Abstract
The LATERAL ORGAN BOUNDARIES (LOB) DOMAIN (LBD) gene family encodes plant-specific transcriptional regulators functioning in organ development. In a screen of Arabidopsis (Arabidopsis thaliana) sequence-indexed transferred DNA insertion mutants, we found disruption of the LOB DOMAIN-CONTAINING PROTEIN20 (LBD20) gene led to increased resistance to the root-infecting vascular wilt pathogen Fusarium oxysporum. In wild-type plants, LBD20 transcripts were barely detectable in leaves but abundant in roots, where they were further induced after F. oxysporum inoculation or methyl jasmonate treatment. Induction of LBD20 expression in roots was abolished in coronatine insensitive1 (coi1) and myc2 (allelic to jasmonate insensitive1) mutants, suggesting LBD20 may function in jasmonate (JA) signaling. Consistent with this, expression of the JA-regulated THIONIN2.1 (Thi2.1) and VEGETATIVE STORAGE PROTEIN2 (VSP2) genes were up-regulated in shoots of lbd20 following treatment of roots with F. oxysporum or methyl jasmonate. However, PLANT DEFENSIN1.2 expression was unaltered, indicating a repressor role for LBD20 in a branch of the JA-signaling pathway. Plants overexpressing LBD20 (LBD20-OX) had reduced Thi2.1 and VSP2 expression. There was a significant correlation between increased LBD20 expression in the LBD20-OX lines with both Thi2.1 and VSP2 repression, and reduced survival following F. oxysporum infection. Chlorosis resulting from application of F. oxysporum culture filtrate was also reduced in lbd20 leaves relative to the wild type. Taken together, LBD20 is a F. oxysporum susceptibility gene that appears to regulate components of JA signaling downstream of COI1 and MYC2 that are required for full elicitation of F. oxysporum- and JA-dependent responses. To our knowledge, this is the first demonstration of a role for a LBD gene family member in either biotic stress or JA signaling.
Plants have evolved inducible defense mechanisms to protect against microbial pathogens, and these include cell wall modifications, the production of antimicrobial metabolites and proteins, and in some instances, hypersensitivity via programmed cell death processes. Several of these host defense responses are transcriptionally regulated via the action of a suite of defense hormones in plants, including salicylic acid (SA), jasmonate (JA), and ethylene (Schenk et al., 2000; Pieterse et al., 2009). In turn, microbial pathogens have evolved mechanisms, such as secreted effector molecules, that avert the activation of these host defense responses (Jones and Dangl, 2006; Boller and He, 2009). Pathogens also reprogram host physiological functions to enhance susceptibility. For example, bacterial and fungal pathogens that enter leaves via stomatal apertures secrete molecules that block stomatal closure (Hok et al., 2010). A further extension of host reprogramming by pathogens is where host processes regulated by growth hormones such as auxins, gibberellins, and cytokinins, are modified by pathogens either by the production of functional hormone analogs by the pathogen itself or via modification of endogenous hormone levels (Robert-Seilaniantz et al., 2007; Grant and Jones, 2009). Alternatively, pathogens can intercept hormone-signaling processes to provide conditions more conducive for infection (Bari and Jones, 2009; Kazan and Manners, 2009). Therefore, to understand the contribution of the host to the final disease outcome, it is necessary to consider pathogen-targeted host processes that may enhance susceptibility in addition to the more commonly studied processes of pathogen perception and the activation of the host defense system.
Fusarium oxysporum is a root-infecting fungal pathogen that causes wilt disease on a broad range of economically important plant species and also the model plant Arabidopsis (Arabidopsis thaliana; Dombrecht et al., 2006; van Hemelrijck et al., 2006; Berrocal-Lobo and Molina, 2008; Michielse and Rep, 2009). This pathogen is soilborne and enters the plant initially through the roots, and subsequently colonizes the vascular tissues and xylem vessels before moving up to stem and foliar tissues (Lagopodi et al., 2002; Czymmek et al., 2007). F. oxysporum is considered to be a hemibiotroph (Thaler et al., 2004) because the initial stages of root infection by this pathogen appear to be biotrophic (Czymmek et al., 2007), while later stages of the infection cycle, particularly the wilting and lesions that occur in foliar tissues, are more typical of the symptoms incited by necrotrophic pathogens. The genomes of pathogen strains of F. oxysporum infecting tomato (Solanum lycopersicum) and Arabidopsis have been sequenced (Ma et al., 2010; Thatcher et al., 2012). Infection by F. oxysporum involves secreted pathogen effector proteins, some of which are encoded on supernumerary pathogenicity chromosomes (Ma et al., 2010). These effectors act as either virulence or avirulence factors, depending on the host genotype (Rep and Kistler, 2010; Takken and Rep, 2010; Thatcher et al., 2012).
The ability to study F. oxysporum interactions on the model host Arabidopsis has opened up diverse genetic- and genomic-based approaches to identify and characterize host factors involved in Fusarium wilt disease development. For example, there is variation in the response to F. oxysporum across Arabidopsis ecotypes and the partial resistance of the commonly studied Columbia-0 (Col-0) ecotype is inherited as a quantitative trait (Diener and Ausubel, 2005). One quantitative trait locus contains the atypical resistance gene RESISTANCE TO FUSARIUM OXYSPORUM1 that encodes WALL-ASSOCIATED KINASE-LIKE KINASE22, but how this gene contributes to resistance is currently unknown (Diener and Ausubel, 2005). One possibility discussed by Diener and Ausubel (2005) is that resistance may be at least partially mediated via SA-regulated defenses. Exogenously applied SA provides increased resistance to this pathogen (Edgar et al., 2006) and transgenic and mutant genotypes that are impaired in SA accumulation have enhanced susceptibility (Diener and Ausubel, 2005; Thatcher et al., 2009).
Recent studies indicate that successful F. oxysporum infection requires the action of diverse host hormonal-signaling pathways, their associated transcriptional regulators, and downstream-regulated response genes. In contrast to SA, some hormone-signaling pathways appear to promote disease susceptibility to F. oxysporum in Arabidopsis. Several components of Arabidopsis auxin-signaling pathways and polar auxin transport processes, but not auxin biosynthesis itself, have been shown to be required for full virulence of F. oxysporum on Arabidopsis (Kidd et al., 2011), suggesting a link between infection and development (Kazan and Manners, 2009). Application of abscisic acid (ABA), a plant hormone usually associated with abiotic stress responses such as water deficit, stimulated increased Fusarium wilt disease development in Arabidopsis while mutations in ABA biosynthesis genes promoted resistance (Anderson et al., 2004). This suggested that ABA signaling may act to prioritize abiotic stress tolerance processes over defense to pathogens like F. oxysporum (Anderson et al., 2004).
The role of the JA-signaling pathway in Arabidopsis-F. oxysporum interactions is of particular interest as it appears to have two opposing effects that either repress or stimulate disease development. The eventual disease outcome reflects the relative balance of these two JA-regulated processes (Thatcher et al., 2009). First, it appears that JA-regulated defenses contribute positively to resistance. For example, the JA-regulated THIONIN2.1 (Thi2.1) gene, which encodes an antimicrobial thionin protein, inhibits infection by F. oxysporum when overexpressed in transgenic plants (Epple et al., 1997; Chan et al., 2005). Overexpression of transcriptional activators (e.g. ETHYLENE RESPONSE FACTOR1 [ERF1]) of JA-responsive defense genes also reduces F. oxysporum infection (Berrocal-Lobo et al., 2002; McGrath et al., 2005). On the other hand, negative transcriptional regulators of JA-responsive defense genes (e.g. ERF4 and MYC2) confer increased F. oxysporum susceptibility (Anderson et al., 2004; McGrath et al., 2005). JAs act in plants by their conjugated form being recognized by the CORONATINE INSENSITIVE1 (COI1) protein (Katsir et al., 2008), and surprisingly the Arabidopsis coi1 mutant shows strong resistance to F. oxysporum despite greatly reduced JA-dependent defenses (Thatcher et al., 2009). Interestingly, coi1 is also nonresponsive to chlorosis-inducing factors present in culture filtrates of F. oxysporum (Thatcher et al., 2009). The strong resistance to F. oxysporum observed in coi1 mutants has been proposed to be due to a reduction of JA-induced senescence, which is exploited by the pathogen to cause disease symptoms such as chlorosis and necrosis at late stages of infection (Thatcher et al., 2009). Similarly analysis of the constitutive expression of pr genes5/hypersenescence1 mutant with constitutively active defenses and enhanced senescence response, shows increased F. oxysporum susceptibility (Schenk et al., 2005). More recently, the MEDIATOR25/PHYTOCHROME AND FLOWERING TIME1 (MED25/PFT1) subunit of the plant mediator complex, which positively regulates the JA-responsive defense genes, has also been shown to act as a F. oxysporum susceptibility gene and mutants show reduced expression of JA-responsive genes but increased F. oxysporum resistance (Kidd et al., 2009).
