Abstract
Background
Acidithiobacillus caldus (A. caldus) is widely used in bio-leaching. It gains energy and electrons from oxidation of elemental sulfur and reduced inorganic sulfur compounds (RISCs) for carbon dioxide fixation and growth. Genomic analyses suggest that its sulfur oxidation system involves a truncated sulfur oxidation (Sox) system (omitting SoxCD), non-Sox sulfur oxidation system similar to the sulfur oxidation in A. ferrooxidans, and sulfur oxygenase reductase (SOR). The complexity of the sulfur oxidation system of A. caldus generates a big obstacle on the research of its sulfur oxidation mechanism. However, the development of genetic manipulation method for A. caldus in recent years provides powerful tools for constructing genetic mutants to study the sulfur oxidation system.
Results
An A. caldus mutant lacking the sulfur oxygenase reductase gene (sor) was created and its growth abilities were measured in media using elemental sulfur (S0) and tetrathionate (K2S4O6) as the substrates, respectively. Then, comparative transcriptome analysis (microarrays and real-time quantitative PCR) of the wild type and the Δsor mutant in S0 and K2S4O6 media were employed to detect the differentially expressed genes involved in sulfur oxidation. SOR was concluded to oxidize the cytoplasmic elemental sulfur, but could not couple the sulfur oxidation with the electron transfer chain or substrate-level phosphorylation. Other elemental sulfur oxidation pathways including sulfur diooxygenase (SDO) and heterodisulfide reductase (HDR), the truncated Sox pathway, and the S4I pathway for hydrolysis of tetrathionate and oxidation of thiosulfate in A. caldus are proposed according to expression patterns of sulfur oxidation genes and growth abilities of the wild type and the mutant in different substrates media.
Conclusion
An integrated sulfur oxidation model with various sulfur oxidation pathways of A. caldus is proposed and the features of this model are summarized.
Introduction
Acidithiobacillus caldus (A. caldus), a gram-negative, acidophilic, obligately chemolithotrophic, moderately thermophilic bacterium [1], [2] and an important member of a consortium of microorganisms used for industrial bioleaching [3], plays key roles together with iron-oxidizing bacteria in bio-leaching processes [4], [5]. A. caldus has the capability of oxidizing elemental sulfur and a wide range of reduced inorganic sulfur compounds (RISCs), but could not oxidize ferrous iron. It uses energy and electrons derived from sulfur oxidation for carbon dioxide fixation and other anabolic processes [1], [2], [6], [7].
Oxidation of elemental sulfur and RISCs happens normally in some chemolithotrophic bacteria and archaea [8]–[10]. The sulfur oxidizing (Sox) enzyme system in lithoautotrophic Paracoccus pantotrophus (P. pantotrophus), responsible for the oxidation of sulfide, elemental sulfur, thiosulfate, and sulfite to sulfate, accompanied by electron transfer to cytochrome c, has been well studied [8]–[12]. It is located in the periplasm and constituted generally by four proteins: SoxYZ, SoxAX, SoxB and Sox(CD)2 [13]. Initially, SoxAX initiates the oxidation of thiosulfate (S2O3 2−) producing SoxY-thiocysteine-S-sulfate (SoxYZ-S-S-SO3 −) [14]; secondly, SoxB hydrolyzes sulfate (SO4 2−) from the thiocysteine-S-sulfate residue (SoxYZ-S-S-SO3 −) producing S-thiocysteine (SoxYZ-S-S−); thirdly, Sox(CD)2 may oxidize the outer sulfur atom of S-thiocysteine producing SoxYZ-cysteine-S-sulfate (SoxYZ-S-SO3 −); finally, sulfate is hydrolyzed and removed by SoxB from SoxYZ-S-SO3 −, and SoxYZ is regenerated. However, Sox(CD)2 is absent in the so-called truncated Sox system of many prototypical α-Proteobacteria [8], [10]. Another sulfur oxidation system based on the sulfur oxygenase reductase (SOR) has been elaborated in several acidophilic and thermophilic archaea [10], [15]. SOR is able to catalyze the disproportionation of elemental sulfur, producing sulfite, thiosulfate, and sulfide [16], [17]. The reaction has several characteristics: (1) it takes place in the cytoplasm; (2) it is dioxygen (O2)-dependent with no external cofactors or electron donors required [16]; (3) sulfur oxidation is not coupled with electron transfer or substrate-level phosphorylation in Acidianus ambivalens (A. ambivalens) [18].
The sulfur oxidation system in acidophilic Acidithiobacillus genus is related to three representative species widely used in bio-leaching, which are A. ferrooxidans, A. thiooxidans and A. caldus. Although, the sulfur metabolic mechanism in A. ferrooxidans has been studied for many years, it is still not completely understood. The elemental sulfur in nature consists of a stable octasulfane ring (S8), which forms orthorhombic crystals insoluble in water [19]. An elemental sulfur activation and oxidation model was proposed in Acidithiobacillus [20], [21]. The S8 is firstly activated to become thiol-bound sulfane sulfur atoms (R-S-SH) and then transported into the periplasm where it is oxidized by sulfur dioxygenase (SDO) [20], [21]. SDO enzyme activity was detected from the crude cell extracts of A. ferrooxidans and A. thiooxidans, but SDO enzyme protein has not been purified and the gene(s) encoding for SDO activity not yet been identified [20], [22]. Another elemental sulfur oxidation enzyme in A. ferrooxidans is the cytoplasmic heterodisulfide reductase complex (HdrABC), but it is a speculation from the genomic and transcriptomic analysis, not from biochemical experimental data [23]. The S4I pathway of thiosulfate oxidation via the form of tetrathionate intermediates exists widely in Acidithiobacillus: the periplasmic thiosulfate is oxidized to tetrathionate by thiosulfate quinine oxidoreduetase (TQO); then, tetrathionate is hydrolyzed by tetrathionate hydrolase (TetH) yielding thiosulfate and other products [10], [24]. In addition, other RISC oxidation enzymes identified in Acidithiobacillus include: sulfide quinone reductase (SQR) being responsible for oxidation of hydrogen sulfide [25], and rhodanese or thiosulfate sulfurtransferase (TST) transferring a sulfur atom from thiosulfate to sulfur acceptors like cyanide and thiol compounds [26], [27]. Recently, a model of sulfur oxidation in A. ferrooxidans was proposed, in which electrons from oxidation of RISCs are transferred via the quinol pool (QH2) to terminal oxidases to produce ATP or to NADH complex I to generate NADPH, coupling the oxidation of RISCs with the generation of energy or reducing power [23].
An unique sulfur oxidation system exists in A. caldus, which is quite different from that of A. ferrooxidans according to comparative genome analysis [28]. The sulfur oxidation system of A. caldus can be classified into three subsystems: the truncated Sox subsystem, non-Sox sulfur subsystem, and SOR subsystem. The truncated Sox subsystem in A. caldus contains two copies of essential soxABXYZ genes but no soxCD genes. The non-Sox sulfur subsystem, similar to the sulfur oxidation system in A. ferrooxidans, contains the sulfur oxidation enzyme genes (tetH, sqr, rhd, and hdrABC) and terminal oxidase genes. The SOR subsystem is characterized by the sulfur oxygenase reductase gene (sor) in A. caldus, which was only found in several acidophilic and thermophilic archaea but not in the two species A. ferrooxidans and A. thiooxidans [10], [29], [30]. Therefore, A. caldus has a complex and integrated sulfur oxidation system. The currently known sulfur oxidation pathways in A. caldus are mainly acquired from the genome sequence analysis, which are not clear and remain unanswered questions: (1) How is the elemental sulfur oxidized and how many elemental sulfur oxidizing pathways exist? (2) How does the truncated Sox subsystem work? (3) How do the various pathways interconnect to complete the sulfur oxidation? Although, a rough model for ISCs metabolism in A. caldus was proposed [31], the above questions are still awaiting clear answers.
Recently, a method for A. caldus mutant construction was developed in our laboratory, which is different from the previous reported gene knock-out method based on the homologous recombination in A. caldus [32]. In our experiments, construction of gene mutant via the transposition insertional mutagenesis of the insertion sequences (IS elements) in A. caldus was discovered for the first time. The first report about IS element transposition in Acidithiobacillus was the ISAfe1 insertional inactivation of resB (a putative cytochrome c-type biogenesis protein) producing the A. ferrooxidans mutant, which lost Fe (II) oxidation ability [33], [34]. IS elements are also widely distributed in A. caldus [35]. The loss of the sor gene resulting from the ISAtc1 transposition in A. caldus SM-1 was proposed according to the comparison of A. caldus SM-1 genome sequence and the sor gene cloned from SM-1 [30], [35].
In this research, A. caldus MTH-04 Δsor mutant is constructed by electroporation of a suicide plasmid. The cell growth of the mutant is measured using S0 and K2S4O6 as the substrates. Then, comparative transcriptome analysis is carried out by whole-genome microarray and quantitative RT-PCR. Finally, an integrated sulfur oxidation model in A. caldus is proposed.
