Abstract
AtPUB18 and AtPUB19 are homologous U-box E3 ubiquitin ligases in Arabidopsis (Arabidopsis thaliana). AtPUB19 is a negative regulator of abscisic acid (ABA)-mediated drought responses, whereas the role of AtPUB18 in drought responses is unknown. Here, loss-of-function and overexpression tests identified AtPUB18 as a negative regulator in ABA-mediated stomatal closure and water stress responses. The atpub18-2atpub19-3 double mutant line displayed more sensitivity to ABA and enhanced drought tolerance than each single mutant plant; therefore, AtPUB18 and AtPUB19 are agonistic. Stomatal closure of the atpub18-2atpub19-3 mutant was hypersensitive to hydrogen peroxide (H2O2) but not to calcium, suggesting that AtPUB18 and AtPUB19 exert negative effects on the ABA signaling pathway downstream of H2O2 and upstream of calcium. AtPUB22 and AtPUB23 are other U-box E3 negative regulators of drought responses. Although atpub22atpub23 was more tolerant to drought stress relative to wild-type plants, its ABA-mediated stomatal movements were highly similar to those of wild-type plants. The atpub18-2atpub19-3atpub22atpub23 quadruple mutant exhibited enhanced tolerance to drought stress as compared with each atpub18-2atpub19-3 and atpub22atpub23 double mutant progeny; however, its stomatal behavior was almost identical to the atpub18-2atpub19-3 double mutant in the presence of ABA, H2O2, and calcium. Overexpression of AtPUB18 and AtPUB19 in atpub22atpub23 effectively hindered ABA-dependent stomatal closure, but overexpression of AtPUB22 and AtPUB23 in atpub18-2atpub19-3 did not inhibit ABA-enhanced stomatal closure, highlighting their ABA-independent roles. Overall, these results suggest that AtPUB18 has a linked function with AtPUB19, but is independent from AtPUB22 and AtPUB23, in negative regulation of ABA-mediated drought stress responses.
Ubiquitin (Ub)-mediated posttranslational protein modifications have ubiquitous functions in eukaryotic cells (Dye and Schulman, 2007; Hunter, 2007). In higher plants, they occur as part of the ubiquitination pathway in physiological processes as diverse as cell cycle progressions, circadian rhythms, environmental stress responses, and hormone and light signaling (Moon et al., 2004; Smalle and Vierstra, 2004; Dreher and Callis, 2007; Vierstra, 2009; Lee and Kim, 2011; Park et al., 2011a). Approximately 6% of the Arabidopsis (Arabidopsis thaliana) proteome is associated with the Ub-26S proteasome system, and over 1,400 different E3 Ub ligase genes exist in the Arabidopsis genome (Smalle and Vierstra, 2004; Vierstra, 2009). E3 proteins possess distinct functional domains, such as those derived from the Homology to E6-AP Carboxyl Terminus, Really Interesting New Gene (RING), and U-box gene families.
Arabidopsis contains at least 64 U-box motif-containing E3 proteins (Mudgil et al., 2004; Wiborg et al., 2008; Yee and Goring, 2009; Lyzenga and Stone, 2012). They have various functions related to biotic and abiotic stress responses (Yang et al., 2006; Cho et al., 2008; Trujillo et al., 2008; Liu et al., 2011), hormonal responses (Luo et al., 2006; Raab et al., 2009), self-incompatibility (Liu et al., 2007; Samuel et al., 2008, 2009), and seed germination (Bergler and Hoth, 2011; Salt et al., 2011). U-box E3s also have cellular functions within a diverse group of monocot and dicot model crops, such as Brassica spp., Medicago spp., rice (Oryza sativa), tobacco (Nicotiana tabacum), and tomato (Solanum lycopersicum) plants (Gu et al., 1998; Stone et al., 1999, 2003; Zeng et al., 2004, 2008; González-Lamothe et al., 2006; Vega-Sánchez et al., 2008; Bos et al., 2010; Mbengue et al., 2010; Park et al., 2011b), suggesting that they are widely conserved in the plant kingdom.
The plant stress hormone abscisic acid (ABA) is a key mediator in drought stress response because it induces stomatal closing to minimize transpirational water loss (Yamaguchi-Shinozaki and Shinozaki, 2006; Cutler et al., 2010; Kim et al., 2010; Raghavendra et al., 2010). ABA turns on various genes necessary for triggering rapid and effective defense programs. ABA-induced genes are diverse and include U-box E3s (Hoth et al., 2002; Cho et al., 2006, 2008); therefore, U-box E3s may be functionally correlated with ABA-mediated stress responses.
Recent studies suggest that Arabidopsis U-box E3s are necessary to induce ABA-mediated responses. PUB44/SAUL1 U-box E3 prevents premature leaf senescence by down-regulating ABA level (Raab et al., 2009). PUB44/SAUL1 inhibits seed germination under stressful conditions, such as high levels of ABA, Glc, salt, or mannitol (Salt et al., 2011), and therefore is a negative regulator of ABA-mediated cell death and seed germination processes. Bergler and Hoth (2011) showed that AtPUB18 and AtPUB19 homologs had negative effects during ABA and salt inhibition of seed germination. AtPUB19 also negatively regulates ABA-mediated drought stress responses (Liu et al., 2011). In contrast, AtPUB22 and AtPUB23 are negative regulators of drought stress responses, but their expression is unaffected by ABA (Cho et al., 2008). AtPUB22 and AtPUB23, in addition to AtPUB24, are involved in the negative regulation of pathogen-associated molecular pattern-triggered immunity (Trujillo et al., 2008), suggesting that AtPUB22 and AtPUB23 play roles in both biotic and abiotic stress responses.
In this study, we investigated AtPUB18 during ABA-mediated drought stress responses. ABA-induced stomatal closure was strongly enhanced in loss-of-function atpub18-2atpub19-3 double mutant plants relative to atpub18-2 and atpub19-3 single mutants and wild-type plants. The progeny of atpub18-2atpub19-3 exhibited even greater drought tolerance than the single mutants. AtPUB18 overexpressors showed inverse phenotypes to knockout plants in all categories examined. These results suggest that AtPUB18 and AtPUB19 have paired functions as negative regulators in ABA-dependent drought stress responses. The stomatal movements of atpub18-2atpub19-3 mutant leaves were hypersensitive to hydrogen peroxide (H2O2) as compared with those of wild-type leaves. In contrast, atpub18-2atpub19-3 displayed wild-type stomatal movement in response to calcium, suggesting that AtPUB18 and AtPUB19 act downstream of H2O2 and upstream of calcium in the ABA-mediated drought response. Our atpub18-2atpub19-3atpub22atpub23 quadruple mutation and reciprocal complementation (35S:AtPUB18/19-atpub22atpub23 and 35S:AtPUB22/23-atpub18-2atpub19-3) tests suggest that AtPUB18 has a combined function with AtPUB19, but is independent from AtPUB22 and AtPUB23, during negative regulation of ABA-mediated drought stress responses.