Despite the importance of root pathogens in plant agriculture and natural ecosystems, much less is known about defense signaling by roots when compared with that of aerial plant organs (Okubara and Paulitz, 2005; Erb et al., 2009). Because F. oxysporum infects via the roots it would be expected that key signaling events determining resistance and susceptibility are initiated by both host and pathogen in root tissues early on during infection. The JA-signaling pathway and downstream responses, but not those of the SA pathway, appear to be activated in both the roots and leaves of Arabidopsis during infection by F. oxysporum (Edgar et al., 2006; Thatcher et al., 2009; Kidd et al., 2011). Activation of JA-regulated genes in leaves was also observed within 24 h after inoculation (Kidd et al., 2011), prior to the invasion of foliar tissues by the fungus, suggesting that systemic signals of host and/or pathogen origin are most likely transmitted from root to shoot during F. oxysporum infection. The critical importance of JA signaling in infected roots was elegantly demonstrated using the coi1 mutant in grafting experiments (Thatcher et al., 2009). It was found that plants consisting of a coi1 rootstock with wild-type scion had strong resistance to F. oxysporum similar to that of plants with coi1 rootstock and coi1 scion. In contrast, plants with a wild-type rootstock and a coi1 scion remained susceptible, indicating that JA-perception and -signaling processes in the roots are critical in determining the eventual disease outcome in Fusarium wilt disease. However, our understanding of the genes involved in JA signaling that determines resistance and susceptibility to F. oxysporum is very limited.
The aim of this study was to identify novel root-expressed genes of Arabidopsis that are required for susceptibility to F. oxysporum, and then to characterize these genes to determine whether they have roles in JA signaling and plant defense regulation or other mechanisms. The approach that we adopted was to initially undertake large-scale unbiased screening of a collection of defined sequence-indexed transferred DNA (T-DNA) insertion mutants of Arabidopsis (O’Malley and Ecker, 2010) and identify mutants that had a reproducible increase in resistance to infection by F. oxysporum when compared with that of the wild type. This was followed by verification of the observed resistant phenotype for the candidate gene by testing a second independent mutant carrying a distinct T-DNA insertion allele in the candidate gene. Because we were particularly interested in genes that function in roots, we then undertook expression analysis of the candidate genes in wild-type plants to test for root expression. In this article, we report on the LATERAL ORGAN BOUNDARIES (LOB) DOMAIN-CONTAINING PROTEIN20 (LBD20) gene that was identified through this process. LBD20 is a member of the plant-specific LBD gene family and we present evidence herein that LBD20 has a novel role as a predominantly root-expressed negative regulator of both resistance to F. oxysporum and a subset of JA responses. The LBD family has previously mainly been studied in regard to plant development with roles in defining the boundaries between organs (Shuai et al., 2002; Majer and Hochholdinger, 2011; Feng et al., 2012). Other LBDs have been shown to have functions in the regulation of nitrogen metabolism and anthocyanin biosynthesis (Rubin et al., 2009). Functions of LBD20 were previously unknown, and our results demonstrate LBD20 is the first member of the LBD family shown to have a role in JA signaling and plant-pathogen interactions.
RESULTS
Large-Scale Screening of Arabidopsis Mutants Identifies LBD20 as an F. oxysporum Susceptibility Gene
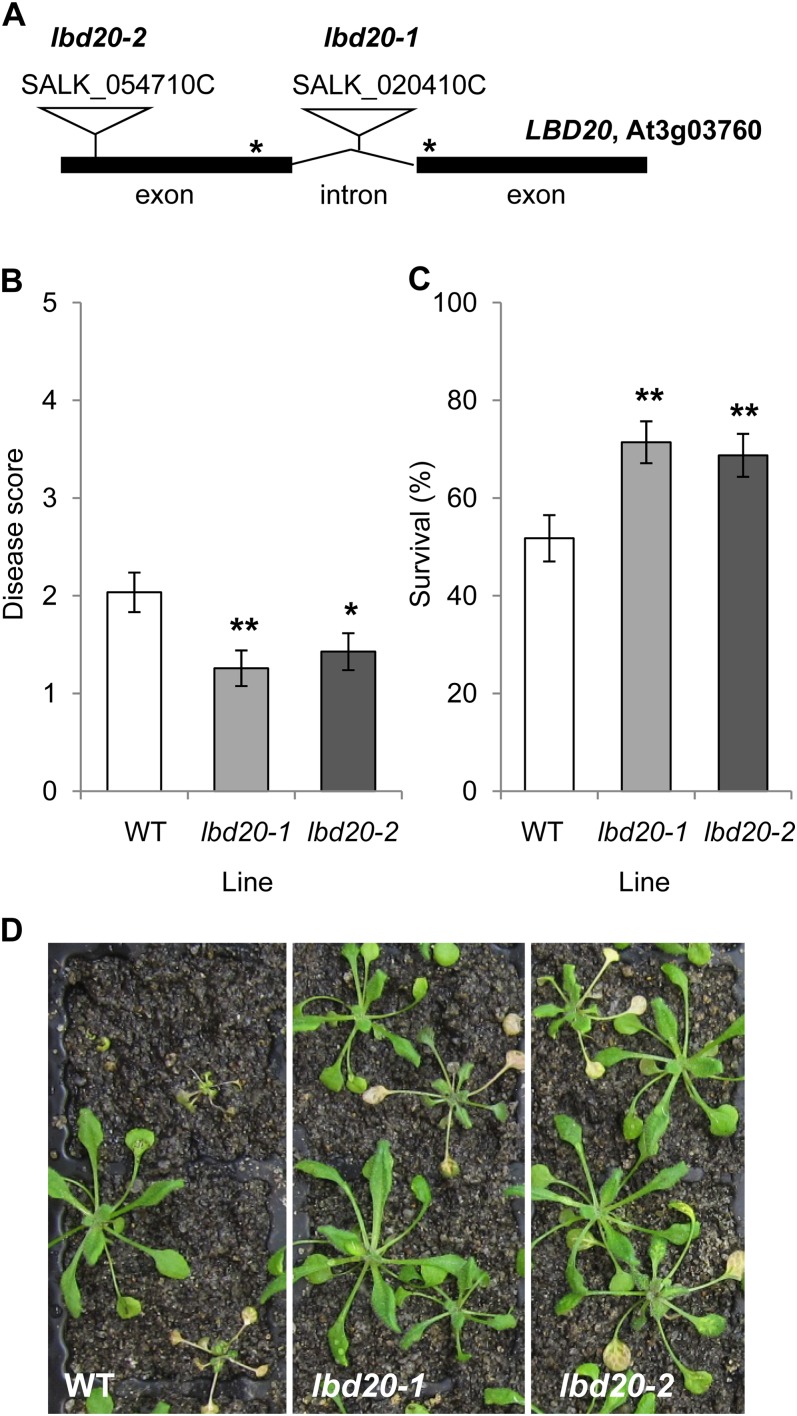
To identify novel genes that affect resistance and susceptibility to F. oxysporum, we systematically screened an Arabidopsis sequence-indexed T-DNA insertion mutant collection (CS27941) consisting of 6,868 T-DNA insertion lines for an altered disease phenotype when compared with that of the wild-type Col-0. Disease phenotypes were determined at 7 and 14 d post inoculation by recording the percentage of diseased plants, survival ratio, and a disease score (rated on a 0–5 scale with 0 being highly resistant and 5 being highly susceptible; see Supplemental Fig. S1). Mutants that showed statistically significant (P < 0.01) disease development compared with Col-0 were selected and rescreened for confirmation of a significantly altered disease phenotype. One of the mutants recovered from this process that showed increased resistance was SALK_020410C (designated here as lbd20-1) and has a T-DNA insertion in the intron of the LBD20 gene. A second homozygous independent mutant line of LBD20 designated as lbd20-2 was obtained (SALK_054710C) with a T-DNA insertion in exon 1 (Fig. 1A). Both lbd20 mutants were confirmed by quantitative reverse transcription (qRT)-PCR to be similarly compromised for LBD20 transcript levels when compared with that of the wild type and are thus expected to be nonfunctional (Supplemental Fig. S2A). Both lbd20 mutants showed significantly increased resistance to F. oxysporum when compared with the wild type both in disease symptoms and plant survival (Fig. 1, B–D). This further indicated a role for LBD20 in susceptibility to Fusarium wilt disease and represents the first case of a LBD gene being implicated in resistance or susceptibility to any plant pathogen, to our knowledge.
Figure 1.
Two independent lbd20 T-DNA mutant lines exhibit increased resistance to F. oxysporum. A, Schematic representation of lbd20-1 and lbd20-2 T-DNA insertion lines. Not drawn to scale. Primer binding sites for determination of LBD20 expression are noted with asterisks. B to d, Disease phenotypes of F. oxysporum inoculated plants with disease score at 14 d post inoculation (B), survival at 21 d post inoculation (C), and representative images of plants 14 d post inoculation (D). The average of three biological replicates consisting of 14 plants each is shown with se. Asterisks indicate values that are significantly different (**P < 0.01, *P < 0.05 Student’s t test) from the wild type (WT).
