SUMMARY
Autophagy is an important intracellular catabolic mechanism that mediates the degradation of cytoplasmic proteins and organelles. We report a potent small molecule inhibitor of autophagy named “spautin-1” for specific and potent autophagy inhibitor-1. Spautin-1 promotes the degradation of Vps34 PI3 kinase complexes by inhibiting two ubiquitin-specific peptidases, USP10 and USP13, that target the Beclin1 subunit of Vps34 complexes. Beclin1 is a tumor suppressor and frequently monoallelically lost in human cancers. Interestingly, Beclin1 also controls the protein stabilities of USP10 and USP13 by regulating their deubiquitinating activities. Since USP10 mediates the deubiquitination of p53, regulating deubiquitination activity of USP10 and USP13 by Beclin1 provides a mechanism for Beclin1 to control the levels of p53. Our study provides a molecular mechanism involving protein deubiquitination that connects two important tumor suppressors, p53 and Beclin1, and a potent small molecule inhibitor of autophagy as a possible lead compound for developing anticancer drugs.
INTRODUCTION
Vps34 is the primordial member of the PI3 kinase family and the only known class III PI3 kinase that can phosphorylate the D-3 position on the inositol ring of phosphatidylinositol (PtdIns) to produce PtdIns3P (Schu et al., 1993). In contrast to class I PI3 kinase, which has been extensively studied, much less is known about the class III PI3 kinase or its regulation in mammalian cells. Emerging evidence indicates a central role of Vps34 PI3K activity and its protein partners in orchestrating both initiation and maturation of autophagosomes (Simonsen and Tooze, 2009). Thus, exploring the mechanisms that regulate the class III PI3 kinase has direct implications in our understanding of these important intracellular mechanisms as well as for developing therapies for treatment of human diseases.
Similar to their homologs in yeast, Vps34 in mammalian cells is present in two complexes: Vps34 complex I and Vps34 complex II (Itakura et al., 2008; Liang et al., 2006; Matsunaga et al., 2009; Zhong et al., 2009). These two complexes share the core components of Vps34, Beclin1 and p150; and in addition, complex I contains Atg14L and complex II contains UVRAG. Interestingly, the stabilities of different components of Vps34 complexes are codependent upon each other as knockdown of one component often reduces the levels of others in the complexes (Itakura et al., 2008).
Beclin1 has been characterized as a tumor suppressor, and its importance is underscored by both the frequent monoallelic loss of beclin1 in human breast, ovarian and prostate tumors, and an increased rate of malignant tumors in BECN1+/− mice (Liang et al., 1999; Qu et al., 2003; Yue et al., 2003). Although autophagy deficiency has been proposed to be the mechanism for the increased tumorigenesis in BECN1+/− mice, a recent study using tissue-specific knockout mice of Atg5 and Atg7 suggests that autophagy deficiency may lead to benign tumors in livers, but not in other tissues (Takamura et al., 2011). Thus, the mechanism of Beclin1 as a tumor suppressor remains as a puzzle.
Small molecule inhibitors are important tools in exploring the cellular mechanisms in mammalian cells. However, the only available small molecule inhibitor of autophagy is 3-methyladenine (3-MA), which has a working concentration of ~10 mM and inhibits multiple forms of PI3 kinases. Therefore, there is an urgent need to develop highly specific small molecule tools that can be used to facilitate the studies of autophagy in mammalian cells. Using an imaging-based screen, we identified a small molecule inhibitor of autophagy and developed it into a highly potent autophagy inhibitor. We named it “spautin-1” for specific and potent autophagy inhibitor-1. We explored the mechanism by which spautin-1 inhibits autophagy and found that it inhibits two ubiquitin specific peptidases, USP10 and USP13, which regulate the deubiquitination of Beclin1 in Vps34 complexes. Using spautin-1 as a tool, we explored the interaction of USP10 and USP13 with Vps34 complexes. Interestingly, we found that Vps34 complexes interact with USP13 and the stabilities of USP10 and USP13 are coordinately regulated with that of Vps34 complexes. Since USP10 is a deubiquitinating enzyme for p53 and regulates the levels of p53 by controlling p53 ubiquitination and degradation (Yuan et al., 2010), regulating the stability of USP10 and USP13 by Vps34 complexes provides a molecular mechanism for class III PI3 kinase to control the levels of p53. Indeed, as predicted by our model, we found that the levels of p53 are reduced in the tissues of BECN1+/− mice, which provide a molecular mechanism for the increased tumorigenesis after monoallelic loss of beclin1. Our results demonstrate that class III PI3 kinase is an important tumor suppressor that can regulate the levels of p53 through controlling its deubiquitination.
RESULTS
Isolation of a Small Molecule Inhibitor of Autophagy by an Image-Based Screen
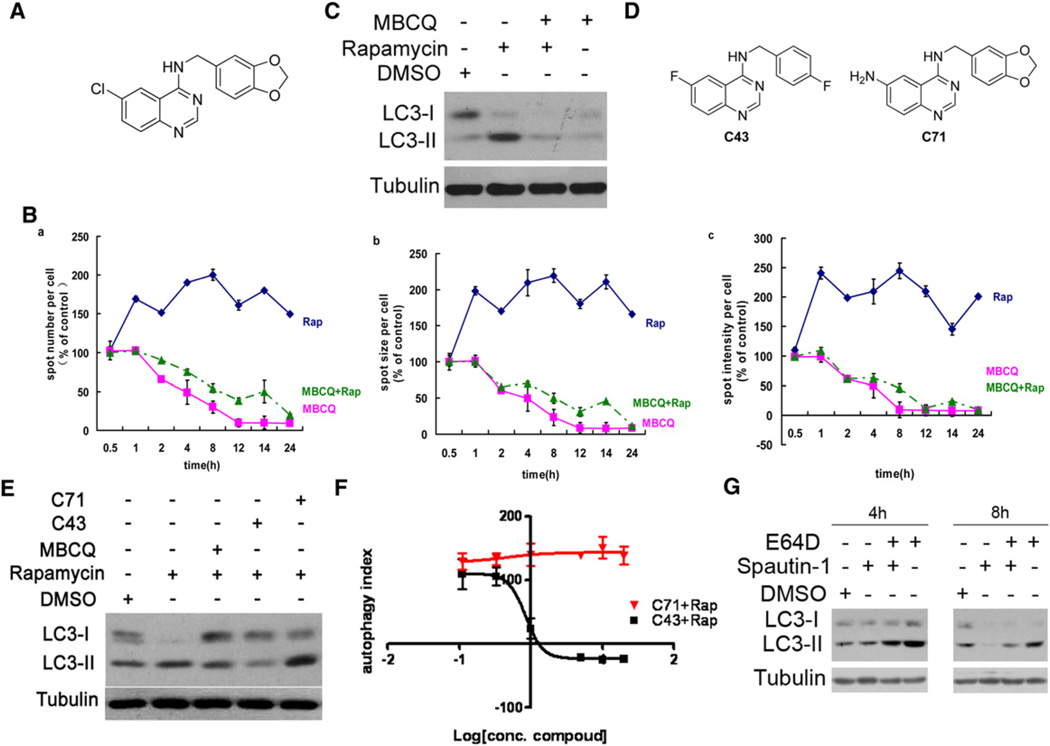
In an imaging-based screen using LC3-GFP as a marker for autophagy (Zhang et al., 2007), we identified a small molecule inhibitor of autophagy, MBCQ, from the ICCB known bioactive library (Figure 1A). MBCQ was previously known as an inhibitor of phosphodiesterase type 5 (PDE5), an enzyme that degrades cGMP by hydrolysis (MacPherson et al., 2006). Stimulation of H4-LC3-GFP cells with rapamycin led to increases in the levels of LC3-GFP as expected. A quantitative analysis of LC3-GFP puncta using high throughput microscopy showed that the treatment of MBCQ reduced the spot numbers as well as spot size and spot intensity of LC3-GFP dots compared to that of control or rapamycin treatment alone (Figure 1B). Thus, the presence of MBCQ inhibited both basal as well as rapamycin induced LC3-GFP autophagic puncta.
Figure 1. Isolation of a Series of Small Molecule Inhibitors of Autophagy.
(A) The structure of MBCQ.
(B) MBCQ reduced the spot numbers (a), spot size (b), and spot intensity (c) of LC3-GFP+ puncta. H4-LC3-GFP cells were treated with rapamycin (0.2 µM) and MBCQ (5 µM) as indicated. The image data are expressed as % of control vehicle treated cells. 1000 cells were analyzed per treatment condition.
