Abstract
The nervous system equips us with capability to adapt to many conditions and circumstances. We rely on an armamentarium of intricately formed neural circuits for many of our adaptive strategies. However, this capability also depends on a well-conserved toolkit of different molecular mechanisms that offer not only compensatory responses to a changing world, but also provide plasticity to achieve changes in cellular state that underlie a broad range of processes from early developmental transitions to life-long memory. Amongst the molecular tools that mediate changes in cellular state, our understanding of post-transcriptional regulation of gene expression is expanding rapidly. Part of the “epigenetic landscape” that shapes the deployment and robust regulation of gene networks during the construction and the remodeling of the brain is the microRNA system controlling both levels and translation of messenger RNA. Here we consider recent advances in the study of microRNA-mediated regulation of synaptic form and function.
Introduction
The success of biological systems depends upon their capacity to adapt to the environment. Over half a century ago, Conrad Waddington proposed that organismal development and reaction to the environment is governed by an “epigenetic system” that sculpts the pathway of embryogenesis (Waddington, 1942, 1959). Waddington’s elegant metaphor of the “epigenetic landscape” illustrated the alternative pathways that a cell might traverse depending on extrinsic influences and adaptive responses; the topology of this landscape being defined by a web of underlying gene networks (Waddington, 1957). Although modern usage of the term epigenetics invokes a rather specific set of chromosomal mechanisms that regulate gene expression, Waddington pondered the relationships between genotype and phenotype before the molecular machinery could be defined. In fact, Waddington described a genetically encoded adaptive mechanism as “a gun which is not only set on a hair trigger but which is aimed to hit the target when it goes off” (Waddington, 1959), anticipating the structure of cellular signaling to regulate downstream target genes (Figure 1A). We now appreciate that cells possess an extensive arsenal of adaptive signaling mechanisms suitable for responses to a wide range of temporal domains and environmental conditions or cellular interactions (Figure 1B). While rapid and local state changes are effectively triggered by conformational, catalytic and posttranslational modification of molecules already available in the cell, sustained adaptive state changes can persist beyond the lifetime of individual molecules, such as the memories stored in neural networks. Mechanisms that link adaptive responses to expression of the genome provide not only the renewable resource of RNA and protein, but can also alter the ‘program’ of the cell via qualitative changes in expression (reviewed by Flavell and Greenberg, 2008). Although transcriptional mechanisms can produce very long-lived state change, they offer limited spatial acuity and thus depend on posttranscriptional processes for regulated delivery of the expressed genome. Spatial constraint is particularly important in the nervous system where extremely complex cell architecture is essential for circuit structure and function. Thus, the topic of translational regulation at the RNA level is an exciting frontier in the context of neurobiology.
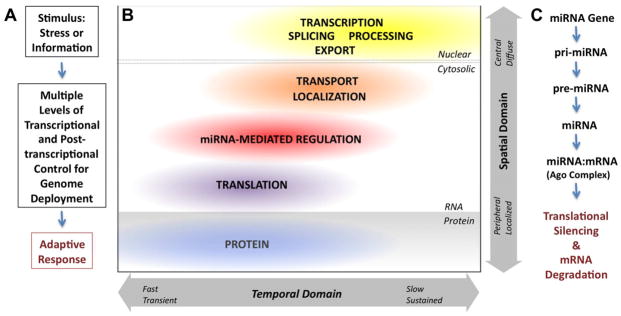
Figure 1. Spatial and Temporal Domains in Genome Expression and Function.
A. Waddington’s adaptive “gun” response triggered by environmental stimuli or information from neirghboring cells can utilize a broad range of molecular mechanisms to mediate changes in the expression of the genome to alter phenotypes or cellular behavior.
B. This diagram represents the relative effective spatial and temporal range of function and adaptive response for different mechanisms in the progression from primary nuclear production of mRNA (transcription, splicing, processing and export; in yellow), to mRNA delivery (transport and localization; in orange), to posttranscriptional miRNA regulation of mRNA (mRNA stability and access to translational machinery; in red), to active translation (in purple) and to the final function of the encoded protein(s) (in blue). While transcriptional mechanisms can be sustained for prolonged periods of cellular and/or organismic lifetime, these processes are slow to respond and have very limited spatial acuity. Posttranscriptional regulation of gene expression offers faster and far more local responses, although conformational change of existing proteins provide the highest spatial and temporal resolution.
C. A simple flow diagram of microRNA biogenesis (from transcriptional production of pri-miRNA to nuclear microprocessor cleavage of pre-miRNA to cytoplasmic cleavage to mature miRNA) and subsequent matching with mRNA targets (in Argonaut [Ago] containing protein complexes), leading to translational silencing and mRNA decay.
Late in his career, Waddington made a somewhat neo-Lamarckian argument that a nervous system capable of learning and teaching was an innovation that freed humans from the arduous process of evolving new genetically encoded capabilities (Waddington, 1959). While the evolution of ideas may be largely uncoupled from the genome, we have learned that memory is quite dependent on gene expression. This was first suggested in 1963 by the memory-blocking effects of the translational inhibitor Puromycin (Flexner et al., 1963). An impressive convergence between the fields of memory and signal transduction research eventually defined highly conserved pathways from cell surface receptors to second messengers to intracellular kinases to transcription factors that link synaptic activity to changes in gene expression (Kandel, 2001). For memory, these pathways showed how short-lived signaling events linked to gene expression could trigger long-lived state changes in a postsynaptic cell, thus coupling adaptive mechanisms across multiple temporal domains. An additional convergence between studies of synaptic plasticity and neurotrophin signaling mechanisms made it clear that signal-dependent deployment of the genome through local protein synthesis was a key to understanding state change at mature synaptic sites (Kang and Schuman, 1996; Martin et al., 1997). It was then discovered that local protein synthesis is also important for multiple stages in the assembly of neural circuits, from axon guidance decisions to synapse formation (reviewed by Jung et al., 2012; Kindler and Kreienkamp, 2012).
The discovery of latent mRNAs that the cell reserves or “masks” for later translation dates back nearly half a century to studies of protein synthesis in sea urchin embryos (e.g. Monroy and Tyler, 1963; Piatigorsky et al., 1967). However, the complexity of mRNA pools that reside in different compartments of developing and mature neurons has been defined only recently with modern genomic technologies, revealing hundreds of candidate transcripts localized in dendrites or axons or even growth cones (Poon et al., 2006; Zhong et al., 2006; Zivraj et al., 2010), many of which may be changing in developmental time (Gumy et al., 2011). Indeed, recent analysis of the hippocampal CA1 neuropil has identified over twenty-five hundred mRNAs in the “local transcriptome” of axons and dendrites (Cajigas et al., 2012). These observations suggest that the ‘RNA space’ subject to posttranscriptional regulation in neurons is substantial. Given their exaggerated morphology, neurons require long-range transport mechanisms to deliver mRNAs along axons and dendrites. Studies of neuronal mRNA transport granules indicate that translation is suppressed en route (Krichevsky and Kosik, 2001), raising intriguing questions regarding the mechanisms that control and activate local translation. While significant progress has been made in defining general components of mRNA transport and storage granules (reviewed by Donnelly et al., 2010), and some exciting insights have been made into signal or state-dependent activation of such players (e.g. Banerjee et al., 2009), a key question is how are individual genes targeted for specific regulation? Although multiple classes of sequence specific RNA regulatory mechanism contribute to shaping the functional landscape, and there are significant interactions between these molecular regulators, we will focus on microRNA (miRNA) mediated control over the maturation and plasticity of neurons and their synaptic connections, highlighting primarily observations made in the past few years.
The Neural miRNA Landscape
miRNAs were first identified based on classical genetics as regulators of developmental timing in Caenorabditis elegans (Lee et al., 1993; Reinhart et al., 2000). These short non-coding RNA were then found in other organisms by virtue of striking sequence conservation across species (Pasquinelli et al., 2000). miRNA genes are transcribed as RNA polymerase II or III transcripts (pri-miRNA) that are processed by specific nuclease cleavage (or RNA splicing for miRtrons) to produce short hairpin RNAs (pre-miRNA) that are transported out of the nucleus and then cleaved once more to generate mature miRNAs that can be loaded into protein complexes that allow binding to specific target mRNA (Figure 1C; reviewed by Bartel and Chen, 2004). Mature miRNA-target mRNA pairs are formed by proteins in the Argonaut (Ago) family together with other components of the RNA-induced silencing complex (RISC; Du and Zamore, 2005). Although there are exceptions, miRNAs inhibit expression for most target genes by reducing steady state message levels (Guo et al., 2010), although this may occur after an initial blockade of translation (Bazzini et al., 2012; Djuranovic et al., 2012).
Many rounds of transcriptome sequence and expression analysis have uncovered a large number of miRNA genes spanning all multi-cellular organisms (see http://www.mirbase.org; Griffiths-Jones et al., 2006). Amongst animal species, the number of miRNA genes has expanded dramatically with increasing organismal complexity (ie. numbers of differentiated cell types), contributing to speculation that despite high conservation in many miRNA families diversification of other miRNA genes has contributed significantly to the evolution of different metazoan bodyplans (Sempere et al., 2006). For example, Cnidarian genomes contain tens of miRNA genes [e.g. 17 in Hydra and 49 in Nematostella], whereas Ecdysozoa have roughly five to ten-fold more [e.g. 223 in C. elegans; 240 in D. melanogaster], and Humans have over fifteen hundred (http://www.mirbase.org). Interestingly, recent comparisons of mRNA and miRNA populations expressed in the brains of different primate species suggest that a subset of developmentally regulated miRNA in prefrontal cortex (PFC) appears to be evolving far more rapidly than other classes of genes including transcription factors (Somel et al., 2011). For example, 19 such developmentally regulated miRNA in PFC were 24-fold more divergent in human than in chimpanzee. Thus, while gene regulatory pathways have long been proposed as a predominant driver of metazoan evolution (see Gerhart and Kirschner, 1997), miRNA may account for a significant part of the expansion in cognitive and intellectual capacity in humans.
Given the cellular and transcriptional complexity of the nervous system, it is not surprising that miRNAs are highly abundant in this tissue (reviewed by Kosik, 2006). Although initial comprehensive profiling of miRNA expression was limited to broad areas of the brain, the advent of new profiling technology makes it clear that the spatial landscape of miRNA expression may be highly complex at the cellular level. For example, by combining immunoprecipitation of tagged, transgenic Ago2 with the cell type specific Cre/Lox system in mouse (a method called “miRAP”; Figure 2A), it has been possible to ascertain the miRNA “finger prints” of different GABAergic interneurons and excitatory pyramidal cells from neocortex or Purkinje cells from cerebellum (He et al., 2012). Nearly half of the over five-hundred miRNA assayed were relatively specific between overall neocortex and cerebellum, and roughly one quarter of the miRNA showed specificity between pyramidal neurons and interneurons, or between two subtypes of interneurons (parvalumin [PV] versus neuropeptide somatostatin expressing [SST]; Figure 2B). For example, six of ten miRNA quantified in follow-up experiments were selectively enriched in PV interneurons, despite the fact that these neurons share many properties with SST interneurons (Figure 2C; He et al., 2012). Thus, while profiling at this single cell-type resolution has just begun, it is clear that the miRNA landscape offers many opportunities to fine-tune the distinct developmental and functional properties of neuronal subpopulations.
