Abstract
Infection with Helicobacter pylori is chronic despite a vigorous mucosal immune response characterized by gastric T-helper type 1 cell expansion and gamma interferon (IFN-γ) production. IFN-γ signals by activation and nuclear translocation of signal transducer and activator of transcription 1 (STAT1); however, the effect of H. pylori infection on IFN-γ-STAT1 signaling is unknown. We infected human gastric (MKN45 and AGS) and laryngeal (HEp-2) epithelial cell lines with type 1 and type 2 H. pylori strains and then stimulated them with IFN-γ. Western blotting of whole-cell protein extracts revealed that infection with live, but not heat-killed, H. pylori time-dependently decreased IFN-γ-induced STAT1 tyrosine phosphorylation. Electrophoretic mobility shift assay of nuclear protein extracts demonstrated that H. pylori infection reduced IFN-γ-induced STAT1 DNA binding. STAT1 was unable to translocate from the cytoplasm to the nucleus in H. pylori-infected HEp-2 cells examined by immunofluorescence, and reverse transcription-PCR showed that IFN-γ-induced interferon regulatory factor 1 expression was inhibited. These effects were independent of the cagA, cagE, and VacA status of the infecting H. pylori strain. Furthermore, neither H. pylori culture supernatants nor conditioned medium from H. pylori-infected MKN45 cells inhibited IFN-γ-induced STAT1 tyrosine phosphorylation, suggesting that inhibition is independent of a soluble epithelial or bacterial factor but is dependent on bacterial contact with epithelial cells. H. pylori disruption of IFN-γ-STAT1 signaling in epithelial cells may represent a mechanism by which the bacterium modifies mucosal immune responses to promote its survival in the human host.
Helicobacter pylori is a gram-negative, spiral-shaped organism that is adapted to survive in the microenvironment of the human stomach. It is estimated that the bacterium chronically colonizes the stomachs of approximately 50% of the human population (41). Most infected individuals develop an asymptomatic chronic gastritis; however, between 3 and 25% develop peptic ulceration (54). There is increasing evidence that H. pylori infection also is a major risk factor for the development of gastric adenocarcinoma (56) and gastric mucosa-associated lymphoid tissue lymphoma (40).
Both microbial and host factors influence the pathogenesis of gastro-duodenal disease. Putative bacterial virulence factors include the presence of the cag pathogenicity island (cag PAI), a horizontally acquired 40-kilobase segment of DNA (6) which defines type 1 (cag PAI+) versus type 2 (cag PAI−) strains. The cag PAI encodes a type and secretion system. The proteins CagA, CagE, and CagF (47) and others that are still uncharacterized. The CagA protein is translocated into host epithelial cells by the type IV secretion system, where it is phosphorylated and induces cytoskeletal reorganization (46), while the CagE protein is integral to the activation of NF-κB and interleukin-8 (IL-8) secretion after infection (12, 24). Non-cag PAI products, such as the vacuolating cytotoxin A (VacA), which activates host signaling molecules Rab7 (39) and Rac1 (28) to cause vacuolation of epithelial cells, may also be important for virulence.
On the host side, evidence from genome-wide linkage studies suggests that gamma interferon (IFN-γ) receptor α chain polymorphisms influence H. pylori colonization of the stomach and the subsequent risk of developing gastric adenocarcinoma (55). Epidemiological studies show that individuals with genetic polymorphisms resulting in increased IL-1β production and who are infected with H. pylori have an increased risk for hypochlorhidria, gastric atrophy, and gastric adenocarcinoma (17). Thus, both microbial and host factors likely contribute to disease progression.
The uninfected stomach has a relatively small T-cell population biased towards a helper T-cell type 1 (Th1) response (27), and this bias is maintained in the inflammatory cell infiltrate in response to H. pylori infection (5). IFN-γ is a characteristic cytokine of the Th1 response (33) and signals largely via signal transducer and activator of transcription 1 (STAT1). This pathway is activated when IFN-γ binds to its receptor α subunit, inducing receptor assembly from two α and two β subunits. Next, Janus kinases 1 and 2 (Jak1 and -2), which are constitutively associated with the α and β subunits, respectively, phosphorylate each other and a functionally critical tyrosine (Y440) residue on the α receptor chains to form a paired set of STAT1 docking sites. This allows two STAT1 molecules to dock and become phosphorylated on a tyrosine (Y701) residue (4). The phosphorylated STAT1 molecules are released and form homodimers that translocate to the nucleus and bind specific DNA sequences to modulate gene transcription (11).
H. pylori can modulate the host immune response; for example, VacA impairs antigen presentation (37), H. pylori infection induces T-cell death through Fas-FasL interactions (60), and H. pylori arginase inhibits macrophage nitric oxide production (25). Since H. pylori elicits and yet evades a Th1 response (5) and since gastric epithelium may play a role in innate and adaptive immunity (14), the aim of this study was to determine whether H. pylori infection disrupted IFN-γ-STAT1 signaling in human epithelial cells.
MATERIALS AND METHODS
Eukaryotic cell culture.
The transformed gastric epithelial cell line MKN45 (58) was cultured in RPMI 1640 medium supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS) and 2.5% penicillin-streptomycin. The transformed human gastric epithelial cell line AGS (American Type Culture Collection [ATCC], Manassas, Va.; CRL-1739) (31) was cultured in Ham's F-12 medium supplemented with 10% FBS, 0.1% sodium bicarbonate, and 1% penicillin-streptomycin. The transformed human laryngeal epithelial cell line HEp-2 (ATCC; CCL-23) (8, 19) was cultured in minimum essential medium supplemented with 10% FBS, 1% sodium bicarbonate, and 1% penicillin-streptomycin (all media and supplements were obtained from Life Technology, GIBCO BRL, Grand Island, N.Y.). Cells were maintained in 75-cm2 tissue culture flasks (Corning Inc., Corning, N.Y.) at 37°C in 5% CO2 and were seeded onto 60-mm-diameter petri dishes (Falcon) for whole-cell or nuclear protein extracts. Prior to bacterial infection and subsequent protein or RNA extraction, cells were incubated in antibiotic-free, reduced-serum (1% FBS) medium overnight at 37°C in 5% CO2.
Bacterial growth and conditions of infection.
The H. pylori strains employed in this study included the clinical isolates LC11 (cagA+ cagE+ VacA+), LC20 (cagA cagE VacA−), and LC14 (cagA+ cagE VacA−) (12, 35); strain 60190 (ATCC 49503) (cagA+ cagE+ VacA+) (32); and the murine-adapted strain SS1 (cagA+ cagE+ VacA−) (13). Bacteria were cultured on Columbia agar plates containing 5% sheep blood and incubated at 37°C under microaerophilic conditions (5% oxygen, 85% nitrogen, and 10% carbon dioxide) for 72 h. The bacteria were then inoculated into brucella broth supplemented with 10% heat-inactivated FBS and grown overnight with shaking (120 rpm) under microaerophilic conditions at 37°C. One milliliter of bacteria from overnight growth in broth was pelleted, washed, and resuspended in phosphate-buffered saline (PBS) to measure growth spectrophotometrically at 550 nm, where an optical density of 1.0 = 2 × 108 bacteria/ml. Prior to infection, bacteria were washed and resuspended in a total volume of 0.05 ml of antibiotic-free tissue culture medium. Epithelial cells were then infected at a multiplicity of infection (MOI) of 100 bacteria to 1 eukaryotic cell for various time periods. After infection, the cell monolayer was rinsed and medium alone or medium containing IFN-γ was added to the remaining adherent cells. Viability of bacteria at the conclusion of experiments was confirmed by visualizing their motility under light microscopy and by microaerobic culture on Columbia blood agar plates. In some experiments H. pylori was heat killed by boiling for 15 min in a sealed sterile tube. Lack of growth on Columbia blood agar plates confirmed heat killing.
