Abstract
A deep-sequencing approach was pursued utilizing 454 and Illumina sequencing methods to discover new genes involved in xyloglucan biosynthesis. cDNA sequences were generated from developing nasturtium (Tropaeolum majus) seeds, which produce large amounts of non-fucosylated xyloglucan as a seed storage polymer. In addition to known xyloglucan biosynthetic genes, a previously uncharacterized putative xyloglucan galactosyltransferase was identified. Analysis of an Arabidopsis thaliana mutant line defective in the corresponding ortholog (AT5G62220) revealed that this gene shows no redundancy with the previously characterized xyloglucan galactosyltransferase, MUR3, but is required for galactosyl-substitution of xyloglucan at a different position. The gene was termed XLT2 for Xyloglucan L-side chain galactosylTransferase position 2. It represents an enzyme in the same subclade of glycosyltransferase family 47 as MUR3. A double mutant defective in both MUR3 (mur3.1) and XLT2 led to an Arabidopsis plant with xyloglucan that consists essentially of only xylosylated glucosyl units, with no further substitutions.
Keywords: seed biology, cell walls, nasturtium, storage polymers, xyloglucan
INTRODUCTION
All plant cells are encased in a wall, a large-scale renewable resource for the production of biofuels and other commodity chemicals (Pauly and Keegstra, 2008). The wall is composed of cellulose, various hemicelluloses, pectic polysaccharides, polyphenols such as lignin, and some glycoproteins (Somerville et al., 2004). However, depending on the plant, the composition of these components can vary, giving rise to the possibility of tailoring the composition of its wall for environmentally sustainable, scalable, and economically competitive processing (Pauly and Keegstra, 2008).
One of the dominant components in growing tissue of plants is the hemicellulose xyloglucan (XyG) (Hayashi, 1989). XyG consists of a β-1,4-linked glucan backbone, which, unlike cellulose, is substituted with α-xylosyl residues on the 6-position. According to the nomenclature for XyG oligosaccharides, the unsubstituted glucosyl residue is abbreviated as ‘G’ and the xylosylated glucosyl residue is abbreviated as ‘X’ (Fry et al., 1993). The xylosylated glucosyl residues can be further decorated, depending on the plant species (Scheller and Ulvskov, 2010). In most dicots such as Arabidopsis thaliana, the xylosyl residue can be further substituted with a galactosyl residue (‘L’), which, in turn, can be O-acetylated (underlined ‘L’) and/or fucosylated (‘F’) (Pauly et al., 2001; Mellerowicz et al., 2008). Due to its structural similarities, XyG binds non-covalently to cellulose microfibrils, thereby connecting them and forming a cellulose-XyG network (Pauly et al., 1999a), which is thought to be the major load-bearing structure of the primary cell wall in dicots (Somerville et al., 2004; Hayashi and Kaida, 2011).
Several glycosyltransferases (GTs) involved in XyG biosynthesis have been identified (Scheible and Pauly, 2004; Lerouxel et al., 2006; Scheller and Ulvskov, 2010). This includes a glucan synthase likely responsible for the XyG glucan backbone, CSLC4 (Cocuron et al., 2007). Moreover, the transferases responsible for adding xylosyl (XXT1, XXT2, XXT5; Faik et al., 2002; Cavalier and Keegstra, 2006), galactosyl (such as MUR3; Madson et al., 2003), and fucosyl residues (Fut1/MUR2; Perrin et al., 1999; Vanzin et al., 2002) have been characterized, as well as a putative XyG O-acetyltransferase (Gille et al., 2011). However, whether these transferases are sufficient to synthesize the described structure of XyG in dicots has not been ascertained.
The synthesis and deposition of many plant cell wall polymers occur simultaneously with other biochemical processes, making the dissection of a synthetic pathway for a specific polysaccharide difficult. However, there are many plants with tissues in the seed or other parts of the plant that deposit only a single type of wall polymer in copious amounts during a specific developmental stage (Pauly and Keegstra, 2010). This fact has been exploited to identify genes involved in the synthesis of the hemicellulose glucogalactomannan in guar seeds (Dhugga et al., 2004; Naoumkina et al., 2007), arabinoxylan in psyllium mucilaginous layers (Jensen et al., 2011), callose in pollen tubes (Doblin et al., 2001), or cellulose in cotton fibers (Pear et al., 1996). XyG is produced by some species as a storage polymer, such as in the parenchyma cells of the cotyledons of nasturtium (Tropaeolum majus; Le Dizet, 1972; Buckeridge et al., 2000). Nasturtium seeds accumulate up to 20% of their dry weight as XyG (Hoth et al., 1986) within 20–25 days post anthesis (dpa) (Desveaux et al., 1998).
The previous identification of 10 000 ESTs of nasturtium cotyledons at a single developmental stage yielded important information leading to the identification of CSLC4 as a putative XyG:glucansynthase (Cocuron et al., 2007). However, the cost of sequencing precluded deep sequencing of large numbers of cDNA libraries required to fully exploit such a XyG production system. The recent introduction of massive parallel sequencing (RNA-Seq) has made it possible to sequence millions of EST for a reasonable cost. This method can yield quantitative representation of transcripts and increased coverage of genes, compared to traditional DNA sequencing (Weber et al., 2007).
Here, we pursued RNA-Seq on developing nasturtium seeds in an attempt to identify and complete the core set of GTs involved in XyG biosynthesis.
