Abstract
Enamel is a hard nanocomposite bioceramic with significant resilience that protects the mammalian tooth from external physical and chemical damages. The remarkable mechanical properties of enamel are associated with its hierarchical structural organization and its thorough connection with underlying dentin. This dynamic mineralizing system offers scientists a wealth of information that allows the study of basic principals of organic matrix-mediated biomineralization and can potentially be utilized in the fields of material science and engineering for development and design of biomimetic materials. This chapter will provide a brief overview of enamel hierarchical structure and properties as well as the process and stages of amelogenesis. Particular emphasis is given to current knowledge of extracellular matrix protein and proteinases, and the structural chemistry of the matrix components and their putative functions. The chapter will conclude by discussing the potential of enamel for regrowth.
Keywords: Amelogenesis, Enamel, Amelogenin, Biomineralization, Tooth, Review
2. INTRODUCTION
The protein components in the extracellular space between ameloblasts and dentin control the initiation, habit, orientation and organization of enamel crystals. The chemical and biological events of organic matrix-mediated biomineralization in enamel are generally common to all mineralized tissues, although some physiological processes distinguish the formation of tooth enamel from that of bone and dentin (1–3). The formation of tooth enamel takes place before the tooth erupts, in a confined extracellular environment situated between the dentin and the overlying layer of ameloblast cells. A series of programmed physiological and chemical events are involved in making enamel, including gene expression, protein secretion, protein folding and assembly, mineral growth, and protein degradation. The extracellular matrix is continuously secreted during enamel formation to result in a “forming mineralized matrix” rather than the “pre-formed matrix” that has been defined for other mineralizing tissues. The matrix is processed in a stepwise and controlled manner during the initial stage when apatite crystals grow in length. The matrix is then is rapidly degraded during the maturation stage when crystals grow mainly in width and thickness, and is eventually removed from the extracellular space to allow completion of mineralization (4, 5). During maturation, massive protein degradation events occur simultaneously with the completion of mineralization. These extracellular events transform a matrix that contains approximately 30% mineral, by weight, with the remainder being organic material and water into a highly organized structure that is almost completely inorganic (>99%) (6, 7).
Enamel is considered a bioceramic, covering and protecting the tooth (8). This biomineralizing system offers a wealth of information that can potentially be utilized in the fields of material science and engineering for development and design of biomimetic materials with potentials for application in biomedicine, dentistry, and industry (9–12). In order to understand how enamel is developed and mineralized, researchers have used animal models such as rat, mouse, pig, and cow. Human and mouse models have been used to characterize enamel mechanical properties and function, as well as the genetics of enamel malformation. Information is rapidly growing in a number of areas related to enamel formation, including the ultrastructure and biomechanics of enamel (8, 13–16), ameloblast cell biology (17), the genetics of enamel malformation (18, 19), the evolutionary aspects of enamel formation (20) and extracellular matrix composition and function (19, 21, 22). Several book chapters and reviews describe in detail different aspects of enamel formation (3, 8, 23–26). The present chapter will provide a brief overview of enamel hierarchical structure and properties, after which a section will describe the process and stages of amelogenesis with a particular emphasis on current knowledge of the structural chemistry of the extracellular matrix components and their putative functions. The chapter will conclude by discussing the potential of enamel for regrowth.
3. ENAMEL BIOCERAMIC AND ITS HIERARCHICAL STRUCTURE
The packing order of mature enamel ranges from microscale level prisms or rods to nanoscale ribbon-like fluoridated carbonatoapatite crystals 50–70 nm in width, 20–25 nm in thickness and with lengths leading to aspect ratios > 500 (Figure. 1). Each enamel prism has about the same diameter as an ameloblast (an enamel-forming cell) and is believed to be the product of a single cell (27). The next level of order is the interwoven arrangement of the prisms (rods) with the interprismatic (inter-rod) enamel. The rods and inter-rod enamel are regarded as the fundamental organization units of mammalian enamel. The packing of the prisms is believed to be the result of an orchestrated receding of the ameloblast cell layer. In human enamel an inter-rod region (interprismatic zone) surrounds each rod, and the orientation of the crystals making the inter-rod region is different from those making up the rods. At the boundary between rod and inter-rod enamel is a narrow space containing organic material known as the “rod sheath” material. This organic material may contribute to the mechanical properties (particularly the fracture toughness) of enamel. At a higher level – measured in hundreds of micrometers – the interwoven structures of the prisms with the interprismatic enamel can be seen with the optical microscope as alternating layers called Hunter-Schreger bands (8). The final product is an approximately 2.5-millimeter thick bioceramic that covers the entire tooth, is translucent and varies in color from yellowish-white to grey-white.
Figure 1.
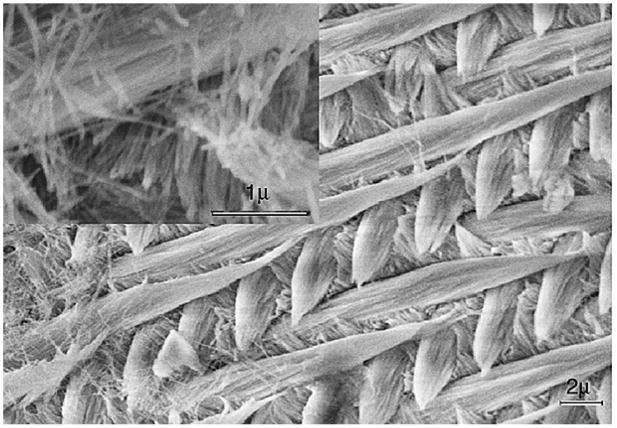
Scanning electron micrograph of etched surface of mouse incisor tooth enamel showing the interwoven arrangement of the prisms and inter-prismatic material. Inset: Higher magnification SEM micrograph showing the calcium fluoridated carbonated apatite crystals arranged in parallel boundless within the prisms.
Dental enamel is the hardest material found in mammals with a high mineral packing density similar to that of bulk hydroxyapatite mineral. When compared to hydroxyapatite mineral, however, enamel has a higher elastic modulus and hardness as well as much better fracture toughness. These mechanical attributes make this natural bioceramic both hard and resistant to fracture and wear. The underlying dentinoenamel junction (DEJ) provides additional mechanical support that prevents enamel deformation, which might otherwise result from the high external forces involved in chewing. Because of noted enamel mechanical properies the whole tooth structure adsorbes considerable damage over time without catastrophic failure (28). In creating a material with such a contradictory combination of mechanical properties, nature has devised a unique assemblage or mineral, water, and organic material, all organized into a structural hierarchy (Figure 1). The key to achieving such a precisely organized architecture lies not only in the cell movements that occur during enamel formation, but also in the highly controlled expression of proteins and enzymes and in the way these organic molecular components interact with each other, with cell surfaces, and most importantly, with the forming mineral (21,24).
4. INITIATION AND STAGES OF AMELOGENESIS
The enamel-making cells, ameloblasts, are of epithelial origin and their differentiation is tightly linked to the differentiation of the dentin-making cells, the odontoblasts, which are of mesenchymal origin (29). In humans, the development of enamel starts in the third trimester of pregnancy and full mineralization is achieved by six months after birth. The three major stages of tooth development are bud, cap, and bell stages. Terminal differentiation of the inner enamel epithelium cells into ameloblasts takes place during the bell stage prior to which odontoblasts secrete the predentin matrix. The basement membrane separating the enamel organ and the dental papilla degrades following the deposition of predentin and before enamel matrix is secreted by the ameloblasts. This is the interface that eventually becomes the dentino-enamel junction. Degradation of the basement membrane allows direct contact between odontoblasts and preameloblasts, an interaction that may function as an inductive signaling between them.
Ameloblasts deposit the enamel matrix onto predentin during the advanced bell stage. The dynamic process of enamel biomineralization occurs in the extracellular space between the presecretory ameloblasts and the mineralized dentin (Figure 2A). The tissue grows continuously, with the secretion of enamel extracellular matrix, until the ameloblasts have secreted the whole thickness of the enamel matrix. During the process of amelogenesis, the ameloblasts pass through a series of differentiation stages that are characterized by changes in cell morphology and function. In general, cell differentiation during enamel development results in pre-secretory, secretory, transitional, and maturation phase ameloblasts. Cell morphology and chemical composition of the enamel extracellular matrix are the basis for definition of the different stages of amelogenesis (4, 6, 30). During the pre-secretory stage, pre-ameloblasts originating from the inner enamel epithelium elongate, polarize, and develop a large capacity for increased protein synthesis. They also play an important role in degradation and resorption of the basal lamina separating them from the predentin (31, 32). Secretory stage ameloblasts are tall columnar epithelial cells that are characterized by histological structures called Tomes’ processes and their main function is a massive production and secretion of enamel proteins. The newly formed enamel in electron micrographs shows a vey irregular wavy boundry at the dentine-enamel junction (DEJ) where processes from dentin and enamel interlock (Figure 2B). The full thickness of enamel is established at the secretory stage following the secretion of the organic matrix and its partial, but not complete, mineralization. Using a constantly growing mouse incisor as a model for early enamel mineralization Beniash et al confirmed that the first mineral phase to form is amorphous calcium phosphate that later transforms to crystalline apatite (33). The size, shape and spatial organization of these amorphous mineral particles and older crystals are the same, suggesting that the mineral morphology and organization in enamel is determined prior to its crystallization.
Figure 2.
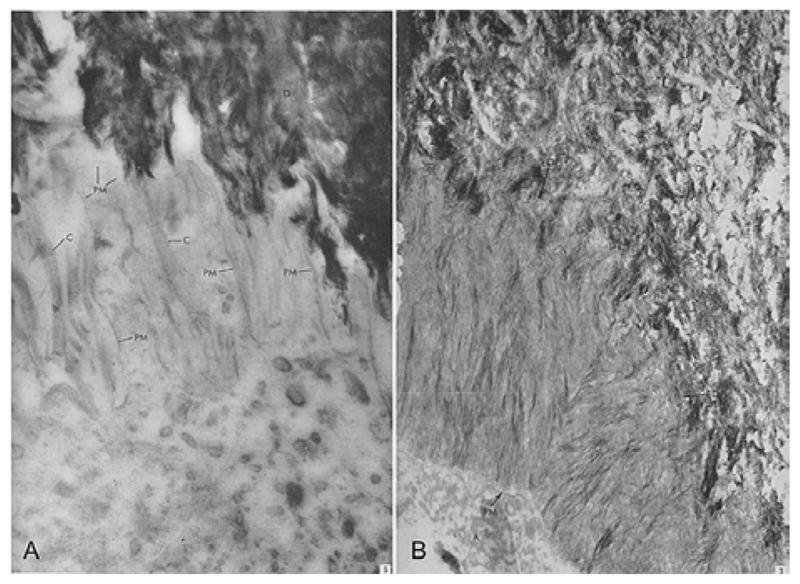
(A) Ameloblast membrane projections on dentin surface in human teeth. Detailed picture of the distal part of the ameloblast cell. Filaments with cross banding typical of collagen are located between the narrow processes extending from the distal surface of the ameloblast cell toward the dentine layer. Magnification=× 67,000. Reproduced with permission from reference # 31; Elsevier through Copyright Clearance Center. (B) The newly formed enamel shows a very irregular wavy boundary at the enamel-dentin junction, EDJ. Processes from enamel and dentine interdigitate, magnification=X 39,000. Reproduced with permission from reference # 32; Elsevier through Copyright Clearance Center. D: dentin, C: collagen, EDJ: Enamel Dentin Junction, A: Ameloblasts, PM: Plasma membrane of the ameloblasts.
Secretory stage ameloblasts, which are connected to each other by junctional complexes and desmosomes, move away from the dentin as they form enamel. It has been reported that in some species they also move laterally and in a twisting motion to create the unique rod/interrod arrangement, and bending patterns, of enamel ulrastructure (34). The transition stage occurs between the secretory and maturation stages when the cells undergo structural changes, becoming shorter and losing their apical cell extension, the Tomes’ process. At this time, their secretory activity is also drastically reduced. The transition stage can be visualized in rat incisor as the boundary between the translucent (less mineralized) and opaque (more mineralized) enamel where the beginning of the maturation stage is occurring. Major changes in cell morphology and function continue to occur during the maturation stage when the cells reorganize to accommodate massive protein degradation and rapid crystal growth. Their secretory activity is down-regulated but not terminated. Maturation stage amelobalsts are characterized by their periodic morphological changes to “ruffle-ended” and “smooth ended” (30). These changes have been associated with the functioning of cells to control ion transport (calcium, phosphate, bicarbonate), endocytosis, and pH (26, 35, 36). Once the enamel is fully mineralized and the organic matrix is degraded and removed, the columnar epithelial ameloblasts cease to function and undergo regression. They shrink dramatically, become cuboidal in shape, and finally form a pellicle on the surface of the tooth enamel that is shed following tooth eruption. Recently, Simmer et al (24) have reviewed five growth parameters, starting from the onset of amelogenesis, that determine the shape of the enamel crown beneath the cusp tips. The five parameters include enamel appositional growth rate, duration of appositional growth, ameloblast extension rate, duration of ameloblast extension, and spreading rate of appositional termination.
5. THE EXTRACELLULAR PROTEIN MATRIX
The extracellular matrix contains proteins and proteases, the timing of expression and secretion of which is well controlled by various genes and signaling pathways (22). The main structural proteins secreted by ameloblasts include amelogenin, enamelin, ameloblastin, amelotin, and the proteinases: matrix metalloproteinase-20 (MMP-20 or enamelysin) and kallikrein-4 (KLK-4) (Table I). Following the isolation of cDNA sequences of the enamel matrix proteins, our understanding of the molecular basis of enamel formation has significantly increased. The chromosomal localizations of the genes encoding these proteins have provided further insight into the origin of the ancestral genes. The genes encoding amelogenin, enamelin and ameloblastin (AMEL, ENAM, and AMBN) belong to the secretory calcium binding phosphoprotein (SCPP) gene family, and all arose from a single ancestor gene (SPARCL1). AMEL arose from AMBN, which itself resulted from a duplication of ENAM (20). Presumably, AMTN (which encodes amelotin) is also derived from ENAM. These genes are all located on the same chromosome except AMEL which is located on chromosome X (Table I). The spatiotemporal expression of each of these enamel proteins has been determined, and experiments using knockout and transgenic animal models have proved their critical function for normal enamel formation. However, what is yet to be elucidated is how these proteins interact with one another, with the growing crystals, and with cells to form one of the most remarkable hierarchical structures in the mammalian body.
Table 1.
Proteins and proteinases of enamel extracellular matrix, their human gene chromosomal localization and proposed function
| Protein and proteinases | Human gene chromosomal localization | Knock out (KO) mousegross enamel phynotype | Proposed function in enamel | Ref. |
|---|---|---|---|---|
| Amelogenin | AMELX; Xp22.3-p22.1, AMELY; Yp11 | Hypoplastic Disorganized (prismless) | Stabilization of amorphous Ca-P phase, control of apatite crystal morphology and organization, control of enamel thickness | 37–40, 45–48 |
| Ameloblastin | AMBN;4q21 | Thin dysplastic mineralized layer is formed on top of dentin | Cell adhesion protein, controls cell differentiation, maintains rod integrity | 114–123 |
| Enamelin | ENAM; 4q21 | No true enamel but focal areas of pathological calcified material | Cooperates with amelogenin to control mineral nucleation and elongated growth | 128–129, 134 |
| Amelotin | AMTN;4q13.3 | ------- | --- | 139–141 |
| KLK-41 | KLK4;19q13 | Delayed maturation and defects at the DEJ | Digests enamel proteins during the maturation stage facilitating their removal and hardening the final layer of enamel | 153–157 |
| Mmp-202 | MMP-20; 11q22.3 | Hypoplastic and hypomineralized enamel with decreased hardness that separates from dentin | Cleaves amelogenin, ameloblastin, and enamelin at the secretory stage to produce stable intermediates with defined functions | 144–148 |
| Ctrc3 | CTRC; 1p36.1 | --- | ---- | 143 |
Abbreviations: Kallikrein-41, Matrix Metalloproteinase -202, Caldecrin3
Ameloblasts express and secrete other proteins and glycoproteins that are not normally considered enamel proteins but which might play important physiological roles during amelogenesis. These include a small proteoglycan called biglycan and dentin sialophosphoprotein (DSPP) (2, 25). The latter is mainly expressed by ameloblasts during the early stages of amelogenesis. In this communication I will focus on the main enamel structural proteins for which in vivo and in vitro data are more available, namely; amelogenin, enamelin, ameloblastin, amelotin and the enamel proteinases.
5.1. Amelogenin
Amelogenin, the major structural protein of the enamel organic matrix, constitutes more than 90% of the enamel’s protein content. In human and cow, the amelogenin gene exists on both X and Y chromosomes (37), while in rodents amelogenin exists only on the X chromosome (38, 39). This makes the mouse a good animal model to study the function of Amelx- (X-chromosomal) encoded amelogenin under conditions that exclude effects of amelogenins encoded from the Y chromosome. The (Amelx) gene is located within an intron of the Arhgap6 gene in human, mouse, cow, and several other species (40). Even in human and cow, the X-chromosome derived amelogenin protein predominates in males. Amelogenin genes on both X and Y chromosomes contain seven exons, the largest being exon 6. The amelogenin primary RNA transcript is subject to extensive alternative splicing in many species with the most abundant amelogenin message including exons 1,2,3,5,6, and 7. In mice, 16 different alternatively spliced products have been identified. In the mouse and rat genomes, additional exons 8 and 9 have been identified and their protein products have been associated with a signaling function of amelogein (41). The N- and C- terminal domains of amelogenin are highly conserved and contain residues that have remained unchanged during 360 (Ma) (million years), supporting the critical function of those residues (42). The central portion contains more variations in sequence but the locations of several PXQ motifs and His residues are conserved. The population of amelogenin polypeptides in developing enamel is heterogeneous because of the presence of alternatively spliced variants and proteolytic activities that produce stable isoforms (43, 44).
