Abstract
cDNA fragments encoding the carboxyltransferase domain of the multidomain plastid acetyl-CoA carboxylase (ACCase) from herbicide-resistant maize and from herbicide-sensitive and herbicide-resistant Lolium rigidum were cloned and sequenced. A Leu residue was found in ACCases from herbicide-resistant plants at a position occupied by Ile in all ACCases from sensitive grasses studied so far. Leu is present at the equivalent position in herbicide-resistant ACCases from other eukaryotes. Chimeric ACCases containing a 1000-aa fragment of two ACCase isozymes found in a herbicide-resistant maize were expressed in a yeast ACC1 null mutant to test herbicide sensitivity of the enzyme in vivo and in vitro. One of the enzymes was resistant/tolerant, and one was sensitive to haloxyfop and sethoxydim, rendering the gene-replacement yeast strains resistant and sensitive to these compounds, respectively. The sensitive enzyme has an Ile residue, and the resistant one has a Leu residue at the putative herbicide-binding site. Additionally, a single Ile to Leu replacement at an equivalent position changes the wheat plastid ACCase from sensitive to resistant. The effect of the opposite substitution, Leu to Ile, makes Toxoplasma gondii apicoplast ACCase resistant to haloxyfop and clodinafop. In this case, inhibition of the carboxyltransferase activity of ACCase (second half-reaction) of a large fragment of the Toxoplasma enzyme expressed in Escherichia coli was tested. The critical amino acid residue is located close to a highly conserved motif of the carboxyltransferase domain, which is probably a part of the enzyme active site, providing the basis for the activity of fop and dim herbicides.
Keywords: wheat, maize, Lolium, Toxoplasma gondii, herbicide resistance
Acetyl-CoA carboxylase (ACCase), a biotin-dependent carboxylase, produces malonyl-CoA by using bicarbonate as a source of the carboxyl group and ATP as a source of energy. The reaction proceeds in two steps: carboxylation of covalently attached biotin (biotin carboxylase activity) followed by transfer of the carboxyl group to acetyl-CoA (carboxyltransferase activity). In prokaryotes, as well as in plastids of some plants, ACCase is a multisubunit enzyme. In eukaryotes, the cytosolic isozyme and in some instances also the plastid isozyme are multidomain enzymes. In these cases, the three major functional domains (biotin carboxylase, biotin carrier, and carboxyltransferase) are present, in that order, in one large polypeptide.
Plastid-localized ACCase contributes to the control of flux through the plant de novo fatty acid biosynthetic pathway (1, 2). This fact is reflected in the response of sensitive plants to herbicides targeting ACCase, which leads to the inhibition of fatty acid biosynthesis to such an extent that the plant dies. Two classes of such herbicides, aryloxyphenoxypropionates (fops) and cyclohexanediones (dims), are very strong inhibitors of the multidomain plastid ACCase found in grasses and some dicot plants. Plants that rely on the prokaryotic type multisubunit plastid ACCase are resistant to these herbicides. Most other eukaryotes, including animals and yeast, are resistant as well. The ACCase subunit structure, the mode of action of fop and dim herbicides, and their use in agriculture, including the emergence of resistance, has been reviewed recently (3–6). (The chemical structures of the herbicides and the International Survey of Herbicide Resistant Weeds are available at www.weedscience.com.) We have shown recently that the multidomain apicoplast ACCase of Toxoplasma gondii is sensitive to fops. The parasite's growth in human cells is inhibited by some of these herbicides, presumably by inhibiting apicoplast-localized fatty acid biosynthesis (7, 8).
We have also shown previously that the herbicide sensitivity determinant in wheat plastid ACCase is located within a 400-aa fragment of the carboxyltransferase domain and that the second ACCase half-reaction is inhibited (9). Wheat cytosolic and cytosolic/plastid chimeric ACCases can be expressed in yeast and can complement a yeast ACC1 null mutation (9, 10). Furthermore, growth of the gene-replacement strains in the presence of ACCase-targeting inhibitors reflects the properties of ACCase. Such gene-replacement yeast strains provide a very convenient system to study ACCase interaction with inhibitors. In this paper, we report that a single amino acid substitution in the carboxyltransferase domain alters the interaction of ACCase with fop and dim herbicides.
Materials and Methods
Plant Material and cDNA Cloning.