RESULTS AND DISCUSSION
An In-Depth Transcript Set Derived from Developing Nasturtium Seeds Producing Storage XyG
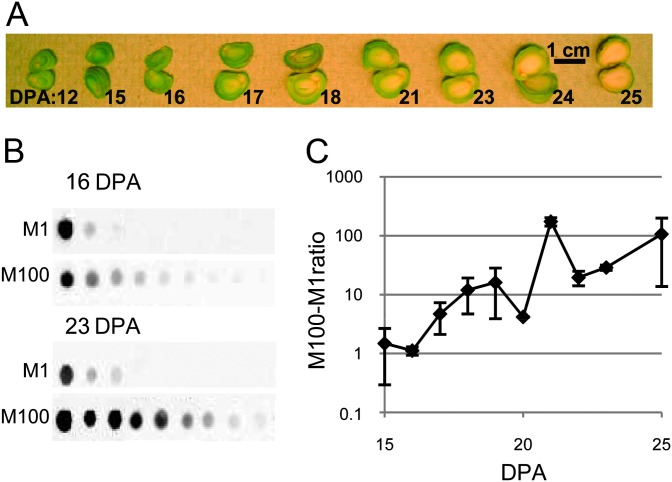
Before probing an expression dataset for genes involved in XyG biosynthesis, the timing of storage XyG deposition in nasturtium seeds needed to be ascertained. Previous efforts to accomplish this goal monitored XyG levels by hot water extraction and iodine staining (Hoth et al., 1986) or by monitoring the expression level of a known XyG biosynthetic gene, a putative XyG:galactosyltransferase TmMUR3, over the time of seed development (Cocuron et al., 2007). These efforts did not take into account that nasturtium cotyledon walls contain two types of XyG: the typical fucosylated XyG present in the cell’s primary walls and the non-fucosylated, non-O-acetylated storage XyG (Desveaux et al., 1998; Marcus et al., 2008). Both types of XyG stain with iodine, and the synthesis of both XyG types require TmMUR3. To distinguish between these two types of XyG, two antibodies were utilized: CCRC-M1, recognizing fucosylated XyG (Puhlmann et al., 1994), and CCRC-M100, exhibiting reactivity only against non-fucosylated XyG (Pattathil et al., 2010). Quantitative dot blot analysis using these carbohydrate-specific antibodies was performed on 4 M KOH extracted material of individual seeds at various dpa and the ratio of the antibody response determined (Figure 1). Based on the results, the ratio of storage XyG to primary wall XyG increased approximately 10-fold at 17–19 dpa. Hence, the time-point for the approximate onset of storage XyG formation was placed just before 17 dpa.
Figure 1.
Deposition of Storage Xyloglucan during Nasturtium Seed Development.
(A) Morphological development of seeds 12–25 d post anthesis (dpa). The cotyledons increase considerably in size from 12 to 18 dpa.
(B) Analysis of xyloglucan deposition by quantitative carbohydrate dot blots. Each successive dot represents a twofold dilution and each set of dot blots in 16 and 23 dpa, respectively, have equal amount of spotted material, so the signal intensities for the two antibodies can be compared. The signal intensities of the individual dots were quantified for each antibody and fitted to a linear regression, hence the slope being representative of antibody reactivity.
(C) Antibody reactivities from (B) represented as total non-fucosylated xyloglucan (CCRC-M100) over fucosylated xyloglucan (CCRC-M1). The carbohydrate dot blot analysis was done on single seeds, two to three seeds for each dpa, and with two technical replicas. Standard error is shown.
Because only very limited sequence information was initially available for nasturtium, sequencing was performed using 454 Life Sciences technology, generating relatively long reads to create a contig scaffold. RNA was prepared from nasturtium seeds at 16, 18, 22, and 25 dpa, spanning the period of XyG deposition from initiation to the completion of seed development. In addition, RNA from 16 and 21 dpa were subjected to a hybridization–normalization procedure (Zhulidov et al., 2004) in an attempt to reduce highly abundant transcripts such as storage proteins (Hoth et al., 1986) and thus increasing the occurrence of lowly expressed transcript in the mRNA pool. 454 sequencing of the non-normalized and normalized cDNA libraries resulted in hundreds of thousands of sequencing reads from each developmental stage with an average length of 215–362 bp (Supplemental Table 1). All 454 sequencing data were combined and assembled using the CLC Genomics Workbench version 4.3.7 resulting in 38 885 total contigs (Supplemental Table 2). Each contig sequence was annotated with its closest TAIR9 Arabidopsis blast hit. To obtain high temporal resolution and robust expression data for lowly expressed genes, Illumina mRNA-Seq was performed and the short sequence reads were mapped onto the 454 contig assemblies. Illumina mRNA-Seq was performed on developing seeds at 13, 16, 17, 18, 21, and 25 dpa with an additional replicate at 16 dpa (Supplemental Table 1). An increase in sequencing reads of about 20-fold was observed with the Illumina data compared to the 454 sequencing. Of the Illumina mRNA-Seq sequence reads, 72% could be matched to a transcript (contig) present in the 454 sequence information (Supplemental Table 2). The remaining 28% of the Illumina sequence reads were ascribed to newly found contigs in the transcriptome or sequencing errors. Of the 454 assembled contigs, 10% were not represented by any Illumina mRNA-Seq read (Supplemental Table 2). Approximately half of these 454 contigs had only one, two, or three reads, and many were found to be concatemers. These artifactual sequences most likely originate from the PCR amplification step of the cDNA libraries prior to 454 FLX sequencing. Another part of the 454 assemblies that lacked Illumina mRNA-Seq consisted of nasturtium rRNA. For further analysis, a combined database was constructed for all contigs with a combined 454 and Illumina sequence read count of at least 5; this database contained 36 120 contigs (Supplemental Tables 2 and 3).