Amelogenin is critical for proper enamel formation as demonstrated by human genetic analyses as well as genetically engineered amelogenin-null mice (45). Three independent strategies have been embraced by investigators to evaluate the loss of function of amelogenin proteins in animal and organ culture models. These include: specific inhibition of amelogenin gene expression by means of a chemically modified ribozyme in newborn mice (46), antisense inhibition of amelogenin translation in organ culture (47), and a null mouse model (45). All these studies have supported the hypothesis that amelogenin functions to control the organization and growth of enamel crystals and that its presence is critical for normal enamel formation. In addition, amelogenin sequence defects lead to defective enamel crystal formation and organization, and deletion of the conserved terminal domains leads to the formation of ill-defined enamel crystals, highlighting the importance of these conserved domains in protein-protein or protein-mineral interactions (48).
Amelogenin possesses signaling activities, as assessed in a variety of cell culture systems (49, 50). This property has in particular been attributed to the small isoform; leucine-rich amelogenin peptide (LRAP) (51). Amelogenin interacts with bone cells and, because of its potential to promote cell differentiation, the protein has been considered for periodontal regenerative therapies. Amelogenin expression in soft tissues has also been reported (52), although no obvious defects were observed in non-dental tissues of amelogenin null-mice. Nor have amelogenin gene defects been directly associated with diseases or abnormalities in tissues other than enamel. In the next several sections I will discuss structural features, some of which may explain the signaling potential and multifinctionality of amelogenin. The rest of the chapter will then focus on amelogenin as the principal protein of enamel extracellular matrix having the main function of controlling enamel thickness and the organized growth of elongated apatite crystals.
5.1.1. The “unstructured” nature of amelogenin
Amelogenin is phosphorylated on a single residue (Ser-16) but not glycosylated (53). The protein sequence has a hydrophobic-hydrophilic polarity in which the core sequence is enriched in hydrophobic amino acids and the majority of the charged amino acids are concentrated in the C-terminal region. The central region of amelogenin is enriched in Pro, His and Gln. The tri-tyrosine motif (PYPSYGYEPMGGW) in the tyrosine-rich amelogenin N-terminal (TRAP) region has been reported to have lectin-like properties. This region might be involved in amelogenin-cell or amelogenin-nonamelogenin interactions (54, 55). The determination of amelogenin secondary structure has been problematic due to both the self-association or aggregation phenomenon, and the intrinsic lability of this protein (56–58).
Several attempts have been made in the past to obtain secondary and tertiary structural information for amelogenin using native proteins isolated from developing enamel. Investigators have reported poorly resolved X-ray diffraction images of developing bovine tooth enamel matrix, possibly showing a beta- or a cross-beta structure (59). CD and NMR studies have led to suggestions that the molecule contains both beta-structures and beta-turns (60, 61), but other work has indicated a highly mobile structure (62).
Our most recent investigations have provided insight into the nature and secondary structure of amelogenin (58, 63, 64). By applying bioinformatic strategies we identified molecular traits that classify amelogenin proteins as members of the family of “intrinsically disordered” proteins (IDPs) or “unstructured proteins” (website http://www.disprot.org). Intrinsically disordered proteins or natively unfolded proteins (IDPs or NUPs) are proteins or large segments of proteins that lack well-structured three dimensional folding and exist as a dynamic ensemble of interconverting structures (65). Despite their lack of a well-defined three dimensional structure, such proteins are indeed functional. The bulk of amelogenin is enriched in Pro (P) and Gln (Q), amino acid residues known to promote disorder in protein structure. Other disorder-promoting residues such as Glu (E), Arg (R) and Lys (K) are all concentrated in the hydrophilic C-terminus. Mammalian amelogenin sequences are highly conserved, and interestingly, all 12 vertebrate amelogenins we examined contained 50–60% disorder-promoting residues (63, 64). Spectroscopic studies on model peptides have confirmed that IDPs have structured regions in their sequence with substantial contribution from a polyproline type II (PPII) structure, PXXP/PXP, where X represents amino acids that have high PPII helix forming propensity (66). The central region of amelogenin contains several PXXP/PXP repeats separated by histidine residues. Based on the PPII propensity scale for individual amino acid residues, we have suggested that PPII is the dominant structure in the central region of amelogenin (67).
Using NMR we confirmed that the recombinant porcine amelogenin isoform rP172, in a monomeric form at pH 3.8 exists in an extended, unfolded state (Figure 3). Although the protein is globally unfolded, the presence of local residual secondary structure (alpha-helix, extended beta strand, turn/loop, and polyproline Type II (PPII) conformation) was detected and these more structured regions may serve several functional roles within the enamel matrix. We then proposed that the extended, labile conformation of rP172 amelogenin is compatible with the known functions of amelogenin in enamel biomineralization, i.e., self-assembly into oligomers, nanospheres, and nanochains; associations with other enamel matrix proteins and with calcium phosphate biominerals; and interaction with cell surfaces. The “unstructured” features in amelogenin sequence may well explain its signaling capability as well as cell adhesion behavior (49, 68). As discussed in the last section, the labile structure of amelogenin also makes it a good candidate for application in synthetic enamel-like biocomposite materials.
Figure 3.
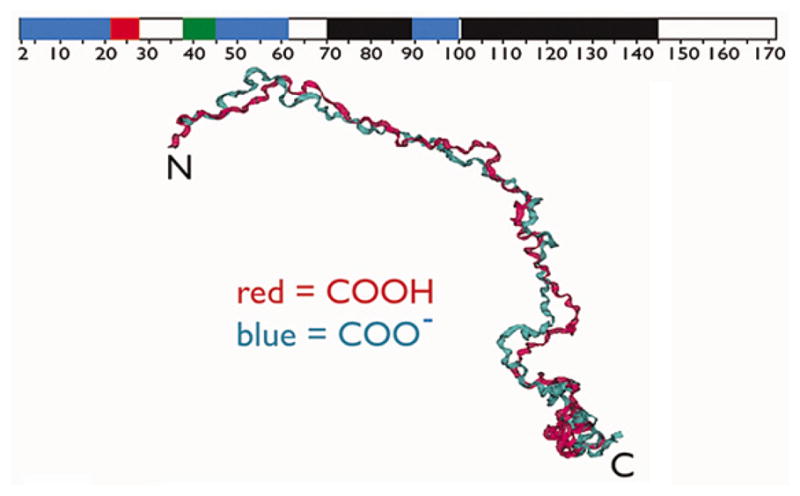
Backbone aligned best structures obtained for the fully protonated (Red) and deprotonated (Blue) forms of recombinant porcine amelogenin (rP172) in monomeric form, showing that the molecule is extended (backbone rmsd = 4.7 Å). The ribbon on the top represents secondary structure assignment based upon nOe and 3J data in this figure; black = PPII, red = alpha-helix, blue = extended beta strand, green = beta turn or loop, and white = random coil or unstructured. Reproduced with permission from (58).
To further characterize the IDP nature of amelogenin, a “solvent engineering” approach was used to simultaneously promote native-like structure in recombinant porcine amelogenin and induce its oligomerization in a manner that allowed identification of intermolecular contacts between amelogenin molecules (69). In the presence of 2,2,2-trifluoroethanol (TFE), significant folding transitions and stabilization occurred primarily within the N- and C-termini, while the polyproline Type II central domain was largely resistant to conformational transitions. TFE induced rP172 self-association via the formation of intermolecular contacts involving the P4-H6, V19-P33, and E40-T58 regions of the N-terminus. It was concluded that the N- and C-termini of amelogenin are conformationally responsive and represent potential interactive sites for amelogenin-target interactions during enamel matrix mineralization. Conversely, the Pro, Gln central domain is resistant to folding and this may have significance in other functions of amelogenin.
5.1.2. In vitro amelogenin self-assembly into oligomers, nanospheres, nanochains
Amelogenin protein assembles into oligomers, nanospheres, nanochains and other elongated assemblies under a variety of different in vitro conditions (70–76). Since the 1960’s it has been well known that amelogenin has a strong tendency towards self-aggregation into large particles (1 to 4 million Dalton) (56, 57). Temperature-dependent reversible aggregation characteristics as well as self-orienting properties characteristic of “liquid crystal mesophases” have also been reported for amelogenin (77).
Nanospheres
Once the recombinant form of amelogenin became available, Fincham and Moradian-Oldak described the formation of “nanospheres” at pH 8 (15–20 nm in radius) resulting from amelogenin self-assembly in vitro by employing dynamic light scattering (DLS), size exclusion chromatography (SEC), atomic force microscopy (AFM) and transmission electron microscopy (78–80). It was then proposed that the nanospheres were formed through intermolecular hydrophobic interactions when the hydrophilic segment of each molecule is exposed on the surface of the nanospheres. Using a two hybrid system two self-assembly domains (A- and B-) were indentified to be invloved in amelogenin-amelogenin interactions during nanosphere formation (81, 82). Later, a model of a dense hydrophobic core with a flexible hydrophilic region exposed on the surface of amelogenin molecules was also proposed based on application of the small-angle X-ray scattering (SAXS) technique, (83).
Oligomers
Dynamic light scattering analyses of amelogenin in different solvents have revealed the presence of monomers and stable discrete oligomers such as dimers, trimers, and hexamers (72). Formation of dodecamers was illustrated by single particle reconstruction of cryo-TEM images of recombinant mouse amelogenin (76). These oligomers were defined as the basic subunits of the typical nanospheres, which have hydrodynamic radii (RH) of about 20 nm. Further insight into the mechanisms of amelogenin oligomerization was obtained in our most recent in vitro studies (70). Our strategy was to change parameters such as pH, temperature and protein concentration to control amelogenin self-association and to understand the driving forces behind assembly and disassembly. Stable oligomers with an average hydrodynamic radius (RH) of 7.5 nm were observed at pH 5.5, and at protein concentrations between 4–10 mg.mL−1. No significant increase in folding upon self-association of the monomers into oligomers was observed indicating that the oligomers are unfolded. Fluorescence experiments with single-tryptophan amelogenins revealed that upon oligomerization at pH 5.5, the C-terminus of amelogenin (around residue W161) is exposed at the surface of the oligomers, while the N-terminal region around W25 and W45 is involved in protein-protein interaction. The truncated rP148 formed similar but smaller oligomers suggesting that the C-terminus is not critical for amelogenin oligomerization. We have separated the interactions that cause monomers to assemble into oligomers from those that cause oligomers to bind and form nanospheres. Specifically, we proposed that oligomer formation is primarily N-terminal mediated (potentially N-terminal to N-terminal), while nanosphere formation is mainly regulated by hydrophobic interactions via the central portion, which are in turn controlled by histidine protonation. Dissociation of nanospheres into oligomers can also be promoted if the nanospheres are exposed to a hydrophilic environment and to charged surfaces, even at pH 8 (71, 84).
Nanochains and other elongated assemblies
Nanochain assemblies formed by amelogenin have been produced in a number of ways, including 1) by slow solvent evaporation (72); 2) by incubation in the presence of calcium (85); 3) by fine-tuning the solution pH around neutral (73) or 4) by incubating a high concentration of amelogenin over an extended period of time together with its proteolytic products (86). Amelogenin nanochain structures were also detected on the surface of crystals grown on fluoroapatite glass ceramic (87) and during an electrodeposition reaction when the local pH was gradually increased (10). Amelogenin can also self-assemble into mircoribbons under slow evaporation conditions (72), and nanoribbons at an oil/water interface and in the presence of calcium phosphate (74). Nanorods of amelogenin can be formed during proteolysis and as the result of its co-assemby with proteolytic products (86, 88).
In our most recent model we propose that nanospheres form via oligomers and we predict that nanospheres will break up to form oligomers in mildly acidic environments, via histidine protonation. The combination of mildly acidic environment during the maturation stage of enamel and the positively charged surface of apatite promotes dissociation of the nanospheres into oligomers. The nano-chains adsorbed on surfaces are therefore the result of the assembly of oligomers and not nanospheres (89, 70, 71, 76). We further suggested that oligomeric structures might be functional components during maturation of enamel apatite.
5.1.3. Amelogenin self-assembly in vivo
Comparisons of the in vitro data on amelogenin assembly with the TEM images of in vivo embedded sections of developing enamel led to reports that amelogenin “nanospheres” appeared as electron-lucent structures after conventional negative staining (47, 90, 91). Following recent data on amelogenin oligomers and careful examination of the sizes of electron lucent spherical particles in some sections of forming enamel, as well as those adsorbed on TEM grids suggest that these structures maybe oligomers and not nanospheres as originally described (47, 71, 79, 80, 90).
Prior to the description of nanospheres Robinson et al. (92) investigated developing rat incisor enamel using a freeze-fracture technique and concluded that “the forming region seems to consist largely of spheres 300–500 Å in diameter”. These spherical particles were proposed to consist of organic matrix and mineral, supporting the theory that the initial nucleation of enamel mineral may start with a co-assembly of the organic matrix (mainly amelogenin) and the mineral phase (initially amorphous calcium phosphate mineral). The recent finding that the initial electron-dense amorphous calcium phosphate mineral in enamel has the form of defined ribbon-like shapes that are separated by chain-like structures of organic matrix (electron-lucent) does not support the theory that the spherical particles at the early stage are a mix of organic/inorganic phase, but supports the notion that amelogenin self-assembly into nanochains may occur at the very early stage of mineral formation, separating the mineral and dictating the morphology of early apatite crystallites (33, 72, 93). It is now well accepted that the amelogenin-based supramolecular assemblies exert control over the organization and directionality of hydroxyapatite crystal growth (Figure 4).
Figure 4.
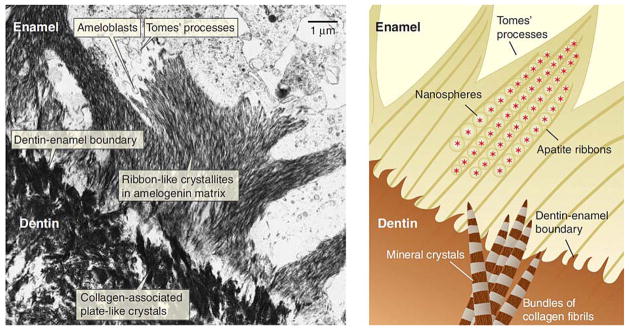
(A) Micrograph of the dentin-enamel boundary in a developing tooth. The apatite of dentin (lower left) forms nanometer-scale plates that initially grow individually and independently in the gap spaces along the collagen fibrils. The ribbon-like apatite crystallites of the enamel (top right) form within the amelogenin-rich enamel matrix. (B) A close-up view (not to scale) of the hypothesized mineral deposition processes in dentin and enamel near the dentin-enamel boundary. In the dentin, plate-like apatite crystals grow in the periodic gap spaces along the collagen fibrils and fibril bundles. The apatite crystal c axis is mostly aligned with the long axis of the collagen fibril. In the enamel, Du et al. (72) propose that linear aggregates of polarized, self-assembled amelogenin nanospheres form a negatively charged template that induces apatite formation. Reproduced with permission from reference # 31; The American Association for the Advancement of Science through Copyright Clearance Center.
Besides spherical structures, the presence of some relatively periodic structures in developing enamel have been previously reported (94). Watson (95) originally described enamel “stippled material” as a finely granular material between the distal end of the secretory ameloblast and the mineralization front. Ronnholm (93) used decalcified embryonic human enamel to demonstrate the presence of a low contrast organic matrix structural pattern. The 80 Å thick “septa”, which originated from the “stippled material,” were parallel to the row of crystallites. Travis and Glimcher (96) used longitudinal sections from developing embryonic bovine enamel to analyze the intra-prismatic organic matrix. They reported many short, swollen, fragmented filaments described as “dense lines approximately 48 Å wide” that were parallel to each other with their long axes parallel to the enamel prisms, and were “separated by approximately 120 Å”. Smales (97) described a helical structure in which “the helices varied in overall width from 15 to 30 nm”.
Additional factors have been suggested to contribute to assembly of amelogenin and the enamel matrix. For example, it has been argued that the assembly of amelogenin may be controlled at some level in vivo by local pH changes (73, 98). Recently we reported that stepwise proteolytic activities in the enamel extracellular matrix can alter amelogenin self-assembly, promoting formation of elongated nanorod-like structures (89). Interactions of amelogenin with other enamel proteins and apatite surfaces are other factors that may control the assembly of amelogein-based matrix (71, 99).
5.1.4. Amelogenin function in controlling mineralization
In order to elucidate the function of amelogenin in controlling mineralization, numerous in vitro experimental strategies have been implemented to study amelogenin-crystal interactions. Availability of recombinant forms of amelogenin has allowed researchers to study the different aspects of calcium phosphate mineralization controlled by amelogenin in a more systematic manner. These strategies have collectively demonstrated that amelogenin has the potential to control calcium phosphate polymorphism, promote parallel crystal organization, and modulate crystal morphology in vitro. Promotion of crystal nucleation by amelogenin in a variety of in vitro model systems has also been reported (100, 101). However, the observation that enamel mineralization still occurs in Amel null-mice does not support a direct nucleating function for amelogenin in vivo (45).