Seeds of herbicide-resistant maize (Zea mays, “SR corn” DK592) were purchased from Dekalb, Dekalb, IL. Seeds of wild-type Lolium (Lolium rigidum) and herbicide-resistant Lolium biotypes (L. rigidum AUS92 and AUS93) were provided by T. Niderman and P. Boutsalis (Syngenta, Basel, Switzerland). RNA from 2-wk-old plants was isolated by using an RNeasy Plant Mini Kit (Qiagen, Chatsworth, CA) according to the manufacturer's protocol. Reverse transcription–PCR was performed by a two-step method by using the 5′RACE System (GIBCO/BRL) according to the manufacturer's protocol. Two gene-specific primers were used for the synthesis of the first cDNA strand: CCTGAACAAACTTCGCTCTCTGAGAG and TAGGAAGAGGTCCACCAATGTTTGC. A 3.2-kb fragment of maize and Lolium plastid ACCase cDNA was PCR-cloned by using the following primers: AGTTGAGGTTATGAAGATGTGCATGC and CAATGTTTGCAGGAACATAGCTGAGC. Plant genomic DNA for PCR was prepared as described previously (11). A 300-bp fragment of the plastid ACCase gene from Lolium biotypes AUS92 and AUS93 was PCR-cloned by using the following primers: ATTAGCTCTTCTGTTATAGCRCA and GCATGTGRGAGCTGTACACTTC. This fragment of the gene had no introns. The High Fidelity PCR System (Roche, Indianapolis, IN) was used for DNA amplification. Multiple clones from each biotype were sequenced.
Chimeric Gene Assembly.
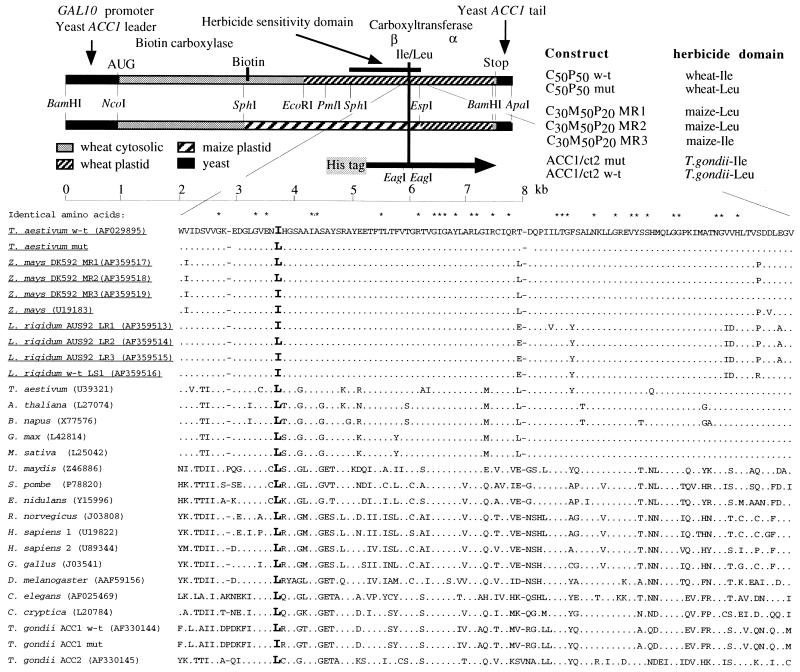
Construction of the C50P50 wheat cytosolic/plastid chimeric ACCase gene consisting of the yeast GAL10 promoter, yeast ACC1 leader, wheat ACCase cDNA, and yeast ACC1 3′-tail was described before (9). Constructs C30M50P20 containing maize sequences were created from C50P50 by replacing the DNA fragment between SphI and EspI sites with maize cDNA fragments PCR-cloned as described above. The C50P50 construct with the Ile (codon ATA) to Leu (codon CTA) substitution was created from C50P50 by replacing the DNA fragment between PmlI and EspI sites with a DNA fragment carrying a single mutation (A to C) introduced by PCR. The yeast shuttle vector pRS426 (12) was used for all of the constructs. The structures of the chimeric genes used in this study are shown in Fig. 1.
Figure 1.