Identification of XyG Glycosyltransferases
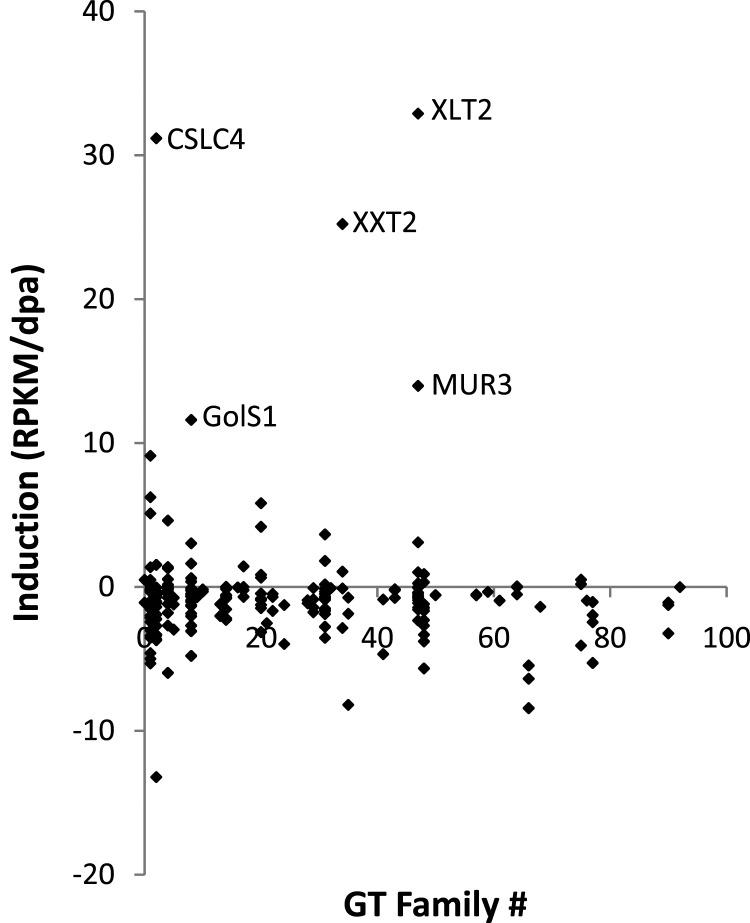
To identify GTs involved in biosynthesis of seed storage XyG in nasturtium, all Arabidopsis GTs present in the CAZy database (www.cazy.org; Cantarel et al., 2009) were compared with the sequences in the nasturtium database using TBLASTN (Altschul et al., 1997). Two hundred and twenty-six contigs were identified corresponding to 179 Arabidopsis genes in 37 GT families (Supplemental Table 4). It is expected that, during the time course of storage XyG production in the developing nasturtium seed, relevant GTs involved in storage XyG biosynthesis will be up-regulated yielding higher transcript levels. The amount of expression induction of the individual 226 GT annotated contigs of each was computed by regression analysis using the slope of a linear best fit for the Illumina expression data (Figure 2 and Supplemental Table 4). Indeed, three of the five most highly induced GT contigs represented putative nasturtium orthologs of Arabidopsis CSLC4, XXT2, and MUR3. The most highly induced GT annotated contig in nasturtium was found to be highly similar to a hitherto uncharacterized Arabidopsis gene, At5g62220. This gene encodes a protein belonging to GT family 47, the same family that contains MUR3 (see phylogenetic tree in Supplemental Figure 1), and hence represents an attractive candidate for a second XyG:galactosyltransferase. The fifth most highly induced contig is highly similar to the Arabidopsis gene GolS1 (At2g47180, GT family 8), which has been shown to contribute to the synthesis of raffinose family oligosaccharides (Liu et al., 1998), the production of which has been shown to increase in later stages of seed development (Kuo et al., 1997). It is thus unlikely that TmGolS1 is involved in XyG biosynthesis.
Figure 2.
Expression Increase of Glycosyltransferases in the Nasturtium Dataset during Seed Development.
Arabidopsis glycosyltransferases from CAZy were used to query the nasturtium database using a TBLASTN to extract glycosyltransferases-related contigs using an e-value cutoff of 10−80. Each identified contig was plotted with the y-axis indicating the increase of expression as measured by the slope of a linear best-fit line of the Illumina expression data, and the x-axis indicating to which glycosyltransferase family the contig belongs (0—not classified in CAZy). Contigs with an induction of less than –20 were omitted for clarity. For the five most induced contigs, the gene name of the putative Arabidopsis ortholog is shown; all other glycosyltransferases contigs can be found in Supplemental Table 4.
These findings confirm that TmCSLC4 likely represents the XyG glucan synthase (Cocuron et al., 2007). While several other CSLCs were identified in the nasturtium database, these all had expression levels several orders of magnitude lower than TmCSLC4 and did not show any temporal induction during seed development (Supplemental Table 4). These data suggest that TmCSLC4 is the only CSL gene involved in synthesizing nasturtium seed storage XyG.
In Arabidopsis, there are seven genes in GT family 34, three of which have been shown to be involved in XyG xylosylation, namely XXT1, XXT2, and XXT5 (Cavalier and Keegstra, 2006; Cavalier et al., 2008; Zabotina et al., 2008). Single mutants containing a disruption of any one of the XXT genes display a 10–50% reduction of xylose levels in XyG, but with no differential impact on the three different positions that are xylosylated in XyG (Cavalier et al., 2008; Zabotina et al, 2008). In the double mutant xxt1 xxt2 there is no detectable XyG (Cavalier et al., 2008) and hence xylosylation at all three positions in the basic XXXG unit appears to have been lost. In vitro activity has not been reported for XXT5, while XXT1 and XXT2 are well characterized, and each protein has been shown to facilitate xylosyl transfer on up to three consecutive positions on a hexameric β-1–4-glucan oligomer (Faik et al., 2002; Cavalier and Keegstra, 2006). In the nasturtium dataset, four members of GT family 34 are present. Only the above-mentioned putative ortholog to Arabidopsis XXT2 displays an expression induction during the time course of seed XyG filling, while the expression of other members including a putative ortholog to Arabidopsis XXT5 remains flat (Supplemental Table 4). While we cannot exclude that any other member of GT family 34 is involved in seed XyG biosynthesis, the data presented here are inconsistent with the hypothesis that more than one XXT is required for XyG xylosylation.
XyG galactosyltransferase activity is needed for galactosylation of XyG. Previously, the MUR3 gene from Arabidopsis has been shown to be a XyG galactosyltransferase responsible for adding galactose at the third position of the repeating XXXG motif (converting XXXG to XXLG) (Madson et al., 2003). However, in Arabidopsis and nasturtium, a galactose can also be found at the second position (XLXG). A gene, AtGT18 (At5g62220), has been hypothesized to be responsible for the occurrence of this structural feature (Li et al., 2004). AtGT18 is a member of GT family 47 like MUR3, and expression analysis using RT–PCR and a promoter–reporter construct are consistent with a co-occurrence of XyG (Li et al., 2004). In addition, a T-DNA knockout line also exhibited a reduction in overall galactose content. However, a role of AtGT18 in XyG synthesis, its activity, and/or substrate specificity, or resulting product characterization was not demonstrated. As mentioned above, two contigs from GT family 47 were found to be highly induced during XyG deposition in the seeds (Figure 2 and Supplemental Table 4). Based on a phylogenetic tree of GT family 47 clade A, these contigs can be considered putative orthologs of Arabidopsis MUR3 and At5g62220 (AtGT18; Supplemental Figure 1). The identification of two GT family 47 genes in nasturtium is consistent with the hypothesis that two separate XyG galactosyltransferases are required to add galactosyl residues to distinct positions on the polymer. However, due to the presence of other members in this GT47 clade in Arabidopsis (Supplemental Figure 1), additional GT47 genes might be involved in XyG galactosylation or substituting xylosyl residues of XyG with other sugar-moieties in this plant species. Nevertheless, the nasturtium expression data provide further support to the hypothesis that AtGT18 might be involved in XyG galactosylation.
