Abstract
The elucidation of breast cancer subgroups and their molecular drivers requires integrated views of the genome and transcriptome from representative numbers of patients. We present an integrated analysis of copy number and gene expression in a discovery and validation set of 997 and 995 primary breast tumours, respectively, with long-term clinical follow-up. Inherited variants (copy number variants and single nucleotide polymorphisms) and acquired somatic copy number aberrations (CNAs) were associated with expression in ~40% of genes, with the landscape dominated by cis- and trans-acting CNAs. By delineating expression outlier genes driven in cis by CNAs, we identified putative cancer genes, including deletions in PPP2R2A, MTAP and MAP2K4. Unsupervised analysis of paired DNA–RNA profiles revealed novel subgroups with distinct clinical outcomes, which reproduced in the validation cohort. These include a high-risk, oestrogen-receptor-positive 11q13/14 cis-acting subgroup and a favourable prognosis subgroup devoid of CNAs. Trans-acting aberration hotspots were found to modulate subgroup-specific gene networks, including a TCR deletion-mediated adaptive immune response in the ‘CNA-devoid’ subgroup and a basal-specific chromosome 5 deletion-associated mitotic network. Our results provide a novel molecular stratification of the breast cancer population, derived from the impact of somatic CNAs on the transcriptome.
Inherited genetic variation and acquired genomic aberrations contribute to breast cancer initiation and progression. Although somatically acquired CNAs are the dominant feature of sporadic breast cancers, the driver events that are selected for during tumorigenesis are difficult to elucidate as they co-occur alongside a much larger landscape of random non-pathogenic passenger alterations1,2 and germline copy number variants (CNVs). Attempts to define subtypes of breast cancer and to discern possible somatic drivers are still in their relative infancy3–6, in part because breast cancer represents multiple diseases, implying that large numbers (many hundreds or thousands) of patients must be studied. Here we describe an integrated genomic/transcriptomic analysis of breast cancers with long-term clinical outcomes composed of a discovery set of 997 primary tumours and a validation set of 995 tumours from METABRIC (Molecular Taxonomy of Breast Cancer International Consortium).
A breast cancer population genomic resource
We assembled a collection of over 2,000 clinically annotated primary fresh-frozen breast cancer specimens from tumour banks in the UK and Canada (Supplementary Tables 1–3). Nearly all oestrogen receptor (ER)-positive and/or lymph node (LN)-negative patients did not receive chemotherapy, whereas ER-negative and LN-positive patients did. Additionally, none of the HER2+ patients received trastuzumab. As such, the treatments were homogeneous with respect to clinically relevant groupings. An initial set of 997 tumours was analysed as a discovery group and a further set of 995 tumours, for which complete data later became available, was used to test the reproducibility of the integrative clusters (described below). An overview of the main analytical approaches is provided in Supplementary Fig. 1. Details concerning expression and copy number profiling, including sample assignment to the PAM50 intrinsic subtypes3,4,7 (Supplementary Fig. 2), copy number analysis (Supplementary Tables 4–8) and validation (Supplementary Figs 3 and 4 and Supplementary Tables 9–11), and TP53 mutational profiling (Supplementary Fig. 5) are described in the Supplementary Information.
Genome variation affects tumour expression architecture
Genomic variants are considered to act in cis when a variant at a locus has an impact on its own expression, or in trans when it is associated with genes at other sites in the genome. We generated a map of CNAs, CNVs (Supplementary Fig. 6, Supplementary Tables 12–15) and single nucleotide polymorphisms (SNPs) in the breast cancer genome to distinguish germline from somatic variants (see Methods), and to examine the impact of each of these variants on the expression landscape. Previous studies8 have shown that most heritable gene expression traits are governed by a combination of cis (proximal) loci, defined here as those within a 3-megabase (Mb) window surrounding the gene of interest, and trans (distal) loci, defined here as those outside that window. We assessed the relative influence of SNPs, CNVs and CNAs on tumour expression architecture, using each of these variants as a predictor (see Methods) to elucidate expression quantitative trait loci (eQTLs) among patients.
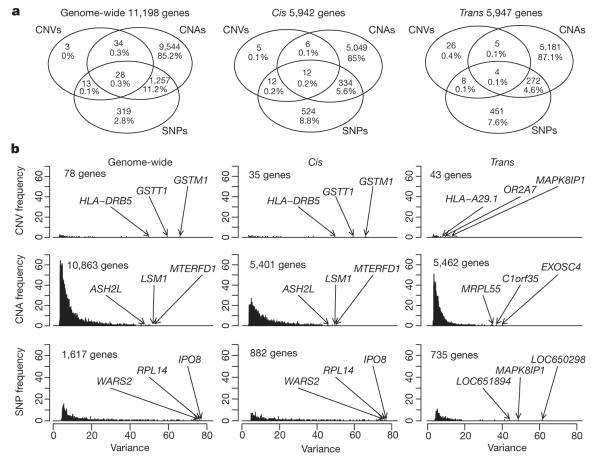
Both germline variants and somatic aberrations were found to influence tumour expression architecture, having an impact on >39% (11,198/28,609) of expression probes genome-wide based on analysis of variance (ANOVA; see Methods), with roughly equal numbers of genes associated in cis and trans. CNAs were associated with the greatest number of expression profiles (Fig. 1, Supplementary Figs 7–13 and Supplementary Tables 16–20), but were rivalled by SNPs to explain a greater proportion of expression variation on a per-gene basis genome-wide, whereas the contribution from CNVs was more moderate (Fig. 1b and Supplementary Table 21). The true ratio of putative trans versus cis eQTLs is hard to estimate9; however, the large sample size used here allowed the detection of small effects, with 5,401 and 5,462 CNAs significantly (Šidák adjusted P value <0.0001) associated in cis or in trans, respectively. Whereas cis-associations tended to be stronger, the trans-acting loci modulated a larger number of messenger RNAs, as described below.