RESULTS
AtPUB18 Is an ABA- and Drought-Inducible Gene
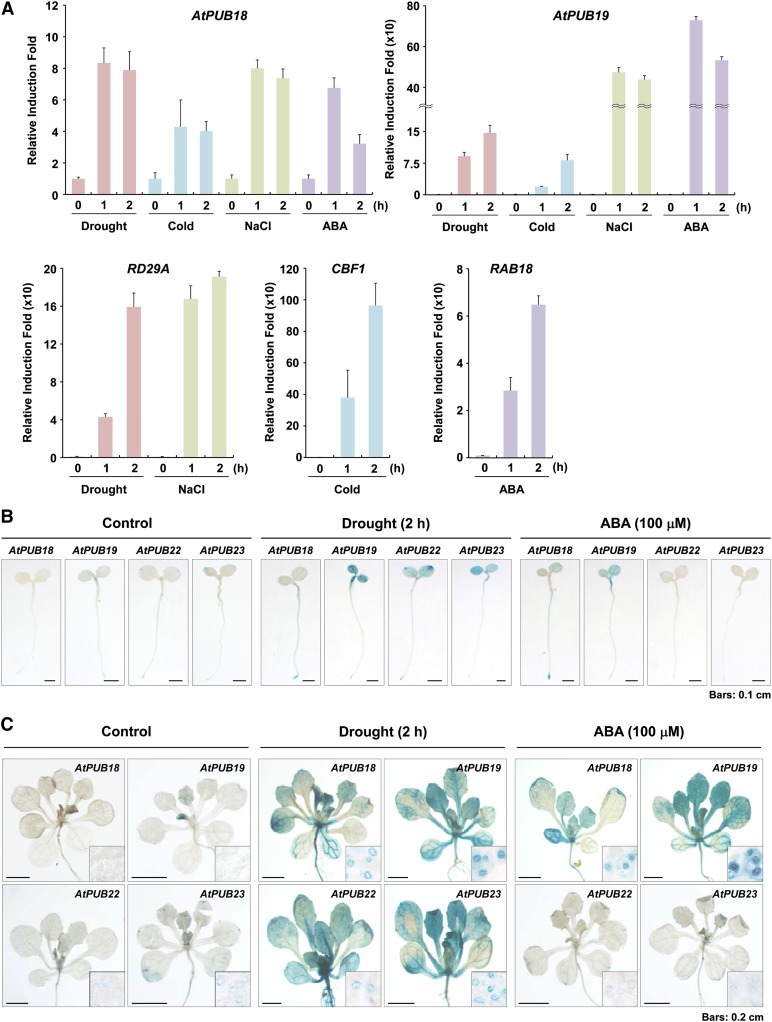
AtPUB18 and AtPUB19 are homologous U-box E3 Ub ligases (Supplemental Fig. S1). Hoth et al. (2002), in a previous genome-scale gene expression study, identified AtPUB18 and AtPUB19 as ABA-induced genes, and Liu et al. (2011) recently confirmed, via RNA gel-blot analysis, the expression of AtPUB19 to be enhanced by ABA and abiotic stress. Here, real-time quantitative reverse transcription (qRT)-PCR showed that AtPUB18 was induced in response to ABA and various abiotic stresses, including NaCl, drought, and coldness (Fig. 1A). Induction kinetics of AtPUB18 were similar to those of both AtPUB19 and marker genes (Responsive to Desiccation29A [RD29A] for drought and salt, C-repeat Binding Factor1 [CBF1] for cold, and Responsive to ABA18 [RAB18] for ABA), while the magnitude of AtPUB18 induction was significantly lower than that of AtPUB19.
Figure 1.
Induction patterns of AtPUB18, AtPUB19, AtPUB22, and AtPUB23 in response to ABA and abiotic stresses, as determined by real-time qRT-PCR and histochemical GUS assays. A, Light-grown, 10-d-old Arabidopsis seedlings were treated with drought, cold, high salinity, and ABA. Total RNA was extracted from the stress-treated whole seedlings and used for qRT-PCR. RD29A, CBF1, and RAB18 were positive controls for drought and high salt, cold, and ABA treatments, respectively. To calculate the relative expression levels of each gene, glyceraldehyde-3-phosphate dehydrogenase C subunit was used as an internal reference gene. Error bars represent sd from three independent experiments. B and C, Promoter (1.0-kb upstream region) activities of AtPUB18, AtPUB19, AtPUB22, and AtPUB23 in response to ABA (100 μm) and dehydration (2 h). B, Histochemical GUS expression patterns in 5-d-old T3 promoter:GUS transgenic seedlings. Bars = 0.1 cm. C, GUS activities in aerial parts and guard cells of 2-week-old transgenic plants. Bars = 0.2 cm. [See online article for color version of this figure.]
A promoter-GUS assay of T3 transgenic lines showed that AtPUB18 and AtPUB19 promoter activities (in the 1.0-kb upstream region) were barely detectable in 5-d-old (Fig. 1B) and 2-week-old (Fig. 1C) plants. This low basal activity was markedly heightened by ABA and drought treatments. In 5-d-old seedlings, drought and ABA inductions of AtPUB18 were predominantly detected in the roots, whereas those of AtPUB19 were found in leaves and upper roots (Fig. 1B). In 2-week-old plants, drought and ABA inductions of both genes were observed throughout the plant tissues, including guard cells (Fig. 1C). These results suggest that AtPUB18 is involved in ABA-mediated stress responses. Promoter activities of AtPUB22 and AtPUB23 were induced by drought but not by ABA treatment (Fig. 1, B and C). Their drought inductions also occurred in guard cells. Overall, gene expression studies indicated that AtPUB18 and AtPUB19 are ABA- and drought-inducible genes, but AtPUB22 and AtPUB23 are only induced by dehydration stress.
Expression of AtPUB18 and AtPUB19, But Not AtPUB22 and AtPUB23, Is Dependent on SnRK
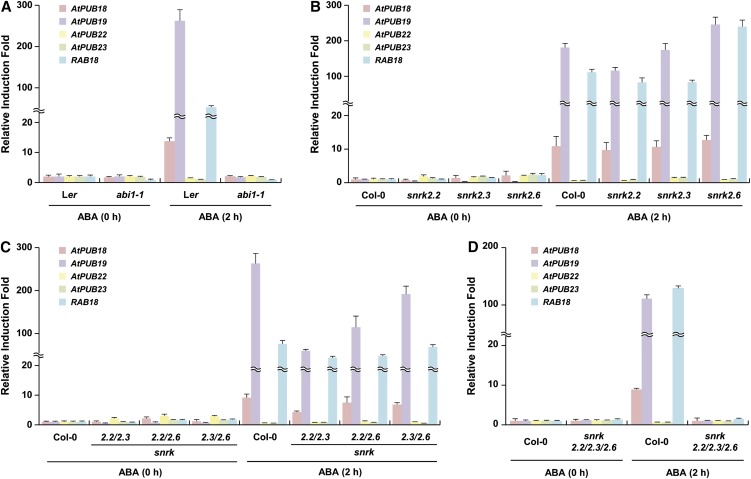
abi1-1 is an ABA-insensitive dominant mutant (Hubbard et al., 2010; Kim et al., 2010; Raghavendra et al., 2010). Figure 2A shows that ABA failed to induce AtPUB18, AtPUB19, and the marker gene, RAB18, in abi1-1. This demonstrates that both AtPUB18 and AtPUB19 are ABA-induced genes. In contrast, AtPUB22 and AtPUB23 expression remained unchanged before and after ABA treatment in wild-type and abi1-1 plants, confirming their ABA-independent expression (Fig. 2A).
Figure 2.
ABA induction profiles of AtPUB18, AtPUB19, AtPUB22, and AtPUB23 in wild-type and abi1-1 and snrk2 mutant plants. Light-grown, 10-d-old wild-type and mutant seedlings were incubated with or without 100 μm ABA for 2 h. Total RNA was isolated and subjected to qRT-PCR. Glyceraldehyde-3-phosphate dehydrogenase C subunit was used as a reference gene for normalization. RAB18 was a marker for the ABA-induced gene. Error bars represent sd from three independent experiments. A, ABA induction profiles of AtPUB18, AtPUB19, AtPUB22, and AtPUB23 in wild-type (Landsberg erecta [Ler]) and abi1-1 mutant plants. B to D, ABA induction profiles of AtPUB18, AtPUB19, AtPUB22, and AtPUB23 in wild-type (Columbia-0 [Col-0]) and various snrk2 single (B), double (C), and triple (D) mutant plants. [See online article for color version of this figure.]