Characterization of a Second Distinct XyG Galactosyltransferase: XyG L-Side Chain GalactosylTranferase Position 2 (XLT2)
Rather than using a generic name, AtGT18, the Arabidopsis gene At5g62220 was renamed XLT2 for XyG L-side chain galactosylTransferase position 2, based on the results described below. To ascertain the function of XLT2, an Arabidopsis T-DNA insertional line (GK-552C10-02168; xlt2) was obtained containing an insertion in the coding sequence of this gene. The xlt2 insertional line was confirmed to be homozygous for the insertion and confirmed to be a true knockout, as no XLT2 transcript could be detected in the plant (Supplemental Figure 2). Leaf wall material from xlt2 was subjected to a xyloglucanase treatment, and the solubilized XyG oligosaccharides were subjected to analysis by High Performance Anion Exchange Chromatography with Pulsed Amperometric Detection (HPAEC–PAD). No compounds eluted at the times for XLXG and XLFG in the xyloglucanase extract of xlt2 (Figure 3 and Table 1). The other galactosylated oligosaccharide, XXLG, was still present, as was its fucosylated version, XXFG. This structural defect is distinct from mur3.1, where XXLG and its fucosylated counterpart are greatly reduced, but XLXG is still present (Figure 3 and Table 1). Further evidence of the XyG structure of the xlt2 mutant comes from oligosaccharide mass profiling (OLIMP; Supplemental Figure 3). In xlt2, oligosaccharide ions corresponding to XLFG and XLFG are not observed. This is again distinct from the phenotype of the mur3.1 mutant, which has greatly reduced XXFG, XXFG, XLFG, and XLFG. To confirm that the observed XyG phenotype in the xlt2 mutant was indeed due to a lack of XLT2-transcript, a genetic complementation was performed by transforming the xlt2 mutant with a wild-type copy of the XLT2 gene. This complementation line, named XLT2_Comp, regained the oligosaccharides containing galactose at the second position as revealed by HPAEC–PAD and OLIMP (Figure 3, Table 1, and Supplemental Figure 3). Analysis of the mur3.1 xtl2 double mutant demonstrated that the XyG of this mutant consists primarily of XXXG subunits with only a residual amount of XXFG (Figure 3 and Supplemental Figure 3). The remaining XXFG may be the result of the mur3.1 allele, which, due to a point mutation, may retain some activity or the presence of another gene with MUR3 activity. The total amount of XyG oligosaccharides released by the xyloglucanase was nearly halved in both of the single mutants and the double mutant compared to the wild-type and the complementation line (Table 1). This indicates either less XyG or that less XyG was accessible to the enzyme (xyloglucanase) in the mutant lines (Pauly et al., 1999a).
Figure 3.
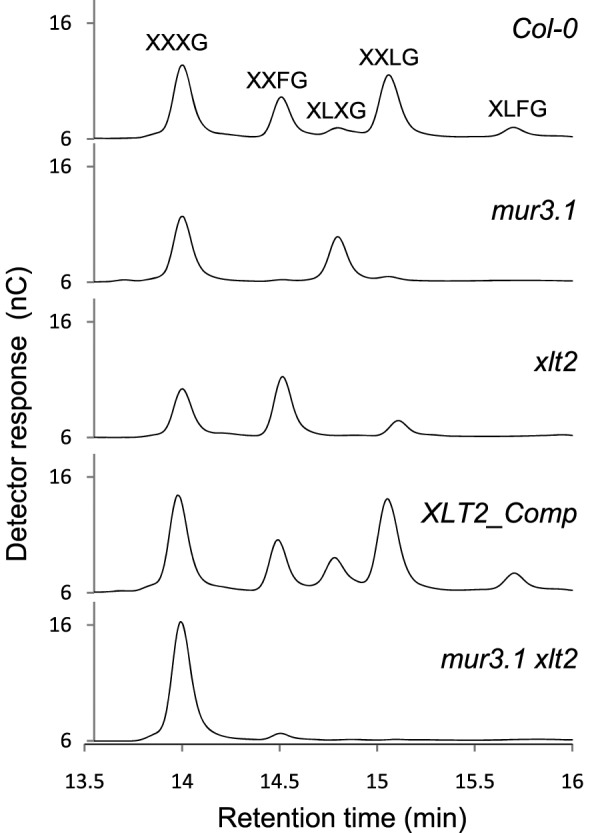
HPAEC–PAD Analysis of Xyloglucan Oligosaccharides Released from AIR Material from Leaves by Xyloglucanase Treatment.
Table 1.
Quantification of Xyloglucanase-Released Xyloglucan Oligosaccharides.
| Genotype | XXXG | XXFG | XLXG | XXLG | XLFG | Gal at 3rda | Total Fb | Total XyG | |
| Wild-type (Col-0) | 0.30 ± 0.04c | 0.12 ± 0.02 | 0.04 ± 0.02 | 0.27 ± 0.05 | 0.04 ± 0.01 | 0.42 ± 0.08 | 0.16 ± 0.03 | 0.76 ± 0.14 | |
| mur3.1 | 0.23 ± 0.04 | 0.00 ± 0.00 | 0.18 ± 0.04 | 0.01 ± 0.01 | n.d. | 0.02 ± 0.01 | 0.00 ± 0.00 | 0.42 ± 0.09 | |
| xlt2 | 0.18 ± 0.05 | 0.22 ± 0.04 | n.d. | 0.05 ± 0.01 | n.d. | 0.28 ± 0.06 | 0.22 ± 0.04 | 0.46 ± 0.11 | |
| XLT2_Comp | 0.27 ± 0.04 | 0.09 ± 0.02 | 0.12 ± 0.02 | 0.31 ± 0.05 | 0.05 ± 0.02 | 0.45 ± 0.09 | 0.14 ± 0.04 | 0.84 ± 0.15 | |
| mur3.1 xlt2 | 0.35 ± 0.06 | 0.01 ± 0.01 | 0.00 ± 0.00 | 0.00 ± 0.00 | n.d. | 0.02 ± 0.01 | 0.01 ± 0.01 | 0.36 ± 0.06 |
Sum of oligosaccharides with galactose at the third position (XXLG, XXFG, XLFG).
