Abstract
Precise classification of tumors is critically important for cancer diagnosis and treatment. It is also a scientifically challenging task. Recently, efforts have been made to use gene expression profiles to improve the precision of classification, with limited success. Using a published data set for purposes of comparison, we introduce a methodology based on classification trees and demonstrate that it is significantly more accurate for discriminating among distinct colon cancer tissues than other statistical approaches used heretofore. In addition, competing classification trees are displayed, which suggest that different genes may coregulate colon cancers.
Targeting specific therapies to pathogenetically distinct tumor types is important for cancer treatment, because it maximizes efficacy and minimizes toxicity (1). Thus, precisely classifying tumors is of critical importance to cancer diagnosis and treatment. Diagnostic pathology has traditionally relied on macro- and microscopic histology and tumor morphology as the basis for tumor classification. Current classification frameworks, however, are unable to discriminate among tumors with similar histopathologic features, which vary in clinical course and in response to treatment (2). Recently, there is increasing interest in changing the basis of tumor classification from morphologic to molecular. In the past decade, microarray technologies have been developed that can simultaneously assess the level of expression of thousands of genes (3–11). Several studies have used microarrays to analyze gene expression in colon, breast, and other tumors and have demonstrated the potential power of expression profiling for classifying tumors (12–14). Gene expression profiles may offer more information than classic morphology and provide an alternative to morphology-based tumor classification systems.
Increasingly detailed information and sensitive data analytic techniques are key ingredients for successful development of tumor classification systems based on gene expression profiles. Most existing statistical and computational methods for gene expression data analysis focus on differential gene expression or cluster analysis (15). The goal of clustering is to group together objects (genes or tissue samples) with similar properties. The hierarchical (16) and k-mean clustering algorithms (17), as well as self-organizing maps (18), have been used for clustering expression profiles (19). Recently, a coupled two-way clustering algorithm was proposed to identify subsets of genes and tissue samples (20). Although clustering techniques will continue to be popular methods for gene expression data analysis, this methodology has the disadvantage that it represents an instance of unsupervised learning. In addition, it is difficult to incorporate prior knowledge about gene expression patterns. Thus, cluster analysis may not be a good statistical framework for diagnosis of disease.
Alternatively, in classification analyses, we are given a training set of observations that contain vectors of gene expressions as well as the labeled (normal or tumor) tissues. These observations are used to induce a classification model. This model can then be applied to predict the class label (normal or tumor) for a set of previously unseen instances (new tissue samples).
To predict tumor type, Golub et al. (1) used supervised learning and derived discriminant decision rules on the basis of magnitude and threshold of prediction strength. However, they did not provide the procedure for selecting a threshold of prediction strength, an essential ingredient for classification. Heuristic rules for selection of the threshold of prediction strength can be used, but with a certain unavoidable level of subjectivity. Brown et al. (21) applied the method of support vector machine to classify genes on the basis of expression data from DNA microarray hybridization experiments and illustrated the method for predicting functional roles of 2,467 uncharacterized genes from yeast Saccharomyces cerevisiae using the expression data. The support vector machine method is based on supervised learning. It can take advantage of prior knowledge by beginning with a set of genes that have a common function, and what is learned from the known genes will be used to discriminate new genes. Although it may not be difficult to assemble a set of training examples from the extant literature and existing databases, the uncertainty that results from the assembled choices is not well-defined. Classifying unknown genes through gene expressions is different from (although related to) classifying tissues through gene expressions. Moler et al. (22) proposed using a naive Bayesian model and support vector machine for tumor classification, both of which achieved comparable classification accuracy. Xiong et al. (23) conducted Fisher's linear discriminant analysis on the data analyzed here for tumor classification. The clustering and classification methods used in the existing literature (1, 12, 21–23) do not have a user-friendly gene selection mechanism and are generally time-consuming when there is a large number of genes to begin with. In particular, the support vector machine makes use of the quadratic programming algorithm and demands even more computational time than other statistical classification methods.