Results and Discussion
Construction of Δsor mutant
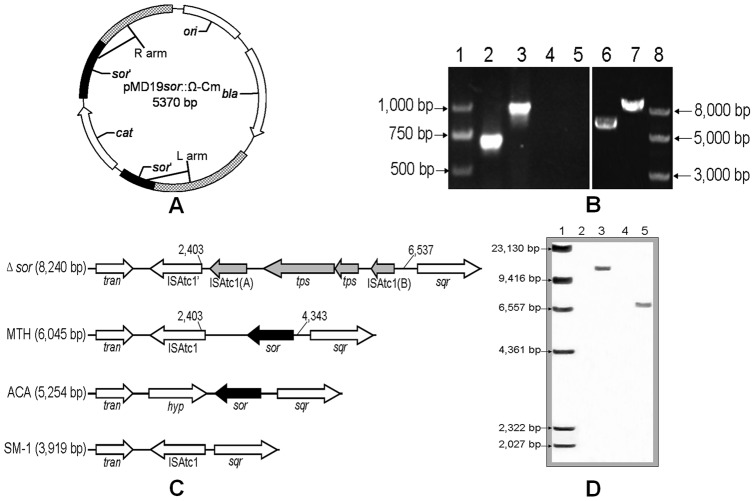
The suicide plasmid pMD19sor::Ω-Cm (Figure 1A) derived from pUC19 is unable to replicate in A. caldus. It carries the homologous sequence of the disrupted sor gene by insertion of chloramphenicol resistance gene (cat, 816 bp) generating two homologous sequences, the L-arm (1,321 bp) and the R-arm (1,332 bp) (Figure 1A). A. caldus MTH-04 was electroporated using the suicide plasmid and the mutants were screened using colony PCR, then the chromosomes of the screened mutants were extracted and analyzed by PCR (Figure 1B) and southern blot analyses (Figure 1D). As shown in Figure 1B, two sets of primers (SorA fwd and SorA rev, SorB fwd and SorB rev) specific to the sor gene and a set of primers (Big fwd and Big rev) specific to the homologous sequence were used to verify the mutant. Fragments of 684 bp (lane 2, Figure 1B) and 971 bp (lane 3, Figure 1B) specific to the sor gene were amplified from the chromosome of the wild type, but not from the mutant. An 8 kbp fragment amplified from the mutant using the primers Big fwd and Big rev (lane 7, Figure 1B) was larger than the fragment of about 6 kbp amplified from the wild type (lane 6, Figure 1B). The fragments of lane 6 and lane 7 in Figure 1B were sequenced for validation. The results are shown in Figure 1C: the sor gene region (from 2,403 bp to 4,343 bp) in A. caldus MTH-04 wild type was different from the corresponding region (from 2,403 bp to 6,537 bp) which is replaced by the IS element (ISAtc1) and the transposase gene in the A. caldus MTH-04 mutant. In order to further confirm the mutant, southern blot analysis was carried out using the sor gene as the probe. As shown in Figure 1D, an EcoR I-fragment (6.7 kbp) was isolated from the wild type chromosome, but not from the mutant. In addition, the analysis of the sor regions in different A. caldus strains is depicted in Figure 1C, ISAtc1 is positioned upstream of the sor gene in the wild type A. caldus MTH-04 genome, similar to A. caldus SM-1 but different from A. caldus ATCC 51765, while the sor gene region in the Δsor mutant was replaced by the IS elements.
Figure 1. Construction of the mutant.
(A) The pMD19sor::Ω-Cm suicide plasmid. The plasmid carries the mutant allele of sor gene disrupted by the chloramphenicol resistance gene (816 bp), generating two homologous sequences L-arm (1,321 bp) and R-arm (1,332 bp). (B) PCR analysis of A. caldus MTH-04 sor mutant. Lanes 4, 5 and 7, the chromosome from the mutant; lanes 2, 3 and 6, the chromosome from the wild type; lanes 2 and 4, primers of SorA fwd and SorA rev; lanes 3 and 5, primers of SorB fwd and SorB rev; lanes 6 and 7, primers of Big fwd and Big rev. (C) Comparison of the regions of the sor gene. Δsor: A .caldus MTH-04 Δsor mutant; MTH: A .caldus MTH-04 wild type; ACA: A .caldus ATCC 51765; SM-1: A .caldus SM-1; tran: ABC transporter ATP-binding protein; ISAtc1: IS elements; tps: transposase; sqr: sulfide quinone reductase; sor: sulfur oxygenase reductase; hyp: hypothetical protein. (D) Southern blot analysis of the wild type and the mutant using the sor probe. Lane 2, negative control with ddH2O; lane 3, positive control with pJRD215-tac-sor (12 kbp) digested by EcoR I; lane 4, the mutant chromosome digested by EcoR I; lane 5, the wild type chromosome digested by EcoR I.
The hypothesis that IS element transposition results in the loss of sor gene in A. caldus SM-1 has been proposed according to sequence analysis [35]. Fortunately, it was confirmed for the first time in our experiments. Firstly, the same result was obtained in the repeated experiments and no mutant was obtained when plasmid pMD19 or pSDU1 was used, which showed that deletion of sor gene was not a natural occurrence. Secondly, no artificial or exogenous IS elements was introduced into the suicide plasmid. Finally, the IS element (ISAtc1) causing the loss of sor gene could be found on A. caldus MTH-04 wild type chromosome. For above reasons, it is clear that the transposition of IS elements in A. caldus MTH-04 chromosome leads to the loss of sor gene. Moreover, it also implies the role of IS elements in gene evolution and metabolic diversity in A. caldus.
Growth curve analysis of the wild type and the Δsor mutant
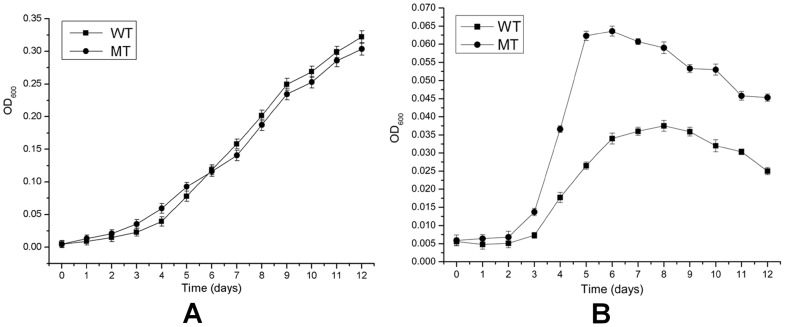
The growth curves of the wild type and the mutant in Starkey-S0 or Starkey-K2S4O6 liquid media are shown in Figure 2. When S0 was utilized as the sole substrate, the cell concentration of the mutant was a little higher than the wild type in the first six days, but was slightly lower for the next six days (Figure 2A). When K2S4O6 was used as the sole substrate, the mutant had an obvious growth advantage compared to the wild and its maximum cell concentration was 70% higher than the wild (Figure 2B). The mutant still had S0 oxidation ability, indicating that SOR is not the sole and determinative elemental sulfur oxidation pathway and there are other additional elemental sulfur oxidation pathways in A. caldus. In addition, the maximum value of OD600 of A. caldus in S0 medium in Figure 2A could reach about 0.35 much higher than that in K2S4O6 medium (OD600≈0.065), so it is suggested that A. caldus has a high efficient elemental sulfur oxidizing ability to enable it grow well.
Figure 2. Growth curves of A. caldus MTH-04 and the Δsor mutant (A) in Starkey-S0 medium and (B) in Starkey-K2S4O6 medium.
Each data point represents triplicate results. The error bars indicate standard deviations.
Comparative transcriptome analysis
Hybridization scheme, microarray experiments, and data analysis are described in materials and methods. Unsupervised hierarchical cluster analysis of sulfur oxidation genes are depicted in Figure 3. It shows the normalized transcription levels of sulfur oxidation genes for each sample compared to the common reference. Several remarkable differences are revealed in this figure: firstly, there was a bright green region related to the genes in the sox operons of the wild but a red color region of the mutant in K2S4O6 medium, suggesting that genes in the sox operons had a low expression level in the wild and a high expression level in the mutant when K2S4O6 was applied; secondly, the bright green color appeared in the row of sor gene of the mutant was resulted from the deletion of sor, moreover the sor gene of the wild displayed red color in K2S4O6 medium and green color in S0 medium, indicating the higher expression level of sor in K2S4O6 medium; thirdly, the green region at the lower right corner of the figure indicated that genes involving rhd, dsrE, and tusA had lower expression levels in the mutant when K2S4O6 was applied.
Figure 3. Hierarchical cluster analysis of genes involved in sulfur oxidation.
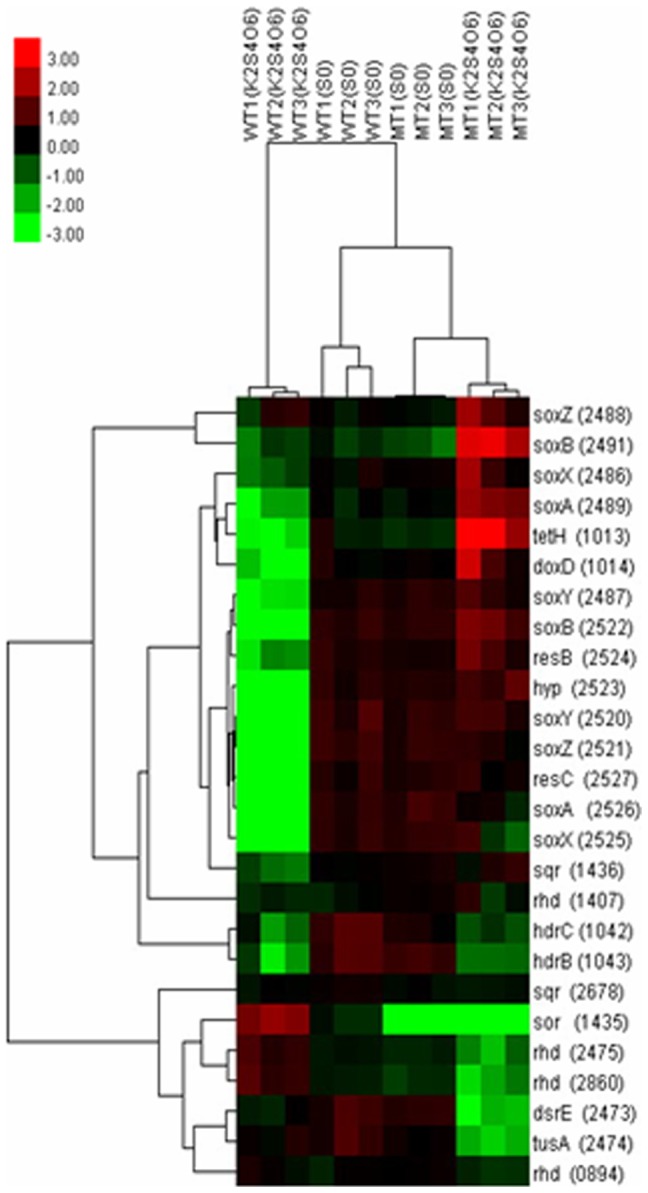
The signals are shown in a red-green color scale, where red represents higher expression and green represents lower expression. Each column stands for a sample and each row stands for a gene.
