Abstract
Previously, a dysfunction of the SMALL ACIDIC PROTEIN1 (SMAP1) gene was identified as the cause of the anti-auxin resistant1 (aar1) mutant of Arabidopsis (Arabidopsis thaliana). SMAP1 is involved in the response pathway of synthetic auxin, 2,4-dichlorophenoxyacetic acid, and functions upstream of the auxin/indole-3-acetic acid protein degradation step in auxin signaling. However, the exact mechanism by which SMAP1 functions in auxin signaling remains unknown. Here, we demonstrate that SMAP1 is required for normal plant growth and development and the root response to indole-3-acetic acid or methyl jasmonate in the auxin resistant1 (axr1) mutation background. Deletion analysis and green fluorescent protein/glutathione S-transferase pull-down assays showed that SMAP1 physically interacts with the CONSTITUTIVE PHOTOMORPHOGENIC9 SIGNALOSOME (CSN) via the SMAP1 F/D region. The extremely dwarf phenotype of the aar1-1 csn5a-1 double mutant confirms the functional role of SMAP1 in plant growth and development under limiting CSN functionality. Our findings suggest that SMAP1 is involved in the auxin response and possibly in other cullin-RING ubiquitin ligase-regulated signaling processes via its interaction with components associated with RELATED TO UBIQUITIN modification.
The plant hormone auxin plays an indispensable role in regulating various morphogenic processes such as root growth, shoot branching, and flower bud formation (Davies, 2004). Understanding the mode of action of auxin has been a major issue in plant physiology ever since the plant hormone concept was developed (Davies, 2004). Recent studies revealed an elegant signaling model for auxin, which is centered on SCFTIR1/AFB ubiquitin E3 ligase (Woodward and Bartel, 2005). The SCF ubiquitin E3 ligase is a multisubunit complex that regulates the ubiquitin-dependent proteolysis of many proteins. This complex consists of S-PHASE KINASE-ASSOCIATED PROTEIN1 (SKP1; ASK1 for Arabidopsis SKP1), CULLIN1 (CUL1), RING H2 finger (RBX1), and substrate-recognition F-box proteins (Woodward and Bartel, 2005). In SCFTIR1/AFB, the F-box protein TRANSPORT INHIBITOR RESISTANT1 (TIR1) or its homologs, the AUXIN-SIGNALING F-BOX (AFB) proteins, function as auxin receptors (Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005). Binding of auxin to the TIR1/AFB receptors facilitates ubiquitin-mediated degradation of auxin/indole-3-acetic acid (Aux/IAA) repressors, which interact with transcriptional factors, AUXIN RESPONSE FACTORs (ARFs), resulting in changes in the patterns of downstream gene expression (Kim et al., 1997; Ulmasov et al., 1997; Tiwari et al., 2001, 2004; Dharmasiri and Estelle, 2004). Another recently discovered F-box protein, SKP2A, directly binds auxin and promotes the degradation of cell cycle transcription factors (Jurado et al., 2010).
The RELATED TO UBIQUITIN (RUB, also known as NEDD8 in mammals) protein is a small conserved protein that covalently binds to several regulatory proteins. One such regulatory protein is CUL, a scaffold protein in CULLIN-RING UBIQUITIN E3 LIGASES (CRLs), including SCF E3 ubiquitin ligase (Dreher and Callis, 2007). This RUB/NEDD8 modification is processed though a series of ATP-dependent steps, similar to the ubiquitin conjugation cascade (Dreher and Callis, 2007). In Arabidopsis (Arabidopsis thaliana), a heterodimer of AUXIN RESISTANT1 (AXR1) and E1 C-TERMINAL-RELATED1 (ECR1) proteins functions as a RUB/NEDD8-activating enzyme (E1) and catalyzes the ATP-dependent formation of a thioester bond between ECR1 and RUB/NEDD8 (del Pozo and Estelle, 1999). Activated RUB/NEDD8 is transferred from ECR1 to RUB-CONJUGATING ENZYME1 (RCE1), which functions as a RUB E2 enzyme and directly binds to RBX1, which is thought to function as a RUB E3 ligase in the CRL complex (Dharmasiri et al., 2003). In the case of RUB modification of CUL, an ε-amino group of a Lys residue in CUL interacts with the thioester of the RUB-E2 conjugate, resulting in the formation of an isopeptide bond between RUB and CUL (Hotton and Callis, 2008). Another protein family, DEFECTIVE IN CULLIN NEDDYLATION1 (DCN1), has also been reported as a RUB E3 ligase in human and yeast (Kurz et al., 2008). Although the biochemical activity of DCN1 proteins in plants is poorly understood, the loss of a DCN1-like gene causes a 2,4-dichlorophenoxyacetic acid (2,4-D)-resistant phenotype in Arabidopsis roots (Biswas et al., 2007). A large body of experimental evidence suggests that RUB modification mechanisms are significant for plant growth and development (Dreher and Callis, 2007).
An evolutionarily conserved complex, the CONSTITUTIVE PHOTOMORPHOGENIC9 (COP9) SIGNALOSOME (CSN), is also required for the activity of CRLs (Schwechheimer et al., 2001). One of the major activities of the CSN is to deconjugate RUB/NEDD8 from RUB/NEDD8-conjugated CULs via the metalloprotease activity of its CSN5 subunit. RUB/NEDD8-associated and -dissociated CRLs are active and inactive forms, respectively (Saha and Deshaies, 2008). A recent model of the RUB/NEDD8 modification cycle suggests that both RUB/NEDD8 conjugation and deconjugation of CUL proteins are required for optimal E3 activity (Cope and Deshaies, 2003; Hotton and Callis, 2008). The stability and efficiency of CRLs depend on the RUB/NEDD8 modification status of CULs, and this status subsequently affects their E3 activity (Schwechheimer and Isono, 2010). Indeed, mutations in either component (i.e. those promoting RUB/NEDD8 conjugation or deconjugation) cause auxin-resistant phenotypes in Arabidopsis (Gray et al., 2001; Schwechheimer et al., 2001; del Pozo et al., 2002).
To better understand auxin signaling mechanisms, we used an inhibitor of auxin action, p-chlorophenoxyisobutylic acid (PCIB), to screen Arabidopsis mutants. This resulted in the isolation of several anti-auxin resistant (aar) mutants (Oono et al., 2003; Biswas et al., 2007). Some aar mutations were located in previously known auxin-related loci such as tir1 and cul1, whereas other mutations were found in unknown loci (Biswas et al., 2007). One of the mutants, aar1-1, showed 2,4-D-specific resistance without any changes in 2,4-D transport or metabolism. The causal gene of aar1 was identified by map-based cloning and designated as SMALL ACIDIC PROTEIN1 (SMAP1) because it encodes a 62-amino acid protein (6.9 kD) with a pI of 3.4. SMAP1 confers the sensitivity of Arabidopsis roots to 2,4-D and PCIB (Rahman et al., 2006). The Arabidopsis genome has another copy of the SMAP gene, SMAP2, which is expressed only in siliques and anthers and potentially mediates the root response to 2,4-D, as SMAP2 overexpression restores the sensitivity of aar1 to 2,4-D (Nakasone et al., 2009). Physiological and genetic analyses of aar1 mutants and the SMAP1 gene suggested that the SMAP1 protein acts upstream of the degradation step of Aux/IAA proteins in auxin signaling (Rahman et al., 2006). Although the SMAP protein has no known functional motifs, there is a highly conserved Phe (F)- and Asp (D)-rich 18-amino acid sequence (F/D region) in the C-terminal region. This region is found in SMAP genes from a wide variety of plants and animals, implying that the SMAP genes are evolutionarily indispensable (Rahman et al., 2006; Nakasone et al., 2009).
Although SMAP1 has been shown to regulate 2,4-D sensitivity and to work upstream of the ubiquitin proteasome pathway, the functional significance of this gene in auxin signaling remains unclear. In this study, we tried to elucidate SMAP1 function by examining the genetic relationships among known auxin-related mutants and attempted to identify SMAP1-interacting proteins. Our results suggested that SMAP1 physically interacts with CSN in Arabidopsis extracts and that its function is linked to the RUB modification components AXR1 and CSN.