SnRK protein kinases are key regulators in the ABA signaling pathway. They mediate ABA-dependent reactive oxygen species production and water and osmotic stress responses (Mustilli et al., 2002; Fujita et al., 2009; Fujii et al., 2011). Furthermore, ABA- and dehydration-induced gene expressions are greatly impaired by triple knockout mutations of SnRK2.2, SnRK2.3, and SnRK2.6 (Fujii and Zhu, 2009; Fujita et al., 2009). ABA induction of AtPUB18 and AtPUB19 in snrk2.2, snrk2.3, and snrk2.6 single and snrk2.2snrk2.3, snrk2.2snrk2.6, and snrk2.3snrk2.6 double knockout mutants was comparable to that in wild-type plants (Fig. 2, B and C). In contrast, ABA induction of both genes was completely abolished in snrk2.2snrk2.3snrk2.6 triple loss-of-function mutant plants (Fig. 2D). Therefore, the redundant functions of SnRK members resulted in ABA induction of AtPUB18 and AtPUB19 in the single and double snrk mutant lines. Similar induction patterns were also observed for RAB18. These results indicate that SnRKs are necessary for ABA induction of AtPUB18 and AtPUB19; however, AtPUB22 and AtPUB23 are not induced by ABA in either wild-type or snkr lines (Fig. 2).
Phenotypic Analyses of atpub18 Single and atpub18atpub19 Double Knockout Mutant Plants in Response to ABA and Drought
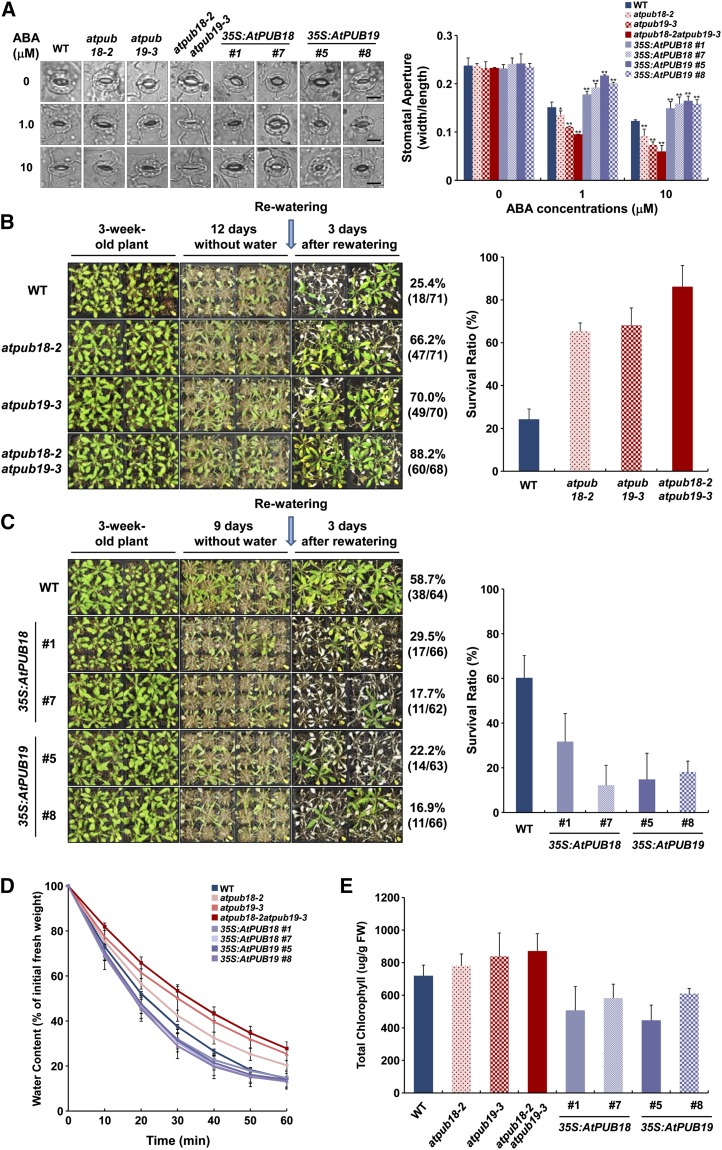
Liu et al. (2011) showed that suppression of AtPUB19 increased ABA sensitivity with regard to stomatal movement. We examined ABA-dependent stomatal behaviors of wild-type, atpub18-2 (SALK_001831) and atpub19-3 (SALK_058791) single mutant, and atpub18-2atpub19-3 double mutant plants (Supplemental Fig. S2). Stomatal apertures (the ratio of width to length) of wild-type and mutant plants were indistinguishable without ABA treatment (Fig. 3A). After treatment with ABA, however, stomatal movement in the atpub18-2 mutant was more markedly enhanced, as compared with that in the wild type, in a dose-dependent manner. The average stomatal apertures of wild-type, atpub18-2, atpub19-3, and atpub18-2atpub19-3 plants were 0.123 ± 0.002, 0.091 ± 0.015, 0.073 ± 0.007, and 0.059 ± 0.013, respectively, in response to 10 μm ABA (Fig. 3A). The atpub18-2 plants, therefore, exhibited hypersensitive phenotypes, relative to the wild type, during ABA-mediated stomatal closure; and the atpub18-2atpub19-3 double mutant was even more sensitive to ABA than each single mutant. In contrast, AtPUB18-overexpressing plants (35S:AtPUB18 transgenic lines 1 and 7) showed hyposensitive stomatal responses toward ABA, which were comparable to those of AtPUB19-overexpressing plants (35S:AtPUB19 lines 5 and 8; Fig. 3A). With 10 μm ABA, stomatal apertures of 35S:AtPUB18 and 35S:AtPUB19 were 0.149 ± 0.014 to 0.159 ± 0.014 and 0.158 ± 0.009 to 0.165 ± 0.010, respectively. Thus, the expression level of AtPUB18 was inversely associated with ABA-regulated stomatal closure, suggesting that AtPUB18 participates in negative regulation of ABA-dependent stomatal movement.
Figure 3.
Phenotypic analyses of wild-type (WT), atpub18-2, atpub19-3, and atpub18-2atpub19-3 knockout mutant, and 35S:AtPUB18- and 35S:AtPUB19-overexpressing transgenic plants in response to ABA and drought treatments. A, ABA-induced stomatal closure. Light-grown, fully expanded leaves were immersed in stomatal opening solution for 2 h and transferred into solution containing various concentrations (0, 1, and 10 µm) of ABA for 2 h. Stomata were photographed using bright-field microscopy. At least 30 stomatal apertures in epidermal peels were measured per replicate. Two replicates were performed for each experiment. Bars = 10 μm. Error bars represent sd (n > 60; *P < 0.05, **P < 0.005, Student’s t test). B, Drought tolerance of wild-type, atpub18-2, atpub19-3, and atpub18-2atpub19-3 plants. Wild-type and mutant plants were grown under normal growth conditions for 3 weeks and then exposed to drought stress by withholding water for 12 d. Surviving plants were counted 3 d after rewatering. C, Drought-sensitive phenotypes of AtPUB18- and AtPUB19-overexpressing plants, which, after 3 weeks, were subjected to water deficit conditions for 9 d. Survival rates were calculated after 3 d of irrigation. D, Water loss rates of detached mature leaves that were incubated at room temperature under dim light. Decreased fresh weights (FW) were recorded at specified time points. Water loss rates were estimated as the percentage of initial to final fresh weights. Data represent means ± sd (n = 12) from four replicates. E, Chlorophyll contents of whole seedlings dehydrated on dry filter paper for 45 min (drought stress). Chlorophyll contents were measured 2 d after rewatering as means ± sd of three replicates. [See online article for color version of this figure.]