Figure 1. Germline and somatic variants influence tumour expression architecture.
a, Venn diagrams depict the relative contribution of SNPs, CNVs and CNAs to genome-wide, cis and trans tumour expression variation for significant expression associations (Šidák adjusted P-value ≤0.0001).
b, Histograms illustrate the proportion of variance explained by the most significantly associated predictor for each predictor type, where several of the top associations are indicated.
Expression outliers refine the breast cancer landscape
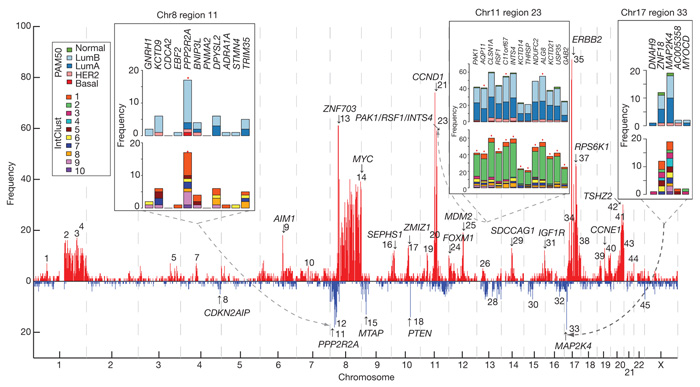
As shown above, ~20% of loci exhibit CNA-expression associations in cis (Supplementary Fig. 14). To refine this landscape further and identify the putative driver genes, we used profiles of outlying expression (see Methods and ref. 10) and the high resolution and sensitivity of the Affymetrix SNP 6.0 platform to delineate candidate regions. This approach markedly reduces the complexity of the landscape to 45 regions (frequency > 5, Fig. 2) and narrows the focus, highlighting novel regions that modulate expression. The full enumeration of regions delineated by this approach and their subtype-specific associations (Supplementary Figs 15 and 16 and Supplementary Tables 22–24) includes both known drivers (for example, ZNF703 (ref. 11), PTEN (ref. 12), MYC, CCND1, MDM2, ERBB2, CCNE1 (ref. 13)) and putative driver aberrations (for example, MDM1, MDM4, CDK3, CDK4, CAMK1D, PI4KB, NCOR1).
Figure 2. Patterns of cis outlying expression refine putative breast cancer drivers.
A genome-wide view of outlying expression coincident with extreme copy number events in the CNA landscape highlights putative driver genes, as indicated by the arrows and numbered regions. The frequency (absolute count) of cases exhibiting an outlying expression profile at regions across the genome is shown, as is the distribution across subgroups for several regions in the insets. High-level amplifications are indicated in red and homozygous deletions in blue. Red asterisks above the bar plots indicate significantly different observed distributions than expected based on the overall population frequency (χ2 test, P < 0.0001).
The deletion landscape of breast cancer has been poorly explored, with the exception of PTEN. We illustrate three additional regions of significance centred on PPP2R2A (8p21, Fig. 2, region 11), MTAP (9p21, Fig. 2, region 15) and MAP2K4 (17p11, Fig. 2, region 33), which exhibit heterozygous and homozygous deletions (Supplementary Figs 15, 17–19 and Supplementary Table 24) that drive expression of these loci. We observe breast cancer subtype-specific (enriched in mitotic ER-positive cancers) loss of transcript expression in PPP2R2A, a B-regulatory subunit of the PP2A mitotic exit holoenzyme complex. Somatic mutations in PPP2R1A have recently been reported in clear cell ovarian cancers and endometrioid cancers14,15, and methylation silencing of PPP2R2B has also been observed in colorectal cancers16. Thus, dysregulation of specific PPP2R2A functions in luminal B breast cancers adds a significant pathophysiology to this subtype.
MTAP (9p21, a component of methyladenosine salvage) is frequently co-deleted with the CDKN2A and CDKN2B tumour suppressor genes in a variety of cancers17 as we observe here (Supplementary Figs 17c and 18). The third deletion encompasses MAP2K4 (also called MKK4) (17p11), a p38/Jun dual specificity serine/threonine protein kinase. MAP2K4 has been proposed as a recessive cancer gene18, with mutations noted in cell lines19. We show, for the first time, the recurrent deletion of MAP2K4 (Supplementary Figs 17d and 19) concomitant with outlying expression (Supplementary Fig. 15) in predominantly ER-positive cases, and verify homozygous deletions (Supplementary Table 9) in primary tumours, strengthening the evidence for MAP2K4 as a tumour suppressor in breast cancer.
