for the normal expression of phototropism in Arabidopsis thaliana seedlings. Physiol. Plant. 1997b;101:278–282. [Google Scholar]
- Jarillo JA, Ahmad M, Cashmore AR. NPH2: A second member of the NPH serine/threonine kinase family of Arabidopsis. Plant Physiol. 1998;117:719. [Google Scholar]
- Jensen PJ, Hangarter RP, Estelle M. Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 1998;116:455–462. doi: 10.1104/pp.116.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- Kagawa T, Sakai T, Suetsugu N, Oikawa K, Ishiguro S, Kato T, et al. Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science. 2001;291:2138–2141. doi: 10.1126/science.291.5511.2138. [DOI] [PubMed] [Google Scholar]
- Kaiserli E, Sullivan S, Jones MA, Feeney KA, Christie JM. Domain swapping to assess the mechanistic basis of Arabidopsis phototropin 1 receptor kinase activation and endocytosis by blue light. Plant Cell. 2009;21:3226–3244. doi: 10.1105/tpc.109.067876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B, Grancher N, Koyffmann V, Lardemer D, Burney S, Ahmad M. Multiple interactions between cryptochrome and phototropin blue-light signalling pathways in Arabidopsis thaliana. Planta. 2008;227:1091–1099. doi: 10.1007/s00425-007-0683-z. [DOI] [PubMed] [Google Scholar]
- Khurana JP, Poff KL. Mutants of Arabidopsis thaliana with altered phototropism. Planta. 1989;178:400–406. [PubMed] [Google Scholar]
- Kim K, Shin J, Lee SH, Kweon HS, Maloof JN, Choi G. Phytochromes inhibit hypocotyl negative gravitropism by regulating the development of endodermal amyloplasts through phytochrome-interacting factors. Proc. Natl Acad. Sci. USA. 2011;108:1729–1734. doi: 10.1073/pnas.1011066108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss JZ, Mullen JL, Correll MJ, Hangarter RP. Phytochromes A and B mediate red-light-induced positive phototropism in roots. Plant Physiol. 2003;131:1411–1417. doi: 10.1104/pp.013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer T, Dümmer M, Landgraf F, Forreiter C. A negative effector of blue light-induced and gravitropic bending in Arabidopsis. Plant Physiol. 2011;156:439–447. doi: 10.1104/pp.110.167411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong SG, Suzuki T, Tamura K, Mochizuki N, Hara-Nishimura I, Nagatani A. Blue light-induced association of phototropin 2 with the Golgi apparatus. Plant J. 2006;45:994–1005. doi: 10.1111/j.1365-313X.2006.02667.x. [DOI] [PubMed] [Google Scholar]
- Lalanne E, Michaelidis C, Moore JM, Gagliano W, Johnson A, Patel R, et al. Analysis of transposon insertion mutants highlights the diversity of mechanisms underlying male progamic development in Arabidopsis. Genetics. 2004;167:1975–1986. doi: 10.1534/genetics.104.030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariguet P, Boccalandro HE, Alonso JM, Ecker JR, Chory J, Casal JJ, et al. A growth regulatory loop that provides homeostasis to phytochrome A signaling. Plant Cell. 2003;15:2966–2978. doi: 10.1105/tpc.014563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariguet P, Fankhauser C. Hypocotyl growth orientation in blue light is determined by phytochrome A inhibition of gravitropism and phototropin promotion of phototropism. Plant J. 2004;40:826–834. doi: 10.1111/j.1365-313X.2004.02256.x. [DOI] [PubMed] [Google Scholar]
- Lariguet P, Schepens I, Hodgson D, Pedmale UV, Trevisan M, Kami C, et al. PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc. Natl Acad. Sci. USA. 2006;103:10134–10139. doi: 10.1073/pnas.0603799103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lascève G, Leymarie J, Olney MA, Liscum E, Christie JM, Vavasseur A, et al. Arabidopsis contains at least four independent blue-light-activated signal transduction pathways. Plant Physiol. 1999;120:605–614. doi: 10.1104/pp.120.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dai X, Cheng Y, Zhao Y. NPY genes play an essential role in root gravitropic responses in Arabidopsis. Mol. Plant. 2011;4:171–179. doi: 10.1093/mp/ssq052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Dai X, Zhao Y. A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiol. 2006;140:899–908. doi: 10.1104/pp.105.070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. Phototropism: mechanisms and outcomes. [Google Scholar]
