Abstract
Analysis of processing, assembly, and function of higher eukaryotic ribosomal RNA (rRNA) has been hindered by the lack of an expression system that enables rRNA to be modified and then examined functionally. Given the potential usefulness of such a system, we have developed one for mammalian 18S rRNA. We inserted a sequence tag into expansion segment 3 of mouse 18S rRNA to monitor expression and cleavage by hybridization. Mutations were identified that confer resistance to pactamycin, allowing functional analysis of 40S ribosomal subunits containing synthetic 18S rRNAs by selectively blocking translation from endogenous (pactamycin-sensitive) subunits. rRNA constructs were suitably expressed in transfected cells, shown to process correctly, incorporate into ≈15% of 40S subunits, and function normally based on various criteria. After rigorous analysis, the system was used to investigate the importance of sequences that flank 18S rRNA in precursor transcripts. Although deletion analysis supported the requirement of binding sites for the U3 snoRNA, it showed that a large segment of the 5′ external transcribed spacer and the entire first internal transcribed spacer, both of which flank 18S rRNA, are not required. The success of this approach opens the possibility of functional analyses of ribosomes, with applications in basic research and synthetic biology.
INTRODUCTION
Ribosomes are macromolecular structures that catalyze protein synthesis in cells. They consist universally of two subunits composed of numerous proteins and several ribonucleic acids (RNAs). Proteins contribute to the structure, stability, and activity of the subunits. However, in general, it has been difficult to assign specific functions to individual ribosomal proteins, as many are not essential for ribosomal function or have additional extraribosomal functions (1,2). By contrast, ribosomal RNAs (rRNAs) are responsible for the overall shape of the ribosomal subunits and for the enzymatic activity that catalyzes protein synthesis. Evidence is accumulating that rRNAs may also affect other aspects of protein synthesis, including mRNA recruitment, regulation of the efficiency of translation of specific mRNAs, and facilitation of ribosomal shunting (3–6).
The ability to study the role of rRNA in ribosome assembly, protein synthesis, and noncanonical aspects of rRNA function requires being able to alter rRNA sequences and monitor the activity of modified ribosomal subunits in vivo (Figure 1a). However, investigations of mammalian rRNAs have lagged behind those of yeast and bacteria because there has been no suitable platform available for expressing modified rRNAs and performing functional analyses. Although previous studies have transcribed portions of mammalian rRNAs, they have not performed downstream analyses of subunits containing correctly processed (mature) rRNAs and could not differentiate between subunits containing synthetic or endogenous rRNAs.
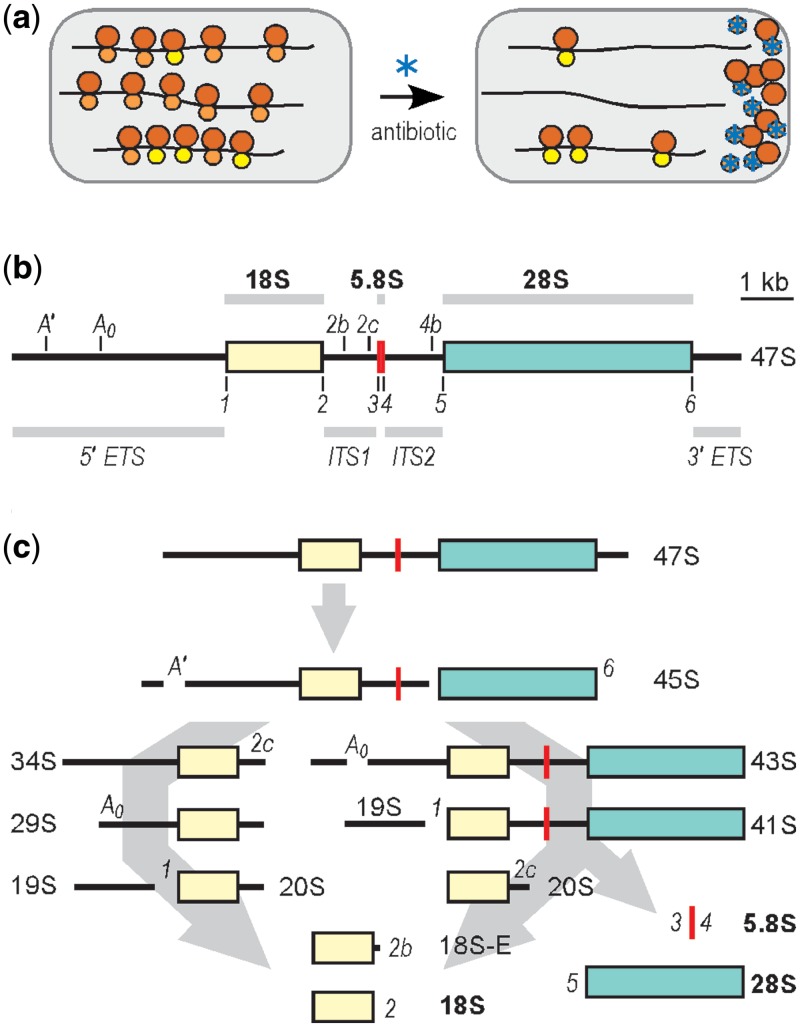
Figure 1.
(a) Schematic representation of an rRNA expression platform. Grey enclosures represent cells and contain polysomes with mRNA indicated as black lines and ribosomal subunits as circles (60S subunits as larger circles; 40S subunits as smaller circles). The sandy brown 40S subunits are antibiotic-sensitive endogenous subunits; the yellow 40S subunits contain synthetic 18 S rRNA and are antibiotic resistant. The different colors are used to indicate that the subunits are physically distinguishable. A cell-permeable antibiotic (blue asterisk) binds to and blocks the activity of endogenous 40S subunits. Synthetic 40S subunits are unaffected and are able to translate under these conditions. (b) Mouse 47S primary rRNA transcript, 47S rRNA is indicated schematically as a horizontal line. The colored sections represent encoded rRNAs: 18 S in light yellow; 5.8S in red; and 28S in turquoise. The 18 S rRNA is the RNA component of 40S ribosomal subunits and the 28S and 5.8S rRNAs are RNA components of 60S subunits. The black lines represent the external transcribed spacer regions (5′ ETS and 3′ ETS) and the internal transcribed spacer regions (ITS1 and ITS2), as indicated. Cleavage sites in the transcribed spacers are indicated as vertical lines and labeled. (c) Processing pathways. Processing of the precursor transcripts involves numerous protein and RNA factors and has been studied extensively in various organisms (44). The mouse 47S precursor rRNA is transcribed in the nucleolus by RNA polymerase I, and is subsequently processed by two possible pathways. Cleavage proceeds in the direction of the arrows from 47S to 18 S, 5.8S, and 28S rRNAs; 45S rRNA can be processed to 18 S rRNA by two pathways as indicated, which generate different intermediate products. Processing sites and major cleavage products resulting from maturation of 18 S rRNA are indicated at each cleavage step. Processing of 5.8S and 28S rRNAs involve additional cleavage steps and intermediate products, which are not indicated. This figure is adapted from ref. (13).
The present study reports the development of a mouse 18S rRNA expression system. This rRNA is the RNA component of the small (40S) ribosomal subunit and is cotranscribed as part of a larger precursor transcript along with the 5.8S and 28S rRNAs, which are components of the large (60S) subunit (Figure 1b). Studies in yeast have supported the feasibility of expressing a mammalian 18S rRNA in isolation (7). To monitor the transcription, processing, and subcellular distribution of 40S subunits containing synthetic rRNAs, we introduced a sequence tag into the 18S rRNA that can be detected by hybridization. In addition, to monitor the translational activity of the ribosomal subunits, we identified mutations in the 18S rRNA that confer resistance to inhibition by the antibiotic pactamycin. Pactamycin is thought to inhibit translation by binding to the E site in 40S ribosomal subunits, blocking translocation of the mRNA-tRNA complex through the ribosome during elongation (8). Therefore, by expressing 18S rRNAs with a pactamycin-resistance mutation in cells, we are able to use the drug to specifically block translation from the cell’s endogenous 40 S subunits and monitor translation from subunits containing the pactamycin-resistant mutation.
The correct processing of suitably modified 18S rRNA in our system provided us with a method to investigate the importance for processing and function of sequences flanking the 18S rRNA: the 5′ external transcribed spacer (ETS) and the first internal transcribed spacer (ITS1). These sequences are expanded in mammalian precursor rRNAs, although many of the principles of rRNA processing seem to be shared amongst species (Figure 1c). For example, mammals have the longest 5′ ETS sequences, with human, mouse, hamster, and rat sequences all longer than other known 5′ ETS sequences by at least 2 kilobases. In addition, mammals have expanded ITS1 sequences, a feature shared with avian species (9).To date, there has been little analysis of the expanded regions contained within the transcribed spacer sequences in mammals. Inasmuch as these expanded sequences represent a significant fraction of the transcription of the most abundant RNAs in cells, expansion may have been selected to benefit ribosomal processing, assembly, or function. Alternatively, these regions may have evolved important secondary functions. Here, we use the rRNA expression system to investigate the importance of sequences in the 5′ ETS and ITS1 for rRNA processing and function.
MATERIALS AND METHODS
rDNA cloning and mutagenesis
18S ribosomal deoxyribonucleic acid (rDNA) and flanking regions were amplified by polymerase chain reaction (PCR) using genomic DNA prepared from mouse Neuro 2a (N2a) cells and oligonucleotide primers rDNA.1 and rDNA.2 (Supplementary Table S1). Cloning into plasmid pRL-cytomegalovirus (CMV) (Promega Corporation, Madison, WI, USA) used NheI and NotI restriction sites that were introduced at the 5′ ends of rDNA.1 and rDNA.2, respectively. All sequencing of PCR products and plasmid constructs was performed using standard Sanger sequencing. To detect synthetic 18S rRNAs, a 24-nt sequence was cloned into the 18S rDNA as a hybridization tag using the SacI site in expansion segment 3. The hybridization tag was generated using a random sequence generator (http://www.faculty.ucr.edu/∼mmaduro/random.htm) and selected based on minimal predicted self-complementarity and secondary structure as determined using Oligo Calc (http://www.basic.northwestern.edu/biotools/OligoCalc.html). Constructs containing the hybridization tag are designated with a “tag” extension, for example, pPol-I (18S-tag).