RESULTS
Genetic Interaction of aar1 and Auxin-Related Mutants
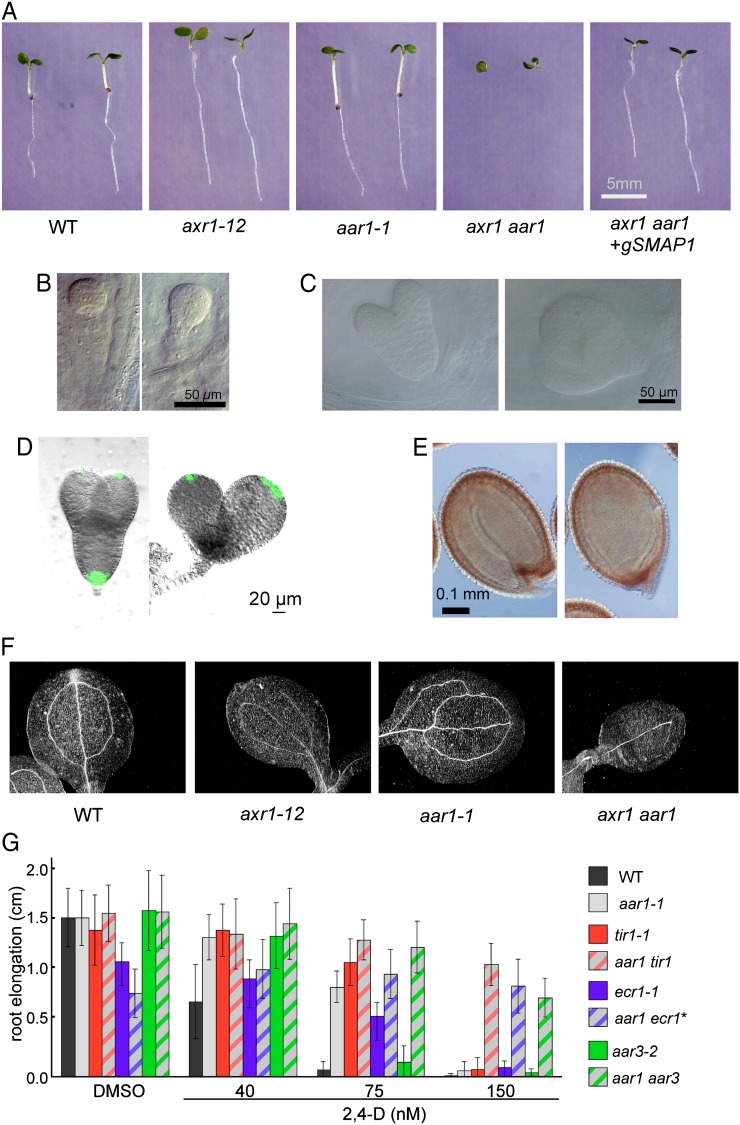
To investigate the relationship between aar1 and other known auxin mutants, we crossed the aar1-1 mutant with various mutants in the auxin signaling pathway: tir1-1, a mutant of an auxin receptor (Dharmasiri et al., 2005a); ecr1-1, a mutant of a subunit of the RUB-activating enzyme E1 (Woodward et al., 2007); axr1-12, a mutant of another subunit of the RUB-activating enzyme E1 (Leyser et al., 1993); and aar3-2, a mutant of a DCN1-like gene (Biswas et al., 2007). Then we attempted to establish double mutants. aar1-1 tir1-1, aar1-1 ecr1-1, and aar1-1 aar3-2 double mutants were successfully obtained. However, the axr1-12 aar1-1 mutant showed severe morphological defects; nearly one-quarter of the offspring from the AXR1/axr1-12 aar1-1 parents did not germinate or died at the early seedling stage without developing roots (Figs. 1, A–F, and 2, A and B). Genotyping with PCR markers confirmed that the rootless siblings were axr1-12 aar1-1 double mutants (data not shown). In normal embryos, the hypophysis divides asymmetrically and forms a quiescent center in the root meristem. Formation of an auxin gradient is required for this process, which can be readily visualized by the authentic auxin reporter DR5rev:GFP (Friml et al., 2003). However, in the abnormal embryos, neither the formation of a quiescent center nor GFP expression was observed at the position where descendants of hypophyseal cells should be located (Fig. 1, B–E), suggesting that axr1-12 aar1-1 lacks normal auxin response. The axr1-12 aar1-1 double mutant also showed severe morphological defects in the aerial parts. For example, venation was poorly developed compared with that in axr1-12 (Fig. 1F), suggesting that the effect of the aar1 mutation is substantial in the axr1-12 background. Similar abnormal seedlings or ungerminated seeds were observed in the descendants of AXR1/axr1-3 aar1-1 or AXR1-12/axr1-12 aar1-2 parental lines (Fig. 2, C–F), suggesting that the abnormal phenotype in double mutants is not allele dependent.
Figure 1.
Genetic interaction between SMAP1 and auxin-related mutants. A, Comparison of the early seedling phenotype in the wild type (WT), axr1-12, aar1-1, axr1-12 aar1-1 double mutant (axr1 aar1), and the double mutant containing the genomic SMAP transgene (axr1 aar1+ gSMAP1). The seedlings were grown for 6 d in GM and photographed. Note the rootless phenotype of axr1 aar1 double mutant and complementation of the phenotype by genomic SMAP1. B and C, Normal (left) and abnormal (right) early globular (B) and torpedo (C) stage embryos in siliques of AXR1/axr1-12 aar1-1 plants. D, Visualization of the auxin response pattern in normal (left) and abnormal (right) torpedo stage embryos in siliques of AXR1/axr1-12 aar1-1 plants containing DR5rev:GFP. GFP expression was absent in the hypophysis of abnormal embryos. E, Mature seeds with normal (left) and abnormal (right) embryos obtained from AXR1/axr1-12 aar1-1 plants. F, Venation patterns in cotyledons of 5-d-old wild type, axr1-12, aar1-1, and axr1 aar1. G, Response of single and double mutants to 2,4-D in a root growth assay. Seedlings were germinated and grown on GM containing 2,4-D. Elongation of roots was measured from day 4 to 7. Data are means ± sd (n = at least 10 seedlings). * The rootless seedlings (see Fig. 2Z) were excluded. DMSO, Dimethyl sulfoxide.
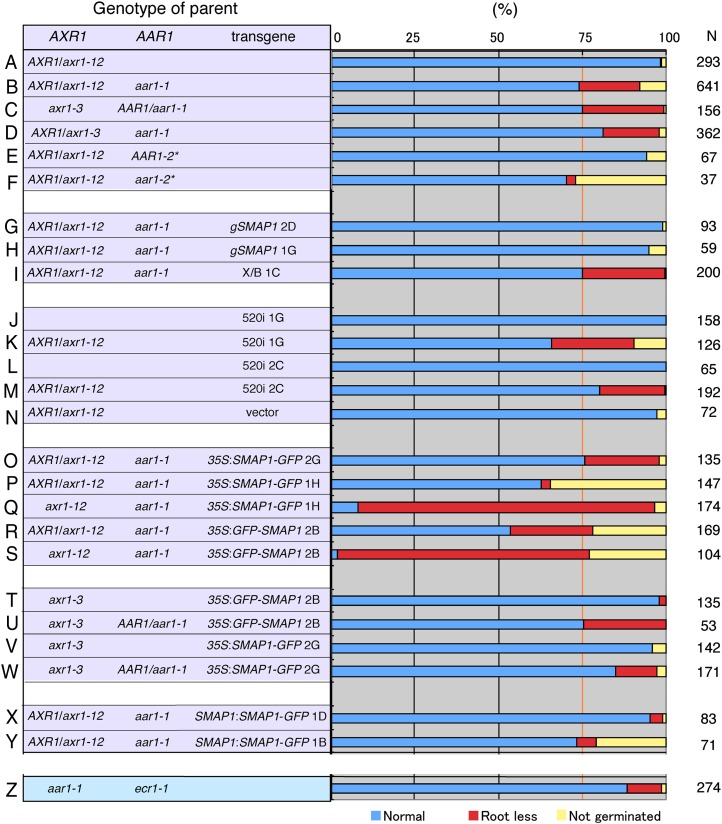
Figure 2.
Frequency of rootless seedlings in various genetic backgrounds. Frequency of normal (blue), rootless (red), and nongerminated (yellow) phenotypes in offspring seedling populations from parent plants of the indicated genotypes is shown. N indicates the number of seedlings observed. The vertical orange line highlights the 3:1 expected ratio for normal:abnormal seedlings from parent lines. Because the axr1 aar1 double mutant was postembryonic lethal, seeds harvested from the AXR1/axr1 heterozygote background were tested in most of the experiments. Transgenes were introduced by crossing. A to F, Phenotype of axr1 aar1 double mutants. * Wassilewskija accession. G to I, Complementation of the rootless phenotype of axr1 aar1 by a genomic fragment containing SMAP1 (gSMAP1). A genomic fragment (X/B) without SMAP1 was used as a control (I). Two independent transgenic lines for gSMAP1 (B/S lines 2D and 1G) and the control line X/B line 1C with the aar1-1 mutation (Rahman et al., 2006) were crossed with axr1-12 mutants to generate parental lines. J to N, Inactivation of SMAP1 by RNAi in the axr1 background. Two independent 520i lines (lines 1G and 2C) or a control line (line F2) transformed with vector pB7GWIWG2(II) (Rahman et al., 2006) were crossed with axr1-12 mutants to generate parental lines. O to W, Expression of the SMAP1∼GFP fusion protein under the control of the 35S promoter in the axr1 aar1 background. Transgenic lines of 35S:SMAP1-GFP/aar1-1 (lines 2G and 1H) or 35S:SMAP1-GFP/aar1-1 line 2B (Supplemental Fig. S1) were crossed with axr1-12 (O–S) or axr1-3 (T–W) mutants. X and Y, Complementation of the rootless phenotype of the double mutant by SMAP1:SMAP1-GFP. Two independent transgenic lines (D4 and B4) of SMAP1:SMAP1-GFP/aar1-1 were crossed with the axr1-12 mutant to generate parental lines. Z, The rootless phenotype was observed in approximately 10% of aar1-1 ecr1-1 seedlings.
The aar1-1 mutation has an approximately 44-kb deletion in chromosome 4; this deleted region contains at least 10 open reading frames, including SMAP1 (Rahman et al., 2006). Therefore, the abnormal phenotype could be caused by the lack of other genes rather than SMAP1. To eliminate this possibility, we crossed axr1-12 with two independent transgenic lines harboring a 3.7-kp BamHI/SacI genomic DNA fragment containing the SMAP1 open reading frame (gSMAP1). As shown in Figures 1A and 2, G to I, the seedling population AXR1/axr1-12 aar1-1 gSMAP1 lines showed no rootless offspring, in contrast to the line harboring the control genomic fragment (X/B). In reverse, the homozygous offspring of the AXR1/axr1-12 heterozygous parental lines harboring the SMAP1 RNA interference (RNAi) construct were rootless (Fig. 2, J–N). Taken together, these results suggest that SMAP1 is necessary for root meristem formation and normal embryo development in the axr1 background.