Structure of chimeric ACCases expressed in yeast. Construct names reflect composition of the encoded proteins: C, wheat cytosolic ACCase; P, wheat plastid ACCase; M, maize plastid ACCase. Numbers indicate approximate contribution (%) of each segment. W-t, wild-type (Ile); mut, mutant (Leu). The origin of ACCase coding sequences and yeast DNA segments is indicated by different patterns. The major functional domains (biotin carboxylase and carboxyltransferase) and the biotin attachment sites are located as described before (17–19). The herbicide sensitivity domain was identified before (9). The approximate position of the T. gondii ACCase fragment (amino acid residues 1,861–2,609 of ACC1, GenBank accession no. AF157612) expressed in E. coli, relative to wheat ACCase is indicated with a horizontal arrow. Locations of key restriction sites used in the constructions are shown. Alignment of amino acid sequences of multidomain ACCases at the site of the critical Ile/Leu residue (in bold) and an approximate location of this domain in ACCase are shown. The fragment shown corresponds to residues 1,752–1,864 of wheat plastid ACCase ORF (AF029895). Names of plastid ACCases of grasses are underlined. Dots indicate residues identical to those in Triticum aestivum (AF029895). Dashes indicate gaps.
The ACC1/ct2 construct encoded amino acid residues 1,861–2,609 of the wild-type T. gondii apicoplast ACCase (ACC1, GenBank accession no. AF157612) fused (at a StuI site) to a 33-aa His tag of vector pProEXHTc (GIBCO/BRL) for expression in E. coli strain DH5α and affinity purification. Details of this construction will be described elsewhere. The ACC1/ct2 construct with the Leu (codon CTT) to Ile (codon ATT) substitution was created by replacing a 0.2-kb EagI fragment with the corresponding DNA fragment carrying a single C to A mutation introduced by PCR. Protein affinity purification was carried out by using Ni-NTA Superflow resin (Qiagen) following the manufacturer's protocol.
Complementation of a yeast ACC1 null mutation and tetrad analysis was performed as described previously (10). Saccharomyces cerevisiae strain W303D-ACC1ΔLEU2 (heterozygous strain ACC1/acc1∷LEU2, Fig. 1) was provided by Dr. S. D. Kohlwein (Technical University, Graz, Austria). All complementation experiments were carried out at 23°C. Asci (20–40) were dissected to test complementation by each of the chimeric genes. Genotypes of the resulting haploid strains were verified by plating on selective media and by PCR by using gene-specific primers for the yeast and maize ACCase genes (not shown). Complementation results and growth properties of the yeast gene-replacement strains are summarized in Table 1.
Table 1.
Properties of chimeric genes and gene-replacement yeast strains
| Construct | Strain | Herbicide domain and its source | Complementation | Doubling time, hr | |
|---|---|---|---|---|---|
| C50P50w-t | a | Wheat-Ile | T. a. w-t | Yes | 8.3 ± 0.6 |
| b | 7.9 ± 0.6 | ||||
| C50P50 mut | a | Wheat-Leu | T. a. mut | Yes | 7.9 ± 0.2 |
| b | 4.3 ± 0.3 | ||||
| c | 4.3 ± 0.3 | ||||
| d | 4.0 ± 0.2 | ||||
| C30M50P20MR1 | NA | Maize-Leu | Z. m. MR1 | No | |
| C30M50P20MR2 | a | Maize-Leu | Z. m. MR2 | Yes | 4.8 ± 0.6 |
| b | 5.2 ± 1.0 | ||||
| C30M50P20MR3 | a | Maize-Ile | Z. m. MR3 | Yes | 5.4 ± 2.0 |
| b | 5.2 ± 0.8 | ||||
Inhibitors.
Haloxyfop and Sethoxydim were purchased from Crescent Chemicals (Hauppauge, NY). Clodinafop and Cethoxydim (our name for CGA215684) were provided by Novartis (now Syngenta, Research Triangle Park, NC).
Growth inhibition of the gene-replacement yeast strains by herbicides was assessed as described previously (10) by measuring culture optical density at 600 nm. One OD unit corresponds to culture density of ≈1.3 × 107 cells per ml (9). Yeast media and growth conditions were as described previously (10). The yeast extract/peptone/raffinose/glucose (YPRG) yeast complex medium for vegetative growth contained 1% Bacto-yeast extract, 2% Bacto-peptone, 0.1% adenine sulfate, 2% galactose, and 2% raffinose. Yeast growth and inhibition by the herbicides was measured at 23°C. Inhibitors were added as 100-fold concentrated stock solutions in DMSO. One-percent DMSO was added to the control medium to measure yeast growth in the absence of herbicides.