A short RNA polymerase I (pol-I) promoter element containing the 5′ SalI-box and a minimal pol-I promoter was cloned from the intergenic spacer of a 45S rDNA gene (−169 to +1) using primers Pol-I.1 and Pol-I.2. A BlgII site in Pol-I.1 and an NheI site in Pol-I.2 were used to clone the PCR fragment into pRL-CMV, replacing the BlgII-NheI fragment, which contains the CMV promoter and a chimeric intron. A pol-I terminator was obtained from the 3′ ETS of 45S rDNA, which contains 10 SalI-box pol-I terminators, using primers Pol-I.3 and Pol-I.4. A NotI site in primer Pol-I.3 and an MfeI site in primer Pol-I.4 were used to clone the PCR fragment into pRL-CMV, replacing a NotI-MfeI fragment that contains the SV40 poly(A) signal.
Three 5′ ETS and two ITS1 fragments were cloned for these studies. The 5′ ETS fragments are the entire 4014-nt sequence, and two shorter fragments that extend 1188 and 703 nucleotides upstream of site 1. The two larger fragments were cloned using NdeI as the 3′ site, which lies immediately 3′ of site 1. The 4014-nt sequence was obtained from a PCR product containing the pol-I promoter and amplified using primers rDNA.3, which contains a 5′ BglII site, and rDNA.4. The 1188-nt fragment was obtained from a PCR product amplified using primers rDNA.4 and rDNA.5 and cloned using an NheI site contained within the 5′ ETS. The 703-nt fragment was cloned together with the 18S rDNA in PCR reactions performed using oligonucleotide primers rDNA.1 and rDNA.2. The two ITS1 fragments are a 1-kb fragment amplified by using primers rDNA.6 and rDNA.7 and a 70-nt fragment cloned with the 18S rDNA. The 1-kb fragment was subcloned using a NotI site that was introduced into rDNA.7 and an EcoNI site present at the 3′ end of the 18S rDNA. The various fragments described earlier were used to generate 6 constructs. The construct containing the full-length 5′ ETS was assembled using the following restriction fragment combination: MfeI-BglII-NdeI-EcoNI-NotI-MfeI. The other constructs were assembled using the restriction fragment combination: MfeI-BglII-NheI-NdeI-EcoNI-NotI-MfeI.
For transcription via the RNA polymerase II (pol-II) promoter, the inserts described earlier, except for the full-length 5′ ETS insert, were transferred into plasmid pRL-CMV, replacing the NheI-NotI fragment. The full-length 5′ ETS insert was first reamplified using oligonucleotide rDNA.12 to introduce an NheI site at the 5′ end. The chimeric intron of pRL-CMV was removed and replaced with a SacI-NheI fragment generated by annealing oligonucleotides rDNA.8 and rDNA.9.
Deletion constructs were generated by digesting plasmid pPol-I(18S-tag) with the following enzymes followed by religation: PvuII-AfeI (p18S.11), PvuII-ZraI (p18S.10), AvrII-NheI (p18S.9), XhoI-XhoI (p18S.8), and AatII (blunt)-BglI (blunt)(p18S.7). p18S.7 was generated using a partial digestion with BglI as the plasmid contains additional sites.
Mutations were generated by PCR, using complementary 45-nt primers containing the mutations and 5′ and 3′ flanking primers located outside of restriction enzyme sites used for cloning. A fragment from each PCR reaction was then subcloned into the expression construct; an NcoI-HindIII fragment of 18S for antibiotic resistance mutations or an AvrII-AatII fragment of 5′ ETS for 5′ hinge mutations.
Reporter genes
pGL4.13 (Promega) was used as a monocistronic control. Dicistronic controls were constructed from pGL4.13 with a double insertion 3′ of the synthetic firefly luciferase gene luc2. The first fragment, an XbaI-NcoI fragment was derived from previous constructs containing a multiple cloning site (MCS), the Encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) or the poliovirus (PV) IRES (10). The second fragment was an NcoI-XbaI fragment from phRG-B (Promega), which contains the humanized Renilla luciferase gene hRluc.
Analysis of rRNA expression and processing
All constructs in these studies were tested in transiently transfected N2a cells. Cells were seeded in 6-well dishes at 100 000 cells/well and transfected the next day using 1-µg DNA per well with 3-µl Fugene 6 (Roche). Approximately 48 hours posttransfection, total RNA was extracted with Trizol reagent (Life Technologies). RNA was quantified, and 2 µg of total RNA and indicated amounts of rRNA in vitro transcripts were electrophoresed on denaturing agarose gels for Northern blot analyses. For each set of samples, gels included a single-stranded RNA marker (1 µg of ssRNA marker; NEB). The positions of the bands were marked on membranes posttransfer using ethidium bromide staining. For Northern blot analyses, membranes were hybridized overnight at 37°C in Ultrahyb solution (Ambion) using 32P- or 33P-5′-labeled oligonucleotide probes. Blots were analyzed using a Molecular Dynamics Phosphorimager system. Different oligonucleotide probes (Supplementary Table S1) were used to detect RNAs containing various sequences. The α-tag probe for RNAs containing the 24-nt hybridization tag; the α-5′ ETS probe for RNAs containing 5′ ETS sequences immediately upstream of site 1, the α-ITS1 probe for RNAs containing ITS1 sequences immediately 3′ of the site 2, and the α-18S rRNA probe for 18S rRNA.
Size control transcripts were generated using an Ambion MEGAscript in vitro transcription kit from PCR-generated fragments amplified using plasmid templates with 5′ primer rDNA.10, which contains the T7 RNA polymerase promoter fused to the first 24 nucleotides of 18S rDNA and 3′ primer rDNA.11 that is complementary to nucleotides at the 3′ end of the 18S rDNA. The resulting transcripts contain 3 additional 5′ guanine nucleotides compared with endogenous 18S rRNAs. The untagged transcript is 1874 nucleotides; the tagged transcript is 1898 nucleotides.
Analysis of synthetic ribosomes
For analyses of protein expression from pactamycin-resistant ribosomes, cells were transfected in 24-well dishes seeded at 20 000 cells/well 24 hours before transfection. 0.5 µg DNA was transfected per well using 1.5-µl Fugene 6 (Roche). Approximately 40 hours post-transfection, cell culture media was switched to starvation medium lacking L-methionine and L-cysteine. Cells were starved for 30 minutes before the media was changed to fresh starvation media containing pactamycin as indicated in the figures, cultured for an additional 30 minutes, and then labeled with 35S-Met/35S-Cys for 4 hours using 1.2 µl of TRAN35S label (MP Biomedicals, Ohio) per well. After labeling, media was removed and cells were washed with ice-cold phosphate buffered saline (PBS). Cells were lysed on ice for 30 minutes on a rocker table with 35-µl ice-cold RIPA buffer (10 mM sodium phosphate pH 7.3, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA) 1% octylphenoxypolyethoxyethanol (IGEPAL) CA-603, 0.1% SDS, 1% sodium deoxycholate, and 1x complete protease inhibitor (Roche)). Cell lysates were centrifuged for 15 minutes at 12 000 x g to remove debris and pellet genomic DNA; 19 µl of lysate was transferred to a new tube with 1-µl 1 M dithiothreitol (DTT) and 4x lithium dodecyl sulfate (LDS) loading buffer (Invitrogen). Samples were heated for 5 minutes at 90°C, placed on ice, and electrophoresed on 4–12% BIS-TRIS NuPage gels using MOPS-SDS buffer (Invitrogen). Gels were soaked in 30% methanol, 5% glycerol for 30 minutes, vacuum dried, and analyzed using a Molecular Dynamics Phosphorimager system or film.
For polysome analysis, cells were seeded at 100 000 per well in 6-well plates, and transfected 24 hour later with an 18S rDNA expression plasmid using 6-µl Fugene 6 per µg DNA. Forty-eight hours post-transfection, samples were processed at 4°C; culture media was aspirated and cells washed with ice-cold PBS containing 0.1 mg/ml cycloheximide. Six wells were scraped with 300 µl of cold lysis buffer A (20 mM Tris-HCl pH 7.5, 15 mM MgCl2, 100 mM KCl, 1% TritonX-100, 0.1 mg/ml cycloheximide, 0.05x Murine RNase inhibitor (NEB), 1x complete protease inhibitor (Roche)). The scraped cells and lysis buffer were then transferred to tubes and incubated on ice for 10 minutes with occasional mixing for passive lysis. Tubes were centrifuged for 10 minutes at 16 000 x g to pellet cellular debris. Approximately 1 OD260 of supernatant was loaded onto a 12-ml, 10–50% sucrose gradient (20 mM Tris-HCl pH 7.5, 15 mM MgCl2, 100 mM KCl, 0.1 mg/ml cycloheximide). Gradients were centrifuged at 35 000 rpm (155 000 x g) in a SW40Ti rotor for 3 hours, and then fractionated using an ISCO fractionator. RNA was ethanol precipitated from these fractions and the resulting pellet was extracted with phenol/chloroform. For each pooled fraction, one-tenth of the total volume was analyzed by primer extension using ddCTP to distinguish between endogenous rRNAs and synthetic rRNAs containing the pactamycin-resistance mutation (11). Controls for the primer extension reactions contained 200 ng of a 1:1 18Swt/18S693mutant transcript mix (Control 1) and 600 ng N2a total RNA (Control 2). RNA sequencing ladders were generated using 200 ng of the same 1:1 transcript mix.
