Abstract
Opitz syndrome (OS) is a human genetic disease characterized by deformities such as cleft palate that are attributable to defects in embryonic development at the midline. Gene mapping has identified OS mutations within a protein called Mid1. Wild-type Mid1 predominantly colocalizes with microtubules, in contrast to mutant versions of Mid1 that appear clustered in the cytosol. Using yeast two-hybrid screening, we found that the α4-subunit of protein phosphatases 2A/4/6 binds Mid1. Epitope-tagged α4 coimmunoprecipitated endogenous or coexpressed Mid1 from COS7 cells, and this required only the conserved C-terminal region of α4. Localization of Mid1 and α4 was influenced by one another in transiently transfected cells. Mid1 could recruit α4 onto microtubules, and high levels of α4 could displace Mid1 into the cytosol. Metabolic 32P labeling of cells showed that Mid1 is a phosphoprotein, and coexpression of full-length α4 decreased Mid1 phosphorylation, indicative of a functional interaction. Association of green fluorescent protein–Mid1 with microtubules in living cells was perturbed by inhibitors of MAP kinase activation. The conclusion is that Mid1 association with microtubules, which seems important for normal midline development, is regulated by dynamic phosphorylation involving MAP kinase and protein phosphatase that is targeted specifically to Mid1 by α4. Human birth defects may result from environmental or genetic disruption of this regulatory cycle.
The Opitz G/BBB syndrome (OS), also known as the hypospadias–dysphagia syndrome or telecanthus with associated abnormalities, is associated with midline abnormalities such as cleft lip, laryngeal cleft, heart defects, hypertelorism, hypospadias, imperforate anus, agenesis of the corpus callosum, and developmental delay (1). It was demonstrated that OS is a heterogeneous disorder, with X linked and autosomal forms. No phenotypic differences between the two linkage types were discerned except that anteverted nares and posterior pharyngeal cleft were seen only in the X linked form (2).
The study (1) of a family in which OS segregated with an X chromosome inversion revealed the position of the gene on Xp22, which was referred to as Mid1 (for midline-1). The Mid1 gene encodes a 667-aa protein that is expressed ubiquitously in both embryonic and adult tissues. The prominent expression of Mid1 in undifferentiated cells in the central nervous, gastrointestinal, and urogenital systems suggested that abnormal cell proliferation and/or migration may underlie the defect in midline development characteristic of OS (3).
The Mid1 protein belongs to a RING finger family of nuclear factors that contain protein–protein interaction domains and have been implicated in fundamental processes such as body-axis patterning and cell transformation. Besides the N-terminal RING finger domain, Mid1 also contains four additional domains: two potential zinc-binding B box domains, a leucine coiled-coil domain characteristic of the “RING-B box-coiled coil” subgroup of RING finger proteins, and a RFP-like C-terminal domain. OS-associated mutations have been identified in both C- and N-terminal regions of Mid1 (2).
Previous studies showed that Mid1 is a microtubule-associated protein that may influence microtubule dynamics in Mid1-overexpressing cells (4, 5). Mid1 is associated with microtubules throughout the cell cycle, colocalizing with cytoplasmic fibers in interphase and with mitotic spindle and midbodies during mitosis and cytokinesis (5). Overexpressed Mid1 proteins harboring C-terminal mutations described in OS patients lack the ability to associate with microtubules, forming cytoplasmic clumps instead (4). Thus, the inability to associate with microtubules is one possible molecular mechanism underlying the OS phenotype. However, the cellular function of Mid1 and the mechanism to control Mid1 subcellular localization are still unknown.
The α4 protein originally was discovered as a phosphoprotein associated with Ig receptor in B cells (6). It shares 28% sequence identity with the yeast Tap42 protein, which functions as part of the yeast target of rapamycin (TOR)-signaling pathway (7–12). Yeast Tap42 binds protein phosphatases Sit4 and Pph21/22 that correspond to mammalian PP6 and PP2A, respectively (7), and binding is independent of Tpd3, the PP2A yeast A subunit (7, 12). Likewise, mammalian α4 associates with the C subunit of PP2A in the absence of A (PR65) subunit (13, 14). In yeast, Tap42-phosphatase association has been shown to be rapamycin-sensitive (7), consistent with a report of TOR-mediated phosphorylation of Tap42 (12). Although this pathway presumably links nutrient status to protein synthesis, the targets of Tap42 phosphatases have not been elucidated. Overexpression of mouse FLAG-α4 in COS cells promoted α4-phosphatase formation and caused dephosphorylation of elongation factor-2, which is activated by dephosphorylation (15). In the present study, we set out to identify proteins that bind directly to α4 and, therefore, might be cellular substrates of this form of phosphatase. Yeast two-hybrid screening with mouse α4 as bait yielded multiple clones of Mid1 as a strong interactor. We demonstrate that the C-terminal 120 residues of α4 are sufficient for binding to Mid1 and expression of FLAG-α4 in cells promotes the dephosphorylation of Mid1 and dissociation of Mid1 from microtubules. This uncovers a novel function for α4 phosphatase in the regulation of Mid1 localization and posits a role for α4 in midline development.