ACCase enzymatic activity was assayed in Sephadex G50-purified protein extracts from yeast strains (10) by measuring incorporation of 14C from bicarbonate into acid-stable malonyl-CoA as described before (13). The reaction mixture contained 100 mM Tris⋅HCl (pH 8.0–9.5), 100 mM KCl, 5 mM MgCl2, 1 mM ATP, 1 mM DTT, 0.5 mM acetyl-CoA, 0.5% DMSO, 1 mM (1.14 μCi per reaction) [14C]NaHCO3 (Amersham Pharmacia), and an aliquot of purified protein. Herbicides were added as 20-fold concentrated stock solutions in 10% DMSO in 0.1 M Tris⋅HCl (pH 8.0). Reaction mixtures (20-μl) were incubated for 30–45 min at 37°C, mixed with 4 μl of ≈12 M HCl, and dried on filter paper at elevated temperature for 10 min before counting radioactivity by using a scintillation mixture.
Enzymatic activity of the carboxyltransferase portion of ACCase was assayed as described in ref. 14 by measuring transfer of the carboxyl group from malonyl-CoA to biotin ester and formation of acid-unstable [14C]acetyl-CoA and carboxybiotin in the reverse carboxyltransferase reaction. The reaction mixture contained 100 mM Tris⋅HCl (pH 8.0), 100 mM KCl, 1 mM MgCl2, 0.5 mg/ml BSA, 0.4 mM DTT, 5 mM d-biotin methyl ester, 1.5% DMSO, 3.6 μM (0.01 μCi per reaction) [2-14C]malonyl-CoA (Amersham Pharmacia), and an aliquot of affinity-purified protein. Reaction mixtures (25-μl) were incubated for 30–45 min at 37°C, mixed with 25 μl of 6 M HCl, and dried in glass scintillation vials at 95°C for 30 min. Biotin ester was added from a 100-fold concentrated stock solution in DMSO, and inhibitors were added as 20-fold concentrated solutions in 10% DMSO in 0.1 M Tris⋅HCl (pH 8.0). The unreacted substrate was dissolved in 100 μl of water, and the residual radioactivity was measured with scintillation mixture. Control reactions in the absence of biotin or inhibitors were supplemented with an appropriate amount of DMSO. The amount of product was calculated by subtracting the residual radioactivity measured in the presence of the enzyme from the residual radioactivity of a minus-enzyme control, treated in the same way. Nonspecific activity in the absence of the biotin ester was negligible.
Results
Carboxyltransferase Domain of ACCases from Herbicide-Resistant Maize and Lolium.
cDNA fragments (3.2-kb) derived from plastid ACCase genes from herbicide-resistant maize were amplified by reverse transcription–PCR and cloned. An approximate location of this fragment in the ACCase ORF is shown in Fig. 1. Polypeptides encoded by MR1 (GenBank accession no. AF359517) and MR2 (AF359518) cDNAs, represented by 3 and 15 clones, respectively, differ by a single one-amino acid deletion. The sequences of MR1 cDNA differed from cDNA for maize plastid ACCase reported earlier (15) by 7 nucleotides, resulting in 5 aa substitutions. These 3 cDNAs most likely represent variants of the same gene copy found in different populations. A single cDNA (MR3, AF359519) representing another plastid ACCase gene was also cloned. Its sequence differs from the cDNA sequences MR1 and MR2 by 5%, resulting in more than 60 aa substitutions, but it has the same three-nucleotide deletion as MR2. MR3 could represent the second copy of the plastid ACCase gene present in cryptotetraploid maize (16).
Corresponding cDNA fragments encoding plastid ACCase were cloned from herbicide-resistant biotype AUS92 and herbicide-sensitive wild-type Lolium. AUS92 is resistant to both aryloxyphenoxypropionates and cyclohexanediones (P. Boutsalis, personal communication). LR2 cDNA from AUS92 (GenBank accession no. AF359514) was represented by multiple clones. Two additional cDNAs, LR1 (AF359513) and LR3 (AF359515), each represented by a single example, were also cloned. Their sequences differed from the sequence of cDNA LR2 more than could be accounted for by PCR errors. Amino acid sequences encoded by the three cDNAs are 99.5% identical. A single cDNA, LS1 (AF359516), represented by five clones, was detected in the herbicide sensitive (wild-type) Lolium. The amino acid sequence encoded by LS1 differed from the sequences encoded by cDNAs LR1, LR2, and LR3 by 1%. Two copies of the plastid ACCase gene were previously detected in L. rigidum (unpublished observations), which is a diploid species.
