GAS STIMULATE PLANT GROWTH AND DEVELOPMENT
GAs are a family of tetracyclic diterpenoid plant hormones that stimulate plant growth and developmental transitions. As sessile organisms, plants rely on developmental plasticity to respond to environmental challenges. Plant hormones regulate developmental responses to diverse environmental stimuli such as changes in light, temperature, moisture, animal feeding, and disease pressure. GAs stimulate seed germination in response to changes in light, temperature, and moisture (Koornneef and van der Veen, 1980; Yamauchi et al., 2004; Seo et al., 2009). GA also stimulates stem elongation and leaf expansion through cell expansion and cell division in response to light or dark (photomorphogenesis and skotomorphogenesis; Ogawa et al., 2003; Alabadí et al., 2008; Feng et al., 2008; de Lucas et al., 2008; Gallego-Bartolomé et al., 2011). GA stimulates developmental transitions from meristematic to shoot growth, from juvenile to adult leaf development, and from vegetative growth to flowering, and also stimulates aspects of flower development (Telfer et al., 1997; Yu et al., 2004; Galinha et al., 2009). The appropriate regulation of these events is essential to the survival of plant species and to successful crop production. GA stimulates many aspects of plant growth and development by lifting DELLA (Asp-Glu-Leu-Leu-Ala) protein repression of these events. This article will review multiple biochemical mechanisms for the regulation of and response to DELLA repression.
Studies using plants with altered GA biosynthesis or catabolism have resulted in a wealth of knowledge of the diverse roles of GA in plant growth and development (for review, see Sun and Gubler, 2004; Yamaguchi, 2008). GA biosynthesis enzyme mutants of dicots and monocots are GA sensitive, showing defects in growth and development that are rescued by GA application. GA-sensitive mutants of rice (Oryza sativa) and barley (Hordeum vulgare) exhibit dwarfism, infertility, and failure to mobilize stored reserves during seed germination through α-amylase induction (Zwar and Chandler, 1995; Chandler and Robertson, 1999; Sakamoto et al., 2004). Similar phenotypes are seen in Arabidopsis (Arabidopsis thaliana) mutants affecting enzymes acting later in the biosynthesis pathway, GA 3-oxidase (GA3ox) and GA20ox (Hedden and Phillips, 2000; Plackett et al., 2012). Since GA3ox and GA20ox belong to multigene families, single mutants are fertile semidwarves. In Arabidopsis and tomato (Solanum lycopersicum), mutants affecting early GA biosynthesis enzymes such as ent-copalyl diphosphate synthase (Sun and Kamiya, 1994) cause failure in seed germination, growth as a dark green dwarf, failure to transition to flowering under short days, and partial to complete infertility (Koornneef and van der Veen, 1980; Karssen et al., 1989; Wilson et al., 1992). Overexpression of the gene encoding the GA catabolic enzyme GA2ox increases GA turnover, leading to reduced grain germination and α-amylase induction in wheat (Triticum aestivum; Appleford et al., 2007) and to failures in seed development and pollen tube growth in Arabidopsis (Singh et al., 2002).
GA signaling is often controlled through direct regulation of hormone accumulation mediated by changes in GA20ox, GA3ox, and GA2ox expression in response to environmental or developmental stimuli. This is logical, as the hormone is the first step in a hormone signaling pathway. Stimulation of Arabidopsis seed germination by red light or cold imbibition and inhibition of germination by far-red light are associated with increased and decreased GA accumulation, respectively (for review, see Seo et al., 2009). Far-red light inhibits seed germination by inducing GA turnover through GA2ox2 and inhibiting the GA biosynthesis GA3ox genes, whereas red light or cold stimulates germination by inducing the biosynthesis genes GA3ox or GA20ox and inhibiting GA2ox expression (Penfield et al., 2006; Oh et al., 2007). The germination of seed imbibing in the cold is stimulated by increased GA levels, but cold acclimation of adult plants is associated with decreased GA. Induction of the C-repeat-binding factor genes by cold acclimation induces the GA turnover GA2ox genes (Achard et al., 2008). Decreased GA levels enhance cold tolerance and suppress plant growth in the cold. GA stimulates the transition from meristematic growth to shoot differentiation. KNOX genes maintain the meristem by repressing the GA biosynthesis GA20ox enzymes and activating the transcript accumulation of the GA turnover GA2ox enzymes (for review, see Galinha et al., 2009). GA20ox expression, and presumably GA accumulation, is high in new shoots but depleted in the meristem.
GA SIGNAL RECEPTION, A CASE OF MOLECULAR GLUE
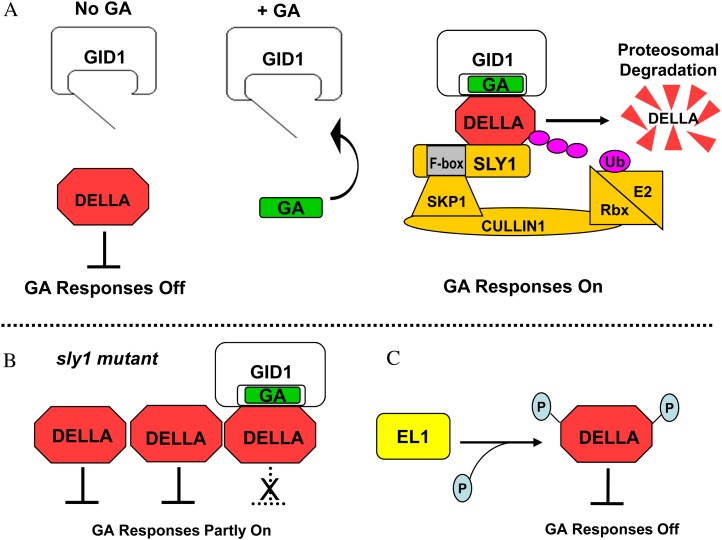
The GA signal is perceived by a soluble receptor protein, GA-INSENSITIVE DWARF1 (GID1). The mechanisms of GA perception are conserved, showing agreement in Arabidopsis and rice, where the signaling pathway has been studied in the greatest detail (Table I). The GID1 gene was identified through map-based cloning of a GA-insensitive mutant in rice, where there is a single copy of the gene (Ueguchi-Tanaka et al., 2005). GA-insensitive GID1 mutants have defined a single barley homolog, GSE1 (Gubler et al., 2002; Chandler et al., 2008), and three Arabidopsis homologs, GID1a, GID1b, and GID1c (Griffiths et al., 2006; Nakajima et al., 2006; Iuchi et al., 2007; Willige et al., 2007). Mutations in the GA receptor result in phenotypes similar to those resulting from severe GA biosynthesis mutations, but they are not rescued by GA application. GID1 protein localizes mainly to the nucleus but also appears to localize to the cytoplasm (Ueguchi-Tanaka et al., 2005; Willige et al., 2007). GID1 encodes a homolog of mammalian hormone-sensitive lipase (Ueguchi-Tanaka et al., 2005). X-ray crystallography demonstrated two key features of the GID1 protein (Murase et al., 2008; Shimada et al., 2008). First, the hormone-sensitive lipase catalytic domain that normally binds a lipid has evolved to bind GAs. Second, the N-terminal “lid” domain of GID1 interacts hydrophobically with the γ-lactone ring of GA4 and upon GA binding folds over the GA-binding pocket (Fig. 1A). This GA-dependent conformational change causes the GID1 N-terminal helical lid domain to behave like “molecular glue” to interact with DELLA repressor target proteins (Ueguchi-Tanaka et al., 2007; Murase et al., 2008). The theme where a small hormone molecule enables a protein-protein interaction is common in plants. For example, the hormones auxin and jasmonate stimulate the interaction of their hormone receptors with the downstream target proteins through direct interactions with both receptor and target proteins (Tan et al., 2007; Sheard et al., 2010). The GID1-GA-DELLA complex binds GA4 with a higher affinity than GID1 alone, indicating that DELLA binding stabilizes the receptor-hormone interaction (Nakajima et al., 2006; Ueguchi-Tanaka et al., 2007). The GID1-GA complex stimulates plant growth and development by down-regulating DELLA repressors through direct protein-protein interaction.