Trans-acting associations reveal distinct modules
We next asked how trans-associated expression profiles are distributed across the genome. We mapped these in the expression landscape by examining the matrices of CNA–expression associations (see Methods). This revealed strong off-diagonal patterns at loci on chromosomes 1q, 7p, 8, 11q, 14q, 16, 17q and 20q (Fig. 3a), including both positive and negative associations, as well as numerous trans-acting aberration hotspots (defined as CNAs associated with >30 mRNAs). Importantly, these aberration hotspots can be grouped into pathway modules, which highlight known driver loci such as ERBB2 and MYC, as well as novel loci associated with large trans expression modules (Supplementary Tables 25 and 26). The T-cell-receptor (TCR) loci on chromosomes 7 (TRG) and 14 (TRA) represent two such hotspots that modulated 381 and 153 unique mRNAs, respectively, as well as 19 dually regulated genes (Supplementary Fig. 20). These cognate mRNAs were highly enriched for T-cell activation and proliferation, dendritic cell presentation, and leukocyte activation, which indicate the induction of an adaptive immune response associated with tumour-infiltrating lymphocytes (Fig. 3b, Supplementary Fig. 20 and Supplementary Tables 27 and 28), as described later.
Figure 3. Trans-acting aberration hotspots modulate concerted molecular pathways.
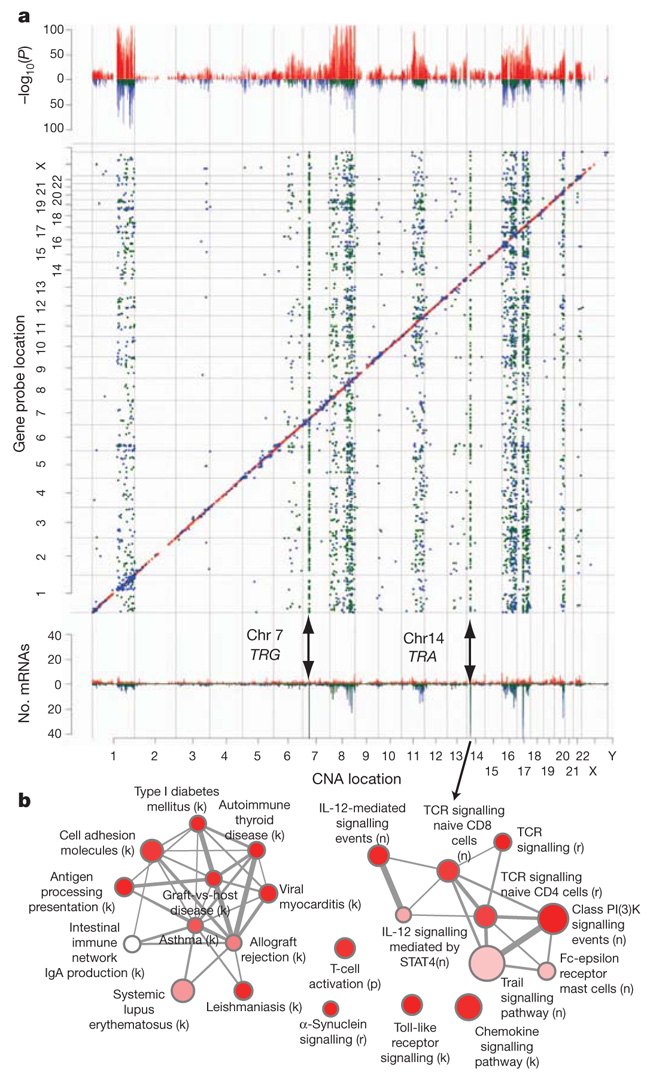
a, Manhattan plot illustrating cis and trans expression-associated copy number aberrations from the eQTL analysis (top panel). The matrix of significant predictor–expression associations (adjusted P-value ≤0.0001) exhibits strong off-diagonal patterns (middle panel), and the frequency of mRNAs associated with a particular copy number aberration further illuminates these trans-acting aberration hotspots (bottom panel). The directionality of the associations is indicated as follows: cis: positive, red; negative, pink; trans: positive, blue; negative, green. b, Enrichment map of immune response modules in the trans-associated TRA network, where letters in parentheses represent the source database as follows: b, NCI-PID BioCarta; c, cancer cell map; k, KEGG; n, NCI-PID curated pathways; p, PANTHER; r, Reactome.
In a second approach, we examined the genome-wide patterns of linear correlation between copy number and expression features (see Methods), and noted the alignment of several off-diagonal signals, including those on chromosome 1q, 8q, 11q, 14q and 16 (Supplementary Fig. 21). Additionally, a broad signal on chromosome 5 localizing to a deletion event restricted to the basal-like tumours was observed (Supplementary Fig. 21), but was not detected with the eQTL framework, where discrete (as opposed to continuous) copy number values were used. This basal-specific trans module is enriched for transcriptional changes involving cell cycle, DNA damage repair and apoptosis (Supplementary Table 29), reflecting the high mitotic index typically associated with basal-like tumours, described in detail below.
