Abstract
Natural RNAs, especially tRNAs, are extensively modified to tailor structure and function diversities. Uracil is the most modified nucleobase among all natural nucleobases. Interestingly, >76% of uracil modifications are located on its 5-position. We have investigated the natural 5-methoxy (5-O-CH3) modification of uracil in the context of A-form oligonucleotide duplex. Our X-ray crystal structure indicates first a H-bond formation between the uracil 5-O-CH3 and its 5′-phosphate. This novel H-bond is not observed when the oxygen of 5-O-CH3 is replaced with a larger atom (selenium or sulfur). The 5-O-CH3 modification does not cause significant structure and stability alterations. Moreover, our computational study is consistent with the experimental observation. The investigation on the uracil 5-position demonstrates the importance of this RNA modification at the atomic level. Our finding suggests a general interaction between the nucleobase and backbone and reveals a plausible function of the tRNA 5-O-CH3 modification, which might potentially rigidify the local conformation and facilitates translation.
INTRODUCTION
Natural RNAs are covalently modified by different functionalities in order to achieve structural and functional specificities as well as diversity (1–3). Many modifications have been found on tRNAs, ribosome RNAs, mRNAs and other functional RNAs (1–4). So far, 109 nucleoside residues with different modifications have been found in natural RNAs (4). However, it is not clear why >94% of the modifications contain nucleobase alterations. tRNA-interaction specificity is required in many biological reactions and processes, including tRNA charging with specific amino acids by aminoacyl tRNA synthetases, tRNA recognition of mRNA through anticodon triplets and selective engagement of amino acid-charged tRNAs with ribosome during protein synthesis. It’s known that the tRNA is the most modified RNA and contains 92 different modifications, 87 of which locate on the nucleobases (4). Interestingly, uracil is the most modified nucleobase in tRNA, and ∼40% of the tRNA nucleobase modifications are on uracil nucleobase of uridine (1, Figure 1 and Supplementary Table S1). The uracil modifications include pseudouracil, 2-thiouracil, 2-selenouracil, etc. Surprisingly, >76% of uracil derivatizations (34 modifications) are located in its 5-position (26 modifications), implying that this position is critical for tRNA functions.
Figure 1.
Chemical structures of uridine 1, 5-modified uridine derivatives (Xo5U, 2), 5-OMe, 5-SMe and 5-SeMe-uracil-containing oligonucleotides (3a, 3b and 3c) and 2′-SeMe-deoxyuridine (4, 2′-SeMe-dU or SedU).
Several natural modifications of uracil are presented in Figure 1. The 5-modified uracil normally contains an exo-5-carbon (e.g. 5-methyluracil) or -oxygen atom (e.g. 5-oxyuridine, 2, Figure 1) to connect with other functionalities (5,6). 5-methoxyuridine (mo5U, 2b) was discovered in many bacterial tRNAs, such as Ala, Thr and Val tRNAs of gram-positive bacteria and Ser and Val tRNAs of Escherichia coli (7). The 5-methoxy (5-O-CH3) modification locates in position 34, the first anticodon nucleotide of tRNA, which interacts with the third nucleotide (the wobble position) of the codon in mRNA. Though it is not very clear how this 5-O-CH3 modification facilitates the interaction between these tRNA anticodons and the mRNA codons, it was hypothesized that the 5-O-CH3 functionality might potentially tune the wobble nucleotide-reading ability of these tRNAs during protein synthesis (8,9). However, it remains elusive how the 5-O-CH3 moiety works.
Though mo5U was studied (10–12), 3D structure of the 5-oxyuracil-containing oligonucleotide has never been investigated. To better understand the role of the 5-O-CH3 modification at the atomic level, we have decided to synthesize an RNA-like system by taking advantage of A-form DNA, which has the same sugar pucker and overall A-form duplex structure as RNA (13,14). To investigate the importance of oxygen, we have also decided to replace the 5-oxygen atom with selenium and sulfur from the same elemental family. Furthermore, we have performed X-ray crystal structure study of the oligonucleotides containing these three modifications. We have discovered for the first time that 5-O-CH3 of the oligonucleotide (3a, Figure 1) forms a unique H-bond with the adjacent 5′-phosphate group. This H-bond was not observed in the corresponding S- and Se-modified oligonucleotides (3b and 3c, Figure 1). Moreover, the computational simulations of these three modified nucleobases have been carried out. Our research results suggest that the electronic effect of oxygen plays unique roles in the function of the 5-O-CH3 modification, particularly in an A-form duplex environment. The H-bond between the nucleobase and phosphate backbone rigidifies the local structure and conformation, which may facilitate the tRNA–mRNA base-pair recognition in protein synthesis.