Comparison of the amino acid sequences of multidomain ACCases within the 400-aa fragment that includes the herbicide sensitivity determinant (9) revealed a correlation between herbicide sensitivity and the presence of Ile at a site within the domain. All ACCases from sensitive grasses have Ile (codon ATA) at this position whereas one ACCase isozyme in resistant maize and Lolium has Leu (codon TTA) at this position (cDNA MR1, MR2, and LR2). ACCases from other eukaryotes, which are all resistant to herbicides, have Leu at this position. The only exception is apicoplast ACCase from T. gondii, which is sensitive to fops but resistant to dims, and has Leu at this position. Alignment of amino acid sequences of multidomain ACCases at the site of the Ile/Leu residue and an approximate location of this 113-aa fragment in ACCase is shown in Fig. 1.
A 300-bp fragment of genomic DNA encoding the plastid ACCase motif including the Ile/Leu residue was PCR-cloned from resistant Lolium biotype AUS92 and AUS93. AUS93 was shown to be resistant to dims and fops (P. Boutsalis, personal communication). In each case, one copy of the gene encoding Leu and one copy of the gene encoding Ile at the critical position were identified. For AUS92, this result confirmed our observations described above. For AUS93, the result was consistent with the suggestion (P. Boutsalis, personal communication) that AUS92 and AUS93 represent the same biotype collected in consecutive years.
Interaction of ACCases with Herbicides.
Chimeric ACCases containing a 1,000-aa fragment of proteins found in herbicide-resistant maize were expressed in a yeast ACC1 null mutant to test herbicide sensitivity of the enzyme in vivo and in vitro. Expression constructs were derived from construct C50P50 prepared previously (9) by exchanging large DNA restriction fragments at conserved sites. Additionally, a single nucleotide mutation changing Ile (codon ATA) to Leu (codon TTA) at the putative herbicide-binding site of the chimeric wheat ACCase encoded by C50P50 was engineered. The structures of these constructs and the chimeric ACCases they encode are shown in Fig. 1. Three of the chimeric ACCases complement the ACC1 null mutation as revealed by tetrad analysis (Table 1). ACCase encoded by C30M50P20 MR1 did not complement (Table 1), even though there are only a few sequence differences between this construct and C30M50P20 MR2, which did complement. The differences include the three-nucleotide gap in MR2 described above and three nonsynonymous changes found in the PCR-cloned cDNA fragment used for the construction. All of these differences are present only in this particular clone and presumably were introduced by PCR.
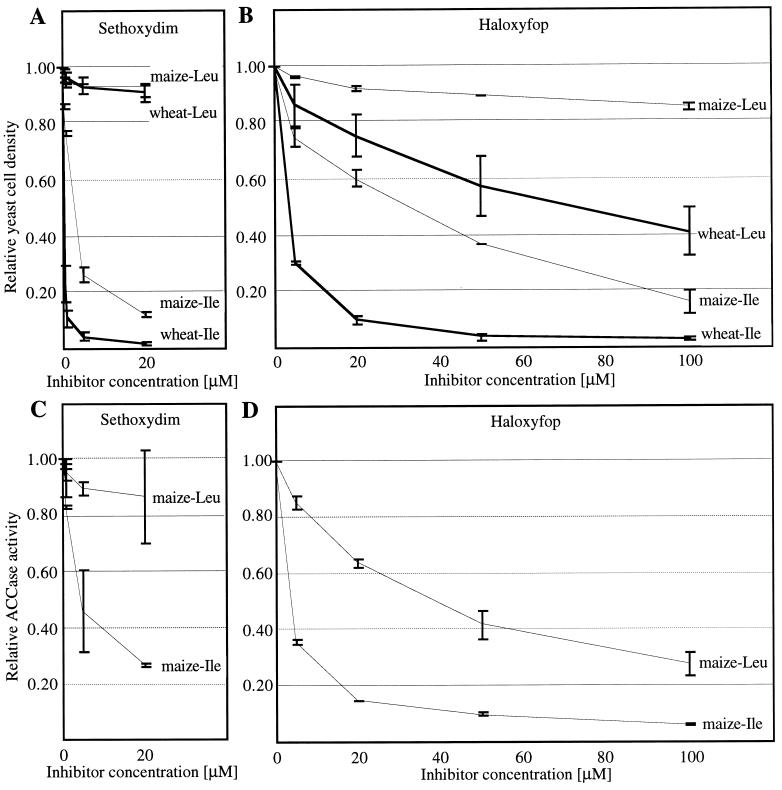
Yeast gene-replacement strains obtained from tetrad dissection grew well, with doubling times similar to that of the parent wild-type yeast diploid strain (Table 1; ref. 9). We have shown previously (9) that growth of such gene-replacement strains in the presence of ACCase-targeting inhibitors reflects the properties of the enzyme. Growth properties of the gene-replacement yeast strains containing 1000-aa fragments of two plastid enzymes from herbicide-resistant maize were significantly different (Fig. 2 A and B). Strains containing ACCase derived from maize variant MR2 were resistant and from maize MR3 were sensitive to herbicides (Fig. 1). Inhibition of the ACCase activity by the same herbicides in partially purified protein extracts from representative yeast strains was