Integrative clustering reveals novel subgroups
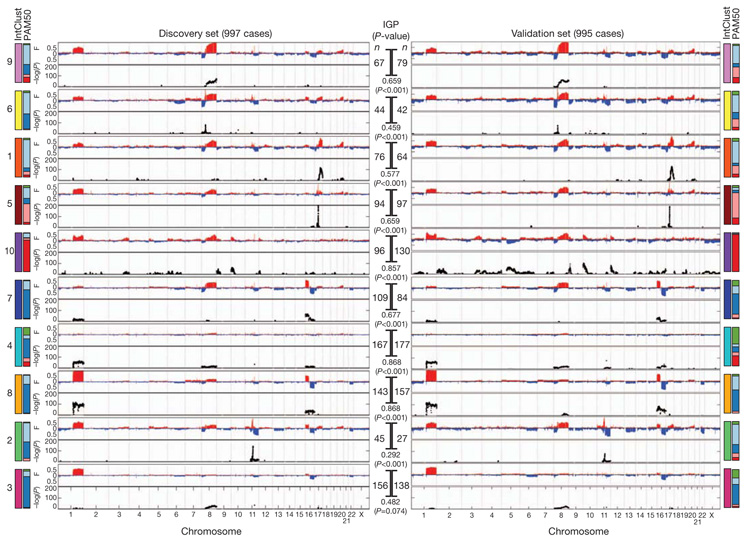
Using the discovery set of 997 breast cancers, we next asked whether novel biological subgroups could be found by joint clustering of copy number and gene expression data. On the basis of our finding that cis-acting CNAs dominated the expression landscape, the top 1,000 cis-associated genes across all subtypes (Supplementary Table 30) were used as features for input to a joint latent variable framework for integrative clustering20 (see Methods). Cluster analysis suggested 10 groups (based on Dunn’s index) (see Methods and Supplementary Figs 22 and 23), but for completeness, this result was compared with the results for alternative numbers of clusters and clustering schemes (see Methods, Supplementary Figs 23–27 and Supplementary Tables 31–33). The 10 integrative clusters (labelled IntClust 1–10) were typified by well-defined copy number aberrations (Fig. 4, Supplementary Figs 22, 28–30 and Supplementary Tables 34–39), and split many of the intrinsic subtypes (Supplementary Figs 31–33). Kaplan–Meier plots of disease-specific survival and Cox proportional hazards models indicate subgroups with distinct clinical outcomes (Fig. 5, Supplementary Figs 34, 35 and Supplementary Tables 40 and 41). To validate these results, we trained a classifier (754 features) for the integrative subtypes in the discovery set using the nearest shrunken centroids approach21 (see Methods and Supplementary Tables 42 and 43), and then classified the independent validation set of 995 cases into the 10 groups (Supplementary Table 44). The reproducibility of the clusters in the validation set is shown in three ways. First, classification of the validation set resulted in the assignment of a similar proportion of cases to the 10 subgroups, each of which exhibited nearly identical copy number profiles (Fig. 4). Second, the groups have substantially similar hazard ratios (Fig. 5b, Supplementary Fig. 35 and Supplementary Table 40). Third, the quality of the clusters in the validation set is emphasized by the in-group proportions (IGP) measure22 (Fig. 4).
Figure 4. The integrative subgroups have distinct copy number profiles.
Genome-wide frequencies (F, proportion of cases) of somatic CNAs (y-axis, upper plot) and the subtype-specific association (−log10 P-value) of aberrations (y-axis, bottom plot) based on a χ2 test of independence are shown for each of the 10 integrative clusters. Regions of copy number gain are indicated in red and regions of loss in blue in the frequency plot (upper plot). Subgroups were ordered by hierarchical clustering of their copy number profiles in the discovery cohort (n = 997). For the validation cohort (n = 995), samples were classified into each of the integrative clusters as described in the text. The number of cases in each subgroup (n) is indicated as is the in-group proportion (IGP) and associated P-value, as well as the distribution of PAM50 subtypes within each cluster.
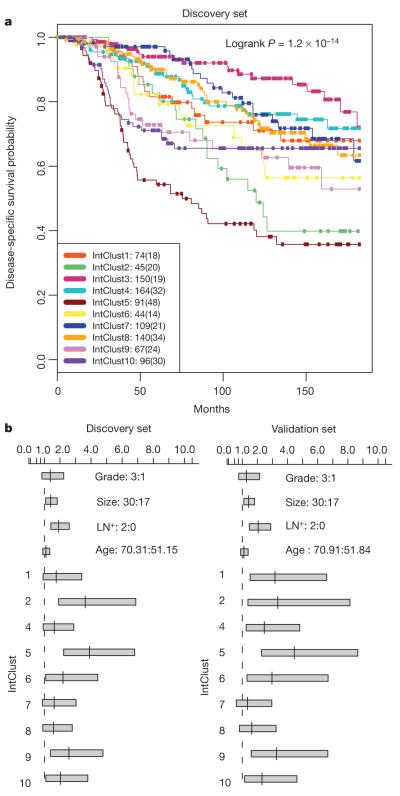
Figure 5. The integrative subgroups have distinct clinical outcomes.
a, Kaplan–Meier plot of disease-specific survival (truncated at 15 years) for the integrative subgroups in the discovery cohort. For each cluster, the number of samples at risk is indicated as well as the total number of deaths (in parentheses). b, 95% confidence intervals for the Cox proportional hazard ratios are illustrated for the discovery and validation cohort for selected values of key covariates, where each subgroup was compared against IntClust 3.