(C) H4-LC3-GFP cells were treated with rapamycin (0.2 µM) and MBCQ (10 µM) as indicated for 2 hr and the cell lysates were analyzed by western blotting using anti-LC3. β-tubulin was used as a control.
(D) An active (C43=spautin-1) and an inactive (C71) derivatives of MBCQ.
(E) MEF cells were treated with DMSO (1‰), rapamycin (0.2 µM) alone, or together with MBCQ (10 µM), C43 (10 µM) or C71 (10 µM) for 4 hr. The cell lysates were analyzed for western blotting using anti-LC3 antibody. β-tubulin was used as a loading control.
(F) Dose-response (in µM) of C43 and inactive C71. H4-LC3-GFP cells were treated with rapamycin (0.2 µM) for 12 hr with C43 or C71 as indicated. The LC3-GFP+ puncta were quantified as in (B). Autophagy index =%{[total LC3-GFP+ spot intensity (compound+rapamycin treated) per cell] – [total LC3-GFP+ spot intensity (DMSO treated) per cell]} / {[total LC3-GFP+ spot intensity (rapamycin treated) per cell] – [total LC3-GFP+ spot intensity (DMSO treated) per cell]}. Rap = rapamycin.
(G) H4-LC3-GFP cells were treated with spautin-1(10 µM) with or without E64D (5 µM) for indicated periods of time. The cell lysates were analyzed by western blotting using anti-LC3 and anti-β-tubulin.
All error bars indicate STD. See also Figure S1 and S2.
This result was further confirmed by LC3 western blot analysis (Figure 1C), and similar results were obtained using mouse embryonic fibroblast cells (MEFs) (Figure S1A). Inhibition of autophagy by MBCQ was rapid (Figure 1B) and dose-dependent with an IC50 of 0.8 µM (Figure S1B), which is significantly more potent than the commonly used class III PI3 kinase inhibitor, 3-methyladenine (3-MA). We have also confirmed the autophagy inhibitory activity of MBCQ by electron microscopic studies. Cells treated with rapamycin showed a large number of autophagosomes with characteristic double membrane, which were conspicuously absent in cells treated with rapamycin and MBCQ (Figure S1C). Finally, the treatment of MBCQ was able to reduce the autophagic puncta of LC3-GFP in the presence of rapamycin, under starvation conditions or with bafilomycin which blocks lysosomal degradation (Figure S1D). Thus, MBCQ is an upstream inhibitor of autophagy.
Autophagy-Inhibiting Activity of MBCQ Can Be Separated from Its PDE5-Inhibiting Activity
One hundred and twelve derivatives of MBCQ were synthesized and analyzed to determine if its activity in inhibiting autophagy could be separated from its inhibition of PDE5 (Table S1 and data not shown). The chemical synthetic schemes are shown in the Extended Experimental Procedures. We selected 9 MBCQ derivatives based on their efficacy in inhibiting autophagy and screened for their activities on PDE5 (Wang et al., 2008). We found that C43 (6-fluoro-N-[4-fluorobenzyl]quinazolin-4-amine), an effective autophagy inhibitor with an IC50 of 0.74 µM (Figure 1D–F), which is comparable to that of MBCQ, has significantly reduced activity toward PDE5 and other PDEs (Figures S2A and S2B and Table S1). Thus, the PDE5 inhibiting activity of MBCQ can be chemically separated from its autophagy inhibiting activity.
Consistent with a separation of PDE5 and autophagy inhibiting activities in MBCQ, there were a number of other known PDE5 inhibitors in the bioactive library that we screened, including MY-5445, dipyridamole, IBMX and sildenafil (Viagra), which were not identified as autophagy inhibitors. To further confirm this conclusion, we treated H4-LC3-GFP cells with rapamycin and other PDE5 inhibitors including MY-5445, dipyridamole, IBMX or sildenafil using MBCQ as a positive control. None of the specific PDE5 inhibitors tested, including the most potent PDE5 inhibitor, sildenafil (Viagra) which has an IC50 of 2.5 nM for PDE5, had any activity on autophagy (data not shown). Thus, we conclude that the autophagy inhibiting activity of MBCQ is not related to its PDE5 inhibiting activity.
To further examine the specificity of C43 in inhibiting autophagy, we treated mouse embryo fibroblasts (MEF) cells with C43 or C71, a negative control, in the presence of rapamycin with the levels of autophagy determined by LC3 western blotting. Treatment with C43, but not a negative control C71, inhibited autophagy induced by rapamycin (Figures 1D–1F) and starvation (Figure S2C). We also confirmed the inhibition of autophagy by C43 using electron microscopy (Figure S2D). Furthermore, the treatment of C43 inhibited autophagy activated in the presence of E64D, a protease inhibitor that increases the accumulation of autophagosome by blocking lysosomal degradation (Figure 1G). Based on these data, we conclude that C43 is a potent inhibitor of autophagy and named it “spautin-1” for specific and potent autophagy inhibitor-1.
Spautin-1 Promotes Cell Death under Starvation Condition and Inhibits Autophagic Cell Death
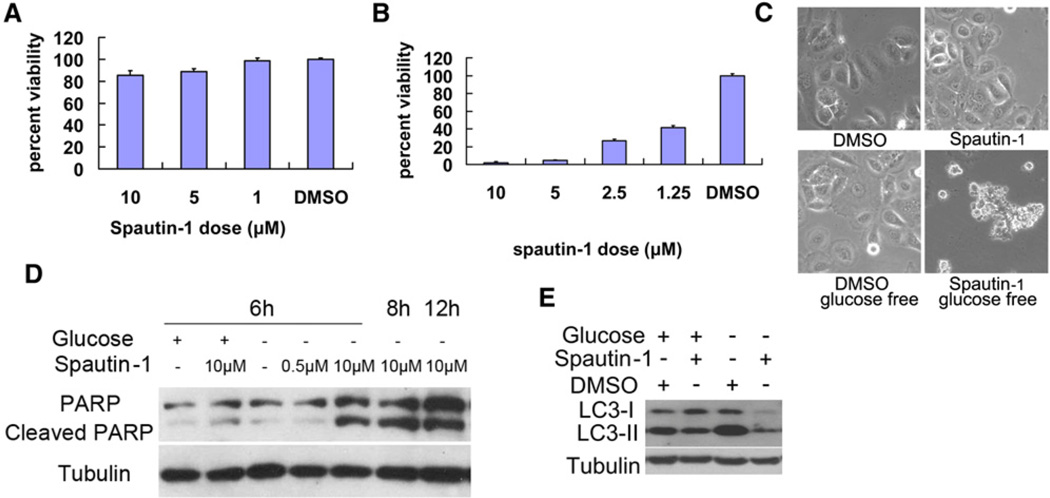
We first characterized the biological effects of spautin-1 at the cellular level in a selected subset of cancer cell lines. Spautin-1 had no effect on the growth and survival of Bcap-37 cells under normal culture conditions (Figure 2A) but dramatically enhanced cell death in glucose-free media (Figure 2B). Bcap-37 cells treated with spautin-1 under glucose-free condition showed apoptotic morphology (Figure 2C) and characteristic PARP cleavage (Figure 2D). Western blotting for LC3 further confirmed that autophagy was induced under glucose-free conditions, which was inhibited by spautin-1 (Figure 2E). Similar results were obtained with MCF-7 and BT549 cells (data not shown). Thus, spautin-1 can sensitize tumor cells to apoptosis under nutritional deprived conditions.
Figure 2. The Biological Effects of Spautin-1 on Cellular Models of Cell Death.
Bcap-37 cells were treated with indicated compounds in normal DMEM with 10% bovine serum (A), glucose free condition (B) or both (C-E) for 48 hr. The cell viability was determined by MTT assay (A), (B), imaged using a phase contrast microscope (C) or the cell lysates were analyzed by western blotting using anti-PARP (D), anti-LC3 and anti-β-tubulin (as a control) (E). All error bars indicate STD. See also Figure S3.
In contrast to the above cancer cell lines analyzed, MDCK cells, a normal cell line derived from the Madin-Darby canine kidney, treatment with spautin-1 under glucose-free conditions did not undergo apoptosis (Figures S3A and S3B). Hs578Bst cells, a myoepithelial cell line established from normal tissue peripheral to a breast cancer, were also not sensitive to the treatment of spautin-1 (Figures S3C and S3D). These results are consistent with the proposal that cancer cells are under increased metabolic pressure and therefore more sensitive to inhibition of autophagy than that of normal cells (Karantza-Wadsworth et al., 2007).