MATERIALS AND METHODS
Synthesis of the 5-SeMe, -SMe and -OMe functionalized DNAs
All the DNA oligonucleotides were chemically synthesized in a 1.0 μmol scale using an ABI392 or ABI3400 DNA/RNA Synthesizer. The normal nucleoside phosphoramidite reagents were used in this work (Glen Research). The concentrations of the 5-modified uridine phosphoramidite were identical to those of the conventional ones (0.1 M in acetonitrile). Coupling was carried out using a 5-(benzylmercapto)-1H-tetrazole (5-BMT) solution (0.25 M) in acetonitrile. The coupling time was 25 s for both native and modified samples. 3% trichloroacetic acid in methylene chloride was used for the 5′-detritylation. Synthesis were performed on control pore glass (CPG-500) immobilized with the appropriate nucleoside through a succinate linker. All oligonucleotides were prepared with 4,4′-dimethoxytrityl group (DMTr)-on form. After synthesis, the DNA oligonucleotides were cleaved from the solid support and fully deprotected by the treatment of concentrated ammonium hydroxide overnight at 55°C. The 5′-DMTr deprotection was performed in a 3% trichloroacetic acid solution for 2 min, followed by neutralization to pH 7.0 with a freshly made aqueous solution of triethylamine (1.1 M) and extraction with petroleum ether to remove DMTr-OH.
High performance liquid chromatography analysis and purification
The modified DNA oligonucleotides were analyzed and purified by reverse-phase high performance liquid chromatography in both DMTr-on and DMTr-off forms. Purification was carried out using a 21.2 × 250 mm Zorbax, RX-C8 column at a flow rate of 6 ml/min. Buffer A consisted of 10 mM triethylammonium acetate (TEAAc, pH 7.1), and buffer B contained 50% aqueous acetonitrile and 10 mM TEAAc, pH 7.1. Similarly, analysis was performed on a Zorbax SB-C18 column (4.6 × 250 mm) at a flow rate of 1.0 ml/min using the same buffer system. The DMTr-on oligonucleotides were eluded with up to 100% buffer B in 20 min in a linear gradient, whereas the DMTr-off oligonucleotides were eluted with up to 70% of buffer B in 20 min in a linear gradient. The collected fractions were lyophilized, the purified compounds were re-dissolved in water, and the pH was adjusted to 7.0 after the final purification.
Crystallization
The purified oligonucleotides (1 mM each) were heated to 70°C for 2 min, and then cooled slowly to room temperature before crystallization. The 2′-SeMe-dU (SedU) was incorporated into the modified DNA for the crystallization facilitation. The Nucleic Acid Mini-Kit (Hampton Research) was applied to screen the crystallization conditions at different temperatures using the hanging-drop method by vapor diffusion.
Data collection
30% glycerol, neat PEG 400 and perfluoropolyether were used as cryoprotectants for the crystal mounting. Data collection was taken under the liquid nitrogen stream at −174°C. All DNA crystal data were collected at beam line X12C in NSLS of Brookhaven National Laboratory. A number of crystals were scanned to find the one with strong anomalous scattering at the K-edge absorption of selenium. The distance of the detector to the crystals was set to 150 mm. The wavelength of 0.9795 Å was chosen for data collection. The crystals were exposed for 10 s/image with 1° oscillation, and 180 images were taken for each data set. All data were processed using HKL2000 and DENZO/SCALEPACK (15).
Structure determination and refinement
The crystal structures of the modified DNAs were solved by molecular replacement with Phaser (16), followed by the refinement of selenium and sulfur atom positions in Refmac 5.0 (17). The refinement protocol includes the simulated annealing, positional refinement, restrained B-factor refinement and bulk solvent correction. The stereo-chemical topology and geometrical restrain parameters of DNA/RNA (18) have been applied. The topologies and parameters for the modified dUs with 2′-SeMe (UMS), 5′-SeMe (USE), 5′-SMe (USM) and 5′-OMe (T5O) were constructed and applied. After several cycles of refinement, a number of highly ordered waters were added. Finally, the occupancies of selenium and sulfur were adjusted. Cross-validation (19) with a 5–10% test set was monitored during the refinement. The σA-weighted maps (20) of the (2 m|Fo| – D|Fc|) and the difference (m|Fo| – D|Fc|) density maps were computed and used throughout the model building.
Molecular modeling calculation
In this work, the density functional theory calculations were performed on the molecules with the GAUSSIAN03W program using the Becke three-parameter and Lee–Yang–Parr hybrid (B3LYP) functions (21,22). The basis set used is 6-31+G(d,p). The energy-minimized structures of given molecules have been determined at the level of B3LYP/6-31+G(d,p). Vibrational spectra were then computed for the structure and obtained within the harmonic approximation. Final geometries of the molecules are confirmed to be stationary points as indicated by the absence of imaginary wavenumbers.