Materials and Methods
Two-Hybrid Screen.
The screen was done by using full-length murine-α4 in pGBT10 vector (BamHI-EcoRI), a derivative of pGBT9 designed and provided by Ian Macara (University of Virginia), that contains the Gal4 DNA-binding domain. Bait was transformed into the yeast strain HF7c by using the lithium acetate method and grown further in synthetic medium lacking tryptophan (16). The library screen was done by using a 9-day embryonic mouse cDNA library cloned into pVP16 (created by Stan Hollenberg, Fred Hutchinson Cancer Center, Seattle). Library transformation was done in a sequential manner, such that 6 × 106 independent clones were tested and plated on synthetic medium containing 10 mM 3-aminotriazole (Sigma) and lacking histidine, leucine, and tryptophan. Positive clones were determined by growth on plates and β-galactosidase filter assays. Clones were rescued by electroporation into Escherichia coli HB101 and grown on M9 plates lacking leucine, which allowed for analysis of positives by transformation tests and DNA sequencing.
Plasmids and Mutagenesis.
Mid1 full-length cDNA was derived from c-Myc/green fluorescent protein (GFP)-Mid1 (5) by PCR amplification. GFP-Mid1 was constructed by placing Mid1 cDNA in-frame in the EcoRI site of pEGFP-C3 vector. FLAG-α4 was constructed as described previously (13). FLAG-α4(1–111), FLAG-α4(1–249), and all mutated forms of GFP-Mid1 were generated by using the Stratagene Quick Change Mutagenesis Kit according to the manufacturer's protocol. FLAG-α4(220–340) was obtained by PCR as described previously (17).
Cell Culture, Transfection, Immunoprecipitation, and Immunoblotting.
COS7 cells were maintained in DMEM (Life Technologies, Gaithersburg, MD) containing 10% newborn calf serum (Life Technologies) at 37°C, 5% CO2. Cells were grown to 50–60% confluency and transfected for 18 h by using FuGene 6 reagent (Roche Diagnostics) according to the manufacturer's instructions. Usually, cells in 60-mm dishes were transfected with 2 μg of plasmid DNA, and 1 μg of DNA was used for cells in 35-mm dishes. Immunoprecipitation with anti-FLAG M2 affinity gel (Sigma) or anti-GFP-conjugated Agarose (Santa Cruz ) and immunoblotting were done as described previously (18).
Metabolic [32P] Labeling.
COS7 cells in 60-mm dishes were transfected for 18 h and then incubated for 3 h in phosphate-free DMEM supplemented with dialyzed newborn calf serum. Cells then were labeled for 1.5 h in the same medium containing 1 mCi/ml [32P]orthophosphate (NEN).
Fluorescence Microscopy.
COS7 cells were grown on fibronectin-coated coverslips in 35-mm dishes. After transfection, cells were washed with 1.2× PEM buffer (120 mM Pipes, pH 7.0/6 mM EGTA/2.4 mM MgCl2) and then fixed with 3.5% paraformaldehyde for 15 min. FLAG-α4 and microtubules were stained with anti-FLAG M2 and anti-β-tubulin antibodies plus a tetramethylrhodamine B isothiocyanate-labeled secondary antibody (Sigma). Nuclei were stained by using the DNA-specific stain 4′,6-diamidino-2-phenylindole (DAPI). Images were acquired on a Nikon Microphot-SA equipped with a Hamamatsu C4742 charge-coupled device camera driven by openlab 2.0.6 (Improvision, Lexington, MA) software. Images were processed in Adobe photoshop 5.5 (Adobe Systems, Mountain View, CA).
GFP-Mid1 Localization in Living Cells.
COS7 cells were plated onto fibronectin-coated 22 × 22-mm coverglasses and transiently transfected with GFP-Mid1. Cultures were incubated overnight at 37°C. Each coverglass was placed into a metal chamber filled with 1 ml of serum-free, phenol red-free DMEM on a warmed stage. Images were captured before and at 10 and 30 min after treatment with either 10 μM U0126 (LC Laboratories, Woburn, MA) or 10 μM PD98059 (Upstate Biotechnology, Lake Placid, NY) by using a Zeiss Axiovert 135TV inverted microscope equipped with a GFP filter set (excitation = 475 nm, emission = 505 nm) from Omega Optical (Brattleboro, VT) and a Hamamatsu (Middlesex, NJ) C4742 charge-coupled device camera driven by innovision (Durham, NC) software. Images were processed in Adobe photoshop 5.5 (Adobe Systems).