Table I. GA signaling genes of Arabidopsis, rice, and barley.
| Plant | GA Receptors | F-Box Proteins | DELLA Repressors | DELLA Targetsa |
|---|---|---|---|---|
| Arabidopsis | GID1a, GID1b, GID1c | SLY1, SNE | GAI, RGA, RGL1, RGL2, RGL3 | JAZ1, PIF3, PIF4, SCL3, ALC,PIF1/PIL5, PIL2, SPT |
| Rice | GID1 | GID2, SNE | SLR1 | – |
| Barley | GSE1 | – | SLN1 | – |
DELLA targets in boldface have been biochemically verified through coimmunoprecipitation or pull-down assay.
Figure 1.
Proteolysis-dependent and -independent GA signaling models. A, The canonical GA signaling model illustrating GA-dependent GID1-DELLA complex formation resulting in DELLA recognition and ubiquitylation by the SCFSLY1 E3. Polyubiquitylation leads to DELLA proteolysis by the 26S proteasome, thereby lifting DELLA repression of GA responses. B, Proteolysis-independent GA signaling in sly1 mutants occurs when GID1-GA-DELLA complex formation blocks DELLA repression of GA responses without DELLA destruction. C, EL1-mediated phosphorylation of DELLA activates DELLA as a repressor of GA responses.
THE DELLA REPRESSORS OF GA SIGNALING ARE REGULATED BY THE UBIQUITIN-PROTEASOME PATHWAY
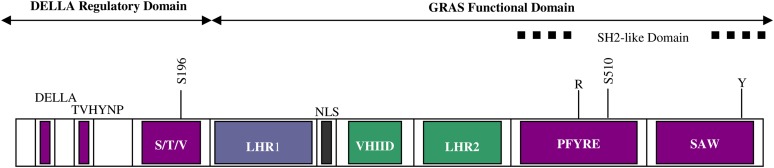
DELLA genes are defined as repressors of GA signaling because gain-of-function mutations in DELLA genes lead to reduced GA signaling (i.e. dwarfism), whereas loss of function leads to increased GA signaling (i.e. tall or slender phenotype). GID1 lifts DELLA repression through direct protein-protein interaction. All DELLA repressors have an N-terminal DELLA regulatory domain containing the conserved amino acid sequence Asp-Glu-Leu-Leu-Ala (DELLA) and a C-terminal GRAS (for GAI, RGA, and SCARECROW) functional domain (Fig. 2; Peng et al., 1997; Silverstone et al., 1998; Pysh et al., 1999; Itoh et al., 2002). The N-terminal DELLA regulatory domain is an intrinsically disordered domain that folds and becomes structured upon GID1 protein binding (Sun et al., 2010). Mutations in the DELLA and TVHYNP regions of the regulatory domain interfere with the ability to bind the GID1 receptor, leading to a semidominant GA-insensitive dwarf phenotype due to the inability to down-regulate the DELLA repressor (Peng et al., 1997; Dill and Sun, 2001; Itoh et al., 2002; Silverstone et al., 2007; Asano et al., 2009). Similar phenotypes have been detected in the PFYRE and SAW regions of the GRAS domain, which form secondary interactions with GID1 (Hirano et al., 2010). The DELLA regulatory domain also contains a Ser/Thr/Val-rich domain believed to be involved in the regulation of DELLA function by phosphorylation, since deletion of both the TVHYNP and Ser/Thr/Val domains blocks DELLA phosphorylation (Itoh et al., 2005a; Silverstone et al., 2007; Dai and Xue, 2010). The DELLA gene of barley is called SLENDER1 (SLN1) and the DELLA gene of rice is called SLENDER RICE1 (SLR1), in reference to the “tall” phenotypes resulting from loss-of-function mutations in the GRAS functional domain (Table I; Chandler et al., 2002; Itoh et al., 2002). The roles of the five Arabidopsis DELLA repressor genes have been determined based on the ability of loss-of-function alleles to rescue phenotypes of the ga1-3 GA biosynthesis mutant. Mutations in REPRESSOR OF GA1-3 (RGA), GA-INSENSITIVE (GAI), and RGA-LIKE (RGL1) rescue plant height; mutations RGA, RGL2, and RGL1 rescue flowering; while mutations in RGL2, RGA, GAI, and RGL3 rescue seed germination (Dill and Sun, 2001; King et al., 2001; Lee et al., 2002; Cheng et al., 2004, Tyler et al., 2004; Cao et al., 2005; Piskurewicz and Lopez-Molina, 2009). DELLA RGL2 has the strongest effect on seed germination, whereas DELLA RGA has the strongest effect on plant height. Promoter-swap experiments have shown that the partially specialized functions of these two Arabidopsis DELLA proteins appear to result mainly from tissue-specific gene expression (Gallego-Bartolomé et al., 2010).
Figure 2.
DELLA protein organization illustrating the conserved domains and subdomains involved in GID1 binding (purple), target binding (blue), nuclear localization (gray), and SLY1/GID2 binding (green). Also depicted are the invariant Arg (R) and Tyr residues (Y) conserved among the STAT-like GRAS protein family and specific Ser residues (Ser-196 and Ser-510) that are sites of EL1-directed phosphorylation.