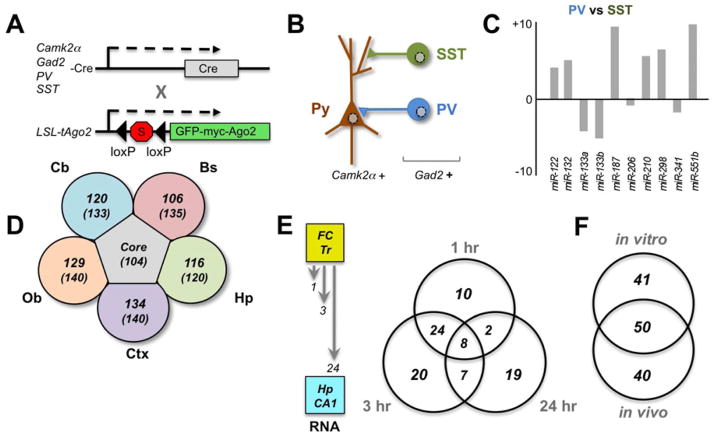
Figure 2. Cellular, Subcellular and Temporal Specificity in Neural miRNA Profiles.
A. Conditional Cre-dependent expression using one of multiple promotors for excitatory Pyramidal neruons (Camk2a) or GABA-ergic interneurons (Gad2, PV or SST) is used to express a GFP-myc tagged-Ago2 fusion (by removal of a stop flanked by loxP sites) to isolate Ago:miRNA:mRNA complexes for cell type-specific immunoprecipitation (miRAP; He et al., 2012).
B. Diagram of three neurons profiled with miRAP: Pyramidal excitatory neurons (Camk2a positive), SST or PV interneurons (both of which are Gad2 positive (adapted from He et al., 2012)
C. Diagram approximates the relative differences in expression for 10 miRNA when PV and SST populations of Gad2+ GABA-ergic interneurons were compared by miRAP (He et al., 2012).
D. A summary of the overlapping sets of miRNA identified by profiling five distinct regions of rodent brain and neurosynaptosomal fractions isolated from these tissues (Pichardo-Casa et al., 2012). Cortex (Ctx), Hippocampus (Hp), Brainstem (Bs), Cerebellum (Cb), and Olfactor buld (Ob) were compared, revealing 104 miRNA common to all five regions. The majority of the total miRNA identified in each tissue (in parenthesis) were also found in synaptosomes from each brain region.
E. Using a fear-conditioning (FC) paradigm, RNA was extracted from dissected hippocampal (Hp) CA1 at three time points (1, 3 and 24 hours) after training (Tr). Subsequent profiling identified overlapping sets of miRNA in each time point whose overall numbers are summarized in the Venn diagram to the right (adapted from Kye et al., 2011).
F. Comparisons of miRNA identified in the in vivo fear conditioning paradigm showed significant overlap with cultured hippocampal neurons subjected to pharmacological stimulation in vitro (Kye et al., 2011).
Even within a single neuron, complex functional architecture offers many compartments that could be regulated by different sets of miRNA. An early comparison between miRNA in the cell bodies and neurites of rodent hippocampal neurons showed a graded distribution across a set of 99 candidates, the extremes of which defined miRNA that are selectively enriched in dendrites versus soma (Kye et al., 2007). This study also examined miRNA copy number and estimated an average of 10,000 copies per cell, a number that is within an order of magnitude of average synapse number per neuron, thus raising the intriguing question of whether synaptic miRNA can be locally effective in very small numbers. Nonetheless, the synaptic compartment appears to contain a large fraction of the neuronal miRNA pool. Recent analysis of miRNA representation in synaptoneurosome fractions from five different rodent brain regions showed that roughly half of the miRNA genes tested were enriched in this synaptic material (Pichardo-Casa et al., 2012). Of roughly 140 miRNA expressed in five regions of the rat brain (cortex, hippocampus, cerebellum, brainstem and olfactory bulb) the majority (79–97%) were also found in synaptosomes from each region (Figure 2D). While a significant number (up to ~25%) of the miRNA detected in the study showed region specificity, the fact that about 100 of the detected miRNA were found in all regions suggests that most miRNA are part of core neural machinery. Interestingly, a small subset of miRNA was exclusively detected in synaptic material in each region (3–9%), implying dedicated synaptic functions. When a subset of the synaptic miRNA were then quantified after kainic acid-induced seizure, the majority (5 out of 6) showed a significant activity-dependent change in the synaptic material even though changes in whole tissue were often not detected (Pichardo-Casa et al., 2012). Of particular interest, several of these activity-dependent miRNA displayed strikingly different changes in different brain regions; for example, miR-150 is increased over 5-fold in cortical synaptosomes, but is reduced about the same fold in hippocampus, whereas miR-125 displays the opposite trend. Although this comparative analysis has only been applied to a handful of synaptic miRNA, it suggests that future functional analysis may reveal many new synaptic functions for miRNA, and that there may be dramatic specificity in these functions in different neural circuits.
If miRNA expression, localization or function can be controlled by neural activity or other influences of neighboring cells and the environment, then miRNA can serve as agents of adaptive state change. Sensory input to the nervous system from the environment appears to trigger significant changes in miRNA stability in the visual system (e.g. Krol et al., 2010). Moreover, from a developmental perspective, a substantial body of evidence shows that miRNA production and activity is controlled by several canonical cell-signaling pathways known to be important for many stages in the construction of neural circuits (reviewed by Saj and Lai, 2011). In addition to hardwiring neural circuits, some of these pathways are also known to link synaptic form and function to neural activity (e.g. Brain-Derived Neurotrophic Factor [BDNF]; Schratt et al., 2006). Multiple studies have surveyed miRNA levels in models of activity-dependent synapse plasticity (reviewed by Olde Loohuis et al., 2012). For example, in hippocampal slices subjected to long-term potentiation (LTP) or depression (LTD) of synaptic output, the majority of detected miRNA (55 of 62) showed more than 2-fold up or down-regulation (Park and Tang, 2009).
The temporal dimension adds another layer of complexity in the adaptive response. For example, a recent profile of hippocampal miRNA levels following contextual conditioning in vivo showed significant changes in miRNA pattern between 1, 3 and 24 hours post-training compared to animals that received NMDA-receptor antagonist prior to training (Kye et al., 2011). Dozens of miRNA were significantly up or down-regulated at each time point, however, the overlap between the initial response at 1 hour and the long-term response at 24 hours was less than 25% (Figure 2E). When cultured hippocampal cells were profiled after pharmacological stimulation in vitro to compare miRNA changes after fear conditioning, just over half of those with detectable changes were found in both the in vitro and in vivo models (Figure 2F). This suggests that while cell culture models for neuronal plasticity can serve as very convenient systems to manipulate miRNA that also provide impressive access to neuronal cell biology, analysis using in vivo models is essential. Interestingly, when downstream target gene mRNAs altered in both in vitro and in vivo were compared (Kye et al., 2011), several components in the miRNA core biosynthetic pathway were found to be part of the adaptive response (including DGCR8, Drosha and Dicer), consistent with other studies suggesting that miRNA processing is actively coupled to neuronal activity in order to propel synaptic plasticity (see below).
Synaptic Functions for the miRNA Pathway
The components of the miRNA biogenesis and processing machinery are well conserved across the animal kingdom. After transcription, pri-miRNA is processed by RNAse III domain-containing protein Drosha in association with the RNA binding protein encoded by DIGeorge syndrome Critical Region gene 8 (DGCR8)/Pasha (reviewed by Du and Zamore, 2005). This “microprocessor” complex binds to the lower stem region of the miRNA self-complementary region (Carthew and Sontheimer, 2009). The double-stranded stem and flanking regions are both important for DGCR8 binding and subsequent Drosha cleavage (Zeng and Cullen, 2006; Han et al., 2006; reviewed by Kim et al., 2009). Processed miRNA precursors (pre-miRNA) are then exported from the nucleus and cleaved by the RNAse III domain-containing protein Dicer. Finally, the remaining duplex is loaded on to the RNA-induced silencing complex (RISC) which is comprised of a set of proteins that mediate mRNA target recognition and suppression, including Ago1, Ago2, Pumilio2 (Pum2) and Moloney leukemia virus (MOV10), (Du and Zamore, 2005).
Pioneering studies of nervous system development using maternal-zygotic mutants of zebrafish dicer revealed gross morphological defects specifically in early brain patterning and morphogenesis (Giraldez et al., 2005). Surprisingly, these dramatic abnormalities are largely rescued by re-introduction of miR-430-family members, suggesting that the complexity of miRNA control over the early stages of neural development may be quite limited. However, detailed studies of later stages in neural development have begun to suggest a more extensive contribution of miRNAs in the formation of synaptic connections, circuit maturation and the activity driven plasticity of these connections. Part of this evidence came from knock out mutations of the miRNA processing genes. For example, a clonal genetic screen in Drosophila identified the miRNA processing proteins Pasha/DGRC8 and Dicer1 as crucial components in the establishment of wiring specificity (Berdnik et al., 2008). Alleles of fly drosha, its dsRBD partner pasha, and novel alleles of dicer-1 were recently identified in another genetic screen in Drosophila. Hypomorphic alleles that gave adult escapers with overtly normal development were identified and shown to exhibit reduced synaptic transmission in the mutant photoreceptor neurons with no accompanying defects in neuronal development or maintenance (Smibert et al., 2011). This suggests that synaptic function is especially sensitive to optimal miRNA pathway function. DGCR8 mutant mice also exhibited abnormalities in synaptic connectivity due to a reduction in the number and size of dendritic spines, reduced synaptic complexity, impaired synaptic transmission and altered short-term plasticity (Stark et al., 2008, Fenelon et al., 2011). Moreover, specific loss of Dgcr8 in pyramidal neurons of the cortex results in a non-cell-autonomous reduction of parvalbumin interneurons in the prefrontal cortex, with a severe deficit in inhibitory synaptic transmission corresponding with a reduction in inhibitory synapses. This research directly implicates miRNAs as functioning in inhibitory synapses and illustrates the global effects cell specific knockdown of miRNAs can impart (Hsu et al., 2012).
Many studies have demonstrated that spatial and temporal specificity is vital to many miRNA roles during neural development. For example, loss of murine dicer in a tissue specific manner revealed a multitude of neuronal abnormalities including impaired neuronal differentiation, reduced neuronal size, neuronal branching deficits and disrupted axonal pathfinding (reviewed by Bian and Sun, 2011). Beyond the morphological changes observed, analysis of downstream elements in the miRNA processing pathway has identified Ataxin-2 as being required for Drosophila long-term olfactory habituation (LTH). Mechanistically, Ataxin-2 binds the DEAD box helicases of the Me31B family, proteins associated with Argonaute (Ago), where it participates as part of the general machinery required for efficient miRNA-mediated translational repression (McCann et al., 2011). However, the requirement for miRNAs in LTH appears to be a very complex one. miRNAs are necessary for the maintenance of neuronal connections as indicated with Ataxin-2 studies, but they are also involved in synaptic remodeling. For example, an inducible deletion of murine dicer 1 decreased expression of specific miRNAs, but demonstrated enhanced memory strength in the CA3 to CA1 synapses (Konopka et al., 2010). Morphologically, the dendritic spines in these dicer 1 mutants displayed an increase in immature filopodia-like dendritic spines. Molecularly the mutants displayed an increase in the translation of synaptic plasticity-related proteins BDNF and MMP-9.
Notably, studies of RISC complex components that mediate the final steps in the core miRNA pathway were some of the first to implicate miRNA in synapse formation. On the other hand, pioneering studies in Drosophila established the importance of the RISC component Armitage in long-lasting memory within the adult olfactory system though analysis of CamKII expression (Ashraf et al., 2006). These studies indicate miRNAs may be acting in both neuronal remodeling and maintenance of neuronal connections in memory and their opposing roles may be due to the spatial temporal specificity of their expression. Zeroing in on the temporal contribution of miRNAs, their role in early hippocampal development was investigated by conditionally ablating dicer at varying embryonic time points. These studies revealed a timing requirement of miRNAs for the formation of specific hippocampal regions (Li Q et al., 2011). As a whole, studies of the core miRNA processing pathway have focused attention on miRNA function in neural circuits, but mechanistic insights into such functions require analysis of individual miRNAs and the target genes they control.