AtPUB19 is negatively involved in drought stress responses (Liu et al., 2011); therefore, we next tested for AtPUB18 function in response to drought. Light-grown, 3-week-old, healthy wild-type, atpub18-2, atpub19-3, and atpub18-2atpub19-3 plants were further grown for 12 d while withholding water and then irrigated for 3 d, at which time their survival rates were recorded, similar to Ryu et al. (2010). The results showed that 25.4% (18 of 71) of wild-type plants resumed their growth after drought stress, whereas survival rates of atpub18-2, atpub19-3, and atpub18-2atpub19-3 plants were 66.2% (47 of 71), 70.0% (49 of 70), and 88.2% (60 of 68), respectively (Fig. 3B). The second alleles of atpub18-1 and atpub19-2 were similarly tolerant to drought stress (Supplemental Fig. S3). In contrast, both AtPUB18- and AtPUB19-overexpressing lines were more susceptible to mild dehydration conditions than wild-type plants. After 9 d of dehydration, the survival rate of wild-type plants was 58.7% (38 of 64; Fig. 3C); however, only 17.7% (11 of 62) to 29.5% (17 of 66) of 35S:AtPUB18 and 16.9% (11 of 66) to 22.2% (14 of 63) of 35S:AtPUB19 survived. Consistent with these results, rosette leaves detached from mutants lost water more slowly during dehydration, and overexpressing plants lost water more quickly, than wild-type leaves during dehydration (Fig. 3D). Finally, leaf chlorophyll contents were higher in mutant leaves than in overexpressing and wild-type leaves after drought (Fig. 3E). Overall, these results indicate that both AtPUB18 and AtPUB19 participate in negative regulation of ABA-dependent drought stress responses.
Stomatal Movement Analysis of atpub18-2atpub19-3 and atpub22atpub23 Double Knockout Mutant Plants in Response to H2O2 and Calcium
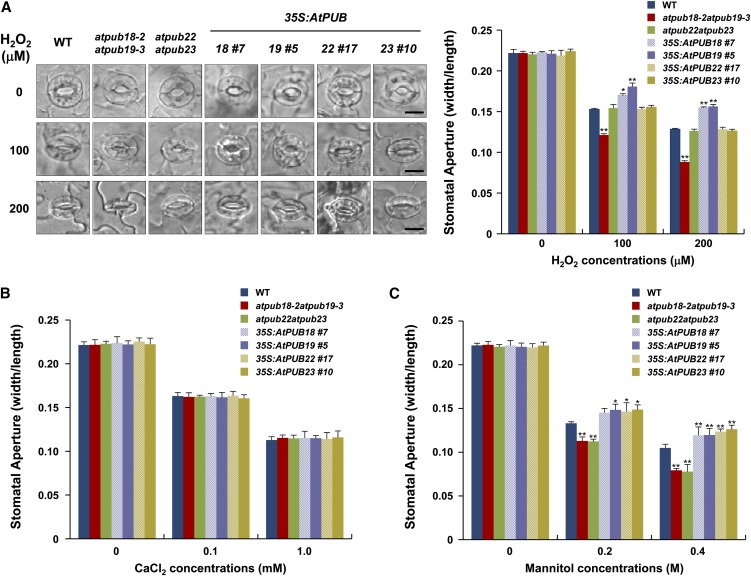
Our results (Figs. 1–3) and Cho et al. (2008) show that four Arabidopsis U-box E3 members, AtPUB18, AtPUB19, AtPUB22, and AtPUB23, participate in negative regulation of drought stress responses. Therefore, we investigated their functions in plant stomatal behavior in response to H2O2 and calcium, both of which are important regulators of ABA-dependent stomatal movement (Huang et al., 2009; Jammes et al., 2009; Kim et al., 2010). After H2O2 treatment, stomatal closure increased in atpub18-2atpub19-3 mutants significantly more than in the wild type. In contrast, H2O2-mediated stomatal movement of atpub22atpub23 mutants was almost identical to that of the wild type. With 100 μm H2O2, stomatal apertures of wild-type, atpub18-2atpub19-3, and atpub22atpub23 plants were 0.153 ± 0.001, 0.121 ± 0.002, and 0.154 ± 0.004, respectively (Fig. 4A). With 200 μm H2O2, the stomatal aperture of atpub18-2atpub19-3 leaves was 0.088 ± 0.002, which was approximately 1.5 times smaller than those of wild-type (0.128 ± 0.001) and atpub22atpub23 (0.126 ± 0.002) plants. These results show that differences in stomatal apertures between atpub18-2atpub19-3 and atpub22atpub23 mutants are more prominent as H2O2 concentrations increase; therefore, knockout mutations of AtPUB18 and AtPUB19 enhance H2O2-mediated stomatal movement, whereas those of AtPUB22 and AtPUB23 result in wild-type stomatal behavior. Furthermore, AtPUB18- and AtPUB19-overexpressing plants displayed H2O2-insensitive stomatal responses relative to the wild type. In contrast, overexpression of AtPUB22 and AtPUB23 could not alter stomatal movement profiles in response to H2O2 treatments, indicating their ABA-independent functions (Fig. 4A). On the other hand, we found endogenous production of H2O2 in wild-type, mutant, and overexpressing plants to be highly similar (Supplemental Fig. S4), as determined by 3,3′-diaminobenzidine (DAB) staining (Cho et al., 2011; Zulfugarov et al., 2011).
Figure 4.
Stomatal closure of wild-type (WT), atpub18-2atpub19-3 and atpub22atpub23 mutant, and 35S:AtPUB18-, 35S:AtPUB19-, 35S:AtPUB22-, and 35S:AtPUB23-overexpressing transgenic plants in response to H2O2, CaCl2, and mannitol. Mature leaves, pretreated with stomatal opening solution, were transferred to solutions containing various concentrations of H2O2 (A), CaCl2 (B), or mannitol (C), each for 2 h. Epidermal peels were mounted on microscope slides, and stomata were observed with bright-field microscopy. Stomatal apertures were measured with three replicates. Error bars represent sd (n > 87; *P < 0.05, **P < 0.005, Student’s t test). Bars = 10 µm. [See online article for color version of this figure.]
Next, we examined calcium-dependent stomatal movements. Neither double mutations nor overexpression of AtPUB18/AtPUB19 changed calcium-dependent stomatal behaviors relative to wild-type plants (Fig. 4B). Therefore, AtPUB18 and AtPUB19 likely acted upstream of calcium and downstream of H2O2 in the ABA-mediated drought responses. As expected, the atpub22atpub23 mutant and AtPUB22/AtPUB23-overexpressing plants displayed wild-type stomatal movement in response to calcium (Fig. 4B).
Finally, stomatal movements of both atpub18-2atpub19-3 and atpub22atpub23 mutant leaves were similarly hypersensitive to mannitol treatment as compared with those of wild-type leaves. In contrast, all of the AtPUB18/AtPUB19- and AtPUB22/AtPUB23-overexpressing lines investigated were hyposensitive to mannitol, confirming their negative roles in drought stress responses (Fig. 4C).