Results and Discussion
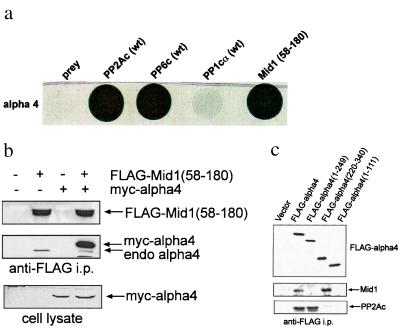
We screened a yeast two-hybrid library derived from mouse embryo by using as bait murine full-length α4, the putative mammalian homologue of yeast Tap 42. From 6 million clones we isolated 200 positives by growth on selective medium, and 65 of these showed expression of β-galactosidase as a reporter. All 65 clones were sequenced, and, of these, 20 were the catalytic subunit of PP2A, 4 were PP4, and another 8 were identical to regions of the Mid1 gene product (1). The Mid1 fragments identified in the α4 screen all contained the region from Thr58 to Asp180, comprising the B box domains, but lacked the N- and C-terminal flanking regions such as Ring finger and coiled-coil domains. We found that Mid1(58–180) alone had no transactivating activity. In the two-hybrid system, pairwise transformation with α4 plus the C subunit of PP2A was strongly positive and Mid1(58–180) gave comparable growth (shown as a darkened area by densitometry in Fig. 1a). We also show (Fig. 1a) that PP6, the mammalian homologue of yeast Sit4, interacted with α4 in a two-hybrid assay, consistent with an earlier report (19). By contrast, PP1c that is about 50% identical in sequence to PP2A but does not bind α4 (13) was a negative control to show specificity of interaction, and, indeed, no growth was observed (Fig. 1a).
Figure 1.
Association of α4 with Mid1 and PP2A. (a) Yeast two-hybrid analysis showed that strains expressing Gal4-DBD-α4 grew on triple-drop out media (trp-, leu-, his-) only when also expressing PP2A, PP6, or Mid1 (residues 58–180), indicative of an interaction with α4. Controls included single transformants and PP1C that did not support growth, because it does not bind α4. The plate was scanned on a densitometer to show growth of yeast colonies. (b) COS7 cells were transfected with FLAG-tagged Mid1(58–180) with or without myc-tagged α4. Cells were lysed and immunoprecipitation was performed by using anti-FLAG M2 affinity gel. FLAG-Mid1(58–180) and α4 were detected in the immunoprecipitates by immunoblotting with anti-FLAG and anti-α4 antibodies. When present, myc-α4 was detected by anti-myc immunoblotting. (c) COS7 cells transfected with vector alone, FLAG-α4 (full-length), FLAG-α4(1–249), FLAG-α4(220–340), or FLAG-α4(1–111) were lysed, and anti-FLAG immunoprecipitation was performed. FLAG-α4 coprecipitated endogenous Mid1 and PP2A-C, detected by anti-FLAG, anti-Mid1, and anti-PP2A-C immunoblotting.
Association of Mid1, α4, and PP2A in Living Cells.
We expressed FLAG-tagged mouse Mid1(58–180) protein with or without myc-tagged α4 in COS7 cells and immunoprecipitated with anti-FLAG antibody (Fig. 1b). FLAG-Mid1(58–180) coprecipitated endogenous α4 (lane 2). Endogenous α4 and, much more, myc-α4 were coprecipitated with FLAG-Mid1(58–180) (lane 4). Conversely, FLAG-α4 was expressed in COS7 cells, and it coprecipitated specifically the endogenous, 80-kDa Mid1 and 36-kDa C subunit of PP2A (Fig. 1c, second lane). The results showed that, in living cells, FLAG-Mid1(58–180) bound α4 and that FLAG-α4 could bind to both endogenous Mid1 and the C subunit of PP2A. Although from these results one cannot tell whether all three proteins, Mid1, α4, and PP2A, are bound together, other evidence (see below) suggests such complexes can assemble in living cells.
We mapped the regions in α4 required for the binding to Mid1 and the C subunit of PP2A (Fig. 1c). Deletion mutants of FLAG-α4 were produced by PCR and expressed in COS7 cells. Analysis of anti-FLAG immunoprecipitates showed that the C-terminal one-third of α4, encompassing only amino acids 220–340, was sufficient for binding to Mid1 but failed to bind the C subunit of PP2A. Conversely, the N-terminal two-thirds of α4, residues 1–249, bound C subunit of PP2A but not Mid1. Further truncation of α4 to a region of residues 1–111 gave a protein that bound no PP2A C subunit, indicating that a region between 111 and 249 of α4 is required for binding to PP2A. These results agree with the previous assignment of the phosphatase-binding site in α4 (14). Our results show that the C-terminal region of α4 binds Mid1. This is the region of α4 most conserved between species. Distinct regions of α4 bind PP2A and Mid1, seemingly independent of one another.