- Liscum E, Briggs WR. Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell. 1995;7:473–485. doi: 10.1105/tpc.7.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Briggs WR. Mutations of Arabidopsis in potential transduction and response components of the phototropic signaling pathway. Plant Physiol. 1996;112:291–296. doi: 10.1104/pp.112.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia E, Klein M, Geisler M, Bovet L, Forestier C, Kolukisaoglu U, et al. Multifunctionality of plant ABC transporters—more than just detoxifiers. Planta. 2002;214:345–355. doi: 10.1007/s004250100661. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl Acad. Sci. USA. 2011;108:18512–18517. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Büttner G, Jürgens G. Apical–basal pattern formation in the Arabidopsis embryo: studies on the role of the gnom gene. Development. 1993;117:149–162. [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- Millar KD, Kumar P, Correll MJ, Mullen JL, Hangarter RP, Edelmann RE, et al. A novel phototropic response to red light is revealed in microgravity. New Phytol. 2010;186:648–656. doi: 10.1111/j.1469-8137.2010.03211.x. [DOI] [PubMed] [Google Scholar]
- Miyazawa Y, Takahashi A, Kobayashi A, Kaneyasu T, Fujii N, Takahashi H. GNOM-mediated vesicular trafficking plays an essential role in hydrotropism of Arabidopsis roots. Plant Physiol. 2009;149:835–840. doi: 10.1104/pp.108.131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev. Biol. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- Möller B, Schenck D, Lüthen H. Exploring the link between auxin receptors, rapid cell elongation and organ tropisms. Plant Signal. Behav. 2010;5:601–603. doi: 10.4161/psb.11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motchoulski A, Liscum E. Arabidopsis NPH3: a NPH1 photoreceptor-interacting protein essential for phototropism. Science. 1999;286:962–964. doi: 10.1126/science.286.5441.961. [DOI] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Křeček P, Bielach A, et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459:1136–1140. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- Muday GK, Brady SR, Argueso C, Deruère J, Kieber JJ, DeLong A. RCN1-regulated phosphatase activity and EIN2 modulate hypocotyl gravitropism by a mechanism that does not require ethylene signaling. Plant Physiol. 2006;141:1617–1629. doi: 10.1104/pp.106.083212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima A, Suzuki G, Uehara Y, Saji K, Furukawa T, Koshiba T, et al. Phytochromes and cryptochromes regulate the differential growth of Arabidopsis hypocotyls in both a PGP19-dependent and a PGP19-independent manner. Plant J. 2008a;53:516–529. doi: 10.1111/j.1365-313X.2007.03358.x. [DOI] [PubMed] [Google Scholar]
- Nagashima A, Uehara Y, Sakai T. The ABC subfamily B auxin transporter AtABCB19 is involved in the inhibitory effects of N-1-naphthyphthalamic acid on the phototropic and gravitropic responses of Arabidopsis hypocotyls. Plant Cell Physiol. 2008b;49:1250–1255. doi: 10.1093/pcp/pcn092. [DOI] [PubMed] [Google Scholar]
- Noh B, Bandyopadhyay A, Peer WA, Spalding EP, Murphy AS. Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature. 2003;423:999–1002. doi: 10.1038/nature01716. [DOI] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP. Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell. 2001;13:2441–2454. doi: 10.1105/tpc.010350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgishi M, Saji K, Okada K, Sakai T. Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl Acad. Sci. USA. 2004;101:2223–2228. doi: 10.1073/pnas.0305984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Shimura Y. Mutational analysis of root gravitropism and phototropism of Arabidopsis thaliana seedlings. Aust. J. Plant Physiol. 1992;19:439–448. [Google Scholar]