For EDTA-dissociated ribosomes, cells were washed 48 hours post-transfection with 1 ml warm PBS, and 18 wells were lysed using 800-µl ice-cold lysis buffer B (20 mM Tris-HCl pH 7.5, 100 mM KCl, 0.3% IGEPAL CA-630, 0.05x Murine RNase inhibitor (NEB), 1x complete protease inhibitor (Roche)). The lysate was further processed using a dounce homogenizer (100 passes) and then spun for 15 minutes at 16 000 x g to pellet debris. The OD260 of each lysate supernatant was determined and one-fifth volume of EDTA buffer (20 mM Tris-HCl pH 7.5, 100 mM KCl, 150 mM EDTA, 0.05 x Murine RNase inhibitor (NEB), 1x complete protease inhibitor (Roche)) was added to adjust the final concentration of EDTA to 30 mM. Approximately 4.5 OD260 of each lysate was loaded onto a 12-ml, 10–35% sucrose gradient in 20 mM Tris-HCl pH 7.5, 100 mM KCl, 30 mM EDTA. Gradients were spun for 5 hours in an SW40Ti rotor at 40 000 rpm (200 000 x g). RNAs from fractionated samples were analyzed, as described earlier, except 1/60th of each pooled fraction was used due to more efficient recovery of RNA by this method.
P100 pellet and S100 supernatant fractions were prepared using mild lysis conditions to minimize lysis of nuclei. Forty-eight hours post-transfection, plates were washed with 1-ml warm PBS, 250-µl ice-cold lysis buffer B with 5 mM MgCl2, and 2-mM DTT was then added to each well. As earlier above, wells were scraped; the lysate was subjected to active lysis and centrifuged to pellet debris. The supernatants were then centrifuged for 3 hours at 100 000g. RNA was isolated from supernatants as described earlier for fractions. The P100 pellet was softened overnight with 20 mM Tris–HCl pH 7.5, 100 mM KCl, 5-mM MgCl2, and RNA was extracted with phenol/chloroform.
For cotransfection experiments using 18S rDNA and luciferase reporter constructs, cells were seeded in 6-well plates and transfected with an 18S rDNA expression construct, as described earlier, using 1-µg vector: 3-µl Fugene 6 per well. For pactamycin treatment, cell media was removed 48 hours post-transfection and replaced with media containing 100-ng/ml pactamycin (generously provided by Dr. Dianna Maar). Cells were cultured for ≈30 minutes and then transfected with reporter constructs using 1-µg plasmid and 2-µl Lipofectamine 2000 (Invitrogen). Cells were cultured overnight, washed with warm PBS, and lysed with passive lysis buffer (Promega). For each independent transfection experiment, an equal volume of cell lysate (20 µl of 250 µl) was assayed using an EG&G Berthold 96 well format dual-injector luminometer.
RESULTS
Identification of an abundant 18S rRNA variant
Mammalian cells contain several hundred rDNA genes that are not identical. These sequence variants provide a source of ribosomal heterogeneity and may have functional consequences (3,4). We began by cloning several sequences from mouse N2a genomic DNA. rDNA fragments were PCR-amplified using primerss rDNA.1 and rDNA.2 (Supplementary Table S1), located ≈700 nucleotides upstream and 70 nucleotides downstream of the 18S rDNA, respectively. Six independent clones were sequenced and found to be >99% identical to each other. However, none of these sequences are completely identical to each other or to two different 18S rDNA sequences in the NCBI nucleotide database (X82564, NR_003278, X00686; sequences X82564 and NR_003278 contain identical 18S sequences). In addition to several unique variations that differ from the published sequences, the sequences identified in this study share a nucleotide difference in helix H41a and two single nucleotide insertions, one in expansion segment 3 and the other in helix H30 (Supplementary Figure S1 (45)).
For our expression system, we decided to start with an 18S rDNA gene sequence that contains the most common sequence variants at each nucleotide position to provide a reference for future studies of rRNA sequences that contain less common sequence variants. To assess how the cloned sequences compare to the population of 18S rDNA sequences, they were compared to sequences obtained using pooled genomic PCR products as templates. Barring significant bias in the PCR reactions, the sequences of the pooled genomic sequences should represent the major genomic variants at each nucleotide. Two independent PCR reactions (PCR1 and PCR2) were performed, and the products were sequenced directly. Comparison of the pooled genomic sequence to those of the clones isolated in this study revealed that one of the clones contains the same sequence as the pooled genomic sequence (see Supplementary Figure S1). This clone (accession #JQ247698) was selected for subsequent experiments. It is 1871 nucleotides long and contains the 3 shared mutations discussed earlier, but no other variations from published sequence X00686.1.
In contrast to the 18S rRNA, sequences from the short region of the 5′ ETS included in this clone showed substantially more variation compared with published sequences. This variability is consistent with previous reports, for example, a report by Tseng et al. (12).
Development of an 18S rRNA expression construct
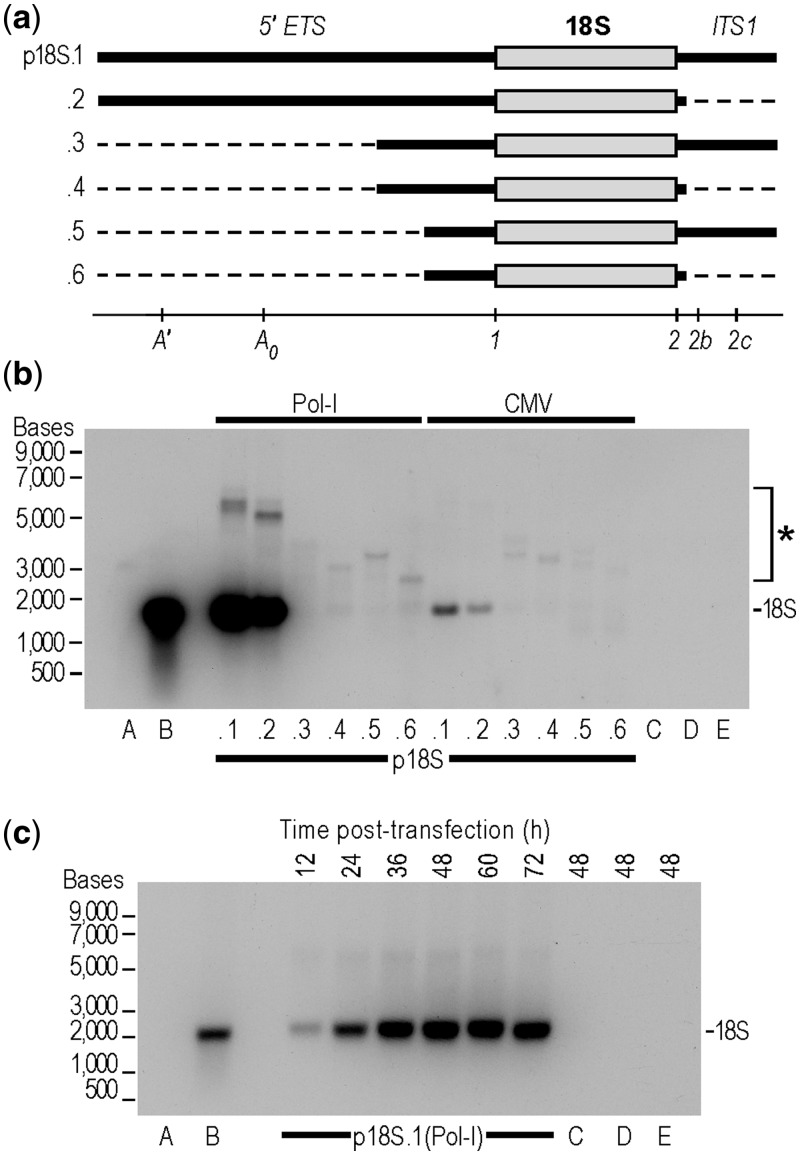
We chose to develop an 18S rRNA expression system to study the role of this RNA in the biogenesis of 40S ribosomal subunits and in the process of translation initiation. Based on previous studies in eukaryotes, we expected that mouse rDNA genomic fragments would require the A′, A0, 1, and 2 sites for proper processing [(13); Figure 1b]. Therefore, we used the 18S rDNA clone identified earlier to generate expression constructs containing the 5′ ETS, 18S rDNA, and ITS1, which include all of these sites (Figure 2a). Constructs were generated using the pol-I promoter and 3′ ETS, or the CMV promoter and an SV40 poly(A) signal. We also tested constructs with deletions at the 5′ end of the 5′ ETS and the 3′ end of ITS1, that is, with less authentic spacer sequence flanking the processed (mature) ends of the 18S rRNA, as it has been reported that cleavage at sites 1 and 2 can occur with minimal flanking sequences in short rRNA mini-gene transcripts (14–16). These various constructs were transfected into N2a cells, and expression of synthetic 18S rRNAs was determined by Northern blotting with an oligonucleotide probe to a 24-nt hybridization tag that was cloned into expansion segment 3 of the 18S rDNA. The results revealed that only constructs with the full-length 5′ ETS, containing sites A′ and A0, yielded a band corresponding to properly processed 18S rRNA (p18S.1 and 18S.2 (Pol-1 and CMV); Figure 2b) confirmed by size comparison to an in vitro 18S RNA transcript. By contrast, ITS1, which contains 3′ processing sites 2b and 2c, does not seem to be required, as construct p18S.2(Pol-1 and CMV) generates a band corresponding to mature 18S rRNA although the majority of ITS1 is deleted in this construct.
Figure 2.