GA lifts DELLA repression of GA responses by targeting DELLA for destruction via the ubiquitin-proteasome pathway. This model was originally based on the observation that rescue of dwarfism due to GA deficiency by GA treatment was associated with DELLA protein disappearance (Silverstone et al., 2001; Itoh et al., 2002). The ubiquitin-proteasome pathway was implicated when it was found that mutations in the F-box genes SLEEPY1 (SLY1) of Arabidopsis and the rice homolog GID2 resulted in a GA-insensitive phenotype associated with an inability to target DELLA for destruction (McGinnis et al., 2003; Sasaki et al., 2003). SLY1/GID2 is the F-box subunit of an SCF (for SKP1, CULLIN, and F-BOX) E3 ubiquitin ligase that catalyzes the polyubiquitylation of DELLA protein (McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004; Gomi et al., 2004; Hussain et al., 2005; Ariizumi et al., 2011; Wang and Deng, 2011). The F-box subunit of an SCF complex confers substrate specificity through a C-terminal protein-protein interaction domain and interacts with the rest of the SCF complex via the N-terminal F-box domain (Smalle and Vierstra, 2004). The SLY1/GID2 F-box requires the C-terminal GGF and LSL domains to bind the DELLA protein VHIID and LHRII domains (Fig. 2; Dill et al., 2004; Fu et al., 2004; Hirano et al., 2010; Ariizumi et al., 2011). SLY1 can bind the Arabidopsis SKP1 protein ASK1 in yeast two-hybrid assays and forms a complex with CULLIN1 that requires the F-box domain in planta (Fu et al., 2004; Ariizumi et al., 2011). Based on a cell-free system, SCFSLY1 catalyzes the formation of polyubiquitin chains covalently attached to DELLA through ubiquitin-Lys-29 rather than through Lys-48 (Wang et al., 2009). GA treatment of the ga1-3 GA biosynthesis mutant results in DELLA destruction through ubiquitylation within 5 h in seeds or 10 min in seedlings (Tyler et al., 2004; Ariizumi and Steber, 2007; Zhang et al., 2011). This disappearance can be blocked by 26S proteasome inhibitors (Fu et al., 2002; Hussain et al., 2005). Thus, GA-stimulated polyubiquitylation of DELLA proteins targets them for destruction via the 26S proteasome.
The formation of the GID1-GA-DELLA complex triggers the protein-protein interaction between DELLA and the F-box protein SLY1/GID2. Yeast three-hybrid analysis demonstrated that GID2 binding to DELLA is stimulated by GID1-GA-DELLA complex formation (Griffiths et al., 2006; Hirano et al., 2010). GA stimulates SLY1 coimmunoprecipitation with GID1, suggesting that GID1-DELLA complex formation also stimulates GID1-DELLA-SLY1 interaction in planta (Ariizumi et al., 2011). DELLA proteins accumulate at higher levels in GA biosynthesis mutants, gid1, and sly1/gid2 mutants, indicating that GA, the GID1 receptor, and components of the SCF complex are required for DELLA proteolysis via the ubiquitin-proteasome pathway (McGinnis et al., 2003; Sasaki et al., 2003; Zhao et al., 2003; Dill et al., 2004; Fu et al., 2004; Gomi et al., 2004; Hussain et al., 2005). GST pull-down and coimmunoprecipitation assays have shown DELLA in complex with SLY1/GID2 in planta (Fu et al., 2004; Ariizumi et al., 2011). Moreover, GA, GID1, SCFSLY1, and ubiquitin are sufficient to polyubiquitylate Arabidopsis DELLAs in a cell-free system (Wang et al., 2009). There is a homolog of SLY1/GID2 both in Arabidopsis and rice called SNEEZY (SNE) or SLY2 (McGinnis et al., 2003; Itoh et al., 2003; Fu et al., 2004; Strader et al., 2004; Cui and Benfey, 2009). While SNE can function in GA signaling, SLY1 is the main regulator of DELLA protein accumulation, given that SNE mutations have a small additive effect in sly1 sne double mutants (Ariizumi et al., 2011; Ariizumi and Steber, 2011).
NONPROTEOLYTIC MECHANISMS FOR GA SIGNALING: VARIATIONS ON THE THEME
If all of GA signaling occurred through DELLA proteolysis, then the level of DELLA protein accumulation should always correlate with the severity of GA-insensitive phenotypes. This is not the case in sly1 and gid2 F-box mutants, which accumulate much more DELLA protein but show less severe GA-insensitive phenotypes than either GA biosynthesis or GA receptor null mutants (McGinnis et al., 2003; Willige et al., 2007; Ueguchi-Tanaka et al., 2008). It appears that DELLA can be inactivated by a nonproteolytic mechanism such that not all of the DELLA protein that accumulates in sly1/gid2 mutants is functional as a repressor of GA signaling (Ariizumi and Steber, 2007; Ariizumi et al., 2008; Ueguchi-Tanaka et al., 2008). A model in which DELLA repression is blocked by GID1-GA-DELLA complex formation alone was proposed based on the following evidence: (1) sly1/gid2 phenotypes are rescued by GID1 gene overexpression without a reduction in DELLA protein levels; and (2) GA signaling in sly1/gid2 requires all of the elements required for GID1-GA-DELLA complex formation, including GA hormone synthesis, the GID1 gene, and the DELLA domain required for DELLA-GID1 protein interaction. In light of this, it appears that GID1 causes GA signaling in sly1 mutants by binding DELLA protein rather than by triggering DELLA proteolysis (Fig. 1B). Unlike Arabidopsis GID1a and GID1c, the GID1b protein binds GA with a higher affinity and shows some ability to interact with DELLA in the absence of GA. This explains why GID1b overexpression better rescues GA-insensitive sly1 mutant phenotypes (Ariizumi et al., 2008; Yamamoto et al., 2010). This GA-independent GID1b-DELLA binding may provide a low level of GA signaling in the absence of GA. Arabidopsis GID1b tends to have low expression levels, suggesting that control of GID1b accumulation may be essential to regulating basal GA responses (Griffiths et al., 2006; Willige et al., 2007). Because DELLA cannot be degraded in the sly1/gid2 F-box mutants, this background provides a tool for examining GA signaling mechanisms that precede DELLA destruction or may occur in situations where DELLA destruction is delayed by ethylene signaling or lack of auxin signaling (Achard et al., 2003; Fu and Harberd, 2003). Determining the contribution of proteolysis-independent DELLA down-regulation to GA signaling without using a sly1/gid2 background requires the ability to directly assay DELLA protein function as well as its accumulation. Phenotypic assays are not useful here, since phenotype is the sum of events over time and can be influenced by multiple signaling pathways. Future work will need to develop a direct assay for DELLA function or use DELLA-regulated promoters as reporters.