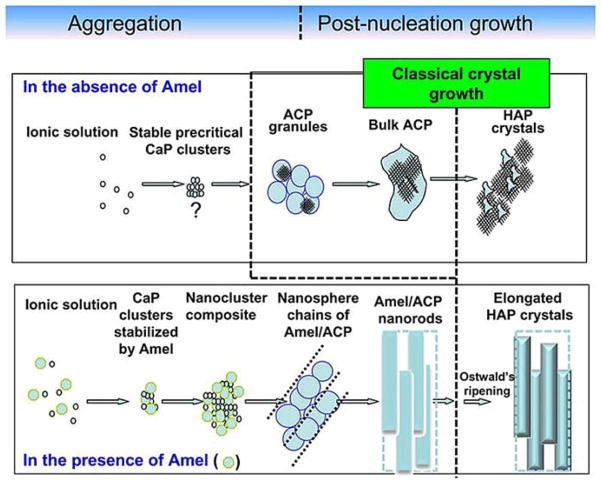
Investigators have utilized in vitro strategies to estimate parameters such as binding affinity and adsorption isotherms of different amelogenin isoforms onto apatite crystal surfaces (102). Other approaches have been used to examine the effect of amelogenin on: 1) the kinetics of seeded apatite crystal growth (103, 104); 2) the morphology (habit) of apatite and octacalcium phosphate crystals grown in solution and in “gel-like” matrices (105); 3) the growth of calcium phosphate coatings on titanium, bioactive surfaces, and glass-ceramics (106), and 4) the regulation of the calcium phosphate mineral phase (76, 107). Specific binding of amelogenin to the side faces of enamel crystals has also been demonstrated (108). The idea of cooperative amelogenin assembly and organized apatite crystal growth was introduced in the past few years by Beniah et al who reported (85) that c-axial co-aligned nanocrystal plates of hydroxyapatite can be formed in mineralization solutions in the presence of amelogenin. The constant composition method (CC) has been applied to demonstrate the promoting effect of amelogenin on the nucleation and growth of apatite crystals. Remarkably, under controlled conditions of supersaturation, elongated ribbon-like crystals similar to enamel crystals were grown in the presence of low concentrations of amelogenin (109). A nucleation model was suggested in which amelogenin was proposed to interact with calcium phosphate in a cooperative manner to form intermediate complex clusters which in turn aggregated along the apatite c-axis (Figure 5). It has been further shown that the complex formed by non-crystalline calcium phosphate and amelogenin plays an important role in cooperative crystallization before the formation of organized crystals (76, 109). By comparing phosphorylated (native) and non-phosphorylated (recombinant) amelogenins Kwak et al. highlighted the importance of phosphate groups in the interaction of amelogenin with mineral and suggested that the phosphate-containing amelogenin proteolytic cleavage product may suppress unwanted rapid mineral formation within the enamel matrix during the secretory stage of amelogenesis. In contrast, presence of the phosphate group did not have a significant effect on amelogenin-self assembly (107).
Figure 5.
Suggested mechanism for in vitro hierarchically organized, elongated HAP microstructures formed by co-assembly of amelogenin and calcium phosphate nanoclusters, based on in vitro experiments Reproduced with permission from (109).
Our understanding of the direct role of amelogenin in controlling mineralization has further improved following in vivo targeted gene knockout strategies that have been used to delete the amelogenin gene (45). The teeth from amelogenin-null mice have a chalky-white discoloration, and a disorganized (prismless) and hypoplastic (thin) enamel (Figure 6B). No amelogenins were expressed in these animal models. The observation that amelogenin- null mice secreted a thin layer of mineralized enamel (10–20% thickness of normal enamel) led the authors to suggest that amelogenin is not required for the initiation of mineralization, but essential for elongation of enamel crystals and achievement of proper enamel thickness. Thus, while the “loss of function” strategy provided important insight into the physiological function of ameloegnin in controlling enamel thickness and prism organization, the in vitro experimental strategies provided clues to the detailed molecular mechanisms of amelogenin-guided mineralization. In vitro and in vivo strategies are complementary, and both are necessary to fully understand the function of amelogenin in enamel formation.
Figure 6.
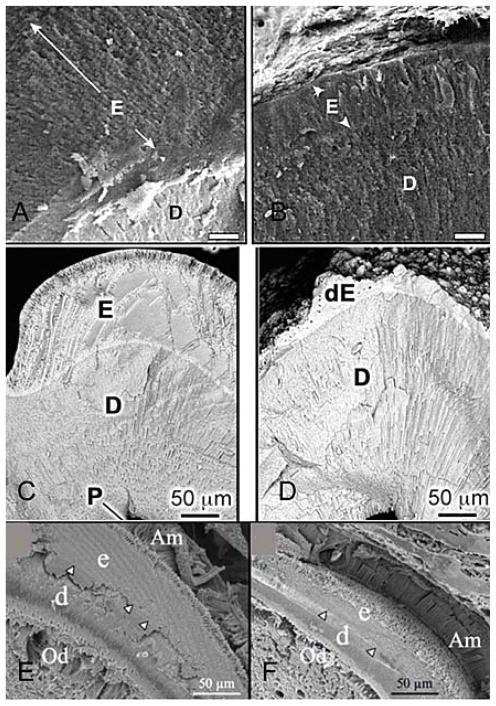
SEM images of fractured incisors showing the defects in enamel formation of amelogenin (Amel), ameloblastin (Ambn), and enamelin (Enam) null-mice. (A) wild type mouse. (B) the enamel from the null mouse does not have a normal prismatic structure and is markedly reduced in thickness compared with that of the wild type mouse shown at the same magnification as A, Bars in A and B = 10 micrometer. Reproduced with permission from reference #45. (C) Heterezygote Ambn+/− and (D) Homozygote Ambn −/− mice revealing a displastic layer with a rough surface over dentin and lack of a prism pattern in D. Reproduced with permission from reference #122. (E) Heterezygote Enam −/+ and (F) Homozygote Enam −/ − mice. The forming enamel in the Enam−/ − mouse (F), is unmineralized, but shows remarkable similarity to normal enamel matrix structure. Reproduced with permission from (131). E: Enamel, D: Dentin, dE: defective enamel, Am: Ameloblasts, Od: Odontoblasts; arrowheads in E and F indicate the dentinoenamel junction.
5. 2. Ameloblastin
While experimental data on amelogenin structure, assembly and function are extensive, such studies are limited for ameloblastin. This is partly due to limitations in expression and purification of its recombinant form. Another reason is that ameloblastin undergoes hydrolysis by enamel proteinase MMP-20 as soon as secreted, making its isolation and purification from in vivo sources challenging (110, 111). This proline-rich protein is expressed by ameloblasts and secreted together with amelogenin in secretory granules, suggesting the existence of a regulatory mechanism for sorting and transport of amelogenin and non-amelogenins, and also a possible functional interaction between the two proteins (112). Amelogenin-ameloblastin interactions have been reported in in vitro experimental assays. The interactions occur through the lectin-like binding domain of amelogenin, although a functional significance for such interactions is still lacking (113). Ameloblastin is the second most abundant protein in enamel, constituting about 5% of the total protein present in the organic matrix. Its expression drops to significantly lower levels during enamel maturation. The Ameloblastin (Ambn) gene is a member of the cluster of secretory calcium-binding phosphoprotein (SCPP) genes located on human chromosome 4q (114) (Table I).
5.2.1. Ameloblastin sequence and processing
Ameloblastin is a glycoprotein (through O-glycosylation only) that contains relatively high percentages of Pro (15.2%), Leu (10.2%) and Gly (9%). Ameloblastin is also phosphorylated and contains hydroxy-prolines (115). Two isoforms from alternatively spliced mRNA transcripts are expressed, and splicing determines the glycosylation state of ameloblastin (116). The N-terminal 86 amino acid domain of ameloblastin is the most conserved region among species, being conserved from amphibians to humans. Interestingly, the sequence of ameloblastin possesses a “bipolar” nature, having a pI of 10.6 for the N-terminal 129 amino acids and a pI of 4.5 for the 66 C-terminal amino acids. These two domains are linked by an “unstructured” region susceptible to proteolysis (117). The major ameloblastin (sheathlin) isoform in pig has 395 amino acids (118). The N-terminal polypeptides appear to be stable while the C-terminal polypeptides degrade rapidly and are removed from the matrix. The inner enamel (119) contains fragments from the N-terminal region, 17 k-Da (amino acids 1–170), 15 k-Da (amino acids 1–130), and 13 k-Da (amino acids 33–130) fragments, and the O-glycolysated 13 and 11 k-Da fragments from the C terminus (120, 121).
5. 2.2. Ameloblastin putative function
It was not possible to learn the direct function of ameloblastin in controlling apatite mineralization from study of “Ambn null mice” animals because of the lack of true enamel layer formation in these mice (Figure. 6D). This Ambn knockout model does not fully abrogate gene and protein expression and only Exons 5 and 6 were deleted in these mice leading to a secreted protein that lacks a highly conserved region at the N-terminus (122, 123). Only a thin layer of hypopalstic mineralized material is deposited onto the dentin. Ameloblastin is considered critical for enamel formation since obvious pathology in the ameloblast layer and defects in the junctional epithelium were noted in these animals (122) (Figure 6D). Ameoblastin shows cell adhesion properties and is believed to control ameloblast cell differentiation (Table I). However, the motif proposed to be responsible for cell attachment (VTLG) is not conserved among species studied so far. Based on immunolocalization studies of ameloblastin N-terminal cleavage products (13- and 17-kDa) appearing in the sheath or inter-prismatic space in porcine enamel, ameloblastins have been hypothesized to be involved in the control of the prismatic structure of enamel (124). While such a pattern of immunolocalization at the sheath space was not observed in rats, “fish-net”-like partitioning that follows the ameloblast outline has also been reported (118, 125, 126). Ameloblastin proteolytic products from the C-terminal region had high affinity for calcium ions, indicating their possible involvement in controlling mineralization (115).
5.3. Enamelin
Enamelin is expressed during the three main stages of enamel formation, but its expression terminates prior to the expression of amelogenin (127). In contrast to the abundant and generally hydrophobic amelogenin, enamelin is much less abundant and hydrophilic in nature. It is the largest known enamel glycoprotein and like amelogenin the gene encoding it belongs to the secretory calcium-binding phosphoprotein (SCPP) gene family. The human enamelin (ENAM) gene, encoding enamelin, maps to chromosome 4q21 (128, 129).
5.3.1. Enamelin sequence and processing
Enamelin sequences from 36 mammalian species are now available (130). Human enamelin contains 9 exons, and is secreted as an 186kDa phosphorylated precursor glycoprotein. Mature enamelin is cleaved from its cysteine-rich carboxy-terminal peptide by proteinases shortly after being secreted and only its cleavage products have been isolated (from pig) and characterized (131). Enamelin cleavage products of 155, 142 and 89kDa apparent molecular weight have been characterized, which retain the original amino terminus. The 89-kDa enamelin accumulates and is further processed to generate the 32 and 25kDa enamelins. In pigs, the most stable and the only studied enamelin cleavage product is the 32kDa form extending from Leu174 to Arg279, and is known to include two phophoserines and three glycosylated asparagines (120, 132). While the Cys-rich carboxy terminal fragment is concentrated at the ameloblast distal membrane, the 32-kDa enamelin is observed throughout the entire thickness of developing porcine enamel and is concentrated in the rod and inter-rod enamel. Evolutionary analysis of enamelin sequences from 36 species representing the mammalian lineages shows high conservation in the region of the 32-kDa segment, supporting a crucial role for this polypeptide (130). The conservation in the 32-kDa region includes phosphorylation, glycolization and proteolytic sites. The 32 kDa enamelin is hydrophilic and acidic, with a pI of 3.2, and is rich in proline (18.8%), glycine (12.3%), threonine (10.4%), and glutamic acid (9.4%) (133).
5.3.2. There is no enamel without enamelin
In Enam null-mice where enamelin protein is not expressed, a true enamel layer is not formed (134). In the heterozygotous animals, initiation of mineral deposition is not observed and only a pathological mineralization of the extracellular matrix occurs leading to the formation of an “undefined” calcified material (Figure 6F). Examination of heterozygous mice reveals that progressive decrease in mineral is associated with reduced enamelin expression. Disorganization and apoptosis of ameloblasts have also been reported in Enam null mice. While utilization of animal models and loss of function strategies provide important clues for elucidating the primary function of enamelin, they do not provide insight into the specific molecular mechanisms of the protein’s interactions with the mineral or of it’s function in controlling mineralization. We have applied in vitro strategies to study the structure and function of enamelin. The 32-kDa enamelin, isolated from developing porcine teeth, was used for these structural and functional studies. The results of our in vitro efforts are summarized in the next few sections:
5.3.3. The 32-kDa enamelin conformation and its interaction with mineral; In vitro models
In vitro studies have highlighted relevant functional properties of enamelin in controlling crystal nucleation or growth. Among different enamelin cleavage products examined, the 32 kDa enamelin was shown to have a relatively high affinity for apatite crystals (115). Using a gelatin gel-containing device we showed that addition of 32 kDa enamelin to amelogenins promoted the nucleation of apatite crystals (135). Based on X-ray diffraction line broadening analysis of d002 of apatite crystals grown in agarose gel, it was demonstrated that the 32-kDa enamelin fragment caused an increase in length of apatite crystals (134). In order to elucidate structural features of enamelin we recently applied circular dichroism (CD) and Fourier transform infrared (FTIR) spectroscopy to study secondary structural preferences of the 32 kDa enamelin isolated from developing porcine enamel. The secondary structure of the 32 kDa enamelin in the absence of calcium had a relatively high content of alpha-helix. We showed that 32 kDa enamelin undergoes conformational changes, with a structural preference for beta-sheet, as a function of calcium ion concentration. We postulated that the increase in beta-sheet conformation in the presence of Ca2+ may allow a preferred interaction of the 32 kDa enamelin with apatite crystal surfaces during enamel biomineralization. The possibility that addition of calcium may induce enamelin aggregation leading to conformational changes cannot be ruled out. The calcium association constant of the 32 kDa enamelin calculated from the fitting curve of ellipticity at 222 nm is Ka = 1.55 (±0.13) × 103 M−1, indicating a relatively low affinity.
5.3.4. Cooperation of amelogenin and enamelin in controlling crystal morphology
The concept of cooperative function between amelogenin and enamelin was first introduced after the observation that, when combined, they promoted the kinetics of apatite nucleation in a gelatin gel (135). One of the first pieces of evidence needed to support the concept of cooperative function was the demonstration that these proteins directly interact. Our biophysical and spectroscopic studies collectively demonstrated that such interactions occur between the 32 kDa enamelin and amelogenin in vitro and that the two proteins might actually co-assemble. Immunoprecipitation studies revealed that the 32 kDa enamelin and amelogenin eluted together from a Protein A column (99). Dynamic light scattering results showed that the 32 kDa enamelin had a profound effect on full-length amelogenin assembly at pH 8.0, causing partial dissociation of the nanospheres in a dose dependent manner. Enamelin stabilized amelogenin oligomers in solution at pH 7.4–8 where nanosphere formation is usually most favorable (99). Interestingly, at lower pH’s (5.8 and 6.5), where monomeric or oligomeric amelogenin particles are favored, addition of the 32 kDa enamelin increased the size of the particles, indicating the occurrence of amelogenin-enamelin co-assembly (136). Conformational changes in amelogenin secondary structure as the result of enamelin addition were evident based on the appearance of an isodichroic point and the shifting and intensity decrease of the ellipticity minima in the circular dichroism spectra of amelogenin following the addition of the 32 kDa enamelin. In the fluorescence spectra, the maximum emission of amelogenin was blue-shifted in the presence of enamelin meaning that the environment around Tyr residues changes as a result of complexation of the two proteins. The above observation supports the concept of cooperation between enamelin and amelogenin in macromolecular self-assembly and in controlling different aspects of crystal formation. Enamelin may also function to: prevent unwanted aggregation of the truncated amelogenin in vivo, act as a chaperone for amelogenin inside the cell (Michael Hubbard, personnal communication), stabilize amelogenin oligomers, enhance the hydrophilicity of the amelogenin oligomers, and affect their interactions with the mineral phase. Amelogenin-enamelin interactions may well be affected by local pH changes that occur during enamel maturation. While a variety of in vitro evidence supports the concept of amelogenin-enamelin interactions, investigating amelogenin-enamelin association in the extracellular matrix in vivo has yet to be explored.
To examine the hypothetical cooperative role of enamelin and amelogenin in controlling the growth morphology to produce enamel crystals with large aspect ratios, we applied a cation-selective membrane system for the growth of octacalcium phosphate (OCP) in a “gel-like” amelogenin matrix with and without addition of the 32kDa enamelin fragment (136, 137). The use of octacalcium phosphate was justified because previous studies of the spontaneous precipitation of ACP, and its subsequent transition into apatite, revealed that the kinetically favored OCP was the first crystalline phase, which formed in very close contact with the ACP particle surface. The ability of OCP to incorporate water molecules and ions other than Ca2+ and PO43− as structural components enables OCP to function as an apatite crystal precursor. Since OCP has been identified as a transient phase for enamel crystals, we have been studying the mechanism of the elongated growth of OCP crystals using a dual membrane experimental device where ionic diffusion is controlled by a cation-selective membrane and a dialysis membrane. We have previously shown that oriented OCP crystals preferentially grow in the c-axis direction on the membrane (105), and that amelogenin increased the length to width ratio (aspect ratio) of OCP crystal through preferential interaction with the side faces of OCP (138).
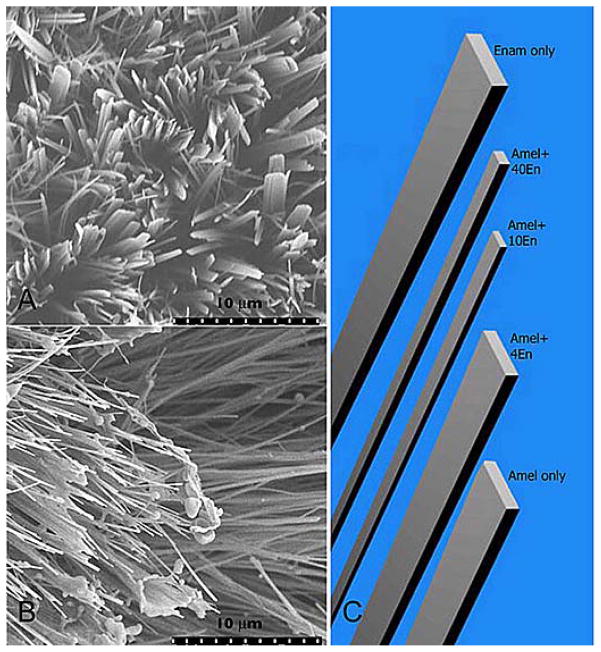
When the 32-kDa enamelin was added to the amelogenin “gel-like matrix”, the aspect ratio of OCP crystal was markedly increased in a dose dependent manner (Figure 7). Interestingly, addition of enamelin to amelogenin enhanced the potential of amelogenin to stabilize the amorphous calcium phosphate (ACP) transient phase. We found that the ratio of enamelin to amelogenin was crucial for stabilization of ACP and the growth of OCP crystals with larger aspect ratios. The cooperative regulatory action of enamelin and amelogenin was attained, presumably, through co-assembling of enamelin and amelogenin.
Figure 7.
SEM images of octacalcium phosphate crystals grown in the cation selective membrane system in the presence of (A) 40 microgram/mL 32-kDa enamelin, (B) 40 microgram/mL 32-kDa enamelin in 10% rP148 porcine amelogenin. Note the increase in crystals aspect ratios in B when compared to A. (C) Computer simulation models for OCP crystals grown in the presence of different amounts of enamelin (Length, width, and thickness values are based on the average values in reference #137).
