Abstract
DNA replication fork stalling poses a major threat to genome stability. This is counteracted in part by the intra-S phase checkpoint, which stabilizes arrested replication machinery, prevents cell-cycle progression and promotes DNA repair. The checkpoint kinase Mec1/ATR and RecQ helicase Sgs1/BLM contribute synergistically to fork maintenance on hydroxyurea (HU). Both enzymes interact with replication protein A (RPA). We identified and deleted the major interaction sites on Sgs1 for Rpa70, generating a mutant called sgs1-r1. In contrast to a helicase-dead mutant of Sgs1, sgs1-r1 did not significantly reduce recovery of DNA polymerase α at HU-arrested replication forks. However, the Sgs1 R1 domain is a target of Mec1 kinase, deletion of which compromises Rad53 activation on HU. Full activation of Rad53 is achieved through phosphorylation of the Sgs1 R1 domain by Mec1, which promotes Sgs1 binding to the FHA1 domain of Rad53 with high affinity. We propose that the recruitment of Rad53 by phosphorylated Sgs1 promotes the replication checkpoint response on HU. Loss of the R1 domain increases lethality selectively in cells lacking Mus81, Slx4, Slx5 or Slx8.
Keywords: intra-S checkpoint, RPA, replication stress, Rad53, Sgs1
Introduction
The accurate replication of DNA and its segregation into daughter cells is aided by the intra-S checkpoint, which is triggered by the single-stranded DNA (ssDNA) that accumulates when DNA polymerases pause, either due to reduced nucleotide concentration or due to the presence of adducts that impair fork progression. Avoidance of fork collapse is mediated both by the Mec1/ATR kinase and by the action of a RecQ helicase, which reverses fold-back structures and resolves strand exchange to suppress inappropriate recombination events. Resumption of replication generally requires that engaged DNA polymerases remain associated with paused forks, which in wild-type yeast cells can persist for many hours (reviewed in Cobb and Bjergbaek, 2006; Tourriere and Pasero, 2007 and Aguilera and Gomez-Gonzalez, 2008).
The checkpoint kinase Mec1-Ddc2 in S. cerevisiae (ATR-ATRIP in humans) plays two critical roles in this event (reviewed in Cimprich and Cortez, 2008 and Friedel et al, 2009). First, Mec1-Ddc2 regulates replisome function and enables the stable retention of replicative polymerases at very early origins like ARS607 (Cobb et al, 2003, 2005; De Piccoli et al, 2012). Second, it modifies and activates Rad53, the downstream checkpoint kinase that in turn retards cell-cycle progression, regulates levels of dNTPs and repair enzymes, represses the firing of late origins, and prevents fork collapse through poorly identified pathways (reviewed in Tourriere and Pasero, 2007; Segurado and Diffley, 2008).
The activation of Mec1/Ddc2 kinase under restricted nucleotide conditions (0.2 M hydroxyurea, HU) most likely stems from the stalling of leading and/or lagging strand DNA polymerases, which generates stretches of ssDNA. These become coated by replication protein A (RPA; Aparicio et al, 1999), which signals the recruitment and activation of Mec1-Ddc2 checkpoint kinase (Zou and Elledge, 2003), not unlike the situation at resected double-strand breaks (Dubrana et al, 2007). In both budding yeast and mammals, RPA contributes to the recruitment of Mec1/ATR to stalled or damaged replication forks, through its cofactor, Ddc2/ATRIP (Melo et al, 2001; Rouse and Jackson, 2002). Intriguingly, RPA is itself a target of Mec1/ATR phosphorylation (Brush et al, 1996; Zou and Elledge, 2003).