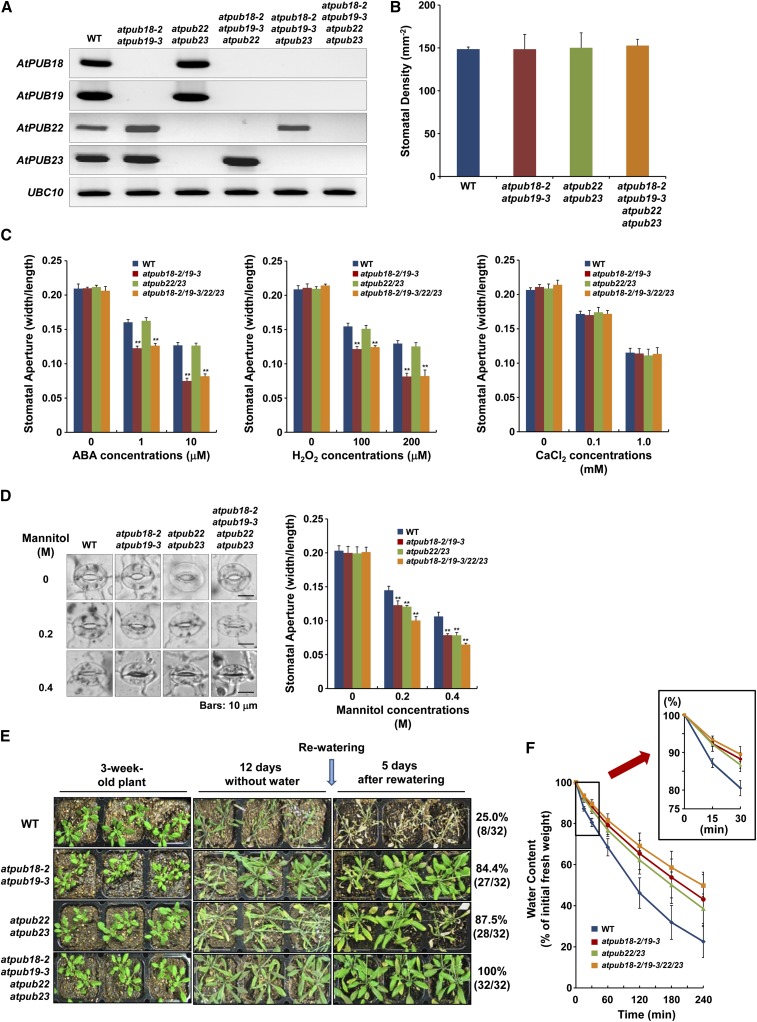
The atpub18-2atpub19-3atpub22atpub23 Quadruple Mutant Line Was Highly Tolerant to Drought Stress, But Their Stomatal Movements Were Similar to Those of atpub18-2atpub19-3 Double Mutants
The atpub18-2atpub19-3atpub22atpub23 quadruple mutant line was generated by crossing the atpub22atpub23 and atpub18-2atpub19-3 double mutant plants (Fig. 5A). The quadruple mutant was phenotypically normal, and their leaf stomatal density was indistinguishable from those of wild-type and double mutant plants (Fig. 5B). As shown in Figure 5C, stomatal movement profiles of the atpub18-2atpub19-3atpub22atpub23 quadruple mutant were highly similar to those of the atpub18-2atpub19-3 double mutant in response to ABA, H2O2, and CaCl2, which is consistent with the ABA-dependent functions of AtPUB18/AtPUB19 and the ABA-independent functions of AtPUB22/AtPUB23, respectively. In contrast, stomatal closure of the quadruple mutant line was even more sensitive than those of atpub18-2atpub19-3 and atpub22atpub23 double mutant plants in response to mannitol treatments (Fig. 5D). These results indicate that quadruple mutations of AtPUB18, AtPUB19, AtPUB22, and AtPUB23 greatly increased tolerance toward drought stress. Consistently, the atpub18-2atpub19-3atpub22atpub23 quadruple mutant was markedly more tolerant to dehydration stress: after 12 d of drought treatment, 100% (32 of 32) of the atpub18-2atpub19-3atpub22atpub23 line survived, whereas the survival rates of wild-type, atpub18-2atpub19-3, and atpub22atpub23 plants were 25.0% (8 of 32), 84.4% (27 of 32), and 87.5% (28 of 32), respectively (Fig. 5E). Detached rosette leaves of the quadruple mutant lost their fresh weight more slowly than each double mutant in the dehydration process (Fig. 5F). These results are consistent with the results that the four U-box E3s participate in the drought stress responses as negative regulators.
Figure 5.
Molecular characterization and phenotype analyses of a series of double, triple, and quadruple homozygous mutant combinations containing atpub18-2, atpub19-3, atpub22, and atpub23. A, Generation of homozygous atpub18-2atpub19-3atpub22 and atpub18-2atpub19-3atpub23 triple and atpub18-2atpub19-3atpub22atpub23 quadruple knockout mutant plants. Semiquantitative RT-PCR analyses were performed to confirm the absence of transcripts. B, Stomatal density (average number of stomata per mm2) of wild-type (WT) and various mutant plants. Abaxial epidermal peels of fully expanded rosette leaves were harvested and observed with a light microscope. This experiment was repeated three times. Error bars represent sd. C, Stomatal movements of the atpub18-2atpub19-3atpub22atpub23 quadruple mutant in response to ABA, H2O2, and CaCl2. Error bars represent sd (n > 87; **P < 0.005, Student’s t test). D, Stomatal movements of the atpub18-2atpub19-3atpub22atpub23 mutant in response to osmotic stress imposed by mannitol. Error bars represent sd (n > 87; **P < 0.005, Student’s t test). E, Drought-tolerant phenotypes of atpub18-2atpub19-3, atpub22atpub23, and atpub18-2atpub19-3atpub22atpub23 plants. Wild-type and mutant plants were grown for 3 weeks with normal irrigation, then subjected to dehydration conditions by interrupting irrigation for 12 d, and supplied with sufficient water for 5 d, after which survival rates were calculated. F, Water loss rates of detached mature leaves, as determined by change in fresh weights (described in Fig. 3D). Data represent means ± sd (n = 5) from three replicates. [See online article for color version of this figure.]
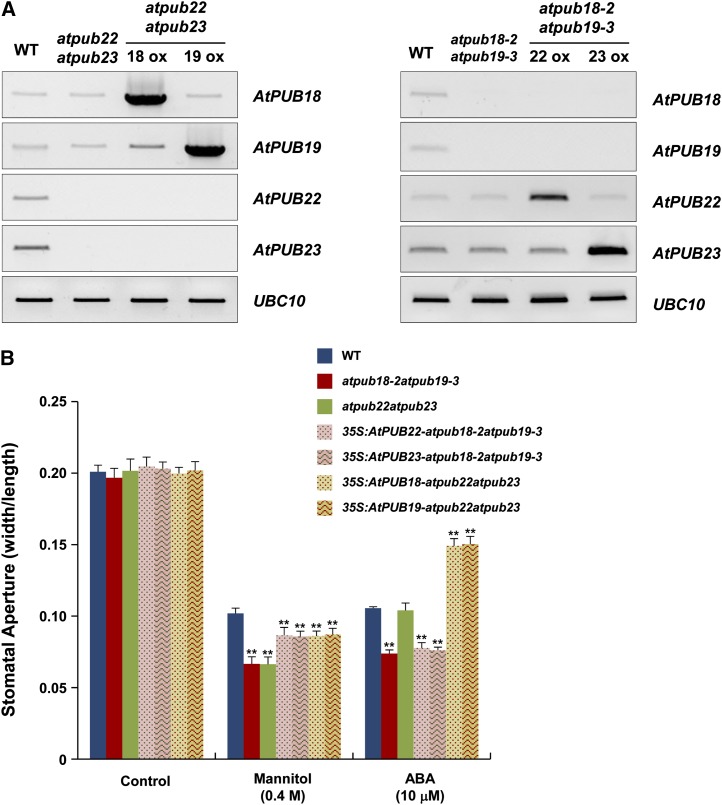
Reciprocal Complementation Analysis of AtPUB18/AtPUB19 and AtPUB22/AtPUB23
The functional relationships between AtPUB18/AtPUB19 and AtPUB22/AtPUB23 were further addressed by reciprocal complementation studies. AtPUB18/AtPUB19 and AtPUB22/AtPUB23 were overexpressed in atpub22atpub23 and atpub18-2atpub19-3 double mutant plants, respectively (Fig. 6A). Complementation transgenic plants (35S:AtPUB18/19-atpub22atpub23 and 35S:AtPUB22/23-atpub18-2atpub19-3) were used for the mannitol- and ABA-dependent stomatal movement tests. The results revealed that ectopic expression of AtPUB18/AtPUB19 and AtPUB22/AtPUB23 in atpub22atpub23 and atpub18-2atpub19-3, respectively, partially inhibited mannitol-dependent stomatal closure, confirming their negative roles in drought stress responses (Fig. 6B). Overexpression of AtPUB18/AtPUB19 in atpub22atpub23 greatly hindered an ABA-mediated stomatal movement, whereas overexpression of AtPUB22/AtPUB23 in atpub18-2atpub19-3 failed to alter the ABA-mediated stomatal movement of atpub18-2atpub19-3 mutant lines (Fig. 6B). On the other hand, AtPUB19 effectively complemented the mutant phenotypes of the atpub18-2 line (Supplemental Fig. S5). Collectively, these results suggest that the U-box E3 Ub ligase AtPUB18 has a combined function with AtPUB19, but is independent from AtPUB22 and AtPUB23, in negative regulation of ABA-mediated drought stress responses.