Analysis of processing of rRNA constructs. (a) Schematic representation of RNAs expressed from constructs p18 S.1-.6. Constructs contain either a pol-I promoter and 3′ ETS, or a CMV promoter and an SV40 poly(A) signal. The 5′ ETS and ITS1 are indicated by thick black lines, the 18 S rRNA by a grey bar and deleted spacer sequences by thin dashed lines. Cleavage sites are indicated. (b) Northern blot analysis of 18 S rRNA processing. N2a cells were transfected with the constructs indicated and RNA analyzed by Northern blots as described in Methods. Nucleotide positions of a single-stranded RNA size ladder are indicated to the left side of the blot. This blot contains the following controls: A: 100-ng 18 S WT transcript, B: 100-ng 18 S-tagged transcript, C: 2-µg total RNA from N2a cells transfected with p18 S.1 untagged (Pol-1), D: 2-µg total RNA from mock transfected N2a cells, E: 2-µg total RNA from N2a cells. Synthetic rRNA was detected by hybridization to an inserted tag sequence using the α-tag probe. The upper bands (asterisk) correspond to full-length and partially processed transcripts. The location of mature 18 S rRNA is indicated. (c) Time course of 18 S rRNA accumulation from construct p18 S.1 (Pol-1). For these experiments, N2a cells were transfected, and RNA was harvested at various times post-transfection as indicated. The controls are the same as in (b).
Expression of mature 18S rRNA was observed to occur with both pol-I and CMV promoters; however, the levels were much higher when transcription was mediated by pol-I (Figure 2b). The results indicate that the processing sites in the 5′ ETS are sufficient for processing of 18S rRNA, and that processing does not require transcription to occur from RNA polymerase I, consistent with results reported in yeast (7). Due to the low yield observed with pol-II transcription of 18S rRNA, we exclusively used the pol-I vector for all subsequent experiments.
Correct processing of the synthetic 18S rRNA precursor transcripts from constructs p18S.1 and p18S.2 was shown by probing blots with the α-tag probe to identify these 18S rRNAs. We then reprobed the same blots with a low specific-activity oligonucleotide probe (α-18S) that recognizes both synthetic and endogenous 18S rRNAs (Supplementary Figure S2a–c). The results show that the mature synthetic and endogenous 18S bands are superimposable. To demonstrate removal of spacer regions from processed RNAs, we performed Northern blots with probes complementary to sequences in the 5′ ETS (α-5′ ETS) and ITS1 (α-ITS1), located immediately 5′ and 3′ of sites 1 and 2, respectively (Supplementary Figure S2d and e). Both blots showed no detectable hybridization with the mature tagged transcript, but hybridized to the longer low abundance precursor transcripts, results consistent with the removal of spacer regions from the mature 18S rRNA. For constructs with deletions in the spacer regions, sizes of the unprocessed and partially processed transcripts are consistent with those expected when compared with pre-rRNA species.
To monitor the relative levels of the synthetic 18S rRNA after transfection, time course experiments were performed. For these experiments, the full-length pol-I construct (p18S.1) that yielded processed 18S rRNA was transfected into cells, and 18S rRNA expression was monitored by harvesting cells at various time points up to 72 hour post-transfection and then probing for the hybridization tag (Figure 2c). For this construct, maximal expression of the synthetic 18S rRNA, as a percentage of total 18S rRNA, was observed 36–48 hours post-transfection. All subsequent experiments were performed 48 hours post-transfection, unless otherwise noted.
To determine whether the processed synthetic 18S rRNAs are associated with ribosomes, we performed Northern blot analyses of whole-cell lysates and of pellet and supernatant fractions of lysates of cells transfected with p18S.1 prepared by centrifugation at 100 000 x g (Supplementary Figure S3a,b). As seen earlier, both unprocessed and processed synthetic 18S rRNAs were present in total RNA prepared from whole-cell lysates (Supplementary Figure S3a,b; right images). However, in the fractionated material, only processed rRNA was seen in the P100 pellet, which contains sedimented ribosomes. Processed RNA was not seen in the supernatant fraction, which contains less dense cytoplasmic material. These fractionated RNA samples were also compared with synthetic in vitro transcribed rRNAs by probing with both α-tag and α-18S probes for determination of relative abundance. Synthetic 18S rRNA in cells 48 hours post-transfection was estimated to be ≈10–15% of total 18S rRNA. Fractionation of lysates from cells transfected with p18S.2(pol-I) yielded similar results (Supplementary Figure S3c), suggesting that synthetic 18S rRNAs lacking ITS1 are incorporated into ribosomal subunits. Taken together, these experiments indicate that the synthetic 18S rRNAs derived from p18S.1 and p18S.2 were correctly processed and incorporated into 40S subunits. They also show that ITS1 is dispensable for formation of mature 18S rRNA.
Analysis of 5′ ETS sequences
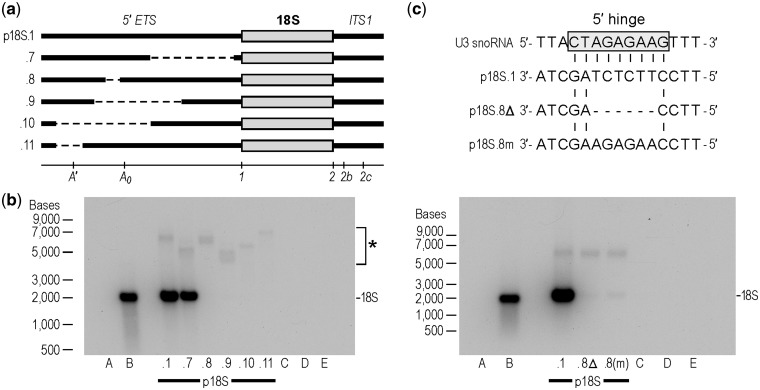
The 5′ region of the 5′ ETS containing the A′ and A0 processing sites seems to be required for processing of 18S rRNA (Figure 1b); however, little is known about the importance of sequences located 3′ of these sites in mammalian rDNA genes. The length of these 3′ sequences in mammalian genes differentiates them from other eukaryotic rDNA genes, which are substantially shorter. To investigate the potential contribution of these additional sequences in mammals, several internal deletions were generated, including deletions to remove the A′ and A0 sites, individually and in combination (Figure 3a; see Methods for construct details).
Figure 3.
Analysis of contribution of 5′ ETS sequences to 18 S rRNA processing. (a) Schematic representation of constructs as in Figure 2. Constructs contain a pol-I promoter and 3′ ETS. (b) Northern blot analysis of 18 S rRNA processing. N2a cells were transfected with the constructs indicated and RNA analyzed by Northern blots as described in Methods. Synthetic rRNA was detected by hybridization to an inserted tag sequence using the α-tag probe. The asterisk indicates full-length and partially processed transcripts. The location of mature 18 S rRNA is indicated. Controls for this blot are as described in Figure 2. (c) Top: Schematic shows comparison of U3 snoRNA 5′ hinge region to p18 S.1, p18 S.8Δ, and p18 S.8m; the complementary sequence match to p18 S.1 is highlighted. Bottom: The Northern blot shows synthetic rRNA expression from N2a cells transfected with the indicated constructs. This blot was hybridized with the α-tag probe, as in panel (b). Controls for this blot are as described in Figure 2.
Northern blots of RNA extracted from cells transfected with the internal deletion constructs were hybridized using an oligonucleotide probe to the hybridization tag. The results confirmed that deletions that remove the A′ or A0 cleavage sites block processing (p18S.9, .10, and .11; Figure 3b); however, deletion of the expanded 3′ region of the 5′ ETS had little to no effect on the maturation of 18S rRNA (p18S.7). This result suggests that this region lacks important cleavage sites and other points of interaction with rRNA processing factors.
Cleavage at A′, A0, and site 1 requires the U3 snoRNA, and in mouse, putative binding sites for the U3 snoRNA have been postulated based on complementary sequence matches to two regions in the snoRNA, termed the 3′ and 5′ hinge regions (13). Based on these predictions and our own observations, deletions in p18S.9, .10, and .11 removed several potential binding sites for the U3 snoRNA 3′ hinge at nucleotides 667-673, 1032-1038, and 1117-1123, whereas deletions in p18S.8, p18S.9, and p18S.10 removed a potential binding site for the 5′ hinge at nucleotides 1552-1560. In all of these constructs, the deletions blocked processing. The deletion in p18S.8 removes a 9-nt putative binding site for the 5′ hinge of the U3 snoRNA, but still contains both the A′ and A0 cleavage sites. To specifically test the requirement of this putative binding site for processing, we deleted or mutated the 9-nt sequence in the 5′ ETS in constructs p18S.8Δ and p18S.8m, respectively (Figure 3c). Northern blot analysis of RNA from cells transfected with these constructs shows that disruption of the 9-nt sequence in both constructs almost completely abolished processing. This result supports that this 9-nt sequence binds to the U3 snoRNA 5′ hinge region as previously proposed (13).
Because A′ cleavage is dependent on U3 snoRNA (17), it is notable that removal of the putative 5′ hinge region in constructs p18S.8 and p18S.9 results in multiple immature rRNA bands, indicating incomplete cleavage at A′ (Figure 3b). It seems that some A′ cleavage can still occur without sequences complementary to the U3 snoRNA 5′ hinge. In contrast, p18S.11, which lacks the A′ site and a potential 3′ hinge binding site, and p18S.10, which lacks binding sites for both hinge regions, yields an uncleaved primary transcript, indicating no discernible cleavage at A0 in the absence of A′, the 3′ hinge region or a combination of these sites.