5.4. Amelotin
Amelotin is the most recently discovered enamel glycoprotein, and its function is not yet clear (139). Amelotin was originally reported to be secreted by maturation stage ameloblasts as a component of the basal lamina between the ameloblasts and the enamel matrix, and to be present in the area around the junctional epithelial cells (140). Recent studies reveal, however, that like other enamel extracellular matrix proteins amelotin is also expressed during the secretory stage of enamel development and its localization is not limited to the basal lamina (141). Alternatively spliced variants of amelotin have also been described. In humans the amelotin gene is localized at the chromosomal locus 4q close to a gene cluster that also includes ameloblastin and enamelin. A sharp increase in expression levels of amelotin occurs at the transition zone from secretory to maturation stage ameloblasts, and this high expression gradually decreases towards the zone of reduced ameloblasts.
5.5. Enamel proteinases
Two proteases, matrix metalloproteinase 20 (MMP-20 or enamelysin) and serine proteinase kallikrein-4 (KLK-4), function to process and degrade amelogenin and other enamel proteins. The two proteases function differentially at different stages of amelogenesis since their peaks of mRNA expression are at different developmental stages of the ameloblast (142). Enamelysin (Mmp-20) is expressed from the onset of enamel matrix secretion through the early maturation stage, while Klk-4 is expressed from the beginning of transition stage and throughout maturation. Mmp-20 processes amelogenin, enamelin, and ameloblastin into stable intermediate products, while Klk-4 functions to completely degrade the extracellular matrix proteins. Recently, a new serine proteinase called caldecrin has been identified and found to be associated with enamel development, although its specific proteolytic activity against enamel proteins still needs to be explored (see 5.5.3., (143)).
5.5.1. Matrix metalloproteinase 20 (MMP-20)
To date, Mmp-20, known also as enamelysin, appears to be the only MMP that is tooth-specific and it is expressed by cells of different developmental origin (i.e. epithelial ameloblasts and mesenchymal odontoblasts) (144, 145). The human matrix metalloproteinase-20 (MMP20) gene contains 10 exons and is part of a cluster of matrix metalloproteinase genes that localize to human chromosome 11q22.3. Enamel in the absence of enamelysin is hypoplastic (thin), contains less mineral (only one-third as much total mineral as wild type), and contains more protein and water (146–148). The enamel layer in enamelysin-null mice is detached from the dentin and has a disorganized prism pattern that apparently results from defective amelogenin processing. The mineral acquisition rates in the mid maturation stage of enamel in Mmp-20 null mice are significantly lower than in normal enamel (148).
It has also been generally thought that Mmp-20 functions to regulate the elongated growth of crystals at the early secretory stage, but a detailed molecular mechanism for such a physiological function is still lacking. In vitro studies have nevertheless provided some insight into the mechanisms of the function of enamelysin. Mmp-20 action on amelogenin in vitro promoted the formation of nanorod structures formed via co-assembly of the parent amelogenin with its proteolytic products (88). Such assembly alteration was proposed to be associated with the elongated growth of apatite crystals. In general, enamelysin functions in enamel to cleave enamel matrix proteins at specific cleavage sites. Enamelysin cleaves amelogenin first at the C-terminal end to produce the truncated “20K” amelogenin lacking the hydrophilic C-terminal region. The N-terminal tyrosine-rich amelogenin polypeptide (TRAP) is cleaved later to produce the central “13” amelogenin (149). Such proteolytic activities affect amelogenin-apatite interactions by producing intermediate products that have less affinity for apatite (150). Removal of TRAP by Mmp-20 may also function to control the assembly or disassembly of amelogenin protein since the N-terminal domain has been defined as a self-assembly domain (151). It has also been shown that the presence of apatite can affect the rate of proteolysis of amelogenin by Mmp-20, making the reaction slower (150). In vitro experiments have provided some clues to the close relationship that exists between proteolysis and the process of mineralization (152). It has been suggested that Mmp-20 activity at the early stage of enamel formation produces protein intermediates that will trigger phase transformation of amorphous calcium phosphate nanopartilces into mineralized hydroxyapatite (107).
5.5.2. Kallikrein-4 (KLK4)
The human gene kallikrein-4 (KLK4) contains 5 exons and is one of a cluster of kallikrein genes located on chromosome 19q13. The gene has also been referred to as “prostase”, since at one time it was believed to demonstrate prostate-restricted expression (153). The primary function of Klk4 is to further digest the proteolystic products of amelogenin, ameloblastin, and enamelin resulting from the Mmp-20 action (154). This digestive function facilitates removal of enamel proteins, resulting in enamel hardening during the maturation stage. The first evidence to support this notion was the observation that the timing of Klk4 expression in the enamel matrix correlates with the disappearance of enamel proteins from the enamel matrix. The finding that the 32-kDa enamelin fragment is resistant to Mmp-20 action, but not to that of Klk4, is further direct evidence for the complementary action of these two proteinases (155). Klk4 digests amelogenin, and unlike Mmp-20 its activity is not significantly affected by the presence of apatite crystals in vitro (156). Klk4 null mice have a less severe phenotype than that of Mmp20 null mice (157). Enamel without Klk4 has normal thickness and typical prismatic structure but there is a delay in hardening and a defect in mineralization near the DEJ is more apparent. The less severe phenotype might well be the result of a compensatory action of a third enzyme (143, 148, 158).
5.5.3. Caldecrin (Ctrc)
Caldecrin is a serine proteinase also known as chymotrypsin C and elastase 4 whose main function is associated with trypsin degradation in the pancreas. The expression pattern of Ctrc in enamel is similar to Klk4, but lower, and Ctrc is predominantly expressed in the maturation stage of amelogenesis in rat and mice. The expression of Ctrc at both the mRNA and protein levels during the maturation stage hints at a possible role in enamel mineralization although its specific function in enamel matrix degradation has yet to be studied (143).
6. EXTRACELLAR MATRIX pH CHANGES AND REGULATION
6.1. pH changes
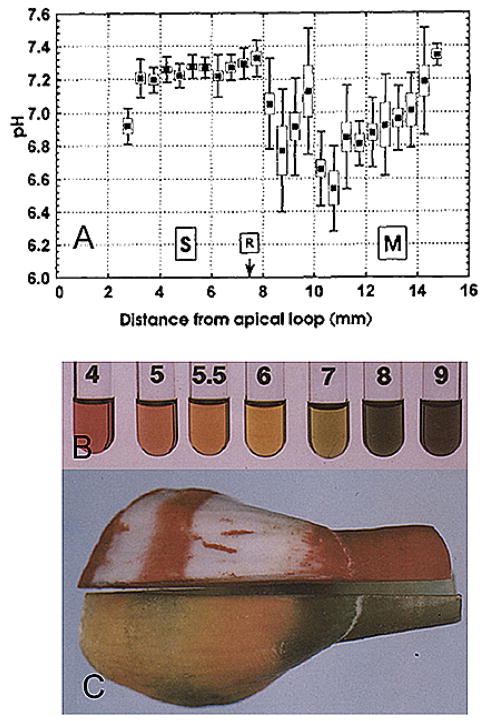
Following the precipitation of carbonated fluoro-hydroxyapatite crystals, significant amounts of protons are generated in the enamel extracellular matrix microenvironment, resulting in a marked decrease in local pH. Changes in pH during amelogenesis have been investigated in incisors from large (cow, pig, dog, monkey) and small (rat and mouse) mammals. The strategies for assessing pH have included: autoradiography (159), direct staining methods using a variety of pH indicators (98, 160), and application of microelectrodes for measurement of enamel homogenates prepared from 0.5 mm freeze-dried enamel strips dissolved in water (161). The acidic and neutral strips are correlated with the zones of “ruffle-ended” and “smooth-ended” ameloblasts (162). Studies on rat incisor strips demonstrated that a sudden drop in pH to mildly acidic conditions occurs after the early maturation stage, while during the secretory stage the pH remains fairly constant (Figure 8A). Staining of un-erupted permanent incisors from 1 year old calves with pH indicator results in a banded color pattern that represents alternate acidic (pH 5.8–6.0) and neutral (pH 7.0–7.2) zones (Figures 8B and 8C). The pH values have been confirmed by measuring the pH of corresponding enamel slices suspended in water using an electrode. Detailed analyses of pH during the development of rat incisors have confirmed similar changes (161). The significance of these morphological features observed on the apical surface of ameloblasts during maturation has been associated with the cells’ function. The “ruffle-ended” ameloblasts have tight junctions and deep membrane infoldings at their apical surfaces associated with their ability to control ion movement. In contrast, “smooth-ended” ameloblasts lack such apical invaginations and their apical junctional complexes are dissociated and “leaky” (163).
Figure 8.
(A) Categorization plot of mean pH (solid squares) ± SD (lines) and SEM (open boxes; n = 10 teeth) for enamel strips cut at 0.5-mm length across the secretory (S) and maturation (M) stages of amelogenesis (R = location of the molar reference as described by Smith et al (161)). The average measured pH of enamel strips remains fairly constant across the secretory stage (S) and into the early maturation stage (M). Reproduced with permission from reference #161. (B and C) Cyclical changes in pH in developing bovine incisor: (B) Universal indicator color standards at different pH values used to stain bovine incisor shown in C. (C) A bovine developing tooth cut in half sagittally. The left half was stained with a pH (Universal) indicator mixture for l-2 min. Alternate stripes of orange correspond to pH 5.5–6.0 and of green to pH 7.0. The right half was stained with GBHA (a calcium chelator dye) solution and rinsed with ethanol. Red stripes of staining correspond to the neutral bands of green staining with the pH indicator and unstained white zones to acidic orange zones. Orange or green coloration occured not only on the forming surface but also in depth on enamel. Reproduced with permission from (98)
6.2. pH regulation
As was pointed out by Smith (4), during amelogenesis, not only are new apatite crystals created, but considerable “post-volumetric” expansion of crystals also occurs after the appositional growth phase of enamel is completed. A buffering function in the system is therefore needed when progressive and rapid growth of enamel crystal thickness occurs, particularly during enamel maturation. Interestingly, while the pH drops at the early maturation stage, a rise in pH value is observed at the end of maturation stage indicating that active mechanisms for control of pH do indeed exist in enamel (Figure 8A). Such control mechanisms may involve programmed expression of carbonic anhydrases (CAs) which catalyze the formation of carbonic acid and production of bicarbonate from water and carbon dioxide. Bicarbonate can be formed either from dissociation of carbonic acid (H2CO3) or through the reaction of hydroxide (OH−) with carbon dioxide (CO2) (equation 1).
| (1) |
Bicarbonate is an essential component of enamel fluid, and has been suggested to be involved in buffering the extracellular environment. Evidence for the presence of bicarbonate ions (HCO3−) in enamel fluid was based on theoretical calculations. The average concentration of free bicarbonate in enamel has been estimated to be 12–27 mM depending on the stage of the life cycle of the ameloblasts (164). Local buffering capacity during amelogenesis could be achieved either by forming bicarbonate ions or by recycling excess carbonic acid back into CO2 and H2O (equation 1).
Two scenarios have been presented to account for the changes in pH conditions impacting the growth of apatite crystals (26). One scenario was proposed to be dominated by apatite nucleation (which may result from protein-ion interactions). In this scenario, formation of apatite unit cells decreases the local pH. The expression of genes involved in bicarbonate transport is then elevated and the cycle back to neutral pH continues. In the second scenario, ameloblasts specifically regulate the level of genes and bicarbonate secretion as a primary step in creating an appropriate pH microenvironment (~8) by minimizing solubility of calcium phosphate and hence inducing crystal nucleation. The release of protons resulting from apatite crystal growth then drops the pH to neutrality, and the pH cycle continues. While there is experimental evidence for cyclical pH changes during enamel development, more studies are needed to address which of the above mechanisms is responsible for the control of initiation and regulation of pH changes.
Ameloblasts in their maturation stage have been shown to express a cytoplasmic isoform of carbonic anhydrase (CA2) (165), and evidence for the expression of a secreted form of CA (CA6) in rat incisor enamel organ during the maturation stage has also been reported (166). Recent studies by Lacruz et al. (26) have demonstrated that pH homeostasis during enamel development is a complex process that might involve more than one or two forms of CA. A systematic screening of the expression of CA isoforms in mouse by RT-PCR showed that most of the 16 CA isoforms are expressed by enamel organ epithelium and ameloblast cells in vivo. The catalytic isozymes CA2, CA6, CA9 (all membrane bound) and CA13 (cytoplasmic) and the acatalytic CA11 isoform have relatively high expression levels. While the post-secretory localization of many of these CAs is not known, it has been proposed that they might have cooperative functions together with anion-exchangers (i.e. AE2) and/or biocarbonate transporters such as the electrogenic bicarbonate cotransporter (NBCe1) (36), which have known sites of localization in developing enamel. It has been proposed that NBCe1-B, which is localized basolaterally, controls the transport of bicarbonate ions into the cytoplasm, while AE2, which is expressed apically, controls the release of bicarbonate ions into the extracellular matrix from the vasculature of the enamel organ (34). CA2 and CA6 are proposed to catalyze the production of bicarbonate in the cytoplasm and in the extracellular matrix, respectively. Both AE2 and NBCe1 are ion transporters expressed by ameloblasts, and mutations in the genes encoding them result in defective enamel formation (167). Both genes belong to the SLC4 gene family with SLC4A2 encoding AE2 and SLC4A4 encoding NBCe1. The lack of Slc4a2 in mice results in edentulism (absence of teeth). Mice lacking the Slc4A4 gene have abnormal enamel formation (36).
6.3. Ion transport
Both passive (simple ion diffusion) and active (carrier-mediated) transport mechanisms have been considered to account for passage of calcium and other ions through the ameloblast layers. The former is a non-energy requiring pathway that is developed in smooth-ended ameloblasts, which are “leakey” because they lack junctional complexes at their apical poles. In the “ruffle-ended” stage, the apical poles of ameloblasts become linked by tight junctions, and ion transport involves the activity of ATPases that actively pump calcium. Even if free diffusion of H+ ions through the smooth-ended ameloblasts is possible, it does not explain the effective neutralization that is seen of the entire acid load of the enamel, since “smooth-ended” ameloblasts cover only 20% of the enamel surface in a 4–5 week old rat. Ameloblast action would not be sufficient to counteract the acidification that occurs at the peak period of mineral growth (163), supporting a mechanism that involves enzymes to control the local pH. Histochemical and immunohistochemical studies have revealed the presence of Ca+2, and Mg+2-ATPase in maturation stage ruffle-ended ameloblasts (168). Pumps for other cations such as Na+ and K+, as well as H+-ATPases have also been identified, which function to balance the osmotic pressure and regulate intracellular pH, respectively (169).
7. DEFECTIVE ENAMEL
A variety of processes can lead to damaged or defective enamel. Even though enamel is the hardest calcified tissue in the human body, being highly anti-abrasive and having a unique combination of wear and fracture resistance (8,28), exposure to acidic pH will lead to its destruction. Extremely acidic conditions in the oral cavity can lead to dental caries and/or dental erosion, conditions that result from the presence of acids in the diet, the presence of acid-forming bacteria, or acid from the stomach due to reflux disorders. While both caries and erosion are the result of enamel mineral loss due to an acidic environment, only caries formation specifically involves the presence of bacteria. Dental erosion, the loss of enamel mineral due to acid exposure not involving bacteria, has been directly associated with disorders such as bulimia nervosa and chronic gastro-oesophageal reflux (170). In addition to the possibility of such damage, the formation of enamel can also be defective from early developmental stages due to mutations in ameloblast gene products.
7.1. Amelogenesis imperfecta
Mutation in any of the genes encoding amelogenin, enamelin, MMP-20 or KLK-4 leads to one of a series of inherited diseases of enamel malformation called amelogenesis imperfecta (AI). The term AI is used to describe hereditary conditions that are clinically heterogeneous and affect dental enamel without the presence of other developmental abnormalities (18, 171). Mutations in genes encoding ion transporters also lead to enamel malformation but are not included in the category of AI because these mutations cause other abnormalities as well (36). FAM83H and WDR72 are two recently discovered genes, with critical cellular functions in enamel development, that have also been associated with AI (172, 173). In general, mutations in ameloblast genes can cause lack of secretion of the gene product, or malformation of the encoded protein or proteinase, leading to its malfunction. The physiological processes that can be affected include: cell adhesion, mineral nucleation and crystal growth, mineral organization, and proteolytic processing of the organic matrix. Depending on the protein affected and the developmental stage involved, the defective enamel could be thin (hypoplastic) or could have normal thickness but abnormal hardness and density due to hypomineralization. The hypomineralization type of AI is subdivided into hypomaturation and hypocalcification phenotypes. Most of the reported cases seen have combined phenotypes. Due to its genetic diversity, the AI trait can be transmitted by autosomal-dominant, autosomal-recessive, or X-linked modes of inheritance (18, 174, 175).
Mutations in the amelogenin AMELX gene correlate with X-linked AI (XAI). To date, there are 14 AMELX-associated AI mutations for which the clinical manifestation, protein expression patterns, and types of mutant amelogenin have been described (176). Genotype-phenotype studies have shown that mutations causing alterations in different functional domains of amelogenin result in diverse and distinct clinical manifestations. Mutations in amelogenin, and their associated phenotypes, fall within three general categories. The first category includes mutations within the signal peptide that result in a premature stop codon or prevent amelogenin secretion. These mutations are mainly associated with hypoplastic (thin) enamel, since they basically result in an enamel extracellular matrix devoid of amelogenin. The second group includes mutations affecting the N-terminal region that are associated with the hypomineralization/hypomaturation type of AI. Point mutations in the N-terminal or the self-assembly A-domain region can hinder proper folding and assembly (177, 178), inhibit lectin binding affinity (54), and disturb proteolytic processing (179). In our most recent studies, we used a series of biophysical techniques, including variable temperature tryptophan fluorescence spectroscopy, circular dichroism, and dynamic light scattering, to investigate amelogenins with point mutations similar to those identified from human DNA sequences in two cases of AI (T21I and P41T) (180). These mutant amelogenins are less stable and exhibit poor refolding ability compared to the wild-type, leading to their pre-mature aggregation. Our studies showed that AI in these cases appears to be due to the destabilization of the secondary structure of amelogenin as a result of the mutations. We postulated that alteration in self-assembly (i.e. premature aggregation) of amelogenins with N-terminal mutations may have profound effects on intra- and extracellular processes such as secretion and proteolysis of amelogenin, and its interactions with non-amelogenin as well as with the forming mineral (178). Finally, mutations that cause the loss of the C-terminal region are mainly associated with hypoplastic enamel (181). Such mutations can affect amelogenin-mineral interactions (182) as well as amelogenin self-assembly (82).
