Abstract
Invasion is generally perceived to be a late event during the progression of human cancer, but to date there are no consistent reports of alterations specifically associated with malignant conversion. We provide evidence that the v-Fos oncogene induces changes in gene expression that render noninvasive normal human diploid fibroblasts highly invasive, without inducing changes in growth factor requirements or anchorage dependence for proliferation. Furthermore, v-Fos-stimulated invasion is independent of the pRb/p16INK4a and p53 tumor suppressor pathways and telomerase. We have performed microarray analysis using Affymetrix GeneChips, and the gene expression profile of v-Fos transformed cells supports its role in the regulation of invasion, independent from proliferation. We also demonstrate that invasion, but not proliferation, is dependent on the activity of the up-regulated epidermal growth factor receptor. Taken together, these results indicate that AP-1-directed invasion could precede deregulated proliferation during tumorigenesis and that sustained activation of AP-1 could be the epigenetic event required for conversion of a benign tumor into a malignant one, thereby explaining why many malignant human tumors present without an obvious premalignant hyperproliferative dysplastic lesion.
The two defining hallmarks of cancer are uncontrolled proliferation and invasion (32). Invasion and subsequent metastasis create difficult problems for the clinical management and treatment of cancer. By the time of diagnosis, many cancers have invaded locally and already metastisized to distant sites (95). It is important to target the invasive component of cancer through identification of the genes that control and mediate invasion and enable metastasis.
Invasion, the migration of cells through tissue boundaries, is, under normal circumstances, a tightly controlled process that is activated in many cell types in response to extracellular signals and ceases once the stimuli are withdrawn. Invasion is complex; the invasive cell must alter its cell-cell and cell-extracellular matrix (ECM) adhesions, control the degradation of the ECM, respond to chemoattractants, rearrange the actin cytoskeleton to facilitate motility, and suppress anoikis (95, 116). The complexity of invasion and tight regulation by extracellular signals, such as growth factors, suggests that it is a transcriptionally regulated multigenic program (13, 46, 71).
Growth factors that bind to receptor tyrosine kinases stimulate proliferation and invasion (13, 34, 53, 54, 61, 113). In human tumors, the growth factor-Ras-Raf-Mek-Erk signal transduction pathway is frequently constitutively activated by mutations in component genes (20). This leads to sustained changes in the activity of transcription factors, which translate the signals into long-term phenotypic responses through changes in gene expression (14, 62, 71, 103). The activator protein 1 (AP-1) family of transcription factors is regulated by the growth factor signal transduction pathway (26) and is important for invasion as well as proliferation (71, 76). Since AP-1 is a downstream effector of the growth factor signal transduction pathway, the sustained stimulation of its activity and subsequent changes in gene expression may be an epigenetic consequence of genetic activation of the Ras pathway by mutations in component proteins (5).
AP-1 denotes a collection of transcription factors composed primarily of heterodimers of Fos and Jun family proteins that recognize a consensus DNA sequence usually in the promoter region of target genes (43). The prototypes of each gene family were originally identified as retroviral oncogenes, thereby highlighting the importance of AP-1 to cellular transformation (18, 60, 106). Transformation by high concentrations of growth factors or oncogenes that function in the growth factor signal transduction pathway is dependent on the sustained increase in expression of AP-1 component proteins and AP-1 activity (71).
Based on the identification of an AP-1 DNA binding site in the collagenase promoter and its role in the degradation of ECM, AP-1 has been implicated in the targeting of genes involved in invasion (2, 17, 114). The epidermal growth factor (EGF)-dependent invasion of human squamous cell carcinoma-derived cell lines is dependent on AP-1 activity (61), as is the conversion of oncogenic Ras-induced hyperproliferative skin lesions into invasive carcinomas (56, 79, 80, 119). Increased expression of AP-1 component proteins, such as Fra-1, rather than mutational activation of AP-1 component proteins, is by far the most frequent means by which AP-1 activity is changed in tumors, and Fra-1 expression is associated with the acquisition of invasive potential by tumors (51, 109, 120).
Fos oncogenes can be used to specifically focus on the contribution of AP-1 to transformation, downstream of the growth factor signal transduction pathway. Transformation of immortal rat 208F fibroblasts with v-FosFBR renders the cells invasive, anchorage independent, and tumorigenic, but they remain dependent on serum for proliferation (34, 64). 208F cells can also be rendered invasive and anchorage independent by growth factor stimulation that induces the expression of AP-1 component proteins and increased AP-1 activity (34, 71). In both cases, invasion is dependent on AP-1 activity (46, 53, 54).
The ability of Fos oncogenes to transform cells is dependent on their functioning as transcription factors (24, 88, 115), which indicates that they mediate transformation through changes in gene expression (34, 46). Interestingly, the changes in expression of AP-1 component proteins following Fos or Ras transformation are similar, emphasizing the importance of AP-1 activation in transformation by oncogenes that function upstream of AP-1 (71). Analysis of genes differentially expressed in v-Fos-transformed 208F fibroblasts indicates that it results in the increased expression of genes that effect invasion (46, 53, 54, 71, 94). However, since v-Fos induces immortality and tumorigenicity in addition to invasion in rodent fibroblasts, it is difficult to determine if differentially expressed genes are specific effectors of invasion or play a wider role in the transformed phenotype (41). Furthermore, others have been unable to find a consistent phenotypic change associated with v-Fos expression in mortal human fibroblasts (1, 50). There is a potential role for tumor suppressors to suppress invasion, since loss or inactivation of tumor suppressors precedes invasion in most tumor model systems.
Herein we provide evidence, based on gene expression profiles, that v-Fos activates a multigenic invasion program in normal human fibroblasts in the presence of active tumor suppressor pathways (p16INK4a and p53) without affecting cell cycle regulation or anchorage-dependent proliferation and suggest that this system can be used to identify genes specifically associated with AP-1 mediated invasion.
MATERIALS AND METHODS
Construction of Myc-tagged v-FosFBJ/R.
Myc-tagged v-FosFBJ/R was generated by PCR, using pBR322 FBJ/R-MSV (65) as a template. The PCR product was ligated into pPCR-Script (Stratagene), and the sequence was verified before being cloned into pLPCX (Clontech Laboratories).
Generation of cell lines.
Telomerase, oncogenic v-Fos, human papillomavirus type 16 (HPV16) E6, and mutant p53 were serially introduced into primary human fetal fibroblasts (hff). The hff strain was established by collagenase digestion of 16-week-old fetal skin. The cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 20 mM HEPES and 20% fetal calf serum (FCS). Early-passage hff were immortalized with the catalytic subunit of telomerase (hTERT) as described previously (68), and the cells were named Tif for “telomerase-immortalized fibroblasts.”
To introduce v-FosFBJ/R into hff and Tif, the amphotropic packaging cell lines Phoenix Ampho (from G. Nolan, Stanford University) or Retropack PT67 (from Clontech Laboratories) were transfected with the retroviral constructs pLPCX (puromycin resistance) or pLPCX FBJ/R expressing Myc-tagged v-FosFBJ/R. The viral supernatants generated were used to infect hff/11 (22 population doublings [P.D.]) and Tif (50 P.D.) using 4 μg of Polybrene per ml. Cells were selected 8 days postinfection in 0.6 μg of puromycin per ml and puromycin-resistant colonies were pooled. Polyclonal populations were used in all experiments to avoid variation due to the clonal heterogeneity of the normal hff and Tif populations. Retroviral infections were repeated independently to confirm phenotypic observations.
p53 was inactivated in late-passage Tif by retroviral infection with either mutant p53 (from K. Ryan, Beatson Institute for Cancer Research) or HPV16 E6 (from A. Phillips, University Medical College of Georgia). Phoenix Ampho cells were transfected with pWZL p53.273H (blasticidin) or pWZL E6 (hygromycin), and the viral supernatants were used to infect Tif. Cells were selected 8 days postinfection by using 10 μg of blasticidin per ml or 100 μg of hygromycin per ml, and pools of drug-resistant colonies were collected.
A mutant form of Fos (aFos) (from R. Hennigan, University of Cincinnati College of Medicine) was expressed in Tif-puro and Tif-Fos by nucleofection. Cells (5 × 105) were resuspended in 100 μl of NHDF-Neo (Amaxa Biosystems) with 3 μg of pEGFP-aFos and nucleofected with program T-20. The cells were incubated for 1 day before being harvested for in vitro invasion assays.
Retrovirally infected cells were screened for virus production by using a C-type reverse transcription assay from Cavidi Tech (Uppsala, Sweden). All cells were routinely tested for mycoplasma contamination by Hoechst 33258 staining (12).
RNA interference.
Transient expression of small interfering RNA (siRNA) was achieved by nucleofection. Tif-Fos cells (5 × 105) were resuspended in 100 μl of NHDF-Neo with 100 nM scrambled siRNA (CGAUCGAUCGAUCGAUCGA), EGF receptor (EGFR) siRNA 1 (CUCUGGAGGAAAAGAAAGU), or EGFR siRNA 2 (CACAGUGGAGCGAAUUCCU) (Dharmacon). Cells were incubated for 48 h before being harvested for Western blotting, growth curve measurement, immunofluorescence, and in vitro invasion assays.
Stable suppression of EGFR expression was achieved using pSUPER.retro (Oligoengine). The puromycin selection cassette was changed to blasticidin resistance, and 64-nucleotide oligonucleotides containing an inhibitory sequence for EGFR (TTCCTATGCCTTAGCAGTC) were annealed and ligated into pSUPER. Tif-Fos cells were retrovirally infected with pSUPER or pSUPER EGFR as described previously, and pools of blasticidin-resistant colonies were obtained.
P.D. and cell counting.
Cells were counted at every passage, and P.D. were calculated from the formula P.D. = 3.32(log10 cells harvested − log10 cells seeded). Cell numbers were determined using a Casy 1 TT cell counter (Schärfe system). For cell growth curves, only attached cells were counted, but for flow cytometric analyses, adherent cells were trypsinized and combined with floating cells from the culture medium to allow analysis of the total cell population.
Western blotting.
Cells were washed with phosphate-buffered saline and lysed for 30 min on ice in buffer containing 50 mM Tris (pH 8.0), 120 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, phosphatase inhibitors (50 mM NaF and 50 μM Na3VO4), protease inhibitors (10 μg of aprotinin per ml, 10 μg of leupeptin per ml, and 1 mM phenylmethylsulfonyl fluoride). The extracts were cleared by centrifugation, and the supernatants stored at −70°C. Protein was quantified by the copper sulfate-bicinchoninic acid method and separated by gel electrophoresis.
Primary antibody incubations were carried out overnight at 4°C with antibodies against the following human proteins: Myc epitope (9E10; Invitrogen), p16INK4a (C-20; Santa Cruz), ERK2 (clone 33; BD Transduction Labs), phospho-p44/42 MAPK (Cell Signaling), p53 (DO-1; Santa Cruz), p21Cip1 (clone 70; BD Transduction Labs), EGFR (clone 13; BD Transduction Labs), and activated EGFR (clone 74; BD Transduction Labs). The blots were washed and probed with horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies (Cell Signaling) and protein detected by enhanced chemiluminescence (Amersham Pharmacia).
Immunofluorescence.
Expression of Myc-tagged v-Fos was detected by immunofluorescence as described previously (97). Cells were grown on glass coverslips for at least 1 to 2 days in DMEM-20% FCS, and v-Fos was detected with 9E10 and tetramethylrhodamine-5-isothiocyanate TRITC-conjugated anti-mouse antibody while F-actin was labeled with fluorescein isothiocyanate FITC-phalloidin.
Wound-healing assay.
Cell motility was assessed using a modification of the wound-healing assay (97). A wound was made in a confluent monolayer of cells by using a 200-μl pipette tip. Three independent areas were marked for identification and photographed at various time points thereafter with a digital camera attached to a phase-contrast microscope. For quantitation of wound closure, the width of each wound at various time points was measured and expressed as a percentage of the original wound size.
In vitro invasion assay.
Invasion was assessed using an in vitro assay (34) with the following modifications. Cells (104) were seeded onto the filter of a polycarbonate chamber containing 100 μl of complete matrigel basement membrane matrix (Becton Dickinson) (in the absence or presence of AG1478). After 3 h of attachment, the cells were washed and placed in serum-free DMEM in the absence or presence of AG1478. DMEM containing 20% FCS with or without EGF in the absence or presence of AG1478 was added above the matrigel as a chemoattractant, and the assay mixture was incubated for 3 days. Invasion was visualized by staining the cells directly with 4 μM calcein acetoxymethyl ester (calcein) (Molecular Probes Europe) in serum-free DMEM for 1 h at 37°C followed by confocal microscopic analysis using a Bio-Rad MRC 600 confocal illumination unit attached to a Nikon Diaphot inverted microscope. Quantitation was performed as described previously (34).
Soft-agar assays.
Anchorage independence was assessed by growth in soft agar. A base layer of 0.5% agar (both Noble and Bacto agars were used independently) in DMEM containing 20% FCS was allowed to set in six-well plates. Cells (104 and 105) were seeded in 0.3% top agar in DMEM-20% FCS. Fresh top agar in DMEM-20% FCS was added once a week, and colonies were analyzed after 3 weeks.
Flow cytometric analysis.
Analysis of the cell cycle phase distribution by propidium iodide staining for DNA content was carried out as follows. Cells were trypsinized and combined with floating cells in retained medium, fixed in ice-cold 70% ethanol, and stored at 4°C. The fixation medium was aspirated after centrifugation and the cells were then stained with 10 μg of propidium iodide per ml and 250 μg of RNase A per ml for 30 min. Cell cycle analyses were made using a Becton Dickinson FACScan. No fewer than 30,000 events were processed per sample, and the bitmap was fixed to discriminate against doublets (cell or nuclear aggregates). Cell cycle phase distributions (expressed as a percentage) were obtained by decomposition of the DNA content histograms by using Cell Quest software.
Microarray analysis with Affymetrix GeneChips.