PHOSPHORYLATION AND O-GLcNAc MODIFICATION OF DELLA PROTEIN ACTIVITY
Evidence also suggests that DELLA activity can be influenced by phosphorylation and O-GlcNAc (for O-linked N-acetylglucosamine) modification. Studies indicate that DELLA is phosphorylated, but the precise role of this modification in controlling DELLA activity or protein degradation is unclear. When DELLA phosphorylation was first observed in rice and in Arabidopsis, it was initially hypothesized that DELLA phosphorylation might increase DELLA affinity for the F-box protein SLY1/GID2, since target protein phosphorylation often stimulates F-box protein target binding in other systems (Sasaki et al., 2003; Fu et al., 2004; Smalle and Vierstra, 2004). Initially, it appeared that rice DELLA SLR1 and Arabidopsis DELLA GAI showed stronger F-box protein binding when phosphorylated (Fu et al., 2004; Gomi et al., 2004). However, kinetic studies of the SLR1-GID2 protein-protein interaction demonstrated no difference in F-box GID2 affinity for phosphorylated or nonphosphorylated DELLA SLR1 (Itoh et al., 2005a). Protein phosphatase inhibitors appear to block the degradation of barley DELLA SLN1 and Arabidopsis DELLAs RGL2 and RGA, suggesting that phosphorylated DELLA is more resistant to degradation (Fu et al., 2002; Hussain et al., 2005; Wang et al., 2009).
Recent evidence suggests that the Ser/Thr protein casein kinase I encoded by rice EARLY FLOWERING1 (EL1) regulates DELLA SLR1 through direct protein phosphorylation (Dai and Xue, 2010). EL1 acts as a negative regulator of GA signaling and a positive regulator of DELLA, since loss of EL1 function leads to increased GA sensitivity associated with early flowering and increased α-amylase expression during seed germination. The el1 phenotypes are associated with a mild increase in the rate of DELLA SLR1 protein degradation following GA treatment, consistent with previous phosphatase inhibitor experiments suggesting that phosphorylated DELLA is more stable. Two lines of evidence suggest that EL1 regulates DELLA by phosphorylation. First, EL1 protein phosphorylates DELLA SLR1 in vitro. Second, Ser-to-Ala mutations in DELLA SLR1 (S196A and S510A), believed to block EL1-dependent phosphorylation, prevented DELLA activation of gene expression. Moreover, Ser-to-Asp phosphomimic mutations (S196D and S510D) led to increased accumulation of DELLA-activated transcripts. This suggests that DELLA may be regulated through phosphorylation at these two sites, one within the DELLA regulatory domain and one within the GRAS functional domain (Fig. 2). Future work will need to demonstrate EL1-directed DELLA phosphorylation at these sites in planta and determine the effect of phosphorylation on DELLA function.
SPINDLY (SPY) encodes an O-GlcNAc transferase that negatively regulates GA signaling in Arabidopsis, rice, and barley (Robertson et al., 1998; Swain et al., 2001; Shimada et al., 2006; Filardo et al., 2009). Loss-of-function mutations in SPY were identified in screens for resistance to the GA biosynthesis inhibitor paclobutrazol and for suppressors of ga1-3 (Jacobsen and Olszewski 1993; Silverstone et al., 1997). In Arabidopsis, spy suppresses the gain-of-function DELLA mutation rga-Δ17. Such suppression is associated not with DELLA disappearance but with apparent DELLA phosphorylation (Silverstone et al., 2007). SPY RNA interference also suppressed the rice gid2 F-box mutant without DELLA proteolysis and resulted in increased DELLA SLR1 protein phosphorylation (Shimada et al., 2006). Since silencing SPY also suppresses rice gid1 and GA biosynthesis mutants, it appears that increased GA signaling in spy requires neither DELLA proteolysis nor GID1-DELLA complex formation. The model proposed suggested that, as in mammals, O-GlcNAcylation competes with phosphorylation for modification of the same Ser/Thr residues and that loss of DELLA O-GlcNAcylation results in decreased DELLA function as a consequence of phosphorylation (Wells et al., 2004; Shimada et al., 2006; Silverstone et al., 2007). We must be cautious in interpreting these data, however, since it has not been clearly demonstrated that DELLA is O-GlcNAcylated. There is an apparent contradiction, since DELLA phosphorylation in spy mutants is thought to cause increased GA signaling, whereas EL1-directed DELLA phosphorylation is believed to cause decreased GA signaling. It is possible that DELLA is activated by phosphorylation as a feedback mechanism to counteract excessive GA signaling due to the spy mutation or that phosphorylation at different amino acid residues may have different effects on DELLA functions. Taken together, these studies suggest that DELLA activity can be modified by posttranslational modification. Future work will need to examine whether DELLA posttranslational modification either alters GID1-DELLA binding affinity or is influenced by the GID1 receptor.
DELLA CONTROLS PLANT GROWTH AND DEVELOPMENT THROUGH INTERACTION WITH MULTIPLE TARGET PROTEINS
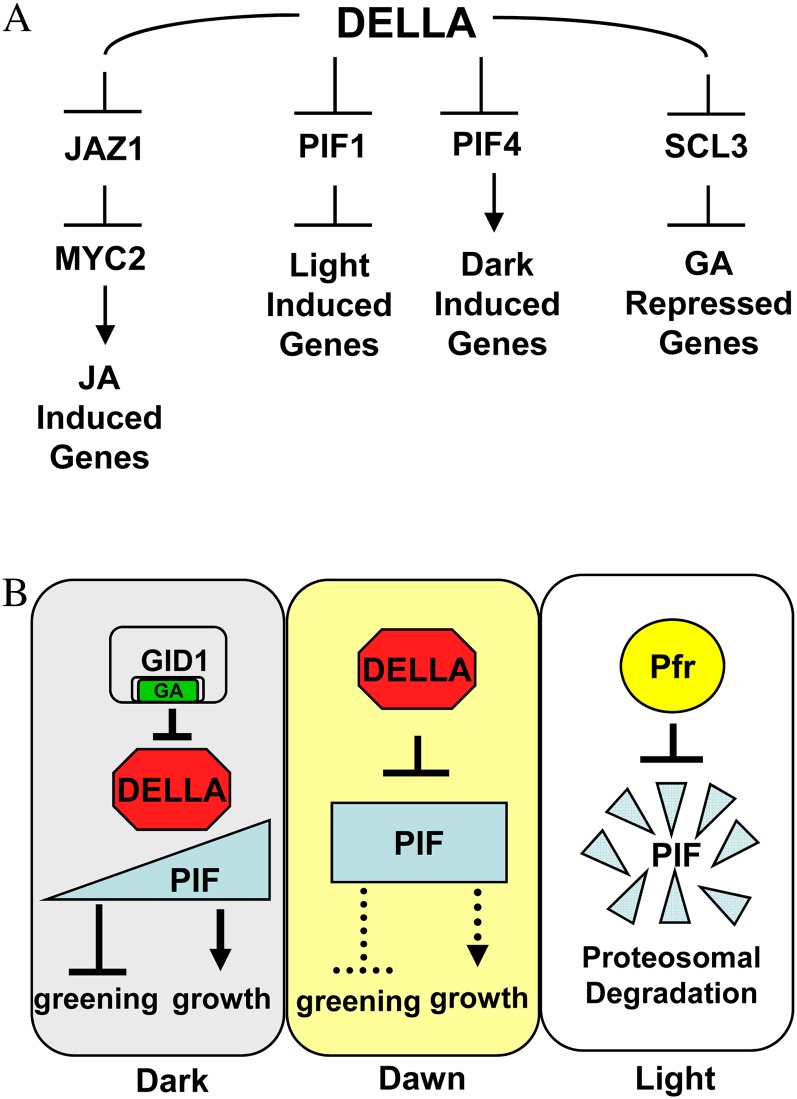
