Abstract
Legionella pneumophila, the gram-negative agent of Legionnaires' disease, possesses type IV pili and a type II protein secretion (Lsp) system, both of which are dependent upon the PilD prepilin peptidase. By analyzing multiple pilD mutants and various types of Lsp mutants as well as performing trans-complementation of these mutants, we have confirmed that PilD and type II secretion genes are required for L. pneumophila infection of both amoebae and human macrophages. Based upon a complete analysis of lspDE, lspF, and lspG mutants, we found that the type II system controls the secretion of protease, RNase, lipase, phospholipase A, phospholipase C, lysophospholipase A, and tartrate-sensitive and tartrate-resistant acid phosphatase activities and influences the appearance of colonies. Examination of the developing L. pneumophila genome database indicated that the organism has two other loci (lspC and lspLM) that are predicted to promote secretion and thus a set of genes that is comparable to the type II secretion genes in other gram-negative bacteria. In contrast to lsp mutants, L. pneumophila pilus mutants lacking either the PilQ secretin, the PspA pseudopilin, or pilin were not defective for colonial growth, secreted activities, or intracellular replication. L. pneumophila dot/icm mutants were also not impaired for type II-dependent exoenzymes. Upon intratracheal inoculation into A/J mice, lspDE, lspF, and pilD mutants, but not pilus mutants, exhibited a reduced ability to grow in the lung, as measured by competition assays. The lspF mutant was also defective in an in vivo kinetic assay. Examination of infected mouse sera revealed that type II secreted proteins are expressed in vivo. Thus, the L. pneumophila Lsp system is a virulence factor and the only type II secretion system linked to intracellular infection.
The gram-negative bacterium Legionella pneumophila is the agent of Legionnaires' disease, a pneumonia which especially affects immunocompromised individuals (28, 89). An inhabitant of freshwater environments, L. pneumophila naturally replicates within protozoan hosts and in biofilms (27, 89). Following inhalation of contaminated aerosols, the bacterium reaches the human respiratory tract. Bacterial multiplication in alveolar macrophages is concomitant with cell death and damage to the lung tissue (89, 100).
In gram-negative bacteria, the PilD prepilin peptidase is necessary for the cleavage and methylation of pilins and pseudopilins that assemble into type IV pili (Tfp) (55, 68, 69, 87). In addition, PilD processes other pseudopilins that are necessary for the biogenesis of a functional type II protein secretion system (8, 13, 55, 71, 86). Accordingly, our previous mutational analysis determined that pilD is required for L. pneumophila piliation and protein secretion (4, 52). In L. pneumophila, Tfp promote attachment to host cells and are involved in competence for DNA transformation (84, 85). The L. pneumophila proteins believed to be secreted via the type II system include a zinc metalloprotease, acid phosphatases, lipases, phospholipases C (PLC), a PLA, and a lysophospholipase A (LPLA) (3-5, 39, 52, 75). In addition to pilD, the loci known to be involved in L. pneumophila type II protein secretion are lspDE, which specifies the outer membrane secretin LspD and the ATPase LspE; lspF, which encodes the inner membrane protein LspF; and lspGHIJK, which encodes the pseudopilins LspGHIJK (39, 75). Previous studies suggested that L. pneumophila pilD, though not required for extracellular replication, is essential for optimal infection of aquatic protozoa and human macrophages (52, 75). Whereas a type IV pilin (pilEL) mutant exhibits intracellular growth identical to wild-type strains (75, 84), mutants lacking either lspDE, lspG, or lspK are critically impaired for replication in Hartmannella vermiformis and Acanthamoeba castellanii amoebae (39, 70, 75). The type II secretion mutants are also defective for growth in human macrophages (70, 75), but an absence of complementation analysis has prevented a definitive conclusion regarding the role of the Lsp system in L. pneumophila infection of mammalian cells. Furthermore, the importance of type II secretion as well as Tfp for L. pneumophila in vivo growth and virulence has remained unknown.
In this study, we constructed and analyzed a large panel of mutants and complemented derivatives in order to confirm the relative roles of PilD, the type II secretion system, and type IV piliation in L. pneumophila extracellular growth, colony formation, protein secretion, natural transformation, and in vitro intracellular infection. Furthermore, to assess the in vivo significance of L. pneumophila type II secretion and pilus biogenesis, we tested the mutants in the A/J mouse model of legionellosis. The results of these studies affirm, among other things, that the type II secretion system promotes the ability of L. pneumophila to infect both protozoan and macrophage hosts and to grow in the mammalian lung. In fact, the data from the animal studies indicate that type II secretion has a greater role in Legionnaires' disease than would have been predicted from the results of in vitro experiments.
(Portions of this work were presented at the 102nd General Meeting of the American Society for Microbiology [O. Rossier and N. P. Cianciotto, Abstr. Gen. Meet. Am. Soc. Microbiol. 2002, abstr. B-207, p. 67, 2002.])
MATERIALS AND METHODS
Bacterial strains and media.
L. pneumophila serogroup 1 strain 130b (ATCC BAA-74), which served as the wild-type strain in this study, and its derivatives NU243, NU258, NU259, BS100, and AA200, which contain stable insertions of a kanamycin resistance (Kmr) gene in pilD, lspDE, lspG, pilEL, and proA, respectively, were described previously (25, 52, 62, 75, 84). Table 1 lists additional strains of L. pneumophila and of Legionella spp. used in this study. Legionellae were cultured at 37°C in buffered yeast extract (BYE) broth or on buffered charcoal yeast extract (BCYE) agar (22). Growth in liquid medium was assessed by measuring the optical density of the culture at 660 nm (OD660). Escherichia coli strains NovaBlue (Novagen, Madison, Wis.) and DH5α (Bethesda Research Laboratories), hosts for recombinant plasmids, were grown at 37°C on LB agar (6). The following antibiotics were added to the media at the indicated final concentrations (μg per ml): ampicillin, 100; chloramphenicol, 6 for L. pneumophila and 30 for E. coli; gentamicin, 2.5; and kanamycin, 25 for L. pneumophila and 50 for E. coli.
TABLE 1.
Wild-type Legionella strains used in this study
| Species and straina | Serogroup | Source | Reference |
|---|---|---|---|
| L. pneumophila | |||
| BAA-74 | 1 | Clinical | 25 |
| 33217 | 1 | Clinical | 15 |
| 33154 | 2 | Clinical | 58 |
| 33155 | 3 | Clinical | 58 |
| 33156 | 4 | Clinical | 58 |
| 33216 | 5 | Clinical | 36 |
| 33215 | 6 | Clinical | 59 |
| 33823 | 7 | Clinical | 10 |
| 35096 | 8 | Clinical | 12 |
| 43736 | 13 | Clinical | 54 |
| 43703 | 14 | Clinical | 9 |
| L. cherrii 35252 | Environmental | 14 | |
| L. feeleii 35072 | 2 | Clinical | 92 |
| L. gormanii 33297 | Environmental | 64 | |
| L. longbeachae 33462 | 1 | Clinical | 57 |
| L. micdadei | |||
| Stanford-R | 1 | Clinical | 74 |
| 33218 | Clinical | 40 | |
| L. parisiensis 35299 | 1 | Environmental | 14 |
| L. spiritensis 35249 | Environmental | 14 |
Except for one L. micdadei isolate, the strain designations refer to ATCC numbers.
DNA isolation, PCR, and sequence analysis.
Genomic DNA was isolated from L. pneumophila as described previously (24). Based on data from the L. pneumophila Philadelphia-I genome database (http://genome3.cpmc.columbia.edu/∼legion/), four pairs of primers were designed for amplifying genes from 130b DNA. Primers F3 and R34 (52) yielded a 1,246-bp fragment containing pilD, OR7lspF (5′-CCTCCAGGATAGTCCGCGTA), and OR2lspH (5′-CTTGTTGTTGAGCCAGGCTT) yielded a 1,972-bp fragment containing lspF and lspG, OR33pilQ (5′-CAACCTGACGGTCAGATTAG) and OR34pilQ (5′-GCCAGCACGCTCTTCAATTA) yielded a 2,857-bp fragment encoding pilQ, and OR29pspA (5′-CATGACGATGTATCTCGGCT) and OR30pspA (5′-GGTAACCGTCTCACACTACT) yielded an 1,830-bp fragment containing pspA. Sequencing reactions were performed using two different PCR amplicons, a series of custom primers, and the BigDye terminator cycle sequencing mix (PE Applied Biosystems, Foster City, Calif.). Automated sequence analysis was done on an ABI Prism 373 DNA sequencer (Applied Biosystems) at the Biotech Facility at Northwestern University. Primers were obtained from Integrated DNA Technologies (Coralville, Iowa). Database searches were performed using programs based on the BLAST algorithm (2), and protein sequences were analyzed for motifs using the PROSITE database (26).
Gene cloning and generation of mutants.
To facilitate construction of L. pneumophila mutants, the PCR fragments encoding pilD, lspF, and pilQ were ligated into pGem-T Easy (Promega, Madison, Wis.), yielding pGD1, pGlspF, and pGQ211, respectively, and the PCR product encoding pspA was cloned into pGem-T (Promega), giving pGpspA. A gentamicin resistance (Gmr) gene was isolated from pX1918GT after HincII and PvuII digestion (82) and was ligated into the single NarI site of the cloned pilD on pGD1, to give pGD::Gm. Plasmid pGlspF was digested with MscI, which cuts in the middle of lspF, and subsequently ligated to a Kmr gene isolated from pMB2190 upon HincII digestion (38), to produce pGlspF::Km. Digestion of pGQ211 with MfeI and ClaI led to the deletion of a 1.5-kb section of pilQ that, following Klenow treatment, was replaced by either the Kmr gene to generate pGQ::Km or the Gmr gene to yield pGQ::Gm. Plasmid pGpspA was digested with MfeI and SspI so as to remove a 0.4-kb fragment from pspA that, following Klenow treatment, was replaced by the Kmr gene producing pGpspA:::Km or by the Gmr gene, yielding pGpspA::Gm. Finally, using our modification to the original L. pneumophila transformation protocol (30, 85), pilD, lspF, pilQ, and pspA mutants were obtained by natural transformation of strain 130b with pGD::Gm, pGlspF::Km, pGQ::Km, pGQ::Gm, pGpspA:::Km, and pGpspA::Gm. Briefly, bacteria were inoculated at an OD660 of 0.3 into 2 ml of BYE broth in a 15-ml polypropylene tube, and then 5 μg of plasmid DNA/ml was added. After the culture was grown for approximately 20 h at 30°C with moderate shaking until mid- to late exponential phase, bacteria were plated on BCYE agar supplemented with kanamycin or gentamicin. Verification of all mutant genotypes was carried out by PCR and Southern hybridization (77). To construct an lspDE pilQ double mutant, the Kmr lspDE mutant NU258 (75) was transformed with plasmid pGQ::Gm, and mutants were selected on BCYE agar supplemented with kanamycin and gentamicin.