Total RNA was isolated from exponentially growing cells by using the RNeasy minikit (Qiagen). RNA (15 μg) was reverse transcribed with T7(dT)24 primer (Helena BioSciences). The double-stranded cDNA was in vitro transcribed with T7 RNA polymerase and biotin-labeled ribonucleotides (Affymetrix). The biotin-labeled cRNA was fragmented at 94°C for 35min. cRNA (15 μg) was hybridized onto Affymetrix GeneChip Human Genome U95Av2 arrays at the Cancer Research UK GeneChip service, Paterson Institute for Cancer Research, Manchester, United Kingdom.
Microarray data analysis.
Using Microarray Suite v5.0, all probe sets on the arrays were scaled to a target intensity of 100 (with scaling factors for each chip within threefold of each other). We excluded genes which were flagged as absent in all samples, and data were normalized either using the Robust Multichip Average method in the Bioconductor “affy” method (6) or by correcting for differences in the mean values for each chip by using GeneSpring (Silicon Genetics). The Robust Multichip Average data were subjected to analysis of variance (ANOVA) (P < 0.05) and significance analysis of microarrays (SAM) (105) with the false discovery rate (FDR) set to 0. The edited Venn diagram selected lists were contained entirely in both the ANOVA and SAM lists (data not shown). Genes were annotated for gene symbol, gene name, and function by using UniGene and LocusLink (National Center for Biotechnology Information) according to the HUGO gene nomenclature committee.
Materials.
Mouse EGF was obtained from Sigma. The EGFR inhibitor tyrphostin AG1478 was purchased from Calbiochem CN Biosciences.
RESULTS
Expression of v-Fos in human fibroblasts.
Expression of Ras (Raf and MEK) oncogenes in human fibroblasts (primary or telomerase immortalized) or human mammary epithelial cells that retain functional pRb/p16INK4a and p53 tumor suppressor pathways results in a permanent growth arrest that is similar to replicative senescence (7, 22, 29, 30, 57, 89, 112, 122). The present study was undertaken to determine if activation of AP-1 using the v-Fos oncogene that functions downstream of the Ras signal transduction pathway could stimulate invasion and/or anchorage independence in the presence of active tumor suppressor pathways in normal human diploid fibroblasts. However, since we expected primary cells to enter into replicative senescence or possibly enter premature oncogene-induced senescence, we also used fibroblasts immortalized with the catalytic subunit of telomerase, and these are used in most of the experiments detailed here.
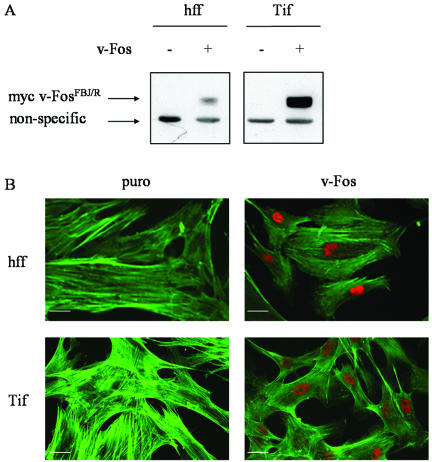
Normal diploid hff and Tif were infected with retroviruses encoding puromycin resistance (puro) or with a Myc epitope-tagged v-FosFBJ/R oncogene (Fos). Colonies of puromycin-resistant cells were pooled, and multiple polyclonal populations of cells were analyzed for the expression of v-Fos. Western analysis, performed using the 9E10 Myc epitope-specific monoclonal antibody to detect exogenous Myc-tagged v-FosFBJ/R protein is shown for representative populations of hff and Tif and demonstrates that only cells infected with the v-Fos virus expressed a protein of the correct molecular mass, 34 kDa for v-FosFBJ/R (65) (Fig. 1A). Immunofluorescence with the Myc antibody revealed that essentially all of the cells in the hff-Fos and Tif-Fos populations expressed the protein in the nucleus of the cell and that none of the control cells, hff-puro and Tif-puro, displayed nuclear staining with anti-Myc (Fig. 1B).
FIG. 1.
Expression of v-Fos and altered morphology in human fibroblasts. Primary hff and telomerase-immortalized hff (Tif) were infected with retroviruses encoding either puromycin resistance (puro) or Myc-tagged v-FosFBJ/R (v-Fos). Colonies of puromycin-resistant cells were pooled and polyclonal populations were analyzed. (A) Expression of v-Fos in the indicated cells was detected by Western blotting for Myc-tagged v-FosFBJ/R with the 9E10 anti-Myc monoclonal antibody. (B) hff-puro, hff-Fos, Tif-puro, and Tif-Fos cells were costained with fluorescence-conjugated antibodies for Myc and phalloidin (TRITC and FITC respectively). Bar, 25 μm.
v-Fos morphologically transforms human fibroblasts.
The morphology of the fibroblasts expressing v-Fos was distinct from that of the puromycin-resistant control cells. At low cell density, the hff-Fos and Tif-Fos cells were more refractile and were easily distinguished from the control cells, hff, Tif, hff-puro, and Tif-puro. At high cell density, the control cells hff-puro and Tif-puro resembled the parental hff and Tif cells, where the cells aligned along their long axes and were arranged into densely packed swirls. The hff-Fos and Tif-Fos cells did not align with each other and were disorganized (data not shown).
Morphological transformation was evident when the actin cytoskeleton was labeled with FITC-phalloidin, demonstrating that the hff-Fos and Tif-Fos cells displayed an equivalent reduction in F-actin stress fibers compared to control cells (Fig. 1B). The effect of v-Fos on the actin cytoskeleton is consistent with previous studies of Fos transformation in other fibroblasts (53).
v-Fos increases the rate of wound healing in human fibroblasts.
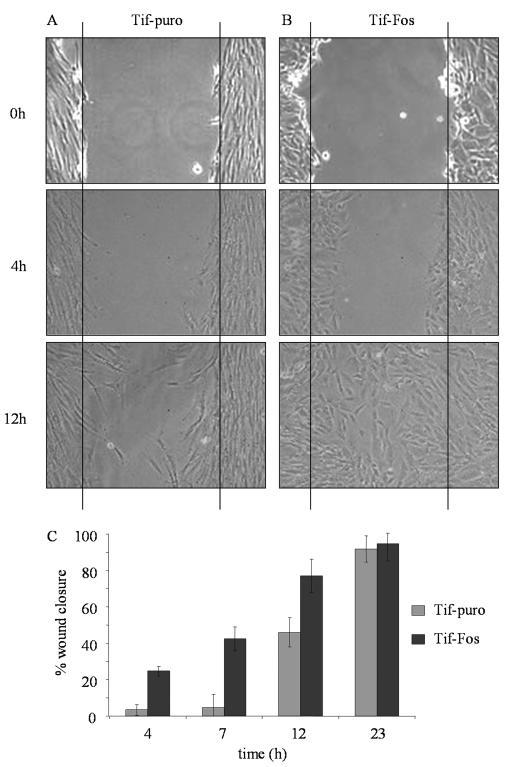
To determine if v-Fos increases cell motility, a wound was made in confluent monolayers of Tif-puro cells (Fig. 2A) and Tif-Fos cells (Fig. 2B) and the closure of the wound was monitored over time. Tif-Fos cells migrated into the wound earlier than did the Tif-puro cells, which displayed a lag phase in wound healing. After only 4 h, Tif-Fos cells displayed 25% wound closure compared to only 3% for Tif-puro cells. By 12 h, wound closure increased to 75 and 45% for Tif-Fos and Tif-puro cells, respectively, and by 23 h, both cell types had essentially closed the wound (Fig. 2C).
FIG. 2.
Motility of telomerase-immortalized fibroblasts expressing v-Fos. (A and B) A wound was made in a confluent monolayer of Tif-puro (A) and Tif-Fos (B) cells, and the area was marked for identification and photographed after 0, 4 and 12 h. (C) Three independent areas of each wound made in Tif-puro (gray bars) and Tif-Fos (black bars) were measured, and the error bars show the standard deviation of the mean percent wound closure after 4, 7, and 23 h.
Human fibroblasts expressing v-Fos are highly invasive.
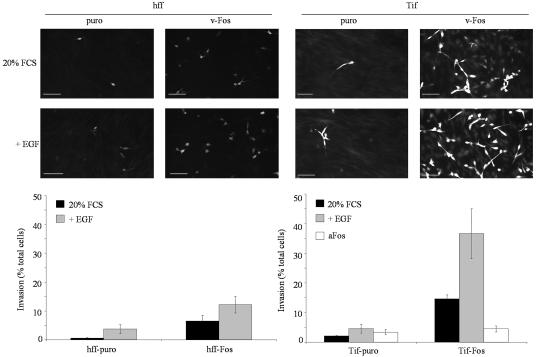
A three-dimensional in vitro inverse invasion assay was used to determine if hff-Fos and Tif-Fos cells were capable of invading through reconstituted basement membrane matrix (34). The assay was configured such that invasion is a measure of a cell's ability to detach from the substrate and to migrate through matrigel. Essentially none of the parental cells (hff or Tif [data not shown]) or control cells (hff-puro or Tif-puro) invade the matrigel, as expected (Fig. 3), although these cells are capable of migrating through the pores of the filter (data not shown). However, v-Fos stimulated the invasion of both hff and Tif, as evidenced by the presence of calcein-stained cells in the matrigel (Fig. 3). Tif-Fos cells are the more invasive of the two, and this difference may be accounted for by the slightly lower expression of v-Fos in hff-Fos cells (Fig. 1A) rather than immortalization by telomerase. Expression of a mutant form of Fos that cannot bind DNA (aFos) does not stimulate invasion in Tif-puro but quite clearly inhibits invasion in Tif-Fos, indicating that v-Fos-stimulated invasion functions through AP-1 activity (Fig. 3).
FIG. 3.
Invasion of human fibroblasts expressing v-Fos. An in vitro assay was used to measure three-dimensional invasion through matrigel reconstituted basement membrane matrix. Primary hff, hff-puro and hff-Fos, and telomerase-immortalized hff, Tif-puro, and Tif-Fos were stained with calcein after 3 days and a Z-series (6 to 30 μm) was captured on a confocal microscope and merged into one image. Bar, 100 μm. Invasion assays were performed in duplicate, and two or three independent fields per sample were analyzed and quantitated. Invasion, in the absence (black bars) or presence of 100 ng of EGF per ml (gray bars) or aFos (white bars) was expressed as a percentage of the total number of cells seeded on the porous filter, and the error bars represent the standard deviation of the mean.
v-Fos does not alter growth parameters or karyotype.
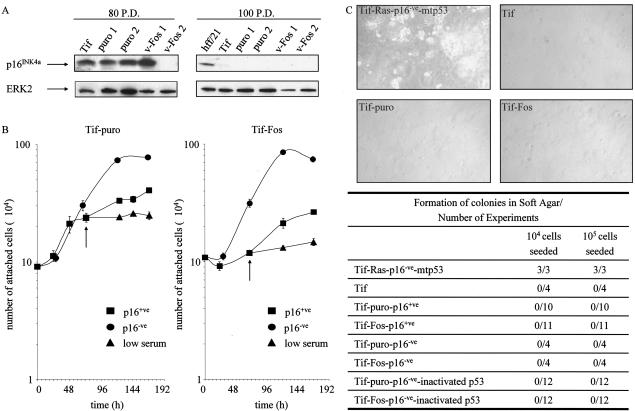
Human fibroblasts expressing Ras pathway oncogenes prematurely enter into a permanent growth arrest that resembles senescence (89). Under the culture conditions described here, the expression of oncogenic v-Fos neither induces a premature senescence-like growth arrest in hff or Tif nor delays or prevents the onset of replicative senescence in hff. The hff-puro, hff-Fos, Tif-puro, and Tif-Fos cells continue to proliferate following puromycin selection with no morphological evidence of senescence or decrease in cell doubling time. As expected, around 7 weeks after retroviral infection, both the hff-puro and hff-Fos enter into replicative senescence and express β-galactosidase, detectable at pH 6.0 (data not shown). Tif-puro and Tif-Fos cells, like parental Tif cells, are immortal and continue to proliferate indefinitely (data not shown). However, after about 80 cumulative P.D., one of the Tif-Fos populations displayed a twofold increase in saturation density, and by 100 P.D., all of the Tif-derived populations also showed a similar increase in saturation density. This was suggestive of loss of a growth-suppressing protein such as p16INK4a (90). To determine if this was the case, Western analysis was performed for p16INK4a with cells that had been passaged for 80 and 100 P.D. All of the cells at 80 P.D., except Tif-Fos 2, expressed p16INK4a, and after 100 P.D., all of the Tif-derived cells had suppressed p16INK4a expression (Fig. 4A). The loss of p16INK4a was not mediated by v-Fos, nor was it a consequence of puromycin selection, since the parental Tif also suppressed p16INK4a expression. The loss of expression of p16INK4a was presumably due to methylation, since p16INK4a expression was restored after exposure to 5-azacytidine (data not shown).
FIG. 4.
v-Fos has no effect on proliferation or serum or anchorage dependence. (A) Telomerase-immortalized fibroblasts. Tif, Tif-puro 1, Tif-puro 2, Tif-Fos 1, and Tif-Fos 2 exhibited an increase in saturation density with each passage after 80 cumulative P.D., which correlated with a loss of p16INK4a expression as demonstrated by Western blotting after 100 P.D. hff/21 (36 P.D.) was used as a positive control for p16INK4a, and equal loading was determined with anti-ERK2. (B) Growth curves of telomerase-immortalized fibroblasts expressing v-Fos. Cells (p16+ve and p16−ve) were grown in 20% serum, and once exponential growth was established after 72 h, the cells were washed and incubated with either 20% serum or a low concentration of serum (0.2%), as indicated by the arrow. Cell proliferation was determined by counting the number of attached cells every day. Each experiment was performed in triplicate, and the error bars show the standard deviation of the mean daily count. (C) Anchorage-dependent growth of telomerase-immortalized fibroblasts with various combinations of v-Fos, loss of p16INK4a (p16−ve), and inactivation of p53 with HPV16 E6 or mutant p53 (273H). For each experiment, 104 or 105 cells were seeded in soft agar. Multiple experiments with multiple polyclonal populations of the cells were performed. During a 3-week period, colony formation was monitored and scored as 0 if there were no soft agar colonies. Tif-Ras that had lost expression of p16INK4a and expressed a mutant form of p53 (mtp53) were used as a positive control for colony formation in soft agar.