RESULTS AND DISCUSSION
DNA oligonucleotide synthesis, crystallization and data collection
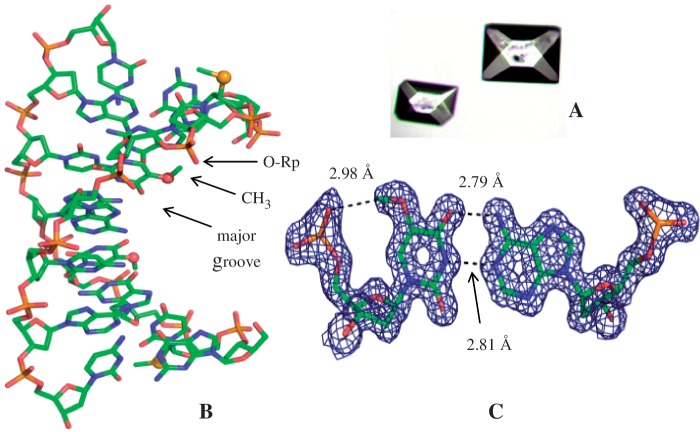
The RNA sequences were examined initially, and unfortunately, satisfactory crystals and diffraction were not obtained. The SedU (4 and Figure 1) has been used to facilitate the crystal growth and maintain the RNA-like conformation, since the 2′-SeMe functionality (2′-Se-facilitator) does not cause structure perturbation. (13,14,23–27) Thus, we have synthesized 3a {5-OMe-DNA: 5′-G(SedU)G(mo5dU)-ACAC-3′; molecular formula: C78H99N30O47P7Se; [M + H]+: measured 2505.6 (calculated 2505.8)}, 3b {5-SMe-DNA: 5′-G(SedU)G(ms5dU)ACAC-3′; molecular formula: C78H99N30O46P7SSe; [M + H]+: measured 2520.6 (calculated 2521.1)} and 3c {5-SeMe-DNA: 5′-G(SedU)G(mSe5dU)ACAC-3′; molecular formula: C78H99N30O46P7Se2; [M + H]+: measured 2568.6 (calculated 2569.0)}. The 5-OMe-, 5-SMe- and 5-SeMe-dU functionalities were introduced into the oligonucleotides by using the corresponding phosphoramidites synthesized from the modified nucleosides (28–30). In addition to its facilitation of crystal growth, the 2′-SeMe functionality can drive the DNA sequences into A-form conformation and structure. Therefore, this RNA-mimic system can serve as a useful model for the structure studies of RNA/RNA interactions and duplexes, which are usually difficult to crystallize and offer high-resolution structures. As expected, these three modified oligonucleotides were crystallized in 3–4 days and with high-diffraction quality, when screening with the Nucleic Acid Mini-kit (Hampton Research). Since the crystallization buffer conditions may affect the crystal packing and molecular interaction, the same buffer for 3a, 3b and 3c was selected for the crystallization. Furthermore, by the diffraction screening, buffer no. 7 of the kit (10% v/v 2-Methyl-2,4-pentanediol (MPD), 40 mM sodium cacodylate, pH 6.0, 12 mM spermine tetra-HCl, 80 mM potassium chloride and 20 mM magnesium chloride) was identified to give the highest resolutions for these three DNA structures. Several large crystals (up to 0.2 × 0.2 × 0.3 mm in size, Figure 2A) from the same buffer were found and mounted for diffraction data collection on X-ray beamline. These DNA molecules crystallized in the same space group (P43212) as the native. The detailed data collection and refinement statistics information for each sequence are presented in Tables 1 and 2.
Figure 2.
X-ray crystal structure of A-form DNA 3a [5-OMe-DNA: 5′-G(SedU)G(mo5dU)ACAC-3′] at 1.3 Å resolution (PDB ID: 3LTR). (A) Crystal images (bigger crystal: 0.2 × 0.2 × 0.3 mm). (B) Duplex structure; the yellow and red balls represent the selenium and oxygen atoms of the modified moieties, respectively. The 5-CH3-O group points to the 5′-phosphate group, instead of the major groove. (C) Electron density map of mo5dU/dA base pair, with δ level = 1.0.
Table 1.
X-ray data collection of 5-OMe, 5-SMe and 5-SeMe-containing DNAs [5′-G(SedU)G(mx5dU)ACAC-3′]
| Data collection | 5-OMe-DNA | 5-SMe-DNA | 5-SeMe-DNA |
|---|---|---|---|
| Space group | P43212 | P43212 | P43212 |
| Unit cell parameters (Å, °) | 43.21, 43.21, 23.69, 90.00, 90.00, 90.00 | 42.88, 42.88, 23.69, 90.00, 90.00, 90.00 | 43.08, 43.08, 23.81, 90.00, 90.00, 90.00 |
| Resolution range (Å) (last shell) | 50–1.30 (1.32–1.30) | 50–1.38 (1.43–1.38) | 50–1.40 (1.45–-1.40) |
| Unique reflections | 5618 (264) | 4616 (324) | 4661 (446) |
| Completeness (%) | 94.5 (70.2) | 94.0 (67.8) | 97.8 (99.3) |
| Rmerge (%) | 3.7 (34.4) | 5.7 (24.5) | 8.7 (13.6) |
| <I/σ(I)> | 38.4 (2.1) | 23.0 (5.9) | 18.9 (14.5) |
| Redundancy | 12.7 (2.0) | 8.4 (5.2) | 23.0 (11.1) |
Rmerge = Σ|I-〈I〉|/ΣI.
Table 2.