We successfully obtained the double mutants aar1-1 tir1-1, aar1-1 ecr1-1, and aar1-1 aar3-2. Most of these double mutants did not show any severe morphological defects compared with their parental lines (data not shown). However, approximately 10% of seedlings from the aar1-1 ecr1-1 parent showed a rootless phenotype similar to that of the axr1 aar1 double mutants (Fig. 2Z). The root elongation assay against 2,4-D revealed that the double mutants aar1-1 tir1-1, aar1-1 ecr1-1, and aar1-1 aar3-2 are more resistant compared with their corresponding single mutants, suggesting an additive relationship between the aar1-1 mutation and the auxin-related mutations tir1-1, ecr1-1, and aar3-2 for regulating the root auxin response (Fig. 1G).
Expression of SMAP1 under the Control of the Cauliflower Mosaic Virus 35S Promoter in the axr1 Background Partially Restored the Wild-Type Phenotype
SMAP1 was fused to GFP and expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter (35S:SMAP1-GFP or 35S:GFP-SMAP1) in aar1-1 plants. The expression of fusion proteins in the transgenic lines restored wild-type sensitivity to PCIB and 2,4-D in the aar1-1 roots, suggesting that the SMAP1-GFP and GFP-SMAP1 fusion proteins are functionally active (Supplemental Fig. S1). The mature 35S:SMAP1-GFP or 35S:GFP-SMAP1 (collectively designated as 35S:SMAP1∼GFP) transgenic Arabidopsis plants were morphologically indistinguishable from nontransgenic plants in either the wild-type or aar1-1 background (data not shown). When 35S:SMAP1∼GFP was introduced into the axr1-12 background (35S:SMAP1-GFP/axr1-12 or 35S:GFP-SMAP1/axr1-12), morphological phenotypes of axr1-12 such as dwarfism, reduced apical dominance, protruding pistils, and low fertility (Lincoln et al., 1990) were partially alleviated (Fig. 3A). In a root growth assay, the axr1-12 mutant is strongly resistant to auxin and other plant hormones such as jasmonic acid (JA; Lincoln et al., 1990; Tiryaki and Staswick, 2002). The introduction of 35S:SMAP1∼GFP also partially restored the 2,4-D sensitivity of axr1-12 roots (Fig. 3B). Furthermore, although the aar1 mutant is reported to be a 2,4-D-specific response mutant (Rahman et al., 2006), the sensitivity of 35S:SMAP1∼GFP/axr1-12 to IAA (Fig. 3C) and methyl jasmonate (Fig. 3D) was intermediate between that of the wild type and axr1-12, suggesting that in the absence of functional AXR1, SMAP1 function is required for normal IAA and JA responses.
Figure 3.
Phenotypic changes resulting from the introduction of 35S:SMAP1∼GFP fusion genes into the axr1-12 mutant. Independent transgenic lines of 35S:SMAP1-GFP (lines 1H and 2G) and 35S:GFP-SMAP1 (line 2B) were crossed with axr1-12, and homozygous lines were established for axr1-12, transgene, and wild-type (WT) AAR1 loci. A, Mature plants (27 d old; top row), inflorescences (middle row), and flowers (bottom row) are shown. Bars = 1 cm (top and middle rows) and 0.5 cm (bottom row). B, Seeds were germinated and grown on GM containing 2,4-D at the indicated concentrations. Increase in root length was measured from day 4 to 7. Data are means ± sd (n > 9 seedlings). DMSO, Dimethyl sulfoxide. C, Seeds were germinated and grown on GM for 4 d, and seedlings were then transferred onto GM containing IAA and grown for a further 3 d. Root elongation after transfer was measured and plotted as a relative value compared with that on medium without chemicals. Data are means ± sd (n = at least 21 seedlings). Mean values (cm) ± sd in the absence of chemicals controlling root elongation were as follows: 1.56 ± 0.25 (wild type), 2.23 ± 0.41 (axr1-12), 1.58 ± 0.37 (35S:SMAP1-GFP/axr1-12 1H), 1.76 ± 0.24 (35S:SMAP1-GFP/axr1-12 2G), and 1.64 ± 0.47 (35S:GFP-SMAP1/axr1-12 2B). D, Seeds were germinated and grown on GM containing 10 µm methyl jasmonate (JA-Me). Root elongation was measured from day 5 to 8 and plotted as a relative value compared with that on medium without chemicals. Data are means ± sd (n = at least 5 seedlings). Mean values (cm) ± sd in the absence of chemicals controlling root elongation were as follows: 2.05 ± 0.26 (wild type), 2.63 ± 0.19 (axr1-12), and 2.19 ± 0.24 (35S:SMAP1-GFP/axr1-12 1H).
Unexpectedly, unlike the SMAP1 genomic fragment, SMAP1∼GFP fusion constructs expressed under the control of the CaMV 35S promoter did not effectively recover the rootless phenotype of the axr1 aar1 double mutant (Fig. 2, O–W). Although we established homozygous axr1-12 aar1-1 35S:SMAP1-GFP (line 1H) and axr1-12 aar1-1 35S:GFP-SMAP1 (line 2B) lines by crossing, many seeds from these lines did not germinate, and most of those that did germinate were rootless (Fig. 2, Q and S). This may be due to the low expression level of SMAP1-GFP at the early stage of embryo development, which could not complement the loss of the SMAP1 gene from hypophyseal initial cells. GFP expression was hardly detected in the globular embryos of the two 35S:SMAP1-GFP lines (Supplemental Fig. S2). The transgene of SMAP1-GFP under the control of the SMAP1 promoter (SMAP:SMAP1-GFP line 1D) that was strongly expressed (as observed by GFP fluorescence) in the lower part of the globular embryo (Supplemental Fig. S2) effectively rescued the morphological defects in the axr1-12 aar1-1 double mutants (Fig. 2X). Another independent line of SMAP1:SMAP1-GFP (line 1B) that showed low-level GFP expression in the globular embryo did not recover the root development in axr1-12 aar1-1 double mutant background (Fig. 2Y; Supplemental Fig. S2). This suggests that strong SMAP1 expression in the globular stage embryo is required to establish root initial organization in the axr1-12 background.
A Conserved F/D-Rich Region Is Significant for SMAP1 Function
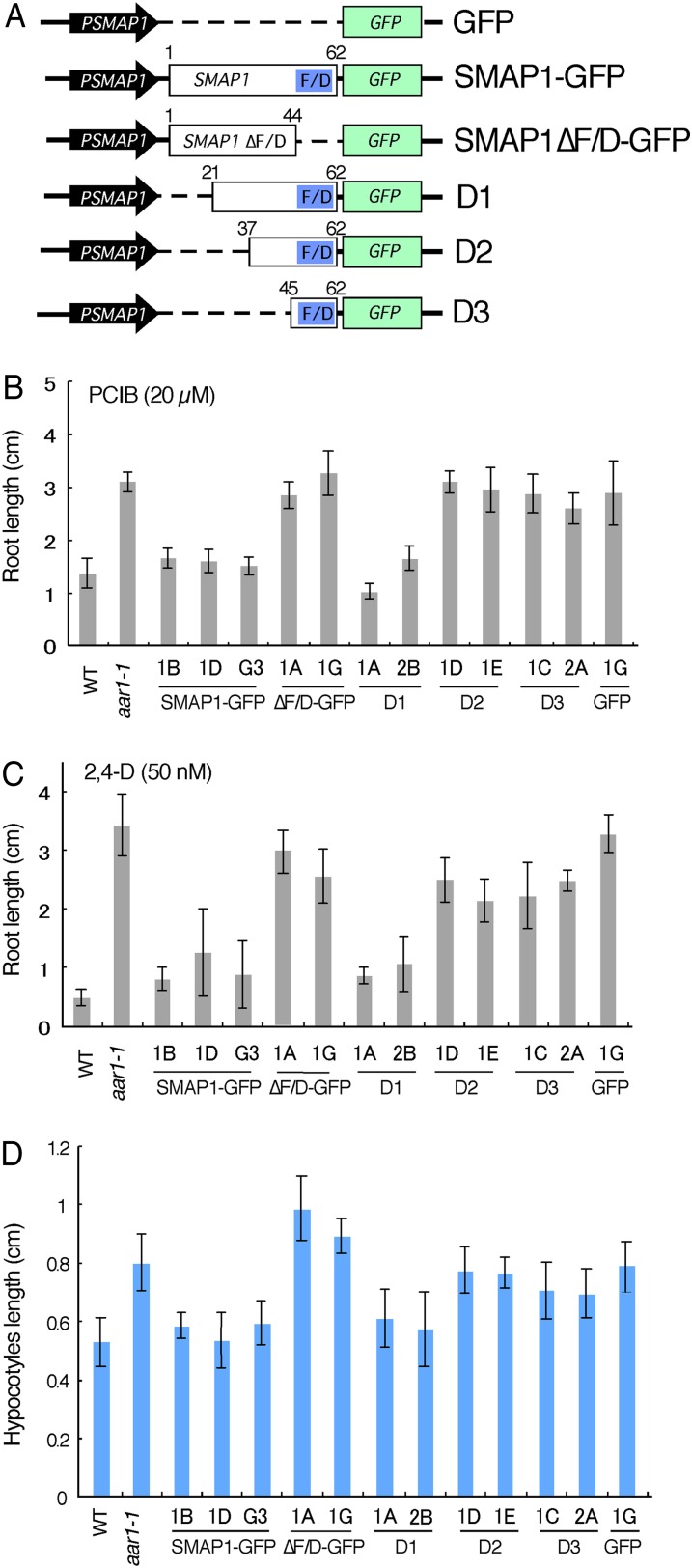
To investigate the functional aspects of the SMAP1 protein, we constructed several versions of SMAP1 derivatives, including those with deleted versions of the conserved F/D region, fused to GFP, under the control of the SMAP1 promoter (approximately 5 kb; Fig. 4A). These constructs were transformed into the aar1-1 mutant, and their ability to complement the aar1 mutation was investigated (Fig. 4, B–D). The GFP fluorescence of the fusion proteins was detected in both nuclei and the cytosol (Supplemental Fig. S3).