We introduce the technique of recursive partitioning (24, 25) for classifying tissues on the basis of gene expression data. This technique has some major advantages over the more traditional methods used by others (1, 12, 21–23). It is very efficient for dealing with a large number of genes. It can classify more than two types of tissues simultaneously, and it automatically selects genes whose expression can distinguish different tissue classes. In addition to this convenience and flexibility, we demonstrate that the classification rules resulting from recursive partitioning can be remarkably precise in comparison to those derived from other methods (1, 12, 21–23).
Materials and Methods
Recursive Partitioning.
Suppose we have data from n units of observations. Each unit contains a vector of feature measurements or covariates (gene expression profiles from a tissue) and a class label (normal or tumor). Recursive partitioning is a technique that builds a classification rule to predict the class membership on the basis of feature information. Unlike Fisher's linear discriminant analysis, which uses linear combinations of the covariates, the recursive partitioning technique extracts homogeneous strata from the data and constructs tree-based classification rules (see Fig. 1, for example). We focus on application of the technique for tissue classification type using gene expression data and refer to Zhang and Singer (24) for a thorough technical description of the method. In essence, the classification tree is constructed through a recursive partitioning process that divides the study sample into smaller and smaller samples (every subsample is called a node) according to whether a particular selected predictor is above a chosen cutoff value. The choices of the selected predictor and its corresponding cutoff value are designed to purify the distribution of the response; namely, separating normal tissues from cancer tissues in the present context. The sample (node) purity is measured by Plog(P) + (1 − P)log(1 − P), where P is the probability of a tissue being normal within the node. This entropy function reaches its maximum when P = 0 or 1 (all tissues are of the same type within the node) and minimum when P = 0.5 (the two types of tissues are equally likely). In general, we have to be careful not to overgrow the tree. A pruning procedure can be used to cut off redundant nodes (24). However, this is less of an issue in the present application, because a relatively small tree can achieve a high precision. Thus, we will not elaborate on the pruning procedure here.
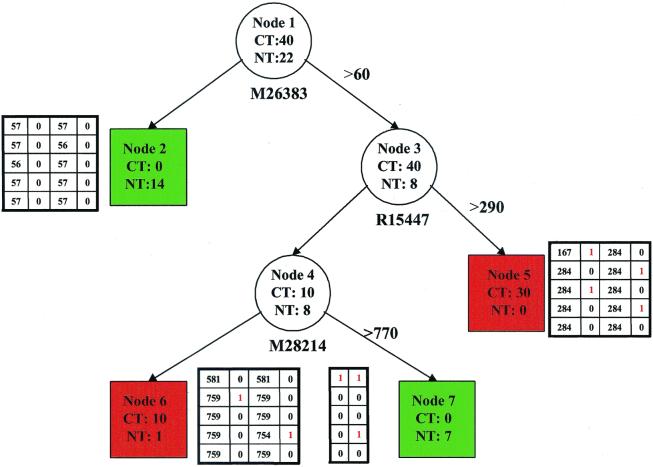
Figure 1.
Classification trees for tissue types by using expression data from three genes (M26383, R15447, M28214). Circles represent internal nodes that are subsequently divided into daughter nodes. The boxes are terminal nodes that do not have further partition and determine the tissue class membership; the red ones contain a total of 40 cancer tissues and 1 normal tissue, and the green ones contain 21 normal tissues. Beneath each internal node is the gene whose expression level is used to split the node, and the cutoff is displayed on the arrow next to the right. The four companion tables provide the information of the predictive precision of the tree based on a cross-validation scheme; see text for details. CT, number of cancer tissues; NT, number of normal tissues.
Gene Expression Data of Tumor and Normal Colon Tissues.
To demonstrate and explain the use of recursive partitioning, we analyze a data set from the expression profiles of 2,000 genes using an Affymetrix oligonucleotide array in 22 normal and 40 colon cancer tissues, which can be retrieved from the website www.sph.uth.tmc.edu/hgc.
Results
Fig. 1 is a classification tree that divides the 62 tissues into 4 groups labeled nodes 2, 5, 6, and 7. Two of them (nodes 2 and 7), shaded in green, contain 21 normal tissues and no cancer tissue. In contrast, the other two nodes (nodes 5 and 6), shaded in red, contain 40 cancer tissues and 1 normal tissue. If we predict the tissue type by the shaded color (green for normal and red for cancer), we misclassify 1 normal tissue out of the total 62 tissues (error rate, 1.6%). This is far more precise than the classification rules using other methods (23).