The clinical manifestation of an enamelin mutation is generally hypoplastic enamel that may either be smooth or have localized horizontal bands of thin enamel. In some cases, open-bite mal-occlusion is also observed. Enamel phenotypes of ENAM mutations may be dose-dependent, with generalized hypoplastic AI segregating as a recessive trait and localized enamel pitting segregating as a dominant trait (183). To date, ten AI-causing mutations in the ENAM gene have been identified (176). The first case of enamelin-associated AI was reported by Rajpar and colleagues who analyzed a family with an autosomal dominant, hypoplastic form of AI (ADAI) (Amelogenesis imperfecta 2, hypoplastic local, AIH2) and showed a mutation in the splice donor site of intron 7 (184). The position of this mutation would have an impact on RNA processing, which in turn would impact protein expression. A second family pedigree has been described with a mutation in the enamelin gene at an exon-intron boundary that also results in AIH2 (185). Mardh and colleagues have described a nonsense mutation in the amino-terminal region of the enamelin gene that effectively blocks the production of enamelin, and this mutation is also responsible for an AIH2 phenotype (186). Other reports of mutations to the enamelin gene resulting in an AIH2 enamel phenotype have been published by Kim and colleagues (187). The first report of autosomal-recessive AI (ARAI) having a “dose dependent” association with an ENAM mutation was described by Hart et al. in 2003 in three probands (188). These researchers found that when the individuals were homozygous for an ENAM mutation on chromosome 4, they showed enamel pitting with an anterior open bite, whereas heterozygosity presented only with enamel pitting. It is interesting to note that in at least four of the mutations identified to date, the 32-kDa fragment is either missing, or affected by mutation or lack of phosphorylation, indicating the important function of this fragment in enamel mineralization (184, 185, 189, 190) (see also 5.3.3).
A single report exists describing an autosomal recessive hypomaturation form of AI in which the offending mutation is within the KLK4 gene locus (191, 192). The resulting protein is truncated and lacks the serine at position 207, which is essential for function of the enzyme. Due to defective enzymatic activity, the apatite crystals grow in length but the growth in thickness is inhibited. Affected individuals from this family show a marked discoloration in both the primary and permanent teeth. Many similar features have been observed when comparing the dental phenotypes of probands bearing mutations in KLK4 and MMP-20. Kim et al have identified a homozygous mutation in the MMP-20 gene in two affected members of a family with autosomal-recessive pigmented hypomaturation AI with the surfaces of the teeth being mottled and rough (193).
Becerik et al evaluated seven Turkish families segregating autosomal recessive AI (ARAI) and did not find any mutations in the major candidate genes discussed above and known to cause AI. Their studies indicated that additional genes are responsible for a significant number of cases of ARAI in the Turkish population (194).
8. REBUILDING ENAMEL
Interest in creation of “synthetic enamel” as a future dental restorative material has been growing among scientists in the fields of dentistry, material science, and engineering (195, 196). The need to develop improved dental restorative materials with mechanical and esthetic attributes close to those of natural enamel is justified because, unlike bone, enamel does not regenerate itself.
8.1. Enamel-like material synthesis
Investigators have applied a few well-known approaches to enamel-like material synthesis. The hydroxyapatite crystals isolated from mature enamel are unusually long and different in shape from those produced under ambient conditions in the laboratory. By applying certain chemical synthetic techniques under extreme laboratory conditions, the synthesis of enamel-like hydroxyapatite nanorods (organized bundles of apatite crystals) is possible. The mineralization-related surfactant or chelate used must have sufficient affinity to hydroxyapatite crystals to prevent the crystal from growing too fast and to guide growth orientation. A hydrothermal method using EDTA-chelated calcium ions has been applied to deposit perfect nano-rod fluoroapatite crystal arrays on a stainless steel surface in a pattern resembling enamel (197). Investigators have attempted to precipitate an apatite coating on an enamel surface by electrolytic deposition ELD at 3–85 °C, but the crystals did not show an enamel-like structure (198). Others have demonstrated that phosphoric acid and fluoroapatite pastes can be used as feasible materials to form a hard repair layer on an enamel surface (198, 199). The layer was found to be organized as parallel fluoroapatite crystals without obvious structural gaps. A very recent report demonstrated that organized fluoroapatite prisms can be directly prepared on an enamel surface at pH 6.0, 37 °C in the presence of a HEDTA chelate and a high concentration (0.1 M) of fluoride (200). Others have reported an etching approach at 80 °C to prepare a nano-HAP array on a calcium silicate glass substrate (201).
8.2. Biomimetic enamel synthesis and remineralization
Although the protein content is low in mature enamel, the presence of proteins is critical and their function in maintaining good fracture toughness of the enamel may be significant. Under such non-physiological conditions as those described in the previous section, however, preparation of protein-containing nanocomposite is not feasible. It is therefore important to develop biomimetic methods that do not require extreme pH and temperature conditions.
Advances in understanding the structure, function, and pattern of expression of ameloblast gene products, together with knowledge of the nature of the mineral phase and its interaction with the organic matrix, allows us to develop techniques for preparing enamel-like material in the laboratory in a cell free system. We now understand that the control of protein self-assembly and simultaneous calcium phosphate crystallization is a basis for the design and development of enamel-like material (196). The key factors would be to carefully control the appropriate conditions (i.e. pH) and the timing of addition of the proteins to the system. A number of in vitro experimental approaches from different laboratories have collectively demonstrated amelogenin’s ability to control particular aspects of calcium phosphate mineralization including stabilization of ACP phases, promotion of nucleation, modulation of crystal morphology, and control of crystal orientation. Understanding these control mechanisms is the key to design and development of biomimetic materials and bio-inspired nanocomposites (202).
A cation-selective membrane system was originally used for synthesis of enamel-like mineral structures under physiological conditions (203). The synthetic octacalcium phosphate and hydroxyapatite formed were in the form of submicron rod-like arrays. Although the resulting material was a purely inorganic crystal, the conditions used in this biomimetic strategy were mild enough to allow incorporation of proteins, and therefore seemed appropriate for preparation of an enamel-like nanocomposite. Following recent advances in biomimetic material synthesis, new strategies have been emerging based on the finding that protein and peptide additives can control critical stages of mineralization such as nucleation and growth organization. One of the first steps towards synthesis of an amelogenin-based composite under biomimetic conditions took advantage of the cation-selective membrane method by applying a dual membrane system and testing the effects of added amelogenin protein on octacalcium phosphate crystal growth. Elongated rodlike crystals were formed, and it was suggested that amelogenin may have bound to the (010) side faces of OCP to promote the formation of these structures. A nanocomposite was assembled in the presence of amelogenin and the nature of the crystals was revealed by AFM, SEM and X-ray powder diffraction (105).
The technique of electrolytic deposition (ELD) has been used in our laboratory to fabricate an enamel-mimicking composite coating from a solution containing calcium, phosphate ions, and soluble recombinant amelogenin proteins, at near physiological pH and ionic strength (10). The advantage of this technique is that local pH near the cathode is increased gradually by continuous electrochemical reactions, resulting in supersaturation of the calcium phosphate mineral phase of interest and crystal nucleation on the surface of the cathode. This increase of pH also induces simultaneous amelogenin self-assembly. In vitro studies showed that solubility and assembly of amelogenin, and even apatite crystal growth in the presence of amelogenin, were dependent on the pH. Fan et al. used recombinant porcine amelogenin (rP172) to prepare an enamel-mimicking composite coating (rP172/CaP) and to study interaction of amelogenin with the growing crystals in the nano-composite. A thicker calcium phosphate coating (300–600 nm) was formed around the Si cathode in the presence of amelogenin compared to the 100 nm thickness formed without amelogenin, indicating that amelogenin promoted mineral deposition. SEM has revealed significant effects of rP172 amelogenin on the surface topography and crystal morphology of the coating when compared to control coatings formed in the absence of protein. The control surface appeared more porous, with numerous cracks, and flaky crystals could be seen on the surface. In contrast, the surface of rP172/CaP appeared to be much denser with fewer cracks. Rod-like crystalline bundles 30 nm in diameter and 100–200 nm in length were observed on the surface of the rP172/CaP under SEM observation. Importantly, the estimated fracture toughness of rP172/CaP (0.47 ± 0.09 MPa.m1/2) was comparable to that of the enamel surface (0.33 ± 0.03 MPa.m1/2) under the same test conditions. Testing using a cubic corner tip showed that a 0.2g load could produce cracks in the surface of mature human enamel, whereas a much higher load of 1.5g was necessary to cause cracks in the rP172/CaP composite coating. Our studies using ELD suggested that full-length amelogenin is essential for the growth of organized structures on the surface of the cathode and that amelogenin self-assembly is the driving force for crystal organization. The next step is to develop techniques for application of amelogenin under conditions relevant to the clinical setting.
A potentially promising clinical application of biomimetic production of enamel-like material in dentistry would be the in situ remineralization of enamel in the presence of amelogenin, enamelin, and related enzymes (152, 196). Developing remineralization techniques would be beneficial and useful for the repair of mineral loss caused by early carious lesions. In the laboratory, we have used a strategy based on application of proteins that are known to control crystal initiation, crystal shape, and packing organization. We have implemented a modified biomimetic approach in the presence of mineralization-modulating amelogenin to rebuild enamel structure. The approach used an acid-etched enamel surface as a model for demineralized enamel (204), and the coating was prepared by simply immersing a tooth slice in the biomimetic calcification solution. We have reported that fluoride had a significant effect on the calcium phosphate crystal morphology in the remineralized enamel. Needle-shaped nanocrystals grew on the acid-etched enamel, and amelogenin (used at optimal concentration of 33 microgram/ml) promoted the formation of oriented bundles of fluoridated hydroxyapatite (Figure 9). The effect of amelogenin on mineral morphology was to facilitate organized crystal formation. In the presence of amelogenin, the minerals formed were mainly OCP in the absence of fluoride and changed to HAP after adding 1 mg/L fluoride. The minimum concentration of recombinant amelogenin rP172 required to mediate organized crystal bundle formation in a calcification system, at pH=7.4–7.6 and physiological temperature, was 33–50 microgram/mL. More recently, it was demonstrated that the concentration of fluoride and the degree of supersaturation of the hydroxyapatite have significant effects on crystal morphology and crystal organization, which varied from plate-like loose crystals to rod-like densely packed nanocrystal arrays (205). Amelogenin promoted the formation of oriented bundles of the needle-like fluoridated hydroxyapatite crystals in a dose dependent manner.
Figure 9.
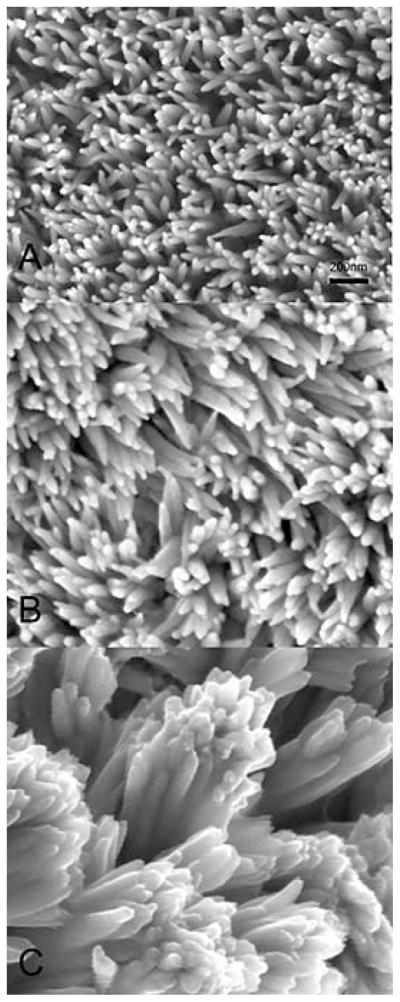
SEM images of fluoridated apatite–amelogenin coatings prepared on enamel surface (longitudinal sections) with 0 (A) 20 (micrograms/mL (B) 33 micrograms/mL (C) of rP172 amelogenin in the mineralization solution and 1 mg/L F. All the images were taken under the same magnification (204).
9. SUMMARY AND PERSPECTIVE
The developmental mechanisms that generate enamel not only provide a strong protective covering for our teeth, but also represent an outstanding biomineralizing system that offers material scientists a wealth of information on the principles and mechanisms of biological mineralization. In the last few decades, with the advances in the understanding of the genetics and proteomics of enamel, together with novel imaging and spectroscopy techniques, our understanding of the mechanisms of enamel formation has greatly improved. We now have more information on the enamel extracellular matrix components, their tissue localization, patterns of expression, chromosomal gene localization, and assembly and degradation. In vitro chemical models and in vivo studies using cell cultures and animal models have taught us that the highly controlled process of enamel biomineralization is the result of programmed expression, secretion, assembly, and degradation of ameloblast gene products (i.e. amelogenins, enamelins, amelobastins, and proteinases). Identification of molecular defects in ameloblast genes and their association with amelogenesis imperfecta (AI) have greatly expanded our knowledge of the importance of enamel proteins in the development of enamel. All of these protein components are critical for normal enamel formation and each may play more than one physiological functional role. Biophysical and computational analyses reveal that many of the extracellular proteins belong to the intrinsically disordered family of proteins. In the case of amelogenin, it is reasonable to propose that the conformational plasticity in the structure of the protein explains its potential to specifically interact with different targets (i.e. mineral, non-amelogenin proteins, cell surfaces, proteinases) and may contribute to its strong tendency to self-assemble into spherical and elongated morphologies. The growth of organized rods and inter-rods in three dimensions is orchestrated and determined by the paths of secretory ameloblasts moving from the DEJ towards the outer tooth surface. The flux of ions into, and from the ameloblasts is tightly controlled by ion transporters and ion channels some of which are also involved in controlling local pH in the extracellular matrix.
There are still gaps with respect to identification and characterization of some intracellular and extracellular components (i.e. ion channels), and in our understanding of the detailed underlying mechanisms by which ameloblast cell products assemble and function to control the processes of nucleation and oriented growth of crystals in this highly mineralized and remarkably organized bioceramic. In particular, information on the structural properties of the non-amelogenin protein components of enamel matrix, and their interactions with each other and with the mineral phase and ameloblast cell surface is yet to be studied.
The knowledge we now have of the genetics and biochemistry of enamel provides a valuable foundation for development of biomaterials with composition and structures similar to enamel. A major challenge remains, however, in efforts to synthesize artificial enamel, and that is the fabrication of enamel’s complex hierarchical prism and interprismatic structures.
Acknowledgments
Research in the laboratory of Prof. J Moradian-Oldak is supported by NIH-NIDCR grants DE-015644, DE-13414, and DE-020099. I would like to thank the students and postdoctoral research associates who worked in my laboratory over the last several years and contributed to the research findings reported in this review.