LBD20 Is Preferentially Expressed in Roots and Responsive to F. oxysporum Infection
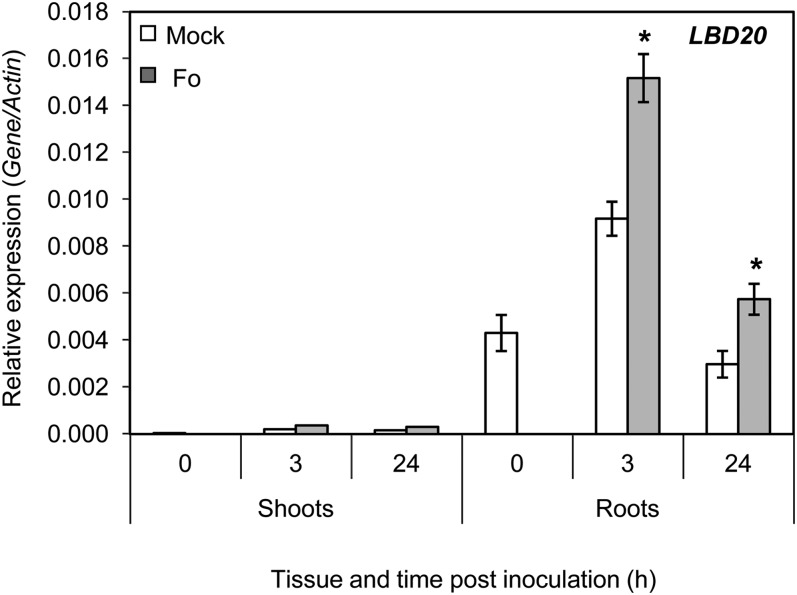
As stated above, we were particularly interested in genes expressed within root tissues where F. oxysporum penetration and the early stages of infection take place, and several LBD genes have previously been shown to be expressed in roots (Shuai et al., 2002; Feng et al., 2012). To test this in further detail for the LBD20 gene, we monitored its expression in wild-type shoot and root tissues before and after F. oxysporum inoculation (Fig. 2). LBD20 expression in shoots could not be reliably detected using qRT-PCR, suggesting it is either very lowly expressed in shoot tissues or expressed within very specific shoot cell types. We did, however, readily measure LBD20 expression in roots just prior to inoculation (time point 0) and after inoculation (Fig. 2). LBD20 expression in 3-h mock-treated samples also increased 2-fold over time point 0 samples, suggesting LBD20 expression is responsive to the inoculation method that involved the potential wounding of roots as they are removed from soil, dipped in water (mock treatment), and repotted. LBD20 expression in F. oxysporum-inoculated samples was significantly higher than those of mock treatments at both 3 and 24 h (Fig. 2). Combined, these results indicate LBD20 is predominantly root expressed and responsive to F. oxysporum infection.
Figure 2.
LBD20 is root expressed and responsive to F. oxysporum infection. LBD20 expression was monitored in wild-type (WT) shoot and root tissue at time 0 h (no treatment) and in mock or F. oxysporum (Fo) challenged plants at 3 and 24 h post inoculation. 0 represents roots and shoots taken from plants just prior to inoculation. The average of three biological replicates consisting of pools of 20 to 30 plants is shown with se. Gene expression levels are relative to the internal control β-actin genes. Asterisks indicate values that are significantly different (*P < 0.05 Student’s t test) from mock treatment at the same time point.
LBD20 Expression Is COI1 and JA Regulated
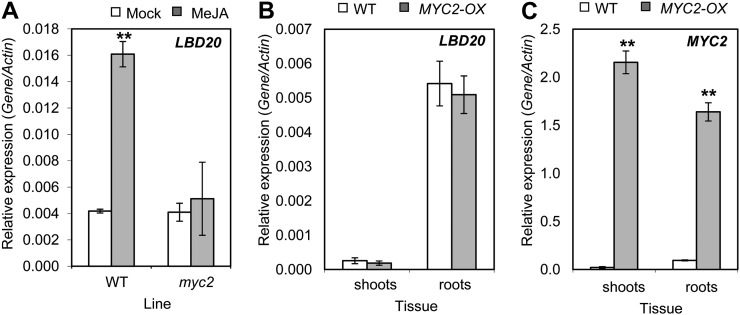
We previously determined that the JA receptor COI1 plays a vital role in susceptibility to F. oxysporum, in particular in root tissues where wild-type scions grafted onto rootstocks in which the COI1 gene has been silenced show complete resistance to F. oxysporum disease symptom development (Thatcher et al., 2009). To determine if LBD20 functions within the framework of the COI1-dependent JA-signaling pathway, we monitored LBD20 expression in wild-type and coi1 roots following F. oxysporum inoculation and found the induced expression of LBD20 to be completely abolished in coi1 (Fig. 3A). This prompted us to examine the potential JA inducibility of LBD20. Expression of LBD20 was induced in root tissues following the transfer of seedlings to methyl jasmonate (MeJA)-containing growth medium (3.8-fold over mock) and this induction was also COI1 dependent (Fig. 3B). These results demonstrate that LBD20 is regulated by the COI1-dependent signaling pathway and response.
Figure 3.
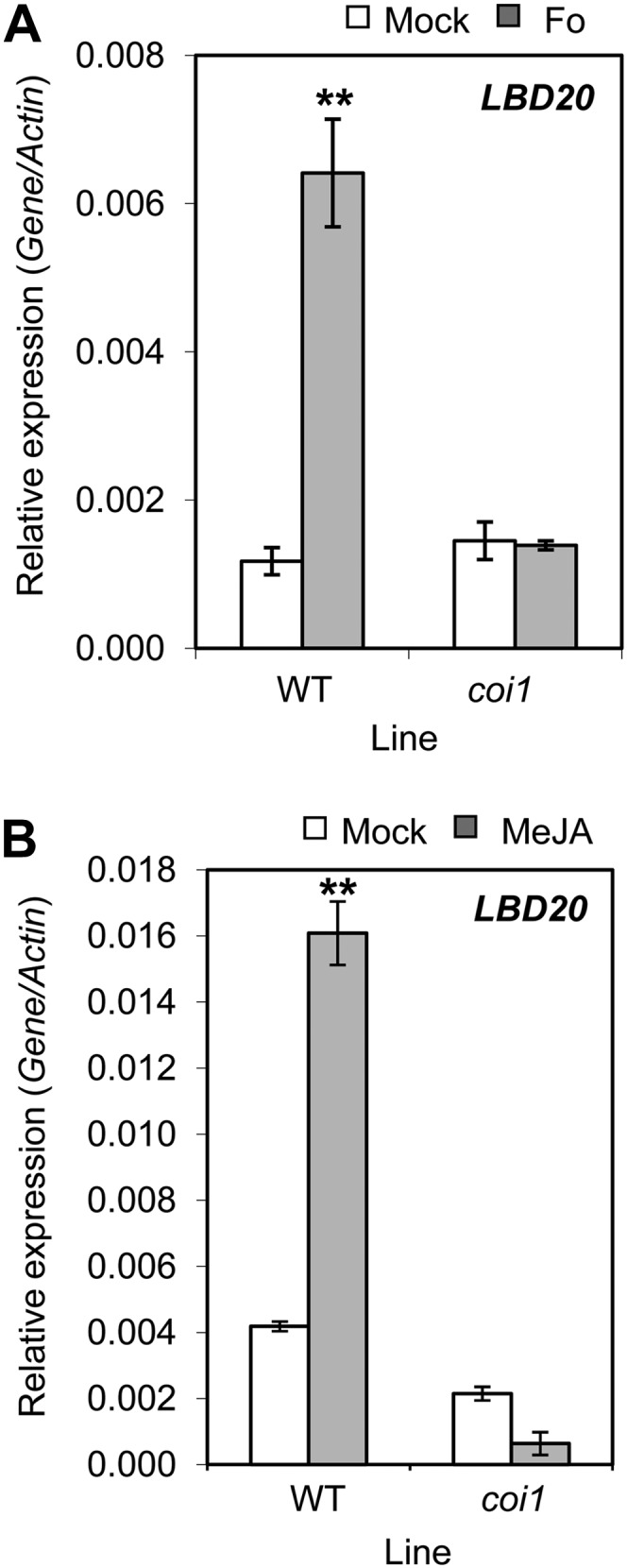
F. oxysporum- and JA-induced LBD20 expression is COI1 dependent. LBD20 expression was monitored in root tissue of mock or F. oxysporum (Fo) challenged wild-type (WT) or coi1 plants at 96 h post infection (A), and in WT and coi1 plants 6 h post mock or MeJA treatment (B). The average of three biological replicates consisting of pools of 10 to 30 plants is shown with se. Gene expression levels are relative to the internal control β-actin genes. Asterisks indicate values that are significantly different (**P < 0.01 Student’s t test) from mock treatment within the same line. Similar results were obtained in an independent experiment.
MeJA-Induced Expression of LBD20 Is Dependent on MYC2
One of the key regulatory genes in early JA-signaling events downstream of COI1 is the transcriptional regulator MYC2 that is thought to act, at least in part, by binding to the G-box cis-element (5′-CACGTG-3′). The LBD20 promoter contains two potential MYC2-binding G-box motifs (−794 to −789, and −338 to −333, relative to the predicted transcription start site). To test whether MYC2 regulates LBD20, we examined LBD20 expression within the myc2 mutant background and found MeJA-induced expression of LBD20 in roots was abolished in myc2 (Fig. 4A). We also examined LBD20 in MYC2-overexpressing (MYC2-OX) plants (35S::MYC2) without providing any MeJA stimulus (Fig. 4, B and C). LBD20 expression, however, did not differ between wild-type and MYC2-OX plants. These findings suggest that LBD20 is part of the JA and MYC2 transcriptional regulon but that up-regulation of MYC2 expression alone is insufficient to stimulate expression of LBD20. Further investigations are required to determine whether these potential MYC2-binding motifs found in the LBD20 promoter are functional and/or positive. Regulation of LBD20 by MYC2 may require JA or pathogen treatment. We also examined MYC2 expression in the lbd20 mutant background and found no difference in F. oxysporum or MeJA induction patterns compared with wild-type plants in either shoot or root tissues (data not shown).
Figure 4.
LBD20 MeJA-induced expression is MYC2 dependent. A, LBD20 expression was examined in wild-type (WT) and myc2 root tissue 6 h post mock or MeJA treatment. B and C, LBD20 and MYC2 expression were examined in shoot and root tissue of WT and 35S::MYC2 (MYC2-OX) plants. MYC2 was examined to confirm its overexpression. Gene expression levels are relative to the internal control β-actin genes. The average of three biological replicates consisting of pools of 30 plants is shown with se. Asterisks indicate values that are significantly different (**P < 0.01 Student’s t test) from mock treatment within the same line (A) or WT within the same tissue (B and C). Similar results were obtained in an independent experiment.