Figure 4.
Construction and analyses of GFP fusion constructs with SMAP1 and SMAP1 deletion series. A, Schematic diagram of constructs. SMAP1 and GFP coding regions are shown by white and green boxes, respectively. The F/D-rich region in SMAP1 is shown by blue boxes. The SMAP1 promoter is shown by black arrows. The nos terminator is not shown. Dotted lines indicate deleted regions in the constructs. SMAP1-GFP contains full-length SMAP1. D1, D2, and D3 are N-terminal deleted constructs. Numbers on white boxes indicate the positions of amino acids within the N and C termini of modified SMAP1. B and C, Transgenic aar1-1 lines, the wild type (WT), and untransformed aar1-1 were planted on GM containing 20 µm PCIB (B) or 50 nm 2,4-D (C), and root length was measured after 10 d. Data are means ± sd (n = at least 13 seedlings). D, Hypocotyl length of 7-d-old seedlings grown on GM in the light. Data are means ± sd (n = at least 15 seedlings). [See online article for color version of this figure.]
The aar1 mutant is resistant to the anti-auxin PCIB and the synthetic auxin 2,4-D and exhibits long hypocotyl when grown in the light (Rahman et al., 2006). As shown in Figure 4, B to D, independent transgenic lines harboring SMAP1-GFP in the aar1 background recovered sensitivities to PCIB (Fig. 4B) and 2,4-D (Fig. 4C) and showed similar hypocotyl length to that of the wild type (Fig. 4D), suggesting that the SMAP1-GFP fusion protein is functional. The construct SMAP1ΔF/D-GFP, which contained a deleted version of the F/D-rich region, did not complement the aar1 mutation, although the fusion protein was expressed in the transgenic lines as confirmed by GFP fluorescence. This finding implies that the F/D region is necessary for SMAP1 function. The N-terminal deletion construct D1, in which one-third of the N-terminal amino acids were deleted, was still able to complement the aar1 phenotype. However, the D2 and D3 constructs, which consisted of 26 and 18 C-terminal amino acids, respectively, did not complement the aar1 phenotype, suggesting that the F/D-rich region alone is not sufficient for SMAP1 function.
SMAP1 Interacts with the COP9 Signalosome
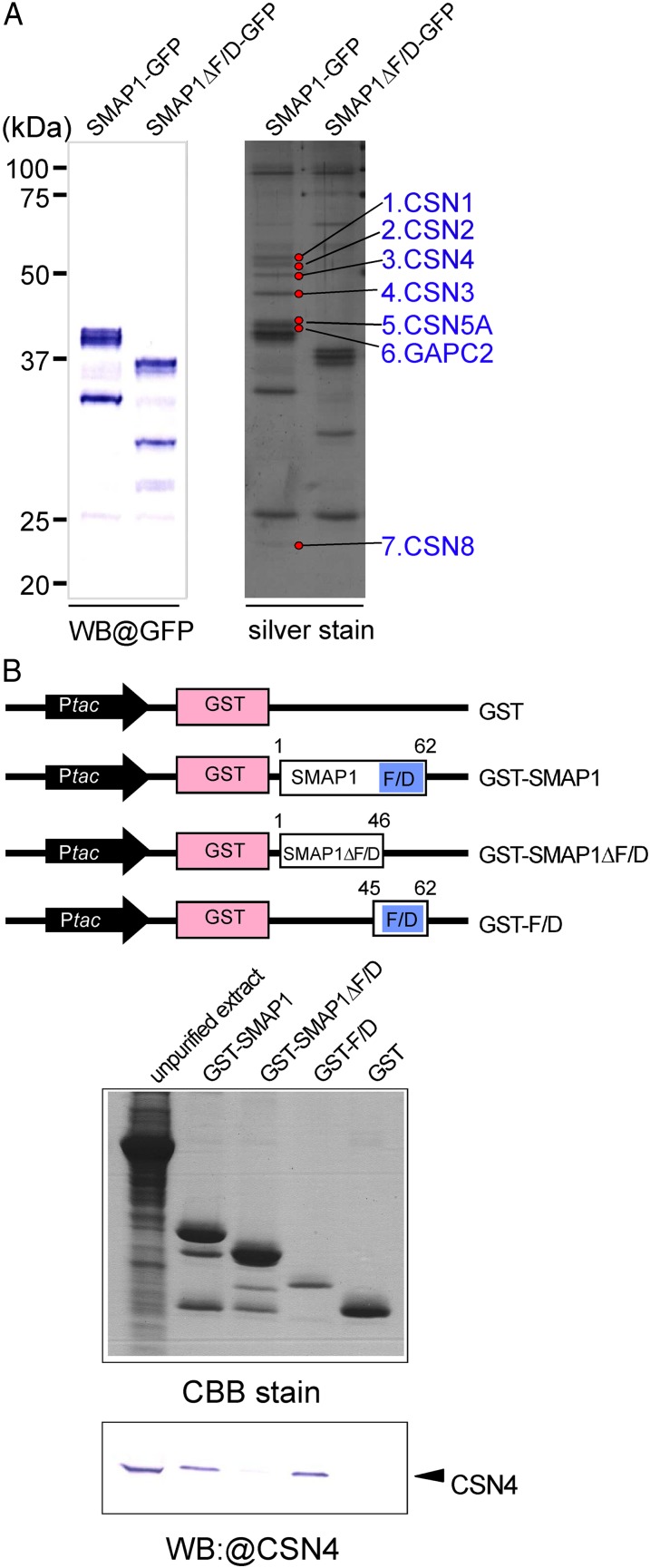
Using the transgenic plants described above, we next attempted to identify SMAP1-interacting proteins in Arabidopsis extracts. Total proteins were extracted from SMAP1-GFP, SMAP1ΔF/D-GFP, and GFP lines using a nondenaturing extraction buffer and subjected to pull-down assays using anti-GFP microbeads. The purified GFP- and/or SMAP1-binding proteins were separated by SDS-PAGE and subjected to silver staining and western blotting with an anti-GFP antibody (Fig. 5A). Silver staining revealed that seven distinct bands are present in the SMAP1-GFP extract but absent in the SMAP1ΔF/D-GFP extract, suggesting that the proteins present in these bands are possibly SMAP1-interacting proteins (Fig. 5A). These proteins were digested by trypsin, eluted from the gel, and analyzed by liquid chromatography (LC)-tandem mass spectrometry (MS/MS). A Mascot database search of the resulting MS/MS spectra identified the proteins as CSN1, CSN2, CSN3, CSN4, CYTOSOLIC GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE2 (GAPC2), CSN5A, and CSN8 (Fig. 5A; Table I). Of all the identified proteins, six out of seven were subunits of the large CSN complex (approximately 321.3 kD) that typically consists of eight subunits (Schwechheimer et al., 2001). Taken together, these results suggest that SMAP1 physically interacts with CSN in Arabidopsis extracts and that this interaction requires the presence of the F/D region.
Figure 5.
Physical interaction between SMAP1 and CSN. A, In vivo pull-down assay using GFP-tagged proteins in aar1-1 transgenic plants. Two transgenic lines, SMAP1-GFP/aar1-1 and SMAP1ΔF/D-GFP/aar1-1, were used in pull-down assays. Pulled down proteins were separated by SDS-PAGE and detected by western blotting (WB) using anti-GFP or by silver staining. Seven protein bands (marked by red circles) that bind to SMAP1-GFP but not to SMAP1ΔF/D-GFP were eluted and identified by LC-MS/MS (Table I). B, In vitro pull-down assay using GST-tagged protein constructs. The top panel shows a schematic diagram of constructs of GST-tagged proteins; GST control, GST-tagged full-length SMAP1 (GST-SMAP1), F/D region-deleted version of SMAP1 (GST-SMAP1ΔF/D), and the F/D region only (GST-F/D). SMAP1 and GST coding regions are shown as white and pink boxes, respectively. Blue boxes indicate the F/D-rich region in SMAP1. Black arrows show the Ptac promoter for expression of the fusion protein in E. coli. The bottom panel shows unpurified total proteins from an aar1-1 plant (left lane) and eluted proteins from glutathione columns (other lanes) separated by SDS-PAGE and detected by Coomassie blue (CBB) or western blot with anti-CSN4 antibody. [See online article for color version of this figure.]
Table I. LC-MS/MS identification of pull-down proteins using SMAP1-GFP protein.
| Band No.a | Protein Name | Theoretical Mass | Mascot Score | Matched Peptides | Sequence Coverage |
|---|---|---|---|---|---|
| kD | % | ||||
| 1 | CSN1 (At3g61140.1) | 50.6 | 400 | 13 | 35 |
| 2 | CSN2 (At2g26990.1) | 51.1 | 142 | 8 | 25 |
| 3 | CSN4 (At5g42970.1) | 45.0 | 864 | 25 | 65 |
| 4 | CSN3 (At5g14250.1) | 47.7 | 370 | 15 | 40 |
| 5 | CSN5A (At1g22920.1) | 39.7 | 288 | 14 | 51 |
| 6 | GAPC2 (At1g13440.1) | 36.9 | 91 | 6 | 24 |
| 7 | CSN8 (At4g14110.1) | 22.5 | 111 | 3 | 18 |
The numbers are shown in Figure 5A.