Complementation analysis.
Although we and others have previously used pMMB207 (63) for trans-complementation of L. pneumophila mutants (56, 73, 96), the conjugal components of the vector can inhibit L. pneumophila growth within macrophages (83). Therefore, in order to facilitate present and future complementation studies, pMMB207 was digested with AgeI and then religated, yielding pMMB2002, which has a 401-bp deletion in the mobA gene (80, 88). After plasmids were electroporated into competent 130b bacteria (52), the stability of pMMB2002 versus pMMB207 was determined by plating serial dilutions of transformants that had been grown without antibiotic selection on plain BCYE agar and BCYE agar supplemented with chloramphenicol. After growth in BYE broth or on BCYE agar to stationary phase, 85 to 100% and 51 to 70% of bacteria retained pMMB2002 and pMMB207, respectively. In addition, whereas 130b(pMMB207) had a 20- to 70-fold decrease in CFU recovery from infected U937 cells relative to 130b, bacteria containing pMMB2002 exhibited intracellular growth that was identical to that of wild type. Thus, to facilitate complementation of the pilD mutants, a 1.2-kb fragment containing only pilD was amplified by PCR using primers F3 and R34 (52) and then cloned under the control of the tac promoter in pMMB2002, yielding pMD1. To assess complementation of an lspF mutant, pMF1 was constructed; i.e., a 1.3-kb fragment containing only lspF was obtained from pGlspF using SstI and SspI and then ligated into pMMB2002 that had been digested with SstI and SmaI.
Analysis of L. pneumophila secreted enzymatic activities.
L. pneumophila protease and lipolytic activities were observed on casein (1%) agar and egg yolk (5%) agar, respectively (4, 30, 93). Filter-sterilized supernatants from cultures in BYE broth in late exponential phase (4) were tested for tartrate-sensitive and tartrate-resistant acid phosphatases as determined by the release of p-nitrophenol (p-NP) from p-NP phosphate in 200 mM sodium acetate (pH 5.5) in the absence or presence of 5 mM tartrate (3). PLC activity was monitored by the release of p-NP from p-NP phosphorylcholine (4). Monoacylglycerol lipase, PLA, and LPLA activities were measured by the ability of supernatants to release free fatty acids from 1-monopalmitoyl glycerol, phosphatidylcholine, and lysophosphatidylcholine, respectively (4, 29, 75). Lipolytic activities were also determined by p-NP palmitate and p-NP caprylate hydrolysis (4). RNase activity was assayed using a protocol adapted from Kar et al. (45). Briefly, 40 μl of culture supernatants was incubated with 400 μl of assay buffer (50 mM Tris, pH 8) containing Baker's yeast type III RNA (10 mg/ml; Sigma Chemical Co., St. Louis, Mo.). At the beginning of the reaction, half of the sample was transferred to a fresh tube, chilled, and precipitated with 2 volumes of ice-cold 10% trichloroacetic acid for 25 min. The remainder was incubated at 37°C for 40 min, at which point the reactions were chilled and precipitated with trichloroacetic acid as described above. Following centrifugation (12 min at 18,000 × g) of the precipitated samples, the absorbance of the supernatants at 260 nm, reflective of released soluble nucleotides, was determined. Bovine pancreas RNase A (Sigma) was used as a positive control.
Intracellular infection by L. pneumophila.
To examine the ability of L. pneumophila to grow within a protozoan host, H. vermiformis was infected as previously described (3, 4, 18, 52). Thus, ca. 104 CFU was added to wells containing 105 amoebae, and then at 0, 24, 48, 72, or 96 h postinoculation, the numbers of bacteria per coculture were determined by plating serial dilutions on BCYE agar, supplemented with the appropriate antibiotics. To quantitate intracellular growth in human macrophages, U937 cells were infected as previously described (3, 4, 52, 75). Briefly, monolayers containing 106 macrophages were inoculated with approximately 105 CFU, incubated for 2 h to allow bacterial entry and then washed three times with media to remove unincorporated bacteria. After 0, 24, or 48 h of incubation, serial dilutions of the lysed monolayers were plated on BCYE agar, supplemented with antibiotics when appropriate, to determine the numbers of bacteria per monolayer.
Pulmonary infection of A/J mice with L. pneumophila.
Pulmonary infection of A/J mice with L. pneumophila is a standard model for Legionnaires' disease (1, 16, 17, 19, 74). Female, 6- to 8-week-old mice (Jackson Laboratory, Bar Harbor, Maine) were anesthetized and then inoculated intratracheally with 25 μl of a bacterial suspension. To assess the virulence of bacterial strains, standard competition assays (31, 50, 60) were performed, as has been described previously (73, 74). Mice were inoculated with 105 CFU of a ca. 1:1 ratio of wild-type and mutant bacteria. At various time points, infected mice (n = 4 to 5) were sacrificed, and lungs were disrupted into 5 to 10 ml of phosphate-buffered saline using a homogenizer Pro 200 equipped with a generator (7 mm by 150 mm; Pro Scientific Inc., Monroe, Conn.). Host cell lysis was achieved by incubation of the tissue sample with 1% saponin for 15 min at 37°C, followed by vortexing. The numbers of viable bacteria and the ratio of wild type to mutant were estimated by plating 10-fold serial dilutions on both standard and antibiotic-supplemented BCYE. To determine the relative ability of strains to replicate and survive in mice lungs, mice (n = 4 or 5) were infected separately with 106 CFU of wild type or mutant, and at various hours postinoculation, the bacterial CFU in the lungs were determined by plating on BCYE agar.
Immunoblot analysis of L. pneumophila culture supernatants.
Filter-sterilized supernatants from BYE cultures in late exponential phase were precipitated with 10% trichloroacetic acid on ice for 30 min. Following centrifugation for 20 min at 4°C, pellets were washed with 70% ethanol, air dried, and resuspended in sample buffer at 1/100 of the supernatant volume (51). Following separation by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE) (51), proteins were transferred onto a nitrocellulose membrane (94) and reacted with a 1:100 dilution of mouse sera harvested 3 weeks after inoculation with 106 CFU of strain 130b (above). After incubation with a 1:1,000 dilution of anti-mouse immunoglobulin G peroxidase-conjugated antibody (Chemicon International, Temecula, Calif.), immunodetection was performed using Enhanced Chemiluminescence Western blotting (Amersham Biosciences) and a charge-coupled device camera (Chemi Imager 5500; Alpha Innotech, San Leandro, Calif.).
Detection of lsp gene sequences in Legionella spp. and serogroups.
Southern blot hybridization was carried out using EcoRI-restricted genomic DNA from strains representing several L. pneumophila serogroups and several Legionella spp. (Table 1). The digoxigenin labeling and detection system (Roche Molecular Biochemicals, Indianapolis, Ind.) was used according to the manufacturer's instructions. Probes were prepared by PCR incorporation using 130b DNA as a template and primers OR70lspC (5′-GTTTAACTGGGATTGGCAAC) and OR71lspC (5′-CAGGCAGCCATCATAGGAAT) for detection of lspC, OR5lspD (5′-TTGATTCTGTCTGGTCGAGC) and OR6lspD (5′-ATCAAGGACTACTACGGAGG) for detection of lspD, OR7lspF and OR2lspH for detection of lspFG, and OR68lspL (5′-GGATTATGCGGCTTCTGGTT) and OR69lspM (5′-AGGTCAGGCTTATAGTCTTG) for detection of lspLM.
Nucleotide sequence accession number.
The nucleotide sequence of L. pneumophila pilQ is deposited in GenBank under accession number AY320053.
RESULTS
Clarification of the role of pilD in L. pneumophila intracellular infection.
In our previous studies of L. pneumophila, the prepilin peptidase mutant NU243 displayed a dramatic reduction in intracellular growth in both amoebae and macrophages (4, 52, 75). Introduction of pMRL13, a derivative of pBBR1MCS expressing pilD, into NU243 restored intracellular growth of the pilD mutant in H. vermiformis to wild-type levels (52). In U937 cells, growth of NU243 was only partially complemented with pMRL13, but since pBBR1MCS derivatives are poorly maintained in strain 130b, we attributed the partial complementation to vector instability (52). With the development of pMMB2002, a vector that is stable in L. pneumophila (see Materials and Methods), we reinvestigated complementation of NU243 in order to definitively assess the importance of pilD for intracellular infection. As expected, NU243 carrying pilD cloned into pMMB2002 (i.e., pMD1) displayed full protease and lipolytic activities on casein agar and egg yolk agar, respectively (data not shown), confirming the role of PilD in L. pneumophila protein secretion. Upon infection of amoebae and macrophages, 130b(pMD1) exhibited the same replication profile as 130b(pMMB2002), demonstrating that expression of multiple copies of pilD does not alter the intracellular growth of wild-type L. pneumophila (Fig. 1). Expression from pMD1 allowed NU243 to effectively replicate in H. vermiformis, with the complementation of the pilD mutation complete after 72 h of coculture (Fig. 1A). NU243(pMD1) also exhibited robust intracellular replication in U937 macrophages, in contrast to NU243(pMMB2002) (Fig. 1B). However, NU243(pMD1)-infected cells always yielded significantly less CFU than did monolayers infected with 130b(pMD1) (Fig. 1B). Since >75% of 130b and NU243 retained pMMB2002 and pMD1 throughout the experiment (data not shown), such partial complementation could not be explained by loss of plasmid alone and suggested that some aspects of the NU243 phenotype are not due to the loss of PilD. Indeed, examination of 130b(pMMB2002), NU243(pMMB2002), and NU243(pMD1) indicated that the grayer color but not the overall morphology of NU243 colonies (52) is associated with the pilD mutation (data not shown).