LBD20 Is a Repressor of a Subset of JA-Regulated Defense Genes in Shoot Tissues
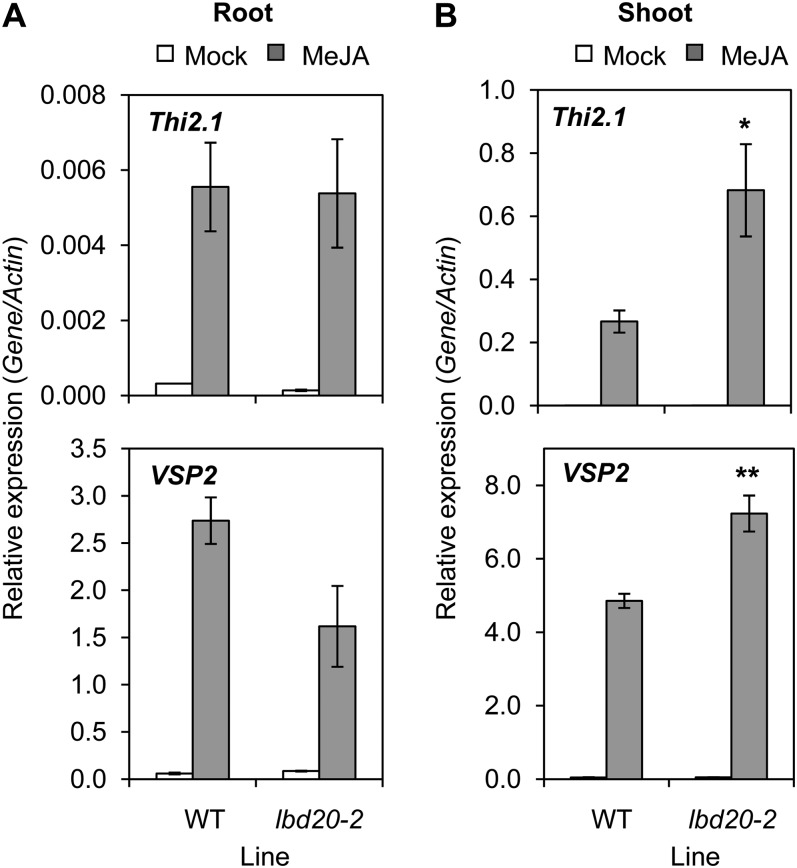
The up-regulation of LBD20 in F. oxysporum- and MeJA-treated wild-type plants and its COI1 and MYC2 dependency prompted us to examine the expression of four well-established marker genes (Thi2.1, VEGETATIVE STORAGE PROTEIN2 [VSP2], PLANT DEFENSIN1.2 [PDF1.2], and PATHOGENESIS RELATED4 [PR4]) for downstream JA-regulated defense responses in the lbd20 mutant. Initially, we examined root tissue following treatment with MeJA. Although transcripts for only Thi2.1, VSP2, and PDF1.2 were induced in root tissue, there was no apparent difference in the induction between the lbd20 mutant and the wild type (Fig. 5A; Supplemental Fig. S3A). However, examination of shoot tissue indicated that all four genes were MeJA induced and that a subset of these JA-response genes were differentially regulated in the lbd20 mutant. For example, expression of the Thi2.1 and VSP2 genes were more strongly induced (P < 0.05) in the lbd20 mutant than the similarly treated wild type (Fig. 5B). In a separate confirmatory experiment, the repressive function on defense gene expression was also confirmed for both lbd20 alleles (e.g. Supplemental Fig. S2B). In contrast, there was no difference detected in the expression of the PDF1.2 and PR4 genes in shoots following MeJA treatment (Supplemental Fig. S3B). We also examined Thi2.1 and VSP2 expression following F. oxysporum inoculation and found the same induction pattern in shoots and roots (Supplemental Fig. S4). These results suggest that LBD20 plays a role in JA signaling and acts as a repressor of a subgroup of JA- and pathogen-induced defense genes in shoots. Given the predominant root expression of the LBD20 gene (Fig. 2), it is possible that LBD20 may either function in a specialized JA-related root-to-shoot signaling process or that very low expression levels in shoots may be sufficient for this regulatory role.
Figure 5.
LBD20 is a repressor of a subset of JA-regulated defense genes. Expression of JA-response genes was examined in wild-type (WT) and lbd20 root (A) or shoot tissue (B) 6 h post mock or MeJA treatment. Gene expression levels are relative to the internal control β-actin genes. The average of three biological replicates consisting of pools of 30 to 40 plants is shown with se. Asterisks indicate values that are significantly different (**P < 0.01, *P < 0.05 Student’s t test) from WT. Similar results were obtained in independent experiments.
F. oxysporum Culture Filtrate-Induced Chlorosis Is Reduced in lbd20
We have previously observed that factors present in F. oxysporum culture filtrates induce a senescence-like chlorotic phenotype in wild-type leaves and this response is absent in the coi1 mutant, which is also highly resistant to F. oxysporum disease symptom development (Thatcher et al., 2009). To determine if the increased F. oxysporum resistance in lbd20 also affects this phenotype, we applied F. oxysporum culture filtrate to wild-type and lbd20 detached leaves, alongside coi1 and myc2 (Fig. 6). We included myc2, as this mutant also exhibits increased resistance to F. oxysporum (Anderson et al., 2004) and regulates LBD20 (Fig. 4), but its response to F. oxysporum culture filtrate was unknown. All three of the mutants tested had reduced lesion development compared with wild-type leaves. Interestingly, the coi1 mutant was the most insensitive to this treatment, with myc2 and lbd20 mutants showing intermediate sensitivity compared with the wild type. Although the secreted F. oxysporum elicitors that may induce the chlorotic phenotype are currently unknown, the nonresponsiveness and reduced responsiveness of coi1 and myc2 leaves, respectively, to F. oxysporum culture filtrate, suggest that possible fungal elicitors act through the JA-signaling pathway. Combined, these results imply LBD20 may also contribute to a JA-signaling-dependent host sensitivity to fungal elicitors of host senescence and chlorosis.
Figure 6.
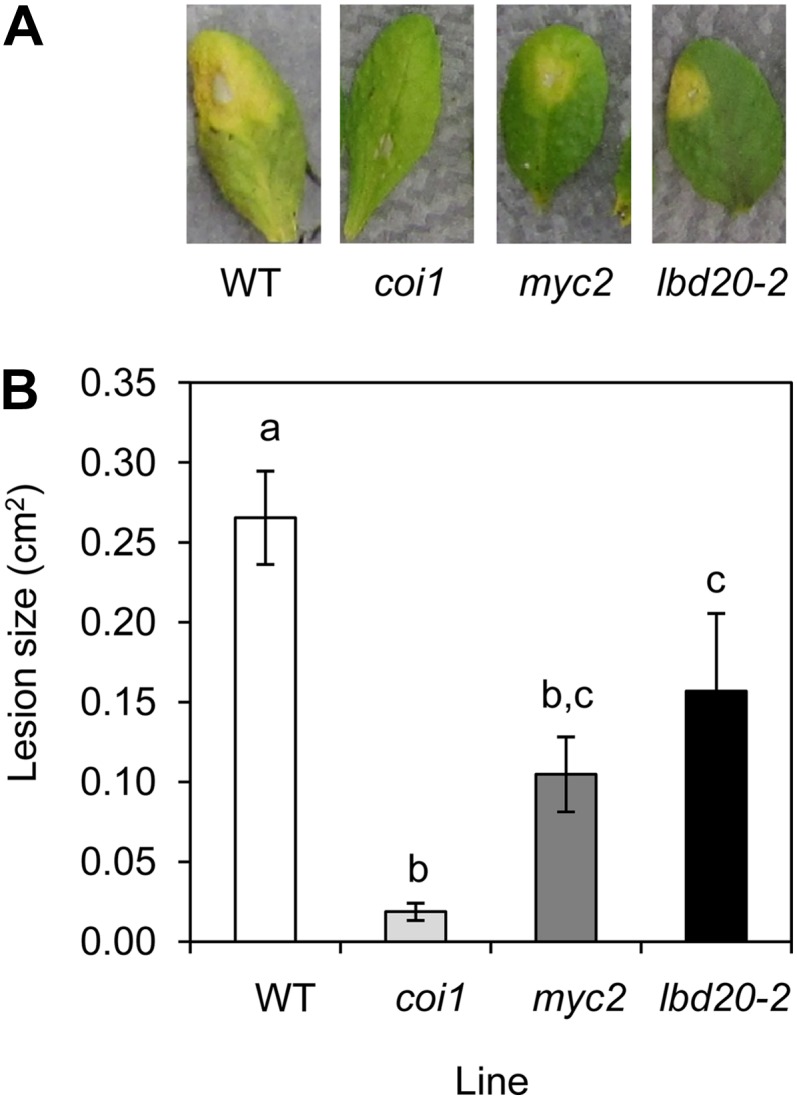
coi1, myc2, and lbd20 leaves are less sensitive to F. oxysporum culture filtrate-induced lesions. A, F. oxysporum culture filtrate initiates a senescence response in wild-type (WT) leaves but lesion size is reduced in coi1, myc2, and lbd20. Representative leaves are shown at 3 d post treatment. Mock treatments of potato dextrose broth (PDB) and water showed no phenotype (data not shown). B, Average lesion size from 15 leaves shown with se. Letters indicate values that are significantly different from each other (P < 0.05, all pairs Student’s t test). Similar results were obtained in an independent experiment.