To further elucidate the interaction between SMAP1 and CSN, we established an in vitro binding assay with glutathione S-transferase (GST)-tagged fusion proteins expressed in Escherichia coli (Fig. 5B). The GST-tagged SMAP1 protein and its derivatives (GST-SMAP1, GST-SMAP1ΔF/D, GST-F/D, and GST) extracted from E. coli were immobilized to glutathione columns. Total proteins extracted from the Arabidopsis aar1-1 mutant were added to these columns. Candidate interacting proteins as well as the GST fusion proteins themselves were eluted from the columns by SDS sample buffer, separated by SDS-PAGE, and subjected to Coomassie blue staining and western-blot analysis with an anti-CSN4 antibody (Fig. 5B). The results showed that GST-SMAP1 and GST-F/D but not GST-SMAP1ΔF/D or GFP pulled down CSN4 protein from the aar1-1 extracts, suggesting that the F/D region of SMAP1 is necessary and sufficient for the interaction between SMAP1 and CSN.
Genetic Interaction of SMAP1 and CSN
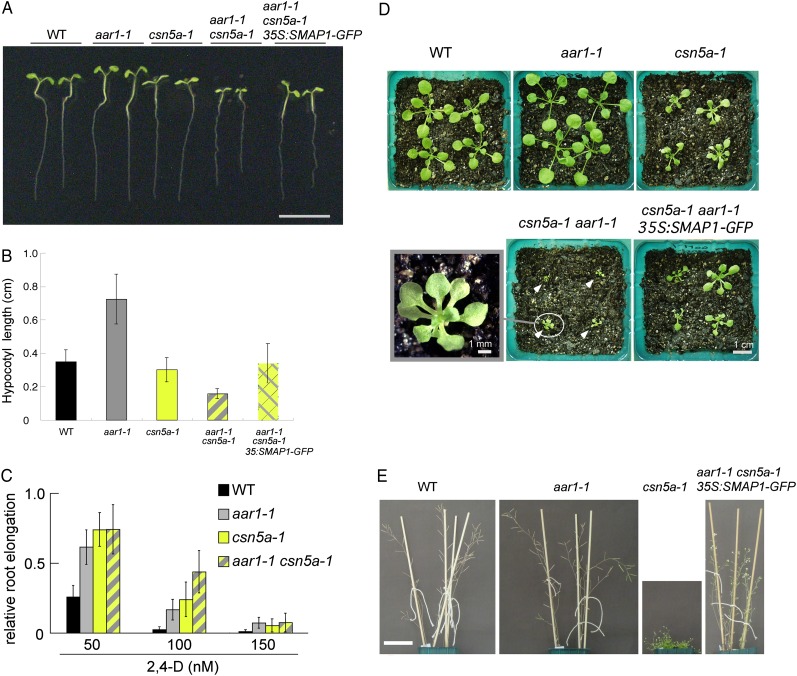
CSN was originally identified through the biochemical characterization of the COP9 protein complex (Wei et al., 1994; Chamovitz et al., 1996), which plays a significant role in regulating photomorphogenesis and postembryo development in Arabidopsis (Kwok et al., 1996). Furthermore, auxin responses are partially impaired in csn mutants (Dohmann et al., 2008). To investigate the interaction between SMAP1 and CSN at the genetic level, we crossed the aar1-1 mutant with the weak csn mutant csn5a-1 and established a line harboring homozygous aar1-1 and heterozygous csn5a-1 (aar1-1 CSN5A/csn5a-1). In the seedling population from the aar1-1 CSN5A/csn5a-1 parental line, both in light- and dark-grown conditions, we observed distinct segregation of dwarf seedlings readily distinguishable from either aar1-1 or csn5a-1 seedlings (70 dwarf seedlings out of 363 light-grown seedlings; χ2 = 6.33, P > 0.01; Fig. 6, A and B; Supplemental Fig. S4B). Genotyping analyses confirmed that these dwarf plants are the aar1-1 csn5a-1 double mutant (data not shown). The longer hypocotyl phenotype of aar1 was suppressed by the csn5a mutation. In fact, the double mutant showed an extreme short hypocotyl phenotype. Similarly, a slower root growth phenotype was found in the double mutant, although the growth of the roots of aar1-1 and csn5a-1 is comparable to the wild type (Fig. 6A; Supplemental Fig. S4A). In addition, the double mutant showed 2,4-D resistance for root growth like its parental lines (Fig. 6C). The aar1-1 csn5a-1 plants at the rosette stage also showed an extremely small seedling phenotype (Fig. 6D), and most of the plants died before producing seeds (data not shown).
Figure 6.
Genetic interaction of CSN5A and SMAP1. We could not obtain a sufficient number of seeds of the aar1-1 csn5a-1 double mutant; therefore, dwarf aar1-1 csn5a-1 seedlings were selected from a seed population from the aar1-1 CSN5A/csn5a-1 parental line for analysis. A and B, Images (A) and root lengths (B) of 7-d-old seedlings grown on GM without growth regulators. Bar in A = 1 cm. Values in B are means ± sd (n = at least 13 seedlings). C, Seeds were germinated and grown on GM for 4 d and then transferred onto GM containing 2,4-D at the indicated concentrations and grown for an additional 3 d. Root elongation after transfer was measured and plotted as a relative value compared with that on medium without chemicals. Values are means ± sd (n = at least 17 seedlings). Mean values (cm) ± sd in the absence of chemicals controlling root elongation were as follows: 1.56 ± 0.18 (wild type [WT]), 1.64 ± 0.22 (aar1-1), 1.71 ± 0.30 (can5a-1), and 1.21 ± 0.22 (aar1-1 csn5a-1). D, Photographs of plants at the rosette stage. Seeds were germinated on GM and grown for 9 d, and seedlings were then transferred to soil and grown for an additional 11 d under 16-h-light/8-h-dark conditions. White arrowheads indicate aar1-1 csn5a-1 double mutants. The photograph at bottom left is an enlarged image of a aar1-1 csn5a-1 double mutant. E, Photographs of adult plants. Seeds were germinated and grown on GM for 9 d, and seedlings were then transferred to soil and further grown for 35 d under 16-h-light/8-h-dark conditions. Bar = 5 cm.
When the 35S:SMAP1-GFP (line 1H) gene was introduced into the aar1-1 csn5a-1 background (35S:SMAP1-GFP/aar1-1csn5a-1) by crossing, normal hypocotyl length was restored (Fig. 6, A and B), suggesting that the lack of the SMAP1 gene is associated with the extreme dwarf hypocotyl phenotype of the aar1-1 csn5a-1 double mutant. When the seedlings were transferred from agar plates and further grown on soil, the morphology of the 35S:SMAP1-GFP/aar1-1csn5a-1 plant was intermediate between that of the wild type and the csn5a-1 mutant (Fig. 6E; Supplemental Fig. S4C), suggesting that the ectopic expression of the SMAP1 gene not only complemented the aar1-1 phenotype but also partially compensated the morphological abnormalities of the csn5a-1 mutant. Taken together, these results suggest that SMAP1 is required for the normal development of seedlings under limiting CSN activity.
RUB Modification Status of CUL1 in the Double Mutants and Transgenic Lines
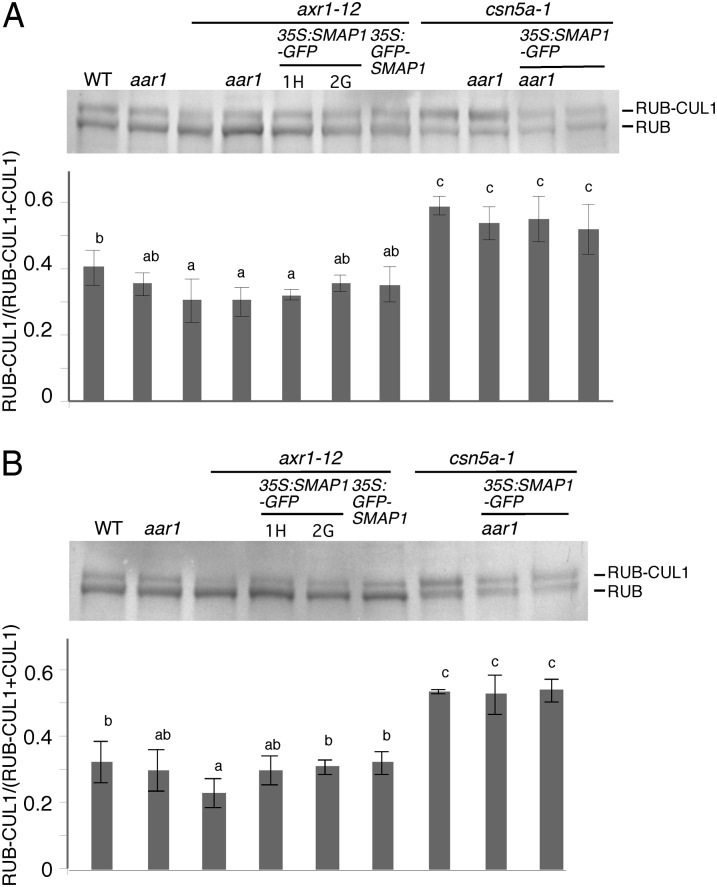
The results from two independent experimental approaches implied that SMAP1 interacts with the RUB modification-related factors AXR1 and CSN. Thus, we next examined the RUB modification status of the CUL1 protein, which is a core subunit of SCF E3 ligase and one of the most characterized RUB-modified proteins, in the double mutants and transgenic lines by western blotting using anti-CUL1 antibody (Fig. 7). As previously published, the ratio of RUB-modified and unmodified CUL1 is lower in axr1-12 and higher in csn5a-1 compared with that in the wild type (Gray et al., 2001; del Pozo et al., 2002; Gusmaroli et al., 2007). No significant difference was observed in RUB modification depending on the presence or absence of the SMAP1 gene, except that a slight increase in RUB-modified CUL1 ratio was detected in the flower extracts of transgenics harboring 35S:SMAP1∼GFP in the axr1-12 background (Fig. 7B).