Identification of a pactamycin resistance mutation and verification of subunit function
The ability in these experiments to selectively monitor translation from subunits containing synthetic 18S rRNAs in vivo requires being able to shut down endogenous subunits to differentiate between the activities of modified and endogenous ribosomal subunits (Figure 1a). This approach is necessary, as quantification of Northern blots of the types shown in Supplementary Figure S3 suggested that ribosomal subunits containing synthetic rRNA only represent a small portion (up to ≈10–15%) of total cellular 40S ribosomal subunits ≈48 hours post-transfection. Therefore, we searched for mutations in 18S rRNA that confer antibiotic resistance. However, there were no reports of functionally tested examples of nucleotide changes conferring antibiotic resistance in higher eukaryotes. By contrast, there are several such mutations in Escherichia coli 16S rRNA, which raised the possibility of targeting regions in the 18S rRNA based on the locations of antibiotic resistance mutations in E. coli 16S rRNA.
A primary concern in selecting an antibiotic for use in mammalian cell cultures is permeability. Although a wide range of antibiotics have the potential to interact structurally with mammalian ribosomes, most have poor cell permeability and are thus unsuitable for use in cell culture. One exception is pactamycin. Although this antibiotic has not been widely used, it has excellent cell permeability and was found to be the most effective antibiotic tested at inhibiting protein synthesis in mouse fibroblast 3T6 cells during a 30-minute incubation (18). This antibiotic affects translation by binding to rRNA in the E site of the small subunit and disrupting the positioning of mRNA at this site. The result of pactamycin binding is a block in elongation and the accumulation of dipeptides resulting from aborted initiation events (19). Importantly, crystal structures of bacterial 30S subunits have revealed specific interactions between pactamycin and 16S rRNA (20). The interaction sites have been mapped to the E site at nucleotides 693, 694, 795, and 796, all of which are conserved in mammalian 18S rRNA. In addition, there is evidence from Halobacterium halobium that spontaneous mutations at three of these residues—694, 795, and 796—confer resistance to pactamycin (21).
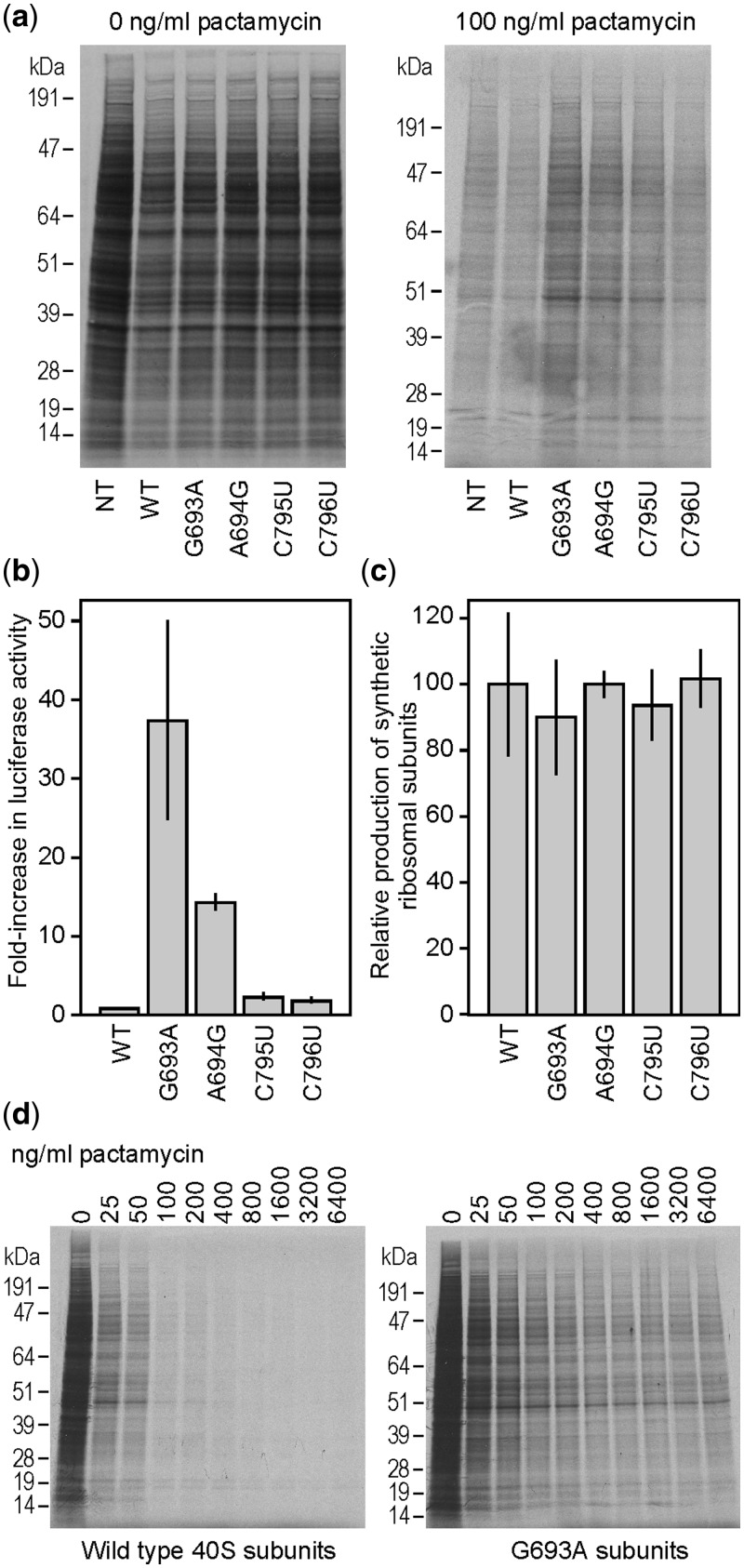
In the present study, we analyzed the effects of mutations at these four residues identified from crystal structures. Mutations were introduced into a construct expressing the untagged mouse 18S rRNA and containing the full-length 5′ ETS and ITS1 spacer regions (p18S.1(Pol-1); Supplementary Figure S4). Pactamycin resistance was assessed by 35S-Met/Cys pulse labeling of transfected cells in the presence of 100 ng/ml antibiotic. At each nucleotide position thought to interact with pactamycin, purine-purine (G693A, A694G), and pyrimidine-pyrimidine (C795U, C796U) transition mutations were generated; three of these mutations were analogous to the spontaneous mutations seen in H. halobium. Note that the numbering of these sites is based on that in E. coli 16S rRNA. For reference, mutations G693, A694, C795, and C796 (E. coli numbering) are located at G963, A964, C1065, and C1066, respectively, in mouse 18S rRNA (accession #JQ247698). Cell lysates were compared from untransfected cells, cells expressing synthetic 18S rRNA, and synthetic mutated 18S rRNAs (Figure 4a) by SDS-PAGE. Mutated ribosomes were also analyzed for their ability to translate a luciferase mRNA in cells transfected with a luciferase reporter plasmid (pGL4.13) and cultured in the presence of 100 ng/ml pactamycin (Figure 4b). The results of both sets of experiments show that all four mutations confer some degree of pactamycin resistance. The G693A and A694G mutations showed the highest levels of resistance, whereas the C795U and C796U mutations gave results that were only slightly above background. The labeling results (Figure 4a) suggest that none of the mutations significantly altered the protein banding pattern compared with wild-type (WT) ribosomes.
Figure 4.
Analysis of pactamycin-resistance mutations in N2a cells. (a) Protein expression in cells expressing WT or mutated 18 S rRNA constructs. Cells were either not transfected (NT) or transfected with constructs expressing WT 18 S rRNA or 18 S rRNAs containing the point mutations indicated. Cells were then 35S pulse-labeled in the absence or presence of 100 ng/ml pactamycin as indicated, cells were lysed, and equal volumes of the cell lysates were analyzed by SDS-PAGE, as described in Methods. Representative autoradiograms are shown. (b) Relative luciferase activities in cell lysates from cells transfected with WT or mutated 18 S rRNAs and with pGL4.13 (Promega). N2a cells were cotransfected with the 18 S rRNA constructs indicated and with a firefly luciferase construct (pGL4.13) and luciferase expression monitored from cell lysates. Luciferase activities are relative to those obtained from cells transfected with the WT construct, which is set to 1.0. Details of the cotransfection and assay methods are described in Methods. (c) Quantification of synthetic rRNA levels from Northern blots for cells transfected with WT or mutated rRNA constructs. Synthetic rRNA was detected by hybridization with the α-tag probe. Signals were quantified using a Molecular Dynamics Phosphorimager system and are represented relative to WT, with WT set to 100%. (d) Cells were transfected with either WT or G693A mutation-containing 18 S rRNA constructs and incubated with various amounts of pactamycin as indicated. Cells were 35S pulse-labeled, and cell lysates analyzed by SDS-PAGE. Representative autoradiograms are shown. In panels (b) and (c) error bars represent standard deviations from 3 independent experiments.
The G693A mutation, which conferred the highest level of pactamycin resistance, was used exclusively for further experiments. We monitored the levels of synthetic 18S rRNA for constructs with each mutation using the hybridization tag and found no substantial differences that could account for the difference in translation between G693A and the other mutations (Figure 4c).
To further characterize the G693A mutation, cells expressing WT or mutated ribosomes were cultured with increasing concentrations of pactamycin. The results showed that translation from WT 40S subunits was substantially blocked by 100 ng/ml pactamycin but could be further blocked at higher concentrations. By contrast, translation from the mutated subunits seemed to be unaffected even at the highest concentration tested of 6400 ng/ml (Figure 4d).