Synaptic Regulatory Functions for Individual miRNA
Much of our knowledge about individual miRNA functions at the synapse was initially informed by studies profiling miRNA expression in the nervous system. Candidate miRNA functions have been frequently explored by initial studies in primary dissociated cell culture models that provide a platform highly accessible to miRNA manipulation through the use of antagomers, “locked nucleic acid” (LNA) oligonucleotides, and over-expression constructs (e.g. Giraldez et al., 2005; Leaman et al., 2005; Krutzfeldt et al., 2005; Lanford et al., 2010). A draw back for use of LNAs to disrupt miRNA is their difficulty of use in in vivo systems. Over-expression models can be easier to execute than in vivo loss-of-function models, but can be misleading due to the very tight expression range in which miRNAs function. As a whole, experiments using both loss and gain-of-function have been very informative in the role miRNAs are playing at the level of individual neurons and neuronal cell biology, but due to the inherent tuning nature of miRNAs and the importance of spatial and temporal control it is important to emphasize that analysis of miRNAs in an intact cellular context at endogenous levels is very important. As we examine recent work in the area of miRNAs at the synapse, two major themes arise (Figure 3). The themes of both the negative and positive regulation of synaptic growth illustrate the balancing and tuning role miRNAs play to facilitate synaptic development and activity driven plasticity.
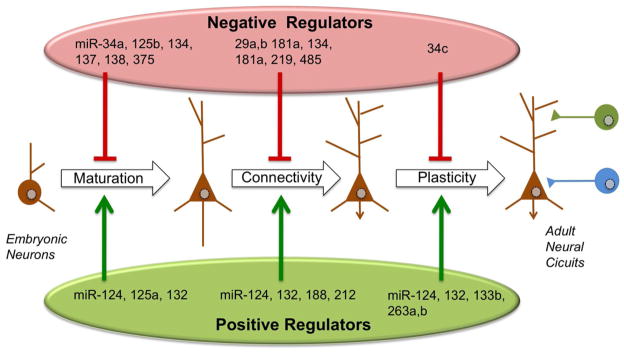
Figure 3. miRNA Involved in Various Aspects of Synaptic Development and Function.
Late stages of neuronal differentiation from process formation (axon and dendrite extension during maturation) to the developmental and continued plasticity required to form higher order circuits. Although comprehensive functional screens will be required to form a complete inventory of functions, several miRNA have been that have been shown to regulate these steps of neuronal and circuit formation or function as either negative regulators (above the timeline) or positive regulators (below the timeline).
Perhaps not surprisingly, negative regulation and suppression of synaptic connections appears to be a primary function of many miRNAs at the synapse (Figure 3). For example, miR-138 is found highly enriched in the brain and localized within dendrites. miR-138 targets acyl protein thioesterase 1 (APT1), an enzyme that defines the palmitoylation status of multiple proteins that are known to function at the synapse including the Gα13 subunits of G proteins negatively regulating the size of dendritic spines in rat hippocampal neurons (Siegel et al., 2009). miR-34a, another miRNA that imparts negative regulation, is controlled by TAp73 (Agostini et al., 2011a). Ultimately, miR-34a negatively regulates both dendritic outgrowth and synaptic function, possibly via targeting the synaptic components synaptotagmin-1 and syntaxin-1 (Agostini et al., 2011a,b), although the relevant target genes have not yet been confirmed. miR-375 on the other hand antagonizes brain-derived neurotrophic factor (BDNF) to inhibit dendritic growth (Abdelmohsen et al., 2010). miR-375’s actions are largely through its target HuD, an RNA binding factor known to control mRNA stability and translation in the nervous system (Deschenes-Furry et al., 2006). As a whole, these observations imply that there are multiple layers of complexity in the regulatory logic of miRNAs in dendritic morphogenesis.
Some miRNAs play different roles at distinct developmental stages. For example, the brain-enriched miR-137 has an early role in neural differentiation: miR-137 regulates CDK6 in cultured mouse neural stem cells, resulting in an increased level of neuronal marker Tuj1 (Silber et al., 2008). miR-137 also controls later steps in developmental plasticity where it is a key regulator in adult neurogenesis (Szulwach et al., 2010) and neuronal maturation (Smrt et al., 2010). However, gain of function studies conducted with miR-137 resulted in decreased dendritic spine growth demonstrating miR-137 was sufficient to negatively regulate synapse morphogenesis. In order to address synaptic function at a late stage of differentiation, miR-137 was suppressed in using an oligo-based technique in cultured primary neurons, and dendritic spine growth was significantly increased. Further study of the mechanism by which dendritic growth regulation occurs revealed that miR-137 elicits changes in synapse morphogenesis largely through regulation of the ubiquitin ligase Mind Bomb-1 (Smrt et al., 2010). Interestingly, a recent genome wide association study (GWAS) has implicated single-nucleotide polymorphisms in the miR-137 gene as being highly associated with schizophrenia (Ripke et al., 2011), and multiple schizophrenia-associated genes including CSMD1, C10orf26, CACNAiC and TCF4 have been confirmed in cell culture to be targets of miR-137 (Kwon et al., 2011). In vivo analysis of miR-137 targets will be an important step in better understanding the role of this miRNA in schizophrenia, a disease where other miRNA genes have been recently implicated.
miRNA regulation at the synapse is not only negative. An example of positive regulation of dendritic spine development is observed with miR-125b. miR-125b and miR-132 (as well as several other miRNA) are associated with Fragile × Mental Retardation Protein (FMRP) in mouse brain. miR-125b over-expression results in longer, thinner processes of hippocampal neurons. FMRP knockdown is shown to ameliorate the effect of over-expressed miR-125b and miR-132 on spine morphology. It has been proposed that miR-125b negatively regulates its target, NR2A, along with FMRP and AGO1 (Edbauer et al., 2010). Recently a mechanism was proposed whereby FMRP phosphorylation provides a reversible switch where AGO2 and miR-125a form and inhibitory complex on PSD-95 mRNA, thus turning off mGluR signaling. However, dephosphorylation of FMRP and subsequent release of Ago2, activates gp1 mGluR signaling (Muddashetty et al., 2011). This switching mechanism could provide the means for temporal and spatial control of translation.
Because some miRNAs can both positively and negatively influence synaptic growth and connections depending on their levels, the concept of miRNAs as fine-tuners of synaptic effector gene networks has long been a popular model for regulation of activity related plasticity. This topic has been extensively reviewed (Siegel et al., 2011; Bredy et al., 2011; Olde et al., 2012); however, we will highlight a few recent advances that illustrate the functional role for miRNAs in this arena. mir-124 is one of the most highly conserved neuronal specific miRNAs and yet gross morphological phenotypes have not been observed in the nervous system in null mutants from multiple species (Miska et al., 2007; Sun et al., 2012). However, when examining the role of miR-124 in activity driven plasticity, we begin to see its functional relevance in the nervous system. miR-124 responds to serotonin in cultured aplysia motor neurons by de-repressing CREB and enhancing serotonin-dependent long-term facilitation (Rajasethupathy et al., 2009). Another miRNA that appears to tune levels of targets in response to activity related plasticity is miR-188. miR-188 was found to be up-regulated with the induction of long-term potentiation (LTP) where it regulated the semaphorin 3F receptor Nrp-2 acting as a negative regulator of spine development and synaptic structure in rat primary hippocampal neuron culture (Lee et al., 2012). These studies continue to illustrate how miRNAs can be playing a very active role in regulation of activity-regulated plasticity.
Pharmacological disruption of neurotransmitter signaling has helped to further elucidate the role of miRNAs in activity driven plasticity. One study disrupted NMDA- mediated glutamate signaling recapitulating behavioral deficits displayed in psychiatric disorders. After blocking glutamate signaling, miR-219 expression was reduced in the prefrontal cortex of mice (Kocerha et al., 2009). A known component of the NMDA receptor signaling cascade, CamKIIγ, was confirmed in cell culture as a miR-219 target. In vivo inhibition of miR-219 was shown to recapitulate the behavioral deficits associated with disruption of the NMDA receptor transmission and treatment with antipsychotic drugs prevented drug induced effects on miR-219 (Kocerha et al., 2009).
Another neurotransmitter pathway examined was dopamine signaling, which is increased with cocaine and amphetamine use. Dopamine signaling was shown to increase the expression of miR-181a in primary neurons. Over-expression and knockdown of miR-181a in primary neurons demonstrated that miR-181a was a negative post-transcriptional regulator of GluA2 surface expression, spine formation and mEPSC frequency in hippocampal neuron cultures establishing a key role for miR-181 in response to neurotransmitters at the synapse (Saba et al., 2011). Furthermore, chronic treatment of cultured hippocampal neurons with nicotine, cocaine or amphetimines also increased miR-29a/b expression, reducing dendritic spines and increased filopodial-like cytoskeleton remodeling. This morphological change, was found to occur through miR29a/b targeting of Arpc3 acting to fine tune structural plasticity through regulation of the actin network branching in mature and developing spines (Lippi et al., 2011).
Neurotransmitters have long been studied as a mechanism of homeostatic neuronal plasticity (reviewed in Pozo and Goda, 2010). Recently, miRNAs have been implicated in neurotransmitter receptor expression. Surface expression of GluR2 as well as PSD-95 clustering and dendritic spine density were negatively altered by miR-485. On a functional level, miR-485 was shown to reduce spontaneous synaptic activity in hippocampal neurons largely through its presynaptic target SV2A (Cohen et al., 2011). This builds on previous studies where miR-485 was found to be dysregulated in neurological disorders such as Huntington and Alzeheimers disease (Packer et al, 2008; Cogswell et al., 2008). These studies build a strong link between miRNAs and neurotransmitter signaling.
Through the study of both negative and positive regulation of synaptic development and remodeling a reoccurring theme of miRNA dysregulation in neuronal disease has come to light. This gives us insight into miRNAs as a very applicable and exciting avenue to follow to better understand neurological diseases and their treatment (Ceman and Saugstad, 2011; Bian and Sun, 2011). Given the importance that miRNAs might play in neuropathology several strategies to manipulate miRNA activity and expression are being pursued as therapeutic models. Ruberti et al., 2012 further discuss these in a recent review. However, dissociated culture models described above lack the context of multicellular environment and global circuitry thus having limitations as disease models. The field is now shifting to in vivo models and gaining the tools necessary to manipulate miRNAs in this context. For a small set of miRNAs we have been able to see the progression of in vivo cell biological data confirmed and studied within the context of in vitro models.
Bridging from In Vitro to In Vivo Models of Investigation
miR-132 and miR-134 are at the vanguard in the study of miRNA function at the synapse. These miRNAs demonstrate the power of studies with neuronal miRNAs in vitro (Vo et al., 2005; Schratt et al., 2006; Wayman et al., 2008) as well as the transition to in vivo models where they clearly demonstrate how miRNAs exert developmental and cellular context-dependent functions. miR-134 was identified in hippocampal neurons as a dendritically localized miRNA and functions to negatively regulate the size of dendritic spines through the inhibition of LimK1, a regulator of actin dynamics. This inhibition was relieved by exposure to stimuli such as BDNF (Schratt et al., 2006). Another layer of complexity was identified for miR-134, as part of the miR378–410 cluster downstream of the transcription factor Mef2. Many members of this cluster were shown in primary culture to be required for activity-dependent dendritic outgrowth of hippocampal cultured neurons. miR-134 regulation of Pumilio2, an RBP involved in miRNA transport and translational inhibition was shown to be key in this activity-dependent dendritic arbor plasticity illustrating a regulatory pathway that couples activity-dependent transcription of miRNA with miRNA-dependent translational control of gene expression in neuronal development (Fiore et al., 2009), suggesting a possible cascade that might alter levels of multiple downstream effector genes.