Figure 7.
RUB modification of CUL1 in double mutants and transgenic lines. Total proteins (10 µg) extracted from 7-d-old light-grown seedlings (A) or flowers (B) were separated by SDS-PAGE and then immunodetected using anti-CUL1 antibody. From left to right, the wild type (WT), aar1-1, axr1-12, axr1-12 aar1-1 double mutant, 35S:SMAP1-GFP/axr1-12 1H, 35S:SMAP1-GFP/axr1-12 2G, 35S:GFP-SMAP1/axr1-12 2B, csn5a-1, aar1-1 csn5a-1 double mutant, 35S:SMAP1-GFP/aar1-1 csn5a-1 1H, and 35S:SMAP1-GFP/csn5a-1 1H were subjected to the analysis for A. We could not obtain flowers for the axr1-12 aar1-1 or aar1-1 csn5a-1 double mutant; therefore, they were excluded from B. Densitometry analyses of the ratio of RUB-modified CUL1 to total CUL1 (RUB-modified plus unmodified CUL1) are presented below the western-blot images that show means and sd derived from four or three (for A or B, respectively) independent experiments. Significant differences (P < 0.05) between mean values are indicated by different letters above the bars.
DISCUSSION
The functional significance of the SMAP1 protein, which confers 2,4-D sensitivity in Arabidopsis, in the auxin signaling pathway remained obscure in spite of the genetic and physiological characterization of the aar1 mutants (Rahman et al., 2006; Nakasone et al., 2009). In this study, using a combinatorial approach of genetics and biochemistry, we demonstrated that the SMAP1 protein interacts with the RUB modification components, AXR1 and CSN, and plays an important role in regulating the growth and development of Arabidopsis seedlings under limiting AXR1 or CSN function. Both AXR1 and CSN have important roles in CRL-mediated signaling processes, including the auxin response, via RUB modification. AXR1 facilitates the RUB modification and CSN functions in deconjugating RUB from RUB-modified proteins. The functional significance of these proteins in auxin signaling has been well demonstrated (Schwechheimer et al., 2001). The genetic and biochemical evidence presented here suggest that SMAP1 associates with the RUB modification cycle, which is consistent with the previous report, where we demonstrated that SMAP1 functions upstream of Aux/IAA protein degradation in Arabidopsis (Rahman et al., 2006).
Our initial characterization of SMAP1 demonstrated that plants with diminished SMAP1 function (aar1 mutants and SMAP1 RNAi lines) show altered responses to 2,4-D but not to IAA or other major plant hormones (Rahman et al., 2006). However, the results in this work suggest that under limited functionality of the RUB modification components AXR1 and CSN, SMAP1 potentially regulates IAA and JA signaling pathways along with 2,4-D. The root phenotypic difference observed between axr1 aar1 and axr1 tir1 double mutants is striking. The axr1 aar1 double mutant showed severe morphological defects with no root meristem formation, while the tir1 aar1 double mutant showed no apparent root developmental phenotype, although they showed increased 2,4-D resistance compared with their respective parental lines. The tir1 axr1 double mutant mimics the tir1 aar1 root phenotype, showing no apparent change in root development and increased auxin resistance (Ruegger et al., 1998). The inability of axr1 aar1 to form a root meristem suggests a strong functional interaction between AXR1 and SMAP1. This idea is further substantiated by the fact that in adult plants, the overexpression of SMAP1 restored a wild-type-like phenotype in the axr1 mutant background, suggesting that SMAP1 functionally cooperates with AXR1 in regulating various biological processes, including embryogenesis, morphology of mature plants, and hormonal responses. A similar genetic feature was reported for the AXR1 homolog gene AXR1-LIKE (AXL). The single mutant of axl did not show any remarkable phenotype, while the axr1 axl double mutant exhibited a rootless phenotype (Dharmasiri et al., 2007). Ectopic expression of AXL under the control of the CaMV 35S promoter in the axr1 background complemented the axr1 phenotype (Dharmasiri et al., 2007). Since both AXR1 and AXL encode a subunit of the RUB E1 enzyme and promote RUB modification to regulate CRL activity, one possible explanation for SMAP function could be its involvement in the RUB modification process.
In vivo GFP and in vitro GST pull-down assays revealed that SMAP1 interacts with CSN. The CSN genes were originally identified as causal genes of cop, deetiolated, or fus mutants, which show defects in photomorphogenesis or embryogenesis (Chamovitz et al., 1996). CSN influences numerous plant hormone signaling pathways, including the auxin, jasmonate, and strigolactone pathways, as well as light signaling, cell cycle progression, and circadian rhythm (Somers and Fujiwara, 2009; Schwechheimer and Isono, 2010). The best-defined function of CSN is to regulate the ubiquitin/proteasome-dependent protein degradation system by removing the covalently conjugated RUB protein from CULs and by stabilizing CRL components, such as TIR1, CUL1, and ASK1 (Stuttmann et al., 2009; Schwechheimer and Isono, 2010). The fact that the functionally significant F/D region is necessary and sufficient for SMAP1 to interact with CSN implies that SMAP1 is involved in the regulation of CRL activity together with CSN.
In our pull-down analysis of SMAP1-GFP, we detected six out of eight CSN subunits but did not detect CSN6 and CSN7 (Table I; Fig. 5A). This could be due to the similar molecular masses of CSN6, CSN7, and GFP-SMAP1, which results in an inadequate separation of these proteins on the gel. Alternatively, SMAP1 may interact with a CSN subcomplex that lacks CSN6 and CSN7. A structural analysis in mammalian cells demonstrated that several CSN subcomplexes exist, for example, CSN1/2/3/8, CSN4/5/6/7, or smaller versions including CSN1/3/8 and CSN4/6/7 (Sharon et al., 2009). Two symmetrical modules, CSN1/2/3/8 and CSN4/5/6/7, are connected by interaction between CSN1 and CSN6 to form an eight-subunit CSN complex. In the CSN1/2/3/8 and CSN4/5/6/7subcomplexes, the most peripheral subunits are CSN2 and CSN5, respectively. Furthermore, subcomplex CSN4/5/6/7 was found to be very stable (Sharon et al., 2009). Therefore, it is difficult to predict that CSN subcomplexes lacking CSN6 or CSN7 are present in plant extracts, although the binding patterns of CSN6 and CSN7 are unknown in plant cells. Further biochemical experiments are required to clarify how SMAP1 physically interacts with CSN. CSN in mammalian cells is involved in various biological responses, including embryonic development, cell cycle progression, T-cell development, signal transduction, oocyte maturation, autophagy, and circadian rhythm (Seeger et al., 1998; Kato and Yoneda-Kato, 2009). Thus, a future challenge is to determine whether SMAP proteins in mammalian cells also interact with mammalian CSN and have a regulatory role in CRL-mediated biological processes.
The strong morphological defects of axr1 aar1 double mutants and the increase of RUB-modified CUL1 in 35S:SMAP1∼GFP/axr1-12 transgenic plants suggest that SMAP1 acts as a positive regulator for RUB modification. Because CSN activates the dissociation of RUB from RUB-modified CULs, one possible explanation for SMAP1 function is that SMAP1 inhibits RUB dissociation activity of CSN by its binding to CSN. Given the marked morphological defects in axr1 and csn mutants, a small change of RUB modification status could be enough to cause the observed morphological changes in the presence or absence of SMAP1. However, we cannot eliminate the possibility that an increase of RUB-modified CUL1 in flower extracts of 35S:SMAP1∼GFP/axr1-12 transgenic plants might be a secondary effect resulting from the morphological changes. Even if this is the case, the morphological defects of the double mutants and the physical interaction between SMAP1 and CSN imply that SMAP1 interacts with the RUB cycle-related regulation of CRL activity. In a recent model in fission yeast, CSN prevents the autocatalytic ubiquitin-mediated degradation of certain F-box proteins that are assembled in the CRL, resulting in maintenance of the CRL complex in the absence of the substrates (Schmidt et al., 2009). The stabilization of CRL components by CSN in Arabidopsis extracts has also been reported (Stuttmann et al., 2009). Thus, it may be possible that SMAP1 affects the stability of CRL components and CRL assembly by binding with CSN. The construction of more sensitive biochemical experimental systems for monitoring RUB modification status, CRL activity, and CRL stability in planta and in vitro would clarify how SMAP1 contributes to the RUB cycle-related regulation of CRL activity.
In conclusion, two independent experimental approaches indicate that the conserved SMAP1 protein interacts with RUB modification-related components. Although the precise mode of action of SMAP1 is still unclear, further biochemical research on the functions and the relationship of SMAP1 with AXR1 and CSN might reveal its biological role in the RUB cycle as well as in the CRL-regulated ubiquitin-proteasome system in plant cells.