Increased LBD20 Expression Correlates with Reduced Thi2.1 and VSP2 Expression and Susceptibility to F. oxysporum
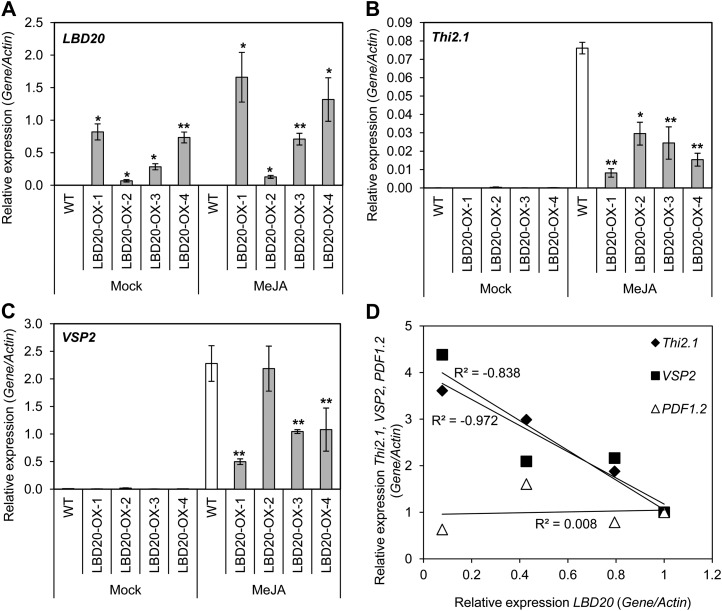
To further characterize the role of LBD20 in defense and JA signaling, we generated LBD20-overexpressing plants (LBD20-OX). We noted that the LBD20-OX plants suffered from varying degrees of lobed leaves, sterility, and termination of development (Supplemental Fig. S5A). Five of the 24 recovered LBD20-OX plants appeared similar to the wild type, nine were sterile, and five died. Similar phenotypes have been observed in plants overexpressing some other LBD genes (Shuai et al., 2002; Nakazawa et al., 2003; Naito et al., 2007). LBD20-OX transformants with milder phenotypes set viable seed and were used in subsequent experiments. From these T2 plants, two lines in either the wild-type Col-0 background or the lbd20 background, and carrying only one LBD20-OX insertion, were analyzed for LBD20 expression (Fig. 7A).
Figure 7.
LBD20 is a repressor of a subset of JA-regulated defense genes. A to C, LBD20, Thi2.1, and VSP2 expression was examined in shoot tissue of wild-type (WT) and LBD20-OX lines 6 h post mock or MeJA treatment. LBD20-OX lines 1 and 2 are in the WT background, while lines 3 and 4 are in the lbd20-2 mutant background. Gene expression levels are relative to the internal control β-actin genes. The average of three biological replicates consisting of pools of 30 plants is shown with se. Asterisks indicate values that are significantly different (**P < 0.01, *P < 0.05 Student’s t test) from WT under the same treatment. No significant differences in expression levels among mock-treated samples were observed for Thi2.1 or VSP2. D, MeJA-induced expression of defense marker genes in LBD20-OX lines was plotted against their internal LBD20 expression. Displayed are trend lines.
To test the hypothesis that LBD20 is a repressor of a subset of JA-regulated defense genes, we analyzed the expression of Thi2.1, VSP2, and PDF1.2 in wild-type and LBD20-OX plants after mock or MeJA treatment. Following MeJA treatment, three of the four LBD20-OX lines exhibited significantly (P < 0.01) lower expression of both Thi2.1 and VSP2 than wild-type plants, while LBD20-OX-2 only had significantly reduced Thi2.1 expression (P < 0.05; Fig. 7, B and C). Of the four LBD20-OX lines tested, LBD20-OX-2 also had the lowest LBD20 levels (Fig. 7A) and exhibited no altered leaf morphology (Supplemental Fig. S5B). This suggests a threshold level of LBD20 expression may be required to observe its effects on plant development. Indeed, we found a strong negative correlation between LBD20 levels and MeJA inducibility of Thi2.1 and VSP2 (Fig. 7D). Consistent with the hypothesis that LBD20 only represses a subset of JA-regulated defense genes, we found no correlation between PDF1.2 and LBD20 expression in the LBD20-OX lines (Fig. 7D).
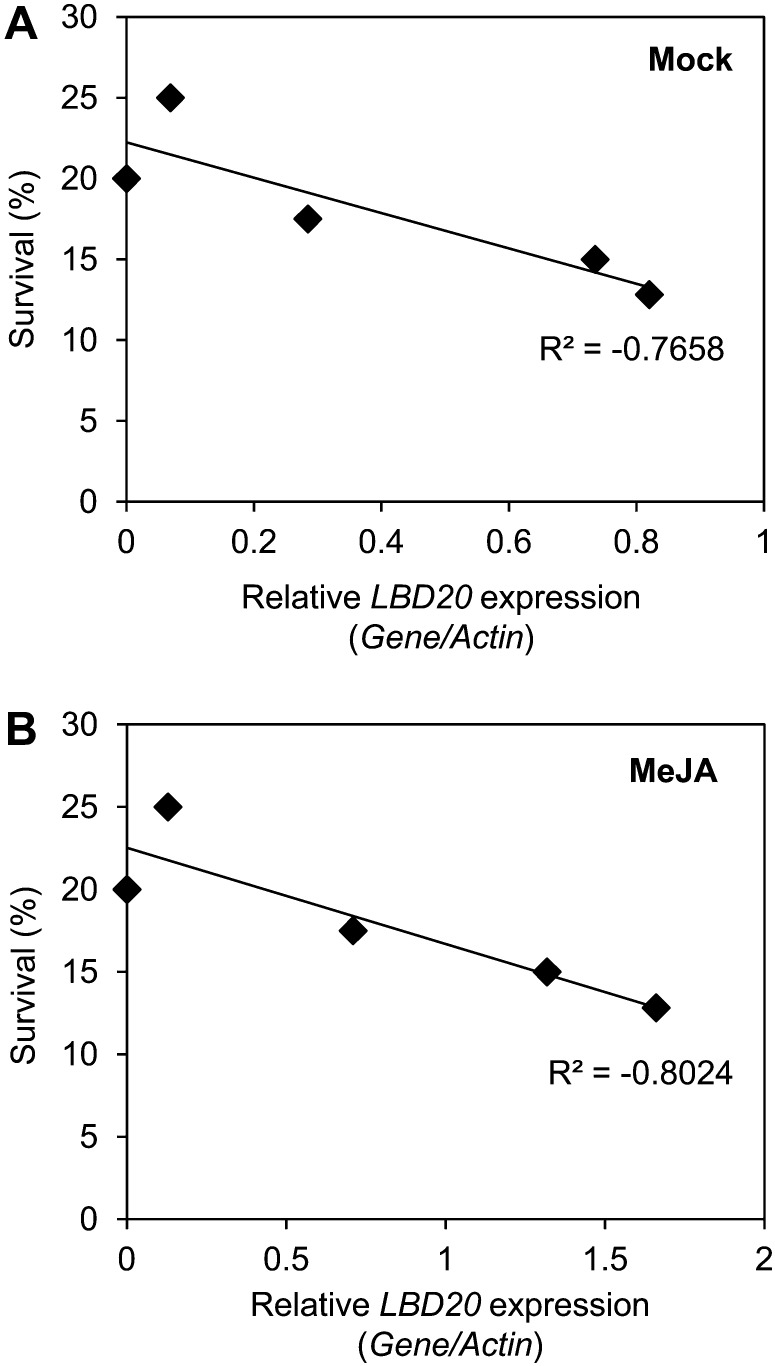
We also inoculated wild-type plants and LBD20-OX lines with F. oxysporum and monitored disease symptom development. There were also strong correlations between both the levels of basal (R2 = −0.77) and JA-induced (R2 = −0.80) LBD20 expression across the LBD20-OX lines and reduced plant survival following inoculation with F. oxysporum (Fig. 8). The diminishing survival of inoculated plants with increasing LBD20 expression strongly supports the conclusion that LBD20 acts as a F. oxysporum susceptibility gene in Arabidopsis.
Figure 8.
Increased LBD20 expression correlates with increased susceptibility to F. oxysporum. Percentage survival of F. oxysporum inoculated wild-type and LBD20-OX plants plotted against their internal LBD20 expression under mock (A) and JA-induced conditions (B). Displayed are trend lines. For F. oxysporum inoculation, the average of two biological replicates consisting of 20 plants each is shown.
DISCUSSION
We initially identified LBD20 as a host-susceptibility gene from a screen of nearly 7,000 independent homozygous sequence-indexed T-DNA insertion lines for increased resistance to the root-infecting pathogen F. oxysporum. Among the mutants identified, lbd20 was selected for further analysis because its increased resistance was confirmed in a second independent homozygous T-DNA insertion line, and because LBD20 was predominantly expressed in roots, the site of primary infection. Even though F. oxysporum infects and colonizes through root tissues, our understanding of host gene functions in root tissue during infection is very limited. LBD20 belongs to the plant-specific LBD family consisting of 42 members in Arabidopsis based on the defining member LOB (Shuai et al., 2002). LOB was initially identified from an enhancer trap screen for genes expressed at the adaxial base of initiating lateral organs and other family members identified based on conservation of the so-called N-terminal LOB domain (Shuai et al., 2002). For most LBD family members the function is not known, but for some, roles have been defined in organ development, anthocyanin, and nitrogen metabolism, as well as in responses to phytohormones such as cytokinin, auxin, and gibberellin (Borghi et al., 2007; Bureau et al., 2010; Majer and Hochholdinger, 2011; Feng et al., 2012). To our knowledge, our findings represent the first functional characterization of LBD20 and the first demonstration of a role for any LBD family member in plant-pathogen interactions.