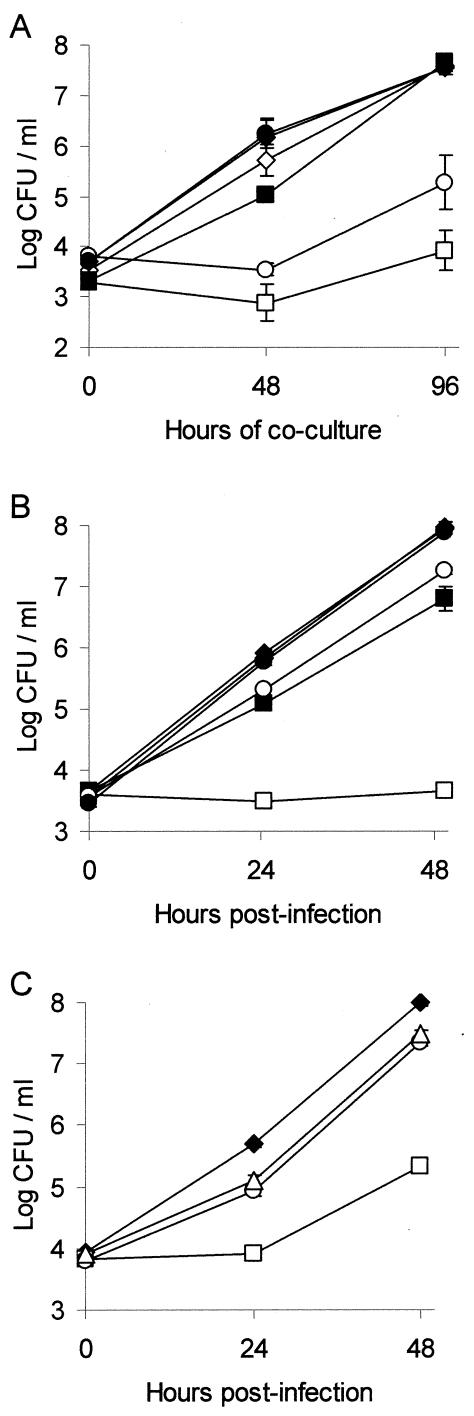
FIG. 1.
Intracellular infection of H. vermiformis amoebae and U937 cell macrophages by L. pneumophila pilD mutants and their complemented derivatives. (A) Wells containing H. vermiformis were inoculated at a multiplicity of infection (MOI) of 0.1 with strains 130b(pMMB2002) (⋄), 130b(pMD1) (♦), NU243(pMMB2002) (□), NU243(pMD1) (▪), NU272(pMMB2002) (○), and NU272 (pMD1) (•), and then the numbers of bacteria in each well were quantified at various times postinoculation by plating on BCYE agar. Results are the means and standard deviations (error bars) of four wells and are representative of two independent experiments. In a third experiment, NU243(pMD1) and NU272(pMD1) also showed full complementation at 72 h postinoculation. (B) U937 cells were infected at an MOI of 0.1 with the same strains indicated above. At various times, the monolayers were lysed, and the total number of CFU in each well was determined. Results are the means and standard deviations (error bars) of triplicate wells and are representative of two independent experiments. The differences in recovery between 130b(pMD1) and both NU243(pMD1) and NU272(pMMB2002) were significant at 24 and 48 h (Student's t test, P < 0.005). (C) Macrophages were infected at an MOI of 0.1 with wild-type 130b (♦), Kmr mutant NU243 (□), Gmr pilD mutant NU272 (○), and lspG mutant NU259 (▵), and then at various times, the total number of CFU in each well was determined. Results are the means and standard deviations (error bars) of triplicate wells and are representative of three independent experiments. At 24 and 48 h postinoculation, significant differences in recovery were obtained between 130b and NU243, 130b and NU259, 130b and NU272, and NU243 and NU272 (Student's t test, P < 0.001).
To explore further the possibility that NU243 has a second site mutation that affects infectivity and colonial growth, we used allelic exchange to isolate new pilD mutants. Three mutants were independently derived from 130b and designated NU272, NU273, and NU274. Each contained a Gmr cassette inserted into the same site in pilD as the Kmr gene in NU243. Since the three new mutants behaved similarly in all experiments, findings will only be presented for NU272. Like NU243, the new mutants grew normally in BYE broth, were unable to secrete type II enzymatic activities (Table 2), and lacked protease and lipolytic activities on casein and egg yolk agar, a phenotype complemented by pilD-containing pMD1 (data not shown). They also had a severe growth defect in H. vermiformis that pMD1 could fully complement (Fig. 1A). However, unlike NU243, the new mutants had a modest growth defect in U937 cells but one that was complemented fully by pMD1 (Fig. 1B) and was comparable to that of the type II secretion (lspG) mutant NU259 (Fig. 1C). Finally, the new mutants' colonies differed from that of wild type and NU243 but were identical to that of the type II secretion mutants (75), and when pMD1 was introduced into NU272, normal colony morphology was restored (data not shown).
TABLE 2.
Secreted activities of L. pneumophila strains
| Strain | Genotype | Proteasea | Activity (%) of supernatantb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Acid phosphatase | Tartrate-resistant acid phosphatase | PLC | PLA | LPLA | Lipase | p-NP caprylate hydrolase | p-NP palmitate hydrolase | |||
| 130b | WT | + | 100 ± 8 | 100 ± 4 | 100 ± 10 | 100 ± 7 | 100 ± 4 | 100 ± 4 | 100 ± 4 | 100 ± 4 |
| NU243 | pilDc | − | 21 ± 1d | 23 ± 2d | 7 ± 4d | 18 ± 1d | 23 ± 1d | 37 ± 1d | 16 ± 1d | 6 ± 1d |
| NU272 | pilD | − | 19 ± 0d | 32 ± 0d | 17 ± 7d | ND | ND | 38 ± 2d | 15 ± 2d | 19 ± 6d |
| NU258 | lspDE | − | 16 ± 1d | 33 ± 4d | 9 ± 3d | 20 ± 0d | 23 ± 2d | 43 ± 4d | 15 ± 1d | 11 ± 2d |
| NU259 | lspG | − | 20 ± 2d | 27 ± 2d | 31 ± 7d | ND | ND | ND | ND | ND |
| NU275 | lspF | − | 17 ± 3d | 25 ± 1d | 23 ± 14d | ND | 45 ± 14d | 34 ± 2d | 13 ± 1d | 27 ± 13d |
| 130b(pMMB2002) | + | 101 ± 5 | 102 ± 5 | 110 ± 11 | ND | 109 ± 11 | 93 ± 5 | 92 ± 9 | 86 ± 7 | |
| 130b(pMF) | + | 103 ± 9 | 92 ± 3 | 117 ± 14 | ND | 83 ± 13 | 108 ± 19 | 99 ± 4 | 101 ± 17 | |
| NU275(pMMB2002) | − | 16 ± 1d | 27 ± 0d | 16 ± 8d | 13 ± 0d | 49 ± 9d | 38 ± 3d | 12 ± 1d | 13 ± 5d | |
| NU275(pMF) | + | 107 ± 7 | 96 ± 4 | 125 ± 11 | 95 ± 13 | 88 ± 12 | 95 ± 12 | 91 ± 6 | 81 ± 5 | |
| AA200 | proA | − | 154 ± 7d | 149 ± 7d | 132 ± 11 | ND | ND | 60 ± 2d | 54 ± 3d | 25 ± 4d |
| BS100 | pilEL | + | 100 ± 7 | 99 ± 4 | 118 ± 3 | 111 ± 3 | 110 ± 3 | ND | 127 ± 3 | 136 ± 10 |
| NU279 | pilQ | + | 94 ± 2 | 93 ± 1 | 99 ± 8 | 92 ± 8 | 97 ± 2 | 109 ± 7 | 95 ± 11 | 110 ± 16 |
| NU280 | pspA | + | 106 ± 3 | 100 ± 7 | 105 ± 9 | 120 ± 9 | 103 ± 0 | 118 ± 11 | 121 ± 5 | 147 ± 16 |
| NU283 | lspDE pilQ | − | 16 ± 0d | 19 ± 0d | 9 ± 5d | 21 ± 1d | 22 ± 1d | 34 ± 3d | 15 ± 0d | 15 ± 6d |
| GG105 | dotA | ND | 109 ± 5 | 108 ± 1 | 123 ± 17 | 98 ± 12 | 99 ± 6 | 104 ± 6 | 99 ± 4 | 102 ± 10 |
| GQ262 | dotDCB | ND | 130 ± 15 | 119 ± 2 | ND | ND | 91 ± 7 | 100 ± 7 | 104 ± 4 | 99 ± 4 |
| GN142 | icmJB | ND | 122 ± 2 | 110 ± 4 | 146 ± 6 | ND | 100 ± 15 | 103 ± 5 | 104 ± 4 | 106 ± 5 |
Presence (+) and absence (−) of clearing on casein agar; ND, not determined.
Culture supernatants (n = 3) were incubated with appropriate substrates. Activities are expressed as percentage of wild-type activity (means ± standard deviations). ND, not determined.
As described in the text, strain NU243 has an unmarked, second-site mutation(s) in addition to the marked mutation in pilD.
Significant differences were obtained between enzymatic activities of the wild-type and the mutant strains (Student's t test, P < 0.005).
In sum, the observation of multiple pilD mutants and their complemented derivatives permits four conclusions. First, L. pneumophila pilD is required for the secretion of multiple enzymatic activities. Second, it promotes L. pneumophila intracellular growth in both amoebae and U937 cells, with its importance being most evident in protozoan hosts. Third, in addition to its pilD mutation, NU243 has a secondary site mutation that also affects colony morphology and intracellular growth in macrophages. Fourth, since the three new pilD mutants behaved identically to lsp mutants, type II secretion appears to be the main subset of the PilD-dependent activities that facilitate intracellular infection.
Clarification of the role of lsp genes in L. pneumophila protein secretion and intracellular infection.