DELLA is able to transmit multiple environmental signals to control plant growth and development by interacting with multiple targets that control gene transcription. The idea that DELLA proteins regulate transcription was originally based on nuclear localization and protein homology. Based on recent studies, DELLA controls gene transcription through protein interaction with specific transcription factor targets. DELLA proteins are nucleus-localized homologs of metazoan signal transducers and activators of transcription (STAT) factors (Darnell, 1997; Peng et al., 1997; Silverstone et al., 1998; Richards et al., 2000; Ikeda et al., 2001). The GRAS functional domain of DELLAs resembles STAT factors in that it contains two Leu heptad repeat domains (LHR1 and LHR2; also called Leu-rich or LZ domains) and a Src Homology2 (SH2)-like domain (Fig. 2; Richards et al., 2000). Additional GRAS subdomains PFYRE and SAW correspond roughly to the SH2-like domain. DELLA regulates transcription, given that most of the GA-regulated transcriptome is DELLA regulated in seeds, seedlings, and flowers (Cao et al., 2006; Zentella et al., 2007; Gallego-Bartolomé et al., 2011). Chromatin immunoprecipitation (ChIP) experiments showed that the Arabidopsis DELLA RGA localizes to the promoter regions of 18 early-DELLA-regulated genes (Zentella et al., 2007). Since all of these genes were GA down-regulated and DELLA up-regulated, it was hypothesized that DELLA functions as a transcriptional activator of repressors of GA signaling, such as the activator of abscisic acid biosynthesis, XERICO (Zentella et al., 2007). That the DELLA domain required to activate transcription in a yeast one-hybrid assay is also required for function as a negative regulator of stem elongation in rice provides circumstantial evidence in favor of this hypothesis (Hirano et al., 2012). Since DELLA showed no more than 3.5-fold enrichment at any early DELLA-regulated promoters, it was proposed that DELLA acts through interaction with DNA-binding proteins (Zentella et al., 2007). Subsequently, DELLA protein was found to interact directly with transcriptional regulators PIF3 (for PHYTOCHROME INTERACTING FACTOR3), PIF4, JAZ1 (for jasmonate ZIM domain1), and SCL3 (for SCARECROW-LIKE3) in planta and is believed to interact with ALCATRAZ (ALC), SPT (for SPATULA), PIF1/PIL5 (for PIF-LIKE), and PIL2 based on yeast two-hybrid assays (de Lucas et al., 2008; Feng et al., 2008; Arnaud et al., 2010; Gallego-Bartolomé et al., 2010; Hou et al., 2010; Heo et al., 2011; Zhang et al., 2011). It is interesting that DELLA protein interaction with JAZ1 and PIF4 requires the LHR1 domain, since mutations in the LHR1 domain result in a gain-of-function GA-insensitive phenotype (Itoh et al., 2002; de Lucas et al., 2008; Wang et al., 2009; Hirano et al., 2010; Hou et al., 2010). In every case where a DELLA-target protein interaction has been well characterized, it appears that DELLA interferes with the ability of a DNA-binding protein to activate or repress a target promoter (Fig. 3A). This does not support the model that DELLA is a transcriptional activator of repressors of GA signaling. Future work will need to (1) examine if DELLA can serve either as a repressor or as a coactivator of gene transcription depending on specific target interactions, and (2) examine how DELLA is able to interact with so many target proteins. Given that the N-terminal region of DELLAs is an intrinsically disordered region, it is possible that the DELLA intrinsically disordered region facilitates multiple protein interactions (Sun et al., 2010). Such disordered regions have structural flexibility enabling specific but low-affinity interactions with multiple proteins (for review, see Sun et al., 2012). Below, the role of DELLA-target protein interactions in the transcriptional control of plant growth and development is discussed with an emphasis on biochemically verified target proteins (Table I).
Figure 3.
A, Downstream DELLA targets negatively regulated through confirmed protein-protein interactions with DELLA. B, Suggested model for DELLA modulation of PIF activity in response to light and the clock. In the night, GA and GID1 levels are highest, thus repressing DELLA’s ability to negatively regulate PIF. PIF mRNA levels rise during the night, leading to rapid hypocotyl growth before morning. At dawn, a drop in GID1 levels results in DELLA stabilization and repression of PIF, leading to greening and slower growth. In the day, red light actives Pfr, which phosphorylates PIF, leading to PIF destruction via the ubiquitin-proteasome pathway. Lack of PIF blocks hypocotyl growth.
DELLA RGA protein negatively regulates the GRAS family transcription factor SCL3. SCL3 is a positive regulator of GA signaling, since loss of scl3 function results in mild GA-insensitive phenotypes, including reduced seed germination, shoot elongation, and root elongation (Heo et al., 2011; Zhang et al., 2011). SCL3 and DELLA appear to act in opposition to each other based on the facts that (1) DELLA RGA binds to SCL3 protein, thereby blocking SCL3 repression of its own promoter, and (2) SCL3 negatively regulates the DELLA-activated genes SCL3, GA20ox2, and GA3ox1 (Zhang et al., 2011). Curiously, SCL3 was originally thought to be a DELLA-activated negative regulator of GA signaling, because ChIP and microarry analyses indicated that the SCL3 promoter is a direct DELLA target showing GA down-regulated/DELLA up-regulated mRNA expression (Zentella et al., 2007). Possibly, microarray data gave a false impression, because the GA down-regulated/DELLA up-regulated expression pattern is actually a negative feedback response. It is interesting that DELLA functions through interaction with the SCL3 GRAS protein, given that rice DELLA SLR1 can interact with itself in yeast two-hybrid analysis and that some DELLA-less GRAS genes can functionally replace DELLA SLR1 function in rice (Itoh et al., 2002, 2005b). Future research will need to investigate whether DELLA functionally interacts with other GRAS proteins.