Sum of fucosylated oligosaccharides (XXFG, XLFG).
XyG oligosaccharide quantification based on HPAEC–PAD using an XXXG standard. Quantities are μg mg−1 of AIR prepared from rosette leaves. n = 4; n.d. - not detected.
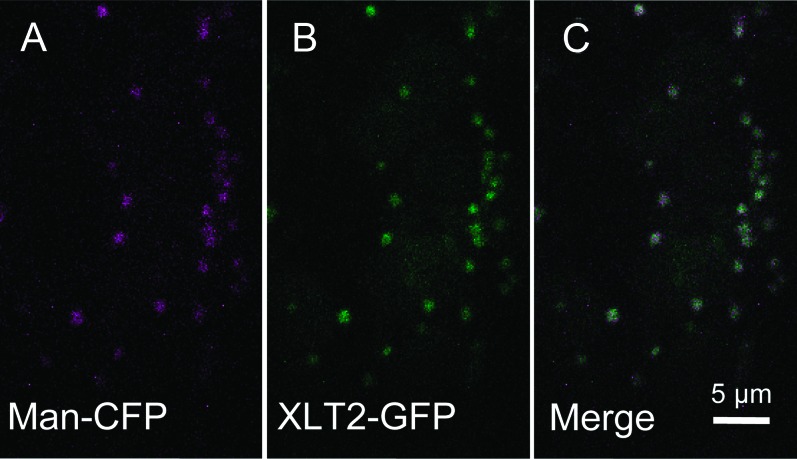
To determine the subcellular localization of XLT2, an XLT2::GFP fusion construct was generated and transiently expressed in Nicotiana benthamiana along with a mannosidase–CFP Golgi-marker (Nelson et al., 2007). The CFP and GFP signals appeared to co-localize in small mobile vesicles (Figure 4). Hence, XLT2 seems to be localized to the Golgi apparatus, consistently with the location of XyG biosynthesis.
Figure 4.
Subcellular Localization of XLT2 using Transient Expression in Nicotiana benthamiana Leaves.
(A) CFP channel colored magenta showing Man–CFP Golgi-marker.
(B) GFP channel colored green showing XLT2–GFP.
(C) Merged image showing co-localization of XLT2–GFP and Man–CFP.
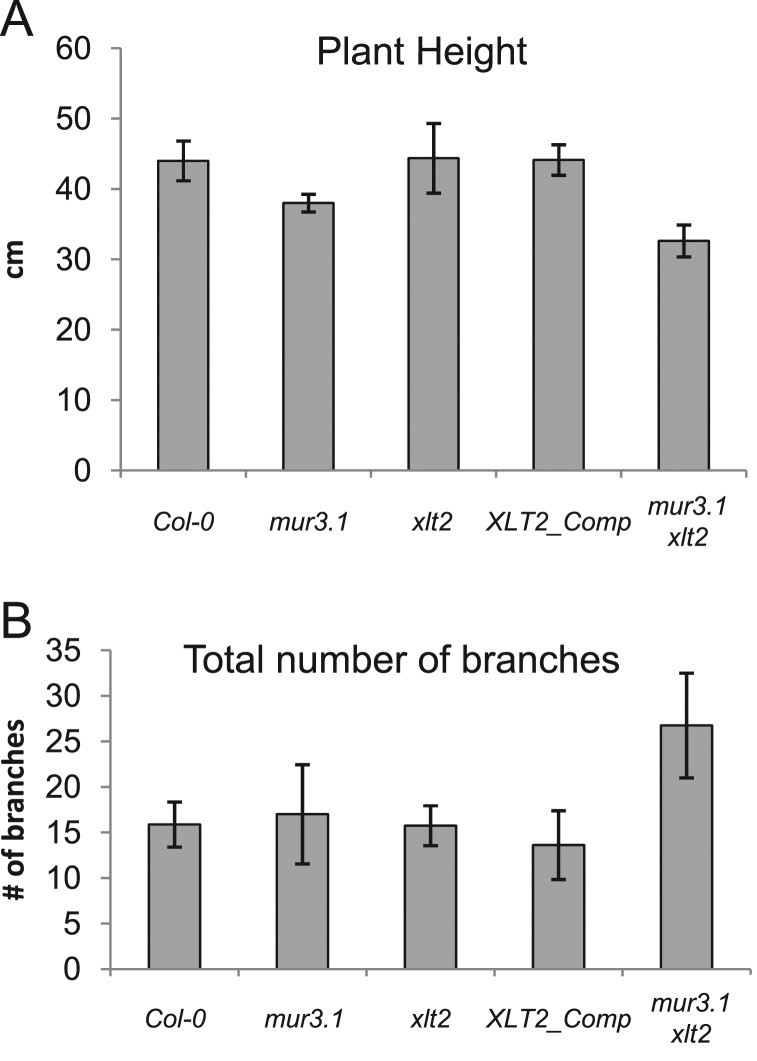
The mur3.1 mutant was, on average, 15% shorter (P < 0.0003) than wild-type plant. This dwarfism was even more pronounced in the mur3.1 xlt2 double mutant (25% shorter than wild-type, P < 10−6; 10% shorter than mur3.1, P < 0.0002; Figure 5A). In addition, the mur3.1 xlt2 double mutant plant had a bushier appearance compared to wild-type or single mutant plants; it had approximately 70% more total stems (including branches, P < 0.001; Figure 5B). The lack of a more severe growth or morphological phenotype of the mur3.1 xlt2 double mutant was unexpected, as a nearly complete lack of XyG substitution beyond the xylosyl residues is likely to have a significant effect on the solubility and binding affinity of this polysaccharide to cellulose. However, the lack of a strong phenotype is consistent with what has been observed for other mutant plants with altered XyG structure (Cavalier et al., 2008; Zabotina et al., 2008).
Figure 5.
Inflorescent Stem Measurements of Wild-Type and Xyloglucan Mutants.