Increased activation of autophagy in apoptosis deficient cells has been shown to mediate cell death (Shimizu et al., 2004). To test this possibility, we treated Bax/Bak double knockout (DKO) cells with etoposide to induce cell death by DNA damage in the presence or absence of spautin-1. We found that spautin-1 inhibited etoposide induced autophagic cell death of Bax-Bak DKO cells (Figures S3E–S3G). Thus, spautin-1 can be used as a tool to explore the requirement of autophagy in cellular processes.
Spautin-1 Selectively Promotes the Degradation of Vps34 Complexes
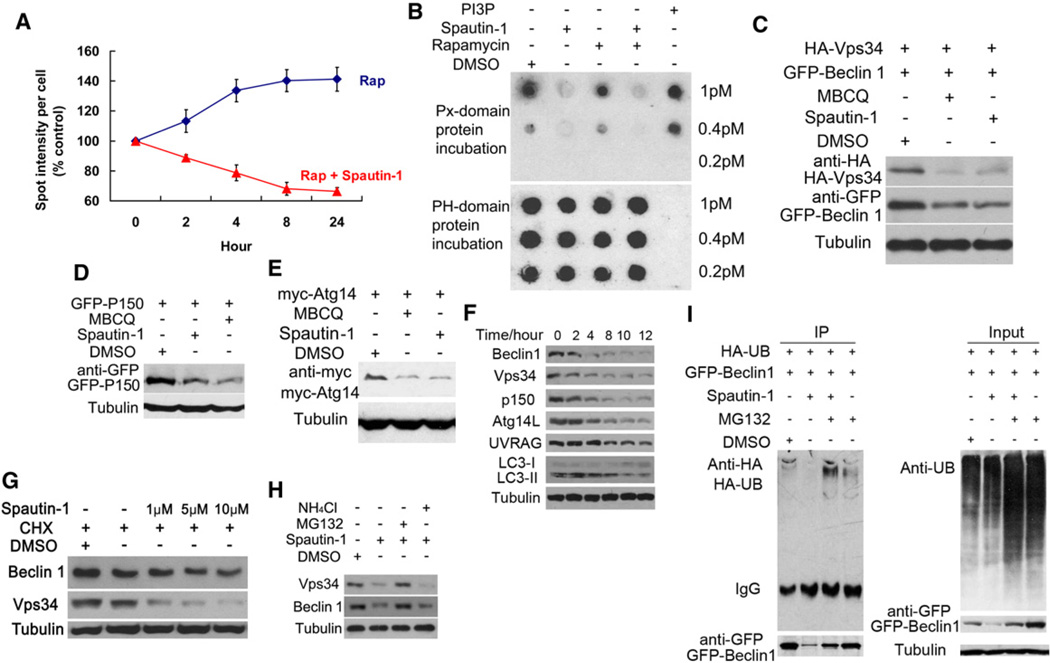
To explore the mechanism by which spautin-1 inhibits autophagy, we first examined the effects of spautin-1 on FYVERFP, an indicator for the activity of class III PI3 kinase, because PtdIns3P, the product of class III PI3 kinase, is important for the formation of autophagosomes (Gaullier et al., 1998; Simonsen and Tooze, 2009). Treatment with spautin-1 (Figure 3A) and MBCQ (Figure S4A) reduced the levels of FYVE-RFP puncta, but had no effect on the protein levels of FYVE-RFP (Figure S4B), suggesting that spautin-1 reduced the levels of PtdIns3P. The reduction of PtdIns3P in spautin-1 treated cells was confirmed using lipid dot blot analysis (Gozani et al., 2003)(Figure 3B). However, spautin-1 does not inhibit the lipid kinase activity of Vps34 in vitro (data not shown). Thus, spautin-1 can reduce the levels of PtdIns3P in cells, but is not a direct inhibitor of class III PI3 kinase activity.
Figure 3. Spautin-1 Reduces the Levels of PtdIns3P by Promoting the Degradation of Vps34 Complexes.
(A) H4-FYVE-RFP cells were treated with rapamycin (0.2 µM) and/or spautin-1 (10 µM) as indicated. The image data are expressed as%of control vehicle treated cells. 1000 cells were analyzed per treatment condition. Rap = rapamycin.
(B) MEF cells were treated with DMSO (1‰), rapamycin (0.2 µM), spautin-1 (10 µM) as indicated for 4 hr. The lipids were extracted and applied onto polyvinylidene fluoride membrane. The commercial PtdIns3P was spotted as indicated for controls. The levels of PtdIns3P were detected using GST-PX-p40 domain protein, which binds to PtdIns3P, and anti-GST antibody (top panel). The levels of PtdIns4P, detected using GST-PH-FAPP-1 domain protein which binds to PtdIns4P, and anti-GST antibody, were used as a loading control (bottom panel).
(C–E) 293T cells were transfected with expression vectors of HA-Vps34 and Flag-Beclin 1 (C), GFP-p150 (D), or myc-Atg14L (E). Twenty-four hours after transfection, cells were treated with DMSO (1‰), MBCQ (10 µM) or spautin-1 (10 µM) as indicated for 24 hr. The cell lysates were analyzed by western blotting using anti-HA, anti-Flag, anti-GFP, anti-myc as indicated or anti-β-tubulin (as a control).
(F) H4-LC3-GFP cells were treated with spautin-1 (10 µM) as indicated, the cell lysates were analyzed by western blotting using indicated antibodies. β-tubulin was used as a control.
(G) H4-LC3-GFP cells were treated with CHX (10 µM) or spautin-1 at indicated concentrations for 12 hr. DMSO (1‰) was used as a negative control. The cell lysates were analyzed by western blotting using anti-Beclin1, anti-Vps34, or anti-β-tubulin (as a control).
(H) H4-LC3-GFP cells were incubated with MG132 (10 µM) or NH4Cl (10 mM) with or without spautin-1 (10 µM) for 6 hr. The cell lysates were analyzed by western blotting using using indicated antibodies. β-tubulin was used as a control.
(I) 293T cells were transfected with GFP-Beclin1 and HA-Ub expression vectors. Twenty-four hours after transfection, cells were treated with spautin-1 (10 µM) for 24 hr and MG132 (5 µM) was added in the last 6 hr. The cell lysates were immunoprecipitated with anti-GFP antibody and the immunocomplexes were analyzed by western blotting using anti-HA antibody. All error bars indicate STD. See also Figure S4.
Interestingly, we noted that the levels of Flag-Beclin1 and HA-Vps34 were considerably lower in spautin-1 treated cells than that of control cells (Figure 3C). In addition, treatment with spautin-1 also reduced the levels of GFP-p150 and Myc-Atg14L (Figures 3D and 3E). On the other hand, the treatment of spautin-1 had no effect on the protein levels of GFP alone, GFP-Arf1, GFP-MT, EGFR, HA-Hrs, HA-Atg3, or GFP-Atg7 (data not shown). Thus, spautin-1 selectively reduces the levels of exogenously expressed components of Vps34 complexes.
To determine if spautin-1 has a similar effect on endogenous Vps34 complexes, we conducted a time course study of H4-LC3-GFP cells treated with spautin-1 by western blotting. We found that the levels of endogenous Beclin1, Vps34, p150, Atg14L, and UVRAG progressively decreased in a time-dependent manner in the presence of spautin-1, and the effect of spautin-1 on the levels of Vps34 complexes was strongly correlated with that of LC3II (Figure 3F). Other active derivatives such as MBCQ have similar activity profiles (data not shown). In contrast, the treatment of 3-MA has no effect on the protein level of Beclin1 (Figure S4C). These data confirm that spautin-1 selectively reduces the levels of Vps34 complexes in mammalian cells.
To explore the mechanism by which spautin-1 reduces the levels of Vps34 complexes, we treated H4-LC3-GFP cells with MBCQ or spautin-1 in the presence or absence of CHX. As shown in Figure 3G, the addition of spautin-1 with CHX reduced the levels of Beclin1 and Vps34 compared to that of CHX alone, suggesting that spautin-1 may promote the degradation of the class III PI3 kinase complexes. To further examine this possibility, we treated H4-LC3-GFP cells with spautin-1 in the presence of MG132 or NH4Cl to inhibit proteasomal or lysosomal degradation, respectively. MG132 but not NH4Cl inhibited the reduction of Beclin1 induced by spautin-1 (Figure 3H). The addition of MG132 restored the levels of Vps34 complexes as well as that of autophagy (Figure S5A). Similar results were found with transfected GFP-Beclin1 in 293T cells (Figure S5B). These results suggest that spautin-1 promotes the degradation of Beclin1 through the proteasomal pathway. Since ubiquitination represents an essential step in mediating proteasomal degradation, we tested if ubiquitination of Beclin1 was increased in cells treated with spautin-1. As shown in Figure 3I, the treatment of spautin-1 promoted the ubiquitination of Beclin1 without an obvious effect on the global levels of ubiquitination. Taken together, we conclude that spautin-1 inhibits autophagy by selectively promoting the degradation of the class III PI3 kinase complexes via the proteasomal pathway.