Thus, v-Fos expression neither induces nor prevents a senescence-like permanent growth arrest. In this regard, human fibroblasts are distinct from rodent fibroblasts, where v-Fos renders the cells immortal (41). The status of the tumor suppressor, p16INK4a, did not affect invasion; hff-Fos cells express p16INK4a and are invasive while the invasiveness of the Tif-Fos cells does not change after the loss of p16INK4a with time in culture (data not shown).
Karyotyping was performed to determine if v-Fos altered chromosomal stability. Analysis of metaphase spreads revealed that hff, hff-puro, and hff-Fos cells remained diploid, 46,XY. The parental Tif cells were 46,XY with a translocation of 14q/14q, and v-Fos did not induce any additional chromosomal changes (data not shown). Hence, v-Fos does not induce gross chromosomal rearrangements.
Tif-puro and Tif-Fos cells retain a functional p53 response to DNA damage.
Mutational inactivation of the p53 tumor suppressor pathway occurs in approximately 50% of human tumors (37), and inactivation of p53 is important for immortalization and transformation of human fibroblasts by oncogenic Ras (22, 29, 30).
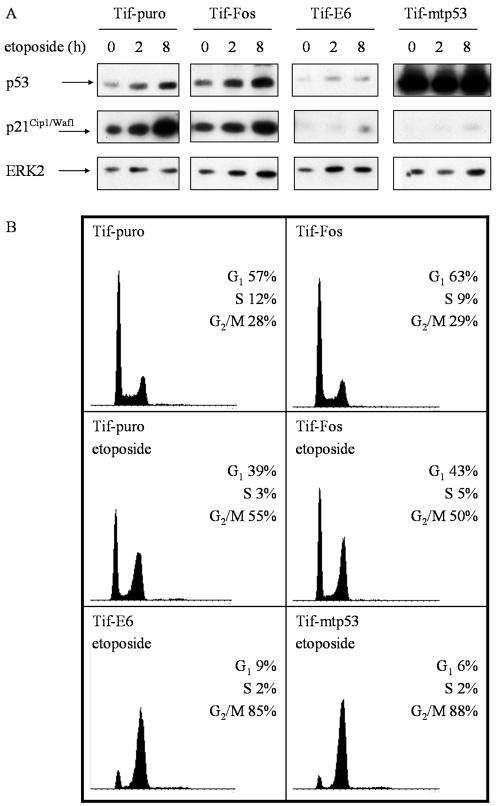
To determine if v-Fos altered p53 function, the cellular response to DNA-damaging agents was assessed. Western blot analysis of p53 expression levels in Tif-puro and Tif-Fos cells revealed slightly elevated levels in Tif-Fos cells (Fig. 5A). However, Tif-puro and Tif-Fos cells both responded to DNA damage induced by etoposide (topoisomerase II inhibitor) by stabilization of p53 protein after 2 to 8 h and a subsequent increase in p21Cip1/Waf1 protein expression (Fig. 5A). Flow cytometric analysis demonstrated that after 20 h, the cells emptied out of S phase and accumulated in both the G1 and G2 phases of the cell cycle (Fig. 5B), which is consistent with a p53-mediated response to DNA damage (8, 47). Similar results were obtained with UV-induced DNA damage, and an enzyme-linked immunosorbent assay specific for mutant p53 (Oncogene Research Products) also confirmed wild-type p53 status (data not shown). These results are consistent with previous reports that telomerase expression does not affect p53 function (42, 66, 108) and further demonstrate that v-Fos does not inactivate p53.
FIG. 5.
v-Fos does not alter the p53 response to DNA damage. (A) Comparison of the expression levels of p53 and p21 in telomerase-immortalized fibroblasts after DNA damage induced by etoposide. Cells were treated with 1 μM etoposide and lysed at the indicated times for Western blot analysis of p53 and p21Cip1/Waf1. p53 was inactivated in Tif cells with HPV16 E6 or mutant p53 (mtp53). The membranes were stripped and reprobed for ERK2 as a loading control. (B) Flow cytometric analysis of the cell cycle phase distribution after DNA damage induced by etoposide in telomerase-immortalized fibroblasts. Exponentially growing cells were incubated in the absence or presence of 1 μM etoposide for 20 h, stained with propidium iodide for DNA content, and analyzed by flow cytometry. The DNA content histograms are shown along with the distribution of cells in the G1, S and G2/M phases of the cell cycle.
To demonstrate that the observed G1 arrest was p53 dependent, p53 was inactivated in Tif, either by degradation induced by the HPV16 E6 oncoprotein (16) or by expression of a dominant negative mutant of p53, 273H (mtp53) (36). Western analysis of Tif-E6 protein extracts revealed a significant reduction in p53 expression, consistent with E6-mediated degradation of p53, while the protein extracts of Tif-mtp53 displayed high-level expression of p53, consistent with overexpression of mutant p53 (Fig. 5A). In contrast to Tif-puro and Tif-Fos cells exposed to etoposide, the majority of Tif-E6 and Tif-mtp53 cells did not display a significant increase in p21Cip1/Waf1 expression (Fig. 5A) and did not arrest in G1 (Fig. 5B), consistent with the inactivation of the endogenous wild-type p53 (107, 108).
v-Fos does not alter the growth regulation of human fibroblasts.
Despite inducing clear morphological transformation and rendering the cells invasive, v-Fos had no effect on growth rate, saturation density, or serum- and anchorage-dependent proliferation. The growth characteristics of the Tif-Fos cells were not radically distinct from those of Tif and Tif-puro cells. Tif-Fos grew to the same saturation density as Tif-puro when grown in 20% serum (Fig. 4B). Growth curves are shown for early-passage p16INK4a-expressing cells and late-passage p16INK4a-negative cells, demonstrating the twofold increase in saturation density observed in both Tif-puro and Tif-Fos cells after the loss of p16INK4a expression. Tif-Fos cells, like Tif-puro cells, were serum dependent for proliferation as judged by the cessation of proliferation on being transferred from 20% serum to a low (0.2%) serum concentration (Fig. 4B).
The Tif-Fos cells also resembled Tif-puro cells in that they remain anchorage dependent for proliferation. In multiple experiments, no colonies formed in soft agar after 3 weeks (Fig. 4C). The Tif-Fos cells were tested for anchorage independence both before and after the loss of p16INK4a expression, confirming that loss of the tumor suppressor p16INK4a was not sufficient to render Fos-transformed human fibroblasts anchorage independent. To determine if additional loss of function of the tumor suppressor protein, p53, altered the anchorage requirements of Tif-Fos cells, p53 was inactivated by expression of either E6 or mtp53 as detailed previously. Tif-Fos-E6 and Tif-Fos-mtp53 cells remained anchorage dependent despite having inactivated both the p16INK4a and p53 tumor suppressor pathways. These results indicate, as expected, that further changes are required to render human fibroblasts anchorage independent (29-31). The additional inactivation of p53 in late-passage (p16-ve) Tif-puro or Tif-Fos cells does not affect the invasiveness of the cells (data not shown). Hence, invasion is independent of the two major tumor suppressor pathways, pRb/p16INK4a and p53.
Gene expression profiling of invasive Tif-Fos cells.
The v-Fos oncogene functions as a component of the AP-1 transcription factor and must mediate its phenotypic consequences, such as morphological transformation, increased motility, and invasiveness, through changes in gene expression. Since the main, if not sole, consequence of v-Fos expression in normal human fibroblasts is to render the cells invasive, analysis of genes differentially expressed in Tif-Fos cells should identify genes that specifically mediate invasion. Microarray analysis was performed with Affymetrix GeneChips using RNA derived from hff, Tif, Tif-puro, and Tif Fos cells. Four independent preparations of RNA from hff and Tif were used to ensure that the differences in gene expression were not due to telomerase expression. To exclude changes in gene expression due to puromycin selection, three independent preparations of RNA were made from two separate populations of Tif-puro. To identify changes in gene expression dependent on v-Fos expression, three independent RNA preparations were made from two separate Tif-Fos populations and a single RNA preparation was made from a third Tif-Fos population.
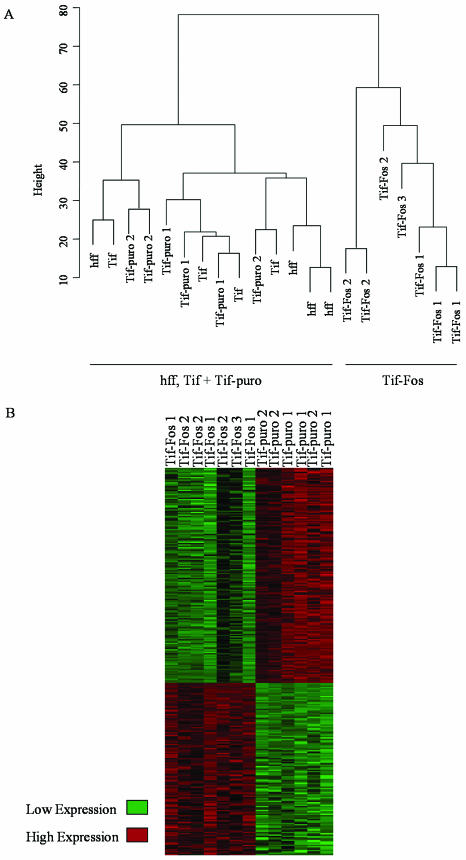
Hierarchical cluster analysis demonstrates that the global expression profiles from hff, Tif, and Tif-puro cells, although marginally distinct from each other, were closely related, while the Tif-Fos populations clustered separately (Fig. 6A). This validates the use of telomerase-immortalized fibroblasts as surrogates for primary fibroblasts for studying v-Fos transformation. It also demonstrates that the changes in gene expression detected in Tif-Fos cells are a consequence of v-Fos.
FIG. 6.
Gene expression profiling. (A) Cluster dendrogram of all samples: hff (×4), Tif (×4), Tif-puro (×6), and Tif-Fos (×7). The metric used was Euclidean distance, and clustering was done by the “complete” method as implemented in the “mva” library in the “R” statistical software. (B) Statistically significant changes in gene expression assessed by SAM for all Tif-puro samples (×6) and all Tif-Fos samples (×7).
We have identified an extremely conservative list of 357 differentially expressed genes with at least a twofold change in gene expression between Tif-puro and Tif-Fos cells (Fig. 6B; Tables 1 and 2). To generate these lists, three replicate RNA samples for a single pool of Tif-puro were compared to three replicate RNA samples for a single pool of Tif-Fos, the genes were filtered for a fold change of twofold, and Venn diagrams were used to select genes that were common to all three lists. To ensure that the lists represented consistent changes in gene expression, each gene was checked for a twofold expression difference in all of the biological replicates not used in the initial pairwise comparison. When a gene was represented by more than one oligonucleotide on the GeneChip, the multiple representation was deleted, resulting in 130 and 227 individually represented genes up- and down-regulated respectively. Similar lists were obtained by ANOVA and SAM (Fig. 6B).
TABLE 1.