Colocalization of α4 and GFP-Mid1 with Microtubules.
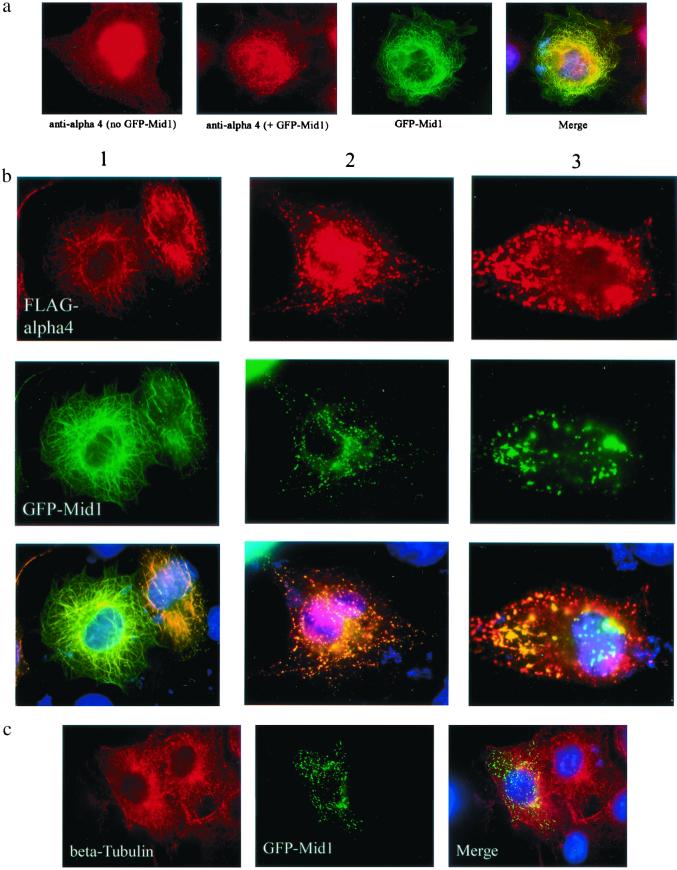
Recent studies have found that a Mid1-GFP fusion protein colocalized with microtubules (4, 5). As expected, this GFP-Mid1 protein expressed in COS cells clearly and predominantly colocalized with the microtubule network (Fig. 2a, green). Endogenous Mid1 gave the same microtubule localization, visualized by indirect immunofluorescence (not shown). Endogenous α4 was stained as diffusely cytoplasmic and in the nucleus by using anti-α4 antibodies (Fig. 2a, first frame), and the same pattern was obtained with anti-FLAG antibodies staining ectopically expressed FLAG-α4 (not shown). However, in cells expressing GFP-Mid1, much of the endogenous α4 became colocalized with microtubules (Fig. 2a). Thus, expression of GFP-Mid1 caused recruitment of cytoplasmic α4 onto microtubules. This also was observed when low levels of FLAG-α4 were coexpressed in COS cells with GFP-Mid1 (Fig. 2b, column 1).
Figure 2.
Codependent distribution of GFP-Mid1 and FLAG-α4. (a) COS7 cells were fixed and stained with anti-α4 antibodies to show the distribution of the endogenous α4 protein in a nontransfected cell (first frame) and in a cell expressing GFP-Mid1 (second frame). The distribution of GFP-Mid1 itself is shown in green, and colocalization of GFP-Mid1 and endogenous α4 is shown in yellow in the merged image (fourth frame). (b) COS7 cells were cotransfected to express both GFP-Mid1 and FLAG-α4. Cells were fixed and GFP-Mid was visualized by direct fluorescence microscopy, and FLAG-α4 was stained as described. From multiple experiments, representative cells with increasing levels of FLAG-α4 (columns 1, 2, and 3, left to right) were compared for localization of FLAG-α4 and GFP-Mid1. In the merged images, nuclei were stained by DAPI. (c) Cells expressing both GFP-Mid1 and FLAG-α4 to give foci of GFP-Mid1 (green) were fixed and stained for β-tubulin (red) and DAPI (blue).
On the other hand, the filamentous pattern of GFP-Mid1 on microtubules was lost as more and more FLAG-α4 was produced in cells by increasing the amount of DNA in transfections and/or the time of expression. Instead, GFP-Mid1 became distributed as numerous, bright cytoplasmic foci (Fig. 2b, column 2) or a few large, fluorescent clumps (see Fig. 2b, column 3). Over this range of expression levels, FLAG-α4 alone was diffusely cytoplasmic (not shown). As seen in the merged images, FLAG-α4 and GFP-Mid1 colocalized, even when their distribution changed. It is important to note that even when essentially all the GFP-Mid1 was accumulated into clumps, removal from microtubules by FLAG-α4 coexpression did not have any apparent effect on the microtubules themselves, based on immunofluorescent staining of β-tubulin in transfected and nontransfected cells (Fig. 2c).