measured to confirm that the enzyme properties determine the observed differences in yeast growth. The two inhibition patterns are similar. However, the response of yeast strains to haloxyfop is weaker than that of the corresponding enzymes. Fifty percent inhibition of the activity of ACCase derived from maize variant MR2 requires 10-fold higher haloxyfop concentration than 50% inhibition of the activity of ACCase derived from maize variant MR3 (Fig. 2D). The less sensitive enzyme (construct C30M50P20 MR2) renders the yeast strain resistant. This result can be explained by the fact that, even at the highest concentration of haloxyfop, 100 μM, the less sensitive enzyme retains 30% of its activity, enough for the yeast strain to grow without significant inhibition (Fig. 2).
Figure 2.
Growth inhibition of yeast gene-replacement strains expressing chimeric ACCases by sethoxydim (A) and haloxyfop (B). The structures of chimeral genes are shown in Fig. 1, and growth properties of the yeast strains are shown in Table 1. The strains are identified by the source of their herbicide-binding ACCase domain. Average values for strains containing the same construct (Table 1), each an average of three measurements for each strain, are shown with error bars. Growth without herbicide is set at 1.00. Inhibition of chimeric ACCases by sethoxydim (C) and haloxyfop (D). Protein extracts were prepared from the yeast gene-replacement haploid strains. One strain of each type was tested for resistance to herbicides (C30M50P20 MR2 strain a and C30M50P20 MR3 strain a, Table 1). Chimeric ACCases are identified by the source of their herbicide-binding domain. Average results of two independent experiments are shown with error bars. Activity without herbicide is set at 1.00.
The results of these experiments (Fig. 2) agree with the correlation derived from amino acid sequence comparisons (Fig. 1). One enzyme from resistant maize (MR2) was resistant/tolerant, and one (MR3) was sensitive to haloxyfop and sethoxydim. The sensitive enzyme has an Ile residue, and the resistant one has a Leu residue at the putative herbicide-binding site. Only one of the two copies of the gene needs to be mutated for the plant to become resistant, if the resistant phenotype is dominant.
Effect of a Single Ile to Leu Amino Acid Substitution on Interaction of the Wheat Plastid ACCase with Haloxyfop and Sethoxydim.
Chimeric wheat ACCase encoded by construct C50P50mut had a single Ile residue, encoded by the wild-type construct C50P50w-t, replaced by Leu (Fig. 1). This mutated chimeric enzyme complemented the yeast ACC1 null mutation, and the haploid yeast gene-replacement strains dependent on this enzyme for growth grew well (Table 1). These strains, however, were resistant to sethoxydim and tolerant (modestly sensitive) to haloxyfop (Fig. 2 A and B). This result is in contrast to comparable strains dependent for growth on the wild-type chimeric ACCase (C50P50w-t), which are sensitive to both dims and fops (Fig. 2 A and B; ref. 9). The tolerant phenotype means that 50% growth inhibition of these strains requires 10 times higher concentration of haloxyfop than for the sensitive strains. This result demonstrates that the single amino acid change dramatically affects interaction between representative fops and dims and their target enzyme, and is consistent with the results described above. Active enzyme could not be isolated from these yeast strains, and therefore in vitro inhibition experiments could not be carried out.