The JA-signaling pathway appears to play two contrasting roles in plant responses to F. oxysporum infection (Thatcher et al., 2009). One JA-regulated pathway promotes susceptibility via the pathogen hijacking JA-induced senescence processes for disease symptom development. Another JA-regulated pathway appears to promote resistance via the expression of JA-induced defenses such as antifungal proteins like thionins, defensins, and chitinases. While it is known that defense gene overexpression or up-regulation in shoot tissues is associated with increased resistance (Epple et al., 1997; Tierens et al., 2002; Anderson et al., 2004; Berrocal-Lobo and Molina, 2004; McGrath et al., 2005; van Hemelrijck et al., 2006), it is not the dominant JA-regulated process in determining the disease outcome during F. oxysporum infection because mutants compromised in defense gene expression such as coi1 and pft1 are still highly resistant to this pathogen (Kidd et al., 2009; Thatcher et al., 2009). To determine the resistance mechanism in the lbd20 mutants, we analyzed JA-mediated defense gene expression following F. oxysporum inoculation. A subset of JA-mediated defense genes were up-regulated following fungal infection in lbd20 shoot tissues compared with wild-type plants, with the same pattern also observed after MeJA treatment (Fig. 5). This defense gene subset included that encoding the antifungal protein Thi2.1 known to reduce disease severity to F. oxysporum (Epple et al., 1997, 1998) and the anti-insect and wound-responsive protein VSP2, while no regulatory effect was observed on the plant defensin marker gene PDF1.2. The chlorosis and senescence-like response elicited by application of F. oxysporum culture filtrate was also reduced in lbd20 (Fig. 6), although not to the degree seen in coi1 that is insensitive to these F. oxysporum elicitors. These results suggest that the increased resistance observed in the lbd20 mutants may be due to a combination of the enhanced production of some JA-regulated antifungal defense proteins such as thionins, as well as a partial reduction in JA-induced chlorosis and senescence processes that are required for symptom development.
The increased expression of some JA-regulated defense genes in the lbd20 mutants indicates that LBD20 has a repressive role for a part of the JA plant-signaling pathway. It is possible that this may be mediated by transcriptional regulation of LBD20 itself as it is also a JA- and F. oxysporum-responsive gene and its transcriptional induction by the fungus and JA is dependent on COI1 and MYC2, the respective JA receptor and a primary transcriptional regulator of JA signaling, respectively. It has been shown that a complex network of transcription factors are regulated downstream of MYC2 and suggested that different branches of this network are responsible for diverse JA-regulated functions (Dombrecht et al., 2007). This model is consistent with the notion that LBD20 regulates a component of JA signaling downstream of COI1 and MYC2. An analysis of public array data (Zimmermann et al., 2004; L. Thatcher, unpublished data) also identified altered JA-defense gene expression in the lbd38 mutant with PDF1.2 and VSP2 expression severalfold higher in lbd38 compared with wild-type plants, suggesting other LBD proteins may act as repressors of JA responses and this warrants a LBD family-wide investigation for this role.
Although it appears that the induction of LBD20 by F. oxysporum infection and JA treatments requires MYC2, regulation downstream of LBD20 differs from that of MYC2. For example, VSP2 induction was attenuated in myc2 following MeJA treatment, while the defensin PDF1.2, PR4 (Hevein-like, encoding a chitin-binding protein), and Thi2.1 were up-regulated (Anderson et al., 2004; Lorenzo et al., 2004; L. Thatcher, unpublished data). Neither PDF1.2 nor PR4 expression was altered in lbd20 compared with the wild type; however, both Thi2.1 and VSP2 were up-regulated. The MYC2 network (Dombrecht et al., 2007) may have repressive and activating branches for genes such as PDF1.2 and VSP2, respectively. In this scenario, LBD20 may occupy a repressive branch or feedback loop that dampens VSP2 expression. The MYC2 regulon is also differentially regulated in shoots and roots temporally during F. oxysporum infection (Anderson et al., 2004; Thatcher et al., 2009; L. Thatcher, unpublished data). In combination with upstream regulators, such as JASMONATE-ZIM-DOMAIN proteins, and downstream regulators, such as LBD20 and ERF1, the JA-dependent response can be finely tuned.
In agreement with published data (Shuai et al., 2002), we found LBD20 expression was virtually absent from leaves, but readily detected in roots (Fig. 2) and floral tissues (data not shown). The highly root-abundant expression of LBD20 compared with that of shoots, taken in combination with the differential expression of JA-regulated defense genes in shoots of the lbd20 mutants, suggests that either LBD20 has a role in a root-to-shoot signaling process that affects specific JA-responsive genes, or that very low levels of transcription of LBD20 in shoots or in specific shoot cells are sufficient for its regulatory activity. Distinguishing these possibilities will require substantially more research, but there are several precedents for the systemic regulation of JA responses from remote tissues. Root inoculation of soybean (Glycine max) with the symbiont Bradyrhizobium japonicum led to activation of JA responses in shoots but not in roots (Kinkema and Gresshoff, 2008), and colonization of roots by certain soil bacteria, in particular Pseudomonas spp. and Bacillus spp., can protect aboveground plant tissues in a JA-dependent manner against different types of pathogens in a process known as induced systemic resistance (for review, see Van der Ent et al., 2009). Treatment with endogenous elicitors or wounding of roots also causes similar systemic effects. For example, wounding or systemin treatment of tomato roots caused induction of systemic responses in leaves and this was dependent on JA signaling (Li et al., 2002), and wounding of Arabidopsis roots caused increased JA biosynthesis in shoots but not roots (Hasegawa et al., 2011). Thus it is possible that LBD20 may be involved in the development of regulatory signals that move from roots to shoots.
Five other LBD genes display similar or root-specific expression patterns like LBD20, with two of these, LBD14 and LBD33, the closest phylogenetically related to LBD20 (Shuai et al., 2002; Matsumura et al., 2009). This suggests subgroups of LBD proteins may have unique roles in roots. In root tissues, the founding member LOB is expressed at the base of lateral roots and at the junction between the primary root and lateral root primordia (Shuai et al., 2002). These points are the preferential site of F. oxysporum colonization and penetration in Arabidopsis (Czymmek et al., 2007; Kidd et al., 2011). Although the cell-specific expression pattern of LBD20 is unknown, it is tempting to speculate that specific root cells with increased levels of LBD20 are more susceptible to F. oxysporum infection. Examination of public array data for LBD genes (26 members with probe sets) responsive to MeJA or other necrotrophic pathogens indicates LBD37 and LBD41 are induced or repressed, respectively, >2-fold in response to Alternaria brassicicola and Botrytis cinerea. This implies other LBD proteins may function in disease responses to fungal pathogens.
LBD function is believed to be mediated, at least in part, through the LOB domain. This domain also determines the unique role of each LBD protein with LOB domains from other members unable to functionally replace each other (Matsumura et al., 2009; Majer and Hochholdinger, 2011). The LOB domain contains a Cys repeat (C motif), a conserved Gly residue, and a Leu-zipper-like motif. The C motif containing four cysteines is conserved in all LBD proteins and is predicted to form a DNA-binding zinc finger, while the less-conserved Leu-zipper-like motif is predicted to form a coiled-coil motif involved in homodimerization or other protein-protein interactions (Shuai et al., 2002; Majer and Hochholdinger, 2011). Nuclear localization and DNA-binding activity shown for some members suggests LBD proteins function as transcription factors (Shuai et al., 2002; Husbands et al., 2007; Naito et al., 2007; Rubin et al., 2009). Indeed, the LOB domain is sufficient for DNA-binding activity, and Husbands et al. (2007) found several LBD members could bind a 6-bp consensus motif (G)CGGC(G), termed the LBD motif, with a broader sequence of A/T C/T GCGGCG C/T/G A/G A/T. Direct promoter targets of LBD transcription factors are not yet known, though the extended LBD motif is present in a diverse set of over 40 genes (L. Thatcher, unpublished data). We found putative LBD motifs in the −1,000-bp Thi2.1 (CTACGGCACTT) and VSP2 promoters (GCACGGCTATG; GTGCGGCGAAT), but not in the PDF1.2 promoter, suggesting LBD20 may directly bind to the Thi2.1 and VSP2 promoters. Further experimental work will be required to confirm this. We also found no significant difference in Thi2.1 or VSP2 expression between root tissue of wild-type and lbd20 plants, suggesting LBD20 either does not bind to these promoters or that other root- and shoot-specific transcription factors are required to mediate their tissue-specific expression.
While knockout mutations of most LBD genes show no obvious phenotypes, their overexpression in many cases results in leaf phenotypes like lobed and curled leaves, along with dwarfing and degrees of infertility (Shuai et al., 2002; Nakazawa et al., 2003; Naito et al., 2007; Mangeon et al., 2011). Similarly, we observed no obvious morphological changes in leaf or root morphology in either lbd20 mutant, however both lines exhibited increased resistance to F. oxysporum accompanied by increased expression of a subset of JA-regulated defense genes. In affirmation, LBD20-OX plants had reduced JA-mediated defense gene expression (Fig. 7), but also suffered from varying degrees of altered leaf morphology, sterility, and development (Supplemental Fig. S5). Overexpression lines with minimal, or no abnormalities were selected for inoculation experiments, and it was shown that a correlation existed between LBD20 expression, reduced Thi2.1 and VSP2 expression, and increasing plant susceptibility, measured as plant survival following inoculation (Figs. 7 and 8). Thus, results from the T-DNA insertion mutants and the overexpression transgenic plants support the notion that LBD20 contributes to susceptibility to F. oxysporum.