Immunoblotting. (i) Whole-cell protein extraction.
Epithelial cell monolayers were washed three times with PBS at 4°C. The remaining adherent cells were scraped with a rubber policeman into 1 ml of PBS and pelleted by centrifugation at 12,000 rpm for 10 s (Microfuge 18; Beckman Coulter). The cell pellet was resuspended in radioimmunoprecipitation assay buffer (1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS] in PBS) supplemented with 50 mM NaF, 150 mM NaCl, 1 mM Na3VO4, 20 μg of phenylmethylsulfonyl fluoride per ml, 15 μg of aprotonin per ml, 2 μg of pepstatin A per ml, and 2 μg of leupeptin per ml (all from Sigma Chemical Co., St. Louis, Mo.) by vortexing. The volume of radioimmunoprecipitation assay buffer was 0.15 ml for MKN45 cells, 0.1 ml for HEp-2 cells, and 0.05 ml for AGS cells. The cells were then left on ice at 4°C for 30 min, followed by centrifugation at 12,000 rpm for 10 min at 4°C (Sorvall SS34 rotor and Sorvall RC-5B centrifuge). Supernatants were stored at −70°C for use as whole-cell protein extracts (10).
(ii) SDS-polyacrylamide gel electrophoresis.
Equal volumes of whole-cell protein extracts were mixed with 2× SDS-polyacrylamide gel electrophoresis loading buffer, boiled for 5 min, and then electrophoresed through a 7.5% Tris-HCl polyacrylamide gel (Bio-Rad) (90 min, 111 V, room temperature). Next, proteins were electrophoretically transferred onto a nitrocellulose membrane (BioTrace NT; Pall Corp., Ann Arbor, Mich.) at 100 V for 60 min at 4 οC and then blocked with 5% milk-0.05% Tween 20 in Tris-buffered saline (TBST-M) for 30 min at room temperature. The membranes were then probed with either polyclonal rabbit anti-human phosphotyrosine (Y701) STAT1 (1:1,000) (Upstate Biotechnology, Lake Placid, N.Y.) or polyclonal rabbit anti-human p84/p91 STAT1 (1:4,000) (Santa Cruz Biotechnology, Santa Cruz, Calif.) in 10 ml of TBST-M at 4οC overnight with shaking. Membranes were then washed three times with TBST, rinsed with distilled water, and incubated with horseradish peroxidase-conjugated donkey anti-rabbit antibody (1:1,000 to 1:4,000) (Santa Cruz) in 10 ml of TBST-M at room temperature for 1 h. Following TBST washes and rinsing in water, membranes were incubated with enhanced chemiluminescence (Western blotting luminol reagent; Santa Cruz) and then developed with Biomax MR film (Kodak, Rochester, N.Y.).
Electrophoretic mobility shift assay (EMSA). (i) Nuclear protein extraction.
Adherent epithelial cells were washed with cold PBS three times, harvested with a rubber policeman in 1 ml of PBS, and pelleted (12,000 rpm, 10 s; Beckman Coulter Microfuge 18). Nuclear proteins were collected by the method of Andrews and Faller (2) with the addition of 10 μg of aprotonin per ml, 2 μg of pepstatin per ml, 2 μg of leupeptin per ml, and 0.5 mM phenylmethylsulfonyl fluoride (all from Sigma) to the extraction buffer (8). The protein concentration of each sample was determined by using the Bio-Rad assay, and extracts were stored at −70°C.
(ii) EMSA.
EMSA was performed as previously described (7). Briefly, binding buffer containing 250 mM Tris-Cl (pH 7.5), 40 mM NaCl, 10 mM EDTA, 2.5 mM dithiothreitol, 10 mM spermidine, 25% glycerol, 1 μg of poly(dI-dC) per μl and 5 μg of calf thymus DNA per μl was mixed with 15 μg of nuclear protein extract. Samples were incubated for 15 min on ice, and approximately 3 × 105 cpm of [α-32P]dCTP end-labeled double-stranded oligonucleotides bearing the STAT1 and STAT3 binding sites (59) was then added to each sample and incubated for a further 20 min at room temperature. Indicator dye (2 μl of 0.25% [wt/vol] bromophenol blue and 5% [wt/vol] glycerol) was added to each sample, which was then electrophoresed through a 5% polyacrylamide gel at 100 V for 3 h at room temperature. Gels were then dried and visualized by autoradiography with Kodak Biomax MR film. Controls included nonradiolabeled double-stranded oligonucleotides as a cold competitor (25 times molar excess) and polyclonal rabbit anti-human p84/p91 STAT1 antibody for supershift, while both an irrelevant DNA probe (STAT6) and an isotype-matched irrelevant polyclonal rabbit anti-human STAT5 antibody (Santa Cruz) served as negative controls.
RNA extraction and reverse transcription-PCR (RT-PCR).
Total RNA was extracted from cells by using the Trizol reagent (Life Technology, Gibco) according to the manufacturer's instructions. Reverse transcription was carried out with 5 ng of total RNA and the oligo(dT) random primers. cDNA products were amplified by using specific primer pairs for interferon regulatory factor 1 (IRF-1) (Primer3 software [http://www-genome.wi.mit.edu/genome_software/other/primer3.html) (IRF-1 sense, 5′-CGA TAC AAA GCA GGG GAA AA-3′; IRF-1 antisense, 5′-TAG CTG CTG TGG TCA TCA GG-3′) and human β-actin (3) (β-actin sense, 5′-GTG GGG CGC CCC AGG CAC CA-3′; β-actin antisense, 5′-CTC CTT AAT GTC ACG CAC GAT TTC-3′). PCR products were resolved by agarose (2%) gel electrophoresis and visualized with UV light after staining with ethidium bromide.
Collection and use of bacterial culture supernatants and conditioned medium.
H. pylori strain LC11 grown in brucella broth overnight was used to inoculate antibiotic-free MKN45 cell medium containing 1% FBS with the same number of bacteria used to infect cells at a MOI of 100:1. After 6 h, bacteria were pelleted (3,000 rpm, 5 min; Beckman GPR centrifuge), and the supernatant was passed twice through 0.2-μm-pore-size sterile syringe filters (Pall) and stored at −70°C until use as bacterial culture supernatants (i.e., containing secreted bacterial factors). Lack of growth on Columbia blood agar plates was used to determine that culture supernatants were sterile. Control culture supernatant was produced by similarly incubating and processing uninfected culture medium. Conditioned medium was obtained from either uninfected or infected (MOI, 100:1; 6 h) confluent MKN45 cell monolayers and centrifuged (3,000 rpm, 5 min; Beckman GPR centrifuge), and the supernatant (i.e., containing secreted bacterial and/or epithelial factors) was passed twice through 0.2-μm-pore-size sterile syringe filters (Pall) and stored at −70°C until use. Confluent MKN45 cell monolayers were then serum starved overnight, incubated with either culture supernatant or conditioned medium (50 to 100% [vol/vol], 6 h), and subsequently stimulated with either medium only or IFN-γ (1 ng/ml, 30 min) before whole-cell protein extraction.
Immunofluorescence.