- Okada K, Shimura Y. Modulation of root growth by physical stimuli. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 665–684. [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–463. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH, Hangarter RP. Phytochrome A regulates red-light induction of phototropic enhancement in Arabidopsis. Plant Physiol. 1996;110:155–162. doi: 10.1104/pp.110.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Dharmasiri PS, Itoh H, Lechner E, et al. Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl Acad. Sci. USA. 2009;106:22540–22545. doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedmale UV, Liscum E. Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. J. Biol. Chem. 2007;282:19992–20001. doi: 10.1074/jbc.M702551200. [DOI] [PubMed] [Google Scholar]
- Petricka JJ, Clay NK, Nelson TM. Vein patterning screens and the defectively organized tributaries mutants in Arabidopsis thaliana. Plant J. 2008;56:251–263. doi: 10.1111/j.1365-313X.2008.03595.x. [DOI] [PubMed] [Google Scholar]
- Poff KL, Janoudi A-K, Rosen ES, Orbović V. The physiology of tropisms. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 639–664. [Google Scholar]
- Rakusová H, Gallego-Bartolomé J, Vanstraelen M, Robert HS, Alabadi D, Blázquez MA, et al. Polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response in Arabidopsis thaliana. Plant J. 2011;67:817–826. doi: 10.1111/j.1365-313X.2011.04636.x. [DOI] [PubMed] [Google Scholar]
- Robert HS, Friml J. Auxin and other signals on the move in plants. Nat. Chem. Biol. 2009;5:325–332. doi: 10.1038/nchembio.170. [DOI] [PubMed] [Google Scholar]
- Robert HS, Offringa R. Regulation of auxin transport polarity by AGC kinases. Curr. Opin. Plant Biol. 2008;11:495–502. doi: 10.1016/j.pbi.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, et al. ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell. 2010;143:111–121. doi: 10.1016/j.cell.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D, Pedmale UV, Morrow J, Sachdev S, Lechner E, Tang X, et al. Modulation of phototropic responsiveness in Arabidopsis through ubiquitination of phototropin 1 by the CUL3-Ring E3 ubiquitin ligase CRL3NPH3. Plant Cell. 2011;23:3627–3640. doi: 10.1105/tpc.111.087999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PR, Smith H. Genetic and transgenic evidence that phytochromes A and B act to modulate the gravitropic orientation of Arabidopsis thaliana hypocotyls. Plant Physiol. 1996;110:211–216. doi: 10.1104/pp.110.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T. NPH3 and RPT2: signal transducers in phototropin signaling pathways. In: Wada M, Shimazaki K, Iino M, editors. Light Sensing in Plants. Tokyo: Springer-Verlag; 2005. pp. 179–184. [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, et al. Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl Acad. Sci. USA. 2001;98:6969–6974. doi: 10.1073/pnas.101137598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Mochizuki S, Haga K, Uehara Y, Suzuki A, Harada A, et al. The WAVY GROWTH 3 E3 ligase family controls the gravitropic response in Arabidopsis roots. Plant J. 2012;70:303–314. doi: 10.1111/j.1365-313X.2011.04870.x. [DOI] [PubMed] [Google Scholar]
- Sakai T, Wada T, Ishiguro S, Okada K. RPT2. A signal transducer of the phototropic response in Arabidopsis. Plant Cell. 2000;12:225–236. doi: 10.1105/tpc.12.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Briggs WR. Cellular and subcellular localization of phototropin 1. Plant Cell. 2002;14:1723–1735. doi: 10.1105/tpc.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon M, Zacherl M, Luff L, Rüdiger W. Exposure of oat seedlings to blue light results in amplified phosphorylation of the putative photoreceptor for phototropism and in higher sensitivity of the plants to phototropic stimulation. Plant Physiol. 1997a;115:493–500. doi: 10.1104/pp.115.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon M, Zacherl M, Rüdiger W. Asymmetric, blue light-dependent phosphorylation of a 116-kilodalton plasma membrane protein can be correlated with the first- and second-positive phototropic curvature of oat coleoptiles. Plant Physiol. 1997b;115:485–491. doi: 10.1104/pp.115.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner AA, Watson JC. The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. Plant J. 2006;45:752–764. doi: 10.1111/j.1365-313X.2005.02641.x. [DOI] [PubMed] [Google Scholar]