Given the equivalent behavior of the new pilD mutants and our previously isolated lsp mutants in macrophage (Fig. 1C) and protozoan (data not shown) infection, we next focused attention on further defining the structure and function of the L. pneumophila type II secretion system. Previously, the lspDE and lspGHIJK loci were shown to be involved in protein secretion (39, 75). As a next step toward identifying genes that promote Legionella type II secretion, we used allelic exchange to insert a Kmr cassette into lspF of strain 130b, generating mutants NU275 and NU276. The lspF gene maps 103 bp upstream of the lspG pseudopilin gene and is believed to encode an inner membrane component of the secretion apparatus (39, 75). NU275 and NU276 grew in BYE broth similarly to wild type (data not shown), indicating that lspF, like the previously studied lsp genes, is not generally required for extracellular replication. The color and morphology of the lspF mutants' colonies were identical to those of the lspDE and lspG mutants (data not shown). The lspF mutants were defective in the secretion of those protease, lipase, PLC, and LPLA activities previously linked to pilD, lspDE, and lspG (Table 2) (4, 75). In addition to a tartrate-sensitive acid phosphatase, there is a tartrate-resistant acid phosphatase in wild-type supernatants that is lacking in pilD-negative NU243 and NU272 (Table 2) (3). Thus, we analyzed the culture supernatants of lsp mutants for acid phosphatase activity in the presence and absence of tartrate. Enzyme activity was always diminished for lspDE, lspF, and lspG mutants (Table 2), suggesting that both of the acid phosphatase activities in L. pneumophila supernatants are dependent upon type II secretion. Prior examination of pilD mutant NU243 revealed a reduction in RNase activity on clear agar matrices impregnated with RNA (4). In order to quantitatively determine the role of lsp genes in RNase secretion, we incubated supernatants with RNA and then measured the release of nucleotides by monitoring increases in absorbance at 260 nm (Table 3). Wild-type supernatants caused an increase in absorbance in the presence but not in the absence of added RNA, confirming that L. pneumophila secretes an RNase. Activity in lspDE and lspF mutant supernatants was reduced by sixfold (Table 3). The introduction of lspF-containing pMF1 into NU275 restored all supernatant activities to wild-type levels (Tables 2 and 3), confirming that the loss of lspF is responsible for the reduced secretion by that strain. Thus, we can now conclude that, in L. pneumophila, the secretion of protease, acid phosphatase, lipase, PLA, PLC, LPLA, and RNase activities is dependent upon a type II secretion pathway.
TABLE 3.
Ribonuclease activitya in L. pneumophila supernatants
| Sample | Increase in A260 (mean ± SD) |
|---|---|
| BYE | 0.013 ± 0.021 |
| 130bb | 0.362 ± 0.040 |
| lspDE mutant NU258 | 0.042 ± 0.023c |
| lspF mutant NU275 | 0.056 ± 0.019c |
| 130b(pMMB2002) | 0.418 ± 0.045 |
| NU275(pMMB2002) | 0.070 ± 0.055c |
| NU275(pMF1) | 0.396 ± 0.058 |
Filtered supernatants from cultures of the indicated bacterial strains or BYE medium were incubated with Baker's yeast RNA, and the release of nucleotides was measured by the increase in A260 over a 40-min period. Values are the means ± standard deviations from three samples.
When RNA was omitted from the 130b sample reaction, the increase in absorbance was only 0.020 ± 0.049, a value that was not different from that obtained with the BYE negative control.
The differences in activity between 130b and this mutant were significant (Student's t test, P < 0.005) and were observed in an additional experiment.
The L. pneumophila zinc metalloprotease ProA appears to be one of the most abundant proteins secreted by the type II pathway (39, 52, 75), and it has been previously observed that the amount of phospholipase A and monoacylglycerol lipase activities in supernatants is reduced in the absence of the protease (30). Therefore, we tested whether other type II-secreted activities are influenced by the metalloprotease by comparing supernatants from 130b and its isogenic proA mutant AA200. The mutant supernatants were reduced in their activity on monoacylglycerol, p-NP caprylate, and p-NP palmitate, suggesting that the protease may promote activation of lipolytic enzymes (Table 2). In contrast, AA200 exhibited a slight increase in secreted acid phosphatase and PLC activity (Table 2), indicating that the protease might partially degrade some secreted enzymes.
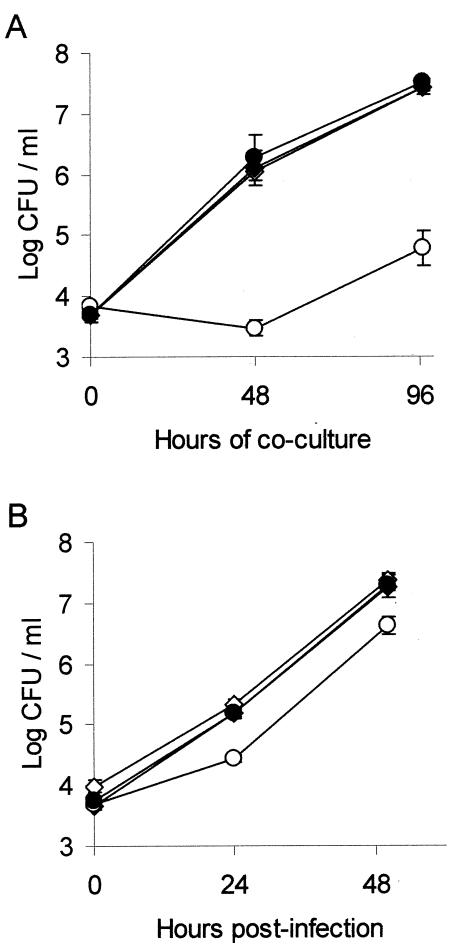
Previously, lspDE, lspG, and lspK mutants of strain 130b were shown to be defective for growth in Hartmannella amoebae and in human macrophages and monocytes (70, 75) (Fig. 1C). Similarly, an lspGH mutant of strain Philadelphia-1 is impaired for growth in acanthamoebae (39). When assessed for its replication in H. vermiformis and U937 macrophages, the lspF-negative mutants exhibited a growth defect similar to that of the other secretion mutants (data not shown). This defect was fully complemented by lspF-containing pMF1 (Fig. 2), proving that lspF is required for intracellular infection. These data represent the first genetic proof for the importance of a type II secretion gene in L. pneumophila infection of both protozoan and human cells. The Legionella Lsp pathway remains the only type II secretion system shown to promote intracellular infection.
FIG. 2.
Intracellular infection of H. vermiformis amoebae and U937 cell macrophages with an L. pneumophila lspF mutant and its complemented derivative. Amoebae (A) and U937 cells (B) were inoculated with strains 130b(pMMB2002) (⋄), 130b(pMF1) (♦), NU275(pMMB2002) (○) and NU275(pMF1) (•) at a multiplicity of infection of 0.1, and then the numbers of bacteria in each well were determined at various times postinoculation. For both panels, results are the means and standard deviations (error bars) of triplicate wells and are representative of two independent experiments. (B) The differences in CFU recovery between 130b(pMMB2002)- versus NU275(pMMB2002)-infected monolayers were significant at both 24 and 48 h postinoculation (Student's t test, P < 0.005).
Given the novel functions associated with L. pneumophila type II secretion, we sought additional lsp genes by examining the developing L. pneumophila Philadelphia-1 genome database (http://genome3.cpmc.columbia.edu/∼legion/). Thus, we performed a BLAST search (2) of the current database using as query sequences the proteins that are conserved in the type II secretory pathways of Pseudomonas aeruginosa, Klebsiella pneumoniae, and Erwinia carotovora. Besides the known lspDE, lspF, and lspGHIJK, we found two loci, designated lspC and lspLM, that encode proteins with greatest homology (49, 43, and 52% similarity) with Aeromonas hydrophila ExeC, Erwinia chrysanthemi OutL, and A. hydrophila ExeM, respectively (42, 46, 53). Thus, L. pneumophila has analogs of most of the genes that have been implicated in type II secretion in other bacteria (78, 79).
The role of Tfp assembly genes in L. pneumophila secretion and intracellular infection.
For several reasons, we wished to explore whether Tfp assembly genes also have a role in L. pneumophila secretion. First, there is similarity between proteins involved in Tfp assembly and those involved in type II secretion; e.g., in P. aeruginosa, the outer membrane PilQ secretin required for piliation is related to the XcpQ secretin involved in secretion, and the PilE, PilVWX, and FimT pseudopilins that help assemble pili are akin to XcpTUVW secretion pseudopilins (67, 76). Second, in enteropathogenic E. coli, the accumulation of ≥11 proteins in culture supernatants depends on the Tfp secretin BfpB (81). Third, in Vibrio cholerae, Tfp assembly genes, including the tcpC secretin gene, control the export of a protein that is not necessary for piliation but appears essential for intestinal colonization (49). Since our previous work showed that the pilEL gene encoding L. pneumophila pilin is not required for secretion of known type II exoproteins (75), we focused the present effort on Tfp secretins and pseudopilins.
Sequencing near the cloned L. pneumophila aroB gene (23) revealed an incomplete open reading frame (ORF) predicted to encode a protein with homology to the PilQ secretin (P. Edelstein, personal communication). Following a BLAST search (2) of the L. pneumophila genome database, we confirmed the identity of that ORF by cloning and completely sequencing the gene from strain 130b. The L. pneumophila pilQ gene encoded a 77-kDa protein with 36% identity and 56% similarity to P. aeruginosa PilQ. BLAST searches of the L. pneumophila database also found an ORF encoding a putative 18-kDa pseudopilin, based upon the presence of a conserved cleavage and methylation site (underlined) in its N-terminal sequence (MRLQLMKITGFTLET [PROSITE PS00409]). We designated this ORF pspA for pseudopilin gene A. In order to examine the function of Legionella PilQ and PspA, we used allelic exchange to isolate 130b mutants containing an antibiotic resistance gene inserted into either pilQ or pspA. Two Kmr pilQ mutants (NU277 and NU278), one Gmr pilQ mutant (NU279), two Kmr pspA mutants (NU280 and NU281), and one Gmr pspA mutant (NU282) were derived. As do Tfp mutants of other gram-negative bacteria (32, 33, 37, 44, 48, 95), the pilQ and pspA mutants were defective for natural transformation (Table 4). Indeed, they were as defective as the L. pneumophila pilEL mutant. An lspG mutant exhibited normal competence (Table 4), indicating that genes involved in type II protein secretion do not influence transformation. All of the new mutants grew in BYE broth similarly to wild type (data not shown), indicating that pilQ and pspA are not required for extracellular replication. When grown on BCYE agar, they appeared as typical colonies (data not shown). All further experiments were done with multiple pilQ and pspA mutants with similar results; however, for simplicity, findings will only be presented for one pilQ mutant (i.e., NU279) and one pspA mutant (i.e., NU280).