Similar to work with other miRNAs, early studies of miR-134 were largely dependent on cultured neurons that lack specific spatial and temporal information that in vivo studies offer. More recent research in mouse models confirmed the negative regulatory role of miR-134 in dendritic arborization of cortical layer V pyramidal neurons (Christensen et al., 2010). Additional in vivo analysis has identified sirtuin1 (SIRT1) as a regulator of miR-134 in synaptic plasticity and memory formation where it acts to limit the expression of miR-134 via a repressor complex containing the transcription factor YY1. In the absence of SIRT1, an increase of miR-134 down regulates CREB resulting in impaired synaptic plasticity (Gao et al., 2010). Additional in vivo studies have identified a functional role for miR-134 in specific periods of neuronal development demonstrating that miR-134 can target Chordin-like 1 and Doublecortin providing stage-specific modulation of cortical development (Gaughwin et al., 2011). miR-134 has also been shown to play a role in neuroprotection and seizure suppression effects in an in vivo mouse model strengthening the need for further study of the implications of miRNAs dysfunction in neuronal disease (Jimenez-Mateos et al., 2012). As a whole, work with miR-134 reinforces the concept that miRNAs exert developmental and cellular context-dependent functions, thus highlighting the need for in vivo models with cell type specific control.
Studies of the miR-132/miR-212 gene cluster indicate that these miRNAs have many diverse functions and targets depending on their spatial and temporal expression, reviewed in Wanet et al., 2012. In the nervous system, miR-132 is a CREB-regulated miRNA that is induced by neuronal activity and neurotrophins, and plays a role in regulating neuronal morphology and cellular excitability (Vo et al., 2005). This links transcriptional regulation of miRNAs furthering the flexibility of the genome in response to selective pressures. In culture, up-regulation of miR-132 increases dendritic outgrowth in an activity-dependent fashion via suppression of a GTPase-activating protein p250GAP translation resulting in activation of the Rac1-PAK actin-remodeling pathway (Vo et al., 2005; Wayman et al., 2008, Impey et al., 2010). In agreement with these studies, over-expression of miR-132 in hippocampal neurons results in stubby and mushroom-shaped spines with an increase in average protrusion width strengthening synaptic transmission (Edbauer et al., 2010). The in vitro work on miR-132 in cultured neurons was confirmed in an in vivo model where the miR-132/miR-212 locus was targeted for deletion in the adult mouse hippocampus. Of these two miRNAs, miR-132 was determined to be the predominately active product in hippocampal neurons and deletion caused a dramatic decrease in dendrite length, arborization and spine density (Magill et al., 2010).
In vitro analysis of miR-132 function not only supports a role for miR-132 in developmental plasticity, but also illustrates a continued role for miR-132 in activity-induced plasticity. miR-132 has been shown to selectively influence short-term plasticity in hippocampal cultures without altering basal synaptic transmission (Lambert et al., 2010). Additionally the induction of long-term potentiation (LTP) in the dentate gyrus of adult rats was coincident with a strong up-regulation of mature miR-212 and miR-132 transcripts. Blocking NMDA receptors enhanced the LTP-dependent induction of these miRNAs where as the blocking of mGlur1 inhibited the enhancement of mature miRNA expression in response to LTP-inducing stimuli (Wibrand et al., 2010). In fact, it was shown that blocking glutamate receptors activates the decay of miR132, whereas glutamate treatment did not have an effect (Krol et al., 2010). These findings suggest specific and fine local regulation through synthesis and degradation in specific synaptic compartments where this cluster is involved in synaptic plasticity modulation.
Because synapse strength and number are scalable properties, the ability of miRNA to fine tune synaptic effector genes is a powerful tool to regulate the functional output of neurons and circuits. The concept of tight regulation and tuning control of miRNAs is illustrated in the research on miR-132 where expression was found to be up-regulated in key layers of the mouse hippocampus after presentation of a spatial learning tasks (Hansen et al., 2012). Furthermore in vivo induction of miR-132 restoring normal endogenous levels significantly enhanced congnitive capacity. In contrast, high levels of miR-132 inhibited learning suggesting that miR-132 must be maintained in a limited range for learning and memory formation. Strict regulation of miR-132 expression is also implicated as the basis of a structural plasticity program in subventricular zone - olfactory bulb postnatal neurogenesis (Pathania et al., 2012). Both of these pieces of data support the role of miRNAs in a tuning capacity to regulate other genes within a specified range of expression.
Beyond implications in morphological change and plasticity, miR-132 has been tied to the pathophysiology of depressive disorders where increased glucocorticoid levels have been shown to down regulate BDNF, which is responsible for normal induction of miR-132 (Kawashima et al, 2010). Recent studies have also implicated miR-132 dysregulation in schizophrenia where miR-132 was found down regulated in schizophrenic subjects. Several key genes, including DNMT3A, GATA2 and DPYSL3 were regulated by miR-132 and exhibited altered expression either during normal neurodevelopment or in tissue from adult schizophrenic subjects (Miller et al., 2012). miR-132 family member, miR-212 has also been suggested to act in adaptive behaviors such as those observed with drug use. miR-212 is believed to act through MECP2 to control the effects of cocaine on striatal BDNF levels (Im et al., 2010; Hollander et al., 2010). For in depth coverage of miR-132 and miR-212 functions, please see recent reviews (Wanet et al., 2012; Tognini and Pizzoruso, 2012).
Overall, work with both miR-134 and miR-132 have demonstrated how complementary work in vitro and in vivo provide a powerful approach to dissect the complex role miRNAs are playing at the synapse. These studies illustrate how miRNAs regulate multiple target genes in different regions and cell types at varied times in development to control both developmental and physiological plasticity.
In Vivo Analysis of Synapse Form and Function in Invertebrates
Much like the in vivo examination in mammalian systems, in vivo analysis in invertebrate systems has helped us understand the spatiotemporal context of miRNA function. The importance of cellular context is clearly demonstrated in the developmental assembly of presynaptic structures, which relies on communication between both neurons and their target cells. At Drosophila neuromuscular junctions retrograde signals from target cells are known to sculpt development of the synapse (reviewed by Collins and DiAntonio, 2007). miR-8, a member of the highly conserved miR-200 family, has been shown to regulate larval morphogenesis of the nerve terminals postsynaptically. This trans-synaptic phenomenon appears to be mediated largely through repression of an actin-binding protein Enabled (Loya et al., 2009). miR-124 provides us with another example of a miRNA requiring trans-synaptic communication between neurons and their targeted tissue. In Drosophila, miR-124 is involved in diversity in dendrite morphology, larval locomotion and synaptic release at the NMJ (Sun et al., 2012). Importantly, components in the retrograde BMP signaling pathway are implicated in the miR-124 presynaptic release phenotype at the NMJ. Interestingly, exosomes have recently emerged as a novel mechanism for the exchange of genetic material between cells. Known to carry small RNA molecules including miRNAs, exosomes have emerged as a likely form of “genetic communication” between the two sides of the synapse (Mittelbrunn and Sanchez-Madrid, 2012). Exosomes have been reported to mediate transsynaptic protein transfer in Drosophila NMJs (Korkut et al., 2009), making the possibility the same mechanism is deployed in the exchange of miRNAs very attractive.
Another group of miRNAs involved at the Drosophila larval neuromuscular junction are the miR-310 cluster, miR310–313, but they appear to be playing an independently presynaptic role not requiring transynaptic communication. Loss of the cluster leads to a significant enhancement of neurotransmitter release, which can be rescued with temporally restricted expression of miR 310–313 in larval presynaptic neurons (Tsurdome et al., 2010). The Kinesin family member Khc-73 is a functional target for the cluster as its expression is increased in cluster mutants and reducing Khc-73 restores normal synaptic function. At later stages of the Drosophila life cycle during periods of tissue remodeling, there is coordinated pre-and postsynaptic expression of another conserved miRNA, let-7 (Caygill and Johnston, 2008; Sokol et al., 2008) Loss of the fly let-7 complex (Let-7, miR-100 and miR-125) prevents the normal maturation of these NMJs as these animals metamorphose to adults, largely via regulation of the muscle transcription factor Abrupt.
Investigation of miRNA function in many contexts indicates that they often act in concert with transcription factors to augment robustness or mediate feedback in the regulation of effector gene networks (reviewed by Pelàez and Carthew, 2012). For example, in the C. elegans neuromuscular system, miR-1 controls both the expression of acetylcholine receptors and the muscle transcription factor MEF-2 (Simon et al., 2008). Interestingly, in this model, MEF-2 is upstream of an unknown trans-synaptic retrograde signal that appears to control presynaptic release properties. This miR-1/MEF-2 pathway highlights the intricate ongoing conversation between neurons and their synaptic partners as miR-1 regulates aspects of both pre-and postsynaptic functions at C. elegans neuromuscular junctions. Further exploration of miRNA-transcripton factor interactions in C. elegans has uncovered a role for miRNA in activity dependent plasticity that is part of normal circuit remodeling during organismal development. In this work, the transcription factor hunchback-like 1 (HBL-1) orthologous to a gene that regulates the timing of neural progenitor fate determination in Drosophila, was found to be specifically expressed in a subset of motor neurons that actively remodel their synaptic connections during larval maturation (Thompson et al., 2012). Interestingly, a change in neural activity induced a corresponding change in HBL-1 expression. In this system, miR-84 was shown to regulate motor neuron plasticity by controlling hbl-1, ultimately allowing for a mechanism of activity regulated circuit refinement (Thompson et al., 2012). Together, these studies have demonstrated the power of in vivo and in vitro models in discovering a functional role for miRNAs in the nervous system providing us with a glimpse of cell contextual roles for miRNAs and a key cooperation with transcription factors.
In Vivo Studies of Learning and Memory
Molecular models of learning and memory have relied heavily on the identification of activity-dependent transcription factors such as c-Fos and CREB (Reviewed Flavell and Greenberg, 2008; Miyamoto, 2006). As mentioned above, extensive studies have identified miR-132 as being regulated by CREB in activity-regulated plasticity. Initial experiments with in the context of learning and memory examined miR-132 expression in response to increased activity in vivo. In these studies, miR132 was rapidly transcribed in the hippocampus following enhanced neuronal activity and contextual fear conditioning (Nudelman et al., 2010). In addition, studies using transgenic mice over-expressing miR-132 in forebrain neurons showed a marked increase in dendritic spine density and impairments in a novel object recognition memory test (Hansen et al., 2010). This functional role for miR-132 in memory formation may at least in part be attributed to the participation of miR-132 in the integration of newborn neurons into the adult dentate gyrus. Expression of miR-132 was increased during neuron differentiation and maturation and knockdown of miR-132 resulted in decreased synapse formation as well as impaired functional integration of newborn neurons (Luikart et al., 2011).
Two recent studies highlight the importance of plasticity mechanisms in the developmental refinement of neural circuits, demonstrating a role for miR-132 in vivo as critical for the formation of ocular dominance (Mellios et al., 2011; Tognini et al., 2011). In this model, one can study the ability to modulate ocular dominance through the reorganization of neuronal connections in response to visual experience. In both papers, visual experience was shown to regulate miR-132 levels in the visual cortex. Interestingly, light exposure increased the presence of multiple histone post-translational modifications within the CRE locus that are important for miR132/212 cluster transcription (Tognini et al., 2011). Both up-regulation of miR-132 through miRNA mimic that caused an increase in the fraction of mature dendritic spines (Tognini et al., 2011), and down-regulation through miRNA sponge technologies which resulted in more immature spines, disrupted optical dominance plasticity (Mellios et al., 2011). Taken together this data indicates that a very tightly regulated balance of miR-132 expression is required in its functional role in plasticity.