Redistribution of OS Mutant Forms of Mid1 by α4.
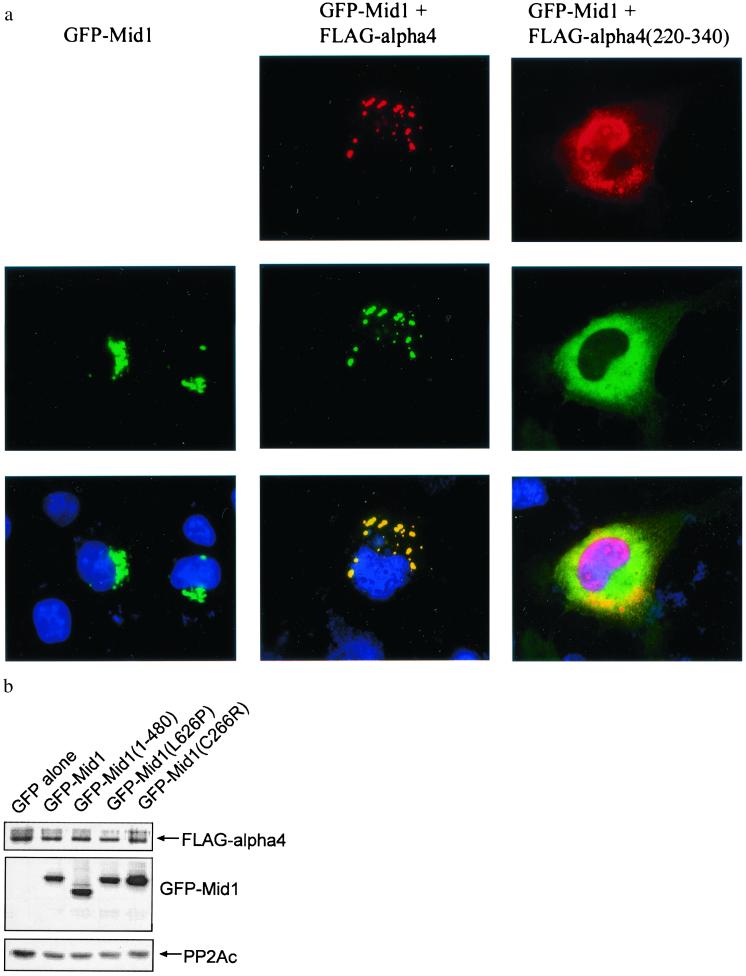
Mutated Mid1 proteins found in OS patients show a tendency to form either cytoplasmic foci or larger clumps (2). We expressed two OS mutant Mid1 proteins, L626P and C226R, as GFP-Mid1 fusion proteins in COS7 cells together with FLAG-α4. As seen in Fig. 3b, in anti-FLAG immunoprecipitates both OS mutant GFP-Mid1 proteins bound FLAG-α4 to the same extent as wild-type GFP-Mid1, so these single-residue mutations did not interfere with the Mid1-α4 interaction. Therefore, the failure of these mutant Mid1 proteins to colocalize with microtubules was not due simply to a loss of binding to α4. Moreover, we produced GFP-Mid1(1–480), a deletion mutant that lacks the entire B30.2 domain, to mimic the Mid1 C-terminal deletions seen in OS. This truncated GFP-Mid1 also coimmunoprecipitated with FLAG-α4 (Fig. 3b). These data showed that various OS mutations in Mid1 did not prevent binding to α4. This conclusion is consistent with these regions not mapping in the α4-binding segment of Mid1, obtained from the yeast two-hybrid screen. Thus, Mid1 has separate sites for binding to microtubules and to α4.
Figure 3.
OS mutants of GFP-Mid1 associate with FLAG-α4. (a) COS7 cells were transfected to express GFP-Mid1(1–480) together with each of the following FLAG constructs: FLAG empty vector, FLAG-α4, and FLAG-α4(220–340). Cells were fixed and GFP-Mid1 was visualized by direct fluorescence microscopy. FLAG-α4 was visualized by anti-α4 immunofluorescence. Nuclei of the cells were visualized by DAPI staining. (b) COS7 cells were cotransfected to express FLAG-α4 and different GFP-Mid1 fusion constructs: GFP alone (as control), GFP-Mid1, GFP-Mid1(1–480), GFP-Mid1(L626P), and GFP-Mid1(C266R). Immunoprecipitation was performed by using anti-FLAG M2 affinity gel and FLAG-α4; GFP-Mid1 fusion proteins and PP2A were detected by immunoblotting with anti-FLAG, anti-GFP, and anti-PP2A. The mutant GFP-Mid1 proteins all bound FLAG-α4.