Among the integrative clusters, we first note an ER-positive subgroup composed of 11q13/14 cis-acting luminal tumours (IntClust 2, n = 45) that harbour other common alterations. This subgroup exhibited a steep mortality trajectory with elevated hazard ratios (discovery set: 3.620, 95% confidence interval (1.905–6.878); validation set: 3.353, 95% confidence interval (1.381–8.141)), indicating that it represents a particularly high-risk subgroup. Several known and putative driver genes reside in this region, namely CCND1 (11q13.3), EMSY (11q13.5), PAK1 (11q14.1) and RSF1 (11q14.1), which have been previously linked to breast13,23 or ovarian cancer24. Both the copy number (Fig. 4) and expression outlier landscapes (Fig. 2) suggest at least two separate amplicons at 11q13/14, one at CCND1 (11q13.3) and a separate peak from 11q13.5-11q14.1 spanning UVRAG–GAB2, centred around PAK1, RSF1, C11orf67 and INTS4, where it is more challenging to distinguish the driver24. Notably, the expression outlier profiles for this region are enriched for samples belonging to IntClust 2 (Fig. 2, inset region 23) and all 45 members of this subgroup harboured amplifications of these genes, with high frequencies of amplification also observed for CCND1 (n = 39) and EMSY (n = 34). In light of these observations, the 11q13/14 amplicon may be driven by a cassette of genes rather than a single oncogene.
Second, we note the existence of two subgroups marked by a paucity of copy number and cis-acting alterations. These subgroups cannot be explained by low cellularity tumours (see Methods). One subgroup (IntClust3, n = 156) with low genomic instability (Fig. 4 and Supplementary Fig. 22) was composed predominantly of luminal A cases, and was enriched for histotypes that typically have good prognosis, including invasive lobular and tubular carcinomas. The other subgroup (IntClust 4, n = 167) was also composed of favourable outcome cases, but included both ER-positive and ER-negative cases and varied intrinsic subtypes, and had an essentially flat copy number landscape, hence termed the ‘CNA-devoid’ subgroup. A significant proportion of cases within this subgroup exhibit extensive lymphocytic infiltration (Supplementary Table 45).
Third, several intermediate prognosis groups of predominantly ER-positive cancers were identified, including a 17q23/20q cis-acting luminal B subgroup (IntClust 1, n = 76), an 8p12 cis-acting luminal subgroup (IntClust 6, n = 44), as well as an 8q cis-acting/20q-amplified mixed subgroup (IntClust 9, n = 67). Two luminal A subgroups with similar CNA profiles and favourable outcome were noted. One subgroup is characterized by the classical 1q gain/16q loss (IntClust 8, n = 143), which corresponds to a common translocation event25, and the other lacks the 1q alteration, while maintaining the 16p gain/16q loss with higher frequencies of 8q amplification (IntClust 7, n = 109). We also noted that the majority of basal-like tumours formed a stable, mostly high-genomic instability subgroup (IntClust 10, n = 96). This subgroup had relatively good long-term outcomes (after 5 years), consistent with ref. 26, and characteristic cis-acting alterations (5 loss/8q gain/10p gain/12p gain).
The ERBB2-amplified cancers composed of HER2-enriched (ER-negative) cases and luminal (ER-positive) cases appear as IntClust 5 (n = 94), thus refining the ERBB2 intrinsic subtype by grouping additional patients that might benefit from targeted therapy. Patients in this study were enrolled before the general availability of trastuzumab, and as expected this subgroup exhibits the worst disease-specific survival at both 5 and 15 years and elevated hazard ratios (discovery set: 3.899, 95% confidence interval (2.234–6.804); validation set: 4.447, 95% confidence interval (2.284–8.661)).