Selected genes up-regulated in invasive Tif-Fos cellsa
| Symbol | Gene name | GenBank accession no. | Fold change |
|---|---|---|---|
| Actin cytoskeleton | |||
| FLNB | Filamin B, beta (ABP-278) | AF042166 | 2.5 |
| GEM | GTP binding protein overexpressed in skeletal muscle | U10550 | 2.9 |
| KRT18 | Keratin 18 | M26326 | 39.6 |
| LPXN | Leupaxin | AF062075 | 3.3 |
| MAPRE2 | Microtubule-associated protein, RP/EB family, 2 | X94232 | 8.6 |
| PPL | Periplakin | AF001691 | 30.2 |
| SDCBP | Syndecan binding protein (syntenin) | AF000652 | 2.7 |
| TNNT1 | Troponin T1, skeletal, slow | AJ011712 | 5.5 |
| TUBB | Tubulin, beta polypeptide | AF035316 | 3.4 |
| Cell motility | |||
| CXCL1 | Chemokine (C-X-C motif) ligand 1 (MGSA) | X54489 | 21.2 |
| CXCL6 | Chemokine (C-X-C motif) ligand 6 (GCP-2) | U81234 | 37.8 |
| LGALS3BP | Lectin, galactoside binding, soluble, 3 binding protein | L13210 | 5.2 |
| ECM | |||
| LAMA3 | Laminin, α3 | L34155 | 5.7 |
| RELN | Reelin | U79716 | 3.8 |
| SPOCK | Sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) | X73608 | 3.2 |
| Proteolysis | |||
| CASP1 | Caspase-1, apoptosis-related cysteine protease (IL1BC) | M87507 | 4.5 |
| CTSC | Cathepsin C | X87212 | 6.4 |
| MMP1 | Matrix metalloproteinase 1 (interstitial collagenase) | M13509 | 5.8 |
| MMP3 | Matrix metalloproteinase 3 (stromelysin 1, progelatinase) | X05232 | 21.1 |
| PLAU | Plasminogen activator, urokinase | X02419 | 6.6 |
| PRSS3 | Protease, serine, 3 (mesotrypsin) | X71345 | 38.7 |
| TFPI | Tissue factor pathway inhibitor (LACI) | M59499 | 7.9 |
| Membrane receptors | |||
| ADORA2B | Adenosine A2b receptor | X68487 | 50.1 |
| CD9 | CD9 antigen (p24) | M38690 | 3.2 |
| EPHA2 | EphA2 | M59371 | 3.6 |
| EPHB6 | EphB6 | D83492 | 8.7 |
| EGFR | EGFR | X00588 | 10.9 |
| FZD2 | Frizzled homolog 2 | L37882 | 3.8 |
| GPRC5B | G-protein-coupled receptor C5B | AC004131 | 11.7 |
| ITGA2 | Integrin, α2 | X17033 | 6.1 |
| ITGA3 | Integrin, α3 | M59911 | 3.0 |
| ITGA6 | Integrin, α6 | S66213 | 12.5 |
| IL1RL1 | Interleukin-1 receptor-like 1 (ST2) | D12763 | 4.2 |
| LIFR | Leukemia inhibitory factor receptor | X61615 | 4.6 |
| MET | Met proto-oncogene (HGFR) | J02958 | 3.5 |
| PTGER4 | Prostaglandin E receptor 4 (subtype EP4) | L25124 | 14.1 |
| PTGFR | Prostaglandin F receptor (FP) | AF004021 | 10.8 |
| RA13 | Retinoic acid induced 3 | AF095448 | 4.2 |
| RAIG-2 | GPRC5B | HG4272-HT4542 | 5.5 |
| SYNGR3 | Synaptogyrin 3 | AJ002309 | 5.3 |
| TM4SF1 | Transmembrane 4 superfamily, 1 | M90657 | 19.8 |
| TM4SF3 | Transmembrane 4 superfamily, 3 | M35252 | 188.9 |
| Signal transduction | |||
| ARL7 | ADP-ribosylation factor-like 7 | AB016811 | 3.9 |
| APBB2 | Amyloid beta (A4) precursor protein binding B2 (Fe65-like) | U62325 | 3.7 |
| ICK | Intestinal cell kinase | AB023153 | 3.7 |
| MAP4K2 | Mitogen-activated protein kinase kinase kinase kinase 2 | U07349 | 4.8 |
| NMU | Neuromedin U | X76029 | 7.0 |
| SHC3 | Neuronal Shc | D84361 | 12.1 |
| NMT2 | N-Myristoyltransferase 2 | AF043325 | 5.5 |
| PPT2 | Palmitoyl-protein thioesterase 2 | AF020543 | 3.1 |
| PIK3CB | Phosphoinositide-3-kinase, catalytic, beta polypeptide | S67334 | 2.7 |
| PIK3CD | Phosphoinositide 3-kinase, catalytic, delta polypeptide | Y10055 | 7.6 |
| PKIA | Protein kinase (cAMP-dependent, catalytic) inhibitor alpha | S76965 | 5.0 |
| PTPRF | Protein tyrosine phosphatase, receptor type, F (LAR) | Y00815 | 3.3 |
| PTPRK | Protein tyrosine phosphatase, receptor type, K | L77886 | 3.1 |
| ARHI | Ras homolog gene family, 1 (NOEY2) | U96750 | 4.7 |
| RRAS2 | Ras-like protein TC21 | HG1111-HT111 | 4.0 |
| RHOBTB3 | Rho-related BTB domain containing 3 | AB020685 | 2.7 |
| STAC | Src homology three (SH3) and cysteine-rich domain | D86640 | 2.8 |
| Transcription factors | |||
| CART1 | Cartilage paired-class homeoprotein 1 | U31986 | 5.0 |
| FOXF2 | Forkhead box F2 | U13220 | 3.5 |
| FOXO1A | Forkhead box O1A (rhabdomyosarcoma) | AF032885 | 2.8 |
| NR2F2 | Nuclear receptor 2F2 | M64497 | 12.4 |
| TFAP2A | Transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | M36711 | 44.3 |
| TP53 | Tumor protein p53 | X02469 | 3.5 |
| Cell cycle and growth | |||
| CDC20 | CDC20 cell division cycle 20 homolog | U05340 | 3.6 |
| CKS2 | CDC28 protein kinase regulatory subunit 2 | X54942 | 3.3 |
| CCNB1 | Cyclin B1 | M25753 | 4.3 |
| CDKN3 | Cyclin-dependent kinase inhibitor 3 | L25876 | 3.8 |
| ORC2L | Origin recognition complex, subunit 2-like | U27459 | 3.1 |
| Apoptosis | |||
| BAX | BCL2-associated X protein | L22475 | 5.4 |
| CARD10 | Caspase recruitment domain family, 10 | AL049851 | 5.2 |
| CLU | Clusterin (CLI, SP-40, TRPM2, APOJ) | M25915 | 9.1 |
| PTTG1 | Pituitary tumor-transforming 1 (securin) | AA203476 | 5.1 |
| TNFRSF10B | Tumor necrosis factor receptor superfamily, 10b (TRAIL-R2) | AF016266 | 3.1 |
| TNFRSF21 | Tumor necrosis factor receptor superfamily, 21 (DR6) | AF068868 | 2.7 |
RNA from Tif-puro (×3) and Tif-Fos (×3) cells was hybridized to Affymetrix GeneChips (human genome U95Av2), and the gene expression profiles were analyzed using GeneSpring. Filtering on fold changes in Tif-Fos cells that are greater than those in Tif-puro cells by a factor of two identified 130 genes up-regulated by v-Fos. The table lists some of the genes that may be important for various aspects of invasion. Fold changes are given as an indication of expression differences and in some cases may be overestimated if the gene is undetectable in Tif-puro cells.
TABLE 2.
Selected genes down-regulated in invasive Tif-Fos cellsa
| Symbol | Gene name | GenBank accession no. | Fold change |
|---|---|---|---|
| Actin cytoskeleton | |||
| ACTC | Actin, alpha, cardiac muscle | J00073 | −61.3 |
| AMPH | Amphiphysin | X81438 | −22.0 |
| CALD1 | Caldesmon 1 | M64110 | −4.1 |
| CNN1 | Calponin 1, basic, smooth muscle | D17408 | −14.9 |
| DSP | Desmoplakin 1 | AL031058 | −140.9 |
| FARP1 | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 | AI701049 | −4.5 |
| FLG | Filaggrin | M60502 | −48.8 |
| HSPB1 | Heat shock 27-kDa protein 1 | Z23090 | −2.4 |
| LMOD1 | Leiomodin 1 (smooth muscle) | X54162 | −6.1 |
| MAP1A | Microtubule-associated protein 1A | W26631 | −4.3 |
| MAP1B | Microtubule-associated protein 1B | L06237 | −3.5 |
| MYO1D | Myosin ID | AB018270 | −4.6 |
| KIAA09921 | Palladin | AB023209 | −7.4 |
| TES | Testis-derived transcript (3 LIM domains) | AL050162 | −8.3 |
| TPM1 | Tropomyosin 1 (alpha) | M19267 | −3.2 |
| Cell adhesion | |||
| FAT | FAT tumour suppressor homolog 1 | X87241 | −2.7 |
| ISLR | Immunoglobulin superfamily containing leucine-rich repeat | AB003184 | −5.7 |
| IGSF4 | Immunoglobulin superfamily, 4 | AL080181 | −19.4 |
| MFGE8 | Milk fat globule-EGF factor 8 protein | U58516 | −6.1 |
| OPCML | Opioid binding protein/cell adhesion molecule-like (OBCAM) | U79251 | −27.3 |
| TRO | Trophinin | AB029037 | −9.6 |
| Cell motility | |||
| CXCL12 | Chemokine (C-X-C motif) ligand 12 (SDF1) | U19495 | −14.3 |
| CTGF | Connective tissue growth factor | X78947 | −21.9 |
| ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) | L35594 | −24.8 |
| EFNB2 | Ephrin-B2 | AI765533 | −3.2 |
| MDK | Midkine (NEGF2) | X55110 | −14.6 |
| SEMA3C | Semaphorin 3C/formerly semaphorin E | AB000220 | −28.8 |
| SEMA7A | Semaphorin 7A/formerly semaphorin L | AF030698 | −5.8 |
| ECM | |||
| DKFZp56411922 | Adlican | AL049946 | −2.5 |
| CRTL1 | Cartilage-linking protein 1 | U43328 | −83.6 |
| CSPG2 | Chondroitin sulfate proteoglycan 2 (versican) | X15998 | −204.5 |
| COL8A1 | Collagen, type VIII, alpha 1 | X57527 | −7.1 |
| COL15A1 | Collagen, type XV, alpha 1 | L25286 | −59.4 |
| COL16A1 | Collagen, type XVI, alpha 1 | M92642 | −5.5 |
| EFEMP1 | EGF-containing fibulin-like ECM protein 1 | U03877 | −9.5 |
| ELN | Elastin (SVAS, WBS) | X52896 | −39.9 |
| FBN1 | Fibrillin 1 (MFS1) | X63556 | −4.2 |
| FMOD | Fibromodulin | U05291 | −5.6 |
| FN1 | Fibronectin 1 | X02761 | −2.9 |
| FBLN1 | Fibulin 1 | U01244 | −13.8 |
| HSPG2 | Heparan sulfate proteoglycan 2 (perlecan) | M85289 | −4.1 |
| HAS2 | Hyaluronan synthase 2 | U54804 | −163.3 |
| LAMA2 | Laminin, α2 (merosin) | Z26653 | −10.1 |
| LAMA4 | Laminin, α4 | S78569 | −6.0 |
| LTBP1 | Latent transforming growth factor beta binding protein 1 | M34057 | −48.0 |
| LTBP2 | Latent transforming growth factor beta binding protein 2 | Z37976 | −11.5 |
| LUM | Lumican | U21128 | −52.8 |
| MAGP2 | Microfibril-associated glycoprotein-2 | U37283 | −17.1 |
| MFAP4 | Microfibrillar-associated protein 4 | L38486 | −10.6 |
| SGCD | Sarcoglycan, delta (35DAG) | X95191 | −30.1 |
| SPP1 | Secreted phosphoprotein 1 (osteopontin, BSP1, ETA-1) | AF052124 | −10.2 |
| SULF1 | Sulfatase 1 | AB029000 | −73.3 |
| THBS1 | Thrombospondin 1 | X14787 | −23.5 |
| Proteolysis | |||
| ADAMTS2 | A disintegrin-like and metalloprotease with thrombospondin type 1 motif 2 | AJ003125 | −19.6 |
| CPE | Carboxypeptidase E | X51405 | −8.3 |
| C1R | Complement component 1, r subcomponent | M14058 | −5.3 |
| C1S | Complement component 1, s subcomponent | J04080 | −4.3 |
| RECK | Reversion-inducing-cysteine-rich protein with kazal motifs | D50406 | −11.5 |
| SERPING1 | Serine (or cysteine) proteinase inhibitor, clade G (C1 inhibitor) 1 | X54486 | −8.5 |
| TFP12 | Tissue factor pathway inhibitor 2 (PP5) | D29992 | −29.6 |
| TIMP1 | Tissue inhibitor of metalloproteinase 1 (EPA, CLG1) | D11139 | −2.9 |
| Membrane receptors | |||
| ACVR2 | Activin A receptor II | D31770 | −3.3 |
| F2RL1 | Coagulation factor II (thrombin) receptor-like I (PAR2) | U67058 | −9.2 |
| EDNRA | Endothelin receptor type A | D11151 | −9.3 |
| FZD1 | Frizzled homolog 1 | AB017363 | −4.4 |
| FZD7 | Frizzled homolog 7 | AB017365 | −3.4 |
| GPR51 | G protein-coupled receptor 51 | AF056085 | −6.8 |
| IL1R1 | Interleukin 1 receptor 1 | M27492 | −2.5 |
| PLXNA2 | Plexin A2 | AB007932 | −4.0 |
| PLXND1 | Plexin D1 | AB014520 | −2.9 |
| PTGER2 | Prostaglandin E receptor 2 (subtype EP2), 53kDa | U19487 | −4.3 |
| Signal transduction | |||
| ADCY9 | Adenylate cyclase 9 | AF036927 | −3.3 |
| BDKRB2 | Bradykinin receptor B2 | M88714 | −4.0 |
| CBLB | cas-Br-M (murine) ecotropic retroviral transforming sequence b | U26710 | −4.7 |
| CD24 | CD24 antigen (small cell lung carcinoma cluster 4 antigen) | L33930 | −134.4 |
| CRABP2 | Cellular retinoic acid binding protein 2 | M97815 | −27.8 |
| DAB2 | Disabled homolog 2, mitogen-responsive phosphoprotein | U53446 | −2.7 |
| DOK1 | Docking protein 1 (p62dok) | U70987 | −2.8 |
| DOK5 | Docking protein 5 | AL050069 | −11.2 |
| EFS | Embryonal Fyn-associated substrate | AB001466 | −8.2 |
| GS3955 | GS3955 protein | D87119 | −11.1 |
| HCK | Hemopoietic cell kinase | M16592 | −3.3 |
| QK1 | Homolog of mouse quaking QK1 (KH domain RNA binding protein) | AL031781 | −4.7 |
| INHBB | Inhibin, beta B (activin AB beta polypeptide) | M31682 | −51.4 |
| MRAS | Muscle Ras oncogene homolog | AF043938 | −3.4 |
| NEK7 | NIMA (never in mitosis gene a)-related kinase 7 | AL080111 | −2.8 |
| PFTK1 | PFTAIRE protein kinase 1 | AB020641 | −3.9 |
| PDE4B | Phosphodiesterase 4B, cAMP-specific (PDE4 dunce homolog) | L20971 | −17.8 |
| PTN | Pleiotrophin (HBGF8, NEGF1) | M57399 | −8.4 |
| PENK | Preproenkephalin | J00123 | −33.1 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 (PGG/HS, COX2) | U04636 | −97.8 |
| PRKG1 | Protein kinase, cGMP-dependent 1 | Y07512 | −4.5 |
| PRKCM | Protein kinase C, mu | X75756 | −3.1 |
| PTPRN | Protein tyrosine phosphatase, receptor type, N | L18983 | −3.0 |
| RAB3B | RAB3B, member RAS oncogene family | M28214 | −2.6 |
| ARHB | Ras homolog gene family, B (RhoB) | M12174 | −6.2 |
| ARHE | Ras homolog gene family, E (RhoE) | S82240 | −3.7 |
| RAC2 | Ras-related C3 botulinum toxin substrate 2 (rho family, small GTPBP Rac2) | M64595 | −6.5 |
| ROR1 | Receptor tyrosine kinase-like orphan receptor 1 | M97675 | −2.8 |
| P164RHOGEF | Rho-specific guanine-nucleotide exchange factor | AB002335 | −5.1 |
| TESK1 | Testis-specific kinase 1 | D50863 | −2.3 |
| TSPAN-2 | Tetraspan 2 | AI924594 | −5.0 |
| TOB1 | Transducer of ERBB2, 1 | D38305 | −2.9 |
| TGFBR1 | Transforming growth factor, beta receptor 1 (ALK-5) | L11695 | −3.4 |
| RALA | v-Ral simian leukemia viral oncogene homolog A (Ras related) | M29893 | −3.4 |
| WASF3 | WAS protein family, 3 (brush-1) | S69790 | −6.1 |
| WNT2 | Wingless-type MMTV integration site 2 | X07876 | −14.9 |
| WNT5A | Wingless-type MMTV integration site 5A | L20861 | −7.7 |
| Transcription factors | |||
| BHLHB2 | Basic helix-loop-helix class B, 2 (DEC1) | AB004066 | −4.5 |
| CUTL1 | Cut-like 1, CCAAT displacement protein | L12579 | −3.9 |
| CKTSF1B1 | Cysteine knot superfamily, 1, BMP antagonist 1 (gremlin/Drm) | AF045800 | −5.5 |
| CRIP1 | Cysteine-rich protein 1 (intestinal) | AI017574 | −54.7 |
| FOXD1 | Forkhead box D1 | U59831 | −4.2 |
| LDB2 | LIM domain binding 2 | AF052389 | −9.4 |
| LITAF | Lipopolysaccharide-induced TNF factor | AF010312 | −4.4 |
| NOTCH3 | Notch homolog 3 | U97669 | −19.8 |
| STAT1 | Signal transducer and activator of transcription 1 | M97935 | −5.1 |
| TCF7 | Transcription factor 7 (T-cell specific, HMG-box) | X59871 | −4.7 |
| TCF21 | Transcription factor 21 | AF047419 | −9.6 |
| Cell cycle and growth | |||
| CCND2 | Cyclin D2 | D13639 | −9.0 |
| DTR | Diphtheria toxin receptor (HBEGF) | M60278 | −10.4 |
| GAS1 | Growth arrest-specific 1 | L13698 | −6.1 |
| GAS6 | Growth arrest-specific 6 | L13720 | −6.0 |
| IGFBP3 | Insulin-like growth factor binding protein 3 | M35878 | −9.3 |
| IGFBP4 | Insulin-like growth factor binding protein 4 | M62403 | −4.5 |
| IGFBP5 | Insulin-like growth factor binding protein 5 | L27560 | −60.6 |
| IGFBP7 | Insulin-like growth factor binding protein 7 (MAC25) | L19182 | −4.3 |
| NRG1 | Neuregulin 1 | M94167 | −5.2 |
| NTF3 | Neurotrophin 3 | X53655 | −10.0 |
| SFRP1 | Secreted frizzled-related protein 1 | AF056087 | −5.2 |
| FGF2 | Fibroblast growth factor 2 (basic) | M27968 | −3.4 |
| FGF5 | Fibroblast growth factor 5 | M37825 | −7.0 |
| FGF7 | Fibroblast growth factor 7 (KGF) | S81661 | −32.7 |
| Apoptosis | |||
| TNFRSF6 | Tumor necrosis factor receptor superfamily, member 6 | X63717 | −2.6 |
| TNFA1P3 | Tumor necrosis factor, alpha-induced protein 3 | M59465 | −5.1 |
RNA from Tif-puro (×3) and Tif-Fos (×3) cells was hybridized to Affymetrix GeneChips (human genome U95Av2), and the gene expression profiles were analyzed using GeneSpring. Filtering on fold changes in Tif-Fos cells that are less than those in Tif-puro cells by a factor of two identified 227 genes down-regulated by v-Fos. The table lists some of the genes that may be important for various aspects of invasion. Fold changes are given as an indication of expression differences and in some cases may be overestimated if the gene is undetectable in Tif-Fos cells.