The B30.2 domain in the C-terminal region of Mid1 can be found in many RING finger proteins as well as other unrelated proteins, but its function is still poorly understood. The potential importance of the B30.2 domain is implied by the concentration of mutations in this region of Mid1 in OS patients. It has been proposed that the RING-B box-coiled coil region is sufficient to mediate homodimerization, and the C-terminal B30.2 domain has a key role in microtubule association (5). Deletion of this domain from Mid1 eliminates microtubule association and results in the protein forming clumps in the cytoplasm (2). We found that FLAG-α4 bound to this truncated form of Mid1 (Fig. 3b, center lane). We transfected COS7 cells to express GFP-Mid1(1–480) and, as expected, observed accumulation of the GFP fusion protein in the perinuclear region (Fig. 3a, left column). Coexpression of FLAG-tagged full-length α4 with the GFP-Mid1(1–480) gave multiple clumps of protein still in the perinuclear region, and, again (as in Fig. 2b), the FLAG-α4 colocalized with the GFP-Mid1(1–480) (Fig. 3a, center column). However, when FLAG-α4(220–340) was coexpressed, the GFP-Mid1(1–480) was redistributed diffusely throughout the cytoplasm (Fig. 3a, right column). The nucleus appears pink in the merged image because there was staining for FLAG-α4(220–340) but not for GFP-Mid1(1–480) in the nucleus. Because FLAG-α4(220–340) binds Mid1 but not the phosphatase catalytic subunits (see Fig. 1b), we expect that FLAG-α4(220–340) competed against endogenous α4 for binding to GFP-Mid1(1–480) and thereby displaced α4-associated phosphatase activity. The results suggest that localization of Mid1 deleted in B30.2 domain is susceptible to regulation by α4-phosphatase, probably involving phosphorylation and dephosphorylation.
Phosphorylation–Dephosphorylation of Mid1.
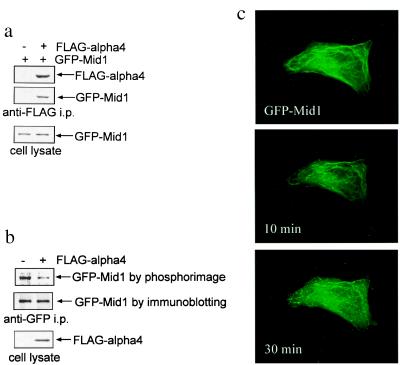
Could Mid1 be a phosphoprotein, and, if so, does binding to the α4-phosphatase reduce its phosphorylation? We transfected COS7 cells to express GFP-Mid1 plus either empty FLAG vector or FLAG-α4. Anti-FLAG antibodies specifically coimmunoprecipitated FLAG-α4 with GFP-Mid1 and PP2A, detected by immunoblotting with anti-GFP (Fig. 4a) and anti-PP2A (not shown). These results are comparable to Fig. 3b but also show that coexpression of FLAG-α4 did not affect the expression level of GFP-Mid1. Separately, cells were transfected the same, labeled with 32Pi, and GFP-Mid1 protein was recovered by anti-GFP immunoprecipitation. PhosphorImager analysis showed that GFP-Mid1 was a 32P-phosphoprotein (Fig. 4b). Coexpression of FLAG-α4 decreased the 32P labeling of GFP-Mid1 by almost half, in two independent experiments, without noticeable effect on the GFP-Mid1 expression level. Our interpretation of these results is that α4 targets phosphatase activity to Mid1, thereby promoting Mid1 dephosphorylation in living cells.
Figure 4.
Dephosphorylation of GFP-Mid1 induced by α4 and MAP kinase activity required for Mid1 association with microtubules. (a) COS7 cells were cotransfected to express GFP-Mid1 together with either FLAG-α4 or FLAG empty vector. Immunoprecipitation was performed by using anti-FLAG M2 affinity gel, and FLAG-α4 and GFP-Mid1 were detected by anti-FLAG and anti-GFP antibodies. (b) COS7 cells were transfected as described and radiolabeled with [32P]orthophosphate for 90 min. GFP-Mid1 was immunoprecipitated with anti-GFP antibodies and was subjected to PhosphorImager analysis after SDS/PAGE. Yield of GFP-Mid1 in the immunoprecipitates was determined by anti-GFP immunoblotting. FLAG-α4 in the cell lysate was detected by anti-FLAG immunoblotting. (c) COS7 cells were transfected to express GFP-Mid1, an initial image was captured (GFP:Mid1), and then the cells were treated with 10 μM UO126 and images were captured at the times indicated.