Abbreviations
- ACP
amorphous calcium phosphate
- AI
amelogenesis imperfecta
- AFM
atomic force microscopy
- Amel
amelogenin
- Ambn
ameloblastin
- Ctrc
caldecrin
- CD
circular dichroism
- DEJ
dentin-enamel junction
- DLS
dynamic light scattering
- Enam
enamelin
- FAP
fluoroapatite
- FTIR
fourier transform infrared
- HAP
hydroxyapatite
- IDP
intrinsically disordered protein
- IEE
inner enamel epithelium
- KLK-4
kallekrin-4
- MMP-20
matrix metalloproteinase 20
- LRAP
Leucine Rich Amelogenin Polypeptide
- OCP
octacalcium phosphate
- OEE
outer enamel epithelium
- rP172
recombinant porcine amelogenin (full-length)
- rP148
recombinant porcine amelogenin (truncated)
- TRAP
tyrosine rich amelogenin polypeptide
- TEM
transmission electron microscopy
- SAXS
small angle X-ray scattering
- SEM
scanning electron microscopy
- XANES
X-ray absorption near edge structure
References
- 1.Lowenstam HA, Weiner S. On Biomineralization. Oxford University Press; New York: 1989. [Google Scholar]
- 2.Moradian-Oldak J, Paine ML. Mammalian Enamel Formation. In: Sigel Astrid, Sigel H, Sigel RKO., editors. Metal Ions In Life Sciences. John Wiley & Sons, Ltd; Chichester: 2008. [Google Scholar]
- 3.Fincham A, Moradian-Oldak J, Simmer J. The structural biology of the developing dental enamel matrix. J Struct Biol. 1999;126(3):270–99. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 4.Smith C. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 5.Robinson C, Kirkham J, Brookes SJ, Bonass WA, Shore RC. The chemistry of enamel development. Int J Dev Biol. 1995;39(1):145–52. [PubMed] [Google Scholar]
- 6.Robinson C, Fuchs P, Deutsch D, Weatherell JA. Four chemically distinct stages in developing enamel from bovine incisor teeth. Caries Res. 1978;12(1):1–11. doi: 10.1159/000260309. [DOI] [PubMed] [Google Scholar]
- 7.Glick PL. Patterns of enamel maturation. J Dent Res. 1979;58(Spec Issue B):883–95. doi: 10.1177/00220345790580024201. [DOI] [PubMed] [Google Scholar]
- 8.Boyde A. Microstructure of Enamel. Dental Enamel; Ciba Foundation Symposium; New York: John Wiley & Sons; 1997. [Google Scholar]
- 9.Sarikaya M. Biomimetics: materials fabrication through biology. Proc Natl Acad Sci USA. 1999;96:13611–13614. doi: 10.1073/pnas.96.25.14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan Y, Sun Z, Wang R, Abbott C, Moradian-Oldak J. Enamel inspired nanocomposite fabrication through amelogenin supramolecular assembly. Biomaterials. 2007;28(19):3034–42. doi: 10.1016/j.biomaterials.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mann S. Molecular tectonics in biomineralization and biomimetic materials chemistry. Nature. 1993;365:499–505. [Google Scholar]
- 12.Palmer LC, Newcomb CJ, Kaltz SR, Spoerke ED, Stupp SI. Biomimetic Systems for Hydroxyapatite Mineralization Inspired By Bone and Enamel. Chemical Reviews. 2008;108(11):4754–4783. doi: 10.1021/cr8004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall GW, Balooch M, Gallagher RR, Gansky SA, Marshall SJ. Mechanical properties of the dentinoenamel junction: AFM studies of nanohardness, elastic modulus, and fracture. J Biomed Mater Res. 2001;54(1):87–95. doi: 10.1002/1097-4636(200101)54:1<87::aid-jbm10>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 14.Habelitz S, Marshall SJ, Marshall GW, Jr, Balooch M. Mechanical properties of human dental enamel on the nanometre scale. Arch Oral Biol. 2001;46(2):173–83. doi: 10.1016/s0003-9969(00)00089-3. [DOI] [PubMed] [Google Scholar]
- 15.White SN, Luo W, Paine ML, Fong H, Sarikaya M, Snead ML. Biological organization of hydroxyapitite crystallites into a fibrous continuum toughens and controls anisotropy in human enamel. J Dent Res. 2001;80:321–326. doi: 10.1177/00220345010800010501. [DOI] [PubMed] [Google Scholar]
- 16.Cui FZ, Ge J. New observations of the hierarchical structure of human enamel, from nanoscale to microscale. Journal of Tissue Engineering and Regenerative Medicine. 2007;1(3):185–191. doi: 10.1002/term.21. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Machule D, Gao C, DenBesten PK. Growth of ameloblast-lineage cells in a three-dimensional Matrigel environment. Eur J Oral Sci. 2006;114(Suppl 1):159–63. doi: 10.1111/j.1600-0722.2006.00308.x. discussion 164-5, 380-1. [DOI] [PubMed] [Google Scholar]
- 18.Stephanopoulos G, Garefalaki ME, Lyroudia K. Genes and related proteins involved in amelogenesis imperfecta. J Dent Res. 2005;84(12):1117–26. doi: 10.1177/154405910508401206. [DOI] [PubMed] [Google Scholar]
- 19.Hu JC, Simmer JP. Developmental biology and genetics of dental malformations. Orthod Craniofac Res. 2007;10(2):45–52. doi: 10.1111/j.1601-6343.2007.00384.x. [DOI] [PubMed] [Google Scholar]
- 20.Sire JY, Davit-Beal T, Delgado S, Gu X. The origin and evolution of enamel mineralization genes. Cells Tissues Organs. 2007;186(1):25–48. doi: 10.1159/000102679. [DOI] [PubMed] [Google Scholar]
- 21.Margolis HC, Beniash E, Fowler CE. Role of macromolecular assembly of enamel matrix proteins in enamel formation. J Dent Res. 2006;85(9):775–93. doi: 10.1177/154405910608500902. [DOI] [PubMed] [Google Scholar]
- 22.Hu JC, Yamakoshi Y, Yamakoshi F, Krebsbach PH, Simmer JP. Proteomics and genetics of dental enamel. Cells Tissues Organs. 2005;181(3–4):219–31. doi: 10.1159/000091383. [DOI] [PubMed] [Google Scholar]
- 23.Robinson C, Kirkham J, Shore R. Dental Enamel: Formation to Destruction. CRC Press; Boca Raton, FL: 1995. [Google Scholar]
- 24.Simmer JP, Papagerakis P, Smith CE, Fisher DC, Rountrey AN, Zheng L, Hu JC. Regulation of dental enamel shape and hardness. J Dent Res. 2010;89(10):1024–38. doi: 10.1177/0022034510375829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartlett J, Ganss B, Goldberg M, Moradian-Oldak J, Paine M, Snead M, Wen X, White S, Zhou Y. 3. Protein-protein interactions of the developing enamel matrix. Curr Top Dev Biol. 2006;74:57–115. doi: 10.1016/S0070-2153(06)74003-0. [DOI] [PubMed] [Google Scholar]
- 26.Lacruz RS, Nanci A, Kurtz I, Wright JT, Paine ML. Regulation of pH During Amelogenesis. Calcif Tissue Int. 2010;86(2):91–103. doi: 10.1007/s00223-009-9326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nanci A. Enamel. Composition, Formation, and Structure. In: Nanci A, editor. Ten Cate’s Oral Histology Development, Structure, and Function. Mosbey Elsevier; St. Louis: 2008. [Google Scholar]
- 28.Chai H, Lee JJ, Constantino PJ, Lucas PW, Lawn BR. Remarkable resilience of teeth. Proc Natl Acad Sci U S A. 2009;106(18):7289–93. doi: 10.1073/pnas.0902466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thesleff I. Homeobox genes and growth factors in regulation of craniofacial and tooth morphogenesis. ACTA Odontol Scand. 1995;53:129–134. doi: 10.3109/00016359509005962. [DOI] [PubMed] [Google Scholar]
- 30.Warshawsky H, Smith CE. Morphological classification of rat incisor ameloblasts. Anat Rec. 1974;179(4):423–46. doi: 10.1002/ar.1091790403. [DOI] [PubMed] [Google Scholar]
- 31.Ronnholm E. An electron microscopic study of the amelogenesis in human teeth. I. The fine structure of the ameloblasts. J Ultrastruct Res. 1962;6:229–48. doi: 10.1016/s0022-5320(62)90055-2. [DOI] [PubMed] [Google Scholar]
- 32.Ronnholm E. The amelogenesis of human teeth as revealed by electron microscopy. II. The development of the enamel crystallites. J Ultrastruct Res. 1962;6:249–303. doi: 10.1016/s0022-5320(62)80036-7. [DOI] [PubMed] [Google Scholar]
- 33.Beniash E, Metzler RA, Lam RSK, Gilbert P. Transient amorphous calcium phosphate in forming enamel. J Stru Biol. 2009;166(2):133–143. doi: 10.1016/j.jsb.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skobe SDNZ, Prostak S. In: Dental Enamel: Formation to Distruction. Robinson KJC, Shore R, editors. CRC Press; Boca Raton: 1995. [Google Scholar]
- 35.Lacruz RS, Hilvo M, Kurtz I, Paine ML. A survey of carbonic anhydrase mRNA expression in enamel cells. Biochem Biophys Res Commun. 2010;393(4):883–7. doi: 10.1016/j.bbrc.2010.02.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lacruz RS, Nanci A, White SN, Wen X, Wang H, Zalzal SF, Luong VQ, Schuetter VL, Conti PS, Kurtz I, Paine ML. The sodium bicarbonate cotransporter (NBCe1) is essential for normal development of mouse dentition. J Biol Chem. 2010;285(32):24432–8. doi: 10.1074/jbc.M110.115188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau EC, Mohandas T, Shapiro LJ, Slavkin HC, Snead ML. Human and mouse amelogenin gene loci are on the sex chromosome. Genomics. 1989;4:162–168. doi: 10.1016/0888-7543(89)90295-4. [DOI] [PubMed] [Google Scholar]
- 38.Snead ML, Zeichner-David M, Chandra T, Robson KJ, Woo SL, Slavkin HC. Construction and identification of mouse amelogenin cDNA clones. Proc Natl Acad Sci U S A. 1983;80(23):7254–8. doi: 10.1073/pnas.80.23.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapman VM, Keitz BT, Disteche CM, Lau EC, Snead ML. Linkage of amelogenin (Amel) to the distal portion of the mouse X chromosome. Genomics. 1991;10:23–28. doi: 10.1016/0888-7543(91)90479-x. [DOI] [PubMed] [Google Scholar]
- 40.Prakash SK, Gibson CW, Wright JT, Boyd C, Cormier T, Sierra R, Li Y, Abrams WR, Aragon MA, Yuan ZA, van den Veyver IB. Tooth enamel defects in mice with a deletion at the Arhgap 6/Amel X locus. Calcif Tissue Int. 2005;77(1):23–9. doi: 10.1007/s00223-004-1213-7. [DOI] [PubMed] [Google Scholar]
- 41.Baba O, Takahashi N, Terashima T, Li W, DenBesten PK, Takano Y. Expression of alternatively spliced RNA transcripts of amelogenin gene exons 8 and 9 and its end products in the rat incisor. J Histochem Cytochem. 2002;50(9):1229–36. doi: 10.1177/002215540205000910. [DOI] [PubMed] [Google Scholar]
- 42.Sire JY, Delgado S, Fromentin D, Girondot M. Amelogenin: lessons from evolution. Arch Oral Biol. 2005;50(2):205–12. doi: 10.1016/j.archoralbio.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Hu CC, Bartlett JD, Zhang CH, Qian Q, Ryu OH, Simmer Cloning JP. cDNA sequence, and alternative splicing of porcine amelogenin mRNAs. J Dent Res. 1996;75(10):1735–41. doi: 10.1177/00220345960750100501. [DOI] [PubMed] [Google Scholar]
- 44.Gibson CW, Golub EE, Abrams WR, Shen G, Ding W, Rosenbloom J. Bovine amelogenin message heterogeneity: alternative splicing and Y-chromosomal gene transcription. Biochemistry. 1992;31(35):8384–8. doi: 10.1021/bi00150a036. [DOI] [PubMed] [Google Scholar]
- 45.Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276(34):31871–5. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 46.Lyngstadaas SP, Risnes S, Sproat BS, Thrane PS, Prydz HP. A synthetic, chemically modified ribozyme eliminates amelogenin, the major translation product in developing mouse enamel in vivo. EMBO J. 1995;14(21):5224–5229. doi: 10.1002/j.1460-2075.1995.tb00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diekwisch TG, David S, Bringas P, Santos V, Slavkin HC. Antisense inhibition of amelogenin translation demonstrates supramolecular controls for enamel HAP crystal-growth during embryonic mouse molar development. Development. 1993;117:471–482. doi: 10.1242/dev.117.2.471. [DOI] [PubMed] [Google Scholar]
- 48.Paine ML, Luo W, Zhu DH, Bringas PJ, Snead ML. Functional domains for amelogenin revealed by compound genetic defects. J Bone Miner Res. 2003;18:466–472. doi: 10.1359/jbmr.2003.18.3.466. [DOI] [PubMed] [Google Scholar]
- 49.Gibson CW. The amelogenin “enamel proteins” and cells in the periodontium. Crit Rev Eukaryot Gene Expr. 2008;18(4):345–60. doi: 10.1615/critreveukargeneexpr.v18.i4.30. [DOI] [PubMed] [Google Scholar]
- 50.Veis A. Amelogenin gene splice products: potential signaling molecules. Cell Mol Life Sci. 2003;60(1):38–55. doi: 10.1007/s000180300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warotayanont R, Zhu D, Snead ML, Zhou Y. Leucine-rich amelogenin peptide induces osteogenesis in mouse embryonic stem cells. Biochem Biophys Res Commun. 2008;367(1):1–6. doi: 10.1016/j.bbrc.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deutsch D, Haze-Filderman A, Blumenfeld A, Dafni L, Leiser Y, Shay B, Gruenbaum-Cohen Y, Rosenfeld E, Fermon E, Zimmermann B, Haegewald S, Bernimoulin JP, Taylor AL. Amelogenin, a major structural protein in mineralizing enamel, is also expressed in soft tissues: brain and cells of the hematopoietic system. Eur J Oral Sci. 2006;114(Suppl 1):183–9. doi: 10.1111/j.1600-0722.2006.00301.x. discussion 201-2, 381. [DOI] [PubMed] [Google Scholar]
- 53.Fincham AG, Moradian-Oldak J. Amelogenin post-translational modifications: carboxy-terminal processing and the phosphorylation of bovine and porcine “TRAP” and “LRAP” amelogenins. Biochem Biophys Res Commun. 1993;197(1):248–55. doi: 10.1006/bbrc.1993.2468. [DOI] [PubMed] [Google Scholar]
- 54.Ravindranath RM, Moradian-Oldak J, Fincham AG. Tyrosyl motif in amelogenins binds N-acetyl-D-glucosamine. J Biol Chem. 1999;274(4):2464–71. doi: 10.1074/jbc.274.4.2464. [DOI] [PubMed] [Google Scholar]
- 55.Paine ML, White SN, Luo W, Fong H, Sarikaya M, Snead ML. Regulated gene expression dictates enamel structure and tooth function. Matrix Biol. 2001;20:273–292. doi: 10.1016/s0945-053x(01)00153-6. [DOI] [PubMed] [Google Scholar]
- 56.Katz EP, Seyer J, Levine PT, Glimcher MJ. The comparative biochemistry of the organic matrix of developing enamel--II. Ultracentrifugal and electrophoretic characterization of proteins soluble at neutral pH. Arch Oral Biol. 1969;14(5):533–9. doi: 10.1016/0003-9969(69)90146-0. [DOI] [PubMed] [Google Scholar]
- 57.Mechanic GL, Katz EP, Glimcher MJ. The Sephadex gel filtration characteristics of the neutral soluble proteins of embryonic bovine enamel. Biochim Biophys Acta. 1967;133(1):97–113. doi: 10.1016/0005-2795(67)90042-6. [DOI] [PubMed] [Google Scholar]
- 58.Delak K, Harcup C, Lakshminarayanan R, Sun Z, Fan Y, Moradian-Oldak J, Evans JS. The tooth enamel protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry. 2009;48(10):2272–81. doi: 10.1021/bi802175a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bonar LC, Glimcher MJ, Mechanic GL. The molecular structure of the neutral-soluble proteins of embryonic bovine enamel in the solid state. J Ultrastruct Res. 1965;13(3):308–17. doi: 10.1016/s0022-5320(65)80079-x. [DOI] [PubMed] [Google Scholar]
- 60.Goto Y, Kogure E, Takagi T, Aimoto S, Aoba T. Molecular conformation of porcine amelogenin in solution: three folding units at the N-terminal, central and C-terminal regions. J Biochem. 1993;113(113):55–60. doi: 10.1093/oxfordjournals.jbchem.a124003. [DOI] [PubMed] [Google Scholar]
- 61.Renugopalakrishnan V, Pattabiraman N, Prabhakaran M, Strawich E, Glimcher MJ. Tooth enamel protein, amelogenin, has a probable beta-spiral internal channel, Gln112-Leu138, within a single polypeptide chain: preliminary molecular mechanics and dynamics studies. Biopolymers. 1989;28(1):297–303. doi: 10.1002/bip.360280130. [DOI] [PubMed] [Google Scholar]
- 62.Termine JD, Torchia DA, Conn KM. Enamel matrix: structural proteins. J Dent Res. 1979;58(Spec Issue B):773–81. doi: 10.1177/00220345790580022901. [DOI] [PubMed] [Google Scholar]
- 63.Moradian-Oldak J, Lakshminarayanan R. Intrinsic disorder in amelogenin. In: Goldberg M, editor. Amelogenins; Multifaceted proteins for dental and bone formation and repair. Bentham Books Ltd; Dubai: 2010. [Google Scholar]
- 64.Lakshminarayanan R, Yoon I, Hegde BG, Fan D, Du C, Moradian-Oldak J. Analysis of secondary structure and self-assembly of amelogenin by variable temperature circular dichroism and isothermal titration calorimetry. Proteins. 2009;76(3):560–9. doi: 10.1002/prot.22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18(6):756–64. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 66.Bienkiewicz EA, Moon Woody A, Woody RW. Conformation of the RNA polymerase II C-terminal domain: circular dichroism of long and short fragments. J Mol Biol. 2000;297(1):119–33. doi: 10.1006/jmbi.2000.3545. [DOI] [PubMed] [Google Scholar]
- 67.Lakshminarayanan R, Fan D, Du C, Moradian-Oldak J. The role of secondary structure in the entropically driven amelogenin self-assembly. Biophys J. 2007;93(10):3664–74. doi: 10.1529/biophysj.107.113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoang AM, Klebe RJ, Steffensen B, Ryu OH, Simmer JP, Cochran DL. Amelogenin is a cell adhesion protein. J Dent Res. 2002;81(7):497–500. doi: 10.1177/154405910208100713. [DOI] [PubMed] [Google Scholar]