DELLA regulates the response to pathogens through direct protein interaction with JAZ1, a repressor of jasmonate (JA) signaling. An initial study suggested that Arabidopsis DELLA genes play a role in defense, because the defense elicitor flagellin 22 appears to block plant growth by stabilizing DELLA RGA-GFP protein and because DELLAs are needed to induce defense-related gene expression in response to methyl-JA (Navarro et al., 2008). This evidence that DELLA is involved in JA and disease responses is interesting in light of the fact that DELLA regulates JA-responsive genes and physically interacts with JAZ1 protein (Hou et al., 2010). JAZ1 repression of JA-responsive genes is lifted when JA binding to the COI1 receptor and E3 ubiquitin ligase facilitates COI1-JAZ protein interaction, leading to JAZ proteolysis via the ubiquitin-proteasome pathway. In the absence of JA, JAZ1 binds the transcriptional activator MYC2, thereby repressing JA-inducible gene expression involved in wound and immune responses as well as the repression of root elongation. In the absence of GA, DELLA binds JAZ1, thereby blocking its ability to bind and inhibit MYC2 (Fig. 3A). This, in turn, leads to shorter roots and the expression of JA and wound-induced genes. Thus, DELLA acts as a positive regulator of JA response by blocking JAZ1 repressor action through protein-protein interaction.
GA signaling modulates growth responses to darkness through the DELLA protein. In the dark, seedlings become etiolated through skotomorphogenesis, growing long hypocotyls with an apical hook and failing to turn green, whereas seedlings grown in light show photomorphogenesis, resulting in short hypocotyls and expanded green cotyledons. GA signaling mediates some developmental responses to dark versus light. The LONG HYPOCOTYL5 (HY5) gene promotes photomorphogenesis in the light, and the PHYTOCHROME INTERACTING FACTOR3 (PIF3) and PIF4 genes promote skotomorphogenesis in the dark. GA signaling appears to negatively regulate HY5 protein stability in the dark, presumably through increased CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) activity (Alabadí et al., 2008). COP1 is an E3 ubiquitin ligase that targets the HY5 protein for destruction. In red light, the basic-helix-loop-helix (bHLH) transcription factors PIF3 and PIF4 interact with the Pfr form of phytochrome, which in turn leads to their destruction through the ubiquitin-proteasome pathway. In the dark, PIF3 and PIF4 activate transcription that stimulates hypocotyl growth and skotomorphogenesis (Huq and Quail, 2002; Park et al., 2004). GA and DELLA modulate this growth in the dark through the DELLA-PIF protein-protein interaction (Fig. 3A; de Lucas et al., 2008; Feng et al., 2008). In the absence of GA, DELLA binds to PIF3 and PIF4, blocking their ability to activate the transcription of genes involved in hypocotyl elongation. Electrophoretic mobility shift assays showed that DELLA can interfere with the ability of PIF4 to bind target promoter DNA (de Lucas et al., 2008). GA stimulates hypocotyl growth in the dark by targeting DELLA for destruction, thereby stimulating PIF protein function.
DELLA and PIFs also appear to have opposing effects on genes involved in greening and photosynthesis (Cheminant et al., 2011). ChIP indicates that in the dark, PIF1 protein directly binds the promoter and represses the expression of CONDITIONAL CHLORINA (CHLH), a gene involved in chlorophyll biosynthesis. Genetic evidence suggests that DELLA proteins block the ability of PIF1 to repress CHLH in the dark. The role of DELLA in controlling gene expression involved in photosynthesis is interesting in light of the fact that GID1, DELLA, and PIF4 are regulated by the circadian rhythm (Nozue et al., 2007; Michael et al., 2008; Arana et al., 2011). GID1a and GID1b mRNA levels are very low at dawn, whereas DELLA protein levels are lowest at night, rise just after dawn as GID1 levels decrease, and are highest in the light, when photosynthesis-related gene expression is needed (Arana et al., 2011). PIF4 and PIF5 protein levels are highest at the end of the night, when seedlings show their fastest growth rate (Nozue et al., 2007). We suggest a variation on the model proposed by Feng et al. (2008), in which DELLA inhibits PIF function at the transition from night to day (Fig. 3B). PIF4 is most active in stimulating seedling growth just before dawn, and GID1 mRNA levels decrease at dawn, leading to increased DELLA protein levels and repression of PIF4, until the ratio of red to far-red light increases sufficiently to trigger PIF4 destruction through interaction with the Pfr form of phytochrome.
DELLA also appears to participate in fruit development through interaction with another bHLH protein, ALC, in Arabidopsis (Arnaud et al., 2010). GA and ALC both stimulate differentiation of the separation layer in the silique value margin. The fact that the DELLA proteins RGA, GAI, and RGL2 interact with the ALC protein in yeast two-hybrid and bimolecular fluorescence complementation assays suggests that DELLA may block the ALC activation of promoters involved in this process by direct protein interaction. According to this model, GA stimulates fruit development in part by lifting DELLA repression of ALC.
GA stimulates and DELLA represses plant growth both through effects on cell expansion and division. GA stimulates the expression of enzymes involved in cell wall loosening and genes controlling cell division and also stimulates microtubule rearrangements associated with cell expansion (Ogawa et al., 2003; Achard et al., 2009; Sambade et al., 2012). GA appears to stimulate cell expansion in part through effects on downstream auxin transport and signaling. PIN-FORMED (PIN) auxin efflux transporter gene expression is repressed by the gene SHY2, which in turn is activated by the ARR1 transcription factor (Moubayidin et al., 2010). DELLA RGA indirectly activates ARR1 transcript accumulation, making DELLA a repressor of PIN. GA stimulates PIN expression through DELLA destruction. Consistent with this idea, Willige et al. (2011) found that GA biosynthesis and signaling are required for efficient auxin transport and appropriate accumulation of PIN1, -2, -3, and -4 proteins. That auxin controls cell expansion downstream of GA signaling is supported by microarray data showing that PIF genes control both GA- and auxin-regulated gene sets (Gallego-Bartolomé et al., 2011; Nozue et al., 2011). GA stimulates cell division in part by lifting the DELLA repression of STUNTED, a receptor-like cytoplasmic kinase that stimulates cell proliferation by repressing cyclin-dependent kinase inhibitor genes (Lee et al., 2012). Thus, GA regulates cell division and expansion through multiple DELLA-regulated mechanisms.