Besides RPA, fork-associated activators of the intra-S phase checkpoint include the 9-1-1 checkpoint clamp and Dbp11/TOPBP1 (Majka et al, 2006; Mordes et al, 2008; Navadgi-Patil and Burgers, 2008), while additional, unidentified co-activators are postulated to exist (Navadgi-Patil and Burgers, 2011). Once recruited Mec1/ATR phosphorylates Mrc1/Claspin, which helps activate the downstream effector kinases Rad53/CHK2, or CHK1 in mammalian cells (reviewed in Tourriere and Pasero, 2007), possibly by facilitating the contact between Mec1 and its target Rad53 (Chen and Zhou, 2009). In mammals, the RecQ helicase BLM was also reported to be a target of ATR/ATM phosphorylation, and to contribute to recovery from replicative stress (Davies et al, 2004; Rao et al, 2005).
Whereas the Rad53 kinase mediates crucial downstream events in the yeast checkpoint response, Mec1/ATR has a distinct role in stabilizing replicative polymerases, particularly at early firing origins, such as the budding yeast ARS607 or ARS305. This was demonstrated by chromatin immunoprecipitation (ChIP) from cells synchronously arrested in S phase: the recovery of DNA polymerases α and ε bound to the replication fork dropped rapidly in cells lacking Mec1, but not in cells lacking Rad53 (Cobb et al, 2003, 2005). Similar separation of function was demonstrated in a study of exo1 deletion effects on viability in rad53 versus mec1 mutants (Segurado and Diffley, 2008). Nonetheless, a loss of Rad53 triggers an accumulation of both ssDNA (Sogo et al, 2002; Tourriere and Pasero, 2007) and recombination intermediates (Lucca et al, 2004). Surprisingly, and in contrast to the situation at early firing origins, it was recently reported that the replisome can be recovered largely intact and associated with later firing origins upon replication stress, in cells lacking either Mec1 or Rad53 (De Piccoli et al, 2012). These checkpoint kinases were proposed to regulate replication fork progression through multiple targets, including Psf1, a component of the replicative Cdc45-MCM-GINS helicase (De Piccoli et al, 2012).
RecQ helicases have also been shown to be important for the stable binding of DNA polymerases at stalled replication forks and for efficient fork restart after exposure to HU or aphidicolin (Cobb et al, 2005; Davies et al, 2007; Bachrati and Hickson, 2008; Pirzio et al, 2008). Loss of Sgs1, the unique RecQ helicase in budding yeast, leads to a reduced recovery of DNA polymerases at early firing origins, a lower survival rate after exposure to HU (Cobb et al, 2003, 2005), and the accumulation of aberrant recombination structures after exposure to MMS (Liberi et al, 2005). Indeed, sgs1 deficient cells display abnormally high levels of recombination (Watt et al, 1996) and spontaneous gross chromosomal rearrangements (GCRs), particularly on HU (Myung and Kolodner, 2002; Schmidt and Kolodner, 2006). The role of RecQ helicases in resistance to replicative stress is conserved: mutations in three human RecQ helicases (BLM, Bloom’s; WRN; Werner’s, and RECQ4) cause syndromes associated with a predisposition to cancer and/or genome instability (reviewed by Bachrati and Hickson, 2008 and Ashton and Hickson, 2010).
Genetic studies argue that Sgs1 acts both in complex with Top3 and Rmi1 (Gangloff et al, 1994; Chang et al, 2005; Mullen et al, 2005) and alone (reviewed in Bernstein et al, 2010). Sgs1 requires Top3 for dissolution of Holliday junctions and for enhancing DNA polymerase at stalled forks (Liberi et al, 2005; Mankouri et al, 2011), while it acts independently of Top3 to activate Rad53 in the presence of HU (Bjergbaek et al, 2005). Sgs1, like BLM and WRN, also binds Rad51 and RPA, and acts both upstream and downstream of Rad51-mediated strand invasion, to prevent and to resolve recombination intermediates. Finally, synthetic lethal screens link Sgs1 not only to recombination enzymes, but also to enzymes and proteins essential for lagging strand synthesis, such as Pol32, RNase H2 and FEN1/Rad27 (Ooi et al, 2003; Tong et al, 2004; Ii and Brill, 2005).