Refinement statistics of 5-OMe-, 5-SMe- and 5-SeMe-containing DNAs [5′-G(SedU)G(mx5dU)ACAC-3′]
| Refinement | 5-OMe-DNA 8-mer | 5-SMe-DNA 8-mer | 5-SeMe-DNA 8-mer |
|---|---|---|---|
| PDB ID: 3LTR | PDB ID: 3IKI | PDB ID: 3LTU | |
| Resolution range (Å) | 30.56-1.30 | 30.32-1.38 | 30.46-1.40 |
| Number of reflections | 5301 | 4373 | 4421 |
| Completeness | 95.11 | 94.04 | 97.83 |
| Rwork (%) | 19.67 | 17.80 | 17.4 |
| Rfree (%) | 21.88 | 19.40 | 20.0 |
| Number of atoms | 201 | 201 | 201 |
| Heavy atom | 1Se | 1Se | 2Se |
| Water | 31 | 37 | 38 |
| Bond length r.m.s. (Å) | 0.006 | 0.007 | 0.004 |
| Bond angle r.m.s. | 1.6 | 1.5 | 1.1 |
| Overall B-factor, (Å2) | 13 | 12 | 9 |
| B11/B22/B33 | −0.09/−0.09/0.17 | 0.12/0.12/−0.24 | 0.17/0.19/−0.34 |
| ESU on R-value, (Å) | 0.055 | 0.061 | 0.059 |
| ESU on Rfree (Å) | 0.056 | 0.060 | 0.061 |
| ESU on max. likelihood (Å) | 0.033 | 0.031 | 1.465 |
ESU = estimated standard uncertainties.
Structure comparison of 5-OMe, 5-SMe and 5-SeMe oligonucleotides
As showed in the duplex structures (Figures 2 and 3), the structures of the native and the 5-MeO-modified DNA (3a) are very similar. We have also observed previously that the 2′-Se-facilitator does not cause significant local and global structure perturbation (13,14,26,31). The 5-MeO locates in the major groove of the A-form helix (Figure 2B). Its methyl group points to the phosphate oxygen (O-Rp), instead of turning around and pointing to a larger space available in the major groove. Moreover, the electron density map of mo5dU/dA pair (Figure 2C) clearly shows that the distance between the methyl carbon and phosphate Rp-oxygen is 2.98 Å. Both the distance and geometry indicate H-bond formation between the 5-CH3-O and 5′-PO4− groups. The H-bond interactions between relatively acidic C-H protons and nearby oxygen or nitrogen atoms have been widely investigated in biological macromolecule systems (32,33). Moreover, the B-factor of the 5′-phosphate of the 5-OMe-dU structure is indeed smaller than that of the 5-Me-dU (i.e. T) structure (PDB ID: 1Z7I). Thus, this unique nucleobase/backbone interaction can reduce the backbone dynamics, significantly rigidifying the local conformation. The 5-OMe modification does not drastically add rigidity to the entire RNA, which is probably expected for this site-specific small modification.
Figure 3.
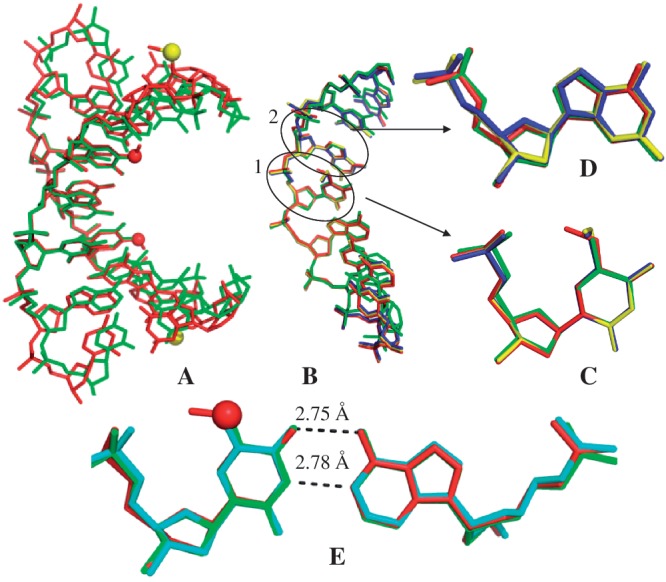
Crystal structure comparisons. The red and yellow balls represent the oxygen and selenium atoms of the modified moieties, respectively. (A) The superimposed duplex structures of 5′-G(SedU)G(mo5dU)ACAC-3′ (3a; in red; PDB ID: 3LTR; 1.30 Å resolution) and the corresponding native (in green; PDB ID: 1DNS; 2.0 Å resolution). (B) The superimposed single-strand structures of 3a (5-OMe-DNA; in red), 3b (5-SMe-DNA; in blue; PDB ID: 3IKI; 1.38 Å resolution), 3c (5-SeMe-DNA; in yellow; PDB ID: 3LTU; 1.40 Å resolution) and the corresponding native (in green; PDB ID: 1DNS). This high-resolution structure of 5-SeMe-DNA (3c), indicating no H-bond formation between the 5-SeMe and 5′-phosphate groups, updates the lower-resolution structure (PDB ID: 3BM0). (C) Superimposed modified dU4 and T4 residues in these four DNAs. (D) Superimposed local structures of dG3 in these four DNAs. (C and D are in the same color code as B). (E) The superimposed structures of mo5dU4/dA base pair of 3a (3LTR; in red), T4/dA of the native (1DNS; in green], and the native T4/dA base pair of 5′-G(SedU)GTACAC-3′ (1Z7I; in cyan; 1.28 Å resolution). The native T4 of 5′-G(SedU)GTACAC-3′ (PDB ID: 1Z7I; 1.28 Å resolution) is displayed, since the resolution of the native structure (1DNS; 2.0 Å resolution) is low and the highly ordered water molecules at T4 are not demonstrated. These two structures (1Z7I and 1DNS) are virtually identical.