- 69.Ndao M, Dutta K, Bromley KM, Lakshminarayanan R, Sun Z, Rewari G, Moradian-Oldak J, Evans JS. Probing the self-association, intermolecular contacts, and folding propensity of amelogenin. Protein Sci. 2011;20(4):724–34. doi: 10.1002/pro.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bromley KM, Kiss AS, Lokappa SB, Laksminarayanan R, Fan D, Ndao M, Evans JS, Moradian-Oldak J. Dissecting amelogenin nanospheres: Characterization of metastable oligomers. J Biol Chem. 2011 Aug 12; doi: 10.1074/jbc.M111.250928. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen C-L, Bromley KM, Moradian-Oldak J, DeYoreo J. In situ AFM Study of Amelogenin Assembly and Dis-assembly Dynamics on Charged Surfaces provides Insights on Matrix Protein Self-Assembly. J Am Chem Soc. 2011 doi: 10.1021/ja206849c. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science. 2005;307(5714):1450–4. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- 73.Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Margolis HC. pH triggered self-assembly of native and recombinant amelogenins under physiological pH and temperature in vitro. J Struct Biol. 2007;160(1):57–69. doi: 10.1016/j.jsb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He X, Wu S, Martinez-Avila O, Cheng Y, Habelitz S. Self-aligning amelogenin nanoribbons in oil-water system. J Struct Biol. 2011;174(1):203–12. doi: 10.1016/j.jsb.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fincham AG, Leung W, Tan J, Moradian-Oldak J. Does amelogenin nanosphere assembly proceed through intermediary-sized structures? Connect Tissue Res. 1998;38(1–4):237–40. doi: 10.3109/03008209809017042. discussion 241-6. [DOI] [PubMed] [Google Scholar]
- 76.Fang PA, Margolis HC, Conway JF, Simmer JP, Dickinson GH, Beniash E. Cryogenic Transmission Electron Microscopy Study of Amelogenin Self-Assembly at Different pH. Cells Tissues Organs. 2011;194(2–4):166–170. doi: 10.1159/000324250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nikiforuk G, Simmons NS. Purification and properties of protein from embryonic bovine enamel. J Dent Res. 1965;44(6 Suppl):1119–22. doi: 10.1177/00220345650440061101. [DOI] [PubMed] [Google Scholar]
- 78.Fincham A, Moradian-Oldak J, Simmer J, Sarte P, Lau E, Diekwisch T, Slavkin H. Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J Struct Biol. 1994;112(2):103–9. doi: 10.1006/jsbi.1994.1011. [DOI] [PubMed] [Google Scholar]
- 79.Moradian-Oldak J. The emergence of “nanospheres” as basic structural components adopted by amelogenin. J Dent Res. 2007;86(6):487–90. doi: 10.1177/154405910708600603. [DOI] [PubMed] [Google Scholar]
- 80.Moradian-Oldak J, Simmer JP, Lau EC, Sarte PE, Slavkin HC, Fincham AG. Detection of monodisperse aggregates of a recombinant amelogenin by dynamic light scattering. Biopolymers. 1994;34(10):1339–47. doi: 10.1002/bip.360341006. [DOI] [PubMed] [Google Scholar]
- 81.Paine CT, Paine ML, Snead ML. Identification of tuftelin- and amelogenin-interacting proteins using the yeast two-hybrid system. Connect Tissue Res. 1998;38(1–4):257–67. doi: 10.3109/03008209809017046. discussion 295-303. [DOI] [PubMed] [Google Scholar]
- 82.Moradian-Oldak J, Paine ML, Lei YP, Fincham AG, Snead ML. Self-assembly properties of recombinant engineered amelogenin proteins analyzed by dynamic light scattering and atomic force microscopy. J Struct Biol. 2000;131(1):27–37. doi: 10.1006/jsbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- 83.Aichmayer B, Margolis HC, Sigel R, Yamakoshi Y, Simmer JP, Fratzl P. The onset of amelogenin nanosphere aggregation studied by small-angle X-ray scattering and dynamic light scattering. J Struct Biol. 2005;151(3):239–49. doi: 10.1016/j.jsb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 84.Tarasevich BJ, Lea S, Bernt W, Engelhard MH, Shaw WJ. Changes in the quaternary structure of amelogenin when adsorbed onto surfaces. Biopolymers. 2009;91(2):103–7. doi: 10.1002/bip.21095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beniash E, Simmer JP, Margolis HC. The effect of recombinant mouse amelogenins on the formation and organization of hydroxyapatite crystals in vitro. J Struct Biol. 2005;149(2):182–90. doi: 10.1016/j.jsb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 86.He X, Li W, Habelitz S. The cooperative self-assembly of 25 and 23kDa amelogenins. J Struct Biol. 2008;164(3):314–21. doi: 10.1016/j.jsb.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Habelitz S, Kullar A, Marshall SJ, DenBesten PK, Balooch M, Marshall GW, Li W. Amelogenin-guided crystal growth on fluoroapatite glass-ceramics. J Dent Res. 2004;83(9):698–702. doi: 10.1177/154405910408300908. [DOI] [PubMed] [Google Scholar]
- 88.Yang X, Sun Z, Ma R, Fan D, Moradian-Oldak J. Amelogenin “nanorods” formation during proteolysis by Mmp-20. J Struct Biol. 2011 Aug 5; doi: 10.1016/j.jsb.2011.07.016. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fincham A, Moradian-Oldak J, Diekwisch T, Lyaruu D, Wright J, Bringas PJ, Slavkin H. Evidence for amelogenin “nanospheres” as functional components of secretory-stage enamel matrix. J Struct Biol. 1995;115(1):50–9. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- 90.Moradian-Oldak J, Goldberg M. Amelogenin supra-molecular assembly in vitro compared with the architecture of the forming enamel matrix. Cells Tissues Organs. 2005;181(3–4):202–18. doi: 10.1159/000091382. [DOI] [PubMed] [Google Scholar]
- 91.Robinson C, Fuchs P, Weatherell JA. The appearance of developing rat incisor enamel using freeze-fracture technique. J Crystal Growth. 1981;53:160–165. [Google Scholar]
- 92.Veis A. A window on biomineralization. Science. 2005;307:1419–1420. doi: 10.1126/science.1109440. [DOI] [PubMed] [Google Scholar]
- 93.Ronnholm E., III The structure of the organic stroma of human enamel during amelogenesis. J Ultrastruct Res. 1962;6:368–89. doi: 10.1016/s0022-5320(62)80041-0. [DOI] [PubMed] [Google Scholar]
- 94.Bonucci E. Crystal ghosts and biological mineralization: fancy spectres in an old castle, or neglected structures worthy of belief? J Bone Miner Metab. 2002;20(5):249–65. doi: 10.1007/s007740200037. [DOI] [PubMed] [Google Scholar]
- 95.Watson ML. The extracellular nature of enamel in the rat. J Biophys Biochem Cytol. 1960;7:489–92. doi: 10.1083/jcb.7.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Travis DF, Glimcher MJ. The Structure And Organization Of, And The Relationship Between The Organic Matrix And The Inorganic Crystals Of Embryonic Bovine Enamel. J Cell Biol. 1964;23:447–97. doi: 10.1083/jcb.23.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smales FC. Structural subunit in prisms of immature rat enamel. Nature. 1975;258(5537):772–4. doi: 10.1038/258772a0. [DOI] [PubMed] [Google Scholar]
- 98.Sasaki S, Takagi T, Suzuki M. Cyclical changes in pH in bovine developing enamel as sequential bands. Arch Oral Biol. 1991;36:227–231. doi: 10.1016/0003-9969(91)90090-h. [DOI] [PubMed] [Google Scholar]
- 99.Fan D, Du C, Sun Z, Lakshminarayanan R, Moradian-Oldak J. In vitro study on the interaction between the 32 kDa enamelin and amelogenin. J Struct Biol. 2009;166(1):88–94. doi: 10.1016/j.jsb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang L, Guan X, Yin H, Moradian-Oldak J, Nancollas GH. Mimicking the Self-Organized Microstructure of Tooth Enamel. J Phys Chem C Nanomater Interfaces. 2008;112(15):5892–5899. doi: 10.1021/jp077105+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tarasevich BJ, Howard CJ, Larson JL, Snead ML, Simmer JP, Paine M, Shaw WJ. The nucleation and growth of calcium phosphate by amelogenin. J Cryst Growth. 2007;304(2):407–415. doi: 10.1016/j.jcrysgro.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bouropoulos N, Moradian-Oldak J. Analysis of hydroxyapatite surface coverage by amelogenin nanospheres following the Langmuir model for protein adsorption. Calcif Tissue Int. 2003;72(5):599–603. doi: 10.1007/s00223-002-1099-1. [DOI] [PubMed] [Google Scholar]
- 103.Aoba T, Moreno EC, Kresak M, Tanabe T. Possible roles of partial sequences at N- and C-termini of amelogenin in protein-enamel mineral interaction. J Dent Res. 1989;68:1331–1336. doi: 10.1177/00220345890680090901. [DOI] [PubMed] [Google Scholar]
- 104.Moradian-Oldak J, Tan J, Fincham AG. Interaction of amelogenin with hydroxyapatite crystals: an adherence effect through amelogenin molecular self-association. Biopolymers. 1998;46(4):225–38. doi: 10.1002/(SICI)1097-0282(19981005)46:4<225::AID-BIP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 105.Iijima M, Moradian-Oldak J. Control of octacalcium phosphate and apatite crystal growth by amelogenin matrices. J Materials Chemistry. 2004;14(14):2189–2199. [Google Scholar]
- 106.Wen HB, Moradian-Oldak J, Zhong JP, Greenspan DC, Fincham AG. Effects of amelogenin on the transforming surface microstructures of Bioglass in a calcifying solution. J Biomed Mater Res. 2000;52(4):762–73. doi: 10.1002/1097-4636(20001215)52:4<762::aid-jbm22>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 107.Kwak SY, Wiedemann-Bidlack FB, Beniash E, Yamakoshi Y, Simmer JP, Litman A, Margolis HC. Role of 20-kDa Amelogenin (P148) Phosphorylation in Calcium Phosphate Formation in vitro. J Biol Chem. 2009;284(28):18972–18979. doi: 10.1074/jbc.M109.020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wallwork ML, Kirkham J, Zhang J, Smith A, Brookes S, Shore RC, Wood SR, Ryu O, Robinson C. Binding of matrix proteins to developing enamel crystals: An atomic force microscope study. Langmuir. 2001;17:2508–2513. [Google Scholar]
- 109.Yang X, Wang L, Qin Y, Sun Z, Henneman ZJ, Moradian-Oldak J, Nancollas GH. How amelogenin orchestrates the organization of hierarchical elongated microstructures of apatite. J Phys Chem B. 2010;114(6):2293–300. doi: 10.1021/jp910219s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Iwata T, Yamakoshi Y, Hu JC, Ishikawa I, Bartlett JD, Krebsbach PH, Simmer JP. Processing of ameloblastin by MMP-20. J Dent Res. 2007;86(2):153–7. doi: 10.1177/154405910708600209. [DOI] [PubMed] [Google Scholar]
- 111.Yamakoshi Y, Hu JC, Zhang H, Iwata T, Yamakoshi F, Simmer JP. Proteomic analysis of enamel matrix using a two-dimensional protein fractionation system. Eur J Oral Sci. 2006;114(Suppl 1):266–71. doi: 10.1111/j.1600-0722.2006.00279.x. discussion 285-6, 382. [DOI] [PubMed] [Google Scholar]
- 112.Zalzal SF, Smith CE, Nanci A. Ameloblastin and amelogenin share a common secretory pathway and are co-secreted during enamel formation. Matrix Biol. 2008;27(4):352–9. doi: 10.1016/j.matbio.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 113.Ravindranath HH, Chen LS, Zeichner-David M, Ishima R, Ravindranath RM. Interaction between the enamel matrix proteins amelogenin and ameloblastin. Biochem Biophys Res Commun. 2004;323(3):1075–83. doi: 10.1016/j.bbrc.2004.08.207. [DOI] [PubMed] [Google Scholar]
- 114.MacDougall M, Simmons D, Gu TT, Forsman-Semb K, Mardh CK, Mesbah M, Forest N, Krebsbach PH, Yamada Y, Berdal A. Cloning, characterization and immunolocalization of human ameloblastin. Eur J Oral Sci. 2000;108(4):303–10. doi: 10.1034/j.1600-0722.2000.108004303.x. [DOI] [PubMed] [Google Scholar]
- 115.Yamakoshi Y, Tanabe T, Oida S, Hu CC, Simmer JP, Fukae M. Calcium binding of enamel proteins and their derivatives with emphasis on the calcium-binding domain of porcine sheathlin. Arch Oral Biol. 2001;46(11):1005–14. doi: 10.1016/s0003-9969(01)00070-x. [DOI] [PubMed] [Google Scholar]
- 116.Kobayashi K, Yamakoshi Y, Hu JC, Gomi K, Arai T, Fukae M, Krebsbach PH, Simmer JP. Splicing determines the glycosylation state of ameloblastin. J Dent Res. 2007;86(10):962–7. doi: 10.1177/154405910708601009. [DOI] [PubMed] [Google Scholar]
- 117.Vymetal J, Slaby I, Spahr A, Vondrasek J, Lyngstadaas SP. Bioinformatic analysis and molecular modelling of human ameloblastin suggest a two-domain intrinsically unstructured calcium-binding protein. Eur J Oral Sci. 2008;116(2):124–34. doi: 10.1111/j.1600-0722.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- 118.Hu CC, Fukae M, Uchida T, Qian Q, Zhang CH, Ryu OH, Tanabe T, Yamakoshi Y, Murakami C, Dohi N, Shimizu M, Simmer JP. Sheathlin: cloning, cDNA/polypeptide sequences, and immunolocalization of porcine enamel sheath proteins. J Dent Res. 1997;76(2):648–57. doi: 10.1177/00220345970760020501. [DOI] [PubMed] [Google Scholar]
- 119.Xu Q, Holder N, Patient R, Wilson SW. Spatially regulated expression of three receptor tyrosine kinase genes during gastrulation in the zebrafish. Development. 1994;120:287–299. doi: 10.1242/dev.120.2.287. [DOI] [PubMed] [Google Scholar]
- 120.Uchida T, Tanabe T, Fukae M, Shimizu M, Yamada M, Miake K, Kobayashi S. Immunochemical and immunohistochemical studies, using antisera against porcine 25kDa amelogenin, 89kDa enamelin and the 13–17kDa nonamelogenins, on immature enamel of the pig and rat. Histochemistry. 1991;96:129–138. doi: 10.1007/BF00315983. [DOI] [PubMed] [Google Scholar]
- 121.Akita H, Fukae M, Shimoda S, Aoba T. Localization of glycosylated matrix proteins in secretory porcine enamel and their possible functional roles in enamel mineralization. Arch Oral Biol. 1992;37(11):953–62. doi: 10.1016/0003-9969(92)90067-i. [DOI] [PubMed] [Google Scholar]
- 122.Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wazen RM, Moffatt P, Zalzal SF, Yamada Y, Nanci A. A mouse model expressing a truncated form of ameloblastin exhibits dental and junctional epithelium defects. Matrix Biol. 2009;28(5):292–303. doi: 10.1016/j.matbio.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Uchida T, Murakami C, Dohi N, Wakida K, Satoda T, Takahashi O. Synthesis, secretion, degradation, and fate of ameloblastin during the matrix formation stage of the rat incisor as shown by immunocytochemistry and immunochemistry using region-specific antibodies. J Histochem Cytochem. 1997;45(10):1329–40. doi: 10.1177/002215549704501002. [DOI] [PubMed] [Google Scholar]
- 125.Nanci A, Zalzal S, Lavoie P, Kunikata M, Chen W, Krebsbach PH, Yamada Y, Hammarstrom L, Simmer JP, Fincham AG, Snead ML, Smith CE. Comparative immunochemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. J Histochem Cytochem. 1998;46(8):911–34. doi: 10.1177/002215549804600806. [DOI] [PubMed] [Google Scholar]
- 126.Snead ML. Enamel biology logodaedaly: Getting to the root of the problem, or “Who’s on first…”. J Bone Min Res. 1996;11:899–904. doi: 10.1002/jbmr.5650110705. [DOI] [PubMed] [Google Scholar]
- 127.Hu JC, Sun X, Zhang C, Simmer JP. A comparison of enamelin and amelogenin expression in developing mouse molars. Eur J Oral Sci. 2001;109:125–132. doi: 10.1034/j.1600-0722.2001.00998.x. [DOI] [PubMed] [Google Scholar]
- 128.Hu CC, Hart TC, Dupont BR, Chen JJ, Sun X, Qian Q, Zhang CH, Jiang H, Mattern VL, Wright JT, Simmer JP. Cloning human enamelin cDNA, chromosomal localization, and analysis of expression during tooth development. J Dent Res. 2000;79:912–919. doi: 10.1177/00220345000790040501. [DOI] [PubMed] [Google Scholar]
- 129.Dong J, Gu TT, Simmons D, MacDougall M. Enamelin maps to human chromosome 4q21 within the autosomal dominant amelogenesis imperfecta locus. Eur J Oral Sci. 2000;108(5):353–8. doi: 10.1034/j.1600-0722.2000.108005353.x. [DOI] [PubMed] [Google Scholar]
- 130.Al-Hashimi N, Sire JY, Delgado S. Evolutionary analysis of mammalian enamelin, the largest enamel protein, supports a crucial role for the 32-kDa peptide and reveals selective adaptation in rodents and primates. J Mol Evol. 2009;69(6):635–56. doi: 10.1007/s00239-009-9302-x. [DOI] [PubMed] [Google Scholar]
- 131.Fukae M, Tanabe T, Uchida T, Yamakoshi Y, Shimizu M. Enamelins in the newly formed bovine enamel. Calcif Tissue Int. 1993;53(4):257–61. doi: 10.1007/BF01320911. [DOI] [PubMed] [Google Scholar]
- 132.Yamakoshi Y, Pinheiro FH, Tanabe T, Fukae M, Shimizu M. Sites of asparagine-linked oligosaccharides in porcine 32 kDa enamelin. Connect Tissue Res. 1998;39(1–3):39–46. doi: 10.3109/03008209809023910. discussion 63-7. [DOI] [PubMed] [Google Scholar]
- 133.Hu JC, Yamakoshi Y. Enamelin and autosomal-dominant amelogenesis imperfecta. Crit Rev Oral Biol Med. 2003;14(6):387–98. doi: 10.1177/154411130301400602. [DOI] [PubMed] [Google Scholar]
- 134.Hu JC, Hu Y, Smith CE, McKee MD, Wright JT, Yamakoshi Y, Papagerakis P, Hunter GK, Feng JQ, Yamakoshi F, Simmer JP. Enamel defects and ameloblast-specific expression in Enam knock-out/lacz knock-in mice. J Biol Chem. 2008;283(16):10858–71. doi: 10.1074/jbc.M710565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bouropoulos N, Moradian-Oldak J. Induction of apatite by the cooperative effect of amelogenin and the 32-kDa enamelin. J Dent Res. 2004;83(4):278–82. doi: 10.1177/154405910408300402. [DOI] [PubMed] [Google Scholar]