Here, we focus on the role of Sgs1 at replication forks stalled by HU, which seems to mimic the situation that ensues when forks encounter tight DNA–protein complexes (reviewed in Aguilera and Gomez-Gonzalez, 2008). Double mutants in budding yeast have been particularly helpful in elucidating this pathway. Whereas the effects of sgs1Δ are relatively mild (Cobb et al, 2003), its combination with mec1-100, an S phase-specific mutation in Mec1, causes extensive fork collapse and a failure of nucleotide incorporation after recovery from acute exposure to HU (Cobb et al, 2005). The mec1-100 mutation compromises the intra-S phase checkpoint response, but is able to modify and activate Rad53, triggering the G2/M checkpoint (Paciotti et al, 2001). Importantly, and in contrast to the effects of mec1-100, deletion of rad53 is not additive with sgs1Δ in GCR or polymerase stability assays. Indeed, neither the loss of checkpoint activity in the rad53-11 mutant nor rad53 deletion coupled with sml1Δ affects polymerase recovery by ChIP at early firing origins (Cobb et al, 2003, 2005).
The fact that Sgs1, Mec1-Ddc2 and DNA pol α all bind RPA, led us to test the hypothesis that Sgs1 influences the association of lagging strand polymerases at stalled forks through its interaction with the ssDNA binding complex. To this end, we mapped the region of Sgs1 that binds RPA and generated a mutant lacking the interaction domain, which we call sgs1-r1. We monitored the status of DNA pol α at stalled forks in mutants lacking the main RPA-interaction domain, with and without Sgs1 helicase activity. We found that the Sgs1 helicase activity and not its RPA interacting domain contributes to the stabilization of engaged DNA pol α/primase at the HU-stalled replication fork. Moreover, we show that Mec1-Ddc2 modifies Sgs1 within the RPA-interaction domain, and that once phosphorylated, Sgs1 has a significant affinity for the FHA1 phospho-threonine binding module of the downstream checkpoint kinase Rad53. We propose that the interaction of Sgs1 and Rad53 contributes to checkpoint kinase activation during replicative stress, independent of the role of Sgs1 helicase activity in stabilizing polymerases at the fork.
Results
Sgs1 interacts with Rpa70 through an acidic region N-terminal of the helicase domain
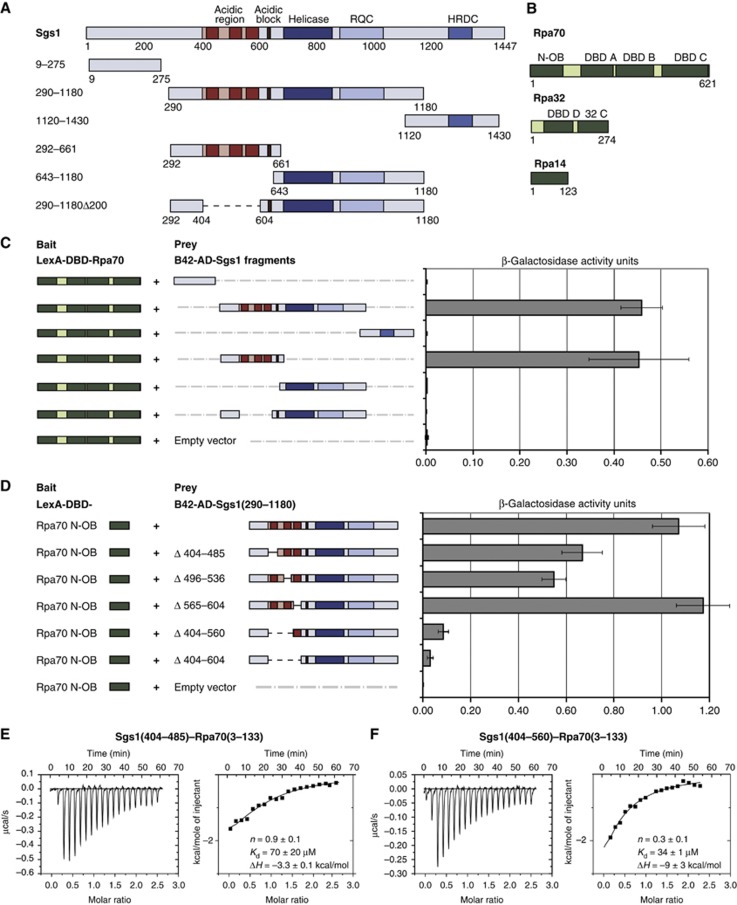