Effect of a Single Leu to Ile Substitution on Interaction of the T. gondii Apicoplast ACCase with Aryloxyphenoxypropionates.
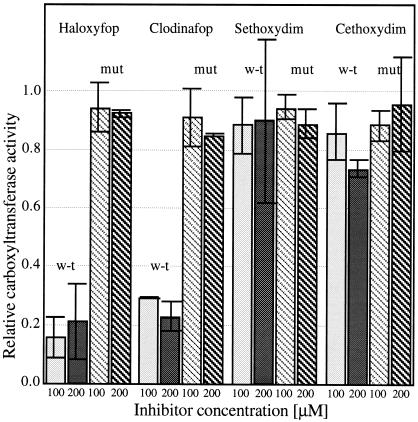
T. gondii apicoplast and cytosolic ACCases (ACC1 and ACC2, respectively, by using the designations for T. gondii) both have a Leu residue at the critical herbicide interaction site identified for plastid ACCases from grasses. This result is consistent with the resistance of both isozymes toward dims and resistance of the cytosolic isozyme toward fops (unpublished results). The apicoplast isozyme (ACC1), however, is sensitive to fops. We have shown recently that the second half-reaction of ACCase (carboxyltransferase activity), measured in reverse, is inhibited by fops if the carboxyltransferase domain is derived from the sensitive T. gondii apicoplast ACCase (unpublished results). A mutated variant of a large fragment of the T. gondii apicoplast ACCase, with the Leu (codon CTT) residue in the putative herbicide-binding site replaced by Ile (codon ATT; Fig. 1), was expressed in E. coli. The expression construct was derived from a similar construct prepared for the expression of the wild-type enzyme. This ACCase fragment, purified by affinity chromatography, had carboxyltransferase activity. The Leu to Ile substitution did not affect interaction with dims: both variants of the enzyme remained resistant. But the substitution made the enzyme resistant to fops (Fig. 3).
Figure 3.
Effect of a single amino acid substitution in T. gondii ACCase on interaction with aryloxyphenoxypropionate and cyclohexanedione inhibitors. Reverse carboxyltransferase activity (ACCase second half-reaction) was measured by using protein fragments expressed in E. coli. Protein structure (ACC1/ct2) and the mutation site are shown in Fig. 1. Average results of two independent experiments, each with replicates, are shown with error bars. Activity without herbicide is set at 1.00. W-t, wild-type (Leu); mut, mutant (Ile).
Discussion
We have shown recently that wheat cytosolic/plastid chimeric ACCases expressed in gene-replacement yeast strains render these strains sensitive to fops and dims and that the herbicide-sensitivity determinant is located in a 400-aa fragment of the carboxyltransferase domain (9). Amino acid sequence comparison of plastid ACCases from herbicide-sensitive and resistant grasses indicated that a single Ile/Leu residue, located within this fragment of the enzyme, appeared to be critical for interaction with dims and fops. This amino acid residue is Ile in sensitive plastid ACCases and Leu in resistant (mutant) plastid ACCases from grasses, and it is Leu in all other ACCases. Our results show that the presence of a Leu residue at the putative herbicide-binding site in plastid ACCase from herbicide-resistant maize coincides with the increased resistance of the enzyme to sethoxydim and haloxyfop. Furthermore, we showed that a single Ile to Leu substitution at the equivalent position in wheat plastid ACCase has the same effect. In both cases, interaction with sethoxydim is affected more strongly than with haloxyfop.
The apicomplexan parasite T. gondii has two ACCases. One of them (ACC1) functions in the apicoplast, a nonphotosynthetic organelle of endosymbiont origin similar to the plant plastid (7, 8). The other isozyme is localized most likely in the cytosol. We have shown recently (unpublished results) that the apicoplast isozyme is sensitive to fops. Its inhibition is most likely responsible for the inhibition of parasite growth in human fibroblasts (8). The cytosolic enzyme is resistant to fops, and both isozymes are resistant to dims (unpublished results). Both proteins have Leu at the putative herbicide-binding site discussed above (Fig. 1). The presence of the Leu residue is consistent with their resistance to dims but not with the sensitivity of the apicoplast ACCase toward fops, which must therefore be determined by other factors. The effect of the Leu to Ile substitution in apicoplast ACCase, yielding an enzyme resistant to representative compounds of both classes of herbicides, is opposite to the effect observed for grass ACCases. This result demonstrates that a small stereochemical change at the herbicide-binding site, including the critical Leu/Ile residue, alters the enzyme–inhibitor interaction. It also indicates that other residues play a role in defining binding specificity and that these were derived by separate evolution of plants and Apicomplexa over a long period. In T. gondii, the Ile residue is no longer compatible with some other structural elements of the inhibitor-binding site.