Pathway deregulation in the integrative subgroups
Finally, we projected the molecular profiles of the integrative subgroups onto pathways to examine possible biological themes among breast cancer subgroups (Supplementary Tables 46 and 47) and the relative impact of cis and trans expression modules on the pathways. The CNA-devoid (IntClust 4) group exhibits a strong immune and inflammation signature involving the antigen presentation pathway, OX40 signalling, and cytotoxic T-lymphocyte-mediated apoptosis (Supplementary Fig. 36). Given that trans-acting deletion hotspots were localized to the TRG and TRA loci and were associated with an adaptive immune response module, we asked whether these deletions contribute to alterations in this pathway. The CNA-devoid subgroup (IntClust 4) was found to exhibit nearly twice as many deletions (typically heterozygous loss) at the TRG and TRA loci (~20% of cases) as compared to the other subtypes (with the exception of IntClust 10), and deletions of both TCR loci were significantly associated with severe lymphocytic infiltration (χ2 test, P < 10−9 and P < 10−8, respectively). Notably, these trans-associated mRNAs were significantly enriched in the immune response signature of the CNA-devoid subgroup (Supplementary Fig. 36) as well as among genes differentially expressed in CNA-devoid cases with severe lymphocytic infiltration (Supplementary Fig. 37). We conclude that genomic copy number loss at the TCR loci drives a trans-acting immune response module that associates with lymphocytic infiltration, and characterizes an otherwise genomically quiescent subgroup of ER-positive and ER-negative patients with good prognosis. These observations suggest the presence of mature T lymphocytes (with rearranged TCR loci), which may explain an immunological response to the cancer. In line with these findings, a recent study27 demonstrated the association between CD8+ lymphocytes and favourable prognosis.
Also among the trans-influenced groups is IntClust 10 (basal-like cancer enriched subgroup), which harbours chromosome 5q deletions (Supplementary Fig. 21). Numerous signalling molecules, transcription factors and cell division genes were associated in trans with this deletion event in the basal cancers, including alterations in AURKB, BCL2, BUB1, CDCA3, CDCA4, CDC20, CDC45, CHEK1, FOXM1, HDAC2, IGF1R, KIF2C, KIFC1, MTHFD1L, RAD51AP1, TTK and UBE2C (Supplementary Fig. 38). Notably, TTK (MPS1), a dual specificity kinase that assists AURKB in chromosome alignment during mitosis, and recently reported to promote aneuploidy in breast cancer28, was upregulated. These results indicate that 5q deletions modulate the coordinate transcriptional control of genomic and chromosomal instability and cell cycle regulation within this subgroup.
In contrast to these subtype-specific trans-associated signatures, the high-risk 11q13/14 subgroup was characterized by strong cis-acting associations. Like the basal cancers, this subgroup also exhibited alterations in key cell-cycle-related genes (Supplementary Fig. 39), which probably have a role in its aggressive pathophysiology, but the nature of the signature differs. In particular, the regulation of the G1/S transition by BTG family proteins, which include CCND1, PPP2R1B and E2F2, was significantly enriched in the 11q13/14 cis-acting subgroup, but not the basal cancers, and this is consistent with CCND1 and the PPP2R subunit representing subtype-specific drivers in these tumours.
Discussion
We have generated a robust, population-based molecular subgrouping of breast cancer based on multiple genomic views. The size and nature of this cohort made it amenable to eQTL analyses, which can aid the identification of loci that contribute to the disease phenotype29. CNAs and SNPs influenced expression variation, with CNAs dominating the landscape in cis and trans. The joint clustering of CNAs and gene expression profiles further resolves the considerable heterogeneity of the expression-only subgroups, and highlights a high-risk 11q13/14 cis-acting subgroup as well as several other strong cis-actingclusters and a genomically quiescent group. The reproducibility of subgroups with these molecular and clinical features in a validation cohort of 995 tumours suggests that by integrating multiple genomic features it may be possible to derive more robust patient classifiers. We show here, for the first time, that subtype-specific trans-acting aberrations modulate concerted transcriptional changes, such as the TCR deletion-mediated adaptive immune response that characterizes the CNA-devoid subgroup and the chromosome 5 deletion-associated cell cycle program in the basal cancers.
The integrated CNA-expression landscape highlights a limited number of genomic regions that probably contain driver genes, including ZNF703, which we recently described as a luminal B specific driver11, as well as somatic deletion events affecting key subunits of the PP2A holoenzyme complex and MTAP, which have previously been under-explored in breast cancer. The CNA-expression landscape also illuminates rare but potentially significant events, including IGF1R, KRAS and EGFR amplifications and CDKN2B, BRCA2, RB1, ATM, SMAD4, NCOR1 and UTX homozygous deletions. Although some of these events have low overall frequencies (<1% patients) (Figs 2, Supplementary Fig. 15 and Supplementary Tables 22–24), they may have implications for understanding therapeutic responses to targeted agents, particularly those targeting tyrosine kinases or phosphatases.
Finally, because the integrative subgroups occur at different frequencies in the overall population, focusing sequencing efforts on representative numbers from these groups will help to establish a comprehensive breast cancer somatic landscape at sequence-level resolution. For example, a significant number (~17%, n = 167 in the discovery cohort) of breast cancers are devoid of somatic CNAs, and are ripe for mutational profiling. Our work provides a definitive framework for understanding how gene copy number aberrations affect gene expression in breast cancer and reveals novel subgroups that should be the target of future investigation.
METHODS SUMMARY
All patient specimens were obtained with appropriate consent from the relevant institutional review board. DNA and RNA were isolated from samples and hybridized to the Affymetrix SNP 6.0 and Illumina HT-12 v3 platforms for genomic and transcriptional profiling, respectively. A detailed description of the experimental assays and analytical methods used to analyse these data are available in the Supplementary Information.