Plant height was measured from the base of the stem to the stretched apex of the longest stem. The total number of branches is the number of stems emerging from the rosette plus the total number of branch points present in these primary stems and all subsidiary branches. Error bars indicate standard deviation. n = 7 for mur3.1; n = 8 for Col-0, xlt2, XLT2_Comp, and mur3.1 xlt2.
CONCLUSION
Plant polysaccharide biosynthesis, as exemplified by XyG, is a complex process involving the coordinated expression and activity of many genes. Co-expression analysis has been used with some success to identify genes involved in the formation of particular polysaccharides. However, in situations in which a cell or tissue is producing many polysaccharides simultaneously, the co-expression signal for any particular polysaccharide can be lost. By moving to a system and tissue, in this case developing nasturtium seeds, that produces large amounts of a particular polysaccharide, one can more easily identify genes involved in the biosynthesis of that polysaccharide and dissect their function. The deep-sequencing approach used here on developing nasturtium seeds allowed the identification of a core set of four GTs involved in seed storage XyG biosynthesis: TmCslC4, TmXXT2, TmMUR3, and TmXLT2. XLT2 was a hitherto uncharacterized XyG galactosyltransferase gene in Arabidopsis. Its effect on XyG structure in planta was established in the corresponding mutants, suggesting that XLT2 is involved in galactosylating XyG at the second position of the XXXG motif to form XLXG.
The expression dataset generated here can be further explored to look for other classes of genes involved in XyG biosynthesis. For example, the most highly expressed and induced glycosylhydrolase identified in the database was most closely related to AtXTH25 (At5g57550), a member of the XyG:endotransglycosylase family (XTH; glycosylhydrolase family 16; Fry et al., 1992; Rose et al., 2002). The expression of TmXTH25 increased more than 400-fold during the time course, whereas other XTHs were lowly expressed and not induced. It is therefore likely that this XTH is involved in assembling nascent XyG polysaccharides into longer chains, and that one XTH is sufficient for this task. Future data mining and biochemical characterization of potential candidate genes, including but not limited to other hydrolases as well as transcription factors, nucleotide sugar interconversion enzymes, and nucleotide sugar transporters, may yield other genes that play a role in XyG biosynthesis.
METHODS
Plant Material and Growth Conditions
Tropaeolum majus seeds were obtained from a local plant nursery and germinated and grown as described by Cocuron et al. (2007). The Arabidopsis mutant mur3.1 (Madson et al., 2003; ABRC stock number CS8566) and the T-DNA line GK-552C10-021687, referred to here as xlt2 and containing an insertion in XLT2 (At5g62220), were obtained from ABRC, Ohio State University, and GABI-KAT (Bielefeld University, Germany; Li et al., 2007), respectively. Soil-grown plant material was grown in long-day conditions (16-h light, 8-h dark) with 130–140 μmol m−2 s−1 light intensity at 22°C.
Quantitative Dot Blot Analysis Using Carbohydrate-Specific Antibodies
Individual pairs of dissected cotelydons were homogenized, extracted with 4 M KOH and analyzed as described by Jensen et al. (2011) using CCRC-M1 (Puhlmann et al., 1994) and CCRC-M100 (Pattathil et al., 2010) as carbohydrate-specific antibodies and goat anti-mouse:HRP (170–6516, Bio-Rad) as secondary antibody used in 1:3000 dilution.
RNA Isolation, 454 FLX Sequencing, and Illumina mRNA-Seq
Total RNA was isolated from individual developing nasturtium seeds as described by Cocuron et al. (2007). 454 FLX sequencing was performed by the Joint Genome Institute (JGI Walnut Creek, CA, USA). cDNA library construction and sequencing are described for the non-normalized libraries in Troncoso-Ponce et al. (2011). Normalized libraries were processed from total RNA at JGI using the kamchatka crab duplex-specific nuclease protocol (Zhulidov et al., 2004). Total RNA (10 μg) was processed and sequenced according to the mRNA-Seq protocol (Illumina). Raw sequence data were deposited at the NCBI Sequence Read Archive (454 FLX sequences: 16 dpa, SRX072972; 16 dpa normalized, SRX015179; 18 dpa, SRX015175; 21 dpa normalized, SRX015180; 22 dpa, SRX072973; 25 dpa, SRX015176–8; Illumina RNA-Seq: SRX108504 to SRX108510 for 13–25 dpa, respectively).
Contig Assembly, Identification, and Annotation
The obtained 454 and Illumina reads were assembled using CLC Genomics Workbench version 4.3.7 (CLC bio, Cambridge, MA, USA) using the de novo assembly algorithm (Parameters: Similarity 0.8; Length fraction 0.5; Insertion cost 3; Deletion cost 3; Mismatch cost 2). The generated consensus sequences were then used as a ‘reference without annotations’ for a batch RNA-Seq experiment resulting in per library RPKM and read counts for the putative transcripts. Each contig has a ‘total members’ value equal to the sum of the read counts from the mappings. The contigs were annotated using BLASTX against the TAIR 9 annotation of the Arabidopsis genome. This annotated assembly with expression values is given in Supplemental Table 3 for all contigs with five or more mapped reads.
Mutant Genotyping
The location of the T-DNA in xlt2 was determined by PCR using JumpStart REDTaq ReadyMix (Sigma-Aldrich) with PCR conditions as recommended. The primers GABI-LB and AtXLT2-RP were used to check for the presence of the T-DNA insertion and AS-124 and AS-125 were used to check for the presence of the native wild-type XLT2 sequence (Supplemental Table 5). Wild-type plants and the XLT2 complementation line were distinguished by performing a PCR reaction with primers AS-120 and M13F, which are specific to the complementation construct. The mur3.1 allele (containing a single nucleotide substitution) was genotyped by amplifying a section of the gene via PCR utilizing primers mur3.1-fwd and mur3.1-rev and digestion of the PCR product with TaqI, a restriction site disrupted in the PCR product resulting from the mur3.1 allele (Neff et al., 2002).