To analyse the role of Sgs1 in stabilizing lagging strand polymerases at stalled forks, we first mapped the Sgs1–RPA interaction site. Sgs1 contains three conserved domains that are characteristic of RecQ helicases: an SF2-type helicase domain, an RQC (RecQ C-terminal) motif and an HRDC (helicase and RNase D C-terminal) domain (Figure 1A). In addition, a region of unknown structure in the N-terminus of Sgs1 (first 158 aa) has been shown to interact with Top3/Rmi1 (Bennett et al, 2000; Fricke et al, 2001; Chen and Brill, 2007; Weinstein and Rothstein, 2008). There is both an acidic block at aa 664 and a larger acidic region located N-terminal of the helicase domain. Although structure prediction suggests that this region is intrinsically disordered in solution, it has been proposed to help prevent or resolve aberrant recombination structures at MMS-treated forks (Bernstein et al, 2009). To see which domain is responsible for binding to RPA, we fused fragments containing the functional domains of Sgs1 to a B42 transactivation domain (B42-AD) and performed yeast two-hybrid (Y2H) analysis with RPA (Figure 1A).
Figure 1.
Mapping the interaction site between Sgs1 and Rpa70. (A) Schematic representation of Sgs1 and its functional domains. Dark and light red—largely disordered acidic region, dark red—sequences that are conserved in close homologues of S. cerevisiae; other domains labelled in figure. RQC=RecQ C-terminal motif, HRDC=helicase and RNase D C-terminal. Below are the Sgs1 domains used in Y2H experiments, which were fused to the B42 activation domain (B42-AD) in pJG46 and expressed under a galactose-inducible promoter. Numbers indicate the boundaries of the Sgs1 domains in aa. (B) Scheme of the RPA subunits with their functional domains. Rpa70 and Rpa32 were fused to the lexA-DNA binding domain (lexA-DBD) in pGAL-lexA, expressed under a galactose-inducible promoter and subjected to Y2H analysis. N-OB=N-terminal OB fold, DBD=DNA binding domain, 32 C=Rpa32 C-terminus. (C) Y2H analysis between Rpa70 fused to lexA-DBD and Sgs1 fragments fused to B42-AD was performed using a quantitative β-galactosidase assay as described in Materials and methods. Error bars indicate standard error of four or more independent transformants. (D) Y2H analysis between Rpa70 N-OB fused to lexA-DBD and Sgs1 fragments fused to B42-AD with different deletions of the three conserved regions within the RPA binding site. (E, F) Isothermal titration calorimetry (ITC) experiment of Rpa70(3–133) with Sgs1(aa 404–485) and Sgs1(aa 404–560). The dissociation constant (Kd), stoichiometry (n) and molar enthalpy (ΔH) are indicated within the figure.