The results of significance analysis of microarrays (SAM) for sulfur oxidation genes under four cases are shown in Table 1 (the numbers without brackets). The expression patterns observed with microarrays were validated by using real-time quantitative RT-PCR (qRT-PCR) for some sulfur oxidation genes (Table 1, the numbers in the brackets). The results from microarrays and qRT-PCR were highly consistent. Genes that met the criterions of fold change ≥1.5 with q-value≤0.05 and fold change ≤0.667 with q-value≤0.05 were up-regulated and down-regulated, respectively. The expression of sulfur oxidation genes showed distinct differences under the four experimental cases (Table 1). In the first case, the comparison was between the mutant and the wild using S0 as sulfur source shown in the column of MT/WT (S0). No obvious difference was found in the expression levels of sulfur oxidation genes except that the expression of sor was not detected in the mutant. The results implied that deletion of sor had no obvious influence on the other sulfur oxidation pathways in S0 medium at the early phase. In the second case, K2S4O6 was used as sulfur source to analyze the differential gene expressions between the mutant and the wild. The loss of sor led to significant gene expression changes in K2S4O6 medium, in particular: (1) the remarkable up-regulation of genes in the two sox operons; (2) the significant up-regulation of genes in tetrathionate hydrolase operon (tetH and doxD); (3) the obvious down-regulation of sulfur transferase genes, such as dsrE (ACAL_2473), tusA (ACAL_2474), and rhd (ACAL_2475, ACAL_2860). The results showed that deletion of sor stimulated the up-regulation of the sox pathway and the S4I pathway in K2S4O6 medium. In the third case, the effect of different sulfur sources on the gene expression of the wild type was showed in the column of S0/K2S4O6 (WT). Sulfur oxidation genes of the two sox operons, tetrathionate hydrolase operon, and heterodisulfide reductase complex operon had higher expression levels of varied degrees when S0 was added, in contrast, sor had a lower expression level. Therefore, the lower expression level of sor gene at the early fast growth phase in S0 medium and the better growth of the mutant at the early days in S0 medium implied that SOR does not play an important role at the early stage of the elemental sulfur oxidation process. The fourth case showed the differential gene expressions of the mutant in the two kinds of media. An obvious change compared to the third case was that the majority of sulfur oxidation genes of sox operons and tetrathionate hydrolase operon had higher expression levels in K2S4O6 medium than that in S0 medium. From above results together with the following facts that the lower expression levels of these genes of the wild type in the K2S4O6 medium (Table 1, column of S0/K2S4O6 (WT)) and the no differential expression of these genes between the mutant and the wild in S0 medium (Table 1, column of MT/WT (S0)), it can be concluded that there was a significant increase of the expression levels of above genes in the mutant compared with that in the wild type in K2S4O6 medium. Above conclusion showed that deletion of SOR pathway stimulated the sox and S4I pathways in K2S4O6 medium.
Table 1. Expression of sulfur oxidation genes under different cases.
| ID | Gene | Function | MT/WT (S0) | MT/WT (K2S4O6) | S0/K2S4O6 (WT) | S0/K2S4O6 (MT) |
| Sox operon I | ||||||
| ACAL_2486 | soxX | cytochrome c class I | NC (1.41±0.02) | 3.86±1.03 (23.88±0.05) | 2.19±0.03 (17.21±0.01) | NC (0.18±0.04) |
| ACAL_2487 | soxY | sulfur covalently binding protein | NC (0.97±0.06) | 12.92±0.08 (14.27±0.04) | 10.89±0.01 (14.40±0.05) | NC (0.17±0.03) |
| ACAL_2488 | soxZ | Sulfur compound chelating protein | NC (1.40±0.03) | 1.96±0.86 (27.43±0.04) | NC (14.68±0.02) | 0.43±0.76 (0.13±0.01) |
| ACAL_2489 | soxA | cytochrome c (diheme) | NC (1.43±0.03) | 13.68±0.14 (6.02±0.04) | 4.35±0.02 (3.32±0.01) | 0.33 ±0.14 (0.14±0.04) |
| ACAL_2491 | soxB | sulfate thiol esterase | NC (1.39±0.03) | 10.64±1.30 (11.15±0.06) | NC (4.90±0.04) | 0.09±1.30 (0.11±0.01) |
| Sox operon II and cytochrome bo3 ubiquinol oxidase | ||||||
| ACAL_2515 | coxB | cytochrome c oxidase, subunit II | NC | 5.40±0.01 | 18.36±0.02 | 3.65±0.02 |
| ACAL_2516 | cox | cytochrome c oxidase | NC | 22.26±0.01 | 53.67±0.05 | 2.36±0.03 |
| ACAL_2517 | cyoC | cytochrome quinol oxidase subunit 3 | NC | 9.61±0.01 | 25.37±0.02 | 2.32±0.01 |
| ACAL_2518 | hyp | hypothetical protein | NC | 11.24±0.01 | 33.64±0.01 | 2.34±0.01 |
| ACAL_2519 | hyp | hypothetical protein | NC | 13.93±0.05 | 10.40±0.01 | NC |
| ACAL_2520 | soxY | sulfur covalently binding protein | NC (1.48±0.01) | 29.24±0.04 (20.59±0.02) | 29.98±0.06 (174.46±0.03) | NC (2.18±0.01) |
| ACAL_2521 | soxZ | sulfur compound chelating protein | NC (1.32±0.01) | 17.12±0.03 (35.92±0.02) | 21.94±0.01 (31.38±0.06) | NC (0.20±0.01) |
| ACAL_2522 | soxB | sulfate thiol esterase | NC (1.66±0.03) | 34.24±0.17 (74.87±0.01) | 21.80±0.01 (180.30±0.04) | 0.61±0.18 (0.70±0.02) |
| ACAL_2523 | hyp | hypothetical protein | NC | 31.41±0.07 | 24.64±0.02 | NC |
| ACAL_2524 | resB | cytochrome c-type maturation protein | NC | 7.04±0.21 | 5.36±0.02 | NC |
| ACAL_2525 | soxX | cytochrome c class I | NC (1.42±0.04) | 10.06±0.21 (15.11±0.05) | 17.66±0.01 (222.40±0.02) | 1.81±0.21 (3.62±0.01) |
| ACAL_2526 | soxA | cytochrome c (diheme) | NC (1.60±0.02) | 22.44±0.02 (117.45±0.05) | 32.03±0.01 (182.13±0.06) | 1.55±0.05 (0.43±0.04) |
| ACAL_2527 | resC | cytochrome c-type maturation protein | NC | 17.33±0.04 | 19.03±0.03 | NC |
| Tetrathionate hydrolase operon | ||||||
| ACAL_1013 | tetH | tetrathionate hydrolase | NC (1.00±0.03) | 57.02±7.02 (83.40±0.02) | 7.73±0.05 (38.06±0.01) | 0.10±0.13 (0.08±0.03) |
| ACAL_1014 | doxD | Thiosuirate quinine oxidoreductase subunit | NC (1.15±0.06) | 18.41±2.74 (14.12±0.02) | 8.91±0.02 (50.22±0.01) | 0.46±0.10 (0.71±0.03) |
| Sulfur oxygenase reductase | ||||||
| ACAL_1435 | sor | sulfur oxygenase reductase | 0.00±0.00 (0.00±0.01) | 0.00±0.08 (0.00±0.04) | 0.26±0.08 (0.51±0.02) | UD (UD) |
| Sulfide-quinone reductase | ||||||
| ACAL_1436 | sqr | sulfide quinone reductase | NC (2.64±0.04) | 2.66±0.06 (1.01±0.05) | 2.24±0.01 (3.06±0.01) | NC (1.40±0.01) |
| ACAL_2678 | sqr | sulfide quinone reductase | 0.83±0.01 | NC | NC | NC |
| Heterodisulfide reductase complex operon | ||||||
| ACAL_1042 | hdrC | heterodisulfide reductase subunit C | 0.65±0.06 (0.94±0.02) | NC (0.42±0.03) | 3.77±0.10 (27.60±0.06) | 2.01±0.02 (10.76±0.01) |
| ACAL_1043 | hdrB | heterodisulfide reductase subunit B | NC (1.08±0.02) | NC (0.59±0.04) | 5.74±0.09 (41.14±0.01) | 3.78±0.01 (1.55±0.03) |
| ACAL_2473 | dsrE | hypothetical protein (sulfur transferase) | NC (0.91±0.04) | 0.20±0.01 (0.21±0.03) | 1.76±0.06 (9.93±0.02) | 7.97±0.00 (7.51±0.01) |
| ACAL_2474 | tusA | hypothetical protein (sulfur transferase) | 0.72±0.01 (1.12±0.02) | 0.20±0.02 (0.14±0.01) | NC (3.86±0.04) | 4.86±0.01 (5.43±0.05) |
| ACAL_2475 | rhd | rhodanese (sulfur transferase) | NC (0.89±0.01) | 0.21±0.04 (0.20±0.03) | 0.56±0.04 (0.85±0.05) | 2.40±0.01 (2.02±0.01) |
| Rhodanese (sulfur transferase) | ||||||
| ACAL_0894 | rhd | rhodanese (sulfur transferase) | NC | NC | NC | NC |
| ACAL_1407 | rhd | Rhodanese (sulfur transferase) | 1.27±0.01 | NC | NC | NC |
| ACAL_2860 | rhd | rhodanese (sulfur transferase) | NC (1.04±0.04) | 0.16±0.05 (0.15±0.01) | 0.50±0.05 (1.66±0.02) | 2.72±0.01 (1.99±0.03) |
| Cytochrome bd ubiquinol oxidase | ||||||
| ACAL_0179 | cydA | cytochrome d ubiquinol oxidase, subunit I | NC | 0.59±0.05 | NC | NC |
| ACAL_0180 | cydB | cytochrome d ubiquinol oxidase, subunit II | NC | 2.09±0.15 | 2.32±0.03 | NC |
| ACAL_1110 | cydB | cytochrome d ubiquinol oxidase, subunit II | 0.61±0.01 | 1.66±0.01 | 3.66±0.01 | NC |
| ACAL_1111 | cydA | cytochrome d ubiquinol oxidase, subunit I | NC | 6.25±0.01 | 6.44±0.02 | NC |
| ACAL_1252 | cydB | cytochrome d ubiquinol oxidase, subunit II | NC | 2.42±0.14 | 2.59±0.03 | NC |