Now, we explain how to read Fig. 1. First, node 1 is split into nodes 2 and 3 in this figure, on the basis of the expression level of gene M26383. The 48 tissues (40 cancer and 8 normal tissues) that have the M26383 level beyond 60 are moved to node 3, and the remaining 14 tissues (all are normal tissues in this case) to node 2. The choices of the M26383 level and its corresponding threshold of 60 are automatically determined by the recursive partitioning algorithm. Briefly, the algorithm examines all of the 2,000 gene expression levels and all possible thresholds for each of the expression levels and selects the combination of gene expression level and threshold that results in the “best” separation of cancer and normal tissues on the basis of the node purity (or impurity) function introduced above. After node 1 is divided into nodes 2 and 3, node 3 is split into nodes 4 and 5 by the same algorithm, while restricting to the 40 tissues in node 3 only. Analogously, node 4 is further partitioned into nodes 6 and 7.
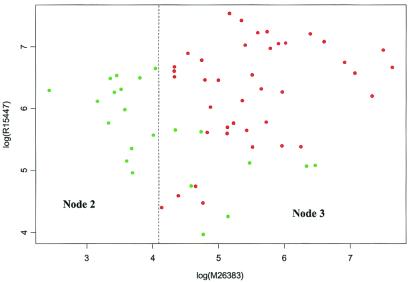
Figs. 2 and 3 are presented to enhance Fig. 1. In Fig. 2, the gene expression from M26383 is plotted against the gene expression from R15447. The 40 points from cancer tissues are labeled in red and the 22 points from normal tissues in green. On the left-hand side of the vertical dotted line are the 14 normal tissues contained in node 2. On the right-hand side are the 48 remaining tissues contained in node 3. Those 48 tissues are plotted again in Fig. 3, although gene expression data from R15447 and M28214 are used this time. The upper left corner in Fig. 3 corresponds to node 7 in Fig. 1. The remaining points are from nodes 5 and 6. The lone green point in the lower left corner is the misclassified tissue in node 6. Furthermore, Fig. 4 is a three-dimensional presentation of Fig. 1 using the gene expressions from M26383, R15447, and M28214 as the three coordinates.
Figure 2.
A scatter plot of expression data from M26383 and R15447. The dots are colored in green and red for normal and cancer tissues, respectively. The dotted line marks the cutoff value for node 1 in Fig. 1, and the two regions are labeled with their corresponding nodes in the same figure.
Figure 3.
A scatter plot of expression data from R15447 and M28214 for a subset of tissues (node 3 in Fig. 1). The dots are colored green and red for normal and cancer tissues, respectively. The dotted lines mark the cutoff values for nodes 3 and 4 in Fig. 1, and the three regions are labeled with their corresponding nodes in the same figure.
Figure 4.
Three-dimensional illustration of gene expressions from M26383, R15447, and M28214, along with tissue types. The 40 points from cancer tissues are labeled in red and the 22 points from normal tissues in green. Because cancer tissues end up in two terminal nodes in Fig. 1 and so are normal tissues, two levels of intensities for each of the red and green colors are highlighted to indicate different terminal node assignments of the same type of tissues.
We have reported the quality of the tree classification on the basis of the number of misclassified tissues. It is important to recall that the tree structure in Fig. 1 was selected to minimize the number of misclassifications. Without adjustments, this selection procedure tends to result in overly optimistic assessments of the tree quality. A commonly used statistical approach is cross-validation (24). Because we have a total of only 62 tissues, the obvious procedure described by Breiman et al. (25) and Zhang and Singer (24) would use very few learning and test samples and produce more uncertainty. Instead, a localized procedure (26) is adopted here to balance the needs of validating the results and retaining as many observations as possible.