RPA is an evolutionarily conserved heterotrimeric protein, consisting of Rpa70, Rpa32 and Rpa14 (names based on molecular weight, or Rpa1, Rpa2 and Rpa3, based on gene names). The smallest subunit, Rpa14, is believed to mediate protein–protein interaction only within the RPA complex, while Rpa70 and Rpa32 were shown to bind other proteins (Binz et al, 2004; Zou et al, 2006). We therefore expressed full-length Rpa70 and Rpa32 fused to the LexA-DNA binding domain (LexA-DBD) under control of a galactose inducible promoter, as bait in the Y2H assay (Figure 1B). Strong interactions were scored between the largest subunit Rpa70 and the large core of the Sgs1 enzyme (aa 290–1180; Figure 1C; Supplementary Figure S1A), while Rpa32 showed a very weak β-galactosidase signal in the Y2H assay with Sgs1 (Supplementary Figure S1B). A 400-aa fragment containing only the acidic region N-terminal of the Sgs1 helicase domain bound Rpa70 as efficiently as a larger fragment (Figure 1C). Within this region, we identified three sequences of 35–41 aa by BLAST and Quick2D analysis, which are conserved among close homologues of the S. cerevisiae enzyme (dark red boxes, Figure 1; aa 404–485, aa 496–536 and aa 565–604). To test the importance of these conserved sequences for the Sgs1–Rpa70 interaction, we removed them by deleting aa 404–604 in the Sgs1-B42-AD construct. Consistently, this deletion abolished Y2H interaction between Rpa70 and Sgs1 (Figure 1C). Similarly, we mapped the interaction site on Rpa70 by fusing Rpa70 subdomains to the LexA-DBD and monitoring their interaction with the Sgs1(290–1180)-B42-AD fusion. The highest β-galactosidase activity was measured for the N-terminal oligonucleotide binding fold (N-OB) of Rpa70 without the linker region (Supplementary Figure S1A).
Sgs1 bears multiple interaction sites for the Rpa70 N-OB fold
To see if each conserved repeat within the Sgs1 acidic domain contributed equally to the interaction, we created three Sgs1-B42-AD deletion constructs each lacking only one of the three conserved sequences (Sgs1(290–1180, Δ404–485), Sgs1(290–1180, Δ496–536) and Sgs1(290–1180, Δ565–604)). These constructs were tested for interaction with the Rpa70 N-OB fold (Rpa70(1–133)) by Y2H analysis (Figure 1D). Deletion of the first or second conserved sequence (Sgs1(290–1180, Δ404–485) or Sgs1(290–1180, Δ496–536)) cut the β-galactosidase signal in half, while deletion of the third conserved sequence (Sgs1(290–1180, Δ565–604)) had no effect. This suggests that Sgs1 binds the RPA70 N-OB fold through two sites, aa 404–485 and aa 496–536. Indeed, deletion of the first two of the three motifs (aa 404–560 in Sgs1-B42-AD), abolished the interaction with the N-OB fold of Rpa70 almost as efficiently as deleting the entire 200 aa region, arguing that two related motifs spanning from aa 404 to 485 and aa 496 to 536, mediate the interaction with Rpa70.
We confirmed that these regions of Sgs1 and Rpa70 interact directly by performing an isothermal titration calorimetry (ITC) assay with purified recombinant proteins (Figure 1E and F). We found that both Sgs1(404–485) and Sgs1(404–560) bound RPA70(3–133) with similar affinity (Kd=70±20 μM and Kd=34±1 μM, respectively). The ITC data suggested differences in the complex stoichiometry (n) and molar enthalpy (ΔH) between the two Sgs1 fragments: it appears that the more N-terminal motif of Sgs1 (aa 404–485) binds one molecule of the RPA70 N-OB fold, while the larger domain (aa 404–560) is able to bind two. This suggests that Sgs1 might be able to bind multiple RPA complexes, possibly leading to RPA delivery and/or removal as Sgs1 unwinds duplex DNA.