CONCLUSION
Many fresh avenues for research on GA signaling have developed since it was first proposed that GA functions by targeting DELLA repressors for destruction, acting as a negative regulator of a negative regulator. Major recent advances have demonstrated that GA stimulates GID1-DELLA protein interaction, proposed GA signaling mechanisms that are independent of DELLA proteolysis, characterized DELLA posttranslational modifications, and identified multiple downstream DELLA target proteins of diverse function. These advances suggest multiple areas for further research. (1) Genetic evidence suggests a functional interaction between DELLA and SPY. It remains to be determined whether DELLA is O-GlcNAc modified and whether this is needed for DELLA repression activity. (2) DELLA repression is stimulated by DELLA phosphorylation, but it is not known if phosphorylation at different residues has different effects on function. (3) Future research will need to address whether posttranslational modification alters the ability of DELLA to bind GID1 or downstream protein targets SCL3, JAZ1, PIF3, and PIF4. It is not known whether the GID1 protein regulates either DELLA posttranslational modification of downstream protein-protein interactions. (4) DELLA controls plant development through diverse factors, including the ZIM domain protein JAZ1 and transcription factors of the bHLH and GRAS families. The mechanisms enabling and controlling such diverse protein interactions remain largely unexplored. (5) The mechanisms by which DELLA interferes with target protein activity need further exploration. DELLA may behave as a coactivator of gene transcription, but characterization of DELLA function as a coactivator of transcription at a natural promoter is still needed. (6) The relative role of proteolytic and nonproteolytic mechanisms for lifting DELLA repression need to be further elucidated.
Acknowledgments
We apologize to all colleagues whose relevant work was not included due to space constraints. We thank members of the Steber laboratory for helpful discussion and reading of the manuscript.
Glossary
- STAT
signal transducers and activators of transcription
- ChIP
chromatin immunoprecipitation
- JA
jasmonate
- bHLH
basic helix-loop-helix
References
- Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P. (2008) The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GTS, Genschik P. (2009) Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol 19: 1188–1193 [DOI] [PubMed] [Google Scholar]
- Achard P, Vriezen WH, Van Der Straeten D, Harberd NP. (2003) Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15: 2816–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadí D, Gallego-Bartolomé J, Orlando L, García-Cárcel L, Rubio V, Martínez C, Frigerio M, Iglesias-Pedraz JM, Espinosa A, Deng XW, et al. (2008) Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J 53: 324–335 [DOI] [PubMed] [Google Scholar]
- Appleford NEJ, Wilkinson MD, Ma Q, Evans DJ, Stone MC, Pearce SP, Powers SJ, Thomas SG, Jones HD, Phillips AL, et al. (2007) Decreased shoot stature and grain α-amylase activity following ectopic expression of a gibberellin 2-oxidase gene in transgenic wheat. J Exp Bot 58: 3213–3226 [DOI] [PubMed] [Google Scholar]
- Arana MV, Marín-de la Rosa N, Maloof JN, Blázquez MA, Alabadí D. (2011) Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci USA 108: 9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Lawrence PK, Steber CM. (2011) The role of two F-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol 155: 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Murase K, Sun T-P, Steber CM. (2008) Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20: 2447–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Steber CM. (2007) Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell 19: 791–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Steber CM. (2011) Mutations in the F-box gene SNEEZY result in decreased Arabidopsis GA signaling. Plant Signal Behav 6: 831–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud N, Girin T, Sorefan K, Fuentes S, Wood TA, Lawrenson T, Sablowski R, Østergaard L. (2010) Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev 24: 2127–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Hirano K, Ueguchi-Tanaka M, Angeles-Shim RB, Komura T, Satoh H, Kitano H, Matsuoka M, Ashikari M. (2009) Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol Genet Genomics 281: 223–231 [DOI] [PubMed] [Google Scholar]
- Cao D, Cheng H, Wu W, Soo HM, Peng J. (2006) Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol 142: 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Hussain A, Cheng H, Peng J. (2005) Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223: 105–113 [DOI] [PubMed] [Google Scholar]
- Chandler PM, Harding CA, Ashton AR, Mulcair MD, Dixon NE, Mander LN. (2008) Characterization of gibberellin receptor mutants of barley (Hordeum vulgare L.). Mol Plant 1: 285–294 [DOI] [PubMed] [Google Scholar]
- Chandler PM, Marion-Poll A, Ellis M, Gubler F. (2002) Mutants at the Slender1 locus of barley cv Himalaya: molecular and physiological characterization. Plant Physiol 129: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler PM, Robertson M. (1999) Gibberellin dose-response curves and the characterization of dwarf mutants of barley. Plant Physiol 120: 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheminant S, Wild M, Bouvier F, Pelletier S, Renou J-P, Erhardt M, Hayes S, Terry MJ, Genschik P, Achard P. (2011) DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell 23: 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J. (2004) Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131: 1055–1064 [DOI] [PubMed] [Google Scholar]
- Cui H, Benfey PN. (2009) Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J 58: 1016–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Xue HW. (2010) Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J 29: 1916–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE., Jr (1997) STATs and gene regulation. Science 277: 1630–1635 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière J-M, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T-P. (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun T-P. (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardo F, Robertson M, Singh DP, Parish RW, Swain SM. (2009) Functional analysis of HvSPY, a negative regulator of GA response, in barley aleurone cells and Arabidopsis. Planta 229: 523–537 [DOI] [PubMed] [Google Scholar]
- Fu X, Harberd NP. (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740–743 [DOI] [PubMed] [Google Scholar]
- Fu X, Richards DE, Ait-Ali T, Hynes LW, Ougham H, Peng J, Harberd NP. (2002) Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14: 3191–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Richards DE, Fleck B, Xie D, Burton N, Harberd NP. (2004) The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16: 1406–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Bilsborough G, Tsiantis M. (2009) Hormonal input in plant meristems: a balancing act. Semin Cell Dev Biol 20: 1149–1156 [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Alabadí D, Blázquez MA. (2011) DELLA-induced early transcriptional changes during etiolated development in Arabidopsis thaliana. PLoS ONE 6: e23918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Marín JA, Prat S, Blázquez MA, Alabadí D. (2010) Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Mol Biol Evol 27: 1247–1256 [DOI] [PubMed] [Google Scholar]
- Gomi K, Sasaki A, Itoh H, Ueguchi-Tanaka M, Ashikari M, Kitano H, Matsuoka M. (2004) GID2, an F-box subunit of the SCF E3 complex, specifically interacts with phosphorylated SLR1 protein and regulates the gibberellin-dependent degradation of SLR1 in rice. Plant J 37: 626–634 [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang Z-L, Powers SJ, Gong F, Phillips AL, Hedden P, Sun T-P, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Chandler PM, White RG, Llewellyn DJ, Jacobsen JV. (2002) Gibberellin signaling in barley aleurone cells: control of SLN1 and GAMYB expression. Plant Physiol 129: 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Heo J-O, Chang KS, Kim IA, Lee M-H, Lee SA, Song S-K, Lee MM, Lim J. (2011) Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc Natl Acad Sci USA 108: 2166–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Asano K, Tsuji H, Kawamura M, Mori H, Kitano H, Ueguchi-Tanaka M, Matsuoka M. (2010) Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell 22: 2680–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Kouketu E, Katoh H, Aya K, Ueguchi-Tanaka M, Matsuoka M. (2012) The suppressive function of the rice DELLA protein SLR1 is dependent on its transcriptional activation activity. Plant J 71: 443–453 [DOI] [PubMed] [Google Scholar]