Supplementary Material
Acknowledgements
The METABRIC project was funded by Cancer Research UK, the British Columbia Cancer Foundation and Canadian Breast Cancer Foundation BC/Yukon. The authors also acknowledge the support of the University of Cambridge, Hutchinson Whampoa, the NIHR Cambridge Biomedical Research Centre, the Cambridge Experimental Cancer Medicine Centre, the Centre for Translational Genomics (CTAG) Vancouver and the BCCA Breast Cancer Outcomes Unit. S.P.S. is a Michael Smith Foundation for Health Research fellow. S.A. is supported by a Canada Research Chair. This work was supported by the National Institutes of Health Centers of Excellence in Genomics Science grant P50 HG02790 (S.T.). The authors thank C. Perou and J.Parkerfor discussions on the use of the PAM50 centroids. Theyalso acknowledge the patients who donated tissue and the associated pseudo-anonymized clinical data for this project.
METABRIC Group
Co-chairs Carlos Caldas1,2, Samuel Aparicio3,4
Writing committee Christina Curtis1,2†, Sohrab P. Shah3,4, Carlos Caldas1,2, Samuel Aparicio3,4
Steering committee James D. Brenton1,2, Ian Ellis5, David Huntsman3,4, Sarah Pinder6, Arnie Purushotham6, Leigh Murphy7, Carlos Caldas1,2, Samuel Aparicio3,4
Tissue and clinical data source sites: University of Cambridge/Cancer Research UK Cambridge Research Institute Carlos Caldas (Principal Investigator)1,2; Helen Bardwell2, Suet-Feung Chin1,2, Christina Curtis1,2†, Zhihao Ding2, Stefan Gräf1,2, Linda Jones8, Bin Liu1,2, Andy G. Lynch1,2, Irene Papatheodorou1,2, Stephen J. Sammut9, Gordon Wishart9; British Columbia Cancer Agency Samuel Aparicio (Principal Investigator)3,4, Steven Chia4, Karen Gelmon4, David Huntsman3,4, Steven McKinney3,4, Caroline Speers4, Gulisa Turashvili3,4, Peter Watson3,4,7; University of Nottingham: Ian Ellis (Principal Investigator)5, Roger Blamey5, Andrew Green5, Douglas Macmillan5, Emad Rakha5; King’s College London Arnie Purushotham (Principal Investigator)6, Cheryl Gillett6, Anita Grigoriadis6, Sarah Pinder6, Emanuele de Rinaldis6, Andy Tutt6; Manitoba Institute of Cell Biology Leigh Murphy (Principal Investigator)7, Michelle Parisien7, Sandra Troup7
Cancer genome/transcriptome characterization centres: University of Cambridge/Cancer Research UK Cambridge Research Institute Carlos Caldas (Principal Investigator)1,2, Suet-Feung Chin (Team Leader)1,2, Derek Chan1, Claire Fielding2, Ana-Teresa Maia1,2, Sarah McGuire2, Michelle Osborne2, Sara M. Sayalero2, Inmaculada Spiteri2, James Hadfield2; British Columbia Cancer Agency Samuel Aparicio (Principal Investigator)3,4, Gulisa Turashvili (Team Leader)3,4, Lynda Bell4, Katie Chow4, Nadia Gale4, David Huntsman3,4, Maria Kovalik4, Ying Ng4, Leah Prentice4
Data analysis subgroup: University of Cambridge/Cancer Research UK Cambridge Research Institute Carlos Caldas (Principal Investigator)1,2, Simon Tavaré (Principal Investigator)1,2,10,11, Christina Curtis (Team Leader)1,2†, Mark J. Dunning2, Stefan Gräf1,2, Andy G. Lynch1,2, Oscar M. Rueda1,2, Roslin Russell2, Shamith Samarajiwa1,2, Doug Speed2,10; Florian Markowetz (Principal Investigator)1,2, Yinyin Yuan1,2; James D. Brenton (Principal Investigator)1,2; British Columbia Cancer Agency Samuel Aparicio (Principal Investigator)3,4, Sohrab P. Shah (Team Leader)3,4, Ali Bashashati3, Gavin Ha3, Gholamreza Haffari3 & Steven McKinney3,4
1Department of Oncology, University of Cambridge, Hills Road, Cambridge CB2 2XZ, UK. 2Cancer Research UK, Cambridge Research Institute, Li Ka Shing Centre, Robinson Way, Cambridge CB2 0RE, UK. 3Department of Pathology and Laboratory Medicine, University of British Columbia, Vancouver, British Columbia V6T 2B5, Canada. 4Molecular Oncology, British Columbia Cancer Research Centre, Vancouver, British Columbia V5Z 1L3, Canada. 5Department of Histopathology, School of Molecular Medical Sciences, University of Nottingham, Nottingham NG5 1PB, UK. 6King’s College London, Breakthrough Breast Cancer Research Unit, London, WC2R 2LS, UK. 7Manitoba Institute of Cell Biology, University of Manitoba, Manitoba R3E 0V9, Canada. 8Cambridge Experimental Cancer Medicine Centre, Cambridge CB2 0RE, UK. 9Cambridge Breast Unit, Addenbrooke’s Hospital, Cambridge University Hospital NHS Foundation Trust and NIHR Cambridge Biomedical Research Centre, Cambridge CB2 2QQ, UK. 10Department of Applied Mathematics and Theoretical Physics, University of Cambridge, Centre for Mathematical Sciences, Cambridge CB3 0WA, UK. 11Molecular and Computational Biology Program, University of Southern California, Los Angeles, California 90089, USA. †Present address: Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, California 90033, USA.