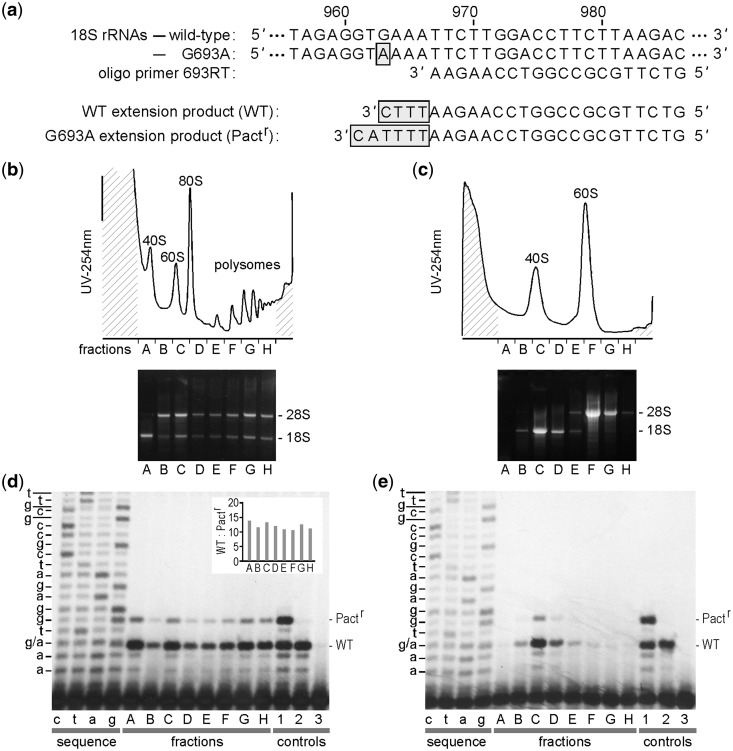
Additional evidence of subunit function was obtained from sucrose density analysis of the distribution of ribosomes containing the G693A mutation. In these experiments, we compared the distribution of ribosomal subunits containing WT (endogenous) and synthetic (G693A) 18S rRNAs by using an oligonucleotide primer (693RT) to hybridize downstream of the G693A mutation and ddCTP as a terminator (Figure 5a). Under these reaction conditions, the mutated 18S rRNA generates a primer extension product that is two nucleotides larger than that generated from the endogenous rRNA. This primer extension reaction enables analysis of the presence and abundance of these rRNAs within the same sample. For these experiments, lysates were prepared from N2a cells transfected with the G693A construct and treated with either cycloheximide or EDTA. Lysates were then fractionated on sucrose density gradients (Figure 5b and c), and the resulting fractions were analyzed by primer extension (Figure 5d and e). Analysis of the distribution of primer extension products through the cycloheximide profile (Figure 5b and d) showed that the mutated rRNA has a relative distribution similar to that of the endogenous 18S rRNA through the polysomes. Quantification of WT and G693A primer extension products and comparison of their relative ratios shows a relatively equal distribution through the gradient (Figure 5d inset), suggesting that ribosomal subunits containing synthetic 18S rRNA are functionally similar to those of endogenous subunits. Likewise, an EDTA-treated lysate confirms the presence of the mutated 18S rRNA in 40S subunits through its colocalization with the 40S peak (Figure 5c and e).
Figure 5.
Sucrose density gradient distributions of ribosomal subunits containing synthetic 18 S rRNAs. (a) ddCTP primer extension assay. Partial sequences of WT and mutated 18 S rRNAs are shown. The position of the pactamycin-resistance mutation (G693A), located at nucleotide 963 in 18 S rRNA, is highlighted. The sequence of oligonucleotide primer 693RT is shown aligned to its complementary match in the 18 S rRNAs. Primer extension reactions performed in the presence of ddCTP will terminate at the first G located upstream of the primer. This will result in 4-nt-extended products from WT 18 S rRNA templates and 6-nt-extended products from G693A-mutated 18 S rRNAs. In each case, the extended nucleotides are highlighted. (b) Top: Lysates from cycloheximide-treated N2a cells transfected with p18 S.1(G693A) were fractionated in a 10–50% (w/v) linear sucrose gradient. Peaks (left to right) represent 40S ribosomal subunits, 60S ribosomal subunits, 80S single ribosomes, and polysomes. The fractions (A-H) that were collected for RNA analysis are indicated. Bottom: RNA prepared from fractions A-H was visualized in an ethidium bromide-stained agarose gel. The 28S and 18 S rRNAs are indicated. (c) Top: EDTA-dissociated ribosomes were fractionated on a 10–35% (w/v) linear sucrose gradient. Peaks (left to right) represent 40S and 60S ribosomal subunits. Bottom: RNA analysis as in panel (b). (d) PAGE analysis of RNA prepared from fractions of the sucrose gradient in panel (b), which were subjected to ddCTP primer extension using oligonucleotide primer 693RT, which was 33-P-labeled. The sequencing reactions (lanes c,t,a,g) used the same primer with total RNA as a template. The upper bands are generated from the synthetic 18 S rRNA (Pactr) and the lower bands are from endogenous 18 S rRNA (WT). Lanes 1-3 are controls. Control 1 is an equimolar mixture of in vitro transcripts that contain or lack the pactamycin resistance mutation. Control 2 is total RNA from untransfected N2a cells, which contains only the WT 18 S rRNA. Control 3 is a no template control. The levels of the primer extension products from the various fractions were quantified from Phosphorimager exposures and the ratios of the two bands (WT:Pactr) are shown in the inset. (e) ddCTP primer extension from fractions of gradient in panel (c).
Analysis of function of 40S subunits derived from precursors with spacer region deletions
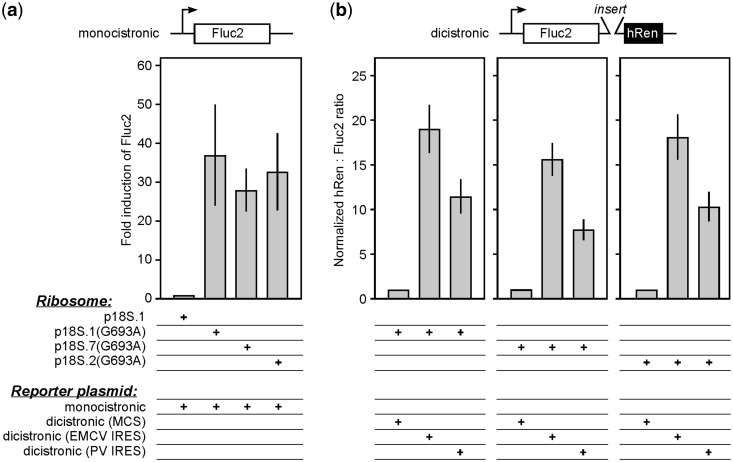
In the present studies, we identified several deletion constructs, which seem to yield properly processed, mature 18S rRNAs based on size (Figures 2 and 3). However, cleavage at the correct sites may not be sufficient to ensure that these rRNAs fold properly and incorporate into functional subunits because interactions between spacer regions and 18S rRNA may be necessary for correct folding. To determine whether rRNAs with deletions in the 5′ ETS and ITS1 can adopt functional conformations in the context of 40S ribosomal subunits, we introduced the G693A mutation into constructs p18S.7 and .2, respectively, and tested them in cells cotransfected with either a monocistronic luciferase reporter construct (Figure 6a) or a dicistronic dual luciferase vector (Figure 6b), as described in Methods. Expression in each case was measured in the presence of pactamycin to inhibit translation from endogenous 40S subunits. We expected that structural alterations resulting from erroneous ribosome formation or folding in the deletion constructs might differentially affect translation of these mRNAs, which represent cap-dependent translation and two classes of IRES-dependent translation that each requires various initiation factors for translation competence. For the monocistronic construct, no significant differences were observed between ribosomes, suggesting that the deletions in the spacer sequences did not disrupt assembly or affect the ability of the subunits to recruit the translation factors associated with cap-dependent translation (Figure 6a). For the dicistronic constructs, we measured expression of the second cistron, facilitated by either the EMCV or PV IRES in the intercistronic region, relative to a control sequence with no known IRES activity. The results showed that the ratio of expression of the two cistrons (hRen/luc2; Figure 6b) was similar between the control and the spacer deletion rRNA constructs, suggesting that the ribosomal subunits in each case are indistinguishable from WT in their abilities to translate mRNAs via these IRESes. These results further support the conclusion that ribosomal subunits derived from the shortened rRNA transcripts are active and do not seem to require the sequences that were deleted from the 5′ ETS and ITS1.
Figure 6.
Analysis of function of synthetic 18 S rRNAs containing deletions in 5′ ETS or ITS1. (a) Reporter assays of N2a cells cotransfected with a monocistronic reporter construct expressing an optimized firefly luciferase (Fluc2) and various 18 S rRNA constructs as indicated. A schematic representation of the monocistronic construct is shown above. The synthetic rRNAs used in each experiment are indicated below the graph: p18 S.1 contains the full 5′ ETS and ITS1, p18 S.7 contains a deletion of the 3′ region of the 5′ ETS, and p18 S.2 lacks ITS1. Luciferase activities were determined as described in Methods. The results of the various transfections are reported as fold induction of Fluc2, which is luciferase activity over background obtained with WT (pactamycin-sensitive) ribosomes (p18 S.1). (b) Reporter assays of N2a cells transfected with dicistronic reporter constructs expressing Fluc2 and an optimized human Renilla luciferase (hRen). The control construct contained an MCS in the intercistronic region; the other constructs contained either the EMCV or PV IRES. The 18 S rRNA constructs and reporter constructs used in each experiment are indicated below the graph. The results are reported as hRen to Fluc2 ratios normalized to the MCS construct, which has no IRES activity. Error bars represent standard deviations from 3 independent experiments.
Effects of 5′ ETS and ITS1 on overall efficiency of subunit formation
Although deletions in the 5′ ETS and ITS1 do not seem to affect the translation competence of 40S subunits, we did not know if these flanking sequences might affect the efficiency of production of ribosomal subunits. Transfection conditions for our studies have been optimized to maximize ribosomal subunit production, potentially masking differences in relative subunit abundance from each construct. Therefore, experiments were performed to reduce expression of synthetic rRNA per cell by transfecting cells with diluted plasmid constructs. These experiments were performed using our standard transfection conditions with the same amount of total plasmid per transfection; however, we varied the amounts of the p18S rRNA expression plasmids (1 µg, 0.1 µg, and 0.01 µg) by using another plasmid (pBS KS) as filler.