The simultaneous effect of the same amino acid substitution on interaction of the sensitive plant ACCase with both classes of herbicides indicates a partial overlap between their binding sites. Our results also indicate that fops bind to an equivalent site in the apicoplast ACCase of T. gondii. The critical amino acid residue (Ile/Leu) is located close to a highly conserved motif of the carboxyltransferase domain, which is probably a part of the active site, providing a basis for the activity of fop and dim herbicides. Amino acid substitution at this site, Ile to Leu or vice versa, alters interaction with inhibitors without inactivating the enzyme and thus provides one venue for creating herbicide-resistant grasses.
Acknowledgments
We are grateful to S. D. Kohlwein for providing yeast strains. We thank T. Niderman and P. Boutsalis (Syngenta) for providing plant material and for valuable discussions. This work was supported by grants from the Consortium for Plant Biotechnology Research and Novartis (Syngenta).
Abbreviations
- ACCase
acetyl-CoA carboxylase
- fop
aryloxyphenoxypropionate
- dim
cyclohexanedione
Footnotes
References
- 1.Ohlrogge J B, Jaworski J G. Annu Rev Plant Physiol Mol Biol. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- 2.Page R A, Okada S, Harwood J L. Biochem Biophys Acta. 1994;1210:369–372. doi: 10.1016/0005-2760(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 3.Devine M D. Pestic Sci. 1997;51:259–264. [Google Scholar]
- 4.Herbert D, Walker K A, Price L J, Cole D J, Pallett K E, Ridley S M, Harwood J L. Pestic Sci. 1997;50:67–71. [Google Scholar]
- 5.Incledon B J, Hall J C. Pestic Biochem Physiol. 1997;57:255–271. [Google Scholar]
- 6.Devine M D, Shukla A. Crop Prot. 2000;19:881–889. [Google Scholar]
- 7.Jelenska J, Crawford M J, Harb O S, Zuther E, Haselkorn R, D S, Roos D S, Gornicki P. Proc Natl Acad Sci USA. 2001;98:2723–2728. doi: 10.1073/pnas.051629998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuther E, Johnson J J, Haselkorn R, McLeod R, Gornicki P. Proc Natl Acad Sci USA. 1999;96:13387–13392. doi: 10.1073/pnas.96.23.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolskaya T, Zagnitko O, Tevzadze G, Haselkorn R, Gornicki P. Proc Natl Acad Sci USA. 1999;96:14647–14651. doi: 10.1073/pnas.96.25.14647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joachimiak M, Tevzadze G, Podkowinski J, Haselkorn R, Gornicki P. Proc Natl Acad Sci USA. 1997;94:9990–9995. doi: 10.1073/pnas.94.18.9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards K, Johnstone C, Thompson C. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 13.Gornicki P, Haselkorn R. Plant Mol Biol. 1993;22:547–552. doi: 10.1007/BF00015984. [DOI] [PubMed] [Google Scholar]
- 14.Guchhait R B, Polakis S E, Dimroth P, Stoll E, Moss J, Lane M D. J Biol Chem. 1974;249:6633–6645. [PubMed] [Google Scholar]
- 15.Egli M, Lutz S, Somers D, Gengenbach B. Plant Physiol. 1995;108:1299–1300. doi: 10.1104/pp.108.3.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devos K M, Gale M D. Plant Mol Biol. 1997;35:3–15. [PubMed] [Google Scholar]
- 17.Gornicki P, Podkowinski J, Scappino L A, DiMaio J, Ward E, Haselkorn R. Proc Natl Acad Sci USA. 1994;91:6860–6864. doi: 10.1073/pnas.91.15.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gornicki P, Faris J, King I, Podkowinski J, Gill B, Haselkorn R. Proc Natl Acad Sci USA. 1997;94:14179–14185. doi: 10.1073/pnas.94.25.14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podkowinski J, Sroga G E, Haselkorn R, Gornicki P. Proc Natl Acad Sci USA. 1996;93:1870–1874. doi: 10.1073/pnas.93.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]