| ACAL_1253 | cydA | cytochrome d ubiquinol oxidase, subunit I | NC | 1.86±0.15 | NC | NC |
| ACAL_2185 | cydA | cytochrome d ubiquinol oxidase, subunit I | UD | UD | UD | UD |
| ACAL_2186 | cydB | cytochrome d ubiquinol oxidase, subunit II | UD | UD | UD | UD |
| ACAL_2017 | cydB | cytochrome d ubiquinol oxidase, subunit II | NC | 2.09±0.04 | 2.02±0.02 | NC |
| Cytochrome bo3 ubiquinol oxidase | ||||||
| ACAL_1757 | cyoB | cytochrome o ubiquinol oxidase, subunit II | NC | 3.31±0.01 | 9.61±0.01 | 2.79±0.01 |
| ACAL_1758 | hyp | hypothetical protein | NC | NC | 3.33±0.11 | 3.69±0.02 |
| ACAL_1759 | cyoA | cytochrome o ubiquinol oxidase, subunit I | NC | 15.28±0.01 | 30.69±0.00 | 1.76±0.01 |
| ACAL_1760 | cyoC | cytochrome o ubiquinol oxidase, subunit III | 0.73±0.08 | 2.16±0.01 | 9.27±0.00 | 3.11±0.01 |
| ACAL_1761 | hyp | hypothetical protein | 0.74±0.05 | 5.96±0.00 | 21.87±0.00 | 2.71±0.02 |
| ACAL_1762 | cyoD | cytochrome o ubiquinol oxidase, subunit IV | NC | 10.49±0.00 | 25.81±0.00 | 2.40±0.01 |
| Cytochrome c protein | ||||||
| ACAL_0446 | resC | cytochrome c-type maturation protein | NC | 20.67±0.21 | 5.34±0.00 | 0.24±0.21 |
| ACAL_1072 | resC | cytochrome c-type maturation protein | NC | 17.35±0.21 | 2.54±0.01 | 0.13±0.09 |
| NADH complex I operon | ||||||
| ACAL_0727 | nuoA | NADH ubiquinone oxidoreductase A subunit | NC | NC | NC | NC |
| ACAL_0728 | nuoB | NADH ubiquinone oxidoreductase B subunit | NC | 0.61±0.08 | 0.67±0.01 | NC |
| ACAL_0729 | nuoC | NADH ubiquinone oxidoreductase C subunit | NC | 0.43±0.06 | 0.35±0.03 | NC |
| ACAL_0730 | nuoD | NADH ubiquinone oxidoreductase D subunit | NC | 0.60±0.04 | NC | 1.79±0.03 |
| ACAL_0731 | nuoE | NADH ubiquinone oxidoreductase E subunit | NC | 0.71±0.00 | NC | 1.57±0.01 |
| ACAL_0732 | nuoF | NADH ubiquinone oxidoreductase F subunit | NC | 2.15±0.02 | 2.88±0.01 | NC |
| ACAL_0733 | nuoG | NADH ubiquinone oxidoreductase G subunit | NC | 3.66±0.03 | 3.68±0.11 | NC |
| ACAL_0734 | nuoH | NADH ubiquinone oxidoreductase H subunit | NC | 3.35±0.11 | 3.15±0.10 | NC |
| ACAL_0735 | nuoI | NADH ubiquinone oxidoreductase I subunit | NC | NC | 0.38±0.54 | ND |
| ACAL_0736 | nuoJ | NADH ubiquinone oxidoreductase J subunit | NC | NC | NC | NC |
| ACAL_0737 | nuoK | NADH ubiquinone oxidoreductase K subunit | NC | NC | 1.88±0.04 | 1.35±0.02 |
| ACAL_0738 | nuoL | NADH ubiquinone oxidoreductase L subunit | ND | ND | ND | ND |
| ACAL_0739 | nuoM | NADH ubiquinone oxidoreductase M subunit | ND | ND | ND | ND |
| ACAL_0740 | nuoN | NADH ubiquinone oxidoreductase N subunit | NC | 6.29±0.15 | 5.53±0.02 | NC |
| ATP synthetase complex operon | ||||||
| ACAL_2147 | atpB | ATP synthase F0, A subunit | NC | 0.34±0.60 | 0.26±0.55 | NC |
| ACAL_2148 | atpE | ATP synthase F0, C subunit | NC | NC | NC | NC |
| ACAL_2149 | atpF | ATP synthase F0, B subunit | NC | NC | NC | 1.53±0.01 |
| ACAL_2150 | atpH | ATP synthase F1, delta subunit | NC | 0.52±0.48 | NC | 1.67±0.01 |
| ACAL_2151 | atpA | ATP synthase F1, alpha subunit | 0.74±0.01 | NC | 2.49±0.23 | 1.37±0.01 |
| ACAL_2152 | atpG | ATP synthase F1, gamma subunit | 0.79±0.01 | NC | 1.54±0.19 | NC |
| ACAL_2153 | atpD | ATP synthase F1, beta subunit | NC | NC | 2.59±0.15 | 1.94±0.05 |
| ACAL_2154 | atpC | ATP synthase F1, epsilon subunit | NC | 4.94±0.03 | 4.56±0.12 | NC |
Fold Change ≥1.5, q-value≤0.05: up-regulation, Fold Change ≤0.667, q-value≤0.05: down-regulation;
NC: not credible, q-value>0.05;
UD: undetected;
Numbers without brackets from microarrays; numbers in brackets from qRT-PCR.
Model of the sulfur oxidation system in A. caldus
The role of SOR in elemental sulfur oxidation
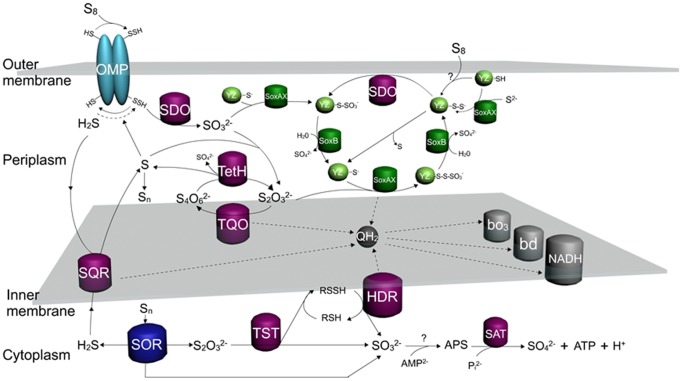
Our experimental data can be explained when SOR is considered a cytoplasmic enzyme oxidizing elemental sulfur in the cytoplasm without coupling with the electron transfer chain or substrate-level phosphorylation in A. caldus. A hypothetical model is shown in Figure 4, sulfur atoms (S) produced from other sulfur oxidation pathways in the periplasm can be accumulated in form of polymeric sulfur (Sn), then S or Sn are transported via an unknown mechanism into the cytoplasm where they are immediately oxidized by SOR. The higher expression level of sor gene in K2S4O6 medium can be explained by cytoplasmic located SOR. K2S4O6 is much easily and quickly entering into the periplasm, where it is hydrolyzed by TetH to produce sulfur atoms and transported into the cytoplasm inducing the expression of sor. While S0 needs to be activated before being transported into the periplasm, which is slower than that of K2S4O6, resulting in the lower expression level of sor in S0 medium. The role of SOR is supported by the facts that the sor (ACAL_1435) gene that encodes a sulfur oxygenase reductase with the ability to oxidize S0, has been cloned and expressed in E. coli and the enzymatic activity of S0 oxidation has been detected without addition of glutathione in our laboratory (unpublished data). The location of SOR in cytoplasm is in agreement with the reports that there are no intracellular sulfur globules in A. caldus, whereas sulfur globules are accumulated in A. ferrooxidans which acks sor gene [36], and SOR is reported existed in the cytoplasm in archaea [10].
Figure 4. Model of sulfur oxidation in A. caldus.
The sulfur oxidation system involves varied sulfur oxidation pathways and the electron transfer system in different cellular compartments. Starting from the extracellular elemental sulfur (S8), it is activated and transported into the periplasmic space as persulfide sulfur (R-SH), and then oxidized by the sulfur dioxygenase (SDO) to produce SO3 2−; SO3 2− can enter into Sox pathway or combine with sulfur atoms to form S2O3 2− via a nonenzymatic reaction; S2O3 2− has two destinies, one is to be oxidized by the Sox pathway, the other is to form S4O6 2− catalyzed by thiosuirate quinine oxidoreductase (TQO); S4O6 2− is hydrolyzed by tetrathionate hydrolase (TetH) producing S2O3 2−, SO4 2−, and S; S produced from hydrolysis of S4O6 2−, oxidation of H2S by sulfide quinone reductase (SQR) or from truncated oxidation of S2O3 2− by the Sox pathway can be accumulated in the form of polymeric sulfur (Sn) in the periplasm and transferred into the cytoplasm; the cytoplasmic elemental sulfur (Sn) is oxidized by sulfur oxygenase reductase (SOR) producing S2O3 2−, SO3 2−, and H2S, which stimulate the cytoplasmic sulfur pathways including the metabolism of S2O3 2− by rhodanese (TST) and heterodisulfidereductase (HDR) and the oxidation of SO3 2− via the APS pathway. Two methods of SoxYZ regeneration are proposed, with one being the sulfur atom is provided from the sulfane intermediate (SoxYZ–S–S−) and the other being oxidation of SoxYZ–S–S− by SDO to complete the Sox sulfur oxidation pathway. Electrons from SQR, TQO, HDR and SoxAX are mediated by the quinol pool in the inner membrane, then are utilized by terminal oxidases bd or bo3 to produce a proton gradient to generate ATP or by the NADH complex I to generate reducing power.
SOR is not coupled with the electron transfer chain or substrate-level phosphorylation. The obvious growth advantage of the Δsor mutant in the first six days in S0 medium or during the whole growth period in K2S4O6 medium (Figure 2) should be caused by the sulfur oxidation pathway shifting from SOR to other pathways coupled with the electron transfer chain. Moreover, it is not an efficient way to produce electrons for sulfur atoms to be oxidized by SOR when sulfur atoms in cytoplasm are insufficient caused by the delay of sulfur activation and transportation at the early growth stage in S0 medium. In contrast, when sulfur atoms are sufficient at the late growth stage in S0 medium, SOR oxidizes the sulfur atoms to produce other sulfur compounds, which enter other sulfur oxidation pathways in the cytoplasm to produce electrons. These could be the reason for the lowered cell growth of the Δsor mutant in S0 medium than the wild type after six days of cultivation (Figure 2A).