The three expression constructs tested in this experiment contain the full-length 5′ ETS and ITS1 sequences (p18S.1)), a deletion in the 5′ ETS (p18S.7), and a deletion in ITS1 (p18S.2). All three constructs contain the hybridization tag for detection of mature 18S rRNA. Cells transfected with the various plasmids at different dilutions showed no substantial differences when compared via Northern blot (Supplementary Figure S5). Taken together, the results indicate that deletion of the flanking spacer regions does not affect translation from or assembly of mature subunits. However, this study does not rule out the possibility that the flanking regions may have more subtle effects. The results also show that a logarithmic dilution of the p18S.1 vector with filler plasmid does not produce a logarithmic decrease in the levels of mature 40S subunits, indicating that the standard conditions used in our experiments (1.0 µg plasmid) are saturating the cell’s ability to produce 40S ribosomal subunits.
DISCUSSION
The mammalian rRNA expression system described here enables synthetic 18S rRNAs to be transcribed, incorporated into functional 40S subunits, and assayed in isolation in vivo. This system can serve as a platform for analyses of any 18S rRNA modification and determination of its contribution to translation, provided such a mutation does not interfere with subunit formation. This system will also allow analysis of aspects of ribosomal function that are unique to multicellular organisms, including ribosomal heterogeneity in different cell types. Such studies can now be performed in cell types that express different ribosomal proteins. Furthermore, the effects of different 18S rRNA primary sequences can be investigated either by mutation or replacement of the current variant.
This mammalian system is comparable with one developed in E. coli, which introduced a spectinomycin resistance mutation into the synthetic 16S rRNA to enable 30S subunits assembled from these rRNAs to function preferentially in the presence of the antibiotic (22). Before the present study, this approach was not directly transferable to mammalian cells (or other eukaryotic cells), as there were no mutations in eukaryotic rRNAs shown to confer antibiotic resistance. It should be noted that rRNA expression systems have also been developed for yeast; however, lack of an antibiotic resistance mutation in the yeast system makes it more difficult to study the subunits in isolation because the translation competence of nonfunctional subunits must be assessed in a background of depleting functional subunits. This is obtained either by disrupting rRNA transcription by using a temperature-sensitive pol-I mutation (7) or by deleting ≈150 tandemly repeated chromosomal rDNA gene copies (23).
The use of antibiotics to differentiate between endogenous and synthetic subunits has been described in bacteria (22), and although numerous antibiotics are effective at blocking translation in various bacteria, a smaller number are effective at blocking translation in mammalian cells. This is because mammalian rRNAs contain nucleotide variations (compared with bacterial rRNAs) that render them resistant to many antibiotics (19). When selected mutations to pactamycin were introduced into mouse 18S rRNA, all gave some level of resistance above background when tested with a reporter construct in the presence of 100 ng/ml of pactamycin. However, mutation G693A gave the highest levels of resistance. Mutations at this nucleotide had previously been proposed to be potentially too damaging to translation to arise spontaneously (20). However, in our studies, ribosomal subunits containing the G693A mutation seemed to function normally as assessed by their codistribution with WT subunits in sucrose gradients. In addition, pulse labeling experiments confirmed that these ribosomal subunits are active in protein synthesis.
All mutations tested in this study were transition mutations, as spontaneous antibiotic resistance mutations are frequently of this type. However, this observation may be due to a higher occurrence of transition vs transversion mutations in nature. Therefore, the possibility that transversion mutations may impart an even higher level of resistance to pactamycin remains untested. In the E. coli 30S/pactamycin crystal structure, the base of nucleotide 693 in the 16S rRNA interacts with pactamycin via 2 hydrogen bonds and one base-stacking interaction. Although the geometry of this interaction may differ in mammalian subunits, these potential additional interactions, when compared with the single hydrogen bonds formed at nucleotides 694, 795, and 796, may explain why this mutation conferred the greatest level of pactamycin resistance in our study. However, at present, we cannot rule out the possibility that the other mutations had additional steric effects.
In eukaryotes, rRNA precursors are transcribed by RNA polymerase I. However, a yeast study showed that it is possible to use RNA polymerase II to drive transcription of synthetic rRNA transcripts (7). An obvious advantage of pol-II is the possibility of using inducible promoters to control expression of synthetic rRNA. In our studies, we, therefore, tested both pol-I and pol-II promoters. Although 18S rRNA was correctly processed in both cases, levels of production of mature 18S rRNA were much higher when transcription was driven by the pol-I promoter. We postulate that these differences may reflect eukaryotic specializations associated with the production of rRNA and mRNA. For example, ribosomal synthesis via pol-I transcription may be efficient because pol-I interacts with other factors involved in ribosomal processing and synthesis, thereby colocalizing rRNA transcription and processing (24). It has been suggested that pol-I transcription and processing of the precursor rRNA are coordinated in a transcription-dependent manner in mammalian cells (25). Likewise, post-transcriptional modifications that occur via factors associated with pol-II may be inhibitory to ribosomal synthesis. For example, factors associated with pol-II mediate the addition of an m7G cap-structure at the 5′ ends of transcripts, and a poly(A) tail at the 3′ ends. Inasmuch as both modifications facilitate RNA export out of the nucleus (26), their presence in precursor rRNA transcripts may interfere with ribosomal processing and subunit assembly in the nucleus. However, it is significant to note that although it was inefficient, subunit processing did occur from a pol-II transcript, showing that transcription and processing are not strictly coordinated.
Analysis of 5′ ETS sequences from various organisms reveals little sequence homology except in regions surrounding the known processing sites (A′ and A0), where common motifs have been identified, including one that is thought to be related to the A′ cleavage site in the 5′ region of 5′ ETS sequences from human, mouse, and Xenopus precursor rRNAs (27). Most of the sequence homologies in the 5′ ETS sequences of mammals are contained within the first 2000 nucleotides; there is little homology in the greatly expanded 3′ half of this spacer sequence. Our deletion analysis of the mouse 5′ ETS confirmed the necessity of the 5′ end of this sequence for the maturation of functional 40S subunits, including a sequence that was previously predicted to be the binding site for the 5′ hinge region of the U3 snoRNA. All of our defective deletion constructs lacked either the proposed 3′ or 5′ U3 snoRNA hinge binding sites, and it is likely that the inability to bind this snoRNA blocked processing, as previous studies have confirmed the necessity of binding for subunit maturation (28–30).
The U3 snoRNA is known to interact with both the 5′ ETS, as well as with a pseudoknot at the 5′ end of the 18S rRNA, which it is suspected to play a role in forming (30). In bacteria, there is no equivalent for the U3 snoRNA; however, it has been shown that regions in the 5′ ETS can base pair to the pseudoknot in 16S rRNA, comparable with the interaction between the U3 snoRNA and 18S rRNA, and possibly mediate formation of the pseudoknot and cleavage (31). The interaction mediated by the U3 snoRNA therefore seems to be conserved throughout evolution. However, several previous studies with partial rRNA transcripts have shown that cleavage at the mature 5′ and 3′ ends of the 18S rRNA (sites 1 and 2; Figure 1b) can occur with only minimal surrounding spacer sequences (14–16). Cleavage at site 1 seems inconsistent with the results of our study, which showed that inhibition of binding of the U3 snoRNA abolishes subsequent maturation of the small subunit. One possibility is that cleavage in vitro does not require the U3 snoRNA, but that this RNA is required for subsequent maturation in vivo, perhaps through its mediation of correct formation of the pseudoknot. If incorrectly folded products are rapidly degraded in cells, we might not detect them on Northern blots. This could explain the lack of partially processed transcripts consistent with cleavage at site 1 (see Figures 2 and 3). We only observed full-length transcripts and what seems to be transcripts cleaved at A′ when synthetic rRNA does not mature into 18S rRNA. Compared with cleavage at site 1, cleavage at site 2 seems to be spontaneous and independent of other factors.
In contrast to the necessity of 5′ sequences for rRNA maturation, a large (≈1.7 kb) region of the 5′ ETS does not seem to be required for formation of functional ribosomal subunits. This result was not completely unexpected, as there is a large degree of sequence divergence in this region of the 5′ ETS between even closely related rodent and human sequences. However, when considering that pre-rRNA transcription accounts for the majority of RNA transcription during rapid cell growth, the large amount of rRNA synthesis required for this region of the 5′ ETS seems to be extravagant if it is not directly involved in ribosomal formation (32). Microscopy and thermodynamic modeling of this region of the 5′ ETS have revealed what seems to be a stable secondary structure (9,33–35). In addition, this fragment of the precursor rRNA is stable enough to be easily detected as a 19S rRNA fragment (13). One possibility is that this RNA has an as yet to be determined extraribosomal function in cells.
Deletions of the ITS1 spacer region revealed that this sequence is not required for the formation of mature 40S subunits. This result may be consistent with the finding that some intermediate pre-rRNAs contain only a short region of the 3′ spacer and that final processing at the 3′ end of the 18S rRNA (site 2; Figure 1c) occurs in the cytoplasm (36). In yeast, it was recently proposed that there is an interaction between ITS1 and helix 44 in 18S rRNA that functions to regulate 40S assembly by inhibiting cleavage at site D (site 2 in mouse) until after cleavage occurs at site A2 (site 2b in mouse) (37). It was suggested that this interaction in yeast serves as an arrest of maturation for quality control and is required for maturation of 18S rRNA. As our shortened constructs do not contain the implicated ITS1 sequences, it is possible that the proposed quality control step is bypassed. Although our results suggest that constructs lacking ITS1 were capable of generating fully functional subunits, additional studies are required to determine whether the absence of ITS1 leads to more subtle effects on the structure or function of 40S subunits.