Figure 6.
Identification and characterization of 35S:AtPUB18/AtPUB19-atpub22atpub23 and 35S:AtPUB22/AtPUB23-atpub18-2atpub19-3 complementation transgenic plants. A, Semiquantitative RT-PCR analysis used to detect ectopic and reciprocal expression of AtPUB18/AtPUB19 and AtPUB22/AtPUB23 in atpub22atpub23 and atpub18-2atpub19-3, respectively. Ubiquitin Conjugating Enzyme10 (UBC10) was used as a loading control. B, ABA- and mannitol-dependent stomatal behaviors in T2 35S:AtPUB18/AtPUB19-atpub22atpub23 and T1 35S:AtPUB22/AtPUB23-atpub18-2atpub19-3 transgenic plants. Fully expanded, light-grown leaves were harvested, and their stomata were observed as described in Figure 3A. Three replicates were performed for each experiment. Error bars represent sd (n = 90; **P < 0.005, Student’s t test). WT, Wild type. [See online article for color version of this figure.]
DISCUSSION
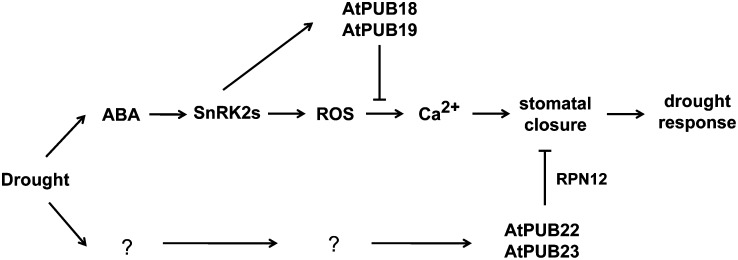
AtPUB19 is a negative regulator of ABA-mediated seed germination and drought response (Bergler and Hoth, 2011; Liu et al., 2011). Although AtPUB18 was recently reported to participate negatively in the ABA- and salt-inhibited seed germination process (Bergler and Hoth, 2011), its role in drought responses was unknown. In this report, AtPUB18 was identified as a drought- and ABA-induced gene and its induction was SnRK dependent (Figs. 1 and 2). Our loss-of-function and overexpression tests revealed that AtPUB18 was a negative regulator in ABA-mediated stomatal closure and water stress responses. Moreover, the atpub18-2atpub19-3 double mutant line displayed even greater sensitivity to ABA and more enhanced drought tolerance than did each single mutant plant (Fig. 3). Therefore, AtPUB18 and AtPUB19 have agonistic functions. Interestingly, the stomatal response of the atpub18-2atpub19-3 mutant was hypersensitive to H2O2, while its response to calcium was comparable to that of wild-type leaves (Fig. 4), suggesting that AtPUB18 and AtPUB19 exert negative effects on the ABA signaling pathway downstream of H2O2 and upstream of calcium (Fig. 7).
Figure 7.
Negative regulation of AtPUB18, AtPUB19, AtPUB22, and AtPUB23 during drought stress responses. Modes of action of the four U-box E3s were divided into two different pathways: AtPUB18 and AtPUB19 were ABA dependent, and AtPUB22 and AtPUB23 were ABA independent. AtPUB18 and AtPUB19 exerted their negative effects on the ABA signaling pathway downstream of H2O2 and upstream of calcium. The cellular mechanism by which AtPUB22 and AtPUB23 regulate drought responses is not known. RPN12a, a non-ATPase subunit of the 19S regulatory particle in the 26S proteasome, is one of the targets of AtPUB22 and AtPUB23, while the possible substrates of AtPUB18 and AtPUB19 are currently unknown. ROS, Reactive oxygen species.
In contrast to clear phenotypic properties in postgermination stages, ABA inhibition of seed germination is relatively weak in atpub18-1atpub19-2 (Bergler and Hoth, 2011). In addition, ABA inhibition of early root growth was also very weak in the mutant lines (Liu et al., 2011; Supplemental Fig. S6); therefore, negative regulation of AtPUB18 and AtPUB19 is likely to mainly occur in postgermination stages. This raises the possibility that other U-box E3 negative regulators that modulate ABA-dependent seed germination processes may exist. On the other hand, a basic leucine zipper (bZIP) ABI5 transcription factor is mainly responsible for the expression of the genes for inhibition of germination and postgermination arrest (Lopez-Molina et al., 2001), whereas other bZIP transcription factors (ABFs/AREBs), which are more highly expressed in mature plants, are likely to function in mature plants (Uno et al., 2000). Therefore, it would be interesting to examine whether various bZIP transcription factors, including ABI5, differentially (negatively or positively) affect the ABA induction of AtPUB18 and AtPUB19 and/or their E3 ligase enzyme activities.
While yeast and humans (Homo sapiens) contain two and eight U-box E3 Ub ligases, respectively, higher plants have more: Arabidopsis has 64 and rice has 77 (Yee and Goring, 2009; Lyzenga and Stone, 2012). Therefore, we speculate that U-box E3s have plant-specific roles and that their cellular functions are coordinated and interconnected as effective defense webs and for concomitant metabolic reprogramming. This is in agreement with other findings that plants have multicombinatorial defensive programs that work coordinately in response to abiotic stresses (Ahuja et al., 2010; Tardieu et al., 2011). In this context, we hypothesize that U-box E3 isoforms also work in a combinatorial pattern to efficiently cope with dehydration stress.
AtPUB22 and AtPUB23 are also negative regulators of drought responses (Cho et al., 2008); therefore, we wanted to elucidate the functional relationships among the four U-box E3s with regard to ABA-dependent drought stress responses. Stomatal behaviors of the atpub18-2atpub19-3atpub22atpub23 quadruple mutant line were highly similar to those of atpub18-2atpub19-3 double mutant plants in response to ABA, H2O2, and calcium (Fig. 5). In contrast, the quadruple mutant exhibited a hypersensitive stomatal response to mannitol and markedly enhanced tolerance to severe drought stress as compared with each atpub18-2atpub19-3 and atpub22atpub23 double mutant plant progeny (Fig. 5). These results strongly suggest that the modes of action of the four U-box E3 negative regulators are divided into two different pathways: AtPUB18 and AtPUB19 are ABA dependent, and AtPUB22 and AtPUB23 are ABA independent. This assumption was further supported by the results of reciprocal complementation studies. Overexpression of AtPUB18 and AtPUB19 in atpub22atpub23 progeny effectively hindered ABA-dependent stomatal closure (Fig. 6). In contrast, overexpression of AtPUB22 and AtPUB23 in the atpub18-2atpub19-3 line failed to inhibit ABA-enhanced stomatal closure. Overall, it is concluded that action mechanisms of AtPUB18 and AtPUB19 are overlapped but separated with those of AtPUB22 and AtPUB23 in ABA-mediated drought stress responses (Fig. 7). These two different drought-responsive pathways, however, are somehow interconnected by an as yet unknown mechanism, since reciprocal complementation partially restored the mannitol-responsive stomatal profiles of atpub18-2atpub19-3 and atpub22atpub23 double mutants (Fig. 6). Mudgil et al. (2004) showed that AtPUB22, AtPUB23, AtPUB18, and AtPUB19 are all related based on the presence of the C-terminal ARM domains in addition to the U-box but that AtPUB18 and AtPUB19 are larger proteins with the additional N-terminal UND domain, indicating different structural relationships between AtPUB18/AtPUB19 versus AtPUB22/AtPUB23 proteins (Supplemental Fig. S1). This is consistent with the possible differences between AtPUB18/AtPUB19 and AtPUB22/AtPUB23 with respect to their functions in drought stress responses.