SOR plays a central role in the cytoplasmic sulfur oxidation pathways. The possible products from the SOR catalyzed reactions are thiosulfate, sulfite and sulfide [17], [37]. Thiosulfate is considered to be catalyzed by rhodanese (TST) in the cytoplasm of A. caldus according to the reasons: (1) TST widely exists in the cytoplasm of both prokaryotes and eukaryotes, and it transfers a sulfur atom from thiosulfate to thiol compounds producing persulfide and sulfite [27], [38]; (2) there is an obvious relevance between the expressions of sor and rhodanese (rhd) genes. As shown in Figure 3, when K2S4O6 was applied, the expressions of sor and rhd (ACAL_2475, ACAL_2860) showed red and dark colors, respectively, for the wild type, while the expressions of the two rhd genes showed bright green for the Δsor mutant; when S0 was applied, the expressions of the two rhd genes showed a little brighter for the Δsor mutant than that for the wild. Above results showed that the expressions of the two rhd genes down-regulated when sor was deleted. Consequently, a hypothetical thiosulfate oxidation pathway was put forward and shown in Figure 4: the thiol proteins (RSH) in the cytoplasm of A. caldus can be used as sulfur atom acceptors for the catalysis of thiosulfate by TST, producing sulfane sulfate (RSSH) which is used as the substrate of the heterodisulfide reductase complex (HDR), thus a cycle is formed, in which RSH obtains a sulfur atom to form RSSH catalyzed by TST and then RSSH is oxidized by HDR to regenerated RSH. Sulfite is toxic to the cell and needs to be oxidized rapidly. An APS (adenosine-5′-phosphosulfate) pathway for sulfite oxidation maybe exists in A. caldus which is similar to the sulfite oxidation pathway in A. ferrooxidans [23]. A putative sulfate adenylyltransferase dissimilatory-type/denylylsulfate kinase (sat) gene was discovered in A. caldus MTH-04 but the APS reductase gene that catalyzed sulfite to adenosine-5′-phosphosulfate (APS) was not determined (see Figure 4). Sulfide could be converted to hydrogen sulfide and then oxidized by sulfide:quinine reductase (SQR) located in the cytoplasmic membrane. Therefore, it is presumed that SOR plays a central role in the cytoplasmic elemental sulfur oxidation in A. caldus.
Other elemental sulfur oxidation pathways
An important elemental sulfur oxidation pathway in the periplasm near the outer membrane of A. caldus is hypothesized as shown in Figure 4: the extracellular elemental sulfur (S8) is activated by thiol groups of special outer-membrane proteins and transported into the periplasmic space as persulfide sulfur (R-SH), and then oxidized by the sulfur diooxygenase (SDO), meanwhile hydrogen sulfide is produced during the activation of S8. Sulfur diooxygenase gene (sdo) in the A. caldus MTH-04 genome was discovered, cloned and expressed in E. coli in our laboratory, and the enzymatic activity of SDO was detected with the addition of glutathione (data to be published). Although several outer-membrane proteins related to sulfur oxidation in A. ferrooxidans have been reported, the thiol-bearing membrane proteins are not yet identified [39], [40]. Another proposed elemental sulfur oxidation pathway locating in the cytoplasm near the inner membrane is shown in Figure 4: heterodisulfide reductase (HDR) catalyzes sulfane sulfate (RSSH) to produce sulfite and regenerate RSH. Two subunits genes (hdrB and hdrC) of HDR were found in the draft genome sequence of A. caldus MTH-04 but all genes (hdrA, hdrB and hdrC) of HDR were found in A. caldus ATCC 51756 [31].
In summary,there are three elemental sulfur oxidation pathways in A. caldus: (1) the pathway based on SDO in the periplasm; (2) the pathway based on the HDR in cytoplasm near the inner membrane; and (3) the pathway based on the SOR in the cytoplasm.
The Sox pathway
The truncated Sox pathway in A. caldus MTH-04 contains two sox gene clusters, sox operon I (soxX-soxY-soxZ-soxA-hyp-soxB) and sox operon II (coxB-cox-cyoC-hyp-hyp-soxY-soxZ-soxB-hyp-resB-soxX-soxA-resC). There is also a strong connection between the expressions of the sox genes and the terminal oxidase genes. As shown in the columns MT/WT (K2S4O6) and S0/K2S4O6 (WT) in Table 1, the expression of terminal oxidase genes, especially the cytochrome bo3 ubiquinol oxidase genes, upregulated significantly when sox genes had high expression levels. The well-studied Sox pathway in P. pantotrophus demonstrated that it couples the sulfur oxidation with the electron transfer [8]. Besides, the sox operon II together with a bo3 ubiquinol oxidase operon make up a big gene cluster on the chromosome, which implies the feasible regulation and control on their transcriptional level. The hypothetical pathway is shown in Figure 4: electrons produced from the Sox system via QH2 were transferred to the terminal oxidases (bd and bo3) and the NADH complex to produce ATP and NADPH, respectively. However, an important question is raised as to how the truncated Sox system works in A. caldus in the absence of SoxCD. Two possible ways to regenerate SoxYZ are put forward. One way is that the sulfur atom of the sulfane intermediate (SoxYZ–S–S−) is dropped from SoxYZ, which has been reported in other bacteria [10], [41], [42]. Another way is that the sulfur atom of the sulfane intermediate is oxidized by sulfur dioxygenase (SDO), because SoxY is actually a kind of thiol-bearing protein that may be used as an oxidation substrate of SDO. In addition, SoxY is presumed to play a role in the activation of S8 for the reasons that elemental sulfur (S8) assembled on the thiol of SoxY via nonenzymatic conjugation has been reported [10], [42], and the expression of soxY gene of the wild type is much higher in S0 medium than that in K2S4O6 medium (Table 1). Therefore, the Sox system in A. caldus has capabilities to oxidize a wide range of sulfur compounds including sulfite, thiosulfate, sulfide and elemental sulfur and couple the process with electron transfer, which make the Sox pathway play a central role in the periplasmic sulfur oxidation system.
The S4I pathway
The reaction cycle linking the hydrolysis of tetrathionate and the oxidation of thiosulfate is an important sulfur oxidation pathway in the periplasm of A. caldus. Tetrathionate hydrolase (TetH), a soluble periplasmic enzyme, is responsible for the hydrolysis of tetrathionate in A. caldus [43]. One of the hydrolysates of tetrathionate catalyzed by TetH is thiosulfate, but the other products are uncertain [43], [44]. The higher expression level of the sor gene in K2S4O6 medium than that in S0 medium (Table 1) implied that sulfur atoms (S) may be one of the products [44]. Thiosulfate quinine oxidoreduetase (TQO), which is constituted of subunits DoxA and DoxD, catalyzes thiosulfate producing tetrathionate. The expression levels of tetH and doxD of the wild type in S0 medium were much higher than that in K2S4O6 medium (Table 1), suggesting that a mass of S4O6 2− and S2O3 2− were produced during elemental sulfur oxidation. Therefore, the pathways are proposed: S2O3 2− is synthesized by nonenzymatic reaction from sulfite and a sulfur atom, then oxidized by TQO, producing S4O6 2− in the cytomembrane (Figure 4). However, only doxD gene was found in all published A. caldus genomes, while doxA was not found. The thiosulfate oxidation process by TQO was illustrated in A. ambivalens: DoxD catalyzes S2O3 2− to S4O6 2− and gains two electrons, meanwhile, DoxA transfers electrons to the quinine [45]. Both subunits are thought to be constituents of the terminal oxidase and the function of DoxA is to transfer electrons, so it is hypothesized that other terminal oxidase in place of DoxA combines with DoxD to constitute TQO in A. caldus. In addition, due to the instability of S2O3 2− and the stability of S4O6 2− in the acidic environment, the produced S2O3 2− in the periplasms is rapidly oxidized by the Sox system or entered into the S4I pathway. For this reason, formation of S4O6 2− via the S4I pathway may be important for sulfur storage.
Electron transfer from sulfur oxidation to respiratory system
Little is known about the electron transfer chain in A. caldus, but it has been well studied in A. ferrooxidans. One of the electron transfer chains in A. ferrooxidans is that electrons from Fe (II) oxidation flow through Cyc2 to rusticyanin, and then reduce oxygen via a cytochrome aa3 complex (downhill pathway) or reduce NAD+ via a bc1/quinone pool/NADH complex (uphill pathway) [23], [46], [47]. The other one is that electrons from RISCs oxidation are transferred via the quinol pool (QH2) either to terminal oxidases bd, bo3 and aa3 to produce a proton gradient, or to NADH complex I to generate reducing power [23], [46]. A. caldus only has the sulfur oxidation system and its genome information shows that four copies of bd ubiquinol oxidase genes (cydAB), two gene clusters encoding bo3 ubiquinol oxidase (coxB and cyoC) and a cytochrome c oxidase gene (cox, ACAL_2516) exist in the A. caldus MTH-04 draft genome. SoxAX, a c-type cytochrome complex that consists of SoxA as a diheme subunit and SoxX as a monoheme [48], can carry two electrons during the catalyzing process in the Sox pathway. The comparative transcriptome analysis showed that once the expression of genes in Sox pathway upregulated, the expression of bd and bo3 oxidases genes were also upregulated (Table 1), which implied that electrons could be transported from the Sox pathway to bd and bo3 oxidases in A. caldus. Therefore, a hypothesized electron transfer chain based on the quinol pool in A. caldus is depicted in Figure 4: electrons from SQR, TQO, HDR and SoxAX are transferred via the quinol pool either directly to terminal oxidases bd or bo3 to produce the proton gradient to generate ATP, or directly to NADH complex I to generate reducing power. Furthermore, there are several features of the electron transfer in A. caldus including: (1) SoxAX connects the Sox pathway with the electron transfer chain, so that makes the Sox pathway become an important sulfur oxidation pathway for producing mass electrons; (2) Quinol terminal oxidases (bd and bo3) are the dominant terminal oxidases as more electrons are produced from S0 oxidation than that from Fe (II) oxidation; (3) The quinol pool (QH2) located in the cytoplasmic membrane may play a regulatory role in the electron transfer chain. Overall, a powerful electron transfer and respiratory system including the quinol pool (QH2), terminal oxidases and NADH complex exist in A. caldus.