To further study the modification, we hypothesized initially that the distance between the methyl C and O-Rp may play a critical role. A larger atom from the same elemental family [such as selenium and sulfur (atomic radii: 1.16 Å and 1.04 Å, respectively) versus oxygen (atomic radius: 0.73 Å)] might present the methyl group closer to the 5′-phosphate and enhance the H-bond. Thus, we replaced the oxygen atom of the 5-O-Me group with a larger selenium or sulfur atom. However, we found that the single Se or S atom replacement disrupts the H-bond and causes local alterations, though the native and modified duplexes have very similar overall structures, which is consistent with the UV-melting study (Supplementary Table S2). In the 5-SeMe or 5-SMe structure (circle 1 in Figure 3B), the methyl group points to the major groove, instead of the 5′-phosphate backbone (Figure 3B and C). Furthermore, the dihedral angle between the 5-CH3-Se-C (or 5-CH3-S-C) and uracil planes is ∼95°, whereas the same dihedral angle in the 5-O-Me modification is approximately zero (0.3°). These observations indicate that the atomic size of the bridging atom is not the key factor for generating the H-bond, instead the electronic effects of 5-oxygen (such as electronegativity and conjugation with uracil) may play critical roles in the H-bond formation. We also attempt to directly compare the differences in rigidity between the OMe, SMe and SeMe RNAs. It is difficult because of the impacts of two factors: one is the H-bond formation in the case of OMe, and the other one is the atomic weights of sulfur and selenium atoms. The H-bond reduces the dynamics (i.e. increasing rigidity) and the higher atomic mass can also reduce the local dynamics. These two competing factors cause the difficulty to evaluate the rigidity order among these modified RNAs, even though we can directly compare the structures of the OMe, SMe, and SeMe RNAs.
Moreover, after the 5-selenium or -sulfur replacement, the 5′-phosphate group of the neighboring 5′-nucleotide (dG3, circle 2 in Figure 3B) rotates ∼110° about its C4′–C5′ bond (Figure 3B and D), compared with the native and 5-O-Me modification. On the contrary, the 5-O-Me does not cause the backbone rotation (Figure 3D), and the 5-O-Me modified and native nucleobases have virtually identical structures (Figure 3E). The backbone rotation caused by the 5-Se-Me and 5-S-Me modifications is due to the alteration of the water networking surrounding the uracil 5-position, which in turn changes the water interactions in the major groove and with the phosphate backbone. Our crystal structures (Figure 4) reveal that two additional highly ordered water molecules (W1 and W2) are recruited to the major groove in the case of the 5-Se and 5-S modifications. The 5-Se or 5-S atom forms a H-bond with W1, considering that the distance between the 5-Se (or -S) atom and W1 is 3.42 Å, Se and S atoms are 0.43 and 0.31 Å, respectively, larger than O atom, and a typical H-bond length is 2.2–3.5 Å (33,34). W1 also forms a H-bond with W2, which subsequently forms H-bonds with W3 and the 5′-phosphate O-Rp of dG3. W4 and W5 remain at almost the same locations in the major groove. W2 is probably the direct cause of the backbone rotation.
Figure 4.
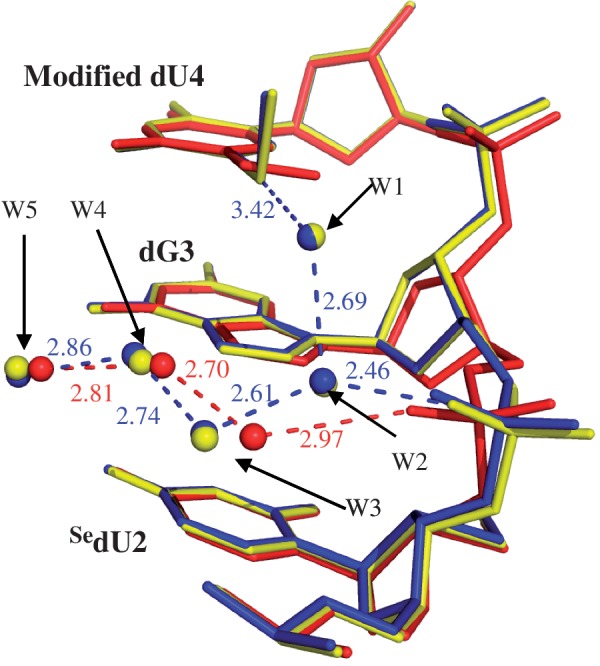
Hydration pattern comparison of the major grooves of SedU2-dG3-dU4. The superimposed local structures of the 5-OMe, 5-SMe and 5-SeMe DNAs are colored as red, blue and yellow, respectively. The H-bonds are shown in red for the 5-OMe DNA, and blue for the 5-S and 5-Se DNAs.