TABLE 4.
Natural transformation frequency of L. pneumophila strains
| Strain | Transformation frequency (mean ± SD)a |
|---|---|
| Wild-type 130b | (8.8 ± 3.2) × 10−6 |
| lspG mutant NU259 | (3.1 ± 0.2) × 10−6 |
| pilEL mutant BS100 | (6.3 ± 0.4) × 10−9 |
| pilQ mutant NU279 | (3.1 ± 5.3) × 10−9 |
| pspA mutant NU280 | (3.3 ± 1.7) × 10−9 |
Following incubation with a 5-μg/ml concentration of either pGD::Gm (see Materials and Methods) or pVA14-1 (5) at 30°C for 20 h, the indicated bacterial suspensions in BYE broth were plated on BCYE agar with the appropriate antibiotics. Values are the means ± standard deviations from three samples. The data presented are representative of at least two independent experiments.
To test whether pilQ or pspA play a role in type II protein secretion, NU279 and NU280 were grown in BYE broth to late exponential phase, and then filtered supernatants were assayed as described previously (4, 75). Both strains had wild-type levels of activities (Table 2), indicating that pilQ and pspA do not have an essential role in the secretion of known activities. We also extended our previous observations (75) indicating that the pilin gene pilEL is not required for the secretion of known type II exoproducts (Table 2). Finally, we explored the possibility that the Tfp secretin and the type II secretory secretin have redundant roles in secretion. Thus, we used allelic exchange to introduce the pilQ mutation into an lspDE mutant. Three double mutants (NU283, NU284, and NU285) were obtained and found to behave similarly. Culture supernatants of these mutants had levels of activity that were comparable to those of the type II secretion mutants (Table 2), confirming that Tfp assembly genes are not involved in L. pneumophila type II secretion.
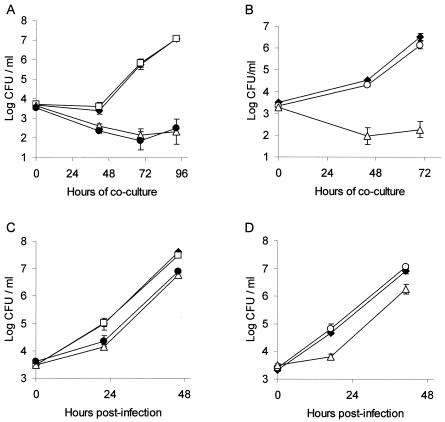
Upon coculture with H. vermiformis, the Tfp mutants grew comparably to wild type, indicating that neither pilQ nor pspA is required for infection of amoebal hosts (Fig. 3A and B). When U937 cells were infected, the mutants continued to behave like wild type (Fig. 3C and D). Finally, in both protozoa and macrophages, the lspDE pilQ double mutants had the same replication defect as the lspDE-negative strain (Fig. 3A and C). These results indicate that pilQ, pspA, and, by extension, the Tfp assembly apparatus, are not required for L. pneumophila intracellular infection in vitro.
FIG. 3.
Intracellular infection of H. vermiformis amoebae and U937 cell macrophages by L. pneumophila pilQ, pspA, and lsp mutants. Amoebae (A and B) and U937 cells (C and D) were infected at a multiplicity of infection of 0.1 with various strains, and then the numbers of bacteria in each well were determined at various times postinoculation. (A and C) Infection profiles for strains 130b (♦), pilQ mutant NU279 (□), lspDE mutant NU258 (•), and lspDE pilQ double mutant NU283 (▵). (B and D) Infectivities of strains 130b (♦), pspA mutant NU280 (○), and lspG mutant NU259 (▵). The results are the means and standard deviations (error bars) of triplicate wells and are representative of two independent experiments. In panels C and D, significant differences in recovery were obtained between 130b and NU258, 130b and NU283, and 130b and NU259 (Student's t test, P < 0.005).
The role of dot/icm genes in L. pneumophila type II protein secretion.
In addition to its type II secretion system, L. pneumophila possesses a type IV secretion system that is involved in intracellular infection (43, 56, 65, 66, 83, 97-99). To test whether this secretion apparatus influences type II protein secretion, we analyzed the enzymatic activities in culture supernatants of strains GG105, GQ262, and GN142, which contain an insertion mutation in dotA, dotDCB, and icmJB, respectively (34). The three mutants secreted normal levels of acid phosphatase, PLC, PLA, LPLA, and lipase (Table 2), indicating that dot/icm genes are not required for Legionella type II secretion.
Virulence of L. pneumophila secretion and piliation mutants in A/J mice.
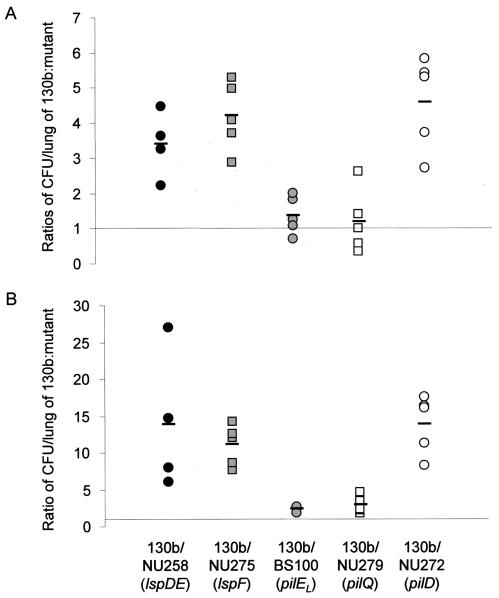
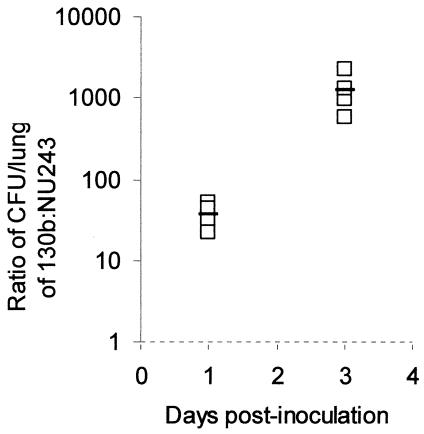
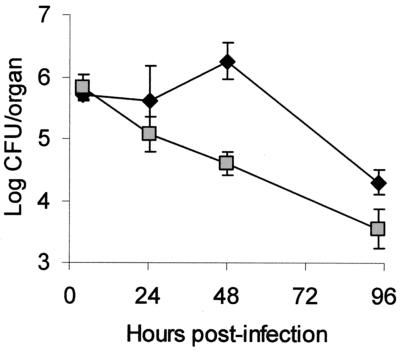
To determine whether PilD, Tfp, and type II secretion are important in vivo, we tested the wild type and isogenic mutants in the A/J mouse model of legionellosis (1, 16, 19, 74). As was done before (73, 74), we performed an in vivo competition assay in which a sublethal dose of bacteria (i.e., 105 CFU) containing a 1:1 mixture of 130b and a mutant are inoculated by intratracheal injection. In such assays, increases in the ratio of wild type to mutant recovered from infected lungs reflect a growth and/or survival defect for the mutant. At 24 h postinoculation, the ratio of 130b to the lspDE mutant NU258 and that of 130b to the lspF mutant NU275 increased to 3.2 and 4.2, respectively (Fig. 4A). By 72 h, the ratios of wild type to lsp mutant had further increased to 11 to 14 (Fig. 4B), showing that type II secretion mutants have a competitive disadvantage in vivo. In contrast, the ratios of 130b to the pilEL mutant BS100 and to the pilQ mutant NU279 did not change significantly at 24 h postinoculation and increased only slightly at 72 h postinoculation (Fig. 4A and B), indicating that L. pneumophila Tfp do not confer a growth advantage in mouse lungs. The pilD mutant NU272 behaved as the type II secretion mutants did (Fig. 4A and 4B), supporting the belief that type II secretion promotes L. pneumophila replication in the A/J lung. Incidentally, strain NU243, which contains a second site mutation in addition to an insertion in pilD, was very defective in the competition assay (Fig. 5). Indeed, the ratio of 130b to NU243 increased to 38 and 1,266 at 24 and 72 h postinoculation, respectively, indicating that the gene(s) affected by the secondary mutation plays a crucial role for in vivo infections. To confirm the importance of type II protein secretion in the mouse lung, we monitored the survival and replication of 130b and NU275 following intratracheal inoculation into separate groups of mice. Whereas the wild type multiplied over the first 48 h postinoculation, as previously observed (16, 20), the lspF mutant exhibited a 17-fold decrease in CFU (Fig. 6). At 4 days postinoculation, there was still a fivefold difference in CFU recovery between the two strains (Fig. 6). In the A/J mouse model, doses of L. pneumophila 130b greater than 107 CFU result in death within 48 h, with this acute lethality being ascribed to a bacterial toxin that is distinct from endotoxin (1, 16). Thus, we challenged groups of mice with 109 CFU of 130b, the pilEL mutant, the lspF mutant NU275, or the lspDE mutant NU258. All mice died acutely and within the same 48-h time frame. Thus, Tfp and type II secretion are not required for rapid killing by high-dose inoculation. However, the results of both the competition assays and the clearance study indicate that type II secretion is critical for the multiplication and survival of L. pneumophila in mammalian lungs.
FIG. 4.