Conclusions
The growth measurements of the wild type and the mutant, and the comparative transcriptome analysis extend our understanding of the complicated sulfur oxidation system in A. caldus. Several distinctive features of the sulfur oxidation system of A. caldus are summarized as follows: (1) The sulfur oxidation system of A. caldus involves the Sox subsystem, a non-Sox sulfur subsystem similar to that in A. ferrooxidans, and SOR which work together to complete the oxidation of elemental sulfur and various sulfur compounds, so that A. caldus has a growth advantage over any other bacteria in the bio-mining reactor [35]. (2) Regional division of the sulfur oxidation system in A. caldus is an obvious characteristic. The first region is the outer membrane of the periplasm where the extracellular elemental sulfur (S8) is activated and transported into the periplasmic space as persulfide sulfur(R-SH). The second region is the periplasm where the Sox pathway, SDO and TetH perform their functions, and this is an important place for sulfur oxidation. Moreover, nonenzymatic reactions producing thiosulfate from sulfite and sulfur atoms and forming polysulfur (Sn) are complementary to the periplasmic sulfur oxidation system. The third region is the cytoplasmic membrane involving SQR, TQO and HDR with a common feature of coupling sulfur oxidation with electron transport. The fourth region is the cytoplasm with an SOR-based sulfur oxidation system. (3) There are complex controls on the sulfur oxidation process. On one hand, the sulfur oxidation related genes belong to different pathways regulated at the transcriptional level to adapt to the production and consumption of various sulfur compounds during the process of elemental sulfur oxidation. On the other hand, metabolic control on the substrate level may be an important regulatory method, as the poly-sulfur (Sn) and poly-thionates (S4O6 2−) accumulated in the periplasm could be the main forms for energy storage.
In summary, an integrated sulfur oxidation model of A. caldus is proposed based on comparative transcriptomic analysis, which provides new insights and guides for the future study of the sulfur oxidation metabolism. In view of the diversity of the RISCs and the complexity of the sulfur oxidation system of A. caldus, many fundamental questions such as identification of sulfur oxidation genes and determination of enzyme activities remain to be resolved. Fortunately, the establishment of genetic manipulation of A. caldus provides effective and powerful tools for elucidation of the sulfur oxidation mechanism of A. caldus [32], [49], [50].
Methods
Bacteria and culture conditions
The bacterial strains and plasmids used are listed in Table 2. The media of Luria broth (LB) or agar plate for E. coli were described in reference [51]. Liquid Starkey-S0 and Starkey-K2S4O6 inorganic medium and solid Starkey-Na2S2O3 medium for cultivation of A. caldus MTH-04 were prepared as described in reference [52]. Elemental sulfur (S0) (boiling sterilized, 20 g/L) and K2S4O6 (membrane filtration, 5 g/L) was added before inoculation. Chloromycetin (Cm) was added to the final concentrations of 34 µg/ml in LB medium, 60 µg/ml in liquid Starkey-S0 medium and solid Starkey-Na2S2O3 medium, respectively. The cultivation temperatures were 37°C for E. coli and 40°C for A. caldus MTH-04. The shaking speed for liquid cultivation of A. caldus MTH-04 was 125 r/min if not specifically stated.
Table 2. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Genotype or description | Source or reference |
| A. caldus MTH-04 | Isolated from Tengchong area, Yunnan province, China | [58] |
| A. caldus MTH-04 Δsor mutant | Δsor | This study |
| E. coli DH5α | F−φ80d lacZΔM15Δ(lacZYA-argF) U169 end A1 recA1 hsdR17(rk −,mk +) supE44λ-thi-1 gyr96 relA1 phoA | TransGen Biotech Corp. China |
| Plasmids | ||
| pSIMPLE19 EcoR V/BAP | Apr; lacZ′; ColE1 replicon; blunt-tailed PCR product cloning vector | TaKaRa Biotechnology Co. China |
| pSIMPLE19hsdM::Ω-Cm | Apr Cmr; suicide plasmid containing the hsdM gene inserted by the chloromycetin resistance gene | Our laboratory |
| pSDU1 | Cmr; IncQ; mob+ | Our laboratory |
| pJRD215-tac-sor | Smr, Kmr; IncQ, Mob+; tac promoter;sor | Our laboratory |
| pMD19sor::Ω-Cm | Apr Cmr; suicide plasmid containing the sor gene inserted by the chloromycetin resistance gene | This study |
Generation of mutant
The suicide plasmid was constructed as follows. The essential part of the suicide plasmid was amplified from pSIMPLE19hsdM::Ω-Cm using the primers of PMD fwd and PMD rev, then digested with Sal I and Not I. A 2.6 kbp homologous sequence arm holding the sor gene was amplified from A. caldus MTH-04 chromosome using the primers of Whol fwd and Whol rev, digested with Sal I and Not I, and ligated to the essential part of the suicide plasmid. The resulting plasmid was named pMD19sor, then linearized in the middle of the sor gene, using the primers of Mid fwd and Mid rev. The linear plasmid pMD19sor was digested with Kpn I and Bgl II, and ligated to the chloramphenicol resistance gene (cat) amplified from plasmid pSDU1 using primers of Cat fwd and Cat rev digested with Kpn I and Bgl II. The generated plasmid carrying the mutant allele of sor disrupted by the cat was the suicide plasmid pMD19sor::Ω-Cm. The suicide plasmid was sequenced by Invitrogen Corp. for sequence confirmation. The sequences of the primers are listed in Table 3. The restriction enzymes and Prime STAR HS Taq were purchased from TaKaRa Corp. The suicide plasmid was electroporated into A. caldus MTH-04 using the methods described in the reference [49].
Table 3. Primers used in constructing the suicide plasmid and the mutant.
| Name | Sequence |
| PMD fwd | ATAAGAATGCGGCCGCGCGGTAATACGGTTATCCAC |
| PMD rev | TCCGGAATTCGCGTCGACAATGGTTTCTTAGACGTCAGGT |
| Wholfwd | TCCGGAATTCGCGTCGACGTGAACACCGTGATCTTGTCC |
| Whol rev | ATAAGAATGCGGCCGCTCACAGTTCCGACGTTTTCAGT |
| Mid fwd | TCGGGGTACCGTTCCGATTTACCCAGGATCA |
| Mid rev | TTGGAAGATCTCTTCGATGACCATATCGAGACAG |
| Cat fwd | CTGAAGATCTTCATGTTTGACAGCT |
| Cat rev | TCGGGGTACCATTCATCCGCTTATTATCACTT |
| Clnfwd | TCCGCTCGAGGGTACCGTCTTAGAGCAAAGCGCCTGT |
| Cln rev | CTAGTCTAGAGCCGTAATCGGCGGAGTTTAT |
| SorAfwd | ATCCCATGGCCGGTCGTTAT |
| SorA rev | TGGTTGTGGATTTGTCGCAG |
| SorBfwd | ATCAGCTTGGAGGCAATGTG |
| SorB rev | ATGGTCTTGGGCTCTTGGTC |
| Big fwd | CGAGTCCCGCCCATTGTT |
| Big rev | TACAGGTAGAAGGCTTCACC |
Colony PCR was used to screen the mutants using the primers of Clnfwd and Cln rev. The mutants were incubated in liquid Starkey-S0 medium, and then collected to extract the chromosome. The chromosomes of the mutants were analyzed using PCR. Firstly, two sets of primers (SorA fwd and SorA rev, SorB fwd and SorB rev) specific to the sor gene were used to verify the mutants. Then, PCR amplification was done using primers of Big fwd and Big rev specific to the mutant allele using LA taq (TaKaRa Corp.) and the amplified fragments were sequenced by Invitrogen Corp. for sequence confirmation. The sequences of the primers used are listed in Table 3.
Southern hybridization was done to further confirm the mutants. Genomic DNA of the wild type and the mutants were extracted using a QIAmp® DNA Mini kit (Qiagen Corp.), digested with EcoRI(NEB Corp.), electrophoresed on agarose gel, and transferred by capillary blotting to positively charged Hybond-N membranes (Roche Corp.). The sor probe was obtained by incorporation of alkali-labile DIG-dUTP (Roche Corp.) during PCR elongation with primers SorAfwd and SorA rev. Hybridization was carried out under stringent conditions as recommended by the manufacturer.
Growth measurements of the wild type and the Δsor mutant
A. caldus MTH-04 and the Δsor mutant were inoculated into 150 ml of fresh Starkey-S0 medium with bacterial loading at 10% (vol/vol). Cells were harvested at the stationary growth phase. The solid sulfur in the cultivation broth was removed by low-speed centrifugation (100× g) before cell harvesting. Then, the cells were harvested by high-speed centrifugation (10,000× g), washed twice using deionized distilled H2O (ddH2O), and diluted to the final concentration with OD600 at 1.0. An aliquot (400 µl) of the diluted cells was inoculated into 150 ml Starkey-S0 or Starkey-K2S4O6 medium and incubated at 40°C, 125 r/min. Each condition was replicated three times and the medium without inoculation was used as the control. An aliquot (250 µl) of each sample was taken, stand for 5 minutes, and then 200 µl was taken from each of the 250 µl sample and used for OD measurement using a Microplate Spectrophotometer (Molecular Devices) at the wavelength of 600 nm. Samples were taken and measured every 24 hours until the 12th day.
Construction of the whole-genome microarray
An A. caldus genome 60 bp oligonucleotide microarray was obtained from CapitalBio Corporation (Beijing, China) [53]. Briefly, the A. caldus genome oligonucleotide set consisting of 5′ amino acid-modified 60 bp probes and representing 3,603 ORFs was synthesized by Biosune Corporation. The 3,603 ORFs consist of 2,972 ORFs from A. caldus MTH-04 and 631 ORFs from A. caldus ATCC 51756 (GenBank: ACVD00000000). Then oligonucleotides were dissolved in EasyArray™ spotting solution (CapitalBio Corp.) at 40 µM concentration, and printed in triplicate on PolymerSlide, in which the surface is covered by a thin layer of aldehyde group modified three-dimensional polymer chain (CapitalBio Corp.). On the slide, there are 48 blocks and each block has 18 columns and 15 rows.
Design of the hybridization scheme on the gene chips
A complex hybridization scheme was designed using two-color platforms and common reference samples to determine the levels of the differential expressions between the group pairs [53]. As shown in Table 4, there were four experimental groups: WT (S0): the wild type in S0 medium, WT (K2S4O6): the wild type in K2S4O6 medium, MT (S0): the mutant inS0 medium, and MT (K2S4O6): the mutant in K2S4O6 medium. Each group contained triplicate biological repeats, so there were totally 12 samples. An aliquot of each sample was taken, mixed and used as common reference. Samples labeled with Cy5-dCTP and common reference labeled with Cy3-dCTP were hybridized on a chip. The gene expression level in the samples was determined based on the common reference. Therefore, there were four cases based on group pairs, which were the gene differential expression in S0 medium between the mutant and the wild type (MT/WT (S0)), the gene differential expression in K2S4O6 medium between the mutant and the wild type (MT/WT (K2S4O6)), the gene differential expression of the wild type between S0 medium and K2S4O6 medium (S0/K2S4O6 (WT)), and the gene differential expression of the mutant between S0 medium and K2S4O6 medium (S0/K2S4O6 (MT)).