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Ch.C. led the analysis, designed experiments and wrote the manuscript. S.P.S. led the HMM-based analyses, expression outlier and TP53 analyses, and contributed to manuscript preparation. S.-F.C. generated data, designed and performed experiments. G.T. generated data, provided histopathology expertise and analysed TP53 sequence data. O.M.R., M.J.D., D.S., A.G.L., S.S., Y.Y., S.G., Ga.H., Gh.H., A.B., R.R., S.M. and F.M. performed analyses. G.T., A.G., E.P., S.P. and I.E. provided histopathology expertise. A.L. performed TP53 sequencing. A.-L.B.-D. oversaw TP53 sequencing. S.P., P.W., L.M., G.W., I.E., A.P., Ca.C. and S.A. contributed to sample selection. J.D.B. and S.T. contributed to study design. S.T. provided statistical expertise. The METABRIC Group contributed collectively to this study. Ca.C. and S.A. co-conceived and oversaw the study, and contributed to manuscript preparation and were responsible for final editing. Ca.C. and S.A. are joint senior authors and project co-leaders.
The authors declare no competing financial interests.
Author Information The associated genotype and expression data have been deposited at the European Genome-Phenome Archive (http://www.ebi.ac.uk/ega/), which is hosted by the European Bioinformatics Institute, under accession number EGAS00000000083. Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Leary RJ, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc. Natl Acad. Sci. USA. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin K, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Chin SF, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teschendorff AE, Naderi A, Barbosa-Morais NL, Caldas C. PACK: Profile analysis using clustering and kurtosis to find molecular classifiers in cancer. Bioinformatics. 2006;22:2269–2275. doi: 10.1093/bioinformatics/btl174. [DOI] [PubMed] [Google Scholar]
- 11.Holland D, et al. ZNF703 is a common Luminal B breast cancer oncogene that differentially regulates luminal and basal progenitors in human mammary epithelium. EMBO Mol. Med. 2011;3:167–180. doi: 10.1002/emmm.201100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 13.Santarius T, Shiply J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nature Rev. Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 14.Jones S, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McConechy MK, et al. Subtype-specific mutation of PPP2R1A in endometrial and ovarian carcinomas. J. Pathol. 2011;223:567–573. doi: 10.1002/path.2848. [DOI] [PubMed] [Google Scholar]
- 16.Tan J, et al. B55β-associated PP2A complex controls PDK1-directed MYC signaling and modulates rapamycin sensitivity incolorectal cancer. Cancer Cell. 2010;18:459–471. doi: 10.1016/j.ccr.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 17.Christopher SA, Diegelman P, Porter CW, Kruger WD. Methylthioadenosine phosphorylase, a gene frequently codeleted with p16 (CDKN2A/ARF), acts as a tumor suppressor in a breast cancer cell line. Cancer Res. 2002;62:6639–6644. [PubMed] [Google Scholar]
- 18.Teng DH, et al. Human mitogen-activated protein kinase kinase 4 as a candidate tumor suppressor. Cancer Res. 1997;57:4177–4182. [PubMed] [Google Scholar]
- 19.Hollestelle A, et al. Distinct gene mutation profiles among luminal-type and basal-type breast cancer cell lines. Breast Cancer Res. Treat. 2010;121:53–64. doi: 10.1007/s10549-009-0460-8. [DOI] [PubMed] [Google Scholar]
- 20.Shen R, Olshen AB, Ladanyi M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics. 2009;25:2906–2912. doi: 10.1093/bioinformatics/btp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl Acad. Sci. USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapp AV, Tibshirani R. Are clusters found in one dataset present in another dataset? Biostatistics. 2007;8:9–31. doi: 10.1093/biostatistics/kxj029. [DOI] [PubMed] [Google Scholar]
- 23.Hughes-Davies L, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 24.Brown LA, et al. Amplification of 11q13 in ovarian carcinoma. Genes Chromosom. Cancer. 2008;47:481–489. doi: 10.1002/gcc.20549. [DOI] [PubMed] [Google Scholar]
- 25.Russnes HG, et al. Genomic architecture characterizes tumor progression paths and fate in breast cancer patients. Sci. Transl. Med. 2010;2:38ra47. doi: 10.1126/scitranslmed.3000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blows FM, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahmoud SMA, et al. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 2011;29:1949–1955. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 28.Daniel J, Coulter J, Woo J-H, Wilsbach K, Gabrielson E. High levels of the Mps1 checkpoint protein are protective of aneuploidy in breast cancer cells. Proc. Natl Acad. Sci. USA. 2011;108:5384–5389. doi: 10.1073/pnas.1007645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.