Genetic Complementation
A 5-kb fragment including the wild-type AtXLT2 gene (At5g62220) was amplified from Arabidopsis Col-0 genomic DNA with Phusion Hi-Fidelity DNA Polymerase (Finnzymes) using the primers AtXLT2-fwd and AtXLT2-rev. The resulting PCR product was TOPO-TA cloned into the pCR8/GW vector (Invitrogen). The restriction enzymes KpnI and XmaI were used to digest this plasmid and clone AtXLT2 into the binary vector pPZP211 (Hajdukiewicz et al., 1994). This plasmid was transformed into Agrobacterium tumefaciens GV3101 and transformed into Arabidopsis using the floral dip method (Clough and Bent, 1998). T1 seeds were obtained and germinated on ½ MS (M5524, Sigma-Aldrich), 1% sucrose plates containing 60 μg ml−1 kanamycin. Resistant plants were transferred to soil and allowed to self. A homozygous line was selected from the T2 generation. The genotype of this line was confirmed to be homozygous for knockout of the native XLT2 and to contain the complementation construct by performing PCR as described above.
Xyloglucan Analysis
OLIMP: the procedure was essentially performed as shown in Gunl et al. (2010). Rosette leaves from 4-week-old Arabidopsis plants were collected and dried. Dried material was frozen in liquid nitrogen and ground in a Retsch Mill Grinder at 25 Hz for 150 s. The ground material was extracted once with 70% aqueous ethanol, three times with 1:1 chloroform:methanol, and dried. The dried material (2 mg) was incubated with 1 ml of 20 mM ammonium formate, pH 4.5, at 37°C overnight and then centrifuged for 10 min at 14 000 rpm to remove buffer-soluble components. The remaining pellet was digested with 0.4 U of a XyG-specific endoglucanase/xyloglucanase (Pauly et al., 1999b) in 100 μl of 20 mM ammonium formate, pH 4.5. Following an overnight incubation at 37°C with shaking, the digest was centrifuged for 10 min at 21000 g. The supernatant was concentrated fivefold by vacuum drying and desalted using approximately 10 BioRex MSZ 501 cation exchange beads. Matrix (2 μl of 10 mg ml−1 2,5-dihydroxybenzoic acid) was spotted onto wells on a MALDI–TOF target plate and dried under vacuum. The desalted concentrated digest (2 μl) was spotted on top of the DHB matrix, incubated for 3 min, and dried under vacuum. An AXIMA Performance (Shimadzu) MALDI–TOF mass spectrometer was used in positive linear mode with an accelerating voltage of 20 000 V to obtain the mass profiles. For separation of XyG oligosaccharides by High Performance Anion Exchange Chromatography with Pulsed Amperometric Detection (HPAEC–PAD), the xyloglucanase digest prepared as described above was diluted four times with water and 25 μl was injected onto a CarboPac PA200 column. 100 mM sodium hydroxide with a gradient of 0–80 mM sodium acetate over 15 min with a flow rate of 0.4 ml min−1 was used to separate the oligosaccharides. Oligosaccharides were quantified using a standard curve generated from commercially available XXXG (Megazymes).
Subcellular Localization
The coding sequence of XLT2 was amplified from the plasmid used for complementation of the xlt2 mutant (XLT2 in pPZP211, described above) using Phusion Hi-Fidelity DNA polymerase (Finnzymes) with the primers AtXLT2CDS-fwd and AtXLT2CDS-rev. The PCR product was gel extracted and a BP reaction was performed with pDONR221 P1-P2 (Invitrogen) to create an entry clone. An LR reaction was subsequently performed with pMDC84 to create an open reading frame consisting of the XLT2 CDS with a C-terminal GFP tag. This plasmid was transformed into A. tumefaciens GV3101. Cultures containing the GFP fusion and a mannosidase-CFP Golgi-marker were grown for 2 d at 30°C, diluted to an OD600 of 0.05, and co-infiltrated into 4-week-old Nicotiana benthamiana leaves (Sparkes et al., 2006). A Zeiss LSM 710 laser scanning confocal microscopy was used to image the epidermal cells. Lasers (405 and 488 nm) were used to excite the fluorescent proteins and band pass filters of 454–481 and 504–598 were used to detect CFP and GFP, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494 to J.K.J., K.K., and C.G.W.), by a National Institutes of Health NRSA Trainee appointment (GM007127 to A.S.), and the Fred Dickinson chair of Wood Science and Technology Endowment to M.P. We thank Christa Pennacchio and Erika Linquist of the US Department of Energy Joint Genome Institute for high-throughput cDNA sequencing, which was supported by the Office of Science of the US Department of Energy under Contract No. (DE-AC02-05CH11231).
Supplementary Material
Acknowledgments
We thank Nick Thrower for providing the bioinformatic expertise clustering of the cDNA libraries; Michael Hahn, CCRC, University of Georgia, for providing the anti-XyG antibodies CCRC-M1 and CCRC-M100; Kirk Schnorr, Novozymes, Bagsvaerd, Denmark, for the xyloglucanase; and Tom Haas, UC Berkeley, for technical assistance. No conflict of interest declared.