MATERIALS AND METHODS
Plant Materials
The Arabidopsis (Arabidopsis thaliana) lines aar1-1 and aar1-2 (Rahman et al., 2006), aar3-2 (Biswas et al., 2007), axr1-3 and axr1-12 (Leyser, et al., 1993), tir1-1 (Ruegger et al., 1998), ecr1-1 (Woodward et al., 2007), csn5a-1 (Gusmaroli et al., 2007), and DR5rev:GFP (Friml et al., 2003) were described previously. The SMAP1 RNAi lines (520i), the transgenic lines transformed with the 3.7-kb SMAP1 genomic fragment (B/S; described as gSMAP1 in this report) in the aar1-1 background, and its control line (X/B) were described by Rahman et al. (2006). 35S:SMAP1-GFP/aar1-1, 35S:GFP-SMAP1/aar1-1, SMAP1:SMAP1-GFP/aar1-1, and their derivative lines were generated in this work as described below. All transgenic and mutant lines were derived from the Columbia accession of Arabidopsis except for aar1-2, which is in the Wassilewskija background. The AAR1 locus was genotyped by PCR using primers T6G15 9538F and T6G15 53634R to detect the DNA deletion in the aar1-1 genome and primers T6G15 24488F and AT4G1 3490F2 to detect the presence of the corresponding wild-type DNA. For aar1-2, the mutant locus was tracked by the kanamycin-resistant phenotype resulting from the NPTII gene in the transgene construct (Rahman et al., 2006). The aar3-2 mutation was genotyped using the primers K5K13 F38890 and K5K13 R39225 to detect the wild-type gene and primers K5K13 F38890 and LB1 to detect the T-DNA. The tir1-1 mutation was detected by PCR amplification using the primers TIR1gF547 and TIR1gR913, and the amplified product was digested with DpnII, which cuts mutant but not wild-type products. The axr1-12 mutation was detected by PCR with the primers AXR1-12F and AXR1-12R, and the amplified product was digested with DraI, which cuts mutant but not wild-type products. For csn5a-1, the primers CSN5A-1F and CSN5A-1R2 were used to detect the wild-type locus and the primers CSN5A-1F and LBb1 were used to detect the mutation. The primer sequences are listed in Supplemental Table S1. The ecr1-1 and axr1-3 mutations were detected by derived cleaved-amplified polymorphic sequence analysis as described by Woodward et al. (2007).
Growth Analyses
Seed sterilization and plant growth conditions were as described by Biswas et al. (2007). Briefly, seeds were plated on germination medium (GM; one-half-strength Murashige and Skoog salts, 1% [w/v] Suc, and 0.5 g L–1 MES, pH 5.8, containing 1× B5 vitamins and 0.8% [w/v] Bacto agar) in rectangular plates, unless otherwise mentioned. For synchronous germination, the seeds were kept in the dark for 2 d at 4°C, and then the plates were transferred to a growth chamber at 23°C. For the hypocotyl and root growth assays, the plants were grown vertically on square culture plates under continuous white light (20–30 µmol m–2 s–1). For transfer assays, seedlings were first grown on GM without plant growth regulators and then transferred after 4 to 5 d to fresh GM containing plant growth regulators. For the IAA root inhibition assay, seedlings were transferred onto GM 1.2 (the same as GM except that the agar concentration is 1.2% to prevent roots from penetrating into the agar). The hypocotyl and root lengths were analyzed using the NIH ImageJ software package (National Institutes of Health) after the plants were photographed with a DP50 digital camera (Olympus). The percentage of root growth inhibition was calculated relative to root growth on medium without growth regulators. To observe the morphology of plants grown on soil, the seedlings were grown in a growth chamber (Biotron; LH-200-RDS, NK system) under a 16-h-light (100–130 µE m–2 s–1)/8-h-dark photoperiod at 23°C.
Plasmid Construction and Plant Transformation
All plasmids were constructed using standard recombinant DNA techniques, and their authenticity was confirmed by DNA sequencing. The SMAP1 DNA fragment was amplified from Arabidopsis bacterial artificial chromosome clone T6G15. The SMAP1:SMAP1-GFP fusion construct and its deleted versions were made by inserting appropriate DNA fragments into the binary vector pEGAD containing GFP and the nos terminator (Cutler et al., 2000). The 35S promoter was removed from pEGAD by digestion with SacI and AgeI, then an approximately 5-kb region of the SMAP1 promoter and the full or deleted version of SMAP1 coding sequences were inserted into the restriction enzyme sites. To generate 35S:SMAP1-GFP and 35S:GFP-SMAP1, SMAP1 coding sequences without or with the stop codon were amplified with the primer sets 13520ATG-topo/13520-TAA and 13520ATG-topo/13520R+TAA (Supplemental Table S1), respectively, and cloned into the pENTR/D-TOPO vector (Invitrogen). The resulting plasmids, 13520-TAA ENTR or 13520+TAA ENTR, were assembled into the Gateway binary vector pK7FWG2 to generate 35S:SMAP1-GFP or into pK7WGF2 to generate 35S:GFP-SMAP1, respectively, by a site-specific recombination reaction between attL and attR sites (Karimi et al., 2005). For the in vitro pull-down assay, GST-SMAP1 and its derivatives were constructed with the pGEX4T-2 expression vector (GE Healthcare). Full details of plasmid construction are available on request.
The resulting constructs in binary vectors were introduced into Agrobacterium tumefaciens GV3101 (pMP90) by electroporation and used to transform Arabidopsis Columbia with the floral-dip method (Clough and Bent, 1998). T1 seeds obtained from infected plants were germinated and selected on GM containing the appropriate antibiotic for selection of the marker gene in the vectors. In the T2 generation, T1 lines that showed a segregation ratio of 1:3 for antibiotic sensitive/antibiotic resistant were selected, and antibiotic-resistant T2 seedlings were grown to harvest T3 seeds. T3 plants were expected to be offspring of T2 plants that were heterozygous or homozygous for the inserted gene. Therefore, T2 lines that gave rise to only antibiotic-resistant T3 plants were selected as homozygous lines.
Observations of Embryo Morphology and GFP Fluorescence
For observations of embryo morphology, siliques at different developmental stages were harvested, dissected using a stereoscope, and cleared in a derivative of Hoyer’s solution (chloral hydrate:glycerol:water, 8:1:2). Cleared ovules were removed from their siliques in a drop of the same clearing solution, mounted whole, and observed with an Olympus BX60 microscope equipped with Nomarski optics. Digital images were captured using an Olympus DP-50 digital camera. To detect GFP accumulation, embryos were dissected in 7% Glc, mounted whole, and then fluorescent signals in roots and embryos were detected using a confocal laser-scanning microscope (Olympus Fluoview FV1000 with digital imaging processing) with a 515- ± 10-nm band-pass filter. For signal localization in embryos, images of GFP and transmitted light channels were electronically overlaid and further processed with Photoshop software (Adobe Systems).
Pull-Down Assay
To detect SMAP1-GFP-binding proteins, we used a µMACS GFP isolation kit (Miltenyi Biotec). Seven-day-old plants grown under continuous light were ground in liquid nitrogen and then ground in ice-cold elution buffer (50 mm Tris-HCl [pH 8.0], 50 mm NaCl, 10% glycerol, 1% Triton X-100, and Protease Inhibitor Complete Mini [Roche Diagnostics]). The extract was centrifuged for 20 min at 13,000g, and the supernatant was filtered through a syringe-driven filter unit with 0.45-µm pore size (Millipore). The solution containing 1 mg of total protein was mixed with magnetic anti-GFP microbeads and then incubated overnight to allow GFP to bind to anti-GFP microbeads. The mixture was transferred to a prewashed column containing magnetic beads, and then the column was washed four times with wash buffer 1 (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 1% Nonidet P-40) and once with wash buffer 2 (20 mm Tris-HCl, pH 8.0) to flush unbound proteins and excess salt. GFP and proteins bound to it were eluted with 50 µL of SDS sample buffer. Eluted proteins were heat denatured and separated by SDS-PAGE on a 12.5% gel. The protein spots on the silver-stained gel were cut out and digested with trypsin (Promega). The digested peptides were analyzed by an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific), and MS/MS spectra were compared against The Arabidopsis Information Resource 8 Arabidopsis genome annotation data using the Mascot server (Matrix Science).
For the GST pull-down assay, Escherichia coli lines harboring the constructs were grown to an optical density at 595 nm of 0.5 to 0.6, and then the expression of fusion proteins was induced by isopropyl β-d-1-thiogalactopyranoside at a final concentration of 0.1 mm for 2 h. Extraction of fusion proteins and their immobilization to the glutathione column were performed using a GST-Spin Trap Purification Module kit (GE Healthcare) according to the manufacturer’s protocol. The columns containing immobilized GST-SMAP1 fusion protein were washed with phosphate-buffered saline and then flushed with ice-cold plant GST extraction buffer (50 mm Tris-HCl, pH 7.0, 10 mm MgCl2, 150 mm NaCl, 10% glycerol, 0.01% Nonidet P-40, 0.5 mm phenylmethylsulfonyl fluoride, and Protease Inhibitor Complete Mini [Roche Diagnostics]). Total protein was prepared from 7-d-old aar1-1 plants as described above for the GFP pull-down assay, except that plant GST extraction buffer was used, and applied to the columns. The columns were incubated at room temperature for 10 min and washed five times with 600 µL of plant GST extraction buffer. The GST and proteins bound to it were eluted with GST elution buffer (50 mm Tris-HCl, pH 7.0, and 10 mm glutathione), and 10 µg of the eluted protein was separated by SDS-PAGE.