In addition to the established roles for miR-132 in learning and memory, novel discoveries are rapidly increasing our understanding of additional miRNAs in these processes. miR-128b is an example of one of these miRNAs. Fear-extinction learning in mice led to increased expression miR-128b disrupting the stability of several plasticity related target genes and regulated formation of fear-extinction memory (Lin et al., 2011). A study of EPAC −/− mice, which demonstrated severe deficits in synaptic transmission, LTP, spatial learning, and social interactions, identified a role for miR-124 in these processes. In this research, EPAC proteins, which act as the guanine nucleotide exchange factors and intracellular receptors for cyclic AMP, were found to activate Rap1, which directly interacts with the regulatory element upstream of miR-124 and restricts miR-124 expression. Further, miR-124 was found to directly bind and inhibit the translation of Zif268, an EGR-family transcription factor. Knock down of miR-124 was found to restore normal levels of Zif268 expression and reverse all aspects of the EPAC−/− phenotypes confirming that EPAC proteins control of miR-124 transcription in the brain is required for processing spatial learning and social interactions (Yang et al., 2012). Large-scale parallel sequencing of mouse hippocampal small RNA libraries identified miR-34c as being highly expressed in the hippocampus relative to the rest of the brain where it acts as a negative constraint during memory consolidation through Sirt1. In the same study, miR-34c was further linked to memory dysfunction because miR-34c levels were found to be elevated in the hippocampus of Alzheimer’s patients and mouse models of Alzheimer’s disease (Zovoilis et al., 2011). Full characterization of miR-34c targets in the hippocampus and in learning and memory remains to be elucidated. Another study used olfaction discrimination training as a learning paradigm for adult mice. After this activity, the hippocampus was profiled for miRNA expression. A significant up regulation of miRNAs was observed indicating that global changes in miRNA expression accompany early stages of learning (Smalheiser et al., 2010).
miRNA Regulation of Other Behaviors
Amongst the many changing conditions that stimulate behavioral adaptation on this planet, cycles of night and day have clearly shaped behaviors that are highly conserved across species. Circadian rhythm is one of these key adaptive mechanisms to manage life in a dynamic world. In mammals, the circadian oscillator is defined by a 25-hour clock controlled by the suprachiasmatic nucleus (SCN), a tiny region of the ventral hypothalamus that contains approximately 20,000 neurons. The timing capacity of the SCN is derived from autonomous neuronal oscillators, which form a pattern of rhythmic neuronal activity to serve as a phasing cue (reviewed in Hansen et al., 2011). Recent work by a number of groups has revealed a role for miRNAs in clock physiology. Initial studies in Drosophila profiled miRNA expression and found oscillations in miR263a and miR-263b that were observed in wild-type flies, but absent in clock mutants (Yang et al., 2008). In a later study, Kadener et al. (2009) found that abrogation of miRNA biogenesis led to both an increase in circadian regulated gene expression and a disruption of circadian regulated behavioral rhythms, revealing a role for miRNA in clock timing.
Recently, miR-279 was also identified in driving rest:activity rhythms in Drosophila through regulation of the JAK/STAT pathway. Over-expression or deletion of miR-279 attenuates rhythms, but oscillations in the clock protein PERIOD were normal indicating miR-279 is downstream of the clock (Luo et al., 2012). The JAK/Stat ligand unpaired (Upd) is a target of miR 279 and knockdown of Upd rescues the behavioral phenotype of miR-279. The central clock neurons were found to project in the vicinity of Upd-expressing neurons and proposed to be a physical connection by which the central clock could regulate Jak/Stat signaling to control rest:activity rhythms. Additionally, a series of in vivo studies has revealed the role of miR-132 in modulating the circadian-clock (Cheng et al., 2007;Alvarez-Saavedra et al., 2010). It was found that exposure to light induces transcription of miR-132 in the SCN in vivo, where it plays a role in regulating entrainment of the circadian clock (Cheng et al., 2007). Further research has indicated that miR-132 acts as a master factor for chromatin remodeling and protein translation in this model enabling the fine-tuned expression of genes involved in the circadian clock regulation (Alvarez-Saavedra et al., 2010).
Sleep and circadian clocks are intimately intertwined so it is not surprising that rhythmic miRNAs have recently been implicated as functioning in sleep behavior. miRNA levels in brain are altered by sleep deprivation and over-expression of miR-132 in vivo decreases duration of non-rapid-eye-movement sleep while simultaneously increasing duration of rapid eye movement sleep during the light phase. Spontaneous cortical levels of miRNA-132 are also lower at the end of the sleep-dominant light period compared the end of the dark period in rats (Davis et al., 2011). This opens up new questions for the implications of miRNAs in sleep that need to be explored.
Social behaviors are some of the most complicated manifestations of neuronal connections. A recent study using the highly socially organized behavior of honey bees has identified miRNAs that are up-regulated in bees that specialize in foraging relative to miRNA levels in bees that specialize in brood care. Evolutionary analysis found the same miRNAs conserved in other eusocial species such as wasps and ants. Interestingly, the up-regulation of specific miRNAs is dependent on social context (Greenberg et al., 2012). This study opens further avenues of study examining miRNAs as regulators of social behaviors and demonstrates the need for functional tools to study miRNAs outside of the traditional model organisms.
Manipulating the In Vivo miRNA Landscape
As true for many developmental regulatory genes, the first in vivo miRNA functions emerged from classical genetic analysis using invertebrate model organisms (Lee et al., 1993; Reinhart et al., 2000; Brennecke et al., 2003). The availability of many defined chromosomal deletions in C. elegans then made it possible to undertake selective screens to map out the miRNA functional landscape for a handful of different phenotypes (Miska et al., 2007; Alvarez-Saavedra and Horvitz, 2010). In screens representing nearly half of the currently known C. elegans miRNAs, the surprising conclusion was drawn that relatively few miRNA are essential for organismal development or simple behaviors (e.g. locomotion, egg-laying and defecation) even when related miRNA families were disrupted. Interestingly, when combinations of miRNA were eliminated in a genetic background compromised for the argonaut-like 1 gene (alg-1), 80% of the mutants displayed defects in viability or development (Brenner et al., 2010), raising the possibility that the sensitized screens feasible in model organisms might overcome functional redundancy built into miRNA target networks. Methods are now available for systematic generation of miRNA deletion mutants in the fly (Chen et al., 2011). Moreover, recent efforts provide effective means for rapid generation of conditional miRNA disruption in the mouse (Park et a., 2012). However, comprehensive in vivo functional screens have not been applied to synaptic development or plasticity phenotypes in these or other species. Elevation of miRNA levels by expression of miRNA mimics (Figure 4) in distinct patterns can be a used an initial assay for potential function (reviewed in Bushati and Cohen, 2007; Dai et al., 2012). For example, large-scale screens have been performed in Drosophila using miRNA misexpression under specific promoters to elicit phenotypes or to probe for genetic interactions (Bejarano et al., 2012; Szuplewski et al., 2012). However, loss-of-function is essential to confirm a functional requirement.
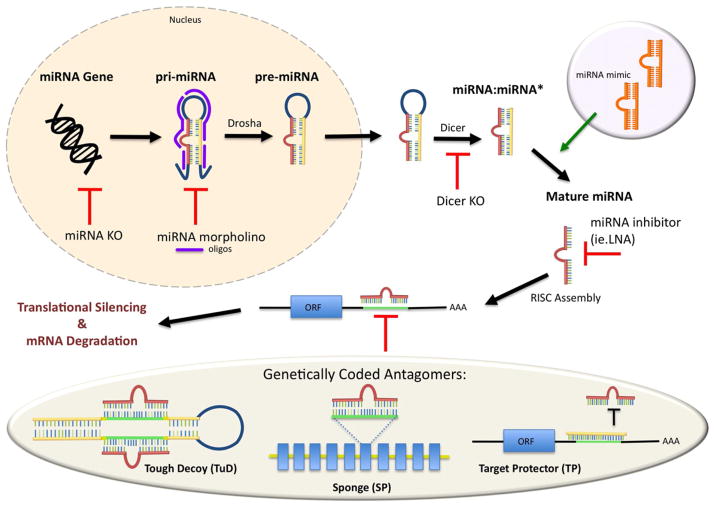
Figure 4. Technologies Available to Manipulate miRNA Levels and Function.
The miRNA biosynthetic and processing pathway is diagrammed to illustrate the stages at which different genetic disruptions can be made. While genetic knock-out (KO) by random or targeted disruption of miRNA eliminates expression completely, such mutations offer conditional loss-of-function only in conjunction with other systems (e.g. mosaic technologies such as Cre-loxP, Flip-FRT, etc.). Antisense oligonucleotides (e.g. LNA morpholino) can block miRNA processing at the pri-miRNA stage to prevent processing to the pre-miRNA form, or later at the level of mature miRNA. Disruption of the Drosha/Pasha microprocessor also prevents formation of pre-miRNA, whereas disruption of Dicer block subsequent formation of mature miRNA. Finally, several genetically encoded antagomer techniques can compete with miRNA or target gene mRNAs to reduce the level of mature miRNAs or the number of miRNA-mRNA complexes. The most widely tested techniques are miRNA sponges (SP), tough decoys (TuD) and target protectors (TP).
Amongst technologies designed to provide spatiotemporal control over miRNA functions in vivo, beyond well-established conditional miRNA gene knockout methods (e.g. Cre-Lox, Flip-FRT; reviewed by), genetically encoded antagomers (called miRNA “sponges” or “decoys”; Figure 4) are promising for analysis of neural development and plasticity (reviewed by Ebert and Sharp, 2012; Ruberti et al., 2012). The miRNA sponge (miR-SP) consists of a DNA construct producing RNAs that bear repeated sequences complementary to a specific miRNA or miRNA family (Ebert et al., 2007). The effect of the sponge is to hybridize with endogenous miRNA and thus win a competition for association of miRNA with their target mRNAs. Sponge constructs were initially shown to be effective and specific in non-neuronal cell culture and xenograft experiments (see Ebert and Sharp, 2012). Placed downstream of promotors to confer spatiotemporal control of miR-SP deployment, transgenic sponges were then tested in Drosophila to recapitulate classical loss-of-function mutations in several miRNA genes (Loya et al., 2009). This first transgenic application of miR-SP technology for analysis of synaptic development in the Drosophila neuromuscular system showed that the technique could distinguish pre- and postsynaptic contributions that matched regulatory effects on a functional target gene.
More recently, miR-SP transgenics have been tested in the mouse. The use of the sponge to inhibit the miR-183/96/182 cluster in retina illustrated not only the effectiveness of this approach to reveal functions in light-dependent neuronal responses, but also the power of miR-SP to simultaneously inhibit miRNA family members with closely related sequences (Zhu et al., 2011). Effective delivery of miR-SP to the CNS has been demonstrated for activity-dependent synaptic plasticity in the mouse visual cortex using a convenient lentiviral system (Mellios et al., 2011). The miR-SP has also been delivered by electroporation to test miR regulation of both early and late stages of neuronal development (de Chevigny et al., 2012; Pathania et al., 2012). Although the miR-SP technology is still being optimized (e.g. Kluivier et al., 2012; Otaegi et al., 2011), current data indicate that it will be a powerful tool that can be generalized to study neural circuit formation and remodeling in many contexts. In addition, improved in vivo inhibition may be achieved by modifications of the approach, including the “tough decoy” (TuD) designed to carry a miRNA seed complement within an overall RNA structure that is resistant to degradation (Haraguchi et al., 2009). The efficacy of TuDs have been recently been compared to miR-SP and one other anti-sense design (miRZips) using an RNA polymerase III promotor in cell culture (Xie et al., 2012). The comparison suggests that under these conditions, TuDs are the most potent genetically encoded antagomer. More importantly, TuDs carried in a DNA parvovirus vector have been validated for in vivo efficacy in the liver by introduction into the bloodstream (Xie et al., 2012), however, they have not been tested in the CNS where access is more limited.