Table 4. The hybridization scheme on the gene chips.
| Number | Hybridization on the chip | Group | Ratio |
| 1 | WT1(S0) vs reference | ||
| 2 | WT2(S0) vs reference | WT(S0) | |
| 3 | WT3(S0) vs reference | ||
| 4 | WT1(K2S4O6) vs reference | ||
| 5 | WT2(K2S4O6) vs reference | WT (K2S4O6) | (1) MT/WT (S0) |
| 6 | WT3(K2S4O6) vs reference | (2) MT/WT (K2S4O6) | |
| 7 | MT1(S0) vs reference | (3) S0/K2S4O6 (WT) | |
| 8 | MT2(S0) vs reference | MT (S0) | (4) S0/K2S4O6(MT) |
| 9 | MT3(S0) vs reference | ||
| 10 | MT1(K2S4O6) vs reference | ||
| 11 | MT2(K2S4O6) vs reference | MT (K2S4O6) | |
| 12 | MT3(K2S4O6) vs reference |
Note: W(S0): the wild type in Starkey-S0 medium with three repeats W1(S0), W2(S0) and W3(S0); W(S4): the wild type in Starkey-K2S4O6 medium with three repeats W1(S4), W2(S4) and W3(S4); M(S0): the Δsor mutant in Starkey-S0 medium with three repeats M1(S0), M2(S0) and M3(S0); M(S4): the Δsor mutant in Starkey-K2S4O6 medium with three repeats M1(S4), M2(S4) and M3(S4); reference:the mixed RNA of the 12 samples.
RNA isolation and microarray experiments
The wild type and the Δsor mutant were cultured as described in the section of growth measurements of the wild type and the Δsor mutant. Cells were collected at exponential growth phase (4th day), washed twice using sterile RNase-free ddH2O and treated with RNA protect Bacteria Reagent (Qiagen Corp.). Total RNA was extracted using RNeasy Mini Kit (Qiagen Corp.) in accordance with the manufacturer's instructions.
Total RNA was purified using Nucleospin® RNA Clean-up (MN Corp.) and tailed with polyA using Poly (A) Polymerase (Ambion Corp.). The tailed RNA was used to produce cy5/cy3-labeled cDNA employing an RNA amplification strategy (CapitalBio eukaryotic RNA Amplification and Labeling Kit, CapitalBio Corp.). Cy5/cy3-labeled cDNA were hybridized with the microarray at 42°C for 16 h. Following hybridization, the arrays were washed using two consecutive solutions (0.2% SDS, 2× SSC at 42°C for 5 min, and 0.2× SSC for 5 min at room temperature). Arrays were scanned using a confocal LuxScan™scanner and the images obtained were then analyzed using LuxScan™ 3.0 software (both from CapitalBio Corp.). For individual channel data extraction, faint spots with intensities below 400 units after background subtraction in both channels (Cy3 and Cy5) were removed. A space- and intensity-dependent normalization based on a LOWESS program was employed [54]. The raw data were Log 2 transformed and median centered by arrays and genes using the Adjust Data function of CLUSTER 3.0 software and then further analyzed using hierarchical clustering with average linkage [55]. To determine the significant differentially expressed genes whose ratio changes ≥1.5 fold with p≤0.05, Significance Analysis of Microarrays (SAM, version 3.02) was performed using two-class unpaired comparison in the SAM procedure [56].
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA was extracted as described above and was treated with RNase-Free DNase Set (Qiagen Corp.) to eliminate the traces of genomic DNA. The total RNA of 2 µg was reversely transcribed using M-MLV Reverse Transcriptase (Invitrogen Corp.) under the following conditions: 25°C for 10 min, 37°C for 60 min, and 70°C for 10 min. RT reaction products of 1 µl were used for PCR amplification using Power SYBR Green PCR Master Mix (Applied Biosystems). The conditions for the PCR reaction were as follows: 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min using a 7900 HT Fast RealTime PCR system (Applied Biosystems). The glyceraldehyde-3-phosphate dehydrogenase gene (gapdh) was used as the reference gene for normalization. The relative expression was calculated using the comparative ΔΔCT method, and the values were expressed as 2−ΔΔCT [57]. Primers used in this study are listed in Table 5.
Table 5. Primers used in qRT-PCR.
| Primers | Sequence | |
| soxX(2486) | Fwd | GGTCGGGCTATTGCCTTTG |
| Rev | TGGTCTGGAACATCTGCTGG | |
| soxY(2487) | Fwd | CGTATCGCCCAAGGTGAAG |
| Rev | CCAGACGGTGCTGACGTAATC | |
| soxZ(2488) | Fwd | GTGGAAGTCCGCTCCCTGA |
| Rev | ACGCTGACGGTCTGAATGAAG | |
| soxA(2489) | Fwd | CAGACCCTCAAAGAATCCCG |
| Rev | CTCCACCTGATGAGTCTTGTTGTC | |
| soxB(2491) | Fwd | AGGACCCATACACCATACCTGAC |
| Rev | TTGAGGACGGAGCCTACGC | |
| soxY(2520) | Fwd | ACAGCATTGGCAAGACCTCC |
| Rev | AACTCACCCTTGTTCGTCCG | |
| soxZ(2521) | Fwd | CAAACCCCTGTGCCATCTG |
| Rev | ATTGTCCTTCCAAGTCATCGC | |
| soxB(2522) | Fwd | GGATTACTACGGCATCAAAGCA |
| Rev | ACCCAACTGTTCACGATAGCG | |
| soxX(2525) | Fwd | GTACAGGCGGGCAATGTTG |
| Rev | CGTCAAGACCTTATCCTTACCAAA | |
| soxA(2526) | Fwd | CTATCAGCAGTATCAAGAGGCAATC |
| Rev | CGGAAAGCAGGACGCATAG | |
| tetH(1013) | Fwd | GGCTTCAACGCCAAAACTG |
| Rev | GGCATCGTAGTCCGAGGTCA | |
| doxD(1014) | Fwd | TGTATGCTGGCGTCATTATCTTTA |
| Rev | CGAAACCATCCCTTCTCCG | |
| sor(1435) | Fwd | ACGGTGTATCGCCCTTGGT |
| Rev | GGATTCGCTCCTAAAGTCGC | |
| sqr(1436) | Fwd | TTTTATCCAGGCGGAAGTGC |
| Rev | TTGTTCCCATAGTAGAAATCGGTG | |
| hdrC(1042) | Fwd | GTACCCTGACGGACTACGAGC |
| Rev | GCCCAAGCGGATGAGGTAG | |
| hdrB(1043) | Fwd | AGCTCGTGCTCAATATCCTCC |
| Rev | TGATGCCAAATTCCACTTCG | |
| drsE(2473) | Fwd | AAGCAAAATGAAGGCGAAGG |
| Rev | CGAAGAGGTCCACCGTCATC | |
| tusA(2474) | Fwd | TCCTGAAAGTCGTTGCCACC |
| Rev | TTCCGCCTGATCCAGAAGC | |
| rhd(2475) | Fwd | CGGGCAGAATTTTCACCTCA |
| Rev | CCGCTCAGACAATAGAACACGA | |
| rhd(2860) | Fwd | GGCTCATCAACGACCACGA |
| Rev | CGGCAGATACTTACCCACCTC | |
| gapdh(2603) | Fwd | ACGTCTCCATCGTCGATCTCA |
| Rev | AGGGCTTGTCGTTGTAGGCA | |
Acknowledgments
The authors would like to thank Dr. Pamela Holt of Shandong University for proofreading the manuscript.