Specifically, we first fix the tree frame in Fig. 1. The same genes will be applied to the same nodes, but the cutoff values for the selected gene profiles can vary. Let us begin with node 1. The 40 cancer tissues were divided randomly into 5 subsamples of 8, and the 22 normal tissues into 5 subsamples of 4, 4, 4, 5, and 5. Four subsamples each from the cancer and normal tissues were used to choose the cutoff values for the three splits. The remaining subsamples were used to count the misclassified tissues as a result of new cutoff values. This 5-fold cross-validation procedure was repeated a second time. The result for node 2 is presented in the 5 × 4 table to the left of the node in Fig. 1. The first column in that table is the cutoff value chosen during each of the five validations, and the second column reports the number of classification errors within node 2. The last two columns are analogous to the first two and report the results for the repeated 5-fold cross-validation.
This cross-validation was then applied to the split of node 3. The table to the right of node 5 displays the result. Likewise, we did the same for the split of node 4. The table to the right of node 6 summarizes the result for node 6, and the table to the left of node 7, for node 7, except that the latter table contains two columns, because the cutoff values are the same as those in the former table and hence are not repeated. In summary, 4 of 62 tissues were misclassified during the first 5-fold cross-validation, and 5 were misclassified the second time. These 6–8% error rates are unbiased estimates. The predictive precision of over 90% is still much better than that obtained by existing analyses.
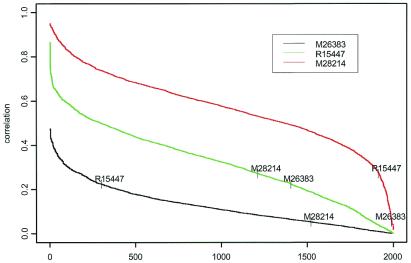
Fig. 1 displays a simple tree constructed from 3 genes that correctly classifies 61 of 62 tissues. It is common that functional expressions from various genes are correlated. Thus, it is interesting to examine the correlation patterns of the expression data between the three selected genes in Fig. 1 with the expression profiles in the remaining genes. Fig. 5 reveals that the correlations among the three selected genes themselves are not high. However, many gene expressions are highly correlated (Pearson's correlation coefficient greater than 0.7) with the expressions from M28214. Fewer gene expressions are highly correlated with the expressions from R15447 and none with expression from M26383. Thus, not only are the expression data from M26383 able to distinguish the tissue types, but they are also relatively unique and may not be easily replaceable, as they are not highly correlated with the other genes. As we move down the tree in Fig. 1, the gene expressions used for classification are more and more replaceable, as highly correlated genes are likely to have a similar performance in classification precision.
Figure 5.
Correlation curves between the three selected gene expressions in Fig. 1 and the remaining expression data. Genes are sorted according to the absolute correlation levels with one of the three selected genes and, obviously, the orders are different among the three selected genes.
The correlation among many gene profiles makes it likely that there exist other competing tree structures that could have similar predictive precision. Using the rtree (http://peace.med.yale.edu) program, we identified another tree, as displayed in Fig. 6, that is based on a different set of three genes. This can be done by selecting competitive node splits rather than the ones that have the best numerical values of node purity as defined through the entropy function. Table 1 shows that there are modest correlations among the new set of three genes and the previous three genes. Thus, different genes may coregulate colon cancer even though their gene expression profiles do not resemble each other very much. The classification prior to the cross-validation is perfect. The tree correctly classifies the 62 tissues. However, if we use the same cross-validation procedure as described above, the error rate is estimated to be between 8 and 11%, slightly higher than the rate for Fig. 1. Similar to Fig. 4, Fig. 7 is used to illustrate Fig. 6. Both of the trees in Figs. 1 and 5 specify high-precision classification rules.
Figure 6.
Classification trees for tissue types by using expression data from three genes (R87126, T62947, X15183). Circles represent internal nodes that are subsequently divided into daughter nodes. The boxes are terminal nodes that do not have further partition and determine the tissue class membership; the red ones contain a total of 40 cancer tissues, and the green ones contain 22 normal tissues. Beneath each internal node is the gene whose expression level is used to split the node, and the cutoff is displayed on the arrow next to the right. The four companion tables provide the information of the predictive precision of the tree based on a cross-validation scheme; see text for details. CT, number of cancer tissues; NT, number of normal tissues.