Once a function has been defined for any specific miRNA, understanding the underlying regulatory mechanism requires one to identify the target genes that are functionally relevant in a specific context. One clever variation of the anti-sense approach was designed to selectively disrupt the access of miRNAs for a specific target gene, thereby relieving that target from endogenous regulation: the “target protector” (TP; reviewed in Staton and Giraldez, 2011). The TP consists of an oligonucleotide (morpholino) designed to be complimentary to sequences within the 3′ UTR of a target mRNA that overlap the miRNA targeting site but extend far enough beyond the miRNA seed complement to ensure specificity to the target (Choi et al., 2007)(see Figure 4). Because the TP should not load into Ago complexes, it will not behave as a miRNA, yet it prevents miRNA access to the transcript by competition for the regulatory site. This technique works well in zebrafish embryos where oligonucleotides can be injected at early blastomere stages (Choi et al., 2007; Staton et al., 2011) and has been introduced by transfection in cell culture (Long and Lahiri, 2011), but has yet to be tested in mammalian or invertebrate models where an adaptation to a transgenic platform would be required for the most versatile applications.
Detecting the location and degree of miRNA regulation for targets in situ is also important because this activity cannot be predicted simply by overlap of miRNA and target gene expression (e.g. Loya et al., 2009), partly due to regulatory interactions that control miRNA function (e.g. Banerjee et al., 2009; Bhattacharyya et al., 2006; Piskounova et al., 2011). For this reason, sensors of miRNA activity have been indispensible for understanding their function in many contexts. However, the majority of miRNA reporters have relied on miRNA downregulation of ubiquitously expressed marker proteins (e.g. luciferase or green fluorescent protein), typically by placing endogenous 3′UTR or synthetic miRNA target sites downstream (e.g. De Pietri Tonelli et al., 2006; reviewed by Van Wynsberghe et al., 2011). Yet, for neurons or other cells deeply embedded in a complex tissue, loss of marker expression in a small subset of cells can be difficult to detect, necessitating future effort to create a robust positive sensor system for in vivo studies.
The Neural MicroRNA Target Landscape
Although the majority of functional analysis for miRNA targets so far has been focused on single genes, many studies using computational sequence predictions and gene or protein profiling techniques show that collectively and individually miRNAs regulate extensive gene networks (reviewed by Bartel, 2009; Peláez and Carthew, 2012). Moreover, amongst related animal species, the target gene sets for miRNA are frequently well conserved (e.g. Grun et al., 2005; Friedman et al., 2009). Consistent with a functional logic within miRNA target networks, genes regulated by miRNA in a given process such as neuronal development and synapse formation have been found to show strong correlation in gene ontogeny (GO) terms assigned based on categories of known function (Manakov et al., 2009; Chen et al., 2011). For these reasons the relatively small number of miRNAs essential for viability and early development in C. elegans (Miska et al., 2007; Alvarez-Saavedra and Horvitz, 2010) or even gross neural patterning in zebrafish (Giraldez et al., 2005) were unexpected. One possible explanation for the discrepancy might be that miRNA functions contribute more frequently to adaptive response mechanisms that are not frequently challenged during embryogenesis in the laboratory setting. The number of miRNA that appear to be involved in the regulation of synaptic plasticity is significant even at an early stage of inquiry before comprehensive in vivo functional screening methods are available beyond C. elegans, suggesting that neural miRNA may play a disproportionate role in circuit formation, refinement and function. Interestingly, recent studies have also implicated miRNA in neuroadaptive responses induced by exposure to substances of abuse (e.g alcohol and cocaine; reviewed by Li and van der Vaart, 2011; Nunez and Mayfield, 2012). While this may simply reflect a central role for miRNA in regulating synaptic biology, as synapse plasticity is thought to be pivotal in addictive behaviors, it reinforces the notion that miRNA contribute to a variety of context-dependent behaviors.
An alternative way of thinking about miRNA function is at the network level where action on single genes may be less informative than the emergent impact of many miRNA on multiple target genes. Even for the most highly conserved miRNA expressed in the nervous system such as miR-9, only a subset of miRNA-target pairings are well-conserved from invertebrates to mammals despite significant conservation in overall function (reviewed by Yuva-Aydemir et al., 2011). Indeed, it has been suggested that the principal features of miRNA that are conserved across the longer evolutionary timeframe are network themes, as opposed to specific target gene relationships (Grun et al., 2005). Thinking globally, beyond first-order regulation of single target genes, it has been suggested that miRNA may collaborate by convergence onto key genes or hubs within networks that require buffering from stochastic noise or onto bottlenecks that link sub-network modules (reviewed by Peláez and Carthew, 2012). The observation that many nodes and bottlenecks are enriched for miRNA regulation is consistent with this idea (Martinez et al., 2008). These miRNA properties can dampen fluctuation at key integrators of convergent information in a network to protect against inappropriate pathway activation, or to set threshold rules for pathway activation. Such dampening will often have a major impact on how a network responds to a change in environmental conditions, making it more robust and reliable. miRNAs are well-known to mediate feedback loops (e.g., Arvanitis et al., 2010), but they also mediate feed-forward systems or can be combined to created coincidence detectors (reviewed by Herranz and Cohen, 2010). Interestingly, since transcription factors tend to concentrate at gene network hubs and are frequently key components to trigger adaptive responses, there is a special relationship between miRNA and transcription factors. Modeling synaptic effector gene networks is an exciting arena for systems biologists given the accumulated molecular and functional data in the field (reviewed by Kotaleski and Blackwell, 2010). The further step of deconvolving the relationship between transcriptional control and post-transcriptional control upstream and downstream of effector gene networks can presumably help define themes and testable hypotheses in the realm of miRNA regulation of synapse development and plasticity. However, putting such hypotheses to the test in vivo will require new techniques and combinations of genetic tools.
Conclusions
Late in his career, Conrad Waddington made efforts to test the possible contribution of “masked” mRNA in the developing Drosophila retina in an attempt to define a latent reservoir of genetic information that might be expressed over the course of developmental events (Waddington and Robertson, 1969). While recent advances in the fields of chromatin structure regulation (reviewed by Margueron and Reinberg, 2010) and post-transcriptional mechanisms such as miRNAs that mediate the complex relationship between genome and phenome would certainly be tremendously exciting to Waddington, one suspects that he would be equally fascinated by the many puzzles that remain. For example, it will be important to complete the process of surveying the ‘map’ of all miRNA functions. For roles in synaptic development and plasticity, profiling data imply that only a small subset of landmarks have been charted so far. Defining the target gene network logic of all these miRNAs will be challenging, and will require new technologies for conditional and combinatorial manipulation of miRNA/target gene function. But other fundamental questions remain. For example, it is not entirely clear how dynamic changes in cellular state are converted into long-lasting and even heritable states, although this process is likely to involve reciprocal interaction between the genome and the RNA space where miRNAs and other non-coding RNAs function. One thing is clear: miRNAs play diverse roles in shaping the neuronal landscape, and we’ve only begun to explore.
Acknowledgments
We express our regrets to many whose work could not be cited due to space constraints. We thank our colleague Dr. Danesh Moazed for thoughtful feedback prior to publication. We also thank Kerry Mojica and Anita Kermode for editorial assistance. This work was supported by grants from NINDS: DVV (R01 NS069695) and EMM (T32 NS007484-12).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelmohsen K, Hutchison ER, Lee EK, Kuwano Y, Kim MM, Masuda K, Srikantan S, Subaran SS, Marasa BS, Mattson MP, et al. miR-375 inhibits differentiation of neurites by lowering HuD levels. Molecular and cellular biology. 2010;30:4197–4210. doi: 10.1128/MCB.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Killick R, Candi E, Sayan BS, Rivetti di Val Cervo P, Nicotera P, McKeon F, Knight RA, Mak TW, et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proceedings of the National Academy of Sciences of the United States of America. 2011a;108:21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, Forsythe ID, Young KW, Ventura A, Concepcion CP, et al. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proceedings of the National Academy of Sciences of the United States of America. 2011b;108:21099–21104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Current biology: CB. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra M, Antoun G, Yanagiya A, Oliva-Hernandez R, Cornejo-Palma D, Perez-Iratxeta C, Sonenberg N, Cheng HYM. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Human Molecular Genetics. 2010;20:731–751. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis DN, Jungas T, Behar A, Davy A. Ephrin-B1 reverse signaling controls a posttranscriptional feedback mechanism via miR-124. Molecular and cellular biology. 2010;30:2508–2517. doi: 10.1128/MCB.01620-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124(1):191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nature reviews Genetics. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome Profiling Shows That miR-430 Reduces Translation Before Causing mRNA Decay in Zebrafish. Science (New York, NY) 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejarano F, Bortolamiol-Becet D, Dai Q, Sun K, Saj A, Chou Y-T, Raleigh DR, Kim K, Ni J-Q, Duan H, et al. A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development (Cambridge, England) 2012 doi: 10.1242/dev.079939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdnik D, Fan AP, Potter CJ, Luo L. MicroRNA processing pathway regulates olfactory neuron morphogenesis. Current biology: CB. 2008;18:1754–1759. doi: 10.1016/j.cub.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Bian S, Sun T. Functions of noncoding RNAs in neural development and neurological diseases. Molecular neurobiology. 2011;44:359–373. doi: 10.1007/s12035-011-8211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Lin Q, Wei W, Baker-Andresen D, Mattick JS. MicroRNA regulation of neural plasticity and memory. Neurobiology of learning and memory. 2011;96:89–94. doi: 10.1016/j.nlm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Brenner JL, Jasiewicz KL, Fahley AF, Kemp BJ, Abbott AL. Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr Biol. 2010;20:1321–1325. doi: 10.1016/j.cub.2010.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annual review of cell and developmental biology. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The Local Transcriptome in the Synaptic Neuropil Revealed by Deep Sequencing and High-Resolution Imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Current biology: CB. 2008;18:943–950. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, Saugstad J. MicroRNAs: Meta-controllers of gene expression in synaptic activity emerge as genetic and diagnostic markers of human disease. Pharmacology & therapeutics. 2011;130:26–37. doi: 10.1016/j.pharmthera.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Pedro MS, Zeller RW. miR-124 function during Ciona intestinalis neuronal development includes extensive interaction with the Notch signaling pathway. Development (Cambridge, England) 2011;138:4943–4953. doi: 10.1242/dev.068049. [DOI] [PubMed] [Google Scholar]
- Chen YW, Weng R, Cohen SM. Protocols for use of homologous recombination gene targeting to produce microRNA mutants in Drosophila. Methods Mol Biol. 2011;732:99–120. doi: 10.1007/978-1-61779-083-6_8. [DOI] [PubMed] [Google Scholar]