In addition to demonstrating that expression of v-Fos in human fibroblasts results in a consistent alteration of the gene expression profile, it identifies known effectors of the invasive phenotype such as genes involved in actin cytoskeleton regulation, cell adhesion and motility, ECM and proteolysis, as well as genes that have not previously been associated with invasion. We propose that up-regulated genes function as effectors of invasion (Table 1) while down-regulated genes function as suppressors of invasion (Table 2). The paucity of cell cycle-associated genes, which supports the data presented here and previous findings that v-Fos does not alter the growth regulation of fibroblasts, is interesting (34, 64). There are a considerable number of genes differentially regulated in Tif-Fos cells that are thought to be involved in oncogenesis despite the cells remaining anchorage and serum dependent for proliferation; these include down-regulated tumor suppressors, membrane receptors, and genes involved in signal transduction, which are discussed in more detail below.
The EGFR is up-regulated by v-Fos in human fibroblasts.
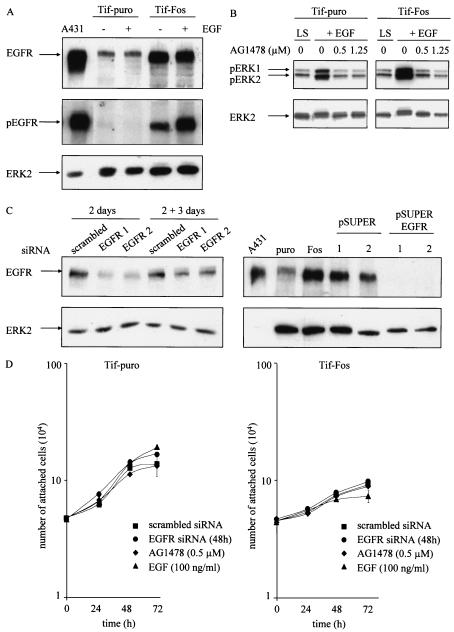
The microarray analysis identified a significant increase in expression of EGFR. EGFR was up-regulated approximately 10-fold at the RNA level in all of the Tif-Fos populations. There was no difference in EGFR expression between the hff, Tif, or Tif-puro cells, indicating that the increase in expression is a consequence of v-Fos expression. Increased EGFR expression was confirmed by Western analysis of Tif-puro and Tif-Fos cells (Fig. 7A). Blotting with an antibody specific for phosphorylated EGFR reveals that the EGFR is constitutively active in the presence of v-Fos and further enhanced by EGF (Fig. 7A).
FIG. 7.
Expression of EGF receptor and EGFR signaling in telomerase-immortalized fibroblasts expressing v-Fos. (A) Western blotting for expression of EGFR and activated EGFR (pEGFR) in Tif-puro and Tif-Fos cells (100 μg of protein) in the absence and presence of EGF (100 ng/ml for 10 min). A431 cells (25 μg of protein) were used as a positive control, and equal loading was determined using anti-ERK2. (B) Tif-puro and Tif-Fos cells incubated in low (0.2%) serum (LS) for 60 h were stimulated for 15 min with EGF (100 ng/ml), and phosphorylation of ERK was detected with the p44/p42 mitogen-activated protein kinase phosphospecific antibody (pERK1 and pERK2). EGFR-mediated signaling to ERK was inhibited by a 10-min pretreatment with the EGFR-specific inhibitor tyrphostin AG1478 (0.5 and 1.25 μM). Total ERK2 is shown as a loading control. (C) Suppression of EGFR expression by RNA interference. Tif-Fos cells were nucleofected with either scrambled siRNA or EGFR siRNA (100 nM). After 2 days, the cells were lysed and EGFR expression was determined by Western blotting. Parallel cultures of cells were trypsinized and incubated in an in vitro invasion assay for 3 days, and EGFR expression was again determined by Western blotting. Tif-Fos cells were also infected with retroviruses encoding pSUPER or pSUPER. EGFR and EGFR expression was determined by Western blotting in two independent polyclonal populations of each. A431 cells were used as a positive control, and ERK2 expression was determined as a loading control. (D) Tif-puro and Tif-Fos cells were nucleofected with siRNA. After 48 h, proliferation was assessed by counting the attached cells in the absence or presence of 100 nM EGFR siRNA, 0.5 μM AG1478, or 100 ng of EGF per ml. (E) Tif-puro and Tif-Fos cells were stained for actin stress fibers with phalloidin-FITC in the absence or presence of EGF (100 ng/ml) for 24 h with or without pretreatment (10 min) with AG1478 (1.25 μM). Fos expression was detected with the 9E10 anti-Myc monoclonal antibody and anti-mouse-TRITC. Bar, 100 μm. (F) Tif-Fos cells transiently expressing scrambled or EGFR siRNA (100 nM for 72h) or stably expressing pSUPER or pSUPER EGFR were stained with phalloidin-FITC in the absence or presence of EGF (100 ng/ml) for 24 h. Fos expression was detected with the 9E10 anti-Myc monoclonal antibody and anti-mouse-TRITC. Bar, 100 μm. (G) Invasion of Tif-Fos in the absence (black bars) or presence of AG1478 (0.5 and 1.25 μM, gray bars) with or without EGF (100 ng/ml) for 3 days. Invasion was quantitated as described in Materials and Methods and normalized with respect to invasion of Tif-Fos alone. The error bars represent the standard deviation of the mean change. (H) Invasion of Tif-Fos cells transiently expressing scrambled (black bars) or EGFR (gray bars) siRNA 2 days after nucleofection, in the absence or presence of EGF (100 ng/ml) for 3 days or stably expressing pSUPER (black bars) or pSUPER EGFR (gray bars). Invasion is normalized with respect to invasion of Tif-Fos alone, and the error bars represent the standard deviation of the mean change.
To determine EGFR tyrosine kinase activation, the response to EGF stimulation was assessed in quiescent Tif-puro and Tif-Fos cells. An increase in the amount of phosphorylated ERK was detected 15 min after treatment with EGF (100 ng/ml) by Western blotting with p44/p42 ERK phosphospecific antibody, indicating that signaling from the EGFR is functional (Fig. 7B). Pretreatment of the Tif-puro and Tif-Fos cells for 10 min with the EGFR-specific inhibitor tyrphostin AG1478 (0.5 and 1.25 μM) inhibited EGF-mediated phosphorylation of ERK, confirming that EGFR signaling was responsible for the stimulation of ERK (Fig. 7B).
To further investigate EGFR function in Tif-Fos cells, expression of the EGFR was knocked down by RNA interference by using either transient expression of EGFR siRNA or stable suppression of endogenous RNA with pSUPER EGFR. Figure 7C demonstrates that EGFR expression was successfully inhibited with siRNA against two different target sequences for EGFR after 2 days, although expression levels were starting to increase after a further 3 days. Expression of pSUPER EGFR completely suppressed EGFR expression in two individual polyclonal populations of Tif-Fos cells (Fig 7C).
To determine whether increased expression of EGFR in Tif-Fos cells resulted in enhanced proliferation, quiescent Tif-puro and Tif-Fos cells were treated with either 10 or 100 ng of EGF per ml. EGF is a potent mitogen for human fibroblasts (9), and after 24 and 48 h, cell counts indicated that Tif-puro and Tif-Fos cells respond mitogenically to EGF in a similar manner (data not shown). However, exponentially growing cells are not dependent on EGF signaling for proliferation, since their growth was not enhanced by treatment with EGF (100 ng/ml) or, conversely, not significantly inhibited by either AG1478 (0.5 μM) or transient expression of EGFR siRNA (Fig. 7D).
EGF treatment of established rodent fibroblast cell lines results in disruption of actin stress fibers followed by morphological transformation (70). Although hff-Fos and Tif-Fos cells have a reduction in actin stress fibers compared to hff-puro and Tif-puro cells, they still have detectable stress fibers (Fig. 1B). Exposure of Tif-Fos cells to EGF (100 ng/ml) for 24 h resulted in a further reduction in actin stress fibers and a more spindle-like cell morphology, while similar treatment of Tif-puro cells with EGF had no effect on the actin cytoskeleton or cell morphology (Fig. 7E). We demonstrate that inhibition of EGFR signaling with AG1478 (1.25 μM) can reverse the loss of stress fibers observed in Tif-Fos cells in the absence or presence of EGF (Fig. 7E). Similar results were obtained for Tif-Fos cells by using either transient EGFR siRNA or pSUPER EGFR (Fig. 7F). This indicates that the consequence of EGF signaling to the actin cytoskeleton in Tif-Fos cells is different from that observed in Tif-puro cells. Of course, other changes in gene expression may contribute to the increased responsiveness of the Tif-Fos cells to EGF, in addition to increased expression of EGFR.
The EGF-induced reduction in the number of actin stress fibers and changes in cell morphology might be expected to enhance the invasiveness of hff-Fos and Tif-Fos cells in response to EGF. Inclusion of EGF (100 ng/ml) in the upper chamber of the in vitro invasion assay resulted in a twofold increase in the number of cells that invade but did not result in a significant increase in the invasiveness of the hff-puro and Tif-puro cells (Fig. 3). The loss of p16INK4a expression and inactivation of p53 did not affect the cells responsiveness to EGF-stimulated invasion (data not shown).
To determine if EGFR signaling was necessary for invasion of Tif-Fos cells, AG1478 was included in the invasion assay at concentrations that inhibit EGF stimulation of ERK phosphorylation (Fig. 7B). AG1478 (0.5 and 1.25 μM) inhibited invasion of Tif-Fos cells in a dose-dependent manner and reduced invasion to a greater extent in the presence of 100 ng of EGF per ml (Fig. 7G). Similarly, transient suppression of EGFR expression using siRNA inhibited invasion in the absence and presence of EGF (Fig. 7H). Furthermore, stable suppression of EGFR expression using pSUPER EGFR completely abolished invasion in Tif-Fos cells (Fig. 7H). This demonstrates that EGFR signaling is required for invasion of Tif-Fos cells. However, overexpression of EGFR is not sufficient to render Tif-puro cells invasive, even in the presence of EGF (data not shown).
Taken together, these results suggest that increased expression of the EGFR in human fibroblasts expressing v-Fos is necessary to maintain the invasive phenotype and validates the EGFR as a component of the AP-1-regulated multigenic invasion program.