HEp-2 cells (104) were seeded onto four-well slide chambers (Nalge Nunc International, Naperville, Ill.) and cultured in antibiotic-free, serum-starved (1% FBS) medium overnight prior to infection with H. pylori (MOI, 100:1; 24 h). HEp-2 cells were then washed twice and incubated with sterile medium or IFN-γ (5 ng/ml, 30 min). The cells were then washed three times with cold PBS, fixed in 10% paraformaldehyde for 20 min, rinsed with PBS, and permeabilized with 0.1% Triton X-100 in PBS for 4 min. Nonspecific binding was blocked with 2% bovine serum albumin in 0.1% Triton X-100-PBS for 30 min, followed by incubation with polyclonal p84/p91 STAT1 antibody (1:250 dilution) in 2% BSA-0.1% Triton X-100-PBS at 4°C overnight. After rinsing with PBS, STAT1 was visualized by treatment with rhodamine-conjugated goat anti-rabbit secondary antibody (1:250 dilution) (Jackson ImmunoResearch, West Grove, Pa.) for 60 min at room temperature. Cells were rinsed with PBS, Vectashield (Vector Labs, Burlingame, Calif.) mounting medium for fluorescence was added, and slides were sealed with coverslips and examined under fluorescence microscopy (Dialux 22; Leica Canada, Willowdale, Ontario, Canada) (8).
RESULTS
H. pylori infection decreases levels of IFN-γ-induced pYSTAT1.
Whole-cell protein extracts from MKN45 gastric epithelial cells analyzed by immunoblotting demonstrated low constitutive levels of tyrosine-phosphorylated STAT1 (pYSTAT1) (Fig. 1), whereas stimulation with IFN-γ increased pYSTAT1 in a dose-dependent manner, with a maximal response at between 1 and 5 ng/ml. Thus, a dose of 1 ng of IFN-γ per ml was chosen for our further experiments with MKN45 cells.
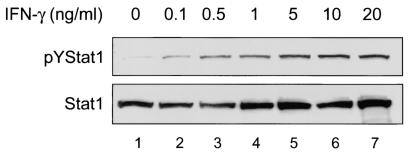
FIG. 1.
IFN-γ activates STAT1 tyrosine phosphorylation in a dose-dependent manner in MKN45 cells. Confluent MKN45 cells were serum starved overnight and stimulated with different doses of IFN-γ for 30 min, and whole-cell protein extracts analyzed by immunoblotting. (Upper panel) Low levels of constitutive pYSTAT1 were present in untreated MKN45 cells (lane 1). Treatment with IFN-γ increased the level of pYSTAT1 in a dose-dependent manner (lanes 2 to 7). (Lower panel) Nonphosphorylated STAT1 levels are similar between samples (n = 2 or 3).
Infection with H. pylori strain LC11 (MOI, 100:1) alone did not increase pYSTAT1 after 2, 4, or 6 h of infection. However, H. pylori infection diminished both constitutive and IFN-γ induced pYSTAT1 after 4 and 6 h, but not 2 h, of infection, indicating a time-dependent mechanism (Fig. 2, upper panel). This effect was not due to decreased expression of nonphosphorylated STAT1 (Fig. 2, lower panel).
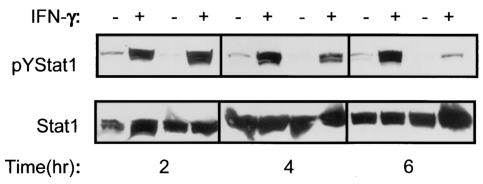
FIG. 2.
H. pylori infection decreases IFN-γ-induced tyrosine phosphorylation of STAT1 in a time-dependent manner. Confluent MKN45 cells were serum starved overnight, infected with H. pylori strain LC11 (MOI, 100:1) for different time periods, rinsed, and stimulated with IFN-γ (1 ng/ml, 30 min), and whole-cell protein extracts were analyzed by immunoblotting. (Upper panels) MKN45 cells displayed low levels of constitutive pYSTAT1. Infection with H. pylori strain LC11 (MOI, 100:1) did not itself increase levels of pYSTAT1 but did decrease the levels of constitutive pYSTAT1 and prevent the IFN-γ-induced increase in pYSTAT1 by a time-dependent mechanism. The effect was absent at 2 h, apparent at 4 h, and well established at 6 h. (Lower panel) Nonphosphorylated STAT1 levels are comparable between samples (n = 1 to 3).
The suppressive effect was reproducible in different epithelial cell lines and with strains of H. pylori differentially expressing CagA, CagE, and VacA. Infection of MKN45 cells with either a type 1 H. pylori strain (LC11) or a type 2 H. pylori strain (LC20) (MOI, 100:1; 6 h) did not increase pYSTAT1 but prevented the IFN-γ-induced (1 ng/ml, 30 min) increase in pYSTAT1 (Fig. 3A). Inhibition of STAT1 signaling was also observed in HEp-2 cells infected with strain LC11, LC20 (Fig. 3B), or LC14 and the mouse-adapted strain SS1 and in MKN45 cells infected with strain 60190 (data not shown). Gastric AGS epithelial cells infected with H. pylori strain LC11 or LC20 yielded equivalent findings (Fig. 3C). These results indicate that suppression of STAT1 signaling was independent of the cag PAI and VacA production and was reproducible in different epithelial cell lines.
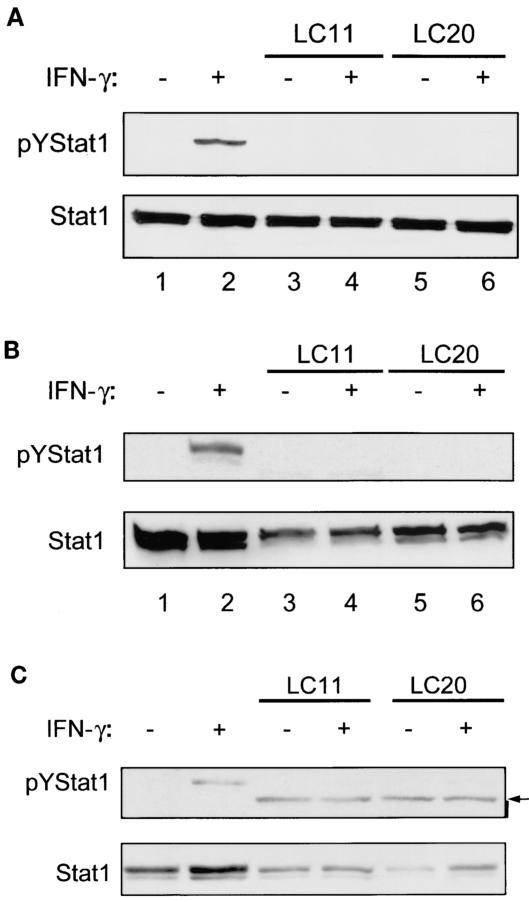
FIG. 3.
H. pylori infection decreases IFN-γ-induced tyrosine phosphorylation of STAT1 in multiple epithelial cell lines. Epithelial cell lines were grown until confluent, serum starved overnight, infected with different H. pylori strains, rinsed, and stimulated with IFN-γ. Whole-cell protein extracts were analyzed by immunoblotting. (A) MKN45 cells. Upper panel, MKN45 cells showed low levels of constitutive pYSTAT1 (lane 1). IFN-γ stimulation (1 ng/ml, 30 min) increased pYSTAT1 (lane 2). Infection of MKN45 cells with H. pylori strains LC11 and LC20 (MOI, 100:1; 6 h) followed by treatment with medium alone did not cause an increase in pYSTAT1 (lanes 3 and 5). However, infection with both strains prevented the IFN-γ-induced increase in pYSTAT1 (lanes 4 and 6 versus lane 2). Lower panel, nonphosphorylated STAT1 levels are similar between samples (n = 3). (B) HEp-2 cells. Upper panel, HEp-2 cells displayed increased pYSTAT1 after IFN-γ stimulation (20 ng/ml, 30 min) (lane 2 [upper band, STAT1 alpha isoform; lower band, STAT1 beta isoform]). Infection with H. pylori strain LC11 or LC20 (MOI, 100:1; 24 h) followed by treatment with medium alone did not increase pYSTAT1 (lane 3 and 5), but infection did prevent the IFN-γ-induced increase in pYSTAT1 (lanes 4 and 6 versus lane 2). Lower panel, nonphosphorylated STAT1 levels are comparable between samples (n = 3). (C) AGS cells. Upper panel, AGS cells lack constitutive pYSTAT1 (lane 1). IFN-γ stimulation (5 ng/ml, 30 min) increased pYSTAT1 (lane 2). Infection with H. pylori strain LC11 or LC20 (MOI, 100:1; 12 h) followed by treatment with medium alone did not increase pYSTAT1 (lanes 3 and 5). Infection with both strains prevented the IFN-γ-induced increase of pYSTAT1 (lanes 4 and 6 versus lane 2). The arrow shows a nonspecific band. Lower panel, nonphosphorylated STAT1 levels are comparable between samples (n = 3).