RPN12a, a non-ATPase subunit of the 19S regulatory particle in the 26S proteasome, is one of the targets of AtPUB22 and AtPUB23, and it was proposed that AtPUB22 and AtPUB23 control a drought signaling pathway by ubiquitinating cytosolic RPN12a (Cho et al., 2008). Ubiquitination and subsequent conformational changes in the 26S proteasome complex may signal negative regulation of drought responses. However, a more detailed mechanism, by which AtPUB22 and AtPUB23 regulate drought responses, remains to be elucidated (Fig. 7). The possible substrates of AtPUB18 and AtPUB19 are currently unknown. Therefore, identification of their target proteins are required to understand how these four U-box E3 negative regulators coordinately mediate a drought stress response in Arabidopsis. Several RING E3 Ub ligases (e.g. AtAIRP1, AtAIRP2, AtRDUF1, and AtRDUF2) positively regulate ABA-dependent drought stress responses (Ryu et al., 2010; Cho et al., 2011; Kim et al., 2012). Thus, functional balances between positive and negative regulators help plants fine-tune their cellular response to dehydration stress, one of the most common environmental stresses of crops. In conclusion, our results suggest that the ubiquitination pathway is critically involved, as a positive and a negative factor, in either ABA-dependent or ABA-independent defensive mechanisms against drought stress in Arabidopsis.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Stress Treatments
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used as the wild type in this study, except for abi1-1, which is in the Landsberg erecta background. The transferred DNA insertion mutants atpub18-1 (SAIL_634_G01; Bergler and Hoth, 2011), atpub18-2 (SALK_001831), atpub19-1 (SALK_035871), atpub19-2 (SALK_152677; Liu et al., 2011), atpub19-3 (SALK_058791), abi1-1 (CS22), snrk2.2 (GABI-Kat 087G04), snrk2.3 (SALK_107315), and snrk2.6 (SALK_008608) were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org). Multiple knockout mutant plants were generated from genetic crosses (Cho et al., 2008). Full-length complementary DNAs (cDNAs) of AtPUB18 and AtPUB19 were cloned downstream of the 35S promoter in the binary vector pBI121 (Arabidopsis Biological Resource Center stock no. CD3-338). The AtPUB22 and AtPUB23 genes were cloned and inserted into a modified pENTR vector (Invitrogen). AtPUB22 and AtPUB23 clones were transferred to pEarlyGate100 destination vectors with a Gateway cloning kit (Invitrogen) and transformed into the Arabidopsis genome using the floral dip method (Ryu et al., 2010). Methods for seed sterilization, growth conditions, and various stress treatments are described by Ryu et al. (2010). All primers used in reverse transcription (RT)-PCR, qRT-PCR, genotyping PCR, and plasmid construction are listed in Supplemental Tables S1 and S2.
RT-PCR and Real-Time qRT-PCR Analyses
Total RNA from ABA- and abiotic stress-treated tissues was extracted using the Easy-Spin IIp plant RNA extraction kit according to the manufacturer’s instructions (Intron Biotechnology). Two micrograms of DNase I-treated total RNA was used for cDNA synthesis using the TOPscript cDNA Synthesis kit (Enzynomics). RT-PCR and real-time qRT-PCR were carried out as described by Cho et al. (2011). Real-time qRT-PCR analyses were performed using an IQ5 light cycler (Bio-Rad). The SYBR Premix Ex Taq II (Takara) was used for the reactions.
Histochemical GUS Assay
For the GUS analysis, mock-, drought-, and ABA-treated 5-d-old seedlings or 2-week-old plants were harvested and fixed with 90% prechilled acetone. After washing with rinsing solution (GUS staining solution without 5-bromo-4-chloro-3-indolyl-β-glucuronic acid A), GUS staining and destaining were performed as described by Cho et al. (2011).
Stomatal Aperture Measurement
Light-grown mature rosette leaves from 4- to 6-week-old plants were immersed in stomatal opening solution (30 mm KCl, 100 μm CaCl2, and 10 mm MES, pH 6.15) for 2 h at 22°C (Kwak et al., 2001). Treated leaves were transferred to a stomatal opening solution containing various concentrations of mannitol (0, 0.2, and 0.4 m), ABA (0, 1, and 10 μm), H2O2 (0, 100, and 200 μm), or CaCl2 (0, 0.1, and 1.0 mm). After incubation for 2 h in white light, 30 stomatal apertures in each epidermal peel were measured and analyzed as described by Ryu et al. (2010). Excel data files of stomatal apertures that were obtained in the experiments are provided in Supplemental Table S3. To measure endogenous leaf H2O2 level, light-grown, 2-week-old seedlings were treated with or without 100 mm ABA for 2 h and transferred to solution containing 100 μg mL−1 DAB as described previously (Cho et al., 2011; Zulfugarov et al., 2011). Chlorophylls of DAB-stained seedlings were removed by boiling in 95% (v/v) ethanol. Reactive oxygen species levels were visualized as a dark brown color.
Drought Phenotype Analysis
Light-grown, 3-week-old wild-type and mutant plants were exposed to water deficit conditions, 9 to 12 d, by withholding irrigation. Three to 5 d after rewatering, survival ratios were calculated as described by Kim et al. (2012). To investigate the water loss rate, aerial parts of 2-week-old seedlings grown on Murashige and Skoog agar plates, or fully expanded rosette leaves detached from soil-grown plants, were placed on glass slides at room temperature. Water loss rate was calculated as the percentage of final to initial fresh weight (Yamaguchi et al., 2007).
Chlorophyll Content Measurement
Light-grown, 2-week-old whole seedlings were collected from Murashige and Skoog agar plates and placed on dry filter papers for 45 min. After dehydration treatment, seedlings were incubated in water for 2 d under white light conditions (Yamaguchi et al., 2007). To extract pigment, each sample was ground in liquid nitrogen and subjected to 80% acetone. Spectrometric analysis of chlorophyll was performed using a DU 800 spectrophotometer (Beckman-Coulter). Chlorophyll contents were determined as described by Fujita et al. (2012).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AtPUB18 (At1g10560), AtPUB19 (At1g60190), AtPUB22 (At3g52450), AtPUB23 (At2g35930), AtPUB8 (At4g21350), AtPUB10 (At1g71020), AtPUB12 (At2g28830), AtPUB13 (At3g46510), AtPUB14 (At3g54850), AtPUB15 (At5g42340), AtPUB16 (At5g01830), AtPUB17 (At1g29340), AtPUB20 (At1g66160), AtPUB21 (At5g37490), AtPUB24 (At3g11840), AtPUB25 (At3g19380), AtPUB26 (At1g49780), AtPUB27 (At5g64660), AtPUB28 (At5g09800), AtPUB29 (At3g18710), and AtPUB31 (At5g65920).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence analysis of AtPUB18 and AtPUB19.
Supplemental Figure S2. Characterization of atpub18, atpub19, and atpub18atpub19 knockout mutant and 35S:AtPUB18 and 35S:AtPUB19 overexpressing transgenic plants.
Supplemental Figure S3. Drought-tolerant phenotypes of atpub18-1 and atpub19-1 second allele mutant plants.
Supplemental Figure S4. ROS accumulation in wild-type, atpub18, atpub19, and atpub18atpub19 knockout mutant, and 35S:AtPUB18 and 35S:AtPUB19 overexpressing transgenic plants in response to ABA.