The 5-Se-Me and 5-S-Me DNAs have virtually identical structures, and the distances between the methyl carbon and phosphate oxygen are 3.66 Å in both the cases (Figure 5 and Supplementary Figure S1), indicating no H-bond formation. The coplanar conformation of the 5-O-Me probably allows maximal conjugation between the oxygen atom and uracil aromaticity, which makes the methyl group most acidic. Unlike the 5-Se-Me and 5-S-Me modifications, the formed H-bond by the 5-O-Me prevents the water networking from disruption and retains the A-form helix structure without significant perturbation. Our research results may provide a plausible reason why oxygen (with proper electronic property) is chosen to modify the uracil 5-position in tRNAs, instead of selenium or sulfur.
Figure 5.
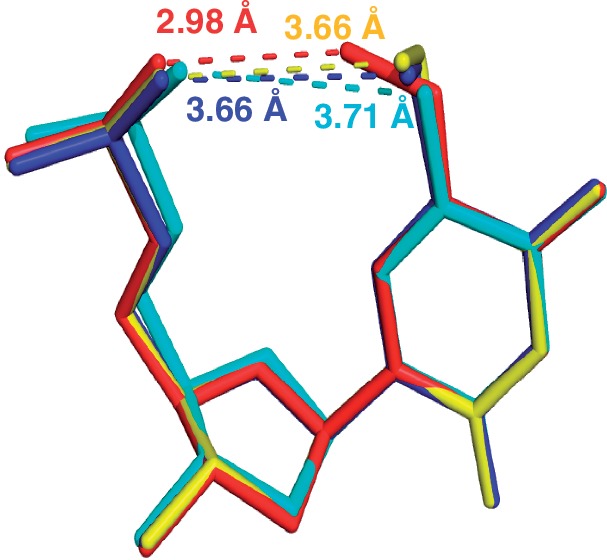
Superimposed structures of the nucleotides. mo5dU4 of 3a [5′-G(SedU)G(mo5dU)ACAC-3′; in red], ms5dU4 of 3b (blue), mSe5dU4 of 3c (yellow), and the native T4 of 5′-G(SedU)GTACAC-3′ (PDB ID: 1Z7I; in cyan). The numbers in the corresponding colors as the lines represent the distances of these atoms in the same structure.
Furthermore, we performed the computation study of the uracils derivatized with the 5-OMe, 5-SMe and 5-SeMe modifications in order to understand the orientation of these 5- functionalities. The computation study indicates that the 5-O-Me, 5-S-Me, and 5-Se-Me moieties form the dihedral angles of 60.2°, 62.7° and 68.7° with the uracil ring, respectively (Figure 6). The theoretical calculation suggests that the out-of-plane conformation is intrinsically favorable to minimize the energy levels of the modified uracils. In the determined crystal structures, the 5-Se-Me and 5-S-Me moieties still prefer the out-of-plane conformation with increased dihedral angles (95°). However, the 5-O-Me moiety forms a small dihedral angle (0.3°) and prefers the in-the-plane conformation. The H-bond formation alters the intrinsic orientation of the 5-OMe, whereas this H-bond is not observed in the 5-SeMe or 5-SMe modification. It is clear that the 5-O makes the methyl hydrogen more acidic than 5-S and 5-Se, and facilitates the H-bond formation in the A-form duplex. Moreover, our high-resolution structure indicates that the coplanar conformation of the 5-OMe with the uracil base enhances the base stacking interaction with the 5′-nucleobase adjacent to the mo5dU. The stabilization gained from the better stacking and H-bonding interactions compensates the destabilization from maintaining the unfavorable coplanar conformation of the 5-OMe, which is consistent with the computation study.
Figure 6.
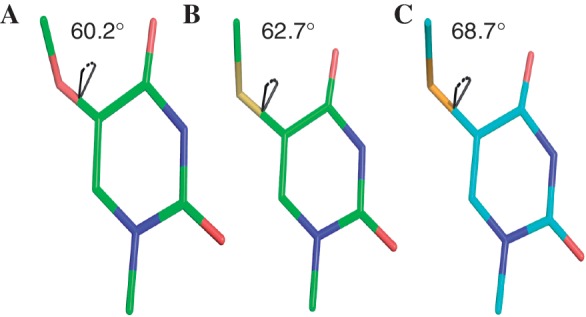
The calculated geometries of the 5-modifications in (A) N-Me-5-OMe-uracil; (B) N-Me-5-SMe-uracil and (C) N-Me-5-SeMe-uracil.
Since the 5-O can make the methyl hydrogen more acidic than 5-S and 5-Se, it is consistent with the H-bond formation observed in the crystal structure containing the 5-OMe modification. Moreover, our high-resolution structure indeed indicates that the coplanar conformation of the 5-OMe with the uracil base enhances the base stacking interaction with the nucleobases adjacent to the mo5dU. The stabilization gained from the better stacking and H-onding interactions is used to compensate the energy required for the unfavorable coplanar conformation, which is indicated by the computation study. According to the theoretical research, the out-of-plane conformation is more stable, thus the 5-SMe and 5-SeMe maintain the out-of-plane conformation, since they cannot form the H-bonding interaction to compensate the coplanar conformation requirement. Furthermore, the melting temperature (Tm) data in Supplementary Table S2 indicate that the overall stabilities of the non-modified and 5-OMe duplexes are virtually identical. The similar stability indicates that the H-bond and stacking of the 5-OMe-U overcome the instability caused by the coplanar conformation of the 5-OMe relative to the uracil base. The structural data is consistent with the results of thermostability and computation studies. On the basis of the computational simulation, the out-of-plane conformation is more stable for all three modifications, including the 5-OMe, 5-SMe and 5-SeMe, which suggests the conjugation is probably less important for the orientation of the 5-OMe relative to the uracil plane. The H-bonding is most likely the major player determining the coplanar conformation of the 5-OMe relative to the uracil base. Consequently, the 5-OMe modification does not affect overall U-pairing stability.