Identification of the Deubiquitinating Enzymes for Vps34 Complexes
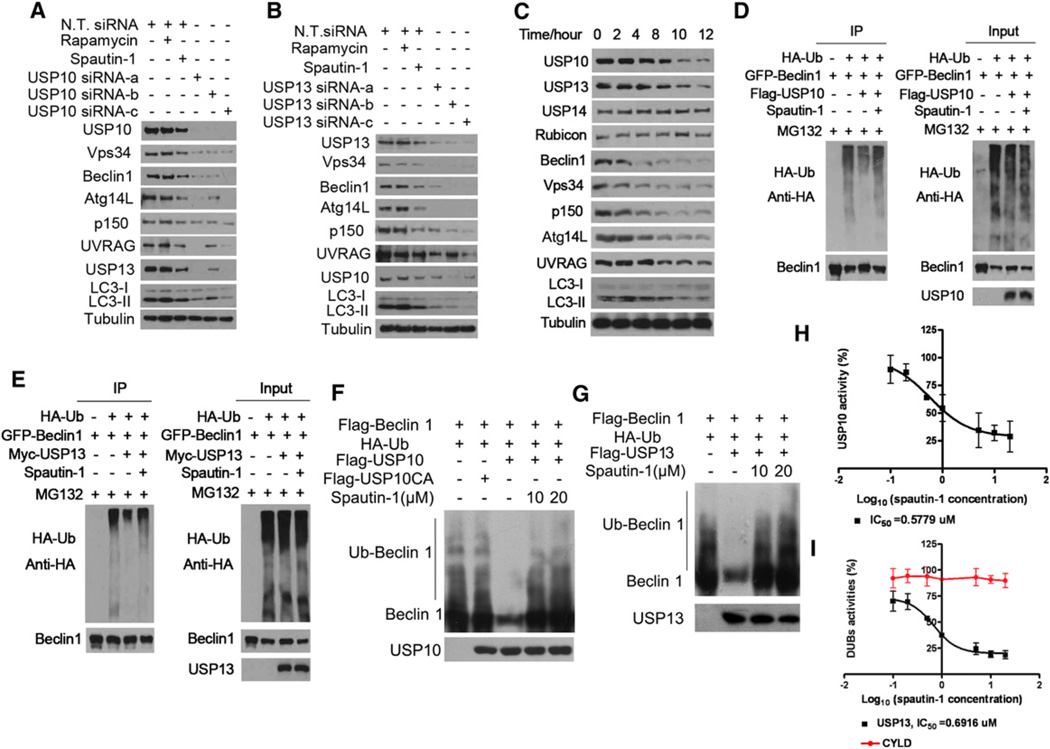
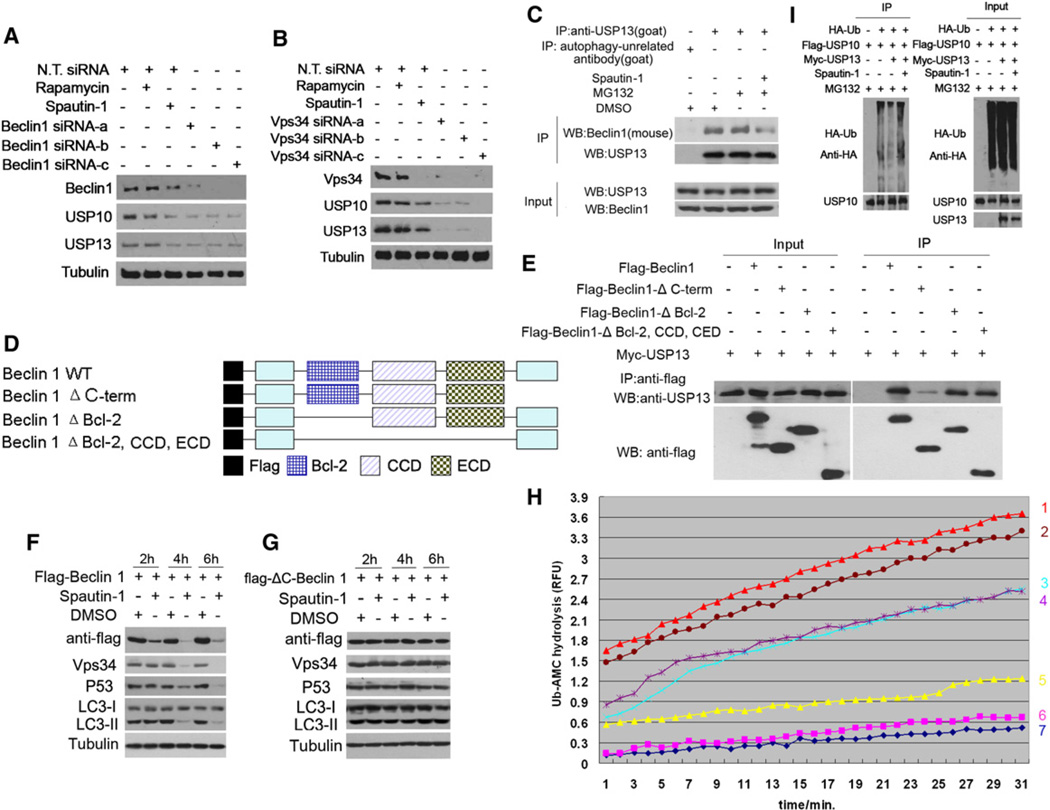
Since ubiquitination of proteins plays a critical role in mediating proteasomal degradation, we hypothesize that spautin-1 targets deubiquitinating enzyme(s) (DUBs) which normally function to negatively regulate the ubiquitination of Vps34 complexes. This follows from the common finding that a small molecule is more likely to be an inhibitor than an activator. To directly test this hypothesis, we screened a collection of 127 siRNAs targeting Human Deubiquitinating Enzymes from the Dharmacon library SMART pools for inhibition of autophagy using H4-LC3-GFP cells as an assay. We found that only knockdown of USP10 or USP13 showed a consistent effect of reducing the levels of endogenous Vps34, Beclin1, Atg14L, p150 and UVRAG (Figures 4A and 4B). Interestingly, the treatment of spautin-1 also reduced the levels of USP10 and USP13, but not USP14, a DUB involved in regulating proteasome function (Lee et al., 2010), or Rubicon, a negative regulator of type III PI3 kinase (Matsunaga et al., 2009; Zhong et al., 2009) (Figure 4C). Similarly, the treatment of MEF cells with spautin-1 also led to a time-dependent reduction in the levels of USP10, USP13, Vps34 complexes and autophagy (Figure S5C). In addition, we compared the effects of spautin-1 on HeLa and Bcap-37 cells under normal culture condition and autophagy induction conditions (Figures S5D and S5E). Interestingly, we found that the reduction in the levels of Vps34 complexes in Bcap-37 cells was significantly stronger under autophagy induction conditions than that under normal culture conditions, where autophagy levels are low.
Figure 4. Spautin-1 Inhibits the Deubiquitination of Vps34 Complexes.
(A) and (B) H4-LC3-GFP cells were transfected with indicated siRNAs for 72 hr or treated with rapamycin (0.25 µM) or spautin-1 (10 µM) as indicated, the cell lysates were analyzed by western blotting using indicated antibodies. β-tubulin was used as a control.
(C) H4-LC3-GFP cells were treated with spautin-1 (10 µM) as indicated, the cell lysates were analyzed by western blotting using indicated antibodies. β-tubulin was used as a control.
(D and E) 293T cells were transfected with indicated expression vectors for 12 hr, incubated with MG132 (10 µM), with or without spautin-1 (10 µM) for 4 hr, the cell lysates were immunoprecipitated with anti-Beclin1 and the immunocomplexes were analyzed by western blotting using anti-HA antibody.
(F) and (G) Ubiquitinated Beclin1 was incubated with immunopurified Flag-USP10, Myc-USP13, or Flag-USP10CA, with or without spautin-1 for 2 hr in vitro in deubiquitinating buffer. The western blot was blotted with anti-Beclin1 antibody.
(H) and (I) Proteins indicated purified from 293T cells and different concentrations of spautin-1 (20 µMto 100 nM) were mixed and incubated for 30 min. Ub-AMC was then added to each well and incubated for another 45 min. The final concentrations of every protein and Ub–AMC were 20 nM and 0.8 µM, respectively.