Only one site of phosphorylation is predicted in Mid1, based on searching for sequence motifs recognized by known kinases (http://expasy.cbr.nrc.ca/tools/). Ser96 in a P-N-S-P sequence is a possible substrate for MAP kinase (http://www.gcg.com/). Xenopus nuclear factor-7 (XNF-7) is a B box protein with sequence similarity to Mid1 (20). XNF-7 functions in Xenopus dorsal–ventral patterning (21), and its nuclear vs. cytoplasmic localization during oocyte maturation is regulated by MAP kinase phosphorylation (20). The phosphorylation site in XNF-7 for MAP kinase aligns with Ser96 in Mid1, so we predicted Mid1 would be a substrate for MAP kinase. To test this hypothesis, we treated cells expressing GFP-Mid1 with UO126 or PD98059 to inhibit MAP kinase activation. Microscopic recording of live cells showed partial redistribution of the GFP-Mid1 from microtubules to a punctate cytosolic localization within 30 min of treatment with UO126 (Fig. 4c). Similar results were obtained with cells treated with PD98059 (not shown). The results show that MAP kinase activity was required to maintain Mid1 association with microtubules.
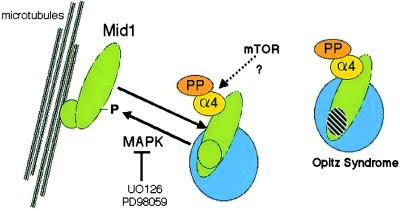
These data support a model (Fig. 5), with phosphorylation of Mid1 regulating its interaction with microtubules and, possibly, with other proteins. The C-terminal B30.2 domain is required for microtubule association, and increased phosphorylation of Mid1 is proposed to enhance the affinity of Mid1 for microtubules or microtubule-associated proteins. Conversely, dephosphorylation of Mid1 by α4-associated phosphatases, probably PP2A, would reduce the association of Mid1 with the microtubule network and result in cytoplasmic localization. Phosphorylation may influence the binding of other partners to Mid1, because even forms of Mid1 deleted in the B30.2 domain and, therefore, not bound to microtubules showed redistribution when coexpressed with wild-type or “phosphatase-deficient” forms of α4.
Figure 5.
Model for regulation of Mid1 association with microtubules by phosphorylation. The Mid1 protein is shown as two domains: a larger, elongated N-terminal domain and a smaller, circular C-terminal domain that is either truncated or contains mutations in OS (cross-hatched). Mid1 binds to other proteins into high Mr complexes in the cytoplasm (large circle) or can bind to microtubules (rods shown on left side). The binding to microtubules is affected by MAP kinase, which is inhibited by compounds UO126 and PD98059, and by binding to α4 (small, yellow circle) that associates with protein phosphatase catalytic subunits (labeled PP) that promote dephosphorylation of Mid1 and its release from microtubules.
Connecting Mid1 to Cellular Signals and Functions.
The cellular function of Mid1 is still unclear. Patients with mutations in Mid1 present with a variable array of malformations that are indicative of a disturbance of primary midline development. Based on this, a possible role for Mid1 has been postulated in the regulation of cell proliferation, programmed cell death, cell migration, or even epithelial–mesenchymal transformation. Our results offer evidence that Mid1 is regulated by a phosphorylation/dephosphorylation mechanism. The potential ability of Mid1 to control cell behavior might be triggered by a pathway that controls the function of α4 and its associated phosphatases during development. Both α4 and its yeast homologue, Tap42, associate with the C subunit of PP2A in the absence of the PP2A PR65 or A subunit. In yeast, Tap42 association with phosphatases is sensitive to the macrocyclic-immunosuppressive antibiotic rapamycin. This is consistent with the report that TOR directly phosphorylates Tap42 and enhances PP2A binding (12). In unpublished work, we have not seen phosphorylation of recombinant α4 by immunoprecipitated mTOR, which otherwise is active in the assay with PHAS-1 as substrate. In addition, α4 showed extremely low levels of 32P labeling in metabolically labeled cells, under a variety of conditions. Whether Tap42 and α4 both are controlled the same, e.g., by phosphorylation, remains an open question. On the other hand, mutation of the phosphorylation (Y307) and methylation (L309) sites in PP2A greatly enhances binding to α4, so modifications of PP2A might govern its association with α4 vs. the A subunit. Recently, yeast TOR was shown to interact with Bik1, a microtubule-associated protein required for microtubule assembly, stability, and function in cell processes such as karyogamy and nuclear migration and positioning (22, 23). Inhibition of TOR by rapamycin caused significant defects in these cellular functions of yeast. Perhaps in yeast, Tap42 and its bound phosphatase connect the TOR-signaling pathway to the regulation of microtubule stability and function. Previous studies showed that overexpression of Mid1 in mammalian cells enhanced the stability of microtubules toward colcemid, a microtubule-depolymerizing drug (4). A cycle of MAP kinase phosphorylation and α4-PP2A dephosphorylation to regulate association of Mid1 with microtubules opens new possibilities for connecting signaling pathways to various functions of microtubules. Learning more about the function of Mid1 and the role of its phosphorylation hopefully will give insights into the developmental defects seen in OS.