DISCUSSION
The initiating event in many tumor progression models results in recognizable benign hyperproliferative lesions, which, through successive steps, acquire an invasive potential that completes the progression from a benign tumor into a malignant one (32). The main finding reported here is that expression of the v-Fos oncogene in human fibroblasts specifically activates the invasive program of the cell. Expression of v-Fos in human fibroblasts (primary or telomerase immortalized) alters morphology, enhances motility, and renders the cells invasive without changing their serum or anchorage requirements for proliferation, inducing a premature senescence-like growth arrest, or, in the case of hff, delaying the onset of replicative senescence. Furthermore, invasion is independent of the p16INK4a and p53 tumor suppressor pathways, since v-Fos stimulates invasion in hff and Tif in which both pathways are intact. The sequential loss of p16INK4a expression and p53 function does not render Tif-puro cells invasive or enhance the invasiveness of Tif-Fos cells. The separation of invasion from proliferation by the v-Fos oncogene indicates that invasion could occur early during tumor progression but may not be recognized since premalignant invasive lesions would be difficult to identify. Early invasiveness could even contribute to proliferation through the loss of growth restraints imposed by the normal architecture of the tissue, the release of growth factors from the degraded ECM, and the repression of anoikis. In this manner, the invasive cells could help to create the environment in which deregulated proliferation and anchorage independence could be selected. This might help to explain why, clinically, many cancers develop without showing evidence of a benign hyperproliferative phase (95).
Gene expression profiles.
Since v-Fos specifically induces invasion following expression in normal human fibroblasts, the changes in gene expression detected in Tif-Fos must account for the invasive phenotype and define the multigenic invasion program. The comparison of gene expression profiles between Tif-Fos and Tif-puro cells revealed consistent significant changes in gene expression that distinguish Tif-Fos cells from the other cells: 130 genes up-regulated and 227 genes down-regulated. The genes that were identified as differentially expressed represent the many cellular functions associated with invasion (Tables 1 and 2). Previous analyses indicate that differentially expressed genes function as part of a program and that the complete program must be intact for the cells to invade (46, 53, 54, 94).
Invasion is dependent on the increased expression of extracellular proteases and a reduction in expression of their inhibitors (15, 40, 110, 111), many of which are AP-1 regulated (17, 114). The gene expression profile of the Tif-Fos cells identifies several extracellular proteases that are up-regulated and several protease inhibitors that are down-regulated. Inhibition of AP-1 activity, which reduces the expression of proteases and increases the expression of their inhibitors, also reduces the invasiveness of the cells (19, 82, 91, 92).
More changes in gene expression in addition to proteases are necessary for AP-1-mediated invasion. Of particular note is the collection of down-regulated genes that are known tumor (oncogene) suppressor genes. Potent inhibitors of fibroblast transformation, lumican (118), fibulin 1 (75, 86), and IGFBP5 (59, 85), are expressed in mouse embryo fibroblasts that are resistant to oncogene-mediated transformation (4). Calponin 1 (100) and DOK1/p62dok (93) are also known suppressors of oncogene-mediated transformation. DAB2 is of particular interest since it suppresses signaling from the growth factor signal transduction pathway to AP-1 (33, 104) and is down regulated in a variety of human tumors (11, 23). Other known tumor suppressor genes, those for thrombospondin 1 (63, 78), RECK (69, 81, 99), TIMP1 (77), IGSF4 (25), gremlin/CKTSF1B1 (10), sFRP1 (49, 98), IGFBP7/MAC25 (96), fibronectin 1 (52, 73, 101), STAT1 (38), and FAT (21), and some purported tumor suppressor genes, those for brush-1/WASF3 (87), TES (102), and WNT5A (39), are all down-regulated in Tif-Fos cells. The number of tumor suppressor genes that are down-regulated in Tif-Fos cells and their breadth of activities (ECM components, transcription regulators, signal transducers, and protease inhibitors) reflect the complexity of the invasion program and the many aspects of cell biology that are engaged during invasion. In support of the role of AP-1 in tumor cell invasion, rather than proliferation, most of the down-regulated tumor suppressors are not direct inhibitors of the cell cycle.
Although it remains to be determined if the down-regulation of each gene is necessary for the invasive phenotype, it demonstrates that multiple suppressors of the invasive and transformed phenotype are expressed in normal fibroblasts. To the extent that each gene is a member of the multigenic invasion program, it seems unlikely that an inactivating mutation in any one of these genes would render the cells invasive or fully transformed. This implies that the down-regulation of most of these genes is the consequence of epigenetic mechanisms such as DNA methylation or histone deacetylation. v-Fos-mediated morphological transformation and invasion are known to be dependent on the activity of DNA methyltransferases and histone deacetylases (3, 71).
Many of the up-regulated genes are also associated with multiple aspects of invasion, such as ECM components and their receptors, proteases, regulators of the actin cytoskeleton, and transcription factors. Again, there are few genes directly associated with the cell cycle. Surprisingly, many known growth factors are down-regulated in Tif-Fos, perhaps accounting for their lack of independence from growth factors. However, several receptors for growth factors, e.g., EGFR (113), met (13), EphA2 (35), and EphB6 (28), that can also affect motility or invasion were significantly up-regulated in the Tif-Fos cells.
The increase in EGFR expression in Tif-Fos cells was unexpected since it has not been demonstrated in diploid human fibroblasts, although it is well established that it is often overexpressed in a variety of human tumors (117). The AP-2 binding site in the promoter region of EGFR may be responsible for phorbol myristate acetate-induced up-regulation of the EGFR (44). The transcription factor AP-2 is notably up-regulated in Tif-Fos. AP-1 also regulates EGFR expression (45, 55, 121). In Tif-Fos, either or both of AP-2 and AP-1 could account for the up-regulation of EGFR expression.
Increased expression of the EGFR is often associated with invasiveness (113). It is known to signal to the actin cytoskeleton and to stimulate cell motility and invasion. In squamous cell carcinoma-derived cell lines that overexpress the EGFR, its activity is required for cellular invasion (61). Tif-Fos cells require EGFR activity for invasion, and inhibition of tyrosine kinase activity dramatically inhibits the invasiveness of the cells in the presence or absence of added ligand. However, EGFR activity in the presence of serum is not necessary for proliferation even though EGF is mitogenic for Tif-puro and Tif-Fos cells. This suggests that increased EGFR is signaling primarily to the invasive phenotype. Contrary to expectation, the known ligands for EGFR are not expressed in Tif-Fos cells. Thus, there is no evidence that an autocrine system for invasion, functioning through ligand binding to the EGFR, occurs in Tif-Fos cells (113). However, there are multiple ways of activating the EGFR through other signaling pathways such as lysophosphatidic acid (27), integrins (67), or prostaglandins (72).
The downstream pathways activated by the EGFR that are important for invasion remain to be elucidated. In the simplest sense, it could be serving as a guidance function and regulating cytoskeleton reorganization for motility (113), in a more complex view, it could be performing that function as well as stimulating changes in gene expression that further enhance invasiveness (53). With regard to a possible role in guidance, it has been demonstrated that in the presence of the EGFR, α6-integrin mediates laminin-mediated chemotaxis in fibroblasts (58, 74). In another system, EGFR activation stimulates the expression of α2-integrin, integrin-mediated adhesion, and motility (83). Both α2- and α6-integrin are up-regulated in Tif-Fos cells according to microarray analysis. The up-regulation of α2-integrin may also contribute to the invasiveness of the cells through the activation of its cytoplasmic domain by R-Ras2, which is also up-regulated in Tif-Fos cells (48). Phosphoinositide-3-kinase catalytic subunit p110delta is up-regulated in Tif-Fos cells and has been shown to regulate EGF-stimulated motility in breast cancer cells by controlling the directionality and speed of migration (84).
The changes in gene expression in human diploid fibroblasts following expression of the v-Fos oncogene must account for the increased invasiveness displayed by Tif-Fos cells. Further functional analysis will establish which differentially expressed genes contribute to the invasive phenotype and comprise the AP-1 regulated invasion program. The lack of changes in expression of genes that regulate cell cycle and the specific down-regulation of genes encoding polypeptide growth factors forms the basis of the activation of the invasive program in the absence of the proliferative program of the cells.
These results establish that activation of AP-1 through v-Fos specifically confers an invasive phenotype upon human fibroblasts that is independent of anchorage independence and cell cycle alterations normally associated with oncogenic transformation (31). The independence of invasion from these cell cycle tumor suppressor genes indicates that the invasion program can be activated in their presence, as would be expected for a normal cellular invasion program.
Acknowledgments
We thank Y. Hey, Cancer Research UK GeneChip Service, Paterson Institute for Cancer Research, Manchester, United Kingdom, and C. Young, Molecular Biology Support Unit, University of Glasgow, Glasgow, United Kingdom for hybridization of microarrays. We also thank S. Kotwaliwale, Genetic Diagnostic Centre, Mumbai, India, for karyotyping.
This work was funded by Cancer Research UK.
REFERENCES
- 1.Alt, M., and R. Grassmann. 1993. Resistance of human fibroblasts to c-fos mediated transformation. Oncogene 8:1421-1427. [PubMed] [Google Scholar]
- 2.Angel, P., M. Imagawa, R. Chiu, B. Stein, R. J. Imbra, H. J. Rahmsdorf, C. Jonat, P. Herrlich, and M. Karin. 1987. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell 49:729-739. [DOI] [PubMed] [Google Scholar]
- 3.Bakin, A. V., and T. Curran. 1999. Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science 283:387-390. [DOI] [PubMed] [Google Scholar]
- 4.Bardeesy, N., M. Sinha, A. F. Hezel, S. Signoretti, N. A. Hathaway, N. E. Sharpless, M. Loda, D. R. Carrasco, and R. A. DePinho. 2002. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature 419:162-167. [DOI] [PubMed] [Google Scholar]
- 5.Bernards, R., and R. A. Weinberg. 2002. A progression puzzle. Nature 418:823. [DOI] [PubMed] [Google Scholar]
- 6.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 7.Brookes, S., J. Rowe, M. Ruas, S. Llanos, P. A. Clark, M. Lomax, M. C. James, R. Vatcheva, S. Bates, K. H. Vousden, D. Parry, N. Gruis, N. Smit, W. Bergman, and G. Peters. 2002. INK4a-deficient human diploid fibroblasts are resistant to RAS-induced senescence. EMBO J. 21:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canman, C. E., A. C. Wolff, C. Y. Chen, A. J. Fornace, Jr., and M. B. Kastan. 1994. The p53-dependent G1 cell cycle checkpoint pathway and ataxia-telangiectasia. Cancer Res. 54:5054-5058. [PubMed] [Google Scholar]
- 9.Carpenter, G., and S. Cohen. 1979. Epidermal growth factor. Annu. Rev. Biochem. 48:193-216. [DOI] [PubMed] [Google Scholar]
- 10.Chen, B., M. Athanasiou, Q. Gu, and D. G. Blair. 2002. Drm/Gremlin transcriptionally activates p21(Cip1) via a novel mechanism and inhibits neoplastic transformation. Biochem. Biophys. Res. Commun. 295:1135-1141. [DOI] [PubMed] [Google Scholar]
- 11.Chen, H., S. Toyooka, A. F. Gazdar, and J. T. Hsieh. 2003. Epigenetic regulation of a novel tumor suppressor gene (hDAB2IP) in prostate cancer cell lines. J. Biol. Chem. 278:3121-3130. [DOI] [PubMed] [Google Scholar]
- 12.Chen, T. R. 1977. In situ detection of mycoplasma contamination in cell cultures by fluorescent Hoechst 33258 stain. Exp. Cell Res. 104:255-262. [DOI] [PubMed] [Google Scholar]
- 13.Comoglio, P. M., and L. Trusolino. 2002. Invasive growth: from development to metastasis. J. Clin. Investig. 109:857-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook, S. J., N. Aziz, and M. McMahon. 1999. The repertoire of Fos and Jun proteins expressed during the G1 phase of the cell cycle is determined by the duration of mitogen-activated protein kinase activation. Mol. Cell. Biol. 19:330-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coussens, L. M., B. Fingleton, and L. M. Matrisian. 2002. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science 295:2387-2392. [DOI] [PubMed] [Google Scholar]