H. pylori infection decreases IFN-γ-induced STAT1 DNA binding.
Nuclear protein extracts from uninfected and unstimulated MKN45 cells demonstrated constitutive DNA binding to STAT1 and STAT3 homodimers and STAT1-STAT3 heterodimers. IFN-γ stimulation (1 ng/ml for 30 min) increased DNA binding to STAT1 (Fig. 4A, lanes 1 and 2). The controls confirmed the specificity of the band as STAT1 by supershift of the IFN-γ-induced STAT1 homodimer band by anti-STAT1 antibody but not by an anti-STAT5 antibody (Fig. 4A, lanes 7 and 8). Abolition of the STAT1 homodimer band following preincubation with a nonradiolabeled STAT1 DNA probe (cold competitor) but not with a STAT6 cold competitor also demonstrated the specificity of the probe (Fig. 4A, lanes 9 and 10).
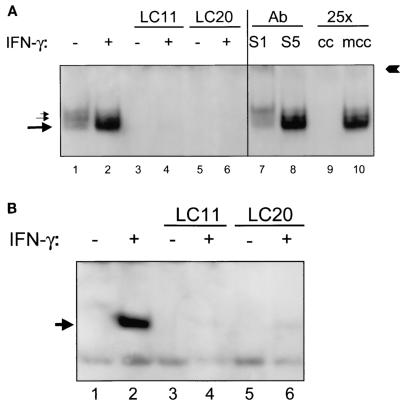
FIG. 4.
H. pylori decreases IFN-γ-induced DNA binding to STAT1 in epithelial cells. Confluent MKN45 and HEp-2 cells were serum starved overnight, infected with H. pylori strains LC11 and LC20, rinsed, and stimulated with IFN-γ. Equal amounts of nuclear protein extracts were then analyzed by EMSA. (A) MKN45 cells exhibited constitutive DNA binding to STAT1 (lower left arrow) and STAT3 (upper left arrow) homodimers and to STAT1-STAT3 (middle left arrow) heterodimers (lane 1). DNA binding to STAT1 homodimers was increased following stimulation with IFN-γ (1 ng/ml, 30 min) (lane 2). Infection with H. pylori strains LC11 and LC20 (MOI, 100:1; 6 h) decreased constitutive (lanes 3 and 5) and IFN-γ induced (lanes 4 and 6) DNA binding to STAT1 (n = 3). The identity of the band as STAT1 was confirmed by supershift of the band with a STAT1 (S1) antibody (Ab) (right arrow) but not with a STAT5 (S5) antibody. The specificity of the radiolabeled double-stranded DNA oligonucleotide was confirmed by competition of the band with a nonradiolabeled STAT1 (25× molar excess) cold competitor (cc) but not with a STAT6 cold competitor (mcc). (B) HEp-2 cells demonstrated inducible STAT1 DNA binding in response to stimulation with IFN-γ (20 ng/ml, 30 min) (arrow, lane 2). Infection with H. pylori strains LC11 and LC20 did not induce STAT1 DNA binding (lanes 3 and 5). IFN-γ-induced DNA binding to STAT1 was decreased by infection with LC11 and LC20 (lanes 4 and 6) (n = 3).
Infection with H. pylori strain LC11 or LC20 (MOI, 100:1; 6 h) alone did not activate DNA binding to STAT1. However, in cells first infected with H. pylori strain LC11 or LC20 and then treated with 1 ng of IFN-γ per ml for 30 min, DNA binding to STAT1 was completely abolished (Fig. 4A, lanes 4 and 6).
Nuclear protein extracts from uninfected and unstimulated HEp-2 cells had no constitutive DNA binding to STAT1 (Fig. 4B, lane 1). IFN-γ stimulation increased DNA binding to STAT1 (Fig. 4B, lane 2). Infection with H. pylori strain LC11 or LC20 inhibited IFN-γ-induced STAT1 DNA binding (Fig. 4B, lanes 4 and 6). Controls confirmed the specificity of the band and the probe (data not shown).
H. pylori inhibits IFN-γ-induced nuclear translocation of STAT1.
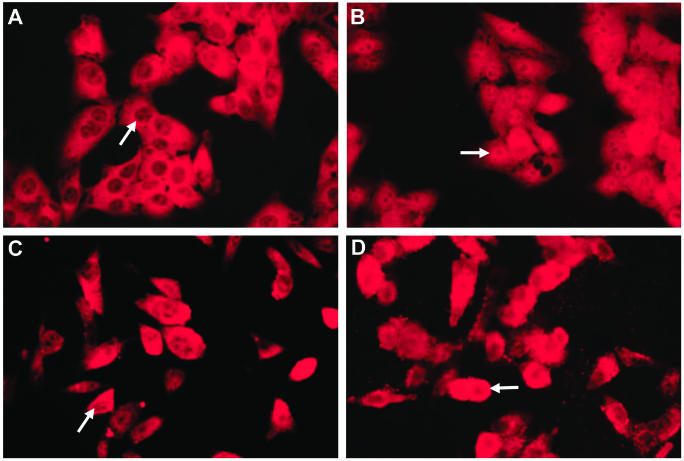
Immunofluorescence analysis of HEp-2 cells was employed to determine the subcellular localization of STAT1 after H. pylori infection. In uninfected HEp-2 cells, STAT1 was distributed throughout the cytoplasm (Fig. 5A) and translocated to the nucleus following IFN-γ stimulation (5 ng/ml, 30 min) (Fig. 5B). Infection with H. pylori strain LC11 (Fig. 5C) alone did not cause redistribution of STAT1, whereas H. pylori infection prevented the IFN-γ-induced nuclear translocation of STAT1 (Fig. 5D). Inhibition of STAT1 nuclear translocation was comparable following infection with H. pylori strains LC20, LC14, and SS1 (data not shown), indicating that the effect was independent of the cag PAI and VacA production.
FIG. 5.
H. pylori inhibits IFN-γ-induced nuclear translocation of STAT1. Arrows indicate nuclei in representative cells for each treatment condition. (A) STAT1 was distributed evenly throughout the cytoplasm but not the nuclei of unstimulated cells. (B) IFN-γ stimulation (5 ng/ml, 30 min) induced translocation of STAT1 into the nucleus. (C) Infection with H. pylori strain LC11 (MOI, 100:1; 24 h) did not affect the distribution of STAT1. (D) Infection with H. pylori strain LC11 followed by IFN-γ stimulation (5 ng/ml, 30 min) inhibited IFN-γ-induced STAT1 nuclear translocation. Approximate magnification, ×400; n = 3.
H. pylori inhibits STAT1-dependent gene expression.
IRF-1 is a gene induced by IFN-γ in a STAT1-dependent manner (48). HEp-2 cells showed constitutive expression of IRF-1 mRNA, as assessed by RT-PCR. Peak IRF-1 mRNA expression occurred after 3 h of IFN-γ stimulation (20 ng/ml). Infection with either H. pylori strain LC11 or strain LC20 alone for 24 h did not increase expression of IRF-1 mRNA. However, the increase in IRF-1 mRNA levels following IFN-γ stimulation was inhibited in H. pylori-infected epithelial cells (Fig. 6). These results indicate that H. pylori infection inhibited IFN-γ-induced gene transcription of a STAT1-dependent gene.