- Cheng HYM, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54(5):813–29. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WYY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Christensen M, Larsen LA, Kauppinen S, Schratt G. Recombinant Adeno-Associated Virus-Mediated microRNA Delivery into the Postnatal Mouse Brain Reveals a Role for miR-134 in Dendritogenesis in Vivo. Frontiers in neural circuits. 2010;3:16. doi: 10.3389/neuro.04.016.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. Journal of Alzheimer’s disease: JAD. 2008;1:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- Cohen JE, Lee PR, Chen S, Li W, Fields RD. MicroRNA regulation of homeostatic synaptic plasticity. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11650–11655. doi: 10.1073/pnas.1017576108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins Ca, DiAntonio A. Synaptic development: insights from Drosophila. Current opinion in neurobiology. 2007;17:35–42. doi: 10.1016/j.conb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Dai Q, Smibert P, Lai EC. Exploiting Drosophila genetics to understand microRNA function and regulation. Curr Top Dev Biol. 2012;99:201–235. doi: 10.1016/B978-0-12-387038-4.00008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CJ, Clinton JM, Taishi P, Bohnet SG, Honn KA, Krueger JM. MicroRNA 132 alters sleep and varies with time in brain. Journal of applied physiology (Bethesda, Md: 1985) 2011;111:665–672. doi: 10.1152/japplphysiol.00517.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chevigny A, Coré N, Follert P, Gaudin M, Barbry P, Béclin C, Cremer H. miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nature Neuroscience. 2012 doi: 10.1038/nn.3142. [DOI] [PubMed] [Google Scholar]
- Deschênes-Furry J, Perrone-Bizzozero N, Jasmin BJ. The RNA-binding protein HuD: a regulator of neuronal differentiation, maintenance and plasticity. BioEssays: news and reviews in molecular, cellular and developmental biology. 2006;28:822–833. doi: 10.1002/bies.20449. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Fainzilber M, Twiss JL. Subcellular communication through RNA transport and localized protein synthesis. Traffic. 2010;11:1498–1505. doi: 10.1111/j.1600-0854.2010.01118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development (Cambridge, England) 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for MicroRNAs in Conferring Robustness to Biological Processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster Ka, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp Pa, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, Greenberg ME, Schratt G. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. The EMBO journal. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling Mechanisms Linking Neuronal Activity to Gene Expression and Plasticity of the Nervous System. Annual review of neuroscience. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flexner JB, Flexner LB, Stellar E. Memory in mice as affected by intracerebral puromycin. Science. 1963;141:57–59. doi: 10.1126/science.141.3575.57. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KKH, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fénelon K, Mukai J, Xu B, Hsu PK, Drew LJ, Karayiorgou M, Fischbach GD, Macdermott AB, Gogos JA. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4447–4452. doi: 10.1073/pnas.1101219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Gräff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cerebral cortex (New York, NY: 1991) 2011;2:1857–1869. doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Kirschner M. Cells, Embryos, and Evolution. Malden: Blackwell Science, Inc; 1997. [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science (New York, NY) 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Greenberg JK, Xia J, Zhou X, Thatcher SR, Gu X, Ament SA, Newman TC, Green PJ, Zhang W, Robinson GE, et al. Behavioral plasticity in honey bees is associated with differences in brain microRNA transcriptome. Genes, Brain and Behavior. 2012 doi: 10.1111/j.1601-183X.2012.00782.x. epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic acids research. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS computational biology. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Yeo GSH, Tung YCCL, Zivraj KH, Willis D, Coppola G, Lam BYH, Twiss JL, Holt CE, Fawcett JW. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17:85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hansen KF, Sakamoto K, Wayman Ga, Impey S, Obrietan K. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PloS one. 2010;5:e15497. doi: 10.1371/journal.pone.0015497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KF, Sakamoto K, Obrietan K. MicroRNAs: a potential interface between the circadian clock and human health. Genome medicine. 2011;3:10. doi: 10.1186/gm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KF, Karelina K, Sakamoto K, Wayman GA, Impey S, Obrietan K. miRNA-132: a dynamic regulator of cognitive capacity. Brain structure & function. 2012 doi: 10.1007/s00429-012-0431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73:35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes & development. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander JA, Im HI, Amelio AL, Kocerha J, Bali P, Lu Q, Willoughby D, Wahlestedt C, Conkright MD, Kenny PJ. Striatal microRNA controls cocaine intake through CREB signalling. Nature. 2010;466:197–202. doi: 10.1038/nature09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu R, Schofield CM, Dela Cruz CG, Jones-Davis DM, Blelloch R, Ullian EM. Dgcr8 is required in pyramidal neurons for normal inhibitory synaptic function. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nature neuroscience. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Davare M, Lasiek A, Fortin D, Ando H, Varlamova O, Obrietan K, Soderling TR, Goodman RH, Wayman GA. An activity-induced microRNA controls dendritic spine formation by regulating Rac1-PAK signaling. Molecular and Cellular Neuroscience. 2010;43:146–156. doi: 10.1016/j.mcn.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Engel T, Merino-Serrais P, McKiernan RC, Tanaka K, Mouri G, Sano T, O’Tuathaigh C, Waddington JL, Prenter S, et al. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nature medicine. 2012 doi: 10.1038/nm.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nature reviews. Neuroscience. 2012;13(5):308–24. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M. A role for microRNAs in the Drosophila circadian clock. Genes & development. 2009;23:2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. The molecular biology of memory storage: a dialog between genes and synapses. Biosci Rep. 2001;21:565–611. doi: 10.1023/a:1014775008533. [DOI] [PubMed] [Google Scholar]
- Kang H, Schuman EM. A Requirement for Local Protein Synthesis in Neurotrophin-Induced Hippocampal Synaptic Plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Kawashima H, Numakawa T, Kumamaru E, Adachi N, Mizuno H, Ninomiya M, Kunugi H, Hashido K. Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience. 2010;165:1301–1311. doi: 10.1016/j.neuroscience.2009.11.057. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews. Molecular cell biology. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kindler S, Kreienkamp H-J. In: Synaptic Plasticity. Kreutz MR, Sala C, editors. Vienna: Springer Vienna; 2012. pp. 285–305. [Google Scholar]
- Kluiver J, Gibcus JH, Hettinga C, Adema A, Richter MK, Halsema N, Slezak-Prochazka I, Ding Y, Kroesen BJ, van den Berg A. Rapid generation of microRNA sponges for microRNA inhibition. PLoS One. 2012;7:e29275. doi: 10.1371/journal.pone.0029275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocerha J, Faghihi MA, Lopez-Toledano MA, Huang J, Ramsey AJ, Caron MG, Sales N, Willoughby D, Elmen J, Hansen HF, et al. MicroRNA-219 modulates NMDA receptor-mediated neurobehavioral dysfunction. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3507–3512. doi: 10.1073/pnas.0805854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka W, Kiryk A, Novak M, Herwerth M, Parkitna JR, Wawrzyniak M, Kowarsch A, Michaluk P, Dzwonek J, Arnsperger T, et al. MicroRNA loss enhances learning and memory in mice. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:14835–14842. doi: 10.1523/JNEUROSCI.3030-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkut C, Ataman B, Ramachandran P, Ashley J, Barria R, Gherbesi N, Budnik V. Trans-synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell. 2009;139:393–404. doi: 10.1016/j.cell.2009.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Kotaleski JH, Blackwell KT. Modelling the molecular mechanisms of synaptic plasticity using systems biology approaches. Nature reviews. Neuroscience. 2010;11:239–251. doi: 10.1038/nrn2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schübeler D, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M, Krutzfeldt J. Silencing of microRNAs in vivo with “antagomirs. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kwon E, Wang W, Tsai L-H. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Molecular psychiatry. 2011 doi: 10.1038/mp.2011.170. [DOI] [PubMed] [Google Scholar]
- Kye MJ, Liu T, Levy SF, Xu NL, Groves BB, Bonneau R, Lao K, Kosik KS. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA. 2007;13:1224–1234. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kye MJ, Neveu P, Lee YS, Zhou M, Steen JA, Sahin M, Kosik KS, Silva AJ. NMDA mediated contextual conditioning changes miRNA expression. PLoS One. 2011;6:e24682. doi: 10.1371/journal.pone.0024682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert TJ, Storm DR, Sullivan JM. MicroRNA132 modulates short-term synaptic plasticity but not basal release probability in hippocampal neurons. PloS one. 2010;5:e15182. doi: 10.1371/journal.pone.0015182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, Marks DS, Sander C, Tuschl T, Gaul U. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Lee K, Kim JH, Kwon OB, An K, Ryu J, Cho K, Suh YH, Kim HS. An Activity-Regulated microRNA, miR-188, Controls Dendritic Plasticity and Synaptic Transmission by Downregulating Neuropilin-2. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2012;32:5678–5687. doi: 10.1523/JNEUROSCI.6471-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Li MD, van der Vaart AD. MicroRNAs in addiction: adaptation’s middlemen? Molecular psychiatry. 2011;16:1159–1168. doi: 10.1038/mp.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Bian S, Hong J, Kawase-Koga Y, Zhu E, Zheng Y, Yang L, Sun T. Timing Specific Requirement of microRNA Function is Essential for Embryonic and Postnatal Hippocampal Development. PloS one. 2011;6:e26000. doi: 10.1371/journal.pone.0026000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Wei W, Coelho CM, Li X, Baker-Andresen D, Dudley K, Ratnu VS, Boskovic Z, Kobor MS, Sun YE, et al. The brain-specific microRNA miR-128b regulates the formation of fear-extinction memory. Nature neuroscience. 2011;14:1115–1117. doi: 10.1038/nn.2891. [DOI] [PubMed] [Google Scholar]
- Lippi G, Steinert JR, Marczylo EL, D’Oro S, Fiore R, Forsythe ID, Schratt G, Zoli M, Nicotera P, Young KW. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. The Journal of cell biology. 2011;194:889–904. doi: 10.1083/jcb.201103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JM, Lahiri DK. MicroRNA-101 downregulates Alzheimer’s amyloid-β precursor protein levels in human cell cultures and is differentially expressed. Biochemical and biophysical research communications. 2011;404:889–895. doi: 10.1016/j.bbrc.2010.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nature methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart BW, Bensen AL, Washburn EK, Perederiy JV, Su KG, Li Y, Kernie SG, Parada LF, Westbrook GL. miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PloS one. 2011;6:e19077. doi: 10.1371/journal.pone.0019077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Sehgal A. Regulation of Circadian Behavioral Output via a MicroRNA-JAK/STAT Circuit. Cell. 2012;148:765–779. doi: 10.1016/j.cell.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroy A, Tyler A. Formation of active ribosomal affrefates (polysomes) upon fertilization and development of sea urchin eggs. Archives of biochemistry and biophysics. 1963;103:431–435. doi: 10.1016/0003-9861(63)90433-8. [DOI] [PubMed] [Google Scholar]
- Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL, Mandel G, Goodman RH. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manakov SA, Grant SGN, Enright AJ. Reciprocal regulation of microRNA and mRNA profiles in neuronal development and synapse formation. BMC genomics. 2009;10:419. doi: 10.1186/1471-2164-10-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Casadio A, Zhu H, EY, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-Specific, Long-Term Facilitation of Aplysia Sensory to Motor Synapses: A Function for Local Protein Synthesis in Memory Storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJM. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes & development. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann C, Holohan EE, Das S, Dervan A, Larkin A, Lee JA, Rodrigues V, Parker R, Ramaswami M. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E655–62. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Sugihara H, Castro J, Banerjee A, Le C, Kumar A, Crawford B, Strathmann J, Tropea D, Levine SS, et al. miR-132, an experience-dependent microRNA, is essential for visual cortex plasticity. Nat Neurosci. 2011;14:1240–1242. doi: 10.1038/nn.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, Zeier Z, Xi L, Lanz TA, Deng S, Strathmann J, Willoughby D, Kenny PJ, Elsworth JD, Lawrence MS, et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3125–3130. doi: 10.1073/pnas.1113793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska Ea, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M, Sánchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nature Reviews Molecular Cell Biology. 2012;13:328–335. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto E. Review Molecular Mechanism of Neuronal Plasticity: Induction and Maintenance of Long-Term Potentiation in the Hippocampus. 2006;442:433–442. doi: 10.1254/jphs.cpj06007x. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudelman AS, DiRocco DP, Lambert TJ, Garelick MG, Le J, Nathanson NM, Storm DR. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 2010;20:492–498. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez YO, Mayfield RD. Understanding Alcoholism Through microRNA Signatures in Brains of Human Alcoholics. Frontiers in genetics. 2012;3:43. doi: 10.3389/fgene.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olde Loohuis NFM, Kos A, Martens GJM, Van Bokhoven H, Nadif Kasri N, Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cellular and molecular life sciences: CMLS. 2012;69:89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaegi G, Pollock A, Sun T. An Optimized Sponge for microRNA miR-9 Affects Spinal Motor Neuron Development in vivo. Front Neurosci. 2011;5:146. doi: 10.3389/fnins.2011.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CS, Tang SJJ. Regulation of microRNA expression by induction of bidirectional synaptic plasticity. J Mol Neurosci. 2009;38:50–56. doi: 10.1007/s12031-008-9158-3. [DOI] [PubMed] [Google Scholar]
- Park CY, Jeker LT, Carver-Moore K, Oh A, Liu HJ, Cameron R, Richards H, Li Z, Adler D, Yoshinaga Y, et al. A Resource for the Conditional Ablation of microRNAs in the Mouse. Cell Reports. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Pathania M, Torres-Reveron J, Yan L, Kimura T, Lin TV, Gordon V, Teng ZQ, Zhao X, Fulga TA, Van Vactor D, et al. miR-132 Enhances Dendritic Morphogenesis, Spine Density, Synaptic Integration, and Survival of Newborn Olfactory Bulb Neurons. PLoS ONE. 2012;7:e38174. doi: 10.1371/journal.pone.0038174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peláez N, Carthew RW. Chapter nine - Biological Robustness and the Role of MicroRNAs: A Network Perspective. In: Biology D, editor. MicroRNAs in Development, E H B T-C T. Academic Press; 2012. pp. 237–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatigorsky J, Ozaki H, Tyler A. RNA- and protein-synthesizing capacity of isolated oocytes of the sea urchin Lytechinus pictus. Developmental Biology. 1967;15:1–22. doi: 10.1016/0012-1606(67)90002-4. [DOI] [PubMed] [Google Scholar]
- Pichardo-Casas I, Goff LA, Swerdel MR, Athie A, Davila J, Ramos-Brossier M, Lapid-Volosin M, Friedman WJ, Hart RP, Vaca L. Expression profiling of synaptic microRNAs from the adult rat brain identifies regional differences and seizure-induced dynamic modulation. Brain Res. 2012;1436:20–33. doi: 10.1016/j.brainres.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Calegari F, Fei JF, Nomura T, Osumi N, Heisenberg CP, Huttner W. Single-cell detection of microRNAs in developing vertebrate embryos after acute administration of a dual-fluorescence reporter/sensor plasmid. BioTechniques. 2006;41:727–732. doi: 10.2144/000112296. [DOI] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon MM, Choi SH, Jamieson CA, Geschwind DH, Martin KC. Identification of process-localized mRNAs from cultured rodent hippocampal neurons. J Neurosci. 2006;26:13390–13399. doi: 10.1523/JNEUROSCI.3432-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo K, Goda Y. Unraveling mechanisms of homeostatic synaptic plasticity. Neuron. 2010;66:337–351. doi: 10.1016/j.neuron.2010.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron. 2009;63:803–817. doi: 10.1016/j.neuron.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin DY, Duan J, Ophoff RA, Andreassen OA, et al. Genome-wide association study identifies five new schizophrenia loci. Nature genetics. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti F, Barbato C, Cogoni C. Exp Neurol. Elsevier Inc; 2012. Targeting microRNAs in neurons: Tools and perspectives. [DOI] [PubMed] [Google Scholar]
- Saba R, Störchel PH, Aksoy-Aksel A, Kepura F, Lippi G, Plant TD, Schratt GM. The dopamine-regulated microRNA, miR-181a, controls GluA2 surface expression in hippocampal neurons. Molecular and cellular biology. 2011:49. doi: 10.1128/MCB.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saj A, Lai EC. Control of microRNA biogenesis and transcription by cell signaling pathways. Curr Opin Genet Dev. 2011;21:504–510. doi: 10.1016/j.gde.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Cole CN, McPeek MA, Peterson KJ. The phylogenetic distribution of metazoan microRNAs: insights into evolutionary complexity and constraint. J Exp Zool B Mol Dev Evol. 2006;306:575–588. doi: 10.1002/jez.b.21118. [DOI] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJL, Kane C, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nature cell biology. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G, Saba R, Schratt G. microRNAs in neurons: manifold regulatory roles at the synapse. Current opinion in genetics & development. 2011;21:491–497. doi: 10.1016/j.gde.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC medicine. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DJ, Madison JM, Conery AL, Thompson-Peer KL, Soskis M, Ruvkun GB, Kaplan JM, Kim JK. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 2008;133:903–915. doi: 10.1016/j.cell.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Lenon AL, Davis JM, Torvik VI, Larson J. Olfactory discrimination training up-regulates and reorganizes expression of microRNAs in adult mouse hippocampus. ASN neuro. 2010;2:e00028. doi: 10.1042/AN20090055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Bejarano F, Wang D, Garaulet DL, Yang J-S, Martin R, Bortolamiol-Becet D, Robine N, Hiesinger PR, Lai EC. A Drosophila genetic screen yields allelic series of core microRNA biogenesis factors and reveals post-developmental roles for microRNAs. RNA (New York, NY) 2011;17:1997–2010. doi: 10.1261/rna.2983511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, Teng ZQ, Luo Y, Peng J, Bordey A, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem cells (Dayton, Ohio) 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes & development. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, Jiang X, Zhang X, Xu G, Xie G, et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9:e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nature genetics. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- Staton AA, Giraldez AJ. Use of target protector morpholinos to analyze the physiological roles of specific miRNA-mRNA pairs in vivo. Nature protocols. 2011;6:2035–2049. doi: 10.1038/nprot.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staton AA, Knaut H, Giraldez AJ. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat Genet. 2011;43:204–211. doi: 10.1038/ng.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Westholm JO, Tsurudome K, Hagen JW, Lu Y, Kohwi M, Betel D, Gao FB, Haghighi AP, Doe CQ, et al. Neurophysiological Defects and Neuronal Gene Deregulation in Drosophila mir-124 Mutants. PLoS genetics. 2012;8:e1002515. doi: 10.1371/journal.pgen.1002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. The Journal of cell biology. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuplewski S, Kugler JM, Lim SF, Verma P, Chen YW, Cohen SM. MicroRNA transgene overexpression complements deficiency-based modifier screens in Drosophila. Genetics. 2012;190:617–626. doi: 10.1534/genetics.111.136689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Peer KL, Bai J, Hu Z, Kaplan JM. HBL-1 Patterns Synaptic Remodeling in C. elegans. Neuron. 2012;73:453–465. doi: 10.1016/j.neuron.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognini P, Putignano E, Coatti A, Pizzorusso T. Experience-dependent expression of miR-132 regulates ocular dominance plasticity. Nature neuroscience. 2011;14(10):1237–9. doi: 10.1038/nn.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognini P, Pizzorusso T. MicroRNA212/132 family: Molecular transducer of neuronal function and plasticity. The international journal of biochemistry & cell biology. 2012;44:6–10. doi: 10.1016/j.biocel.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Tsurudome K, Tsang K, Liao EH, Ball R, Penney J, Yang JS, Elazzouzi F, He T, Chishti A, Lnenicka G, et al. The Drosophila miR-310 cluster negatively regulates synaptic strength at the neuromuscular junction. Neuron. 2010;68:879–893. doi: 10.1016/j.neuron.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. Endeavour. 1942;1:18–20. [Google Scholar]
- Waddington CH. The Strategy of the Genes; a Discussion of Some Aspects of Theoretical Biology. London: Allen and Unwin; 1957. [Google Scholar]
- Waddington CH. Evolutionary systems; animal and human. Nature. 1959;183:1634–1638. doi: 10.1038/1831634a0. [DOI] [PubMed] [Google Scholar]
- Waddington CH, Robertson E. Determination, activation and actinomycin D insensitivity in the optic imaginal disk of Drosophila. Nature. 1969;221:933–935. doi: 10.1038/221933a0. [DOI] [PubMed] [Google Scholar]
- Wanet A, Tacheny A, Arnould T, Renard P. miR-212/132 expression and functions: within and beyond the neuronal compartment. Nucleic acids research. 2012 doi: 10.1093/nar/gks151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HYM, Marks D, Obrietan K, Soderling TR, Goodman RH, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibrand K, Panja D, Tiron A, Ofte ML, Skaftnesmo KOO, Lee CS, Pena JTG, Tuschl T, Bramham CR. Differential regulation of mature and precursor microRNA expression by NMDA and metabotropic glutamate receptor activation during LTP in the adult dentate gyrus in vivo. Eur J Neurosci. 2010;31:636–645. doi: 10.1111/j.1460-9568.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Chan S-P, Slack FJ, Pasquinelli AE. Chapter 8 - Analysis of microRNA Expression and Function. In: Biology C, editor. Caenorhabditis elegans: Molecular Genetics and Development, J H R and A S B T-M. Academic Press; 2011. pp. 219–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Ameres SL, Friedline R, Hung JH, Zhang Y, Xie Q, Zhong L, Su Q, He R, Li M, et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat Methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Lee JE, Padgett RW, Edery I. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC genomics. 2008;9:83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Shu X, Liu D, Shang Y, Wu Y, Pei L, Xu X, Tian Q, Zhang J, Qian K, et al. EPAC Null Mutation Impairs Learning and Social Interactions via Aberrant Regulation of miR-124 and Zif268 Translation. Neuron. 2012;73:774–788. doi: 10.1016/j.neuron.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuva-Aydemir Y, Simkin A, Gascon E, Gao FB. MIcroRNA-9:functional evolution of a conserved small regulatory RNA. RNA Biol. 2011;8:557–564. doi: 10.4161/rna.8.4.16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR. Recognition and cleavage of primary microRNA transcripts. Methods in molecular biology (Clifton, NJ) 2006;342:49–56. doi: 10.1385/1-59745-123-1:49. [DOI] [PubMed] [Google Scholar]
- Zhong J, Zhang T, Bloch LM. Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci. 2006;7:17. doi: 10.1186/1471-2202-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Sun W, Okano K, Chen Y, Zhang N, Maeda T, Palczewski K. Sponge transgenic mouse model reveals important roles for the microRNA-183 (miR-183)/96/182 cluster in postmitotic photoreceptors of the retina. J Biol Chem. 2011;286:31749–31760. doi: 10.1074/jbc.M111.259028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivraj KH, Tung YC, Piper M, Gumy L, Fawcett JW, Yeo GS, Holt CE. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovoilis A, Agbemenyah HY, Agis-Balboa RC, Stilling RM, Edbauer D, Rao P, Farinelli L, Delalle I, Schmitt A, Falkai P, et al. microRNA-34c is a novel target to treat dementias. The EMBO journal. 2011;30:4299–4308. doi: 10.1038/emboj.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]