Ub–AMC hydrolysis was measured.
All error bars indicate STD. See also Figure S5.
Because the reductions in the levels of USP10 and USP13 in H4-LC3-GFP cells treated with spautin-1 appeared later than the reductions in the levels of Vps34 complexes and autophagy (Figure 4C), the reduced levels of USP10 and USP13 are unlikely to be the primary reason for the ability of spautin-1 to reduce the levels of PtdIns3P and inhibit autophagy. Since the treatment with spautin-1 increases the ubiquitination levels of Beclin1 and knockdown of USP10 or USP13 reduces the levels of Vps34 complexes, we considered the possibility that spautin-1 targets USP10 and USP13 mediated the deubiquitination of Vps34 complexes. We first examined the ability of USP10 and USP13 to mediate the deubiquitination of Vps34 complexes. We found that the overexpression of USP10 was highly effective in reducing the levels of ubiquitinated Beclin1, and this effect was inhibited in the presence of spautin-1 (Figure 4D). Similarly, the overexpression of USP13 reduced the levels of ubiquitinated Beclin1 which was inhibited by spautin-1 (Figure 4E). On the other hand, overexpression of USP10 or USP13 had no obvious effects on the ubiquitination levels of overexpressed Vps34, Atg14L, p150, and UVRAG (Figures S6A–S6H). Taken together, these results suggest that Beclin1 is the primary target of USP10 and USP13.
To directly test if spautin-1 can inhibit the deubiquitinating activity of USP10 and USP13, we tested the activity of isolated USP10 and USP13 on ubiquitinated Beclin1 in vitro. As shown in Figures 4F and 4G, the coincubation of ubiquitinated Beclin1 with USP10 or USP13 but not a catalytically inactive USP10 mutant reduced the levels of Beclin1 ubiquitination. Furthermore, the presence of spautin-1 inhibited the deubiquitination of Beclin1 mediated by USP10 and USP13. In contrast, spautin-1 had no effect on CYLD-mediated deubiquitination of RIP1 in vitro (data not shown). To further confirm this result, we developed an in vitro deubiquitination assay using Ub-AMC (the C-terminal derivatization of ubiquitin with 7-amino-4-methylcoumarin), which is a fluorogenic substrate for deubiquitinating enzymes (DUBs) (Dang et al., 1998). Using this assay, we found that spautin-1 inhibited USP10 and USP13 with IC50 of ~0.6–0.7 µM while having no inhibitory activity toward CYLD which is also a member of ubiquitin specific peptidase family (Figures 4H and 4I). Thus, spautin-1 is an inhibitor of the deubiquitinating activity of USP10 and USP13. Our results suggest that inhibition of USP10 and USP13 by spautin-1 promotes the ubiquitination and degradation of Vps34 complexes which in turn leads to a reduction in the levels of PtdIns3P and consequent inhibition of autophagy.
Regulation of USP10 and USP13 by Vps34 Complexes
Unexpectedly, we found that the knockdown of Beclin1 or Vps34 could also reduce the endogenous levels of USP10 and USP13 (Figures 5A and 5B). This suggests that Vps34 complexes may be able to regulate their own levels by stabilizing their cognate deubiquitinating enzymes including USP10 and USP13. This effect is not likely mediated through PtdIns3P, the product of Vps34 complexes, as the treatment of 3-MA which inhibits the kinase activity of class III PI3 kinase had no effect on the levels of USP10 or USP13 (data not shown). Thus, Vps34 complexes have the surprising role of regulating the stability of USP10 and USP13.
Figure 5. Regulation of USP13 by Vps34 Complexes.
(A and B) H4-LC3-GFP cells were transfected with indicated siRNAs for 72 hr or treated with rapamycin (0.25 µM) or spautin-1 (10 µM) for 4 hr, the cell lysates were analyzed by western blotting using indicated antibodies. β-tubulin was used as a control.
(C) H4-LC3-GFP cells were treated with MG132 (10 µM) and spautin-1 (10 µM) for 6 hr. The cell lysates were immunoprecipitated with anti-USP13 antibody and the immunocomplexes were analyzed by western blotting using anti-Beclin1 antibody.
(D) A schematic diagram of Beclin1 truncation mutants used in (E).
(E) 293T cells were transfected with Myc-USP13, Flag-Beclin1, Flag-Beclin1-ΔC-term, Flag-Beclin1-ΔBD, Flag-Belin1-ΔBD, CCD, CED as indicated for 24 hr. The cell lysates were immunoprecipitated with anti-flag antibody and the immunocomplexes were analyzed by western blotting using anti-USP13 antibody.
(F and G) 293T cells were transfected with flag-Beclin1 or flag-ΔC-Beclin1 for 24 hr, and then treated with spautin-1(10 µM) as indicated. The cell lysates were assayed by anti-flag, anti-Vps34, anti-p53, anti-LC3, β-tubulin (loading control) as indicated.
(H) Flag-Beclin1, Flag-USP10 and Myc-USP13 proteins were isolated from 293T cells individually transfected with the relevant expression constructs by immunoprecipitation followed by extensive washing (12×) and elution with tag peptides. Deubiquitinating activities of indicated proteins were analyzed using Ub-AMC assay. Line 1: Myc-USP13, Flag-USP10 and Flag-Beclin1; Line 2: Myc-USP13 and Flag-Beclin1; Line 3: Flag-USP10 and Flag-Beclin1; Line 4: Myc-USP13 and Flag-USP10; Line 5: Myc-USP13; Line 6: Flag-USP10; Line 7: Flag-Beclin1.
(I) 293T cells were transfected with indicated expression vectors for 12 hr, incubated with MG132 (10 µM) in the presence or absence of spautin-1 (10 µM) for an additional 4 hr. The cell lysates were immunoprecipitated with anti-USP10 antibody and the immunocomplexes were analyzed by western blotting using anti-HA antibody.
See also Figure S6.
To determine the mechanism by which Vps34 complexes regulate the stability of USP13 and USP10, we examined the possibility that Beclin1 may interact with USP10 and USP13. We found that endogenous Beclin1 can interact with USP13 and the interaction was reduced in the presence of spautin-1 (Figure 5C). However, the interaction of Beclin1 and USP10 was considerably weaker (data not shown). These data suggest that Beclin1 may closely interact with USP13, whereas its interaction with USP10 is indirect or transient in nature.
To further characterize the interaction of USP13 with Beclin1, we determined the domains of Beclin1 that interact with USP13 (Figure 5D & E). Different truncation mutants of Beclin1 were coexpressed with USP13 in 293T cells and the interaction of USP13 with different Beclin1 mutants was analyzed by coimmunoprecipitation. A C-terminal deletion mutant of Beclin1 (ΔC-Beclin1) showed significantly reduced binding with USP13, suggesting the C terminus of Beclin1 is important for the interaction. We further compared the effect of spautin-1 in 293T cells expressing ΔC-Beclin1 mutant or full length Beclin1 (Figures 5F and 5G). Interestingly, we found that not only was the ΔC-Beclin1 mutant resistant to spautin-induced degradation, but the expression of ΔC-Beclin1 mutant significantly blocked the effect of spautin-1 in inhibiting autophagy and inducing the degradation of Vps34. Since ΔC-Beclin1 can bind to Vps34 (Furuya et al., 2010) but not USP13, this experiment suggests that the interaction of Beclin1 and USP13 is critically important for regulating the stability of Vps34 complexes in response to spautin-1 treatment.
To directly examine the mechanism by which Beclin1 regulates USP10 and USP13, we determined the effects of their interaction on the deubiquitinating activities in vitro using Ub-AMC as a substrate. As shown in Figure 5H, the DUB activities of USP10 or USP13 were comparatively low when incubated alone with Ub-AMC. Interestingly, the DUB activities were significantly increased when USP13 and USP10 coincubated together or with Beclin1 or all 3 proteins together, suggesting the DUB activity can be significantly enhanced when USP13 interacts with its substrate Beclin1 or USP10. Thus, reduced levels of USP10 and USP13 in the presence of spautin-1 or with beclin1 knockdown may be due to their increased ubiquitination and degradation through the proteasome pathway. Consistent with this possibility, the effect of spautin-1 on the levels of USP10 and Vps34 complexes can be fully restored in the presence of MG132 (Figure S5A). Furthermore, the effect of Beclin1 knockdown on reduced levels of USP10 and USP13 can also be inhibited by MG132 (Figure S7B).