Another transcription factor that represses JA responses and functions within the LBD family framework is the MYB transcription factor MYB91/ASYMMETRIC LEAVES1 (AS1; Nurmberg et al., 2007). MYB91/AS1 binds to LBD6/AS2 to repress the expression of class 1 KNOTTED-like homeobox genes (for review, see Moon and Hake, 2011). In as1/myb91 or lbd6/as2 mutants the ectopic misexpression of KNOTTED-like homeobox genes results in plants with strongly lobed leaves. MYB91/AS1 also acts as a negative regulator of JA-inducible genes such as Thi2.1 and VSP1, and as a susceptibility gene to the fungal necrotrophic pathogens B. cinerea and A. brassicicola (Nurmberg et al., 2007). Nurmberg and colleagues (2007) also showed MYB91/AS1 could bind to the promoters of over 30 genes responsive to B. cinerea, JA, or ethylene, including many involved in defense responses (e.g. ERF, NUCLEOTIDE BINDING SITE-LEUCINE RICH REPEAT, GLUTATHIONE S-TRANSFERASE TAU7, CONSTITUTIVE EXPRESSION OF VSP1, Thaumatin family gene). While MYB91/AS1-LBD6/AS2 binding implies LBD6 may be involved in defense responses, increased fungal resistance and up-regulation of JA defenses is not evident in lbd6/as2 (Nurmberg et al., 2007). Some pathogens also actively target AS1 to induce disease symptom development. The tomato yellow leaf curl China virus effector βC1 competes with LBD6/AS2 for AS1 binding to selectively repress JA-responsive defense genes including PDF1.2, PR4, and VSP1 (Yang et al., 2008). The F. oxysporum effector Fo5176-SECRETED IN XYLEM4 also promotes increased disease symptom development through a mechanism yet to be discovered, but it is suggested it may act with other Fo5176 effectors to activate components of the JA-signaling pathway (Thatcher et al., 2012). Other isolates of F. oxysporum and Pseudomonas syringae seem to target host JA signaling by secreting oxylipins or coronatine that mimic the host’s endogenous JA signal (Miersch et al., 1999; Kloek et al., 2001; Katsir et al., 2008; Thatcher et al., 2009). These studies detail a common theme where pathogens selectively target host susceptibility genes to cause disease.
CONCLUSION
In summary, we identified LBD20 as a novel highly root-expressed negative regulator of a subset of JA responses and as a susceptibility gene for Fusarium wilt disease. It will now be interesting to determine the role of other LBD family members in plant-pathogen interactions, to determine the active targets of LBD proteins, and to determine cell- and tissue-specific LBD20 expression and function to explore the potential of LBD20 to regulate root-to-shoot signaling processes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Unless otherwise specified, all experiments were conducted with the wild-type Arabidopsis (Arabidopsis thaliana) Col-0 ecotype grown in soil under a short-day light regime (8-h ligh/16-h dark) at 21°C as described previously (Campbell et al., 2003; Edgar et al., 2006). For growth of seedlings on Murashige and Skoog salt plates, seeds were surface sterilized and plated on one-half-strength Murashige and Skoog (supplied with 3% Suc, 0.8% Bacto Agar, pH 7.2), stratified at 4°C, and incubated under the same conditions as soil-grown plants. The T-DNA insertion mutants (Alonso et al., 2003) lbd20-1 (SALK_020410C), lbd20-2 (SALK_054710C), myc2 (SALK_061267C), and coi1 (SALK_035548) were obtained from the Arabidopsis Biological Resource Centre. T-DNA mutants were confirmed for correct loci insert and homozygous state using the iSct Primers tool at http://signal.salk.edu/cgi-bin/tdnaexpress. lbd20-1 plants were used for initial gene expression experiments under F. oxysporum infection. For all other experiments, the lbd20-2 line was used. Homozygous coi1 plants were selected on Murashige and Skoog plates containing 50 μm MeJA. 35S::MYC2 plants are described in Dombrecht et al. (2007). For generation of plants expressing the LBD20 (35S::LBD20), the LBD20 CDS was amplified using LBD20-HindIII-F 5′-GTTTAAGCTTAACAATGGCTGATCAGCAGCGAG-3′ that includes an ATG start codon, and LBD20-EcoRI-R 5′-GGTAGAATTCTCATCTCCGGTGAAAATCC-3′. The resulting amplicon cloned into HindIII/EcoRI digested binary vector pKEN (McGrath et al., 2005) and confirmed by sequencing. 35S::LBD20 pKEN was mobilized into Agrobacterium tumefaciens AGL1 and transformed into Arabidopsis Col-0 and lbd20-2 as per McGrath et al. (2005). Transgenic plants were selected based on their resistance to 10 mg/L Pestanal (glufosinate ammonium; Riedel-de Haen) and resulting T2 and T3 lines were used in subsequent experiments.
F. oxysporum Inoculation
The F. oxysporum isolate used in this study was strain Fo5176 obtained from Dr. Roger Shivas, Queensland Plant Pathology Herbarium, Queensland Department of Primary Industries and Fisheries, Brisbane, Australia. Root-dip inoculations on 3- to 4-week-old plants with a 1 × 106 cell/mL spore suspension were performed as described (Campbell et al., 2003; Edgar et al., 2006; Thatcher et al., 2009).
qRT-PCR
qRT-PCR experiments were performed on tissue collected after mock, F. oxysporum, or MeJA treatment. For analysis of root and shoot tissues, plants were cut at the top of the root, just below the crown, so shoot tissue consisted of the hypocotyl and aerial tissue. Three biological replicates were taken in all experiments consisting of tissue samples pooled separately from 10 to 30 plants grown and treated at the same time in the same environment. For gene expression under MeJA treatment, 14-d-old plants were germinated on Murashige and Skoog plates then gently lifted into a mock medium (Murashige and Skoog broth) or 100 μm MeJA medium (Murashige and Skoog medium plus MeJA) such that the roots were submerged, and left for 6 h before harvesting. For experiments using coi1, homozygous coi1 plants that are JA insensitive were selected on Murashige and Skoog agar plates containing 50 μm MeJA and at 7 d of age transferred to Murashige and Skoog-only medium. For all experiments, plants were gently lifted from the soil or broth, rinsed in water, blotted on filter paper, frozen in liquid nitrogen, and stored at −80°C. RNA extraction, complementary DNA synthesis, and qRT-PCR were conducted as described by McGrath et al. (2005) using an Applied Biosystems 7900HT fast real-time PCR system. Absolute gene expression levels relative to the previously validated (Anderson et al., 2004; Kidd et al., 2009; Thatcher et al., 2009) reference gene mix β-actin2, β-actin7, and β-actin8 (At1g49240, At3g18780, and At5g09810, respectively) were used for each complementary DNA sample using the equation: relative ratio gene of interest/actin = (Egene−Ct gene)/(Eactin−Ct actin) where Ct is the cycle threshold value. The β-actin mix contains reverse primers for each of the three β-actin genes and a universal forward primer. The mean expression range of the reference gene was found to be within ±1 Ct across all samples. The gene-specific primer sequences have mostly been previously published (Anderson et al., 2004; McGrath et al., 2005; Edgar et al., 2006; Kidd et al., 2009; Thatcher et al., 2009) and are also listed in Supplemental Table S1.
F. oxysporum Culture Filtrate Assay
F. oxysporum culture filtrate assays were performed as per Thatcher et al. (2009) on 15 leaves per line. Lesion size was measured at 3 d post inoculation using the ImageJ freeware package (http://rsb.info.nih.gov/ij/).
Sequence data from this article can be found in the The Arabidopsis Information Resource data libraries under accession numbers At3g03760 (LBD20), At1g32640 (MYC2), At1g72260 (Thi2.1), At3g04720 (PR4), At5g24770 (VSP2), and At5g44420 (PDF1.2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. F. oxysporum disease score ratings system.
Supplemental Figure S2. Analysis of lbd20 T-DNA mutants.
Supplemental Figure S3. PDF1.2 and PR4 expression in wild-type versus lbd20 plants following MeJA treatment.
Supplemental Figure S4. LBD20 is a repressor of a subset of JA-regulated defense genes following F. oxysporum infection.
Supplemental Figure S5. Plants overexpressing LBD20 have altered leaf morphology and fertility.
Supplemental Table S1. qRT-PCR primers used in gene expression analyses.
Supplementary Material
Acknowledgments
We thank Roger Shivas for the F. oxysporum (strain Fo5176) and the Arabidopsis Biological Resource Centre for seeds of Arabidopsis mutants. We also thank Anca Rusu for excellent technical assistance and Emma Campbell, Emily Christoffels, Cameron Edgar, Karri Hartley, Andrew Kettle, Lyeta Payet, Amber Stephens, Fiona Soper, Mucella Tekeoglu, and Shi Zhuge for technical assistance in F. oxysporum inoculation experiments. We also thank Dr. Brendan Kidd for critical reading of the manuscript and useful discussions.