Acknowledgments
We thank Mary Foley for constant encouragement, members of the lab for support and discussions, and Christine Palazzolo for assistance in preparing the manuscript. We gratefully acknowledge grant support from the U.S. Public Health Service–National Cancer Institute (CA77584 to D.L.B.), the W. M. Keck Foundation (to D.L.B.), and the March of Dimes Birth Defects Foundation (1-FY00-465 to G.M.).
Abbreviations
- OS
Opitz syndrome
- Mid1
midline-1
- GFP
green fluorescent protein
- TOR
yeast target of rapamycin
References
- 1.Quaderi N A, Schweiger S, Gaudenz K, Franco B, Rugarli E I, Berger W, Feldman G J, Volta M, Andolfi G, Gilgenkrantz S, et al. Nat Genet. 1997;17:285–291. doi: 10.1038/ng1197-285. [DOI] [PubMed] [Google Scholar]
- 2.Cox T C, Allen L R, Cox L L, Hopwood B, Goodwin B, Haan E, Suthers G K. Hum Mol Genet. 2000;9:2553–2562. doi: 10.1093/hmg/9.17.2553. [DOI] [PubMed] [Google Scholar]
- 3.Dal Zotto L, Quaderi N A, Elliott R, Lingerfelter P A, Carrel L, Valsecchi V, Montini E, Yen C H, Chapman V, Kalcheva I, et al. Hum Mol Genet. 1998;7:489–499. doi: 10.1093/hmg/7.3.489. [DOI] [PubMed] [Google Scholar]
- 4.Schweiger S, Foerster J, Lehmann T, Suckow V, Muller Y A, Walter G, Davies T, Porter H, van Bokhoven H, Lunt P W, et al. Proc Natl Acad Sci USA. 1999;96:2794–2799. doi: 10.1073/pnas.96.6.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cainarca S, Messali S, Ballabio A, Meroni G. Hum Mol Genet. 1999;8:1387–1396. doi: 10.1093/hmg/8.8.1387. [DOI] [PubMed] [Google Scholar]
- 6.Inui S, Kuwahara K, Mizutani J, Maeda K, Kawai T, Nakayasu H, Sakaguchi N. J Immunol. 1995;154:2714–2723. [PubMed] [Google Scholar]
- 7.Di Como C J, Arndt K T. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 8.Zaragoza D, Ghavidel A, Heitman J, Schultz M C. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt A, Beck T, Koller A, Kunz J, Hall M N. EMBO J. 1998;17:6924–6931. doi: 10.1093/emboj/17.23.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis P B, Fumagalli S, Thomas G. Curr Opin Genet Dev. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- 11.Beck T, Hall M N. Nature (London) 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Y, Broach J R. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murata K, Wu J, Brautigan D L. Proc Natl Acad Sci USA. 1997;94:10624–10629. doi: 10.1073/pnas.94.20.10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inui S, Sanjo H, Maeda K, Yamamoto H, Miyamoto E, Sakaguchi N. Blood. 1998;92:539–542. [PubMed] [Google Scholar]
- 15.Chung H, Nairn A C, Murata K, Brautigan D L. Biochemistry. 1999;38:10371–10376. doi: 10.1021/bi990902g. [DOI] [PubMed] [Google Scholar]
- 16.Zhu L. Methods Mol Biol. 1997;63:173–196. doi: 10.1385/0-89603-481-X:173. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Kleiner U, Brautigan D L. Biochemistry. 1996;35:13858–13864. doi: 10.1021/bi961669e. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Brautigan D L. J Biol Chem. 2000;275:26074–26081. doi: 10.1074/jbc.M003843200. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Peterson R T, Schreiber S L. Biochim Biophys Acta. 1998;247:827–832. doi: 10.1006/bbrc.1998.8792. [DOI] [PubMed] [Google Scholar]
- 20.El-Hodiri H M, Che S, Nelman-Gonzalez M, Kuang J, Etkin L D. J Biol Chem. 1997;272:20463–20470. doi: 10.1074/jbc.272.33.20463. [DOI] [PubMed] [Google Scholar]
- 21.El-Hodiri H M, Shou W, Etkin L D. Dev Biol. 1997;190:1–17. doi: 10.1006/dbio.1997.8692. [DOI] [PubMed] [Google Scholar]
- 22.Choi J H, Adames N R, Chan T F, Zeng C, Cooper J A, Zheng X F. Curr Biol. 2000;10:861–864. doi: 10.1016/s0960-9822(00)00599-6. [DOI] [PubMed] [Google Scholar]
- 23.Bonatti S, Simili M, Galli A, Bagnato P, Pigullo S, Schiestl R H, Abbondandolo A. Chromosoma. 1998;107:498–506. doi: 10.1007/s004120050335. [DOI] [PubMed] [Google Scholar]