Deubiqutination of USP10 by USP13
Since the treatment of spautin-1 also led to reduced levels of USP10, which was inhibited by the addition of MG132 (Figure S5A), it is likely that the levels of USP10 and USP13 are also regulated by ubiquitination. Interestingly, knockdown of either USP10 or USP13 led to reductions in the levels of the other (Figures 4A and 4B). Thus, we considered the possibility that USP10 and USP13 may regulate deubiquitination of each other. Consistent with this possibility, the ubiquitination levels of USP10 were reduced when cells were cotransfected with an expression vector of USP13 and the addition of spautin-1 inhibited the deubiquitination of USP10 by USP13 (Figure 5I). On the other hand, coexpression of USP10 with USP13 has a much less pronounced effect on ubiquitination of USP13 (data not shown). These results suggest that USP13 may directly regulate the deubiquitination of USP10; however, USP10 may regulate USP13 indirectly perhaps by affecting the levels of Vps34 complexes. Since USP10 mediates the deubiquitination of Beclin1 and reduced levels of USP10 leads to increased ubiquitination and degradation of Vps34 complexes, reduced levels of Vps34 complexes as a result of USP10 reduction may in turn lead to destabilization of USP13.
Our data supports an interactive regulatory relationship of USP10 and USP13 with Vps34 complexes. We propose that USP10 and USP13 mediate the deubiqutination of Vps34 complexes to regulate the levels of class III PI3 kinase. Furthermore, Beclin1 also interacts with USP13 and regulates the stability of USP13. Since USP13 can also deubiquitinate USP10, regulating the stability of USP13 by Beclin1 provides a mechanism for Beclin1 to control the stability of USP10. Thus, our data suggest that the levels of Vps34 complexes may be coupled to the levels of USP10 and USP13.
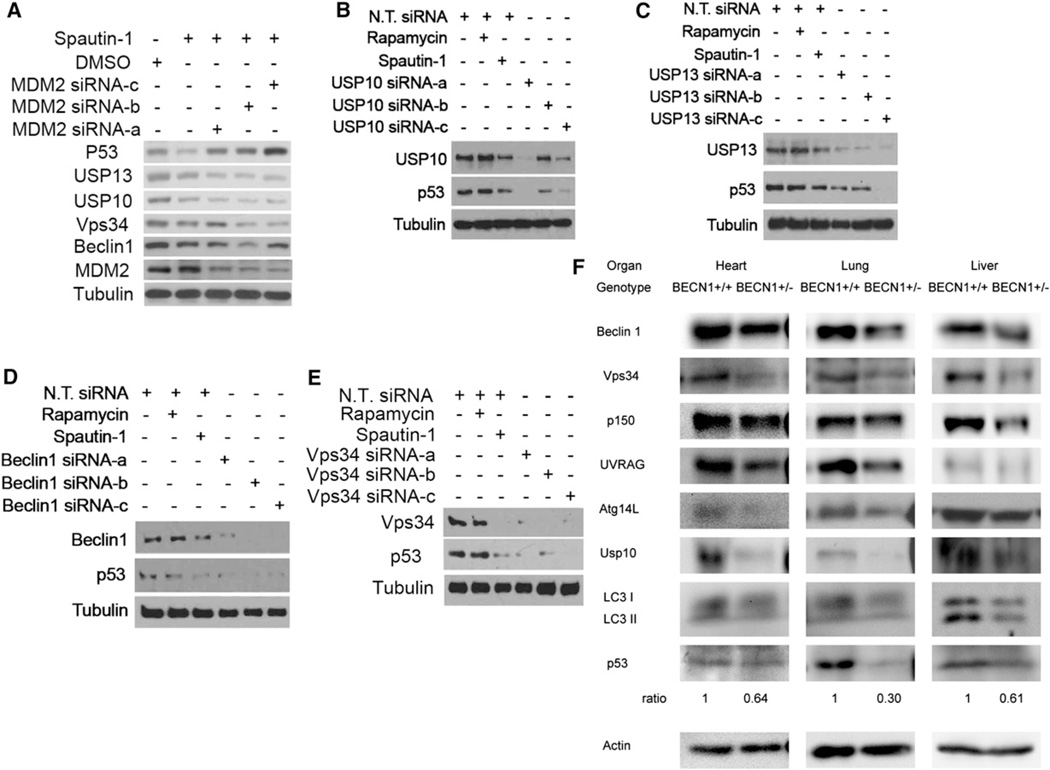
Regulation of p53 via Vps34 Complexes and Deubiquitination
Since USP10 is known as a deubiquitinating protease of p53 (Yuan et al., 2010), inhibition of USP10 by spautin-1 may promote the degradation of p53. Consistent with this possibility, the treatment of spautin-1 led to a reduction in the levels of p53 that was inhibited in the presence of MG132 (Figure S5A). Furthermore, spautin-1 induced reduction in the levels of p53 was inhibited with knockdown of MDM2, the major E3 ubiquitin ligase for p53 (Figure 6A). On the other hand, knockdown of Mdm2 had no effect on spautin-1 induced reduction of USP10, USP13, Vps34 or Beclin1. In addition, we found that knockdown of USP10, USP13, Beclin1, Vps34, p150, UVRAG, Atg14L all led to reduction in the levels of p53 (Figures 6B–6E; Figure S7A). Thus, the cellular levels of p53 may be coordinately regulated with that of Vps34 complexes via deubiquitinating enzymes such as USP10 and USP13. Finally, consistent with Beclin1 being the primary target of USP10 and USP13, the expression of ΔC Beclin1, which behaves as a dominant negative in inhibiting the loss of Vps34 complexes and autophagy, also inhibited the reduction of p53 induced by spautin-1 (Figure 5F and 5G).
Figure 6. Regulation of p53 by Vps34 Complexes, USP10 and USP13.
(A–E) H4-LC3-GFP cells were transfected with indicated siRNAs for 72 hr and treated with rapamycin (0.25 µM) or spautin-1 (10 µM) for 4 hr. The cell lysates were analyzed by western blotting using indicated antibodies. Anti-β-tubulin is a loading control.
(F) Heart, lung and liver tissues of newborn BECN1+/+ and BECN1+/− mice were isolated and analyzed by western blotting using indicated antibodies. Anti-actin was used as a loading control.
Also see Figure S7.
Since our model predicts that the levels of class III PI3 kinase should be correlated with that of p53, we examined the levels of p53 in BECN1+/− mice. As shown in Figure 6F, the levels of Beclin1 in newborn BECN1+/− mice are approximately half of that in wt mice. Consistent with a coordinated regulation of Vps34 complex components, the levels of Vps34, Atg14L, p150 and UVRAG are also significantly reduced in BECN1+/− tissues. Interestingly, as predicted by our model, the levels of USP10 and p53 in the heart, lung and liver of newborn BECN1+/− mice are correspondingly reduced. The levels of USP13 could not be examined currently due to a lack of antibody that can recognize murine USP13. The levels of LC3II in BECN1+/− liver are reduced compared to that of wt. On the other hand, the reduction of LC3II in heart and lung of BECN1+/− mice is not as obvious as that in liver. The reduced levels of p53 provide an important molecular mechanism contributing to the increased tumorigenesis in BECN1+/− mice.
To further characterize the effect of spautin-1 on p53, we examined the effect of spautin-1 on the cytoplasmic and the nuclear levels of p53 and found that the treatment of spautin-1 can reduce both nuclear and cytoplasmic p53 (Figure S7C). Furthermore, we found that the effects of spautin on the levels of Vps34 complexes and autophagy could still be observed in SKOV-3 ovarian cancer cell line which is null for p53 (Figure S7D). These results are consistent with the target of spautin-1 being upstream and independent of p53.