Our studies indicate that the 5-Me group orientation and the H-bond formation are unique features of the 5-O-CH3 modification in A-form helix. The rigidified local structure and conformation may facilitate the base recognition in tRNA–mRNA interaction and translation. So far, the 5-OMe-U modification has only been found on 34 position of tRNA, which is the first nucleotide in anticodon and pairs with the third nucleobase in codon. It was reported that the mnm5 (5-CH2NHCH3) modification reduces the flexibility of the anticodon and contributes to ‘pre-organize’ the anticodon into an A-form structure ready to interact with the codon (9). Similarly, by forming the H-bond, the 5-OMe functionality can make the anticodon pre-organized for interacting with codon. Moreover, it was reported (8) that the reading efficiency of tRNA containing the 5-OMe-U modification (UGA) is higher than that of its non-modified counterpart (i.e. tRNA containing native UGA) when reading UCU and UCG codons (the wobble reading). Our observations of the H-bond formation and the local rigidification shed new light on the roles of the 5-OMe in anticodon/codon interaction and mRNA reading function. Although the context of the sequence is different from tRNA, the formation of the duplexes, including A-form duplex, is sequence-independent, as long as the sequences are complementary to each other. Our observation reveals rather general H-bond formation of the OMe modification in the A-form duplex. Therefore, our crystal structure study suggests that the H-bond may form when the OMe-modified tRNA encounters the condon in mRNA.
CONCLUSION
In summary, via crystal structure determination of an A-form helix and atom-specific substitution, we have studied the uracil 5-O-CH3 functionality, which is a natural tRNA modification. The A-form DNA duplex was used as a mimic of RNA duplex, and the 2′-Se-moiety was used to ensure the A-form conformation and facilitate growth of oligonucleotide crystals with high diffraction quality. The 5-O-Me modified structure is virtually identical to the corresponding native structure. This is the first observation of the interaction between a nucleobase and its 5′-phosphate group. This interaction might rigidify the local backbone and conformation. The 5-O-Me modification promotes this interaction by forming the H-bond between the CH3 and 5′-phosphate Rp-oxygen. Our computational result is also consistent with the H-bond formation. The atom-specific replacement of the 5-O with Se or S abolishes the H-bond formed by the methyl group, and these 5-modifications do not significantly alter the duplex stability. Furthermore, the 5-O-Me moiety does not cause local structure perturbation, whereas the 5-Se-Me and 5-S-Me cause the local backbone rotation by disrupting the water networking in the major groove. Our studies reveal the unique features of the 5-O-CH3 modification of the tRNA uracil, suggesting a general interaction between modified nucleobases and backbones. The H-bond formation and local rigidification by the 5-OMe might help explaining the facilitation of the codon wobble reading. Moreover, this atom-specific replacement demonstrates a useful methodology to study RNA modifications.
ACCESSION NUMBERS
PDB IDs: 3LTR, 3IKI, 3LTU.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figure 1.
FUNDING
USA National Science Foundation [MCB-0824837]; Georgia Cancer Coalition Distinguished Cancer Clinicians and Scientists Award and the US National Institute of Health [GM095086]. Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
We thank the mail-in program and PXrr in Brookhaven National Laboratory (BNL) for the X-ray crystal diffraction data collection. The National Synchrotron Light Source (NSLS) at BNL is funded by NIH’s National Center for Research Resources and DOE’s Office of Biological and Environmental Research. The structure pictures were prepared by PyMol (DeLano Scientific LLC, http://www.pymol.org).