- 16.Crook, T., J. A. Tidy, and K. H. Vousden. 1991. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell 67:547-556. [DOI] [PubMed] [Google Scholar]
- 17.Curran, S., and G. I. Murray. 2000. Matrix metalloproteinases: molecular aspects of their roles in tumour invasion and metastasis. Eur. J. Cancer 36:1621-1630. [DOI] [PubMed] [Google Scholar]
- 18.Curran, T., G. Peters, C. Van Beveren, N. M. Teich, and I. M. Verma. 1982. FBJ murine osteosarcoma virus: identification and molecular cloning of biologically active proviral DNA. J. Virol. 44:674-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong, Z., H. C. Crawford, V. Lavrovsky, D. Taub, R. Watts, L. M. Matrisian, and N. H. Colburn. 1997. A dominant negative mutant of jun blocking 12-O-tetradecanoylphorbol-13-acetate-induced invasion in mouse keratinocytes. Mol. Carcinog. 19:204-212. [DOI] [PubMed] [Google Scholar]
- 20.Downward, J. 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3:11-22. [DOI] [PubMed] [Google Scholar]
- 21.Dunne, J., A. M. Hanby, R. Poulsom, T. A. Jones, D. Sheer, W. G. Chin, S. M. Da, Q. Zhao, P. C. Beverley, and M. J. Owen. 1995. Molecular cloning and tissue expression of FAT, the human homologue of the Drosophila fat gene that is located on chromosome 4q34-q35 and encodes a putative adhesion molecule. Genomics 30:207-223. [DOI] [PubMed] [Google Scholar]
- 22.Elenbaas, B., L. Spirio, F. Koerner, M. D. Fleming, D. B. Zimonjic, J. L. Donaher, N. C. Popescu, W. C. Hahn, and R. A. Weinberg. 2001. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 15:50-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fazili, Z., W. Sun, S. Mittelstaedt, C. Cohen, and X. X. Xu. 1999. Disabled-2 inactivation is an early step in ovarian tumorigenicity. Oncogene 18:3104-3113. [DOI] [PubMed] [Google Scholar]
- 24.Funk, M., B. Poensgen, W. Graulich, V. Jerome, and R. Muller. 1997. A novel, transformation-relevant activation domain in Fos proteins. Mol. Cell. Biol. 17:537-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomyo, H., Y. Arai, A. Tanigami, Y. Murakami, M. Hattori, F. Hosoda, K. Arai, Y. Aikawa, H. Tsuda, S. Hirohashi, S. Asakawa, N. Shimizu, E. Soeda, Y. Sakaki, and M. Ohki. 1999. A 2-Mb sequence-ready contig map and a novel immunoglobulin superfamily gene IGSF4 in the LOH region of chromosome 11q23.2. Genomics 62:139-146. [DOI] [PubMed] [Google Scholar]
- 26.Greenberg, M. E., and E. B. Ziff. 1984. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature 311:433-438. [DOI] [PubMed] [Google Scholar]
- 27.Gschwind, A., N. Prenzel, and A. Ullrich. 2002. Lysophosphatidic acid-induced squamous cell carcinoma cell proliferation and motility involves epidermal growth factor receptor signal transactivation. Cancer Res. 62:6329-6336. [PubMed] [Google Scholar]
- 28.Gurniak, C. B., and L. J. Berg. 1996. A new member of the Eph family of receptors that lacks protein tyrosine kinase activity. Oncogene 13:777-786. [PubMed] [Google Scholar]
- 29.Hahn, W. C., C. M. Counter, A. S. Lundberg, R. L. Beijersbergen, M. W. Brooks, and R. A. Weinberg. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464-468. [DOI] [PubMed] [Google Scholar]
- 30.Hahn, W. C., S. K. Dessain, M. W. Brooks, J. E. King, B. Elenbaas, D. M. Sabatini, J. A. DeCaprio, and R. A. Weinberg. 2002. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22:2111-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahn, W. C., and R. A. Weinberg. 2002. Rules for making human tumor cells. N. Engl. J. Med. 347:1593-1603. [DOI] [PubMed] [Google Scholar]
- 32.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 33.He, J., E. R. Smith, and X. X. Xu. 2001. Disabled-2 exerts its tumor suppressor activity by uncoupling c-Fos expression and MAP kinase activation. J. Biol. Chem. 276:26814-26818. [DOI] [PubMed] [Google Scholar]
- 34.Hennigan, R. F., K. L. Hawker, and B. W. Ozanne. 1994. Fos-transformation activates genes associated with invasion. Oncogene 9:3591-3600. [PubMed] [Google Scholar]
- 35.Hess, A. R., E. A. Seftor, L. M. Gardner, K. Carles-Kinch, G. B. Schneider, R. E. Seftor, M. S. Kinch, and M. J. Hendrix. 2001. Molecular regulation of tumor cell vasculogenic mimicry by tyrosine phosphorylation: role of epithelial cell kinase (Eck/EphA2). Cancer Res. 61:3250-3255. [PubMed] [Google Scholar]
- 36.Hinds, P. W., C. A. Finlay, R. S. Quartin, S. J. Baker, E. R. Fearon, B. Vogelstein, and A. J. Levine. 1990. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: a comparison of the “hot spot” mutant phenotypes. Cell Growth Differ. 1:571-180. [PubMed] [Google Scholar]
- 37.Hollstein, M., M. Hergenhahn, Q. Yang, H. Bartsch, Z. Q. Wang, and P. Hainaut. 1999. New approaches to understanding p53 gene tumor mutation spectra. Mutat. Res. 431:199-209. [DOI] [PubMed] [Google Scholar]
- 38.Huang, S., C. D. Bucana, M. Van Arsdall, and I. J. Fidler. 2002. Stat1 negatively regulates angiogenesis, tumorigenicity and metastasis of tumor cells. Oncogene 21:2504-2512. [DOI] [PubMed] [Google Scholar]
- 39.Huguet, E. L., K. Smith, R. Bicknell, and A. L. Harris. 1995. Regulation of Wnt5a mRNA expression in human mammary epithelial cells by cell shape, confluence, and hepatocyte growth factor. J. Biol. Chem. 270:12851-12856. [DOI] [PubMed] [Google Scholar]
- 40.Janulis, M., S. Silberman, A. Ambegaokar, J. S. Gutkind, and R. M. Schultz. 1999. Role of mitogen-activated protein kinases and c-Jun/AP-1 trans-activating activity in the regulation of protease mRNAs and the malignant phenotype in NIH 3T3 fibroblasts. J. Biol. Chem. 274:801-813. [DOI] [PubMed] [Google Scholar]
- 41.Jenuwein, T., D. Muller, T. Curran, and R. Muller. 1985. Extended life span and tumorigenicity of nonestablished mouse connective tissue cells transformed by the fos oncogene of FBR-MuSV. Cell 41:629-637. [DOI] [PubMed] [Google Scholar]
- 42.Jiang, X. R., G. Jimenez, E. Chang, M. Frolkis, B. Kusler, M. Sage, M. Beeche, A. G. Bodnar, G. M. Wahl, T. D. Tlsty, and C. P. Chiu. 1999. Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat. Genet. 21:111-114. [DOI] [PubMed] [Google Scholar]
- 43.Jochum, W., E. Passegue, and E. F. Wagner. 2001. AP-1 in mouse development and tumorigenesis. Oncogene 20:2401-2412. [DOI] [PubMed] [Google Scholar]
- 44.Johnson, A. C. 1996. Activation of epidermal growth factor receptor gene transcription by phorbol 12-myristate 13-acetate is mediated by activator protein 2. J. Biol. Chem. 271:3033-3038. [PubMed] [Google Scholar]
- 45.Johnson, A. C., B. A. Murphy, C. M. Matelis, Y. Rubinstein, E. C. Piebenga, L. M. Akers, G. Neta, C. Vinson, and M. Birrer. 2000. Activator protein-1 mediates induced but not basal epidermal growth factor receptor gene expression. Mol. Med. 6:17-27. [PMC free article] [PubMed] [Google Scholar]
- 46.Johnston, I. M., H. J. Spence, J. N. Winnie, L. McGarry, J. K. Vass, L. Meagher, G. Stapleton, and B. W. Ozanne. 2000. Regulation of a multigenic invasion programme by the transcription factor, AP-1: re-expression of a down-regulated gene, TSC-36, inhibits invasion. Oncogene 19:5348-5358. [DOI] [PubMed] [Google Scholar]
- 47.Kastan, M. B., O. Onyekwere, D. Sidransky, B. Vogelstein, and R. W. Craig. 1991. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51:6304-6311. [PubMed] [Google Scholar]
- 48.Keely, P. J., E. V. Rusyn, A. D. Cox, and L. V. Parise. 1999. R-Ras signals through specific integrin alpha cytoplasmic domains to promote migration and invasion of breast epithelial cells. J. Cell Biol. 145:1077-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ko, J., K. S. Ryu, Y. H. Lee, D. S. Na, Y. S. Kim, Y. M. Oh, I. S. Kim, and J. W. Kim. 2002. Human secreted frizzled-related protein is down-regulated and induces apoptosis in human cervical cancer. Exp. Cell Res. 280:280-287. [DOI] [PubMed] [Google Scholar]
- 50.Kolettas, E., A. Evangelou, and E. S. Gonos. 2001. v-FBR-fos oncogene fails to rescue mammalian cells from growth arrest but affects the responses of human fibroblasts to heparin. Anticancer Res. 21:435-444. [PubMed] [Google Scholar]
- 51.Kustikova, O., D. Kramerov, M. Grigorian, V. Berezin, E. Bock, E. Lukanidin, and E. Tulchinsky. 1998. Fra-1 induces morphological transformation and increases in vitro invasiveness and motility of epithelioid adenocarcinoma cells. Mol. Cell. Biol. 18:7095-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ladeda, V., P. Frankel, L. A. Feig, D. A. Foster, E. Bal de Kier Joffe, and J. A. Aguirre-Ghiso. 2001. RalA mediates v-Src, v-Ras, and v-Raf regulation of CD44 and fibronectin expression in NIH3T3 fibroblasts. Biochem. Biophys. Res. Commun. 283:854-861. [DOI] [PubMed] [Google Scholar]
- 53.Lamb, R. F., R. F. Hennigan, K. Turnbull, K. D. Katsanakis, E. D. MacKenzie, G. D. Birnie, and B. W. Ozanne. 1997. AP-1-mediated invasion requires increased expression of the hyaluronan receptor CD44. Mol. Cell. Biol. 17:963-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lamb, R. F., B. W. Ozanne, C. Roy, L. McGarry, C. Stipp, P. Mangeat, and D. G. Jay. 1997. Essential functions of ezrin in maintenance of cell shape and lamellipodial extension in normal and transformed fibroblasts. Curr. Biol. 7:682-688. [DOI] [PubMed] [Google Scholar]
- 55.Li, G., C. Gustafson-Brown, S. K. Hanks, K. Nason, J. M. Arbeit, K. Pogliano, R. M. Wisdom, and R. S. Johnson. 2003. c-Jun is essential for organization of the epidermal leading edge. Dev. Cell 4:865-877. [DOI] [PubMed] [Google Scholar]
- 56.Li, J. J., Y. Cao, M. R. Young, and N. H. Colburn. 2000. Induced expression of dominant-negative c-jun downregulates NFkappaB and AP-1 target genes and suppresses tumor phenotype in human keratinocytes. Mol. Carcinog. 29:159-169. [DOI] [PubMed] [Google Scholar]
- 57.Lin, A. W., M. Barradas, J. C. Stone, L. van Aelst, M. Serrano, and S. W. Lowe. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 12:3008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin, M. L., and P. J. Bertics. 1995. Laminin responsiveness is associated with changes in fibroblast morphology, motility, and anchorage-independent growth: cell system for examining the interaction between laminin and EGF signaling pathways. J. Cell. Physiol. 164:593-604. [DOI] [PubMed] [Google Scholar]
- 59.Lin, S. C., C. P. Wang, Y. M. Chen, S. Y. Lu, M. J. Fann, C. J. Liu, S. Y. Kao, and K. W. Chang. 2002. Regulation of IGFBP-5 expression during tumourigenesis and differentiation of oral keratinocytes. J. Pathol. 198:317-325. [DOI] [PubMed] [Google Scholar]
- 60.Maki, Y., T. J. Bos, C. Davis, M. Starbuck, and P. K. Vogt. 1987. Avian sarcoma virus 17 carries the jun oncogene. Proc. Natl. Acad. Sci. USA 84:2848-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malliri, A., M. Symons, R. F. Hennigan, A. F. Hurlstone, R. F. Lamb, T. Wheeler, and B. W. Ozanne. 1998. The transcription factor AP-1 is required for EGF-induced activation of rho-like GTPases, cytoskeletal rearrangements, motility, and in vitro invasion of A431 cells. J. Cell Biol. 143:1087-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mechta, F., D. Lallemand, C. M. Pfarr, and M. Yaniv. 1997. Transformation by ras modifies AP1 composition and activity. Oncogene 14:837-847. [DOI] [PubMed] [Google Scholar]
- 63.Mettouchi, A., F. Cabon, N. Montreau, P. Vernier, G. Mercier, D. Blangy, H. Tricoire, P. Vigier, and B. Binetruy. 1994. SPARC and thrombospondin genes are repressed by the c-jun oncogene in rat embryo fibroblasts. EMBO J. 13:5668-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miao, G. G., and T. Curran. 1994. Cell transformation by c-fos requires an extended period of expression and is independent of the cell cycle. Mol. Cell. Biol. 14:4295-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller, A. D., I. M. Verma, and T. Curran. 1985. Deletion of the gag region from FBR murine osteosarcoma virus does not affect its enhanced transforming activity. J. Virol. 55:521-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morales, C. P., S. E. Holt, M. Ouellette, K. J. Kaur, Y. Yan, K. S. Wilson, M. A. White, W. E. Wright, and J. W. Shay. 1999. Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat. Genet. 21:115-118. [DOI] [PubMed] [Google Scholar]
- 67.Moro, L., L. Dolce, S. Cabodi, E. Bergatto, E. B. Erba, M. Smeriglio, E. Turco, S. F. Retta, M. G. Giuffrida, M. Venturino, J. Godovac-Zimmermann, A. Conti, E. Schaefer, L. Beguinot, C. Tacchetti, P. Gaggini, L. Silengo, G. Tarone, and P. Defilippi. 2002. Integrin-induced epidermal growth factor (EGF) receptor activation requires c-Src and p130Cas and leads to phosphorylation of specific EGF receptor tyrosines. J. Biol. Chem. 277:9405-9414. [DOI] [PubMed] [Google Scholar]
- 68.Munro, J., K. Steeghs, V. Morrison, H. Ireland, and E. K. Parkinson. 2001. Human fibroblast replicative senescence can occur in the absence of extensive cell division and short telomeres. Oncogene 20:3541-3552. [DOI] [PubMed] [Google Scholar]
- 69.Oh, J., R. Takahashi, S. Kondo, A. Mizoguchi, E. Adachi, R. M. Sasahara, S. Nishimura, Y. Imamura, H. Kitayama, D. B. Alexander, C. Ide, T. P. Horan, T. Arakawa, H. Yoshida, S. Nishikawa, Y. Itoh, M. Seiki, S. Itohara, C. Takahashi, and M. Noda. 2001. The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell 107:789-800. [DOI] [PubMed] [Google Scholar]