- Sauer M, Kleine-Vehn J. AUXIN BINDING PROTEIN1: the outsider. Plant Cell. 2011;23:2033–2043. doi: 10.1105/tpc.111.087064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck D, Christian M, Jones A, Lüthen H. Rapid auxin-induced cell expansion and gene expression: a four-decade-old question revisited. Plant Physiol. 2010;152:1183–1185. doi: 10.1104/pp.109.149591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer GFE. AUXIN-BINDING-PROTEIN1, the second auxin receptor: what is the significance of a two-receptor concept in plant signal transduction? J. Exp. Bot. 2011;62:3339–3357. doi: 10.1093/jxb/err033. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C. Understanding gibberellic acid signaling—are we there yet? Curr. Opin. Plant Biol. 2008;11:9–15. doi: 10.1016/j.pbi.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, et al. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinitz B, Ren Z, Poff KL. Blue and green light-induced phototropism in Arabidopsis thaliana and Lactuca sativa L. seedlings. Plant Physiol. 1985;77:248–251. doi: 10.1104/pp.77.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, et al. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;286:316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, et al. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 2011;23:3961–3973. doi: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoelzle S, Kagawa T, Wada M, Hedrich R, Dietrich P. Blue light activates calcium-permeable channels in Arabidopsis mesophyll cells via the phototropin signaling pathway. Proc. Natl Acad. Sci. USA. 2003;100:1456–1461. doi: 10.1073/pnas.0333408100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone BB, Stowe-Evans EL, Harper RM, Celaya RB, Ljung K, Sandberg G, et al. Disruptions in AUX1-dependent auxin influx alter hypocotyl phototropism in Arabidopsis. Mol. Plant. 2008;1:129–144. doi: 10.1093/mp/ssm013. [DOI] [PubMed] [Google Scholar]
- Stowe-Evans EL, Luesse DR, Liscum E. The enhancement of phototropin-induced phototropic curvature in Arabidopsis occurs via a photoreversible phytochrome A-dependent modulation of auxin responsiveness. Plant Physiol. 2001;126:826–834. doi: 10.1104/pp.126.2.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Thomson CE, Kaiserli E, Christie JM. Interaction specificity of Arabidopsis 14-3-3 proteins with phototropin receptor kinases. FEBS Lett. 2009;583:2187–2193. doi: 10.1016/j.febslet.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Sullivan S, Thomson CE, Lamont DJ, Jones MA, Christie JM. In vivo phosphorylation site mapping and functional characterization of Arabidopsis phototropin 1. Mol. Plant. 2008;1:178–194. doi: 10.1093/mp/ssm017. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Dhonukshe P, Brewer PB, Friml J. Spatiotemporal asymmetric auxin distribution: a means to coordinate plant development. Cell Mol. Life Sci. 2006;63:2738–2754. doi: 10.1007/s00018-006-6116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, et al. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell. 2004;16:379–393. doi: 10.1105/tpc.018630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomann A, Brukhin V, Dieterle M, Gheyeselinck J, Vantard M, Grossniklaus U, et al. Arabidopsis CUL3A and CUL3B genes are essential for normal embryogenesis. Plant J. 2005;43:437–448. doi: 10.1111/j.1365-313X.2005.02467.x. [DOI] [PubMed] [Google Scholar]