REFERENCES
- 1.Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agris PF, Vendeix FA, Graham WD. tRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 3.Gustilo EM, Vendeix FA, Agris PF. tRNA’s modifications bring order to gene expression. Curr. Opin. Microbiol. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishimura S. Minor components in transfer RNA: their characterization, location and function. Prog. Nucleic Acid Res. Mol. Biol. 1972;12:49–85. [PubMed] [Google Scholar]
- 6.Albani M, Schmidt W, Kersten H, Geibel K, Luderwald I. 5-methoxyuridine, a new modified constituent in tRNAs of Bacillaceae. FEBS Lett. 1976;70:37–42. doi: 10.1016/0014-5793(76)80721-1. [DOI] [PubMed] [Google Scholar]
- 7.Murao K, Ishikura H, Albani M, Kersten H. On the biosynthesis of 5-methoxyuridine and uridine-5-oxyacetic acid in specific procaryotic transfer RNAs. Nucleic Acids Res. 1978;5:1273–1281. doi: 10.1093/nar/5.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takai K, Okumura S, Hosono K, Yokoyama S, Takaku H. A single uridine modification at the wobble position of an artificial tRNA enhances wobbling in an Escherichia coli cell-free translation system. FEBS Lett. 1999;447:1–4. doi: 10.1016/s0014-5793(99)00255-0. [DOI] [PubMed] [Google Scholar]
- 9.Takai K, Yokoyama S. Roles of 5-substituents of tRNA wobble uridines in the recognition of purine-ending codons. Nucleic Acids Res. 2003;31:6383–6391. doi: 10.1093/nar/gkg839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egert E, Lindner HJ, Hillen W, Buhm MC. Influence of substituents at the 5-position on the structure of uridine. J. Am. Chem. Soc. 1980;102:3707–3713. [Google Scholar]
- 11.Egert E, Lindner H. Crystal structure and conformation of 2-thio-5-methylaminomethyluridine dihydrate. Acta. Crystallogr. B. 1979;35:122–125. [Google Scholar]
- 12.Egert E, Lindner H. Crystal structure and conformation of 5-aminouridine. Acta. Crystallogr. B. 1978;34:2204–2208. [Google Scholar]
- 13.Sheng J, Jiang J, Salon J, Huang Z. Synthesis of a 2′-Se-thymidine phosphoramidite and its incorporation into oligonucleotides for crystal structure study. Org. Lett. 2007;9:749–752. doi: 10.1021/ol062937w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang J, Sheng J, Carrasco N, Huang Z. Selenium derivatization of nucleic acids for crystallography. Nucleic Acids Res. 2007;35:477–485. doi: 10.1093/nar/gkl1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 16.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta. Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 18.Parkinson G, Vojtechovsky J, Clowney L, Brunger AT, Berman HM. New parameters for the refinement of nucleic acid-containing structures. Acta. Crystallogr. D Biol. Crystallogr. 1996;52:57–64. doi: 10.1107/S0907444995011115. [DOI] [PubMed] [Google Scholar]
- 19.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 20.Read RJ. Improved fourier coefficients for maps using phases from partial structures with errors. Acta. Crystallogr. A. 1986;42:140–149. [Google Scholar]
- 21.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 22.Becke A. Density functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 23.Salon J, Sheng J, Gan J, Huang Z. Synthesis and crystal structure of 2′-Se-modified guanosine containing DNA. J. Org. Chem. 2010;75:637–641. doi: 10.1021/jo902190c. [DOI] [PubMed] [Google Scholar]
- 24.Hassan AE, Sheng J, Zhang W, Huang Z. High fidelity of base pairing by 2-selenothymidine in DNA. J. Am. Chem. Soc. 2010;132:2120–2121. doi: 10.1021/ja909330m. [DOI] [PubMed] [Google Scholar]
- 25.Salon J, Sheng J, Jiang J, Chen G, Caton-Williams J, Huang Z. Oxygen replacement with selenium at the thymidine 4-position for the Se base pairing and crystal structure studies. J. Am. Chem. Soc. 2007;129:4862–4863. doi: 10.1021/ja0680919. [DOI] [PubMed] [Google Scholar]
- 26.Sheng J, Huang Z. Selenium derivatization of nucleic acids for X-ray crystal-structure and function studies. Chem. Biodivers. 2010;7:753–785. doi: 10.1002/cbdv.200900200. [DOI] [PubMed] [Google Scholar]
- 27.Sheng J, Salon J, Gan J, Huang Z. Synthesis and crystal structure study of 2′-Se-adenosine-derivatized DNA. Sci. China Chem. 2010;53:78–85. [Google Scholar]
- 28.Zhang W, Sheng J, Hassan AE, Huang Z. Synthesis of 2′-Deoxy-5-(methylselenyl)cytidine and Se-DNAs for structural and functional studies. Chem. Asian J. 2012;7:476–479. doi: 10.1002/asia.201100898. [DOI] [PubMed] [Google Scholar]
- 29.Hassan AE, Sheng J, Jiang J, Zhang W, Huang Z. Synthesis and crystallographic analysis of 5-Se-thymidine DNAs. Org. Lett. 2009;11:2503–2506. doi: 10.1021/ol9004867. [DOI] [PubMed] [Google Scholar]
- 30.Stout MG, Robins RK. The synthesis of some 5-methoxypyrimidine nucleosides. J. Heterocycl. Chem. 1972;9:545–549. [Google Scholar]
- 31.Lin L, Sheng J, Huang Z. Nucleic acid X-ray crystallography via direct Selenium derivatization. Chem. Soc. Rev. 2011;40:4591–4602. doi: 10.1039/c1cs15020k. [DOI] [PubMed] [Google Scholar]
- 32.Jiang L, Lai L. CH•••O hydrogen bonds at protein–protein interfaces. J. Biol. Chem. 2002;277:37732–37740. doi: 10.1074/jbc.M204514200. [DOI] [PubMed] [Google Scholar]
- 33.Castellano RK. Progress toward understanding the nature and function of C–H•••O interactions. Curr. Org. Chem. 2004;8:845–865. [Google Scholar]
- 34.Emsley J. Very strong hydrogen bonding. Chem. Soc. Rev. 1980;9:91–124. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.