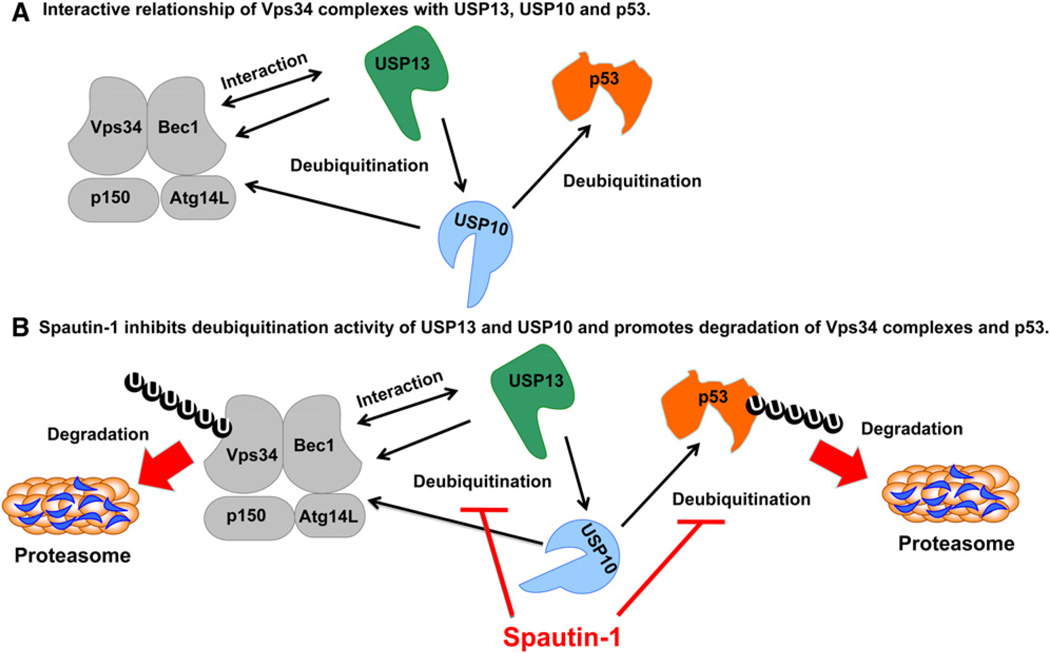
Taken together, our data suggest a model of regulatory relationship between class III PI3 kinase and p53 via protein interaction and deuqibuitination and the mechanism by which the treatment with spautin-1 leads to the reduced levels of Vps34 complexes and p53 (Figure 7).
Figure 7. A Model: USP10 and USP13 Mediate the Deubiquitination of Vps34 Complexes an p53.
(A) In cells treated with spautin-1, USP10 and USP13 are inhibited which leads to increased ubiquitination and degradation of Beclin1 in Vps34 complexes and p53.
(B) USP13 interacts with Beclin1 in Vps34 complexes which provides a mechanism for Vps34 complexes to regulate the deubiquitination activity of USP13. USP13 also mediates the deubiquitination of USP10 which explains why knockdown of USP13 also leads to increased degradation of USP10. On the other hand, knockdown of USP10 leads to the loss of Vps34 complexes which might in turn destabilizes USP13.
DISCUSSION
In this study, we describe a potent small molecule inhibitor of autophagy, named spautin-1, that targets the deubiquitination activity of USP10 and USP13. Using spautin-1 as a tool, we demonstrate that the ubiquitination and degradation of Vps34 complexes are regulated by two ubiquitin-specific peptidases, USP10 and USP13. Inhibiting deubiquitination of Vps34 complexes by spautin-1 leads to increased ubiquitination and degradation of class III PI3 kinase complexes through the proteasomal pathway. Furthermore, our study demonstrates a physiological mechanism for regulating the class III PI3 kinase via protein deubiquitination. Since class III PI3 kinase plays an important role in regulating multiple intracellular vesicular trafficking events including autophagy and endocytosis, the ability of USP10 and USP13 to regulate the stability of Vps34 complexes provides a molecular mechanism for ubiquitination and proteasomal degradation to control intracellular vesicular trafficking. Unlike that of class I and class II PI3 kinases which need to be activated through receptor signaling, the activity of class III PI3 kinase is believed to be constitutive (Lindmo and Stenmark, 2006). Thus, regulating the protein levels of class III PI3 kinase might provide an important mechanism for controlling the constitutively active class III PI3 kinase and intracellular levels of PtdIns3P. Our recent genome-wide siRNA screen on autophagy demonstrated a prominent role of class III PI3 kinase in regulating autophagy (Lipinski et al., 2010). It will be interesting in future to examine if regulating deubiquitination and ubiquitination of Vps34 complexes provides a general mechanism for controlling the constitutively active class III PI3 kinase activities under different physiological conditions. Consistent with the regulation of this deubiquitination mechanism, we found that the effect of spautin-1 on the levels of Vps34 complexes are dramatically enhanced in Bcap-37 cells under glucose-free condition, suggesting that the deubiquitination of Vps34 complexes may be under the control of nutritional availability. Thus, inhibiting the deubiquitination of Vps34 complexes might provide a strategy for developing autophagy inhibitors as an anticancer therapy.
USPs are cysteine proteases containing conserved regions in their amino acid sequence surrounding the Cys, His and Asp/Asn residues that form the catalytic triad. From the structural studies of a number of USP family members, it has been noted that the USP catalytic domains are often not appropriately aligned without binding to their substrates (Komander, 2010). That is, the catalytic Cys in USP7 shifts from catalytically inactive position to an active position where it interacts with the catalytic His only when binding to ubiquitin. On the other hand, although the catalytic machineries of USP14 and USP8 are properly aligned for catalysis in the absence of ubiquitin, the ubiquitin binding sites are blocked by the ubiquitin-binding surface loops (Hu et al., 2005). In addition, the Fingers domain of USP8, which is important for binding to ubiquitin, folds inward which blocks the ubiquitin binding site when not interacting with its substrates. Although no structural information for USP13 and USP10 is currently available, the enhanced DUB activity when USP13 interacts with USP10 or when they interact with Beclin1 suggest that the interaction of USP13 and USP10 with each other or with their substrates can lead to changes in their conformation which may be critical for the catalytic activities. This provides a possible model and mechanism for the close interactive regulatory relationship between USP13/USP10 with Vps34 complexes to explain why knockdown of USP13/USP10 or Vps34 complexes lead to reduced levels of the others.
We demonstrate that USP10 and USP13 can both mediate the deubiquitination of Beclin1. Since the stabilities of the core components of Vps34 complexes are codependent upon each other (Itakura et al., 2008), regulating deubiquitination of Beclin1 may be sufficient to control the levels of whole complex. On the other hand, our study also demonstrates that Vps34 complexes can regulate their own levels by a feedback control of USP13. Since the interaction of USP13 and Beclin1 is detectable by coimmunoprecipitation, we propose that Beclin1 may have close interaction with USP13. On the other hand, the interaction of Beclin1 and USP10 is consistent with that of enzyme/substrate which is expected to be weak and transient in nature. Most DUBs with resolved structures show that the enzymes are in an unproductive conformation before binding to the substrates. These inactive states might result from the blocking of the active site by loops or a misalignment of catalytic triads. Thus, binding to Beclin1 may trigger a major change in the conformation of USP13 to allow catalysis.
Our study demonstrates that class III PI3 kinase is an important tumor suppressor. The role of beclin1 as a haploid-insufficient tumor suppressor is well-established; however, it has been unclear how might a reduction in beclin1 expression can have such a dramatic impact on genomic instability and tumorigenesis (Karantza-Wadsworth et al., 2007). Our study demonstrates that a reduction of beclin1 expression leads to a reduced p53 level by increasing its ubiquitination, providing an important molecular mechanism contributing to the role of beclin1 as a haploid-insufficient tumor suppressor that is frequently monoallelically lost in human breast, ovarian, and prostate cancers (Liang et al., 1999; Qu et al., 2003; Yue et al., 2003).
Recently, using mutant mice with tissue-specific Atg5 or Atg7 deficiency, Takamura et al. showed that multiple benign tumors developed from autophagy deficient liver, but not in other tissues (Takamura et al., 2011). Thus, it is unlikely that increased rate of tumorigenesis in BECN1+/−mice is due to autophagy deficiency as assumed originally. Since Beclin1 has been shown to interact with Bcl-2, it has also been proposed that decreased levels of Beclin1 in BECN1+/− cells may promote the activity of Bcl-2 to increase cell survival which in turn promotes tumorigenesis (Pattingre et al., 2005). While our model does not rule out of a potential contribution of Bcl-2 or autophagy deficiency from promoting tumorigenesis in BECN1+/− mice, the reduced levels of p53 as a result of reduction in Beclin1 might provide a mechanism to promote genomic instability which in turn leads to tumorigenesis. The ability of Beclin1 to regulate the levels of p53 provides a mechanism underlying the observations that monoallelic loss of beclin1 is sufficient to lead to DNA damage and genomic instability via gene amplification (Karantza-Wadsworth et al., 2007).
Consistent with the contribution of p53 deficiency to increased tumorigenesis in beclin1 heterozygous background, the tumor spectra of TP53+/− mice and BECN1+/− mice strongly overlap: the highest frequencies of tumors in both TP53+/− and BECN1+/− mice are lung carcinoma, hepatoma and lymphoma (Jacks et al., 1994; Qu et al., 2003). Furthermore, the beclin1 gene is frequently monoallelically deleted in human sporadic ovarian, prostate and breast cancers similar to that of p53 mutations (http://www-p53.iarc.fr). The similarities in tumor spectra of BECN1+/− mice and TP53+/− mice suggest that reduced p53 levels play an important role in promoting tumorigenesis in BECN1+/− mice. Since the stability of the components of Vps34 complexes are largely codependent upon each other, reduced expression of other components of Vps34 complexes, including Vps34, p150, Atg14L and UVRAG, also leads to reduced levels of p53. Thus, our data suggest that all components of Vps34 complexes can regulate the levels of p53.