Potential applications
Applications of the vector system described in this study include analyses of ribosome processing and assembly but are not limited to such studies. The 40S subunit is involved in the earliest steps of translation initiation, and the ability to modify the 40S subunit will enable analysis of the contributions of 18S rRNA to this level of gene regulation, including the possible role of various mRNA-rRNA base pairing interactions during translation initiation, reinitiation, and shunting. The pactamycin-resistance mutation identified for this expression system now enables the study of mutations in isolation that would otherwise be fatal to cells as protein synthesis can be monitored from deficient ribosomes representing only a subset of the total ribosome population. One application of the synthetic rRNA expression system is the study of specific pseudouridylation and methylation sites in 18S rRNA. For example, there have been several studies that have correlated a lack of rRNA pseudouridylation with generation of cancerous phenotypes (38). The ability to block specific modifications provides an improvement over current approaches that inhibit global pseudouridylation or methylation by blocking or removing snoRNAs, particularly because snoRNAs have targets in cells other than rRNAs (39). Another application of the synthetic rRNA expression system is to investigate the importance of expansion segments in 18S rRNA, which are discrete sequence elements that are found in eukaryotic rRNA but absent in bacteria (40).
In addition to fundamental analyses of ribosomal structure and translation, the 18S rRNA expression system has applications for both synthetic biology and biotechnology. This includes the development of ribosomes with altered function and specialized ribosomes, which preferentially translate specific mRNAs more efficiently than endogenous ribosomes (22,41–43). The ability to develop such orthogonal ribosome mRNA pairs may have immediate practical applications for bioproduction in mammalian cells.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–5.
FUNDING
Funding for open access charge: National Institutes of Health [GM078071]; Promosome, LLC [SFP 1539].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs. Gerald M. Edelman and Daiki Matsuda for critical discussions and reading of the manuscript and Dr. Maurille Fournier for advice regarding these studies.
REFERENCES
- 1.Stelzl U, Connell S, Wittman-Liebold B. Encyclopedia of Life Sciences. John Wiley & Sons, Ltd; 2001. Ribosomal proteins: role in ribosomal functions; pp. 1–12. [Google Scholar]
- 2.Wool IG. Extraribosomal functions of ribosomal proteins. Trends. Biochem. Sci. 1996;21:164–165. [PubMed] [Google Scholar]
- 3.Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc. Natl Acad. Sci. USA. 2002;99:12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mauro VP, Edelman GM. The ribosome filter redux. Cell Cycle. 2007;6:2246–2251. doi: 10.4161/cc.6.18.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luttermann C, Meyers G. The importance of inter-and intramolecular base pairing for translation reinitiation on a eukaryotic bicistronic mRNA. Genes. Dev. 2009;23:331–344. doi: 10.1101/gad.507609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang WQ, Fournier MJ. Synthesis of functional eukaryotic ribosomal RNAs in trans: development of a novel in vivo rDNA system for dissecting ribosome biogenesis. Proc. Natl. Acad. Sci. USA. 1997;94:2864–2878. doi: 10.1073/pnas.94.7.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinos G, Wilson DN, Teraoka Y, Szaflarski W, Fucini P, Kalpaxis D, Nierhaus KH. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site RNA binding. Mol. Cell. 2004;13:113–124. doi: 10.1016/s1097-2765(04)00002-4. [DOI] [PubMed] [Google Scholar]
- 9.Schibler U, Wyler T, Hagenbuchle O. Changes in size and secondary structure of the ribosomal transcription unit during vertebrate evolution. J. Mol. Biol. 1975;94:503–517. doi: 10.1016/0022-2836(75)90217-x. [DOI] [PubMed] [Google Scholar]
- 10.Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl Acad. Sci. USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sigmund CD, Ettayebi M, Borden A, Morgan EA. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- 12.Tseng H, Chou W, Wang J, Zhang X, Zhang S, Schultz RM. Mouse ribosomal RNA genes contain multiple differentially regulated variants. PLoS One. 2008;3:e1843. doi: 10.1371/journal.pone.0001843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kent T, Lapik YR, Pestov DG. The 5′ external transcribed spacer in mouse ribosomal RNA contains two cleavage sites. RNA. 2009;15:14–20. doi: 10.1261/rna.1384709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannon GJ, Maroney PA, Branch A, Benenfield BJ, Robertson HD, Nilsen TW. Accurate processing of human pre-rRNA in vitro. Mol. Cell. Biol. 1989;9:4422–4431. doi: 10.1128/mcb.9.10.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hadjiolova KV, Normann A, Cavaille J, Soupene E, Mazan S, Hadjiolov AA, Bachellerie JP. Processing of truncated mouse or human rRNA transcribed from ribosomal minigenes transfected into mouse cells. Mol. Cell. Biol. 1994;14:4044–4056. doi: 10.1128/mcb.14.6.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shumard CM, Torres C, Eichler DC. In vitro processing at the 3′-terminal region of pre-18S rRNA by a nucleolar endoribonuclease. Mol. Cell. Biol. 1990;10:3868–3872. doi: 10.1128/mcb.10.8.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kass S, Tyc K, Steitz JA, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 18.Contreras A, Vazquez D, Carrasco L. Inhibition, by selected antibiotics, of protein synthesis in cells growing in tissue cultures. J. Antibiot. 1978;31:598–602. doi: 10.7164/antibiotics.31.598. [DOI] [PubMed] [Google Scholar]
- 19.Wilson DN. Protein Synthesis and Ribosome Structure. In: Nierhaus KH, Wilson DN, editors. Antibiotics and the Inhibition of Ribsome Function. Wiley-VCH, Weinheim, Germany: 2004. pp. 449–527. [Google Scholar]
- 20.Brodersen DE, Clemons WM, Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 21.Mankin AS. Pactamycin resistance mutations in functional sites of 16 S rRNA. J. Mol. Biol. 1997;274:8–15. doi: 10.1006/jmbi.1997.1387. [DOI] [PubMed] [Google Scholar]
- 22.Hui A, de Boer HA. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1987;84:4762–4766. doi: 10.1073/pnas.84.14.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wai HH, Vu L, Oakes M, Nomura M. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 2000;28:3524–3534. doi: 10.1093/nar/28.18.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis JD, Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- 25.Kopp K, Gasiorowski JZ, Chen D, Gilmore R, Norton JT, Wang C, Leary DJ, Chan EK, Dean DA, Huang S. Pol I transcription and pre-rRNA processing are coordinated in a transcription-dependent manner in mammalian cells. Mol. Biol. Cell. 2007;18:394–403. doi: 10.1091/mbc.E06-03-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartshorne T, Toyofuku W, Hollenbaugh J. Trypanosoma brucei 5′ETS A′-cleavage is directed by 3′-adjacent sequences, but not two U3 snoRNA-binding elements, which are all required for subsequent pre-small subunit rRNA processing events. J. Mol. Biol. 2001;313:733–749. doi: 10.1006/jmbi.2001.5078. [DOI] [PubMed] [Google Scholar]
- 28.Borovjagin AV, Gerbi SA. An evolutionary intra-molecular shift in the preferred U3 snoRNA binding site on pre-ribosomal RNA. Nucleic Acids Res. 2005;33:4995–5005. doi: 10.1093/nar/gki815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savino R, Gerbi SA. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes JM. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 31.Dennis PP, Russell AG, Moniz De Sa M. Formation of the 5′ end pseudoknot in small subunit ribosomal RNA: involvement of U3-like sequences. RNA. 1997;3:337–343. [PMC free article] [PubMed] [Google Scholar]
- 32.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 33.Bourbon H, Michot B, Hassouna N, Feliu J, Bachellerie JP. Sequence and secondary structure of the 5′ external transcribed spacer of mouse pre-rRNA. DNA. 1988;7:181–191. doi: 10.1089/dna.1988.7.181. [DOI] [PubMed] [Google Scholar]
- 34.Michot B, Bachellerie JP. Secondary structure of the 5′ external transcribed spacer of vertebrate pre-rRNA. Presence of phylogenetically conserved features. Eur. J. Biochem. 1991;195:601–609. doi: 10.1111/j.1432-1033.1991.tb15743.x. [DOI] [PubMed] [Google Scholar]
- 35.Wellauer PK, Dawid IB, Kelley DE, Perry RP. Secondary structure maps of ribosomal RNA. II. Processing of mouse L-cell ribosomal RNA and variations in the processing pathway. J. Mol. Biol. 1974;89:397–407. doi: 10.1016/0022-2836(74)90527-0. [DOI] [PubMed] [Google Scholar]
- 36.Rouquette J, Choesmel V, Gleizes PE. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamanna AC, Karbstein K. An RNA conformational switch regulates pre-18S rRNA cleavage. J. Mol. Biol. 2010;405:3–17. doi: 10.1016/j.jmb.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 39.Bratkovic T, Rogelj B. Biology and applications of small nucleolar RNAs. Cell Mol. Life Sci. 2011;68:3843–3851. doi: 10.1007/s00018-011-0762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerbi SA. Ribosomal RNA structure, evolution, processing, and function in protein biosynthesis. CRC Press; 1996. Expansion segments: regions of variable size that interrupt the universal core secondary structure of ribosomal RNA; pp. 71–87. [Google Scholar]
- 41.Rackham O, Chin JW. Cellular logic with orthogonal ribosomes. J. Am. Chem. Soc. 2005;127:17584–17585. doi: 10.1021/ja055338d. [DOI] [PubMed] [Google Scholar]
- 42.Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 2007;25:770–777. doi: 10.1038/nbt1314. [DOI] [PubMed] [Google Scholar]
- 43.An W, Chin JW. Orthogonal gene expression in Escherichia coli. Methods Enzymol. 2011;497:115–134. doi: 10.1016/B978-0-12-385075-1.00005-6. [DOI] [PubMed] [Google Scholar]
- 44.Connolly K, Culver G. Deconstructing ribosome construction. Trends Biochem. Sci. 2009;34:256–263. doi: 10.1016/j.tibs.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.