In vivo competition between wild-type L. pneumophila and mutants lacking type II protein secretion and Tfp. A mixture of 130b and mutant were introduced into the lungs of A/J mice by intratracheal inoculation. At 1 day (A) and 3 days (B) postinoculation, the ratio of 130b to mutant in infected lungs was determined and normalized to the ratio of wild type to mutant in the inoculum. Data are representative of actual values obtained per mouse (n = 4 to 6), and the solid bars represent the mean values. Strains tested in competition with wild type were from left to right: lspDE mutant NU258 (black circles), lspF mutant NU275 (gray squares), pilEL mutant BS100 (gray circles), pilQ mutant NU279 (white squares), and pilD mutant NU272 (white circles). For the trials done with NU275, BS100, NU279, and NU272, significant differences were obtained between ratios observed at day 1 and day 3 (Student's t test, P < 0.05).
FIG. 5.
In vivo competition between wild-type L. pneumophila and mutant NU243. A mixture of 130b and the Kmr mutant NU243 was introduced into the lungs of A/J mice by intratracheal inoculation. At 1 and 3 days postinoculation, the ratio of 130b to mutant in infected lungs was determined. Data are representative of the actual values obtained per mouse (n = 4), and a solid bar represents the mean value. The 130b to mutant ratio increased significantly between days 1 and 3 (Student's t test, P < 0.05).
FIG. 6.
Growth and survival of wild-type and lspF mutant L. pneumophila in the lungs of infected A/J mice. Mice were intratracheally inoculated with equal numbers of either 130b (black diamonds) or lspF mutant NU275 (gray squares), and then at various time points, the CFU in infected lungs were determined by plating. Data are the means and standard deviations (error bars) obtained for four to six infected animals. Significant differences were obtained between the CFU recovered from mice infected with 130b and those infected with NU275, at 48 and 96 h postinoculation (Student's t test, P < 0.01).
Detection of antibodies against type II-secreted proteins in the antisera of L. pneumophila-infected mice
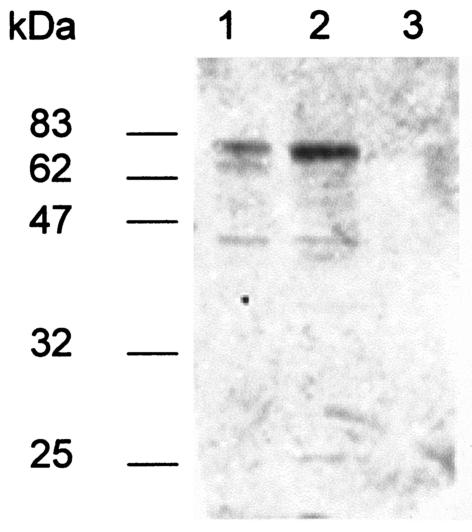
To confirm that the L. pneumophila type II secretion is operating during in vivo growth, we investigated whether A/J mice infected with wild-type 130b developed antibodies that reacted with type II exoproteins. Sera were obtained from two infected animals 3 weeks after their inoculation with a sublethal dose of 106 CFU and were incubated with blots containing concentrated culture supernatants from strain 130b and its lspF-negative derivative NU275, as well as ATCC 33155, a representative of L. pneumophila serogroup 3 (Fig. 7). The sera reacted with at least five proteins in the 130b supernatants whose sizes were ca. 72, 65, 55, 42, and 27 kDa. The 65- and 42-kDa proteins, as well as a 74-kDa species, were detected in the supernatants of strain 33155. All reactive proteins were absent in the supernatant of the lspF mutant. Thus, L. pneumophila-infected mice develop antibodies against multiple type II-secreted proteins, and therefore, we conclude that Legionella type II secretion is functioning during bacterial infection of the mammalian lung.
FIG. 7.
Immunodetection of L. pneumophila type II-secreted proteins by infected mouse sera. Concentrated supernatants from cultures of L. pneumophila strains 33155 (lane 1) and 130b (lane 2) and lspF mutant NU275 (lane 3) were separated by SDS-PAGE, transferred to a nitrocellulose membrane and reacted with the diluted serum of a 130b-infected A/J mouse. Similar results were obtained using the serum of a second infected mouse.
Distribution of lsp genes in L. pneumophila serogroups and other Legionella spp.
Given the importance of type II secretion for serogroup 1 strains of L. pneumophila, we sought to evaluate the distribution of lsp genes in other L. pneumophila serogroups as well as other Legionella spp. Thus, Southern hybridizations were performed using probes derived from lspC, lspD, lspFG, and lspLM of 130b. Under high-stringency conditions (10% base pair mismatch allowed), sequences homologous to all of the probes were observed in all L. pneumophila strains tested, i.e., representatives of serogroups 1 to 8, 13, and 14 (Table 1). Under low-stringency conditions (30% bp mismatch allowed), sequences homologous to all of the probes were detected in all Legionella species tested, i.e., L. cherrii, L. feeleii, L. gormanii, L. longbeachae, L. micdadei, L. parisiensis, and L. spiritensis (Table 1). Thus, type II secretion genes seem to be distributed throughout the genus Legionella. With the exception of L. cherrii, the Legionella spp. were tested for protease and lipolytic activities on casein and egg yolk agar (data not shown). Most strains produced both types of activities. However, the L. micdadei strains failed to show any activity, even though they contained the lsp genes. The L. spiritensis strain did not produce clearing on the egg yolk agar, suggesting that it does not secrete a PLA. Finally, the L. feeleii isolate did not produce iridescence on the egg yolk agar, indicating that it lacks a secreted lipase. Taken together, these data indicate that a type II secretion system is present and operative in most type of legionellae.
DISCUSSION
Using thorough complementation analysis, we have now formally established that the Lsp type II secretion system promotes infection of amoebae and macrophages, the two host cells most relevant to L. pneumophila ecology and pathogenesis. To our knowledge, this L. pneumophila pathway remains the only type II secretion system to be implicated in intracellular infection. In a previous study, we had demonstrated that protease, acid phosphatase, PLC (p-NP phosphorylcholine hydrolase), multiple lipase, PLA, and LPLA activities were lacking in the supernatants of lspG and lspDE mutants (75). With additional mutant constructions and a corresponding complementation analysis, we have confirmed that the L. pneumophila type II secretion system governs the secretion of all of these activities as well as tartrate-resistant and tartrate-sensitive acid phosphatase and an RNase. Thus, L. pneumophila lsp genes promote the secretion of at least eight extracellular enzymatic activities, a workload that matches if not exceeds that of other known type II systems (79). SDS-PAGE analysis of bacterial supernatants confirms that the L. pneumophila type II system controls the secretion of multiple protein species, some of which may facilitate yet-to-be-defined enzymatic activities (39, 52). L. pneumophila genes encoding protease (proA), tartrate-sensitive acid phosphatase (map), lipase (lipA and lipB), LPLA (plaA), and PLC (plcA) activities have been identified (3, 5, 30, 62, 90), although the corresponding mutants are not impaired in intracellular infection. Thus, it will be important to continue to pursue the genes encoding the PLA, RNase, tartrate-resistant acid phosphatase, as well as the lipase and PLC activities not associated with lipA, lipB, or plcA. Besides its influence on the release of proteins and enzymes into supernatants, the Lsp system had appeared to affect the morphology of L. pneumophila colonies (75). This supposition has also now been confirmed, suggesting that the L. pneumophila type II system may also mediate the localization of cell envelope proteins. It is possible that the type II-dependent factors that promote intracellular infection include outer membrane proteins.
In addition to pilD, lspDE, and lspFGHIJK, we have now identified in the L. pneumophila genome three more genes, lspC, lspL, and lspM, which encode conserved components of type II secretion pathways. Although components associated with some type II systems (e.g., homologs of V. cholerae EpsA, EpsB, and EpsN) have not yet been identified in the L. pneumophila genome, the Legionella organism does have a set of genes that matches the xcp genes of P. aeruginosa (78, 79). Unlike P. aeruginosa (7), the Philadelphia-1 database does not reveal a second type II secretion system. In L. pneumophila, the 12 lsp genes are located in five loci scattered throughout the chromosome, a feature rarely seen in other bacteria, where most type II secretion genes are clustered in one or two loci (76, 78, 79). Further studies are needed in order to establish whether the additional lsp genes are required for protein secretion.
Stone and Abu Kwaik had established that the L. pneumophila pilEL gene is required for Tfp-mediated natural transformation (85). We have now identified two additional genes, pilQ and pspA, that are necessary for DNA transformation. No cross talk between Tfp and type II protein secretion was evident, since disruption of lspG did not abolish transformation and loss of pilEL, pilQ, or pspA and did not influence the secretion of enzymatic activities. In extracellular gram-negative pathogens, Tfp are often important virulence factors that promote adherence to and colonization of the host (11, 41, 91). Although an L. pneumophila pilEL-negative strain exhibits a 50% reduction in adherence to mammalian and protozoan cells (84), it replicates like the wild type within these cells in vitro (75, 84). Similarly, the pilQ and pspA mutants showed no replication defect in U937 cells or H. vermiformis. When tested in vivo, strains lacking pilEL and pilQ strains were only slightly outcompeted by the wild type in A/J mouse lungs, suggesting that Tfp are not a significant virulence determinant in the mammalian host. However, it is possible that Tfp have a role that cannot be easily monitored with the A/J mouse model. Nonetheless, Tfp appear to be quite significant for L. pneumophila persistence in the environment; i.e., pilus mutants are defective for colonization of aquatic biofilms (C. E. Lucas, E. Brown, T. S. Forster, R. Murga, R. M. Donlan, N. P. Cianciotto, Y. Abu Kwaik, and B. S. Fields, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol 2002, abstr. Q-258, p. 422, 2002).
In contrast to the Tfp mutants, type II secretion mutants exhibited a severe defect in the competition assay in A/J mouse lungs. That the pilD mutant NU272 had a defect similar to that of the lsp mutants implies that the importance of the prepilin peptidase in vivo is due to its role in promoting protein secretion. By monitoring the kinetics of bacterial growth in vivo, we firmly established that the lspF mutant is impaired for replication and survival in the animal lungs. Thus, we strongly believe that type II secretion genes have a significant role in L. pneumophila pathogenesis. The in vivo relevance of type II secretion was also evidenced by the presence of antibodies specific to Lsp-dependent proteins in the sera of infected mice. This finding is compatible with a previous study in which the ProA metalloprotease was shown to be expressed in the lungs of guinea pigs infected with L. pneumophila (21). Moreover, individuals diagnosed with Legionnaires' disease have antibodies specific to the protease, indicating that factors secreted by the type II pathway are also expressed during human infection (47, 72).