Table 1.
| M26383 | R15447 | M28214 | R87126 | T62947 | X15183 | |
|---|---|---|---|---|---|---|
| M26383 | 1 | 0.224 | −0.053 | −0.315 | 0.300 | 0.333 |
| (0.08) | (0.68) | (0.01) | (0.02) | (0.01) | ||
| R15447 | 1 | 0.271 | −0.143 | 0.605 | 0.489 | |
| (0.03) | (0.27) | (<0.001) | (<0.001) | |||
| M28214 | 1 | 0.318 | 0.363 | 0.390 | ||
| (0.01) | (0.004) | (<0.001) | ||||
| R87126 | 1 | −0.144 | −0.002 | |||
| (0.26) | (0.99) | |||||
| T62947 | 1 | 0.412 | ||||
| (<0.001) |
Pearson's correlations and their P values are displayed.
Figure 7.
Three-dimensional illustration of the gene expressions from X15183, R87126, and T62947, along with tissue types.
Discussion
The simultaneous monitoring of the expression of thousands of genes holds great promise for a better understanding of cancer biology and for development of accurate tumor classification schemes. However, the very large amount of gene expression information provided by contemporary microarray technology leads to difficulties for both basic research and clinical applications. Gene expression analyses for tumor classification requires cost-effective and streamlined methodology. In particular, if a handful of genes provide the basis for an accurate tumor classification scheme, the cost and complexity of monitoring the expression of thousands of genes will not be necessary in a clinical setting. In this report, we have demonstrated the use of recursive partitioning tree-based rules for tumor classification.
The selection of an optimal subset of genes poses two related problems: determining the number of genes to be selected and determining which genes belong to the set. Recursive partitioning is able to incorporate feature (gene) selection as a part of its learning algorithm and thus to simultaneously address these two issues. Using recursive partitioning, we have analyzed available expression data from 2,000 genes in 22 normal and 40 colon cancer samples and found that using three genes, IL-8 (M26383), CANX (R15447), and RAB3B (M28214), we can achieve 98% classification accuracy. These three genes are related to tumors. It was reported that IL-8 is correlated with the stage of colon cancer (27), the migration of human colonic epithelial cell lines (28), and metastasis of bladder cancer (29). The expression of the molecular chaperone CANX was found to be decreased in HT-29 human colon adenocarcinoma cells (30) and to be involved in apoptosis in human prostate epithelial tumor cells (31). RAB3B is a member of the RAS oncogene family. It is associated with significant increase of the mRNA expression in a human leukemia cell line (32).
This result is appealing and may have profound implications for clinical applications. It bodes well for the following scenario. Initially, basic research and clinical trials will monitor the expression of thousands of genes by using microarrays to identify a handful of genes providing optimal tumor classification information. Clinical applications will then require monitoring of only this small subset of genes, thus avoiding the cost and complexity of large-scale gene expression arrays. Of course, the number of selected genes and the optimal set of genes will likely differ according to tumor type, and thus clinical laboratories will still need the capability of monitoring a variety of genes.
Some of the gene expression levels across tissue samples are correlated and may form clusters. As a result, it is likely that the information contained in a large number of genes can be captured by a smaller number without significant loss of information. This is a direct result of the fact that clusters of genes are similarly regulated and hence play a similar role in tumor classification. The precision of classification exhibited herein by recursive partitioning—in comparison, for example, with linear discriminant analysis (23)—is critically important for such clinical applications. Furthermore, our results imply that gene expression on the basis of tumor classification systems not only provides an informative supplement to morphology-based classification systems but also possibly represents an improved alternative to them.
Acknowledgments
This research was supported in part by National Institutes of Health Grants DA12468 and AA12044.