References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckeridge M, dos Santos H, Tine M. Mobilisation of storage cell wall polysaccharides in seeds. Plant Physiol. Bioch. 2000;38:141–156. [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier DM, Keegstra K. Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. J. Biol. Chem. 2006;281:34197–34207. doi: 10.1074/jbc.M606379200. [DOI] [PubMed] [Google Scholar]
- Cavalier DM, et al. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell. 2008;20:1519–1537. doi: 10.1105/tpc.108.059873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cocuron JC, et al. A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc. Natl Acad. Sci. U S A. 2007;104:8550–8555. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Faik A, Maclachlan G. Fucosyltransferase and the biosynthesis of storage and structural xyloglucan in developing nasturtium fruits. Plant Physiol. 1998;118:885–894. doi: 10.1104/pp.118.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga K, et al. Guar seed beta-mannan synthase is a member of the cellulose synthase super gene family. Science. 2004;303:363–366. doi: 10.1126/science.1090908. [DOI] [PubMed] [Google Scholar]
- Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM. Pollen tubes of Nicotiana alata express two genes from different beta-glucan synthase families. Plant Physiol. 2001;125:2040–2052. doi: 10.1104/pp.125.4.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Price NJ, Raikhel NV, Keegstra K. An Arabidopsis gene encoding an alpha-xylosyltransferase involved in xyloglucan biosynthesis. Proc. Natl Acad. Sci. U S A. 2002;99:7797–7802. doi: 10.1073/pnas.102644799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry S, et al. An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol. Plantarum. 1993;89:1–3. [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 1992;282:821–828. doi: 10.1042/bj2820821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, et al. O-acetylation of Arabidopsis hemicellulose xyloglucan requires AXY4 or AXY4L, proteins with a TBL and DUF231 domain. Plant Cell. 2011;23:4041–4053. doi: 10.1105/tpc.111.091728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenl M, Gille S, Pauly M. OLIgo mass profiling (OLIMP) of extracellular polysaccharides. J. Vis. Exp. 2010;40:2046. doi: 10.3791/2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Hayashi T. Xyloglucans in the primary cell wall. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989;40:139–168. [Google Scholar]
- Hayashi T, Kaida R. Functions of xyloglucan in plant cells. Mol. Plant. 2011;4:17–24. doi: 10.1093/mp/ssq063. [DOI] [PubMed] [Google Scholar]
- Hoth A, Blaschek W, Franz G. Xyloglucan (amyloid) formation in the cotyledons of Tropaeolum-majus L seeds. Plant Cell Rep. 1986;5:9–12. doi: 10.1007/BF00269706. [DOI] [PubMed] [Google Scholar]
- Jensen JK, Kim H, Cocuron J-C, Orler R, Ralph J, Wilkerson CG. The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis . Plant J. 2011;66:387–400. doi: 10.1111/j.1365-313X.2010.04475.x. [DOI] [PubMed] [Google Scholar]
- Kuo TM, Lowell CA, Smith PT. Changes in soluble carbohydrates and enzymic activities in maturing soybean seed tissues. Plant Science. 1997;125:1–11. [Google Scholar]
- Le Dizet P. Analysis of the structure of amyloid from nasturtiums. Carbohydrate Research. 1972;24:505–509. doi: 10.1016/s0008-6215(00)85085-5. [DOI] [PubMed] [Google Scholar]
- Lerouxel O, Cavalier DM, Liepman AH, Keegstra K. Biosynthesis of plant cell wall polysaccharides: a complex process. Curr. Opin. Plant Biol. 2006;9:621–630. doi: 10.1016/j.pbi.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Li X, Cordero I, Caplan J, Molhoj M, Reiter WD. Molecular analysis of 10 coding regions from Arabidopsis that are homologous to the MUR3 xyloglucan galactosyltransferase. Plant Physiol. 2004;134:940–950. doi: 10.1104/pp.103.036285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Rosso MG, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: an Arabidopsis thaliana T-DNA mutant database with detailed information for confirmed insertions. Nucleic Acids Res. 2007;35:D874–D878. doi: 10.1093/nar/gkl753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJJ, Krenz DC, Galvez AF, de Lumen BO. Galactinol synthase (GS): increased enzyme activity and levels of mRNA due to cold and desiccation. Plant Science. 1998;134:11–20. [Google Scholar]
- Madson M, et al. The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell. 2003;15:1662–1670. doi: 10.1105/tpc.009837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus SE, et al. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008;8:60. doi: 10.1186/1471-2229-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz EJ, Immerzeel P, Hayashi T. Xyloglucan: the molecular muscle of trees. Ann. Bot. 2008;102:659–665. doi: 10.1093/aob/mcn170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina M, et al. Analysis of cDNA libraries from developing seeds of guar (Cyamopsis tetragonoloba (L.) Taub) BMC Plant Biol. 2007;7:62. doi: 10.1186/1471-2229-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Turk E, Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002;18:613–615. doi: 10.1016/s0168-9525(02)02820-2. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Pattathil S, et al. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 2010;153:514–525. doi: 10.1104/pp.109.151985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008;54:559–568. doi: 10.1111/j.1365-313X.2008.03463.x. [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K. Plant cell wall polymers as precursors for biofuels. Curr. Opin. Plant Biol. 2010;13:305–312. doi: 10.1016/j.pbi.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Pauly M, Albersheim P, Darvill A, York WS. Molecular domains of the cellulose/xyloglucan network in the cell walls of higher plants. Plant J. 1999a;20:629–639. doi: 10.1046/j.1365-313x.1999.00630.x. [DOI] [PubMed] [Google Scholar]
- Pauly M, Eberhard S, Albersheim P, Darvill AG, York WS. Effects of the mur1 mutation on xyloglucans produced by suspension-cultured Arabidopsis thaliana cells. Planta. 2001;214:67–74. doi: 10.1007/s004250100585. [DOI] [PubMed] [Google Scholar]
- Pauly M, et al. A xyloglucan-specific endo-beta-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification and characterization of the recombinant enzyme. Glycobiology. 1999b;9:93–100. doi: 10.1093/glycob/9.1.93. [DOI] [PubMed] [Google Scholar]
- Pear JR, Kawagoe Y, Schreckengost WE, Delmer DP, Stalker DM. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl Acad. Sci. U S A. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, et al. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science. 1999;284:1976–1979. doi: 10.1126/science.284.5422.1976. [DOI] [PubMed] [Google Scholar]
- Puhlmann J, et al. Generation of monoclonal antibodies against plant cell-wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal alpha-(1→2)-linked fucosyl-containing epitope. Plant Physiol. 1994;104:699–710. doi: 10.1104/pp.104.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002;43:1421–1435. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- Scheible W-R, Pauly M. Glycosyltransferases and cell wall biosynthesis: novel players and insights. Curr. Opin. Plant Biol. 2004;7:285–295. doi: 10.1016/j.pbi.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. Hemicelluloses. Annu. Rev. Plant Biol. 2010;61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- Somerville C, et al. Toward a systems approach to understanding plant cell walls. Science. 2004;306:2206–2211. doi: 10.1126/science.1102765. [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006;1:2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- Troncoso-Ponce MA, et al. Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 2011;68:1014–1027. doi: 10.1111/j.1365-313X.2011.04751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD. The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc. Natl Acad. Sci. U S A. 2002;99:3340–3345. doi: 10.1073/pnas.052450699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber APM, Weber KL, Carr K, Wilkerson C, Ohlrogge JB. Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol. 2007;144:32–42. doi: 10.1104/pp.107.096677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, et al. Arabidopsis XXT5 gene encodes a putative alpha-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. Plant J. 2008;56:101–115. doi: 10.1111/j.1365-313X.2008.03580.x. [DOI] [PubMed] [Google Scholar]
- Zhulidov PA, et al. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 2004;32:e37. doi: 10.1093/nar/gnh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.