The reduced survival of the lsp mutants in the A/J mouse lung is likely due, at least in part, to diminished growth in alveolar macrophages, since the mutants are also defective for in vitro infection of U937 cells. However, the number of lspF mutant CFU did not increase within the mouse lungs, whereas they did, albeit not optimally, in vitro. Several factors may be responsible for this difference. First, the mouse alveolar macrophages and the U937 cell macrophages may provide different responses to L. pneumophila infection. On the one hand, mouse and human macrophages may have significant differences in permissiveness, and on the other, resident macrophages may be much more restrictive than a macrophage cell line; e.g., an lspK mutant exhibits a modest defect in U937 cells but is unable to replicate in human blood monocyte-derived macrophages (70). Second, since L. pneumophila can also infect lung epithelial cells (35, 61), it is possible that type II secretion is needed for effective intracellular spread beyond the alveolar macrophage. Third, the lsp mutants may be defective for extracellular processes that are operative in the lungs. In attempting to reconcile in vitro and in vivo findings, our experience with the lsp mutants is somewhat reminiscent of observations made with protease mutants, i.e., a proA (msp) mutant, while not defective for in vitro intracellular infection (62, 90), elicits less necrosis and more macrophage infiltration in the lungs of guinea pigs inoculated by the intratracheal route (62). In light of our results with the lspF mutant, all type II effector mutants should be examined in the animal model of disease, rather than just pursuing those that display intracellular growth defects in vitro.
Acknowledgments
We are grateful to Paul Edelstein for sharing the partial sequence of pilQ; Alan Hauser for providing pX1918GT; and Yousef Abu Kwaik for providing strains BS100, GG105, GQ262, and GN142. For technical assistance with the competition assays, we thank Kimberly Allard, Virginia Aragon, Antje Flieger, Joseph Garonski-Salerno, and Marianne Robey. We also thank Bethany Boardman for assistance with generating the lspF mutant. We are extremely grateful to past and present members of the laboratory for helpful discussions and comments.
This work was supported by NIH grant AI43987 awarded to N.P.C.
Editor: D. L Burns
REFERENCES
- 1.Alli, O. A., L. Y. Gao, L. L. Pedersen, S. Zink, M. Radulic, M. Doric, and Y. Abu Kwaik. 2000. Temporal pore formation-mediated egress from macrophages and alveolar epithelial cells by Legionella pneumophila. Infect. Immun. 68:6431-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Aragon, V., S. Kurtz, and N. P. Cianciotto. 2001. The Legionella pneumophila major acid phosphatase and its role in intracellular infection. Infect. Immun. 69:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aragon, V., S. Kurtz, A. Flieger, B. Neumeister, and N. P. Cianciotto. 2000. Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila. Infect. Immun. 68:1855-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aragon, V., O. Rossier, and N. P. Cianciotto. 2002. Legionella pneumophila genes that encode lipase and phospholipase C activities. Microbiology 148:2223-2231. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. Wiley, New York, N.Y.
- 7.Ball, G., E. Durand, A. Lazdunski, and A. Filloux. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43:475-485. [DOI] [PubMed] [Google Scholar]
- 8.Bally, M., A. Filloux, M. Akrim, G. Ball, A. Lazdunski, and J. Tommassen. 1992. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol. Microbiol. 6:1121-1131. [DOI] [PubMed] [Google Scholar]
- 9.Benson, R. F., W. L. Thacker, H. W. Wilkinson, R. J. Fallon, and D. J. Brenner. 1988. Legionella pneumophila serogroup 14 isolated from patients with fatal pneumonia. J. Clin. Microbiol. 26:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bibb, W. F., P. M. Arnow, D. L. Dellinger, and S. R. Perryman. 1983. Isolation and characterization of a seventh serogroup of Legionella pneumophila. J. Clin. Microbiol. 17:346-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 12.Bissett, M. L., J. O. Lee, and D. S. Lindquist. 1983. New serogroup of Legionella pneumophila, serogroup 8. J. Clin. Microbiol. 17:887-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bleves, S., R. Voulhoux, G. Michel, A. Lazdunski, J. Tommassen, and F. A. 1998. The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX (GspK family). Mol. Microbiol 27:31-40. [DOI] [PubMed] [Google Scholar]
- 14.Brenner, D. J., A. G. Steigerwalt, G. W. Gorman, H. W. Wilkinson, W. F. Bibb, M. Hackel, R. L. Tyndall, J. Campbell, J. C. Feeley, W. L. Thacker, P. Skaliy, W. T. Martin, B. J. Brake, B. S. Fields, H. V. McEachern, and L. K. Corcoran. 1985. Ten new species of Legionella. Int. J. Syst. Bacteriol. 35:50-59. [Google Scholar]
- 15.Brenner, D. J., A. G. Steigerwalt, and J. E. McDade. 1979. Classification of the Legionnaires' disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann. Intern. Med. 90:656-658. [DOI] [PubMed] [Google Scholar]
- 16.Brieland, J., P. Freeman, R. Kunkel, C. Chrisp, M. Hurley, J. Fantone, and C. Engleberg. 1994. Replicative Legionella pneumophila lung infection in intratracheally inoculated A/J mice. A murine model of human Legionnaires' disease. Am. J. Pathol. 145:1537-1546. [PMC free article] [PubMed] [Google Scholar]
- 17.Brieland, J., M. McClain, L. Heath, C. Chrisp, G. Huffnagle, M. LeGendre, M. Hurley, J. Fantone, and C. Engleberg. 1996. Coinoculation with Hartmannella vermiformis enhances replicative Legionella pneumophila lung infection in a murine model of Legionnaires' disease. Infect. Immun. 64:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cirillo, J. D., S. L. Cirillo, L. Yan, L. E. Bermudez, S. Falkow, and L. S. Tompkins. 1999. Intracellular growth in Acanthamoeba castellanii affects monocyte entry mechanisms and enhances virulence of Legionella pneumophila. Infect. Immun. 67:4427-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirillo, S. L., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conlan, J. W., A. Williams, and L. A. Ashworth. 1988. In vivo production of a tissue-destructive protease by Legionella pneumophila in the lungs of experimentally infected guinea-pigs. J. Gen. Microbiol. 134:143-149. [DOI] [PubMed] [Google Scholar]
- 22.Edelstein, P. H. 1981. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J. Clin. Microbiol. 14:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edelstein, P. H., M. A. Edelstein, F. Higa, and S. Falkow. 1999. Discovery of virulence genes of Legionella pneumophila by using signature tagged mutagenesis in a guinea pig pneumonia model. Proc. Natl. Acad. Sci. USA 96:8190-8195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engleberg, N. C., C. Carter, D. R. Weber, N. P. Cianciotto, and B. I. Eisenstein. 1989. DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect. Immun. 57:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engleberg, N. C., D. J. Drutz, and B. I. Eisenstein. 1984. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect. Immun. 44:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 28.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flieger, A., S. Gong, M. Faigle, M. Deeg, P. Bartmann, and B. Neumeister. 2000. Novel phospholipase A activity secreted by Legionella species. J. Bacteriol. 182:1321-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flieger, A., B. Neumeister, and N. P. Cianciotto. 2002. Characterization of the gene encoding the major secreted lysophospholipase A of Legionella pneumophila and its role in detoxification of lysophosphatidylcholine. Infect. Immun. 70:6094-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francis, M. S., and C. J. Thomas. 1997. Mutants in the CtpA copper transporting P-type ATPase reduce virulence of Listeria monocytogenes. Microb. Pathog. 22:67-78. [DOI] [PubMed] [Google Scholar]
- 32.Friedrich, A., C. Prust, T. Hartsch, A. Henne, and B. Averhoff. 2002. Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl. Environ. Microbiol. 68:745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fussenegger, M., T. Rudel, R. Barten, R. Ryll, and T. F. Meyer. 1997. Transformation competence and type-4 pilus biogenesis in Neisseria gonorrhoeae—a review. Gene 192:125-134. [DOI] [PubMed] [Google Scholar]
- 34.Gao, L. Y., O. S. Harb, and Y. Abu Kwaik. 1997. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect. Immun. 65:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao, L. Y., B. J. Stone, J. K. Brieland, and Y. Abu Kwaik. 1998. Different fates of Legionella pneumophila pmi and mil mutants within macrophages and alveolar epithelial cells. Microb. Pathog. 25:291-306. [DOI] [PubMed] [Google Scholar]
- 36.Garrity, G. M., E. M. Elder, B. Davis, R. M. Vickers, and A. Brown. 1982. Serological and genotypic diversity among serogroup 5-reacting environmental Legionella isolates. J. Clin. Microbiol. 15:646-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graupner, S., V. Frey, R. Hashemi, M. G. Lorenz, G. Brandes, and W. Wackernagel. 2000. Type IV pilus genes pilA and pilC of Pseudomonas stutzeri are required for natural genetic transformation, and pilA can be replaced by corresponding genes from nontransformable species. J. Bacteriol. 182:2184-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grindley, N. D., and C. M. Joyce. 1980. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc. Natl. Acad. Sci. USA 77:7176-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hales, L. M., and H. A. Shuman. 1999. Legionella pneumophila contains a type II general secretion pathway required for growth in amoebae as well as for secretion of the Msp protease. Infect. Immun. 67:3662-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hebert, G. A., C. W. Moss, L. K. McDougal, F. M. Bozeman, R. M. McKinney, and D. J. Brenner. 1980. The rickettsia-like organisms TATLOCK (1943) and HEBA (1959): bacteria phenotypically similar to but genetically distinct from Legionella pneumophila and the WIGA bacterium. Ann. Intern. Med. 92:45-52. [DOI] [PubMed] [Google Scholar]
- 41.Herrington, D. A., R. H. Hall, G. Losonsky, J. J. Mekalanos, R. K. Taylor, and M. M. Levine. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howard, S. P., J. Critch, and A. Bedi. 1993. Isolation and analysis of eight exe genes and their involvement in extracellular protein secretion and outer membrane assembly in Aeromonas hydrophila. J. Bacteriol. 175:6695-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joshi, A. D., S. Sturgill-Koszycki, and M. S. Swanson. 2001. Evidence that Dot-dependent and -independent factors isolate the Legionella pneumophila phagosome from the endocytic network in mouse macrophages. Cell. Microbiol. 3:99-114. [DOI] [PubMed] [Google Scholar]