References
- 1.Golub T R, Slonim D K, Tamayo P, Huard C, Gaasenbeek M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 2.Stephenson J. J Am Med Assoc. 1999;282:927–928. [Google Scholar]
- 3.Tlsty T D, Margolin B H, Lum K. Proc Natl Acad Sci USA. 1989;86:9441–9445. doi: 10.1073/pnas.86.23.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Theillet C. Nat Med. 1998;4:767–768. doi: 10.1038/nm0798-767. [DOI] [PubMed] [Google Scholar]
- 5.Strausberg R L, Austin M J F. Physiol Genomics. 1999;1:25–32. doi: 10.1152/physiolgenomics.1999.1.1.25. [DOI] [PubMed] [Google Scholar]
- 6.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 7.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, et al. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 8.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Nat Biotechnol. 1997;15:1539–1567. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 9.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang G P, Ross D T, Kuang W W, Brown P O, Weigel R J. Nucleic Acids Res. 1999;27:1517–1523. doi: 10.1093/nar/27.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 12.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Raffeld M, et al. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- 13.Alon U, Barkai N, Notterman D A, Gish K, Ybarra S, Mack D, Levine A J. Proc Natl Acad Sci USA. 1999;96:6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perou C M, Jeffrey S S, van de Rijn M, Rees C A, Eisen M B, Ross D T, Pergamenschikov A, Williams C F, Zhu S X, Lee J C F, et al. Proc Natl Acad Sci USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Butte A J, Tamayo P, Slonim D, Golub T R, Kohane I S. Proc Natl Acad Sci USA. 2000;97:12182–12186. doi: 10.1073/pnas.220392197. . (First Published October 10, 2000; 10.1073/pnas.220392197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tavazoie S, Hughes J D, Campbell M J, Cho R J, Church G M. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 18.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander E S, Golub T R. Proc Natl Acad Sci USA. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brazma A, Vilo J. FEBS Lett. 2000;480:17–24. doi: 10.1016/s0014-5793(00)01772-5. [DOI] [PubMed] [Google Scholar]
- 20.Getz G, Levine E, Domany E. Proc Natl Acad Sci USA. 2000;97:12079–12084. doi: 10.1073/pnas.210134797. . (First Published October 17, 2000; 10.1073/pnas.210134797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown M P S, Grundy W N, Lin D, Cristianini N, Sugnet C W, Furey T S, Ares M, Jr, Haussler D. Proc Natl Acad Sci USA. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moler E J, Chow M L, Mian I S. Physiol Genomics. 2000;4:109–126. doi: 10.1152/physiolgenomics.2000.4.2.109. [DOI] [PubMed] [Google Scholar]
- 23.Xiong M M, Jin L, Li W, Boerwinkle E. BioTechniques. 2000;29:1264–1270. doi: 10.2144/00296bc02. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H P, Singer B. Recursive Partitioning in the Health Sciences. New York: Springer; 1999. [Google Scholar]
- 25.Breiman L, Friedman J, Stone C, Olshen R. Classification and Regression Trees. Monterey, CA: Wadsworth; 1984. [Google Scholar]
- 26.Zhang H P, Holford T, Bracken M. Stat Med. 1995;15:37–50. doi: 10.1002/(SICI)1097-0258(19960115)15:1<37::AID-SIM144>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 27.Fox S H, Whalen G F, Sanders M M, Burleson J A, Jennings K, Kurtzman S, Kreutzer D. J Surg Oncol. 1998;69:230–234. doi: 10.1002/(sici)1096-9098(199812)69:4<230::aid-jso7>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 28.Toshina K, Hirata I, Maemura K, Sasaki S, Murano M, Nitta M, Yamauchi H, Nishikawa T, Hamamoto N, Katsu K. Scand J Immunol. 2000;52:570–575. doi: 10.1046/j.1365-3083.2000.00815.x. [DOI] [PubMed] [Google Scholar]
- 29.Inoue K, Slaton J W, Karashima T, Shuin T, Sweeney P, Millikan R, Dinney C P. Clin Cancer Res. 2000;6:4866–4873. [PubMed] [Google Scholar]
- 30.Yeates L C, Powis G. Biochem Biophys Res Commun. 1997;238:66–70. doi: 10.1006/bbrc.1997.7238. [DOI] [PubMed] [Google Scholar]
- 31.Nagata K, Okano Y, Nozawa Y. Thromb Haemostasis. 1997;77:368–375. [PubMed] [Google Scholar]
- 32.Prasad S C, Soldatenkov V A, Kuettel M R, Thraves P J, Zou X, Dritschilo A. Electrophoresis. 1999;20:1065–1074. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<1065::AID-ELPS1065>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]