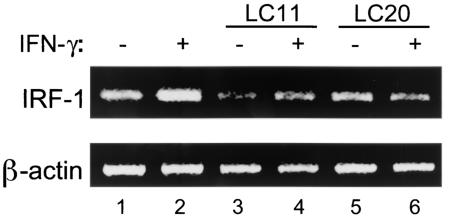
FIG. 6.
H. pylori infection inhibits transcription of the IFN-γ-induced, STAT1-dependent IRF-1 gene. HEp-2 cells display constitutive expression of IRF-1 mRNA (lane 1) that is increased by IFN-γ stimulation (20 ng/ml for 3 h) (lane 2). Infection with H. pylori strain LC11 or LC20 (MOI, 100:1; 24 h) decreased constitutive IRF-1 mRNA levels (lanes 3 and 5 versus lane 1). The increase in expression of IRF-1 mRNA following IFN-γ stimulation was inhibited following H. pylori infection (lanes 4 and 6 versus lane 2). The β-actin gene was employed as a housekeeping gene (LC11, n = 2; LC20, n = 1).
Decrease in pYSTAT1 requires infection with viable H. pylori.
MKN45 cells infected with live H. pylori strain LC11 (MOI, 100:1; 6 h) showed decreased levels of constitutive and IFN-γ-induced pYSTAT1, but this did not occur following incubation with heat-killed H. pylori strain LC11 (Fig. 7A). Similarly, AGS cells infected with heat-killed H. pylori strain LC11 or LC20 had levels of IFN-γ-induced pYSTAT1 comparable to those in uninfected IFN-γ-treated cells (Fig. 7B). These findings indicate that a heat-labile, lipopolysaccharide-independent H. pylori-derived factor is required to achieve the inhibitory effect on STAT1 signaling.
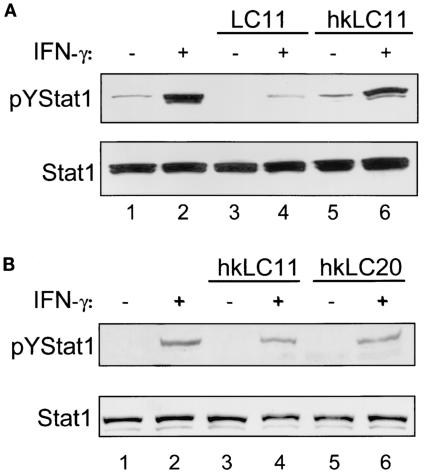
FIG. 7.
Heat-killed H. pylori does not decrease IFN-γ-induced pYSTAT1. H. pylori was heat killed by boiling for 15 min and incubated with (MOI, 100:1) confluent MKN45 and AGS cells after serum starvation overnight. The cells were then rinsed and stimulated with IFN-γ. Whole-cell protein extracts were analyzed by immunoblotting. (A) MKN45 cells. Upper panel, MKN45 cells incubated for 6 h with H. pylori strain LC11 exhibited decreased levels of constitutive and IFN-γ-stimulated (1 ng/ml, 30 min) pYSTAT1 (lanes 3 and 4) compared to uninfected controls (lanes 1 and 2). Incubation with heat-killed LC11 (hkLC11) did not decrease levels of pYSTAT1 (lanes 5 and 6) compared to uninfected controls. Lower panel, nonphosphorylated STAT1 levels are similar between samples (n = 3). (B) AGS cells. Upper panel, AGS cells incubated for 12 h with heat-killed H. pylori strains LC11 (lanes 3 and 4) and LC20 (lanes 5 and 6) showed no reduction of IFN-γ-induced pYSTAT1 compared to noninfected cells (lanes 1 and 2). Lower panel, nonphosphorylated STAT1 levels are similar between samples.
Inhibition of IFN-γ-induced STAT1 activation is independent of a soluble factor.
To determine if a secreted bacterial factor was responsible for disrupting STAT1 activation, MKN45 cells were incubated with culture supernatant prepared from H. pylori strain LC11 (100%, 6 h) followed by stimulation with IFN-γ (1 ng/ml, 30 min). Figure 8A shows that culture supernatant did not inhibit STAT1 activation, suggesting that a soluble bacterial factor is unlikely to mediate the disruption of STAT1 signaling.
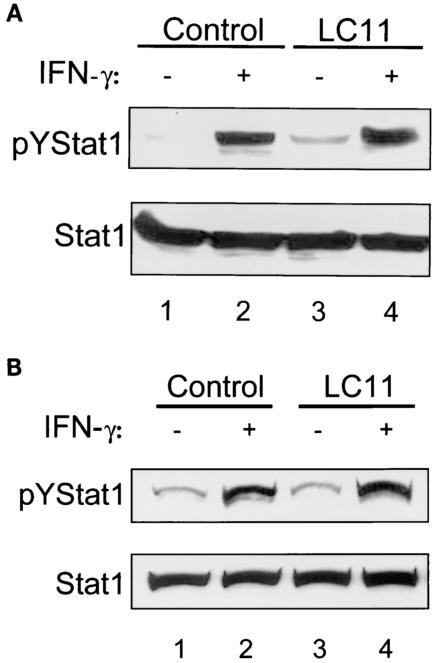
FIG. 8.
Neither H. pylori culture supernatant nor conditioned medium decreases levels of IFN-γ-induced pYSTAT1. Confluent MKN45 cells were serum starved overnight and incubated for 6 h with either H. pylori strain LC11 culture supernatant (A) or conditioned medium from MKN45 cells infected with H pylori strain LC11 (B). Monolayers were then rinsed and stimulated with IFN-γ (1 ng/ml, 30 min), and whole-cell protein was extracted and analyzed by immunoblotting. Upper panels, MKN45 cells incubated with culture supernatant or conditioned medium did not change the levels of constitutive or IFN-γ induced pYSTAT1 compared to those incubated with control culture supernatant or conditioned medium. Lower panels, nonphosphorylated STAT1 levels are comparable between samples (n = 3).
To determine whether a soluble factor produced as a result of interactions between bacteria and epithelial cells was involved, MKN45 cells were incubated with bacterium-free conditioned medium (100%, 6 h) collected from MKN45 cells with or without H. pylori infection (MOI, 100:1; 6 h). Figure 8B shows that conditioned medium did not inhibit STAT1 activation, indicating that a soluble factor released after bacterium-epithelial cell interaction is unlikely to explain the disruption of STAT1 signaling. Taken together with the lack of inhibition with heat-killed H. pylori, these results suggest that host epithelial cell interactions with viable bacteria are required to inhibit IFN-γ STAT1 signaling.
DISCUSSION
This study demonstrates the activation of IFN-γ-STAT1 signaling in human epithelial cell lines (gastric MKN45 and AGS cells and laryngeal HEp-2 cells) by using complementary techniques, including immunoblotting, EMSA, immunofluorescence, and RT-PCR. Furthermore, we determined, for the first time, that H. pylori infection inhibits this signaling pathway in epithelial cells. Inhibition of STAT1 activation was time dependent, was independent of a soluble bacterial or epithelial factor, and required eukaryotic cell interaction with live bacteria. This disruption may represent a bacterial adaptation to modulate the host gastric mucosal immune response to promote bacterial survival.
Studies of knockout mice confirm the importance of IFN-γ-STAT1 signaling in the host response to microbial infections. For example, studies with both IFN-γ receptor and STAT1 gene knockout mice show that IFN-γ-mediated protection is important for the control of infection with microbial pathogens such as Mycobacterium species (57), Chlamydia pneumoniae (44), and Listeria monocytogenes (36). Furthermore, humans with IFN-γ receptor α or β chain deficiencies are particularly vulnerable to disseminated mycobacterial infections (15). Two infants with a homozygous STAT1 mutation developed disseminated infection after Mycobacterium bovis bacillus Calmette-Guérin (BCG) vaccination (16). Thus, alteration of the IFN-γ response in the host significantly affects the outcome of microbial infections.