- 44.Kang, Y., H. Liu, S. Genin, M. A. Schell, and T. P. Denny. 2002. Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46:427-437. [DOI] [PubMed] [Google Scholar]
- 45.Kar, S., L. Soong, M. Colmenares, K. Goldsmith-Pestana, and D. McMahon-Pratt. 2000. The immunologically protective P-4 antigen of Leishmania amastigotes. A developmentally regulated single strand-specific nuclease associated with the endoplasmic reticulum. J. Biol. Chem. 275:37789-37797. [DOI] [PubMed] [Google Scholar]
- 46.Karlyshev, A. V., and S. MacIntyre. 1995. Cloning and study of the genetic organization of the exe gene cluster of Aeromonas salmonicida. Gene 158:77-82. [DOI] [PubMed] [Google Scholar]
- 47.Keen, M. G., and P. S. Hoffman. 1989. Characterization of a Legionella pneumophila extracellular protease exhibiting hemolytic and cytotoxic activities. Infect. Immun. 57:732-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kennan, R. M., O. P. Dhungyel, R. J. Whittington, J. R. Egerton, and J. I. Rood. 2001. The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosus is essential for virulence, protease secretion, and natural competence. J. Bacteriol. 183:4451-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirn, T. J., N. Bose, and R. K. Taylor. 2003. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol. Microbiol. 49:81-92. [DOI] [PubMed] [Google Scholar]
- 50.Krogfelt, K. A., M. Hjulgaard, K. Sorensen, P. S. Cohen, and M. Givskov. 2000. rpoS gene function is a disadvantage for Escherichia coli BJ4 during competitive colonization of the mouse large intestine. Infect. Immun. 68:2518-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 52.Liles, M. R., P. H. Edelstein, and N. P. Cianciotto. 1999. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol. Microbiol. 31:959-970. [DOI] [PubMed] [Google Scholar]
- 53.Lindeberg, M., and A. Collmer. 1992. Analysis of eight out genes in a cluster required for pectic enzyme secretion by Erwinia chrysanthemi: sequence comparison with secretion genes from other gram-negative bacteria. J. Bacteriol. 174:7385-7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindquist, D. S., G. Nygaard, W. L. Thacker, R. F. Benson, D. J. Brenner, and H. W. Wilkinson. 1988. Thirteenth serogroup of Legionella pneumophila isolated from patients with pneumonia. J. Clin. Microbiol. 26:586-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marsh, J. W., and R. K. Taylor. 1998. Identification of the Vibrio cholerae type 4 prepilin peptidase required for cholera toxin secretion and pilus formation. Mol. Microbiol. 29:1481-1492. [DOI] [PubMed] [Google Scholar]
- 56.Matthews, M., and C. R. Roy. 2000. Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect. Immun. 68:3971-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKinney, R. M., R. K. Porschen, P. H. Edelstein, M. L. Bissett, P. P. Harris, S. P. Bondell, A. G. Steigerwalt, R. E. Weaver, M. E. Ein, D. S. Lindquist, R. S. Kops, and D. J. Brenner. 1981. Legionella longbeachae species nova, another etiologic agent of human pneumonia. Ann. Intern. Med. 94:739-743. [DOI] [PubMed] [Google Scholar]
- 58.McKinney, R. M., L. Thacker, P. P. Harris, K. R. Lewallen, G. A. Hebert, P. H. Edelstein, and B. M. Thomason. 1979. Four serogroups of Legionnaires' disease bacteria defined by direct immunofluorescence. Ann. Intern. Med. 90:621-624. [DOI] [PubMed] [Google Scholar]
- 59.McKinney, R. M., H. W. Wilkinson, H. M. Sommers, B. J. Fikes, K. R. Sasseville, M. M. Yungbluth, and J. S. Wolf. 1980. Legionella pneumophila serogroup six: isolation from cases of legionellosis, identification by immunofluorescence staining, and immunological response to infection. J. Clin. Microbiol. 12:395-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merrell, D. S., and A. Camilli. 1999. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34:836-849. [DOI] [PubMed] [Google Scholar]
- 61.Mody, C. H., R. Paine, 3rd, M. S. Shahrabadi, R. H. Simon, E. Pearlman, B. I. Eisenstein, and G. B. Toews. 1993. Legionella pneumophila replicates within rat alveolar epithelial cells. J. Infect. Dis. 167:1138-1145. [DOI] [PubMed] [Google Scholar]
- 62.Moffat, J. F., P. H. Edelstein, D. P. Regula, Jr., J. D. Cirillo, and L. S. Tompkins. 1994. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig pneumonia model. Mol. Microbiol. 12:693-705. [DOI] [PubMed] [Google Scholar]
- 63.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 64.Morris, G. K., A. Steigerwalt, J. C. Feeley, E. S. Wong, W. T. Martin, C. M. Patton, and D. J. Brenner. 1980. Legionella gormanii sp. nov. J. Clin. Microbiol. 12:718-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 66.Nagai, H., and C. R. Roy. 2001. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J. 20:5962-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nunn, D. 1999. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell. Biol. 9:402-408. [DOI] [PubMed] [Google Scholar]
- 68.Nunn, D. N., and S. Lory. 1991. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc. Natl. Acad. Sci. USA 88:3281-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pepe, C. M., M. W. Eklund, and M. S. Strom. 1996. Cloning of an Aeromonas hydrophila type IV pilus biogenesis gene cluster: complementation of pilus assembly functions and characterization of a type IV leader peptidase/N-methyltransferase required for extracellular protein secretion. Mol. Microbiol. 19:857-869. [DOI] [PubMed] [Google Scholar]
- 70.Polesky, A. H., J. T. Ross, S. Falkow, and L. S. Tompkins. 2001. Identification of Legionella pneumophila genes important for infection of amoebas by signature-tagged mutagenesis. Infect. Immun. 69:977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pugsley, A. P., and B. Dupuy. 1992. An enzyme with type IV prepilin peptidase activity is required to process components of the general extracellular protein secretion pathway of Klebsiella oxytoca. Mol. Microbiol. 6:751-760. [DOI] [PubMed] [Google Scholar]
- 72.Quinn, F. D., M. G. Keen, and L. S. Tompkins. 1989. Genetic, immunological, and cytotoxic comparisons of Legionella proteolytic activities. Infect. Immun. 57:2719-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robey, M., and N. P. Cianciotto. 2002. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 70:5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robey, M., W. O'Connell, and N. P. Cianciotto. 2001. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect. Immun. 69:4276-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rossier, O., and N. P. Cianciotto. 2001. Type II protein secretion is a subset of the PilD-dependent processes that facilitate intracellular infection by Legionella pneumophila. Infect. Immun. 69:2092-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279:485-499. [DOI] [PubMed] [Google Scholar]
- 77.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 78.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 79.Sandkvist, M. 2001. Type II secretion and pathogenesis. Infect. Immun. 69:3523-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scherzinger, E., R. Lurz, S. Otto, and B. Dobrinski. 1992. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 20:41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmidt, S. A., D. Bieber, S. W. Ramer, J. Hwang, C. Y. Wu, and G. Schoolnik. 2001. Structure-function analysis of BfpB, a secretin-like protein encoded by the bundle-forming-pilus operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:4848-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 83.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stone, B. J., and Y. Abu Kwaik. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect. Immun. 66:1768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stone, B. J., and Y. Abu Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Strom, M. S., D. Nunn, and S. Lory. 1991. Multiple roles of the pilus biogenesis protein pilD: involvement of pilD in excretion of enzymes from Pseudomonas aeruginosa. J. Bacteriol. 173:1175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Strom, M. S., D. N. Nunn, and S. Lory. 1993. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc. Natl. Acad. Sci. USA 90:2404-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sturgill-Koszycki, S., and M. S. Swanson. 2000. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 192:1261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 90.Szeto, L., and H. A. Shuman. 1990. The Legionella pneumophila major secretory protein, a protease, is not required for intracellular growth or cell killing. Infect. Immun. 58:2585-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tang, H., M. Kays, and A. Prince. 1995. Role of Pseudomonas aeruginosa pili in acute pulmonary infection. Infect. Immun. 63:1278-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thacker, W. L., H. W. Wilkinson, B. B. Plikaytis, A. G. Steigerwalt, W. R. Mayberry, C. W. Moss, and D. J. Brenner. 1985. Second serogroup of Legionella feeleii strains isolated from humans. J. Clin. Microbiol. 22:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thorpe, T. C., and R. D. Miller. 1981. Extracellular enzymes of Legionella pneumophila. Infect. Immun. 33:632-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Villar, M. T., R. L. Hirschberg, and M. R. Schaefer. 2001. Role of the Eikenella corrodens pilA locus in pilus function and phase variation. J. Bacteriol. 183:55-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Viswanathan, V. K., S. Kurtz, L. L. Pedersen, Y. Abu-Kwaik, K. Krcmarik, S. Mody, and N. P. Cianciotto. 2002. The cytochrome c maturation locus of Legionella pneumophila promotes iron assimilation and intracellular infection and contains a strain-specific insertion sequence element. Infect. Immun. 70:1842-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 98.Watarai, M., I. Derre, J. Kirby, J. D. Growney, W. F. Dietrich, and R. R. Isberg. 2001. Legionella pneumophila is internalized by a macropinocytotic uptake pathway controlled by the Dot/Icm system and the mouse Lgn1 locus. J. Exp. Med. 194:1081-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wiater, L. A., K. Dunn, F. R. Maxfield, and H. A. Shuman. 1998. Early events in phagosome establishment are required for intracellular survival of Legionella. Infect. Immun. 66:4450-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Winn, W. C., Jr. 1988. Legionnaires disease: historical perspective. Clin. Microbiol. Rev. 1:60-81. [DOI] [PMC free article] [PubMed] [Google Scholar]