Curiously, although monoallelic loss of beclin1 is frequently observed in breast, ovarian and prostate cancers, the loss of heterozygocity of beclin1 was not commonly observed (Liang et al., 1999; Qu et al., 2003; Yue et al., 2003). Thus, beclin 1 might not represent a “conventional” tumor suppressor such as Rb that satisfies the “Knudson two-hit hypothesis” criteria for classification as a tumor suppressor gene which indicates that it is necessary to demonstrate loss of both alleles, via either deletion or the presence of inactivating mutations (Knudson, 1971). Since TP53−/− mice are viable while BECN1−/− mice are early embryonic lethal, Vps34 complexes must provide a wider range of vital cellular functions than controlling p53 protein levels. Thus, while a reduction in beclin1 expression might promote tumorigenesis by reducing the levels of p53, a complete loss of beclin1 might negatively impact the development of certain tumors at least as a complete loss of beclin1 leads to early embryonic lethality (Qu et al., 2003; Yue et al., 2003) and might be required for cell viability at least for certain cell types. Thus, unlike “conventional” tumor suppressor, bi-allelic loss of beclin1 may not promote tumorigenesis and may lead to cell death. Consistent with this possibility, in contrast to that of normal cells, selected cancer cell lines demonstrate an increased sensitivity toward spautin-1 under starvation condition, suggesting that spautin-1 may be used to synergize with selected chemotherapeutic agents to induce cancer cell death. Spautin-1 might therefore provide a potential lead compound for developing a class of autophagy inhibitors as anticancer therapy.
EXPERIMENTAL PROCEDURES
High-Throughput Image Analysis
Cells were fixed with 4% paraformaldehyde (Sigma) and stained with 3 µg/ml DAPI (Sigma). Images data were collected with an ArrayScan HCS 4.0 Reader with a 203 objective (Cellomics ArrayScan VTI) for DAPI-labeled nuclei and GFP/RFP-tagged intracellular proteins.
Cell Lines and Culture Conditions
293T, MEF, HeLa, and Bcap-37 cells were cultured in DMEM media with 10% NCS. H4-LC3-GFP, H4-FYVE-RFP and MDCK cells were cultured in DMEM supplemented with 10% FBS and 1 X Na pyruvate (Invitrogen). Hs578Bst cells were cultured in Hybri-Care Medium (ATCC), supplemented with 30 ng/ml mouse EGF and 10% FBS. For starvation experiments, cells were cultured in DMEM supplemented with 10% serum without Glucose (GIBCO).
Antibodies
Rabbit polyclonal antibody anti-USP10, anti-USP13, anti-UVRAG, anti-Vps34 and anti-p53, were from Abcam. Rabbit polyclonal antibody anti-LC3B and mouse monoclonal antibody anti-β-tubulin were from sigma. Monoclonal antibodies anti-flag, anti-Myc and anti-HA were from Abmart. Rabbit polyclonal antibody anti-Beclin1 was from Santa Cruz. Polyclonal antibody anti-Atg14L was from MBL.
Protein-Lipid Blot Assay
Protein-lipid blot assays were carried out as reported (Dowler et al., 2002; Gozani et al., 2003). Briefly, lipids extracted from a 100 mm plate was spotted onto Hybond C-extra membrane (Amersham) and allowed to dry overnight in the dark. The membrane was incubated with lipid blocking buffer (1% BSA in TBST) for 1 hr, washed once in TBST for 30 min, and incubated with protein buffer (1 µg GST-tagged protein per 1 ml TBST with 1% BSA) overnight at 4C. Then the membrane was washed again in TBST for four times at 30 min each, incubated with anti-GST (Sigma) in 1% BSA buffer for 4 hr, washed in TBST for four changes with 5 min each, incubated with secondary antibody for 1 hr, and washed in TBST for four changes with 5 min each. All incubations were at roomtemperature unless noted otherwise. The signals were visualized with ECL.
In Vitro Deubiquitination Assay
In vitro deubiquitination assay was carried out using a similar protocol as described in (Yuan et al., 2010). Ubiquitinated Beclin1 was isolated from 293T cells transfected with expression vectors for HA-UB and FLAG-Beclin1. After Incubation with proteasome inhibitor MG132 (25 µM) and a pan DUB inhibitor G5 (25 µM) for 6 hr, ubiquitinated Beclin1 was purified from the cell extracts with anti-FLAG-affinity column in FLAG-lysis buffer (50 mM Tris-HCl [pH 7.8], 137mM NaCl, 10mM NaF, 1mM EDTA, 1% Triton X-100, 0.2% Sarcosyl, 1mM DTT, 10% glycerol and fresh proteinase inhibitors). After extensive washing with the FLAG-lysis buffer, the proteins were eluted with FLAG-peptides (Sigma). The recombinant Flag-USP10 and USP10CA were expressed in 293T cells and purified using FLAG affinity column and eluted with FLAG-peptide. For in vitro deubiquitination assay, ubiquitinated Beclin1 protein was incubated with recombinant USP10 in the deubiquitination buffer (50 mM Tris-HCl [pH 8.0], 50mM NaCl, 1mM EDTA, 10mM DTT, 5% glycerol) for 2 hr at 37°C.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dan Finley, Bruce Yankner, Zhujun Yao, Dana Christofferson, Dimitry Ofengeim, and Bénédicte Py for comments on the manuscript; Dr. Xin Xie of the National Center for Drug Screening in Shanghai for help with the original compound screen for autophagy regulators; Dr. Wade Harper for providing expression vectors of USP10 and USP13; Dr. Zhenkun Lou for mutant expression vector for USP10; Dr. Caroline Shamu (the director of the ICCB screening facility); and David Wrobel and Stewart Rudnicki for helps with siRNA screening. This work was supported in part by a NIH Director’s Pioneer Award US (to J.Y.), grants from the Chinese Academy of Sciences (KGCX2-SW-209 and KJCX2-YW-H08 [to D.M.]), the National Natural Science Foundation of China (21020102037 [to D.M.], and 90813007 [to L.Z.]) and the National Institute on Aging US (R37 AG012859 and PO1 AG027916 [to J.Y.]). M.K. is a recipient of Samsung Scholarship from South Korea. H.V.N. is supported in part by a fellowship form the Swedish Society for Medical Research (SSMF).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, seven figures, and one table and can be found with this article online at doi:10.1016/j.cell.2011.08.037.
REFERENCES
- Dang LC, Melandri FD, Stein RL. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry. 1998;37:1868–1879. doi: 10.1021/bi9723360. [DOI] [PubMed] [Google Scholar]
- Dowler S, Kular G, Alessi DR. Protein lipid overlay assay. Sci. STKE. 2002:pl6. doi: 10.1126/stke.2002.129.pl6. 2002. [DOI] [PubMed] [Google Scholar]
- Furuya T, Kim M, Lipinski M, Li J, Kim D, Lu T, Shen Y, Rameh L, Yankner B, Tsai LH, et al. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol. Cell. 2010;38:500–511. doi: 10.1016/j.molcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier JM, Simonsen A, D’Arrigo A, Bremnes B, Stenmark H, Aasland R. FYVE fingers bind PtdIns(3)P. Nature. 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO. J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. Mechanism, specificity and structure of the deubiquitinases. Subcell Biochem. 2010;54:69–87. doi: 10.1007/978-1-4419-6676-6_6. [DOI] [PubMed] [Google Scholar]
- Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Hoffman G, Ng A, Zhou W, Py BF, Hsu E, Liu X, Eisenberg J, Liu J, Blenis J, et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev. Cell. 2010;18:1041–1052. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson JD, Gillespie TD, Dunkerley HA, Maurice DH, Bennett BM. Inhibition of phosphodiesterase 5 selectively reverses nitrate tolerance in the venous circulationJPharmacol. Exp. Ther. 2006;317:188–195. doi: 10.1124/jpet.105.094763. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yan Z, Yang S, Cai J, Robinson H, Ke H. Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity. Biochemistry. 2008;47:12760–12768. doi: 10.1021/bi801487x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, Yu AD, Xie X, Ma D, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc. Natl. Acad. Sci. USA. 2007;104:19023–19028. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.