H. pylori infection of IFN-γ knockout mice results in reduced gastric inflammation and increased H. pylori colonization in comparison with wild-type mice. Also, a more diverse array of H. pylori strains are able to colonize IFN-γ knockout mice compared with wild-type littermates (45, 49). Furthermore, H. pylori-infected IFN-γ knockout mice show reduced expression of proinflammatory mediators such as macrophage inflammatory protein 2, the mouse homologue of human IL-8, and the inducible form of nitric oxide synthase (iNOS) in comparison to controls (38). These studies indicate that inhibition of IFN-γ-STAT1 signaling in response to H. pylori infection may result in increased bacterial colonization while decreasing mucosal inflammation.
IFN-γ stimulates Jak1 and -2-dependent phosphorylation of tyrosine residues on the cytoplasmic tail of the IFN-γ receptor α chain, which allows docking of monomeric STAT1 protein and tyrosine phosphorylation by Jaks, with subsequent STAT1 dimerization and translocation to the nucleus to bind DNA and modulate transcription (4). In the present study, disruption of this signaling pathway in epithelial cells by H. pylori was demonstrated at multiple levels. First, immunoblotting showed decreased levels of pYSTAT1 without a decrease in expression of nonphosphorylated STAT1. As tyrosine phosphorylation is a prerequisite for nuclear translocation and DNA binding of STAT1, nuclear protein extracts were next examined by EMSA, and decreased IFN-γ-induced STAT1 DNA binding was observed. Fluorescence microscopy studies corroborated this finding by showing inhibition of STAT1 translocation to the nucleus following H. pylori infection. A functional consequence of STAT1 inhibition was confirmed by using RT-PCR to show decreased transcription of IRF-1 mRNA following H. pylori infection.
Inhibition of the IFN-γ-STAT1 pathway is also observed in macrophages infected with L. monocytogenes (51) and in epithelial cells infected with enterohemorrhagic Escherichia coli O157:H7 but not enteropathogenic E. coli (8). Possible mechanisms of inhibition include decreased expression of signaling cascade components, such as the decrease in Jak1 and Jak2 expression caused by membrane vesicles of Porphyromonas gingivalis (50), or the induction of inhibitory proteins, such as suppressor-of-cytokine-signaling proteins (51).
The gut epithelium plays an important role in both innate and adaptive immune responses to microbes (14). IFN-γ alters human epithelial cell function by increasing expression of major histocompatibility complex class II (20), intercellular adhesion molecule 1 (9), IRF-1 (43), and iNOS (22); therefore, the modulation of IFN-γ signaling by H. pylori infection likely has biological significance. This study demonstrates an inhibitory effect of a number of H. pylori strains on IFN-γ-STAT1 signaling in two gastric epithelial cell lines. However, other in vitro studies demonstrated that H. pylori infection and IFN-γ act synergistically, for example, to induce apoptosis (29) and increase major histocompatibility complex class II (20) and iNOS (34) expression. One possible explanation for this discrepancy is that in other models IFN-γ was introduced prior to or simultaneously with bacterial infection (20, 34) rather than, as in this study, after a period of infection. Whether the effect of H. pylori infection on IFN-γ-dependent cell functions in vivo is synergistic or inhibitory ultimately may also depend on the balance between STAT1 and non-STAT1 IFN-γ signaling (23, 42) and on the influence of other cytokines, such as tumor necrosis factor alpha and IL-1β, secreted by immune cells in the gastric mucosa (61).
Neither heat-killed H. pylori, culture supernatant, nor conditioned medium inhibited IFN-γ-STAT1 signaling, suggesting that direct interaction between bacteria and eukaryotic cells, independent of lipopolysaccharide, is required to inhibit signal transduction events. The inhibition occurs independent of the presence of the cag PAI, ruling out as a mechanism the translocation of prokaryotic effector molecules into epithelial cells via the bacterial type IV secretion system.
Adherence of H. pylori to host epithelial cells is a critical step in virulence (18). H. pylori binds to a variety of host molecules, including phosphatidylethanolamine (26), Lewisb antigen (53), major histocompatibility class II antigens (21), and Toll-like receptor 4 (52). However, it is not clear what events occur downstream of these interactions. On the bacterial side, genome sequencing has identified genes encoding a large family of outer membrane proteins (1). Functional studies have defined the roles of some of the outer membrane proteins; for example, BabA appears to be involved in mediating adherence (30). The product of the HP0638 gene (outer inflammatory protein [oipA] gene) induces an increased epithelial IL-8 response (62) and is expressed in both type1 and type 2 H. pylori, consistent with a cag PAI-independent factor. Our data, which demonstrate that a heat-labile and contact-dependent factor inhibits IFN-γ-STAT1 signaling, suggest that an outer membrane protein could be a candidate to consider. Future studies should define the role of H. pylori adhesins in the modulation of IFN-γ-STAT1 signaling.
In summary, this study demonstrates, for the first time, that H. pylori infection disrupts IFN-γ-induced STAT1 signaling in host epithelial cells. Such disruption of a key signal transduction cascade could represent bacterial adaptation for modulating the host mucosal immune response to promote bacterial survival in the stomach.
Acknowledgments
D.J.M. and H.Q.H. contributed equally to the planning and execution of this work and the preparation of this paper.
This research was supported by operating grants from the Canadian Institutes of Health Research (CIHR). D.J.M. and H.Q.H. were recipients of CIHR/University Industry (Canadian Association of Gastroenterology/AstraZeneca) fellowships. P.J.M.C. is the recipient of a CIHR/Canadian Digestive Health Foundation doctoral award and is also a Cell Signals CIHR Strategic Training Fellow (STP-53877). N.L.J. is the recipient of an American Digestive Health Foundation Research Scholar Award. P.M.S. holds a Canada Research Chair in Gastrointestinal Disease.
We thank J. L. Watson and D. M. McKay (McMaster University), who provided the primer sequence for IRF-1 mRNA and control IRF-1 PCR product for IRF-1 RT-PCR.