- Hou X, Lee LYC, Xia K, Yan Y, Yu H. (2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH. (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A, Cao D, Cheng H, Wen Z, Peng J. (2005) Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J 44: 88–99 [DOI] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. (2001) slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Matsuoka M, Steber CM. (2003) A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trends Plant Sci 8: 492–497 [DOI] [PubMed] [Google Scholar]
- Itoh H, Sasaki A, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Hasegawa Y, Minami E, Ashikari M, Matsuoka M. (2005a) Dissection of the phosphorylation of rice DELLA protein, SLENDER RICE1. Plant Cell Physiol 46: 1392–1399 [DOI] [PubMed] [Google Scholar]
- Itoh H, Shimada A, Ueguchi-Tanaka M, Kamiya N, Hasegawa Y, Ashikari M, Matsuoka M. (2005b) Overexpression of a GRAS protein lacking the DELLA domain confers altered gibberellin responses in rice. Plant J 44: 669–679 [DOI] [PubMed] [Google Scholar]
- Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M. (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Suzuki H, Kim Y-C, Iuchi A, Kuromori T, Ueguchi-Tanaka M, Asami T, Yamaguchi I, Matsuoka M, Kobayashi M, et al. (2007) Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J 50: 958–966 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karssen CM, Zagorski S, Kepczynski J, Groot SPC. (1989) Key role for endogenous gibberellins in the control of seed germination. Ann Bot (Lond) 63: 71–80 [Google Scholar]
- King KE, Moritz T, Harberd NP. (2001) Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH. (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Lee LYC, Hou X, Fang L, Fan S, Kumar PP, Yu H. (2012) STUNTED mediates the control of cell proliferation by GA in Arabidopsis. Development 139: 1568–1576 [DOI] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J. (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun T-P, Steber CM. (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, Chory J. (2008) A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. (2010) The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr Biol 20: 1138–1143 [DOI] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun T-P, Hakoshima T. (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim Y-C, Park S-H, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JDG. (2008) DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Nozue K, Harmer SL, Maloof JN. (2011) Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol 156: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee H-S, Sun T-P, Kamiya Y, Choi G. (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Kim J, Lee Y, Shin J, Oh E, Chung W-I, Liu JR, Choi G. (2004) Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol 45: 968–975 [DOI] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Lopez-Molina L. (2009) The GA-signaling repressor RGL3 represses testa rupture in response to changes in GA and ABA levels. Plant Signal Behav 4: 63–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett ARG, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, Seo M, Jikumaru Y, Benlloch R, Nilsson O, Ruiz-Rivero O, et al. (2012) Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 24: 941–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18: 111–119 [DOI] [PubMed] [Google Scholar]
- Richards DE, Peng J, Harberd NP. (2000). Plant GRAS and metazoan STATs: one family? BioEssays 22: 573–577 [DOI] [PubMed] [Google Scholar]
- Robertson M, Swain SM, Chandler PM, Olszewski NE. (1998) Identification of a negative regulator of gibberellin action, HvSPY, in barley. Plant Cell 10: 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambade A, Pratap A, Buschmann H, Morris RJ, Lloyd C. (2012) The influence of light on microtubule dynamics and alignment in the Arabidopsis hypocotyl. Plant Cell 24: 192–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong D-H, An G, Kitano H, Ashikari M, et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Seo M, Nambara E, Choi G, Yamaguchi S. (2009) Interaction of light and hormone signals in germinating seeds. Plant Mol Biol 69: 463–472 [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M. (2008) Structural basis for gibberellin recognition by its receptor GID1. Nature 456: 520–523 [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M. (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J 48: 390–402 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T. (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP. (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PY, Martínez EC, Sun TP. (1997) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Tseng T-S, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun T-P. (2007) Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 143: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DP, Jermakow AM, Swain SM. (2002) Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell 14: 3133–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Strader LC, Ritchie S, Soule JD, McGinnis KM, Steber CM. (2004) Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc Natl Acad Sci USA 101: 12771–12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Gubler F. (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y. (1994) The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6: 1509–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Jones WT, Harvey D, Edwards PJB, Pascal SM, Kirk C, Considine T, Sheerin DJ, Rakonjac J, Oldfield CJ, et al. (2010) N-terminal domains of DELLA proteins are intrinsically unstructured in the absence of interaction with GID1/gibberellic acid receptors. J Biol Chem 285: 11557–11571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Jones WT, Rikkerink EHA. (2012) GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. Biochem J 442: 1–12 [DOI] [PubMed] [Google Scholar]
- Swain SM, Tseng TS, Olszewski NE. (2001) Altered expression of SPINDLY affects gibberellin response and plant development. Plant Physiol 126: 1174–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. (2007) Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS. (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654 [DOI] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun T-P. (2004) DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow T-Y, Hsing Y-IC, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Hirano K, Hasegawa Y, Kitano H, Matsuoka M. (2008) Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20: 2437–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Hongyu X, Ashikari M, Kitano H, Yamaguchi I, et al. (2007) Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19: 2140–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Deng XW. (2011) Plant ubiquitin-proteasome pathway and its role in gibberellin signaling. Cell Res 21: 1286–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan L-M, Deng XW. (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L, Kreppel LK, Comer FI, Wadzinski BE, Hart GW. (2004) O-GlcNAc transferase is in a functional complex with protein phosphatase 1 catalytic subunits. J Biol Chem 279: 38466–38470 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EMN, Maier A, Schwechheimer C. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Isono E, Richter R, Zourelidou M, Schwechheimer C. (2011) Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell 23: 2184–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR. (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Hirai T, Yamamoto E, Kawamura M, Sato T, Kitano H, Matsuoka M, Ueguchi-Tanaka M. (2010) A rice gid1 suppressor mutant reveals that gibberellin is not always required for interaction between its receptor, GID1, and DELLA proteins. Plant Cell 22: 3589–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S. (2004) Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ito T, Zhao Y, Peng J, Kumar P, Meyerowitz EM. (2004) Floral homeotic genes are targets of gibberellin signaling in flower development. Proc Natl Acad Sci USA 101: 7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Zhang Z-L, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al. (2007) Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z-L, Ogawa M, Fleet CM, Zentella R, Hu J, Heo J-O, Lim J, Kamiya Y, Yamaguchi S, Sun T-P. (2011) Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc Natl Acad Sci USA 108: 2160–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Ni W, Feng B, Han T, Petrasek MG, Ma H. (2003) Members of the Arabidopsis SKP1-like gene family exhibit a variety of expression patterns and may play diverse roles in Arabidopsis. Plant Physiol 133: 203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwar JA, Chandler PM. (1995) α-Amylase production and leaf protein synthesis in a gibberellin-responsive dwarf mutant of ‘Himalaya’ barley (Hordeum vulgare L.). Planta 197: 39–48 [Google Scholar]