Antibodies and Western-Blot Analyses
We used antibodies against GFP (1181446001; Roche Diagnostics) and CSN4 (PW8360; Enzo Life Sciences). For the Arabidopsis CUL1 antibody, the N-terminal region of CUL1 protein (380 amino acid residues of the N terminus of CUL1) was inserted into the pET16b vector (Novagen) and expressed in the E. coli strain BL21(DE3) (Novagen). The insoluble protein fraction was solubilized in His-binding buffer (20 mm Tris-HCl, pH 7.9, 0.5 m NaCl, 5 mm imidazole, and 6 m guanidine-HCl), bound to nickel-nitrilotriacetic acid agarose (Qiagen), washed with washing buffer (20 mm Tris-HCl, pH 7.9, 0.5 m NaCl, 60 mm imidazole, and 6 m guanidine-HCl), and eluted with elution buffer (20 mm Tris-HCl, pH 7.9, 0.5 m NaCl, 250 mm imidazole, and 6 m guanidine-HCl). The eluted protein was precipitated by the addition of 80% cold acetone, and then proteins were separated by SDS-PAGE and purified by electroelution from the gel. The anti-CUL1 antibody was prepared from a rabbit injected with the purified CUL1 protein. For RUB modification analysis of CUL1, Arabidopsis seedlings or flowers were ground in lysis buffer (125 mm Tris-HCl, pH 8.5, 50 mm NaS2O3, 1% SDS, and 10% glycerol). The extracts were centrifuged at 13,000g for 10 min, and the protein concentration in the supernatant diluted 1:9 with water was determined by the Bradford assay (Bio-Rad Laboratories). Protein samples were boiled in SDS-PAGE sample buffer, run on SDS-PAGE gels, and blotted onto polyvinylidene difluoride membranes. For detection, alkaline phosphatase-conjugated anti-rabbit IgG (A3687; Sigma-Aldrich) as secondary antibody and nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate solution (Roche Diagnostics) were used. For densitometry analyses, the digital images of the blots were acquired by scanning the blots, and the peak intensity of the bands was determined by Quantity One software (Bio-Rad Laboratories). The values of the ratio of RUB-modified CUL1 to total CUL1 (RUB-modified plus unmodified CUL1) were analyzed by Fisher’s lsd test in KaleidaGraph software (Synergy Software).
Sequence data for this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SMAP1 (At4g13520), AXR1 (At1g05180), TIR1 (At3g62980), AAR3 (At3g28970), ECR1 (At5g19180), CSN1 (At3g61140), CSN2 (At3g26990), CSN3 (At5g14250), CSN4 (At5g42970), CSN5A (At1g22920), CSN8 (At4g14110), GAPC2 (At1g13440), and CUL1 (At4g02570).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Complementation of the aar1-1 phenotype with SMAP1 and GFP fusion protein derived by 35S promoter.
Supplemental Figure S2. Expression of SMAP1-GFP in early embryonic stage.
Supplemental Figure S3. Cellular localization of GFP fluorescence in root tip epidermal cells of 7-d-old plants.
Supplemental Figure S4. Additional data for genetic interaction of the CSN5A and SMAP1.
Supplemental Table S1. List of primers used in this study for genotyping of mutant and transgenic lines.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center, the Salk Institute Genomic Analysis Laboratory, and Dr. Bonnie Bartel (Rice University) for providing biological materials used in this work. We also thank Chihiro Suzuki (Japan Atomic Energy Agency) for technical assistance and Drs. Tomoki Chiba (Tsukuba University) and Tomohiro Kiyosue (Gakusyuin University) and members of Ion Beam Mutagenesis Research Group at the Japan Atomic Energy Agency for helpful discussions. We are grateful to Dr. William Gray (University of Minnesota) for critical reading of the manuscript.
Glossary
- 2,4-D
2,4-dichlorophenoxyacetic acid
- GST
glutathione S-transferase
- AUX
auxin
- IAA
indole-3-acetic acid
- PCIB
p-chlorophenoxyisobutylic acid
- RNAi
RNA interference
- CaMV
cauliflower mosaic virus
- JA
jasmonic acid
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
References
- Biswas KK, Ooura C, Higuchi K, Miyazaki Y, Van Nguyen V, Rahman A, Uchimiya H, Kiyosue T, Koshiba T, Tanaka A, et al. (2007) Genetic characterization of mutants resistant to the antiauxin p-chlorophenoxyisobutyric acid reveals that AAR3, a gene encoding a DCN1-like protein, regulates responses to the synthetic auxin 2,4-dichlorophenoxyacetic acid in Arabidopsis roots. Plant Physiol 145: 773–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng XW. (1996) The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86: 115–121 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cope GA, Deshaies RJ. (2003) COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114: 663–671 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ. (2004) Plant Hormones. Biosynthesis, Signal Transduction, Action! Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M. (2002) AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14: 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M. (1999) The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc Natl Acad Sci USA 96: 15342–15347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. (2005a) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Karunarathna N, Jurgens G, Estelle M. (2007) AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J 52: 114–123 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M. (2005b) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Estelle M. (2004) Auxin signaling and regulated protein degradation. Trends Plant Sci 9: 302–308 [DOI] [PubMed] [Google Scholar]
- Dharmasiri S, Dharmasiri N, Hellmann H, Estelle M. (2003) The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J 22: 1762–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmann EMN, Levesque MP, Isono E, Schmid M, Schwechheimer C. (2008) Auxin responses in mutants of the Arabidopsis CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome. Plant Physiol 147: 1369–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K, Callis J. (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Gusmaroli G, Figueroa P, Serino G, Deng XW. (2007) Role of the MPN subunits in COP9 signalosome assembly and activity, and their regulatory interaction with Arabidopsis Cullin3-based E3 ligases. Plant Cell 19: 564–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotton SK, Callis J. (2008) Regulation of cullin RING ligases. Annu Rev Plant Biol 59: 467–489 [DOI] [PubMed] [Google Scholar]
- Jurado S, Abraham Z, Manzano C, López-Torrejón G, Pacios LF, Del Pozo JC. (2010) The Arabidopsis cell cycle F-box protein SKP2A binds to auxin. Plant Cell 22: 3891–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P. (2005) Modular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kato JY, Yoneda-Kato N. (2009) Mammalian COP9 signalosome. Genes Cells 14: 1209–1225 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A. (1997) Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci USA 94: 11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T, Chou Y-C, Willems AR, Meyer-Schaller N, Hecht M-L, Tyers M, Peter M, Sicheri F. (2008) Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell 29: 23–35 [DOI] [PubMed] [Google Scholar]
- Kwok SF, Piekos B, Misera S, Deng XW. (1996) A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol 110: 731–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser HMO, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. (1993) Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161–164 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasone A, Kawai-Yamada M, Kiyosue T, Narumi I, Uchimiya H, Oono Y. (2009) A gene encoding SMALL ACIDIC PROTEIN 2 potentially mediates the response to synthetic auxin, 2,4-dichlorophenoxyacetic acid, in Arabidopsis thaliana. J Plant Physiol 166: 1307–1313 [DOI] [PubMed] [Google Scholar]
- Oono Y, Ooura C, Rahman A, Aspuria ET, Hayashi K-i, Tanaka A, Uchimiya H. (2003) p-Chlorophenoxyisobutyric acid impairs auxin response in Arabidopsis root. Plant Physiol 133: 1135–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Nakasone A, Chhun T, Ooura C, Biswas KK, Uchimiya H, Tsurumi S, Baskin TI, Tanaka A, Oono Y. (2006) A small acidic protein 1 (SMAP1) mediates responses of the Arabidopsis root to the synthetic auxin 2,4-dichlorophenoxyacetic acid. Plant J 47: 788–801 [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M. (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell 32: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MW, McQuary PR, Wee S, Hofmann K, Wolf DA. (2009) F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Mol Cell 35: 586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Isono E. (2010) The COP9 signalosome and its role in plant development. Eur J Cell Biol 89: 157–162 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng X-W. (2001) Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292: 1379–1382 [DOI] [PubMed] [Google Scholar]
- Seeger M, Kraft R, Ferrell K, Bech-Otschir D, Dumdey R, Schade R, Gordon C, Naumann M, Dubiel W. (1998) A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J 12: 469–478 [PubMed] [Google Scholar]
- Sharon M, Mao H, Boeri Erba E, Stephens E, Zheng N, Robinson CV. (2009) Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure 17: 31–40 [DOI] [PubMed] [Google Scholar]
- Somers DE, Fujiwara S. (2009) Thinking outside the F-box: novel ligands for novel receptors. Trends Plant Sci 14: 206–213 [DOI] [PubMed] [Google Scholar]
- Stuttmann J, Lechner E, Guérois R, Parker JE, Nussaume L, Genschik P, Noël LD. (2009) COP9 signalosome- and 26S proteasome-dependent regulation of SCFTIR1 accumulation in Arabidopsis. J Biol Chem 284: 7920–7930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiryaki I, Staswick PE. (2002) An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol 130: 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Wang X-J, Hagen G, Guilfoyle TJ. (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Wei N, Chamovitz DA, Deng XW. (1994) Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 78: 117–124 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Ratzel SE, Woodward EE, Shamoo Y, Bartel B. (2007) Mutation of E1-CONJUGATING ENZYME-RELATED1 decreases RELATED TO UBIQUITIN conjugation and alters auxin response and development. Plant Physiol 144: 976–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.