Editor: B. B. Finlay
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai, Y., Y. Ohyama, K. Gen, and T. Ogawa. 2001. Bacterial fimbriae and their peptides activate human gingival epithelial cells through Toll-like receptor 2. Infect. Immun. 69:7387-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bach, E. A., M. Aguet, and R. D. Schreiber. 1997. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 15:563-591. [DOI] [PubMed] [Google Scholar]
- 5.Bamford, K. B., X. Fan, S. E. Crowe, J. F. Leary, W. K. Gourley, G. K. Luthra, E. G. Brooks, D. Y. Graham, V. E. Reyes, and P. B. Ernst. 1998. Lymphocytes in the human gastric mucosa during Helicobacter pylori have a T helper cell 1 phenotype. Gastroenterology 114:482-492. [DOI] [PubMed] [Google Scholar]
- 6.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceponis, P. J., F. Botelho, C. D. Richards, and D. M. McKay. 2000. Interleukins 4 and 13 increase intestinal epithelial permeability by a phosphatidylinositol 3-kinase pathway. Lack of evidence for STAT 6 involvement. J. Biol. Chem. 275:29132-29137. [DOI] [PubMed] [Google Scholar]
- 8.Ceponis, P. J., D. M. McKay, J. C. Ching, P. Pereira, and P. M. Sherman. 2003. Enterohemorrhagic Escherichia coli O157:H7 disrupts Stat1-mediated gamma interferon signal transduction in epithelial cells. Infect. Immun. 71:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y. J., M. J. Holtzman, and C. C. Chen. 2002. Interferon-gamma-induced epithelial ICAM-1 expression and monocyte adhesion. Involvement of protein kinase C-dependent c-Src tyrosine kinase activation pathway. J. Biol. Chem. 277:7118-7126. [DOI] [PubMed] [Google Scholar]
- 10.Ching, J. C., N. L. Jones, P. J. Ceponis, M. A. Karmali, and P. M. Sherman. 2002. Escherichia coli Shiga-like toxins induce apoptosis and cleavage of poly(ADP-ribose) polymerase via in vitro activation of caspases. Infect. Immun. 70:4669-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 12.Day, A. S., N. L. Jones, J. T. Lynett, H. A. Jennings, C. A. Fallone, R. Beech, and P. M. Sherman. 2000. cagE is a virulence factor associated with Helicobacter pylori-induced duodenal ulceration in children. J. Infect. Dis. 181:1370-1375. [DOI] [PubMed] [Google Scholar]
- 13.Day, A. S., N. L. Jones, Z. Policova, H. A. Jennings, E. K. Yau, P. Shannon, A. W. Neumann, and P. M. Sherman. 2001. Characterization of virulence factors of mouse-adapted Helicobacter pylori strain SS1 and effects on gastric hydrophobicity. Dig. Dis. Sci. 46:1943-1951. [DOI] [PubMed] [Google Scholar]
- 14.Didierlaurent, A., J. C. Sirard, J. P. Kraehenbuhl, and M. R. Neutra. 2002. How the gut senses its content. Cell Microbiol. 4:61-72. [DOI] [PubMed] [Google Scholar]
- 15.Dupuis, S., R. Doffinger, C. Picard, C. Fieschi, F. Altare, E. Jouanguy, L. Abel, and J. L. Casanova. 2000. Human interferon-gamma-mediated immunity is a genetically controlled continuous trait that determines the outcome of mycobacterial invasion. Immunol. Rev. 178:129-137. [DOI] [PubMed] [Google Scholar]
- 16.Dupuis, S., E. Jouanguy, S. Al-Hajjar, C. Fieschi, I. Z. Al-Mohsen, S. Al-Jumaah, K. Yang, A. Chapgier, C. Eidenschenk, P. Eid, A. Al Ghonaium, H. Tufenkeji, H. Frayha, S. Al-Gazlan, H. Al-Rayes, R. D. Schreiber, I. Gresser, and J. L. Casanova. 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33:388-391. [DOI] [PubMed] [Google Scholar]
- 17.El-Omar, E. M., M. Carrington, W. H. Chow, K. E. McColl, J. H. Bream, H. A. Young, J. Herrera, J. Lissowska, C. C. Yuan, N. Rothman, G. Lanyon, M. Martin, J. F. Fraumeni, Jr., and C. S. Rabkin. 2000. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature 404:398-402. [DOI] [PubMed] [Google Scholar]
- 18.Evans, D. J., Jr., and D. G. Evans. 2000. Helicobacter pylori adhesins: review and perspectives. Helicobacter 5:183-195. [DOI] [PubMed] [Google Scholar]
- 19.Fahey, J. W., X. Haristoy, P. M. Dolan, T. W. Kensler, I. Scholtus, K. K. Stephenson, P. Talalay, and A. Lozniewski. 2002. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc. Natl. Acad. Sci. USA 99:7610-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan, X., S. E. Crowe, S. Behar, H. Gunasena, G. Ye, H. Haeberle, N. Van Houten, W. K. Gourley, P. B. Ernst, and V. E. Reyes. 1998. The effect of class II major histocompatibility complex expression on adherence of Helicobacter pylori and induction of apoptosis in gastric epithelial cells: a mechanism for T helper cell type 1-mediated damage. J. Exp. Med. 187:1659-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan, X., H. Gunasena, Z. Cheng, R. Espejo, S. E. Crowe, P. B. Ernst, and V. E. Reyes. 2000. Helicobacter pylori urease binds to class II MHC on gastric epithelial cells and induces their apoptosis. J. Immunol. 165:1918-1924. [DOI] [PubMed] [Google Scholar]
- 22.Fu, S., K. S. Ramanujam, A. Wong, G. T. Fantry, C. B. Drachenberg, S. P. James, S. J. Meltzer, and K. T. Wilson. 1999. Increased expression and cellular localization of inducible nitric oxide synthase and cyclooxygenase 2 in Helicobacter pylori gastritis. Gastroenterology 116:1319-1329. [DOI] [PubMed] [Google Scholar]
- 23.Gil, M. P., E. Bohn, A. K. O'Guin, C. V. Ramana, B. Levine, G. R. Stark, H. W. Virgin, and R. D. Schreiber. 2001. Biologic consequences of Stat1-independent IFN signaling. Proc. Natl. Acad. Sci. USA 98:6680-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glocker, E., C. Lange, A. Covacci, S. Bereswill, M. Kist, and H. L. Pahl. 1998. Proteins encoded by the cag pathogenicity island of Helicobacter pylori are required for NF-kB activation. Infect. Immun. 66:2346-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gobert, A. P., D. J. McGee, M. Akhtar, G. L. Mendz, J. C. Newton, Y. Cheng, H. L. Mobley, and K. T. Wilson. 2001. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc. Natl. Acad. Sci. USA 98:13844-13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gold, B. D., M. Huesca, P. M. Sherman, and C. A. Lingwood. 1993. Helicobacter mustelae and Helicobacter pylori bind to common lipid receptors in vitro. Infect. Immun. 61:2632-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haeberle, H. A., M. Kubin, K. B. Bamford, R. Garofalo, D. Y. Graham, F. El-Zaatari, R. Karttunen, S. E. Crowe, V. E. Reyes, and P. B. Ernst. 1997. Differential stimulation of interleukin-12 (IL-12) and IL-10 by live and killed Helicobacter pylori in vitro and association of IL-12 production with gamma interferon-producing T cells in the human gastric mucosa. Infect. Immun. 65:4229-4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hotchin, N. A., T. L. Cover, and N. Akhtar. 2000. Cell vacuolation induced by the VacA cytotoxin of Helicobacter pylori is regulated by the Rac1 GTPase. J. Biol. Chem. 275:14009-14012. [DOI] [PubMed] [Google Scholar]
- 29.Houghton, J., R. M. Korah, M. R. Condon, and K. H. Kim. 1999. Apoptosis in Helicobacter pylori-associated gastric and duodenal ulcer disease is mediated via the Fas antigen pathway. Dig. Dis. Sci. 44:465-478. [DOI] [PubMed] [Google Scholar]
- 30.Ilver, D., A. Arnqvist, J. Ogren, I. M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Boren. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373-377. [DOI] [PubMed] [Google Scholar]
- 31.Jones, N. L., A. S. Day, H. A. Jennings, and P. M. Sherman. 1999. Helicobacter pylori induces gastric epithelial cell apoptosis in association with increased Fas receptor expression. Infect. Immun. 67:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keates, S., A. C. Keates, M. Warny, R. M. Peek, Jr., P. G. Murray, and C. P. Kelly. 1999. Differential activation of mitogen-activated protein kinases in AGS gastric epithelial cells by cag+ and cag− Helicobacter pylori. J. Immunol. 163:5552-5559. [PubMed] [Google Scholar]