Funding Statement
This research was supported by the National Basic Research Program (http://www.973.gov.cn)(2010CB630902, 2004CB619202), the National Natural Science Foundation (http://www.nsfc.gov.cn)(31070034), and the Knowledge Innovation Program of CAS (http://www.cas.cn)(2AKSCX2-YW-JS401) of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hallberg KB, Lindström EB (1994) Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology 140: 3451–3456. [DOI] [PubMed] [Google Scholar]
- 2. Hallberg KB, Lindström EB (1996) Multiple serotypes of the moderate thermophile Thiobacillus caldus, a limitation of immunological assays for biomining microorganisms. Appl Environ Microbiol 62: 4243–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Okibe N, Gericke M, Hallberg KB, Johnson DB (2003) Enumeration and characterization of acidophilic microorganisms isolated from apilot plant stirred-tank bioleaching operation. Appl Environ Microbiol 69: 1936–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rawlings DE (1998) Industrial practice and the biology of leaching of metals from ores. J Ind Microbiol Biotechnol 20: 268–274. [Google Scholar]
- 5. Dopson M, Lindström EB (1999) Potential role of Thiobacillus caldus in arsenopyrite bioleaching. Appl Environ Microbiol 65: 36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kamimura K, Okayama T, Murakami K, Sugio T (1999) Isolation and characterization of a moderately thermophilic sulfur-oxidizing bacterium. Microbios 99: 7–18. [Google Scholar]
- 7. Edwards KJ, Bond PL, Banfield JF (2000) Characteristics of attachment and growth of Thiobacillus caldus on sulphide minerals: A chemotactic response to sulphur minerals? Environ Microbiol 2: 324–332. [DOI] [PubMed] [Google Scholar]
- 8. Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J (2005) Prokaryotic sulfur oxidation. Curr Opin Microbiol 8: 253–259. [DOI] [PubMed] [Google Scholar]
- 9. Frigaard N, Dahl C (2009) Sulfur metabolism in phototrophic sulfur bacteria. Adv Microb Physiol 54: 103–200. [DOI] [PubMed] [Google Scholar]
- 10. Ghosh W, Dam B (2009) Biochemistry and molecular biology of lithotrophic sulfur oxidation by taxonomically and ecologically diverse bacteria and archaea. FEMS Microbiol Rev 33: 999–1043. [DOI] [PubMed] [Google Scholar]
- 11. Rainey FA, Kelly DP, Stackebrand E, Burghard J, Hiraishi A, et al. (1999) A re-evaluation of the taxonomy of Paracoccus denitrificans and a proposal for the combination Paracoccus pantotrophus comb. nov. Int J Syst Bacteriol 2: 645–651. [DOI] [PubMed] [Google Scholar]
- 12. Friedrich CG, Rother D, Bardischewsky F, Quentmeie A, Fischer J (2001) Oxidation of Reduced Inorganic Sulfur Compounds by Bacteria:Emergence of a Common Mechanism? Appl Environ Microbiol 67: 2873–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Friedrich CG, Quentmeier A, Bardischewsky F, Rother D, Kraft R, et al. (2000) Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J Bacteriol 182: 4677–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quentmeier A, Friedrich CG (2001) The cysteine residue of the SoxY protein as the active site of protein-bound sulfur oxidation of Paracoccus pantotrophus GB17. FEBS Lett 503: 168–172. [DOI] [PubMed] [Google Scholar]
- 15. Kletzin A, Urich T, Müller F, Bandeiras TM, Gomes CM (2004) Dissimilatory Oxidation and Reduction of Elemental Sulfur in Thermophilic Archaea. J Bioenerg Biomembr 36: 77–91. [DOI] [PubMed] [Google Scholar]
- 16. Kletzin A (1989) Coupled enzymatic production of sulfite,thiosulfate, and hydrogen sulfide from sulfur: purification and properties of a sulfur oxygenase reductase from the facultatively anaerobic archaebacterium Desulfurolobus ambivalens . J Bacteriol 171: 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kletzin A (1992) Molecular characterization of the sor gene,which encodes the sulfur oxygenase/reductase of the thermoacidophilic Archaeum Desulfurolobus ambivalens . J Bacteriol 174: 5854–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Urich T, Bandeiras TM, Leal SS, Rachel R, Albrech T, et al. (2004) The sulphur oxygenase reductase from Acidianus ambivalens is a multimeric protein containing a low-potential mononuclear non-haem iron centre. Biochem J 381: 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steudel R (2000) The chemical sulfur cycle. London: IWA Publishing. 1–31 p. [Google Scholar]
- 20. Rohwerder T, Sand W (2003) The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp . Microbiology 149: 1699–1709. [DOI] [PubMed] [Google Scholar]
- 21. Rohwerder T, Sand W (2007) Oxidation of inorganic sulfur compounds in acidophilic prokaryotes. Eng Life Sci 7: 301–309. [Google Scholar]
- 22.Sugio T, Ochi K, Muraoka T, Takeuchi F, Kanao T (2005) Isolation and some properties of sulfur dioxygenase from Acidithiobacillus thiooxidansNB1-3. In: Harrison STL, Rawlings DE, Petersen, editors. Proceedings of the 16th international biohydromet. Symposium, cape town, South Africa. compress, pp 25–29. [Google Scholar]
- 23. Quatrini R, Appia-Ayme C, Denis Y, Jedlicki E, Holmes DS, et al. (2009) Extending the models for iron and sulfur oxidation in the extreme Acidophile Acidithiobacillus ferrooxidans . BMC Genomics 10: 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hallberg KB, Dopson M, Lindstrom EB (1996) Reduced sulfur compound oxidation by Thiobacillus caldus . J Bacteriol 178: 6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wakai S, Kikumoto M, Kanao T, Kamimura K (2004) Involvement of sulfide:quinone oxidoreductase in sulfur oxidation of an acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans NASF-1. Biosci Biotechnol Biochem 68: 2519–2528. [DOI] [PubMed] [Google Scholar]
- 26. Gardner MN, Rawlings DE (2000) Production of rhodanese by bacteria present in bio-oxidation plants used to recover gold from arsenopyrite concentrates. J Appl Microbiol 89: 185–190. [DOI] [PubMed] [Google Scholar]
- 27. Schlesinger P, Westley J (1974) An expanded mechanism for rhodanese catalysi. J Biol Chem 249: 780–788. [PubMed] [Google Scholar]
- 28. Valdés J, Pedroso I, Quatrini R, Holmes DS (2008) Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: Insights into their metabolism and ecophysiology. Hydrometallurgy 94: 180–184. [Google Scholar]
- 29. Valdés J, Quatrini R, Hallberg K, Dopson M, Valenzuela P, et al. (2009) Genome announcement: draft genome sequence of the extremely acidophilic bacterium Acidithiobacillus acldus ATCC 51756 reveals metabolic versatility in the genus Acidithiobacillus . J Bacteriol 191: 5877–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen ZW, Liu YY, Wu JF, She Q, Jiang CY, et al. (2007) Novel bacterial sulfur oxygenase reductases from bioreactors treating gold-bearing concentrates. Appl Microbiol Biotechnol 74: 688–698. [DOI] [PubMed] [Google Scholar]
- 31. Mangold S, Valdés J, Holmes DS, Dopson M (2011) Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front Microbiol 2: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leonardo JvZ, Jolanda MvM, Rawlings DE (2008) Construction of arsB and tetH Mutants of the sulfur-oxidizing bacterium Acidithiobacillus caldus by Marker Exchange. Appl Environ Microbiol 74: 5686–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cabrejos M-E, Zhao HL, Guacucano M, Bueno S, Levican G, et al. (1999) IST1 insertional inactivation of the resB gene: implications for phenotypic switching in Thiobacillus ferrooxidans . FEMS Microbiol Lett 175: 223–229. [DOI] [PubMed] [Google Scholar]
- 34. Schrader JA, Holmes DS (1988) Phenotypic Switching of Thiobacillus ferrooxidans . J Bacteriol 170: 3915–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. You XY, Guo X, Zheng HJ, Zhang MJ, Liua LJ, et al. (2011) Unraveling the Acidithiobacillus caldus complete genome and its central metabolisms for carbon assimilation. J Genet Genomics 38: 243–252. [DOI] [PubMed] [Google Scholar]
- 36.Huan H (2009) The study on the speciation and transformation of elemental sulfur mediated by typical acidophilic sulfur-oxidizing microbes [D]. Changsha: Central South University. [Google Scholar]
- 37. Sun CW, Chen ZW, He ZG (2003) Purification and properties of the sulfur oxygenase/reductase from the acidothermophilic archaeon Acidianus strain S5. Extremophiles 7: 131–134. [DOI] [PubMed] [Google Scholar]
- 38. Billaut-Laden I, Allorge D, Crunelle-Thibaut A, Rat E, Cauffiez C, et al. (2006) Evidence for a functional genetic polymorphism of the human thiosulfate sulfurtransferase (Rhodanese), a cyanideand H2S detoxification enzyme. Toxicology 225: 1–11. [DOI] [PubMed] [Google Scholar]
- 39. Ramírez P, Guiliani N, Valenzuela L, Beard S, Jerez CA (2004) Differential protein expression during growth of Acidithiobacillus ferrooxidans on ferrous iron, sulfur compounds, or metal sulfides. Appl Environ Microbiol 70: 4491–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buonfiglio V, Polidoro M, Flora L, Citro G, Valenti P, et al. (1993) Identification of two outer membrane proteins involved in the oxidation of sulphur compounds in Thiobacillus ferrooxidans . FEMS Microbiol Rev 11: 43–50. [Google Scholar]
- 41. Dahl C, Prange A (2006) Bacterial Sulfur Globules: Occurrence, Structure and Metabolism. Microbiol Monogr 1: 21–51. [Google Scholar]
- 42. Sauvé VR, Bruno S, Berks BC, Hemmings A (2007) The SoxYZ Complex Carries Sulfur Cycle Intermediates on a Peptide Swinging Arm. J Biol Chem 282: 23194–23204. [DOI] [PubMed] [Google Scholar]
- 43. Bugaytsova Z, Lindström EB (2004) Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus . Eur J Biochem 271: 272–280. [DOI] [PubMed] [Google Scholar]
- 44. Meulenberg R, Pronk JT, Hazeu W, Bos P, Kuenen JG (1992) Oxidation of reduced sulphur compounds by intact cells of Thiobacillus acidophilus . Arch Microbiol 157: 161–168. [Google Scholar]
- 45. Mueller FH, Bandeiras TM, Urich T, Teixeira M, Gomes CM, et al. (2004) Coupling of the pathway of sulphur oxidation to dioxygen reduction: characterization of a novel membrane-bound thiosulphate: quinone oxidoreductase. Mol Microbiol 53: 1147–1160. [DOI] [PubMed] [Google Scholar]
- 46. Valdés J, Pedroso I, Quatrini R, Dodson RJ, Tettelin H, et al. (2008) Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9: 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Castelle C, Guiral M, Malarte G, Ledgham F, Leroy G, et al. (2008) A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes isolated from the extreme acidophile Acidithiobacillus ferrooxidans . J Biol Chem 283: 25803–25811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dambe T, Quentmeier A, Rother D, Friedrich C, Scheidig AJ (2005) Structure of the cytochrome complex SoxXA of Paracoccus pantotrophus, a heme enzyme initiating chemotrophic sulfur oxidation. J Struct Biol 152: 229–234. [DOI] [PubMed] [Google Scholar]
- 49. Chen L, Lin J, Li B, Lin J, Liu X (2010) Method Development for Electrotransformation of Acidithiobacillus caldus . J Microbiol Biotechnol 20: 39–44. [PubMed] [Google Scholar]
- 50. Liu XM, Lin JQ, Zhang Z, Bian J, Zhao Q (2007) Construction of conjugative gene transfer system between E. coli an moderately thermophilic, extremely acidophilic Acidithiobacillus caldus MTH-04. J Microbiol Biotechnol 17: 162–167. [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual., 2nd ed.; Harbor CS, editor. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- 52. Jin SM, Yan WM, Wang ZN (1992) Transfer of IncP plasmids to extremely acidophilic Thiobacillus thiooxidans . Appl Environ Microbiol 58: 429–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Patterson TA, Lobenhofer EK, Fulmer-Smentek SB, Collins PJ, Chu TM, et al. (2006) Performance comparison of one-color and two-color platforms within the MicroArray Quality Control (MAQC) project. Nat Biotechnol 24: 1140–1150. [DOI] [PubMed] [Google Scholar]
- 54. Yang YH, Dudoit S, Luu P, Lin DM, Peng V, et al. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. ProcNatl Acad Sci 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Livaka KJ, Schmittgenb TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta] CT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 58. Liu Y, Qi FJ, Lin JQ, Tian KL, Yan WM (2004) Isolation and phylogenetic analysis of a moderately thermophilic acidophilic sulfur oxidizing bacterium. Acta Microbiol Sin 44: 382–385. [Google Scholar]