Glossary
- MeJA
methyl jasmonate
- JA
jasmonate
- SA
salicylic acid
- Col-0
Columbia-0
- ABA
abscisic acid
- qRT
quantitative reverse transcription
- T-DNA
transferred DNA
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16: 3460–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JD. (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A. (2004) Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol Plant Microbe Interact 17: 763–770 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A. (2008) Arabidopsis defense response against Fusarium oxysporum. Trends Plant Sci 13: 145–150 [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29: 23–32 [DOI] [PubMed] [Google Scholar]
- Boller T, He SY. (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324: 742–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi L, Bureau M, Simon R. (2007) Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell 19: 1795–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau M, Rast MI, Illmer J, Simon R. (2010) JAGGED LATERAL ORGAN (JLO) controls auxin dependent patterning during development of the Arabidopsis embryo and root. Plant Mol Biol 74: 479–491 [DOI] [PubMed] [Google Scholar]
- Campbell EJ, Schenk PM, Kazan K, Penninckx IA, Anderson JP, Maclean DJ, Cammue BP, Ebert PR, Manners JM. (2003) Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiol 133: 1272–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YL, Prasad V, Sanjaya, Chen KH, Liu PC, Chan MT, Cheng CP. (2005) Transgenic tomato plants expressing an Arabidopsis thionin (Thi2.1) driven by fruit-inactive promoter battle against phytopathogenic attack. Planta 221: 386–393 [DOI] [PubMed] [Google Scholar]
- Czymmek KJ, Fogg M, Powell DH, Sweigard J, Park SY, Kang S. (2007) In vivo time-lapse documentation using confocal and multi-photon microscopy reveals the mechanisms of invasion into the Arabidopsis root vascular system by Fusarium oxysporum. Fungal Genet Biol 44: 1011–1023 [DOI] [PubMed] [Google Scholar]
- Diener AC, Ausubel FM. (2005) RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171: 305–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Kazan K, Manners JM (2006) Improved resistance to Fusarium wilt through genetic engineering of defense signaling pathways. In JA Teixeira da Silva, ed, Floriculture, Ornamental and Plant Biotechnology (Global Science Books), pp 388–398 [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar CI, McGrath KC, Dombrecht B, Manners JM, Schenk PM, Maclean DJ, Kazan K. (2006) Salicylic acid mediates resistance to the vascular wilt pathogen Fusarium oxysporum in the model host Arabidopsis thaliana. Australas Plant Pathol 35: 581–591 [Google Scholar]
- Epple P, Apel K, Bohlmann H. (1997) Overexpression of an endogenous thionin enhances resistance of Arabidopsis against Fusarium oxysporum. Plant Cell 9: 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple P, Vignutelli A, Apel K, Bohlmann H. (1998) Differential induction of the Arabidopsis thaliana Thi2.1 gene by Fusarium oxysporum f. sp. matthiolae. Mol Plant Microbe Interact 11: 523–529 [DOI] [PubMed] [Google Scholar]
- Erb M, Lenk C, Degenhardt J, Turlings TC. (2009) The underestimated role of roots in defense against leaf attackers. Trends Plant Sci 14: 653–659 [DOI] [PubMed] [Google Scholar]
- Feng Z, Sun X, Wang G, Liu H, Zhu J. (2012) LBD29 regulates the cell cycle progression in response to auxin during lateral root formation in Arabidopsis thaliana. Ann Bot (Lond) 110: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant MR, Jones JDG. (2009) Hormone (dis)harmony moulds plant health and disease. Science 324: 750–752 [DOI] [PubMed] [Google Scholar]
- Hasegawa S, Sogabe Y, Asano T, Nakagawa T, Nakamura H, Kodama H, Ohta H, Yamaguchi K, Mueller MJ, Nishiuchi T. (2011) Gene expression analysis of wounding-induced root-to-shoot communication in Arabidopsis thaliana. Plant Cell Environ 34: 705–716 [DOI] [PubMed] [Google Scholar]
- Hok S, Attard A, Keller H. (2010) Getting the most from the host: how pathogens force plants to cooperate in disease. Mol Plant Microbe Interact 23: 1253–1259 [DOI] [PubMed] [Google Scholar]
- Husbands A, Bell EM, Shuai B, Smith HMS, Springer PS. (2007) LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res 35: 6663–6671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. (2009) Linking development to defense: auxin in plant-pathogen interactions. Trends Plant Sci 14: 373–382 [DOI] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K. (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BN, Kadoo NY, Dombrecht B, Tekeoglu M, Gardiner DM, Thatcher LF, Aitken EA, Schenk PM, Manners JM, Kazan K. (2011) Auxin signaling and transport promote susceptibility to the root-infecting fungal pathogen Fusarium oxysporum in Arabidopsis. Mol Plant Microbe Interact 24: 733–748 [DOI] [PubMed] [Google Scholar]
- Kinkema M, Gresshoff PM. (2008) Investigation of downstream signals of the soybean autoregulation of nodulation receptor kinase GmNARK. Mol Plant Microbe Interact 21: 1337–1348 [DOI] [PubMed] [Google Scholar]
- Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN. (2001) Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J 26: 509–522 [DOI] [PubMed] [Google Scholar]
- Lagopodi AL, Ram AF, Lamers GE, Punt PJ, Van den Hondel CA, Lugtenberg BJ, Bloemberg GV. (2002) Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol Plant Microbe Interact 15: 172–179 [DOI] [PubMed] [Google Scholar]
- Li L, Li C, Lee GI, Howe GA. (2002) Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc Natl Acad Sci USA 99: 6416–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma LJ, van der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ, Di Pietro A, Dufresne M, Freitag M, Grabherr M, Henrissat B, et al. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer C, Hochholdinger F. (2011) Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci 16: 47–52 [DOI] [PubMed] [Google Scholar]
- Mangeon A, Bell EM, Lin WC, Jablonska B, Springer PS. (2011) Misregulation of the LOB domain gene DDA1 suggests possible functions in auxin signalling and photomorphogenesis. J Exp Bot 62: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y, Iwakawa H, Machida Y, Machida C. (2009) Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J 58: 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. (2005) Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol 139: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse CB, Rep M. (2009) Pathogen profile update: Fusarium oxysporum. Mol Plant Pathol 10: 311–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch O, Bohlmann H, Wasternack C. (1999) Jasmonates and related compounds from Fusarium oxysporum. Phytochemistry 50: 517–523 [Google Scholar]
- Moon J, Hake S. (2011) How a leaf gets its shape. Curr Opin Plant Biol 14: 24–30 [DOI] [PubMed] [Google Scholar]
- Naito T, Yamashino T, Kiba T, Koizumi N, Kojima M, Sakakibara H, Mizuno T. (2007) A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci Biotechnol Biochem 71: 1269–1278 [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Ichikawa T, Ishikawa A, Kobayashi H, Tsuhara Y, Kawashima M, Suzuki K, Muto S, Matsui M. (2003) Activation tagging, a novel tool to dissect the functions of a gene family. Plant J 34: 741–750 [DOI] [PubMed] [Google Scholar]
- Nurmberg PL, Knox KA, Yun BW, Morris PC, Shafiei R, Hudson A, Loake GJ. (2007) The developmental selector AS1 is an evolutionarily conserved regulator of the plant immune response. Proc Natl Acad Sci USA 104: 18795–18800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Ecker JR. (2010) Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J 61: 928–940 [DOI] [PubMed] [Google Scholar]
- Okubara PA, Paulitz TC. (2005) Root defense responses to fungal pathogens: a molecular perspective. Plant Soil 274: 215–226 [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Rep M, Kistler HC. (2010) The genomic organization of plant pathogenicity in Fusarium species. Curr Opin Plant Biol 13: 420–426 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JDJ. (2007) Pathological hormone imbalances. Curr Opin Plant Biol 10: 372–379 [DOI] [PubMed] [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Rusu AG, Manners JM, Maclean DJ. (2005) The SEN1 gene of Arabidopsis is regulated by signals that link plant defence responses and senescence. Plant Physiol Biochem 43: 997–1005 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM. (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97: 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Peña CG, Springer PS. (2002) The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol 129: 747–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken F, Rep M. (2010) The arms race between tomato and Fusarium oxysporum. Mol Plant Pathol 11: 309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JS, Owen B, Higgins VJ. (2004) The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiol 135: 530–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher LF, Gardiner DM, Kazan K, Manners JM. (2012) A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Mol Plant Microbe Interact 25: 180–190 [DOI] [PubMed] [Google Scholar]
- Thatcher LF, Manners JM, Kazan K. (2009) Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J 58: 927–939 [DOI] [PubMed] [Google Scholar]
- Tierens KF, Thomma BP, Bari RP, Garmier M, Eggermont K, Brouwer M, Penninckx IA, Broekaert WF, Cammue BP. (2002) Esa1, an Arabidopsis mutant with enhanced susceptibility to a range of necrotrophic fungal pathogens, shows a distorted induction of defense responses by reactive oxygen generating compounds. Plant J 29: 131–140 [DOI] [PubMed] [Google Scholar]
- Van der Ent S, Van Wees SC, Pieterse CM. (2009) Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 70: 1581–1588 [DOI] [PubMed] [Google Scholar]
- van Hemelrijck W, Wouters PFW, Brouwer M, Windelinckx A, Goderis IJWM, De Bolle MFC, Thomma BPHJ, Cammue BPA, Delauré SL. (2006) The Arabidopsis defense response mutant esa1 as a model to discover novel resistance traits against Fusarium diseases. Plant Sci 171: 585–595 [Google Scholar]
- Yang JY, Iwasaki M, Machida C, Machida Y, Zhou X, Chua NH. (2008) betaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev 22: 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.