- 136.Fan D, Iijima M, Bromley KM, Yang X, Mathew S, Moradian-Oldak J. The Cooperation of Enamelin and Amelogenin in Controlling Octacalcium Phosphate Crystal Morphology. Cells Tissues Organs. 2011;194(2–4):194–8. doi: 10.1159/000324208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Iijima M, Fan D, Bromley KM, Sun Z, Moradian-Oldak J. Tooth enamel proteins enamelin and amelogenin cooperate to regulate the growth morphology of octacalcium phosphate crystals. Cryst Growth Des. 2010;10(11):4815–4822. doi: 10.1021/cg100696r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Iijima M, Moradian-Oldak J. Interactions of amelogenins with octacalcium phosphate crystal faces are dose dependent. Calcif Tissue Int. 2004;74(6):522–31. doi: 10.1007/s00223-002-0011-3. [DOI] [PubMed] [Google Scholar]
- 139.Iwasaki K, Bajenova E, Somogyi-Ganss E, Miller M, Nguyen V, Nourkeyhani H, Gao Y, Wendel M, Ganss B. Amelotin--a Novel Secreted, Ameloblast-specific Protein. J Dent Res. 2005;84(12):1127–32. doi: 10.1177/154405910508401207. [DOI] [PubMed] [Google Scholar]
- 140.Moffatt P, Smith CE, St-Arnaud R, Simmons D, Wright JT, Nanci A. Cloning of rat amelotin and localization of the protein to the basal lamina of maturation stage ameloblasts and junctional epithelium. Biochem J. 2006;399(1):37–46. doi: 10.1042/BJ20060662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gao Y, Wang W, Sun Y, Zhang J, Li D, Wei Y, Han T. Distribution of amelotin in mouse tooth development. Anat Rec (Hoboken) 2010;293(1):135–40. doi: 10.1002/ar.21022. [DOI] [PubMed] [Google Scholar]
- 142.Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP. Functions of KLK4 and MMP-20 in dental enamel formation. Biol Chem. 2008;389(6):695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lacruz RS, Smith CE, Smith SM, Hu P, Bringas P, Jr, Sahin-Toth M, Moradian-Oldak J, Paine ML. Chymotrypsin C (Caldecrin) Is Associated with Enamel Development. J Dent Res. 2011;19(10):1228–33. doi: 10.1177/0022034511418231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bartlett JD, Simmer JP, Xue J, Margolis HC, Moreno EC. Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene. 1996;183:123–128. doi: 10.1016/s0378-1119(96)00525-2. [DOI] [PubMed] [Google Scholar]
- 145.Fukae M, Tanabe T. Degradation of enamel matrix proteins in porcine secretory enamel. Connect Tissue Res. 1998;39(1–3):123–9. doi: 10.3109/03008209809023918. discussion 141-9. [DOI] [PubMed] [Google Scholar]
- 146.Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, Birkedal-Hansen H, Bartlett JD. Enamelysin (MMP-20) deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2002;277:49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- 147.Bartlett JD, Beniash E, Lee DH, Smith CE. Decreased mineral content in MMP-20 null mouse enamel is prominent during the maturation stage. J Dent Res. 2004;83(12):909–13. doi: 10.1177/154405910408301204. [DOI] [PubMed] [Google Scholar]
- 148.Smith CE, Richardson AS, Hu Y, Bartlett JD, Hu JC, Simmer JP. Effect of Kallikrein 4 Loss on Enamel Mineralization: Comparison With Mice Lacking Matrix Metalloproteinase 20. J Biol Chem. 2011;286(20):18149–60. doi: 10.1074/jbc.M110.194258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ryu OH, Fincham AG, Hu CC, Zhang C, Qian Q, Bartlett JD, Simmer JP. Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J Dent Res. 1999;78:743–750. doi: 10.1177/00220345990780030601. [DOI] [PubMed] [Google Scholar]
- 150.Sun Z, Fan D, Fan Y, Du C, Moradian-Oldak J. Enamel proteases reduce amelogenin-apatite binding. J Dent Res. 2008;87(12):1133–7. doi: 10.1177/154405910808701212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sun Z, Abbott C, Moradian-Oldak J. Control of amelogenin assembly and dis-assembly by enamelysin (MMP-20). The 8th International Conference on Chemistry and Biology of Mineralized Tissue; University of Toronto Press Inc., Banff, Canada. 2005. [Google Scholar]
- 152.Uskokovic V, Kim MK, Li W, Habelitz S. Enzymatic processing of amelogenin during continuous crystallization of apatite. J Mate Res. 2008;23(12):3184–3195. doi: 10.1557/JMR.2008.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nelson PS, Gan L, Ferguson C, Moss P, Gelinas R, Hood L, Wang K. Molecular cloning and characterization of prostase, an androgen-regulated serine protease with prostate-restricted expression. Proc Natl Acad Sci U S A. 1999;96:3114–3119. doi: 10.1073/pnas.96.6.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ryu O, Hu JC, Yamakoshi Y, Villemain JL, Cao X, Zhang C, Bartlett JD, Simmer JP. Porcine kallikrein-4 activation, glycosylation, activity, and expression in prokaryotic and eukaryotic hosts. Eur J Oral Sci. 2002;110(5):358–65. doi: 10.1034/j.1600-0722.2002.21349.x. [DOI] [PubMed] [Google Scholar]
- 155.Yamakoshi Y, Hu JC, Fukae M, Yamakoshi F, Simmer JP. How do enamelysin and kallikrein 4 process the 32-kDa enamelin? Eur J Oral Sci. 2006;114(Suppl 1):45–51. doi: 10.1111/j.1600-0722.2006.00281.x. discussion 93-5, 379-80. [DOI] [PubMed] [Google Scholar]
- 156.Sun Z, Carpiaux W, Fan D, Fan Y, Lakshminarayanan R, Moradian-Oldak J. Apatite reduces amelogenin proteolysis by MMP-20 and KLK4 in vitro. J Dent Res. 89(4):344–8. doi: 10.1177/0022034509360660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC. Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J Biol Chem. 2009;284(28):19110–21. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tye CE, Lorenz RL, Bartlett JD. Lysosomal protease expression in mature enamel. Cells Tissues Organs. 2009;189(1–4):111–4. doi: 10.1159/000151431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lyman GE, Waddell WJ. pH gradients in the developing teeth of young mice from autoradiography of (14C)DMO. Am J Physiol. 1977;232(4):F364–7. doi: 10.1152/ajprenal.1977.232.4.F364. [DOI] [PubMed] [Google Scholar]
- 160.Takano Y, Crenshaw MA, Reith EJ. Correlation of 45Ca incorporation with maturation ameloblast morphology in the rat incisor. Calcif Tissue Int. 1982;34:211–213. doi: 10.1007/BF02411236. [DOI] [PubMed] [Google Scholar]
- 161.Smith CE, Issid M, Margolis HC, Moreno EC. Developmental changes in the pH of enamel fluid and its effects on matrix-resident proteinases. Adv Dent Res. 1996;10(2):159–69. doi: 10.1177/08959374960100020701. [DOI] [PubMed] [Google Scholar]
- 162.Smith CE, Nanci A. Overview of morphological changes in enamel organ cells associated with major events in amelogenesis. Int J Dev Biol. 1995;39:153–161. [PubMed] [Google Scholar]
- 163.Smith CE, McKee MD, Nanci A. Cyclic induction and rapid movement of sequential waves of new smooth-ended ameloblast modulation bands in rat incisors as visualized by polychrome fluorescent labeling and GBHA-staining of maturing enamel. Adv Dent Res. 1987;1(2):162–75. doi: 10.1177/08959374870010020401. [DOI] [PubMed] [Google Scholar]
- 164.Aoba T, Moreno EC. The enamel fluid in the early secretory stage of porcine amelogenesis: chemical composition and saturation with respect to enamel mineral. Calcif Tissue Int. 1987;41(2):86–94. doi: 10.1007/BF02555250. [DOI] [PubMed] [Google Scholar]
- 165.Toyosawa S, Ogawa Y, Inagaki T, Ijuhin N. Immunohistochemical localization of carbonic anhydrase isozyme II in rat incisor epithelial cells at various stages of amelogenesis. Cell Tissue Res. 1996;285(2):217–25. doi: 10.1007/s004410050639. [DOI] [PubMed] [Google Scholar]
- 166.Smith CE, Nanci A, Moffatt P. Evidence by signal peptide trap technology for the expression of carbonic anhydrase 6 in rat incisor enamel organs. Eur J Oral Sci. 2006;114(Suppl 1):147–53. doi: 10.1111/j.1600-0722.2006.00273.x. discussion 164-5, 380-1. [DOI] [PubMed] [Google Scholar]
- 167.Lacruz RS, Smith CE, Moffatt P, Chang EH, Bromage TG, Bringas P, Jr, Nanci A, Baniwal SK, Zabner J, Welsh MJ, Kurtz I, Paine ML. Requirements for ion and solute transport, and pH regulation, during enamel maturation. J Cell Physiol. 2011 Jul 15; doi: 10.1002/jcp.22911. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Crenshaw MA, Takano Y. Mechanisms by which the enamel organ controls calcium entry into developing enamel. J Dent Res. 1982;(Spec No):1574–9. [PubMed] [Google Scholar]
- 169.Garant PR, Sasaki T, Colflesh PE. Na-K-ATPase in the enamel organ: localization and possible roles in enamel formation. Adv Dent Res. 1987;1(2):267–75. doi: 10.1177/08959374870010021601. [DOI] [PubMed] [Google Scholar]
- 170.Nunn JH. Prevalence of dental erosion and the implications for oral health. Eur J Oral Sci. 1996;104(2):156–61. doi: 10.1111/j.1600-0722.1996.tb00064.x. [DOI] [PubMed] [Google Scholar]
- 171.Hart PS, Hart TC, Simmer JP, Wright JT. A nomenclature for X-linked amelogenesis imperfecta. Arch Oral Biol. 2002;47:255–260. doi: 10.1016/s0003-9969(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 172.Kim JW, Lee SK, Lee ZH, Park JC, Lee KE, Lee MH, Park JT, Seo BM, Hu JC, Simmer JP. FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta. Am J Hum Genet. 2008;82(2):489–94. doi: 10.1016/j.ajhg.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.El-Sayed W, Parry DA, Shore RC, Ahmed M, Jafri H, Rashid Y, Al-Bahlani S, Al Harasi S, Kirkham J, Inglehearn CF, Mighell AJ. Mutations in the beta propeller WDR72 cause autosomal-recessive hypomaturation amelogenesis imperfecta. Am J Hum Genet. 2009;85(5):699–705. doi: 10.1016/j.ajhg.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Karrman C, Backman B, Dixon M, Holmgren G, Forsman K. Mapping of the locus for autosomal dominant amelogenesis imperfecta (AIH2) to a 4-Mb YAC contig on chromosome 4q11-q21. Genomics. 1997;39:164–170. doi: 10.1006/geno.1996.4485. [DOI] [PubMed] [Google Scholar]
- 175.Wright JT, Hart PS, Aldred MJ, Seow K, Crawford PJ, Hong SP, Gibson CW, Hart TC. Relationship of phenotype and genotype in X-linked amelogenesis imperfecta. Connect Tissue Res. 2003;44(Suppl 1):72–78. [PubMed] [Google Scholar]
- 176.Wright JT. Consequences of amelogenin mutations: Implications in Amelogenesis Imperfecta. In: Goldberg M, editor. Amelogenins; Multifaceted proteins for dental and bone formation and repair. Bentham Books Ltd; Dubai: 2010. [Google Scholar]
- 177.Lakshminarayanan R, Bromley KM, Lei YP, Snead ML, Moradian-Oldak J. Perturbed amelogenin secondary structure leads to uncontrolled aggregation in amelogenesis imperfecta mutant proteins. J Biol Chem. 2010;285(52):40593–603. doi: 10.1074/jbc.M110.131136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bromley KM, Lakshminarayanan R, Lei YP, Snead ML, Moradian-Oldak J. Folding, Assembly, and Aggregation of Recombinant Murine Amelogenins with T21I and P41T Point Mutations. Cells Tissues Organs. 2011;194(2–4):284–90. doi: 10.1159/000324342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li W, Gibson CW, Abrams DW, Andrews DW, DenBesten PK. Reduced hydrolysis of amelogenin may result in X-linked amelogenesis imperfecta. Matrix Biology. 2001;19:755–760. doi: 10.1016/s0945-053x(00)00121-9. [DOI] [PubMed] [Google Scholar]
- 180.Collier PM, Sauk JJ, Rosenbloom J, Yuan ZA, Gibson CW. An amelogenin gene defect associated with human x-linked amelogenesis imperfecta. Archs Oral Biol. 1997;42(3):235–242. doi: 10.1016/s0003-9969(96)00099-4. [DOI] [PubMed] [Google Scholar]
- 181.Pugach MK, Li Y, Suggs C, Wright JT, Aragon MA, Yuan ZA, Simmons D, Kulkarni AB, Gibson CW. The amelogenin C-terminus is required for enamel development. J Dent Res. 2010;89(2):165–9. doi: 10.1177/0022034509358392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Moradian-Oldak J. Amelogenins: assembly, processing and control of crystal morphology. Matrix Biol. 2001;20(5–6):293–305. doi: 10.1016/s0945-053x(01)00154-8. [DOI] [PubMed] [Google Scholar]
- 183.Ozdemir D, Hart PS, Firatli E, Aren G, Ryu OH, Hart TC. Phenotype of ENAM mutations is dosage-dependent. J Dent Res. 2005;84(11):1036–41. doi: 10.1177/154405910508401113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rajpar MH, Harley K, Laing C, Davies RM, Dixon MJ. Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta. Hum Mol Genet. 2001;10:1673–1677. doi: 10.1093/hmg/10.16.1673. [DOI] [PubMed] [Google Scholar]
- 185.Kida M, Ariga T, Shirakawa T, Oguchi H, Sakiyama Y. Autosomal-dominant hypoplastic form of amelogenesis imperfecta caused by an enamelin gene mutation at the exon-intron boundary. J Dent Res. 2002;81(11):738–42. doi: 10.1177/0810738. [DOI] [PubMed] [Google Scholar]
- 186.Mardh CK, Backman B, Holmgren G, Hu JC, Simmer JP, Forsman-Semb K. A nonsense mutation in the enamelin gene causes local hypoplastic autosomal dominant amelogenesis imperfecta (AIH2) Hum Mol Genet. 2002;11:1069–1074. doi: 10.1093/hmg/11.9.1069. [DOI] [PubMed] [Google Scholar]
- 187.Kim JW, Seymen F, Lin BP, Kiziltan B, Gencay K, Simmer JP, Hu JC. ENAM mutations in autosomal-dominant amelogenesis imperfecta. J Dent Res. 2005;84(3):278–82. doi: 10.1177/154405910508400314. [DOI] [PubMed] [Google Scholar]
- 188.Hart PS, Wright JT, Savage M, Kang G, Bensen JT, Gorry MC, Hart TC. Exclusion of candidate genes in two families with autosomal dominant hypocalcified amelogenesis imperfecta. Eur J Oral Sci. 2003;111(4):326–31. doi: 10.1034/j.1600-0722.2003.00046.x. [DOI] [PubMed] [Google Scholar]
- 189.Gutierrez SJ, Chaves M, Torres DM, Briceno I. Identification of a novel mutation in the enamalin gene in a family with autosomal-dominant amelogenesis imperfecta. Arch Oral Biol. 2007;52(5):503–6. doi: 10.1016/j.archoralbio.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 190.Chan HC, Mai L, Oikonomopoulou A, Chan HL, Richardson AS, Wang SK, Simmer JP, Hu JC. Altered enamelin phosphorylation site causes amelogenesis imperfecta. J Dent Res. 2010;89(7):695–9. doi: 10.1177/0022034510365662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wright JT. The molecular etiologies and associated phenotypes of amelogenesis imperfecta. Am J Med Genet A. 2006;140(23):2547–55. doi: 10.1002/ajmg.a.31358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, Wright JT. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41(7):545–9. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, Hu JC. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J Med Genet. 2005;42(3):271–5. doi: 10.1136/jmg.2004.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Becerik S, Cogulu D, Emingil G, Han T, Hart PS, Hart TC. Exclusion of candidate genes in seven Turkish families with autosomal recessive amelogenesis imperfecta. Am J Med Genet A. 2009;149A(7):1392–8. doi: 10.1002/ajmg.a.32885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mann S. The biomimetics of enamel: a paradigm for organized biomaterials synthesis. John Wiley & Sons Ltd; Chichester: 1997. [PubMed] [Google Scholar]
- 196.Moradian-Oldak J. The regeneration of tooth enamel. Dimens Dent Hyg. 2009;7(8):12–15. [PMC free article] [PubMed] [Google Scholar]
- 197.Chen JD, Wang YJ, Wei K, Zhang SH, Shi XT. Self-organization of hydroxyapatite nanorods through oriented attachment. Biomaterials. 2007;28(14):2275–80. doi: 10.1016/j.biomaterials.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 198.Lei C, Liao Y, Feng Z. Kinetic model for hydroxyapatite precipitation on human enamel surface by electrolytic deposition. Biomed Mater. 2009;4(3):035010. doi: 10.1088/1748-6041/4/3/035010. [DOI] [PubMed] [Google Scholar]
- 199.Wang X, Xia C, Zhang Z, Deng X, Wei S, Zheng G, Chen H. Direct growth of human enamel-like calcium phosphate microstructures on human tooth. J Nanosci Nanotechnol. 2009;9(2):1361–4. doi: 10.1166/jnn.2009.c157. [DOI] [PubMed] [Google Scholar]
- 200.Yin Y, Yun S, Fang J, Chen H. Chemical regeneration of human tooth enamel under near-physiological conditions. Chem Commun (Camb) 2009;(39):5892–4. doi: 10.1039/b911407f. [DOI] [PubMed] [Google Scholar]
- 201.Hayakawa S, Li Y, Tsuru K, Osaka A, Fujii E, Kawabata K. Preparation of nanometer-scale rod array of hydroxyapatite crystal. Acta Biomater. 2009;5(6):2152–60. doi: 10.1016/j.actbio.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 202.Moradian-Oldak J, Fan D. Tooth-Inspired Nanocomposites. In: Kumar C, editor. Nanomaterials for the life sciences : Biomimetic and bioinspired nanomaterials. Willey-VCH; Weinheim: 2010. [Google Scholar]
- 203.Iijima M, Moriwaki Y, Takagi T, Moradian-Oldak J. Effects of bovine amelogenins on the crystal morphology of octacalcium phosphate in a model system of tooth enamel formation. J of Crystal Growth. 2001;222(3):615–626. [Google Scholar]
- 204.Fan Y, Sun Z, Moradian-Oldak J. Controlled remineralization of enamel in the presence of amelogenin and fluoride. Biomaterials. 2009;30(4):478–83. doi: 10.1016/j.biomaterials.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Fan Y, Nelson JR, Alvarez JR, Hagan J, Berrier A, Xu X. Amelogenin-assisted ex vivo remineralization of human enamel: Effects of supersaturation degree and fluoride concentration. Acta Biomater. 2011;7(5):2293–302. doi: 10.1016/j.actbio.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]