- 70.Ozanne, B., R. J. Fulton, and P. L. Kaplan. 1980. Kirsten murine sarcoma virus transformed cell lines and a spontaneously transformed rat cell-line produce transforming factors. J. Cell. Physiol. 105:163-180. [DOI] [PubMed] [Google Scholar]
- 71.Ozanne, B. W., L. McGarry, H. J. Spence, I. Johnston, J. Winnie, L. Meagher, and G. Stapleton. 2000. Transcriptional regulation of cell invasion: AP-1 regulation of a multigenic invasion programme. Eur. J. Cancer 36:1640-1648. [DOI] [PubMed] [Google Scholar]
- 72.Pai, R., B. Soreghan, I. L. Szabo, M. Pavelka, D. Baatar, and A. S. Tarnawski. 2002. Prostaglandin E2 transactivates EGF receptor: a novel mechanism for promoting colon cancer growth and gastrointestinal hypertrophy. Nat. Med. 8:289-293. [DOI] [PubMed] [Google Scholar]
- 73.Perez, S., E. Vial, H. van Dam, and M. Castellazzi. 2001. Transcription factor ATF3 partially transforms chick embryo fibroblasts by promoting growth factor-independent proliferation. Oncogene 20:1135-1141. [DOI] [PubMed] [Google Scholar]
- 74.Pouliot, N., L. M. Connolly, R. L. Moritz, R. J. Simpson, and A. W. Burgess. 2000. Colon cancer cells adhesion and spreading on autocrine laminin-10 is mediated by multiple integrin receptors and modulated by EGF receptor stimulation. Exp. Cell Res. 261:360-371. [DOI] [PubMed] [Google Scholar]
- 75.Qing, J., V. M. Maher, H. Tran, W. S. Argraves, R. W. Dunstan, and J. J. McCormick. 1997. Suppression of anchorage-independent growth and matrigel invasion and delayed tumor formation by elevated expression of fibulin-1D in human fibrosarcoma-derived cell lines. Oncogene 15:2159-2168. [DOI] [PubMed] [Google Scholar]
- 76.Reichmann, E., H. Schwarz, E. M. Deiner, I. Leitner, M. Eilers, J. Berger, M. Busslinger, and H. Beug. 1992. Activation of an inducible c-FosER fusion protein causes loss of epithelial polarity and triggers epithelial-fibroblastoid cell conversion. Cell 71:1103-1116. [DOI] [PubMed] [Google Scholar]
- 77.Rigg, A. S., and N. R. Lemoine. 2001. Adenoviral delivery of TIMP1 or TIMP2 can modify the invasive behavior of pancreatic cancer and can have a significant antitumor effect in vivo. Cancer Gene Ther. 8:869-878. [DOI] [PubMed] [Google Scholar]
- 78.Rodriguez-Manzaneque, J. C., T. F. Lane, M. A. Ortega, R. O. Hynes, J. Lawler, and M. L. Iruela-Arispe. 2001. Thrombospondin-1 suppresses spontaneous tumor growth and inhibits activation of matrix metalloproteinase-9 and mobilization of vascular endothelial growth factor. Proc. Natl. Acad. Sci. USA 98:12485-12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rutberg, S. E., T. L. Adams, A. Glick, M. T. Bonovich, C. Vinson, and S. H. Yuspa. 2000. Activator protein 1 transcription factors are fundamental to v-rasHa-induced changes in gene expression in neoplastic keratinocytes. Cancer Res. 60:6332-6338. [PubMed] [Google Scholar]
- 80.Saez, E., S. E. Rutberg, E. Mueller, H. Oppenheim, J. Smoluk, S. H. Yuspa, and B. M. Spiegelman. 1995. c-fos is required for malignant progression of skin tumors. Cell 82:721-732. [DOI] [PubMed] [Google Scholar]
- 81.Sasahara, R. M., C. Takahashi, M. C. Sogayar, and M. Noda. 1999. Oncogene-mediated downregulation of RECK, a novel transformation suppressor gene. Braz. J. Med. Biol. Res. 32:891-895. [DOI] [PubMed] [Google Scholar]
- 82.Sato, T., L. Koike, Y. Miyata, M. Hirata, Y. Mimaki, Y. Sashida, M. Yano, and A. Ito. 2002. Inhibition of activator protein-1 binding activity and phosphatidylinositol 3-kinase pathway by nobiletin, a polymethoxy flavonoid, results in augmentation of tissue inhibitor of metalloproteinases-1 production and suppression of production of matrix metalloproteinases-1 and -9 in human fibrosarcoma HT-1080 cells. Cancer Res. 62:1025-1029. [PubMed] [Google Scholar]
- 83.Sawhney, R. S., G. H. Zhou, L. E. Humphrey, P. Ghosh, J. I. Kreisberg, and M. G. Brattain. 2002. Differences in sensitivity of biological functions mediated by epidermal growth factor receptor activation with respect to endogenous and exogenous ligands. J. Biol. Chem. 277:75-86. [DOI] [PubMed] [Google Scholar]
- 84.Sawyer, C., J. Sturge, D. C. Bennett, M. J. O'Hare, W. E. Allen, J. Bain, G. E. Jones, and B. Vanhaesebroeck. 2003. Regulation of breast cancer cell chemotaxis by the phosphoinositide 3-kinase p110delta. Cancer Res. 63:1667-1675. [PubMed] [Google Scholar]
- 85.Schenker, T., and B. Trueb. 1998. Down-regulated proteins of mesenchymal tumor cells. Exp. Cell. Res. 239:161-168. [DOI] [PubMed] [Google Scholar]
- 86.Schiemann, W. P., G. C. Blobe, D. E. Kalume, A. Pandey, and H. F. Lodish. 2002. Context-specific effects of fibulin-5 (DANCE/EVEC) on cell proliferation, motility, and invasion. Fibulin-5 is induced by transforming growth factor-beta and affects protein kinase cascades. J. Biol. Chem. 277:27367-27377. [DOI] [PubMed] [Google Scholar]
- 87.Schott, D. R., J. N. Chang, G. Deng, W. Kurisu, W. L. Kuo, J. Gray, and H. S. Smith. 1994. A candidate tumor suppressor gene in human breast cancers. Cancer Res. 54:1393-1396. [PubMed] [Google Scholar]
- 88.Schuermann, M., G. Hennig, and R. Muller. 1993. Transcriptional activation and transformation by chimaeric Fos-estrogen receptor proteins: altered properties as a consequence of gene fusion. Oncogene 8:2781-2790. [PubMed] [Google Scholar]
- 89.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 90.Sharpless, N. E., N. Bardeesy, K. H. Lee, D. Carrasco, D. H. Castrillon, A. J. Aguirre, E. A. Wu, J. W. Horner, and R. A. DePinho. 2001. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413:86-91. [DOI] [PubMed] [Google Scholar]
- 91.Shiratsuchi, T., H. Ishibashi, and K. Shirasuna. 2002. Inhibition of epidermal growth factor-induced invasion by dexamethasone and AP-1 decoy in human squamous cell carcinoma cell lines. J. Cell. Physiol. 193:340-348. [DOI] [PubMed] [Google Scholar]
- 92.Simon, C., M. J. Hicks, A. J. Nemechek, R. Mehta, B. W. O'Malley, Jr., H. Goepfert, C. M. Flaitz, and D. Boyd. 1999. PD 098059, an inhibitor of ERK1 activation, attenuates the in vivo invasiveness of head and neck squamous cell carcinoma. Br. J. Cancer 80:1412-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Songyang, Z., Y. Yamanashi, D. Liu, and D. Baltimore. 2001. Domain-dependent function of the rasGAP-binding protein p62Dok in cell signaling. J. Biol. Chem. 276:2459-2465. [DOI] [PubMed] [Google Scholar]
- 94.Spence, H. J., I. Johnston, K. Ewart, S. J. Buchanan, U. Fitzgerald, and B. W. Ozanne. 2000. Krp1, a novel kelch related protein that is involved in pseudopod elongation in transformed cells. Oncogene 19:1266-1276. [DOI] [PubMed] [Google Scholar]
- 95.Sporn, M. B. 1997. The war on cancer: a review. Ann. N. Y. Acad. Sci. 833:137-146. [DOI] [PubMed] [Google Scholar]
- 96.Sprenger, C. C., M. E. Vail, K. Evans, J. Simurdak, and S. R. Plymate. 2002. Over-expression of insulin-like growth factor binding protein-related protein-1(IGFBP-rP1/mac25) in the M12 prostate cancer cell line alters tumor growth by a delay in G1 and cyclin A associated apoptosis. Oncogene 21:140-147. [DOI] [PubMed] [Google Scholar]
- 97.Stapleton, G., A. Malliri, and B. W. Ozanne. 2002. Downregulated AP-1 activity is associated with inhibition of protein kinase C-dependent CD44 and ezrin localisation and upregulation of PKC theta in A431 cells. J. Cell Sci. 115:2713-2724. [DOI] [PubMed] [Google Scholar]
- 98.Suzuki, H., E. Gabrielson, W. Chen, R. Anbazhagan, M. van Engeland, M. P. Weijenberg, J. G. Herman, and S. B. Baylin. 2002. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet. 31:141-149. [DOI] [PubMed] [Google Scholar]
- 99.Takahashi, C., Z. Sheng, T. P. Horan, H. Kitayama, M. Maki, K. Hitomi, Y. Kitaura, S. Takai, R. M. Sasahara, A. Horimoto, Y. Ikawa, B. J. Ratzkin, T. Arakawa, and M. Noda. 1998. Regulation of matrix metalloproteinase-9 and inhibition of tumor invasion by the membrane-anchored glycoprotein RECK. Proc. Natl. Acad. Sci. USA 95:13221-13226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takeoka, M., T. Ehara, J. Sagara, S. Hashimoto, and S. Taniguchi. 2002. Calponin h1 induced a flattened morphology and suppressed the growth of human fibrosarcoma HT1080 cells. Eur. J. Cancer 38:436-442. [DOI] [PubMed] [Google Scholar]
- 101.Taylor, G. A., M. Jeffers, C. P. Webb, H. M. Koo, M. Anver, K. Sekiguchi, and G. F. Vande Woude. 1998. Decreased fibronectin expression in Met/HGF-mediated tumorigenesis. Oncogene 17:1179-1183. [DOI] [PubMed] [Google Scholar]
- 102.Tobias, E. S., A. F. Hurlstone, E. MacKenzie, R. McFarlane, and D. M. Black. 2001. The TES gene at 7q31.1 is methylated in tumours and encodes a novel growth-suppressing LIM domain protein. Oncogene 20:2844-2853. [DOI] [PubMed] [Google Scholar]
- 103.Treinies, I., H. F. Paterson, S. Hooper, R. Wilson, and C. J. Marshall. 1999. Activated MEK stimulates expression of AP-1 components independently of phosphatidylinositol 3-kinase (PI3-kinase) but requires a PI3-kinase signal to stimulate DNA synthesis. Mol. Cell. Biol. 19:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tseng, C. P., B. D. Ely, R. C. Pong, Z. Wang, J. Zhou, and J. T. Hsieh. 1999. The role of DOC-2/DAB2 protein phosphorylation in the inhibition of AP-1 activity. An underlying mechanism of its tumor-suppressive function in prostate cancer. J. Biol. Chem. 274:31981-31986. [DOI] [PubMed] [Google Scholar]
- 105.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Van Beveren, C., F. van Straaten, T. Curran, R. Muller, and I. M. Verma. 1983. Analysis of FBJ-MuSV provirus and c-fos (mouse) gene reveals that viral and cellular fos gene products have different carboxy termini. Cell 32:1241-1255. [DOI] [PubMed] [Google Scholar]
- 107.Vaziri, H., and S. Benchimol. 1996. From telomere loss to p53 induction and activation of a DNA-damage pathway at senescence: the telomere loss/DNA damage model of cell aging. Exp. Gerontol. 31:295-301. [DOI] [PubMed] [Google Scholar]
- 108.Vaziri, H., J. A. Squire, T. K. Pandita, G. Bradley, R. M. Kuba, H. Zhang, S. Gulyas, R. P. Hill, G. P. Nolan, and S. Benchimol. 1999. Analysis of genomic integrity and p53-dependent G1 checkpoint in telomerase-induced extended-life-span human fibroblasts. Mol. Cell. Biol. 19:2373-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vial, E., E. Sahai, and C. J. Marshall. 2003. ERK-MAPK signaling coordinately regulates activity of Rac1 and RhoA for tumor cell motility. Cancer Cell 4:67-79. [DOI] [PubMed] [Google Scholar]
- 110.Wang, I. K., S. Y. Lin-Shiau, and J. K. Lin. 2000. Suppression of invasion and MMP-9 expression in NIH 3T3 and v-H-Ras 3T3 fibroblasts by lovastatin through inhibition of ras isoprenylation. Oncology 59:245-254. [DOI] [PubMed] [Google Scholar]
- 111.Wang, Y. 2001. The role and regulation of urokinase-type plasminogen activator receptor gene expression in cancer invasion and metastasis. Med. Res. Rev. 21:146-170. [DOI] [PubMed] [Google Scholar]
- 112.Wei, S., and J. M. Sedivy. 1999. Expression of catalytically active telomerase does not prevent premature senescence caused by overexpression of oncogenic Ha-Ras in normal human fibroblasts. Cancer Res. 59:1539-1543. [PubMed] [Google Scholar]
- 113.Wells, A., J. Kassis, J. Solava, T. Turner, and D. A. Lauffenburger. 2002. Growth factor-induced cell motility in tumor invasion. Acta Oncol. 41:124-130. [DOI] [PubMed] [Google Scholar]
- 114.Westermarck, J., and V. M. Kahari. 1999. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 13:781-792. [PubMed] [Google Scholar]
- 115.Wisdon, R., and I. M. Verma. 1993. Transformation by Fos proteins requires a C-terminal transactivation domain. Mol. Cell. Biol. 13:7429-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Woodhouse, E. C., R. F. Chuaqui, and L. A. Liotta. 1997. General mechanisms of metastasis. Cancer 80:1529-1537. [DOI] [PubMed] [Google Scholar]
- 117.Yarden, Y. 2001. The EGFR family and its ligands in human cancer; signalling mechanisms and therapeutic opportunities. Eur. J. Cancer 37 (Suppl. 4):S3-S8. [DOI] [PubMed] [Google Scholar]
- 118.Yoshioka, N., H. Inoue, K. Nakanishi, K. Oka, M. Yutsudo, A. Yamashita, A. Hakura, and H. Nojima. 2000. Isolation of transformation suppressor genes by cDNA subtraction: lumican suppresses transformation induced by v-src and v-K-ras. J. Virol. 74:1008-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Young, M. R., J. J. Li, M. Rincon, R. A. Flavell, B. K. Sathyanarayana, R. Hunziker, and N. Colburn. 1999. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc. Natl. Acad. Sci. USA 96:9827-9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zajchowski, D. A., M. F. Bartholdi, Y. Gong, L. Webster, H. L. Liu, A. Munishkin, C. Beauheim, S. Harvey, S. P. Ethier, and P. H. Johnson. 2001. Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells. Cancer Res. 61:5168-5178. [PubMed] [Google Scholar]
- 121.Zenz, R., H. Scheuch, P. Martin, C. Frank, R. Eferl, L. Kenner, M. Sibilia, and E. F. Wagner. 2003. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev. Cell 4:879-889. [DOI] [PubMed] [Google Scholar]
- 122.Zhu, J., D. Woods, M. McMahon, and J. M. Bishop. 1998. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 12:2997-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]