- 33.Kelso, A. 1995. Th1 and Th2 subsets: paradigms lost? Immunol. Today 16:374-379. [DOI] [PubMed] [Google Scholar]
- 34.Kim, J. M., J. S. Kim, H. C. Jung, I. S. Song, and C. Y. Kim. 2002. Up-regulation of inducible nitric oxide synthase and nitric oxide in Helicobacter pylori-infected human gastric epithelial cells: possible role of interferon-gamma in polarized nitric oxide secretion. Helicobacter 7:116-128. [DOI] [PubMed] [Google Scholar]
- 35.Loeb, M., P. Jayaratne, N. Jones, A. Sihoe, and P. Sherman. 1998. Lack of correlation between vacuolating cytotoxin activity, cagA gene in Helicobacter pylori, and peptic ulcer disease in children. Eur. J. Clin. Microbiol. Infect. Dis. 17:653-656. [DOI] [PubMed] [Google Scholar]
- 36.Meraz, M. A., J. M. White, K. C. Sheehan, E. A. Bach, S. J. Rodig, A. S. Dighe, D. H. Kaplan, J. K. Riley, A. C. Greenlund, D. Campbell, K. Carver-Moore, R. N. DuBois, R. Clark, M. Aguet, and R. D. Schreiber. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84:431-442. [DOI] [PubMed] [Google Scholar]
- 37.Molinari, M., M. Salio, C. Galli, N. Norais, R. Rappuoli, A. Lanzavecchia, and C. Montecucco. 1998. Selective inhibition of Ii-dependent antigen presentation by Helicobacter pylori toxin VacA. J. Exp. Med. 187:135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obonyo, M., D. G. Guiney, J. Harwood, J. Fierer, and S. P. Cole. 2002. Role of gamma interferon in Helicobacter pylori induction of inflammatory mediators during murine infection. Infect. Immun. 70:3295-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papini, E., B. Satin, C. Bucci, M. de Bernard, J. L. Telford, R. Manetti, R. Rappuoli, M. Zerial, and C. Montecucco. 1997. The small GTP binding protein rab7 is essential for cellular vacuolation induced by Helicobacter pylori cytotoxin. EMBO J. 16:15-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parsonnet, J., S. Hansen, L. Rodriguez, A. B. Gelb, R. A. Warnke, E. Jellum, N. Orentreich, J. H. Vogelman, and G. D. Friedman. 1994. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 330:1267-1271. [DOI] [PubMed] [Google Scholar]
- 41.Pounder, R. E., and D. Ng. 1995. The prevalence of Helicobacter pylori infection in different countries. Aliment. Pharmacol. Ther. 9(Suppl. 2):33-39. [PubMed] [Google Scholar]
- 42.Ramana, C. V., M. P. Gil, Y. Han, R. M. Ransohoff, R. D. Schreiber, and G. R. Stark. 2001. Stat1-independent regulation of gene expression in response to IFN-gamma. Proc. Natl. Acad. Sci. USA 98:6674-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reis, L. F., H. Harada, J. D. Wolchok, T. Taniguchi, and J. Vilcek. 1992. Critical role of a common transcription factor, IRF-1, in the regulation of IFN-beta and IFN-inducible genes. EMBO J. 11:185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rottenberg, M. E., A. Gigliotti Rothfuchs, D. Gigliotti, M. Ceausu, C. Une, V. Levitsky, and H. Wigzell. 2000. Regulation and role of IFN-gamma in the innate resistance to infection with Chlamydia pneumoniae. J. Immunol. 164:4812-4818. [DOI] [PubMed] [Google Scholar]
- 45.Sawai, N., M. Kita, T. Kodama, T. Tanahashi, Y. Yamaoka, Y. Tagawa, Y. Iwakura, and J. Imanishi. 1999. Role of gamma interferon in Helicobacter pylori-induced gastric inflammatory responses in a mouse model. Infect. Immun. 67:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segal, E. D., J. Cha, J. Lo, S. Falkow, and L. S. Tompkins. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA 96:14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seydel, A., E. Tasca, D. Berti, R. Rappuoli, G. Del Giudice, and C. Montecucco. 2002. Characterization and immunogenicity of the CagF protein of the cag pathogenicity island of Helicobacter pylori. Infect. Immun. 70:6468-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sims, S. H., Y. Cha, M. F. Romine, P. Q. Gao, K. Gottlieb, and A. B. Deisseroth. 1993. A novel interferon-inducible domain: structural and functional analysis of the human interferon regulatory factor 1 gene promoter. Mol. Cell. Biol. 13:690-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smythies, L. E., K. B. Waites, J. R. Lindsey, P. R. Harris, P. Ghiara, and P. D. Smith. 2000. Helicobacter pylori-induced mucosal inflammation is Th1 mediated and exacerbated in IL-4, but not IFN-gamma, gene-deficient mice. J. Immunol. 165:1022-1029. [DOI] [PubMed] [Google Scholar]
- 50.Srisatjaluk, R., G. J. Kotwal, L. A. Hunt, and D. E. Justus. 2002. Modulation of gamma interferon-induced major histocompatibility complex class II gene expression by Porphyromonas gingivalis membrane vesicles. Infect. Immun. 70:1185-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoiber, D., S. Stockinger, P. Steinlein, J. Kovarik, and T. Decker. 2001. Listeria monocytogenes modulates macrophage cytokine responses through STAT serine phosphorylation and the induction of suppressor of cytokine signaling 3. J. Immunol. 166:466-472. [DOI] [PubMed] [Google Scholar]
- 52.Su, B., P. J. Ceponis, S. Lebel, H. Huynh, and P. M. Sherman. 2003. Helicobacter pylori activates Toll-like receptor 4 expression in gastrointestinal epithelial cells. Infect. Immun. 71:3496-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su, B., P. M. Hellstrom, C. Rubio, J. Celik, M. Granstrom, and S. Normark. 1998. Type I Helicobacter pylori shows Lewis(b)-independent adherence to gastric cells requiring de novo protein synthesis in both host and bacteria. J. Infect. Dis. 178:1379-1390. [DOI] [PubMed] [Google Scholar]
- 54.Suerbaum, S., and P. Michetti. 2002. Helicobacter pylori infection. N. Engl. J. Med. 347:1175-1186. [DOI] [PubMed] [Google Scholar]
- 55.Thye, T., G. D. Burchard, M. Nilius, B. Muller-Myhsok, and R. D. Horstmann. 2003. Genomewide linkage analysis identifies polymorphism in the human interferon-gamma receptor affecting Helicobacter pylori infection. Am. J. Hum. Genet. 72:448-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uemura, N., S. Okamoto, S. Yamamoto, N. Matsumura, S. Yamaguchi, M. Yamakido, K. Taniyama, N. Sasaki, and R. J. Schlemper. 2001. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345:784-789. [DOI] [PubMed] [Google Scholar]
- 57.van den Broek, M. F., U. Muller, S. Huang, R. M. Zinkernagel, and M. Aguet. 1995. Immune defence in mice lacking type I and/or type II interferon receptors. Immunol. Rev. 148:5-18. [DOI] [PubMed] [Google Scholar]
- 58.Wada, A., K. Ogushi, T. Kimura, H. Hojo, N. Mori, S. Suzuki, A. Kumatori, M. Se, Y. Nakahara, M. Nakamura, J. Moss, and T. Hirayama. 2001. Helicobacter pylori-mediated transcriptional regulation of the human beta-defensin 2 gene requires NF-kappaB. Cell Microbiol. 3:115-123. [DOI] [PubMed] [Google Scholar]
- 59.Wagner, B. J., T. E. Hayes, C. J. Hoban, and B. H. Cochran. 1990. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 9:4477-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, J., E. G. Brooks, K. B. Bamford, T. L. Denning, J. Pappo, and P. B. Ernst. 2001. Negative selection of T cells by Helicobacter pylori as a model for bacterial strain selection by immune evasion. J. Immunol. 167:926-934. [DOI] [PubMed] [Google Scholar]
- 61.Wilson, M., R. Seymour, and B. Henderson. 1998. Bacterial perturbation of cytokine networks. Infect. Immun. 66:2401-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamaoka, Y., D. H. Kwon, and D. Y. Graham. 2000. A M(r) 34, 000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 97:7533-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]