Supplemental Figure S5. Characterization of 35S:AtPUB19-atpub18 complementation transgenic plants and their stomatal movements in response to mannitol and ABA treatments.
Supplemental Figure S6. ABA-induced root growth inhibition of wild-type, atpub18, atpub19, and atpub18atpub19 knockout mutant, and 35S:AtPUB18 and 35S:AtPUB19 overexpressing transgenic plants.
Supplemental Table S1. Primer sequences used for RT-PCR and qRT-PCR.
Supplemental Table S2. Primer sequences used for genotyping PCR and constructions.
Supplemental Table S3. Excel data files of stomatal apertures that were obtained in experiments.
Supplementary Material
Glossary
- ABA
abscisic acid
- H2O2
hydrogen peroxide
- Ub
ubiquitin
- qRT
quantitative reverse transcription
- RT
reverse transcription
- DAB
3,3′-diaminobenzidine
- bZIP
basic leucine zipper
- cDNA
complementary DNA
References
- Ahuja I, de Vos RCH, Bones AM, Hall RD. (2010) Plant molecular stress responses face climate change. Trends Plant Sci 15: 664–674 [DOI] [PubMed] [Google Scholar]
- Bergler J, Hoth S. (2011) Plant U-box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol (Stuttg) 13: 725–730 [DOI] [PubMed] [Google Scholar]
- Bos JIB, Armstrong MR, Gilroy EM, Boevink PC, Hein I, Taylor RM, Zhendong T, Engelhardt S, Vetukuri RR, Harrower B, et al. (2010) Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc Natl Acad Sci USA 107: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Chung HS, Ryu MY, Park MJ, Lee MM, Bahk Y-Y, Kim J, Pai HS, Kim WT. (2006) Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-box E3 ubiquitin ligase homolog. Plant Physiol 142: 1664–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Ryu MY, Seo DH, Kang BG, Kim WT. (2011) The Arabidopsis RING E3 ubiquitin ligase AtAIRP2 plays combinatory roles with AtAIRP1 in abscisic acid-mediated drought stress responses. Plant Physiol 157: 2240–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Ryu MY, Song C, Kwak JM, Kim WT. (2008) Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20: 1899–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dreher K, Callis J. (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BT, Schulman BA. (2007) Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct 36: 131–150 [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu J-K. (2011) Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci USA 108: 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu J-K. (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Iuchi S, Yamada K, Kobayashi Y, Urano K, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. (2012) Natural variation in a polyamine transporter determines paraquat tolerance in Arabidopsis. Proc Natl Acad Sci USA 109: 6343–6347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, et al. (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- González-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JD. (2006) The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18: 1067–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TS, Mazzurco M, Sulaman W, Matias DD, Goring DR. (1998) Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc Natl Acad Sci USA 95: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth S, Morgante M, Sanchez J-P, Hanafey MK, Tingey SV, Chua N-H. (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Huang X-Y, Chao D-Y, Gao J-P, Zhu M-Z, Shi M, Lin H-X. (2009) A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev 23: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. (2010) Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev 24: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell 28: 730–738 [DOI] [PubMed] [Google Scholar]
- Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D, Cho D, Lee S, Giordo R, Sritubtim S, et al. (2009) MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci USA 106: 20520–20525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Ryu MY, Kim WT. (2012) Suppression of Arabidopsis RING-DUF1117 E3 ubiquitin ligases, AtRDUF1 and AtRDUF2, reduces tolerance to ABA-mediated drought stress. Biochem Biophys Res Commun 420: 141–147 [DOI] [PubMed] [Google Scholar]
- Kim T-H, Böhmer M, Hu H, Nishimura N, Schroeder JI. (2010) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Murata Y, Baizabal-Aguirre VM, Merrill J, Wang M, Kemper A, Hawke SD, Tallman G, Schroeder JI. (2001) Dominant negative guard cell K+ channel mutants reduce inward-rectifying K+ currents and light-induced stomatal opening in Arabidopsis. Plant Physiol 127: 473–485 [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Kim WT. (2011) Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol Cells 31: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Sherman-Broyles S, Nasrallah ME, Nasrallah JB. (2007) A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr Biol 17: 734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-C, Wu Y-R, Huang X-H, Sun J, Xie Q. (2011) AtPUB19, a U-box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Mol Plant 4: 938–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua N-H. (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Shen G, Yan J, He C, Zhang H. (2006) AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J 46: 649–657 [DOI] [PubMed] [Google Scholar]
- Lyzenga WJ, Stone SL. (2012) Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot 63: 599–616 [DOI] [PubMed] [Google Scholar]
- Mbengue M, Camut S, de Carvalho-Niebel F, Deslandes L, Froidure S, Klaus-Heisen D, Moreau S, Rivas S, Timmers T, Hervé C, et al. (2010) The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22: 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Parry G, Estelle M. (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16: 3181–3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y, Shiu SH, Stone SL, Salt JN, Goring DR. (2004) A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol 134: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli A-C, Merlot S, Vavasseur A, Fenzi F, Giraudat J. (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Park HC, Lee SY, Bohnert HJ, Yun D-J. (2011a) Ubiquitin and ubiquitin-like modifiers in plants. J Plant Biol 54: 275–285 [Google Scholar]
- Park J-J, Yi J, Yoon J, Cho L-H, Ping J, Jeong HJ, Cho SK, Kim WT, An G. (2011b) OsPUB15, an E3 ubiquitin ligase, functions to reduce cellular oxidative stress during seedling establishment. Plant J 65: 194–205 [DOI] [PubMed] [Google Scholar]
- Raab S, Drechsel G, Zarepour M, Hartung W, Koshiba T, Bittner F, Hoth S. (2009) Identification of a novel E3 ubiquitin ligase that is required for suppression of premature senescence in Arabidopsis. Plant J 59: 39–51 [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Ryu MY, Cho SK, Kim WT. (2010) The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol 154: 1983–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt JN, Yoshioka K, Moeder W, Goring DR. (2011) Altered germination and subcellular localization patterns for PUB44/SAUL1 in response to stress and phytohormone treatments. PLoS ONE 6: e21321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Chong YT, Haasen KE, Aldea-Brydges MG, Stone SL, Goring DR. (2009) Cellular pathways regulating responses to compatible and self-incompatible pollen in Brassica and Arabidopsis stigmas intersect at Exo70A1, a putative component of the exocyst complex. Plant Cell 21: 2655–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Mudgil Y, Salt JN, Delmas F, Ramachandran S, Chilelli A, Goring DR. (2008) Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol 147: 2084–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Stone SL, Anderson EM, Mullen RT, Goring DR. (2003) ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15: 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Arnoldo M, Goring DR. (1999) A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286: 1729–1731 [DOI] [PubMed] [Google Scholar]
- Tardieu F, Granier C, Muller B. (2011) Water deficit and growth: co-ordinating processes without an orchestrator? Curr Opin Plant Biol 14: 283–289 [DOI] [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K. (2008) Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr Biol 18: 1396–1401 [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Sánchez ME, Zeng L, Chen S, Leung H, Wang GL. (2008) SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice. Plant Cell 20: 1456–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wiborg J, O’Shea C, Skriver K. (2008) Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem J 413: 447–457 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Takahashi Y, Berberich T, Imai A, Takahashi T, Michael AJ, Kusano T. (2007) A protective role for the polyamine spermine against drought stress in Arabidopsis. Biochem Biophys Res Commun 352: 486–490 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yang CW, González-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O’Donnell E, Jones JD, Sadanandom A. (2006) The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18: 1084–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D, Goring DR. (2009) The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot 60: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Zeng LR, Park CH, Venu RC, Gough J, Wang GL. (2008) Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol Plant 1: 800–815 [DOI] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL. (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulfugarov IS, Tovuu A, Kim J-H, Lee C-H. (2011) Detection of reactive oxygen species in higher plants. J Plant Biol 54: 351–357 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.