- Titapiwatanakun B, Blakeslee JJ, Bandyopadhyay A, Yang H, Mravec J, Sauer M, et al. ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J. 2009;57:27–44. doi: 10.1111/j.1365-313X.2008.03668.x. [DOI] [PubMed] [Google Scholar]
- Titapiwatanakun B, Murphy AS. Post-transcriptional regulation of auxin transport proteins: cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J. Exp. Bot. 2009;60:1093–1107. doi: 10.1093/jxb/ern240. [DOI] [PubMed] [Google Scholar]
- Tromas A, Paponov I, Perrot-Rechenmann C. AUXIN BINDING PROTEIN 1: functional and evolutionary aspects. Trends Plant Sci. 2010;15:436–446. doi: 10.1016/j.tplants.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Tseng TS, Briggs WR. The Arabidopsis rcn1-1 mutation impairs dephosphorylation of phot2, resulting in enhanced blue light responses. Plant Cell. 2010;22:392–402. doi: 10.1105/tpc.109.066423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida-Mayama T, Nakano M, Uehara Y, Sano M, Fujisawa N, Okada K, et al. Mapping of the phosphorylation sites on the phototropic signal transducer, NPH3. Plant Sci. 2008;174:626–633. [Google Scholar]
- Tsuchida-Mayama T, Sakai T, Hanada A, Uehara Y, Asami T, Yamaguchi S. Role of the phytochrome and cryptochrome signaling pathways in hypocotyl phototropism. Plant J. 2010;62:653–662. doi: 10.1111/j.1365-313X.2010.04180.x. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- Vitha S, Zhao L, Sack FD. Interaction of root gravitropism and phototropism in Arabidopsis wild-type and starchless mutants. Plant Physiol. 2000;122:453–462. doi: 10.1104/pp.122.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Signaling mechanisms of higher plant photoreceptors: a structure–function perspective. Curr. Top. Dev. Biol. 2005;68:227–261. doi: 10.1016/S0070-2153(05)68008-8. [DOI] [PubMed] [Google Scholar]
- Watahiki MK, Tatematsu K, Fujihira K, Yamamoto M, Yamamoto KT. The MSG1 and AXR1 genes of Arabidopsis are likely to act independently in growth–curvature responses of hypocotyls. Planta. 1999;207:362–369. doi: 10.1007/s004250050493. [DOI] [PubMed] [Google Scholar]
- Went FW, Thimann KV. New York: Macmillan; 1937. Phytohormones. [Google Scholar]
- Whippo CW, Hangarter RP. Second positive phototropism results from coordinated co-action of the phototropins and cryptochromes. Plant Physiol. 2003;132:1499–1507. doi: 10.1104/pp.102.018481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whippo CW, Hangarter RP. Phytochrome modulation of blue-light-induced phototropism. Plant Cell Environ. 2004;27:1223–1228. [Google Scholar]
- Whippo CW, Hangarter RP. A brassinosteroid-hypersensitive mutant of BAK1 indicates that a convergence of photomorphogenic and hormonal signaling modulates phototropism. Plant Physiol. 2005;139:448–457. doi: 10.1104/pp.105.064444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whippo CW, Hangarter RP. Phototropism: bending towards enlightenment. Plant Cell. 2006;18:1110–1119. doi: 10.1105/tpc.105.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005;43:118–130. doi: 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, et al. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl Acad. Sci. USA. 2011;108:18518–18523. doi: 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Liu H, Klejnot J, Lin C. The cryptochrome blue light receptors. Arabidopsis Book. 2010;8:e0135. doi: 10.1199/tab.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žádníková P, Petrášek J, Marhavý P, Raz V, Vandenbussche F, Ding Z, et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development. 2010;137:607–617. doi: 10.1242/dev.041277. [DOI] [PubMed] [Google Scholar]
- Zažímalová E, Murphy AS, Yang H, Hoyerová K, Hošek P. Auxin transporters. Why so many? Cold Spring Harb. Perspect. Biol. 2010;2:a001552. doi: 10.1101/cshperspect.a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu X, Foo E, Symons GM, Lopez J, Bendehakkalu KT, et al. A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiol. 2007;145:106–118. doi: 10.1104/pp.107.099838. [DOI] [PMC free article] [PubMed] [Google Scholar]