The Sgs1–RPA interaction site is structurally isolated from the helicase domain and its deletion does not affect protein stability
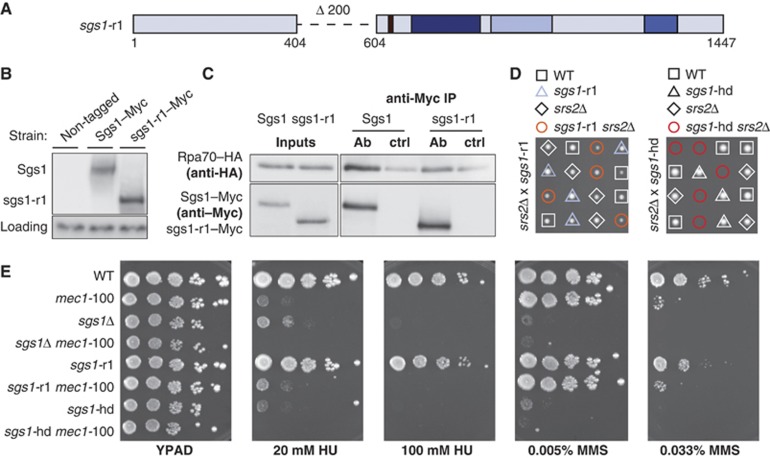
To determine whether this region of Sgs1 is also important for interaction with RPA in vivo, we deleted amino acids 404–604 within the SGS1 chromosomal locus using a PCR-based allele-replacement technique. The resulting allele (sgs1-r1; Figure 2A), C-terminally tagged by 13Myc, is expressed when tested by western blot analysis on whole cell extracts (Figure 2B and C, inputs). The signal for sgs1-r1–13Myc is slightly stronger than for wild-type Sgs1–13Myc, which may either reflect a better blotting efficiency or slightly improved stability. This deletion leaves intact the aa 664 shown to be important for resolving recombination intermediates, and is distinct from the previously published AR2 deletion, which reduced Sgs1 levels (Bernstein et al, 2009).
Figure 2.
Loss of the acidic region in Sgs1 impairs Sgs1–RPA interaction in vivo. (A) Schematic representation of sgs1-r1: a new allele generated by deleting the acidic region (aa 404–604) at the endogenous SGS1 locus. The deleted region is indicated by a dashed line. Black=acidic block, dark blue=helicase domain, light blue=RQC domain, blue=HRDC domain. (B) Wild type and sgs1-r1 were C-terminally fused to 13Myc epitopes at the endogenous SGS1 locus (Sgs1–13Myc; GA-5311, sgs1-r1–13Myc; GA-5313) and expression levels were analysed by western blot with anti-Myc antibody. Non-tagged strain (GA-7249) was used as a negative control. Anti-actin was used to detect Act1 as a loading control. (C) Co-IP of exponentially growing 13Myc-tagged Sgs1 (GA-1759) or sgs1-r1 (GA-5316) with 3HA-tagged Rpa70. Exponentially growing cells were collected for IP using Dynabeads either coupled to monoclonal anti-Myc (AB) or not (ctrl). Western blots were probed with anti-Myc (9E10) for Sgs1 or sgs1-r1 and anti-HA (F-7) for Rpa70. (D) sgs1-r1 (GA-4848) and srs2Δ (GA-1805), as well as sgs1-hd (GA-5445) and srs2Δ (GA-5334) mutants were crossed and sporulated and tetrad analysis was performed. (E) Ten-fold serial dilutions of the following strains were plated onto YPAD, ±20 mM or 100 mM HU, 0.005% or 0.033% MMS: GA-1981 (WT), GA-4978 (mec1-100), GA-5457 (sgs1Δ), GA-4967 (sgs1Δ mec1-100), GA-5076 (sgs1-r1), GA-5077 (sgs1-r1 mec1-100), GA-5445 (sgs1-hd) and GA-5447 (sgs1-hd mec1-100).
To monitor the interaction of the sgs1-r1 mutant protein with RPA in vivo, we performed co-immunoprecipitation (co-IP) experiments with appropriately tagged proteins. Strains expressing Rpa70–3HA and either 13Myc-tagged Sgs1 or sgs1-r1 were released from G1 phase for 20 min to allow cells to accumulate in S phase. Rpa70–3HA and Sgs1-13Myc were efficiently precipitated as a complex using either anti-Myc antibody (Figure 2C) or anti-HA (Supplementary Figure S2). In the sgs1-r1–13Myc precipitation, the efficiency of Rpa70 recovery was reduced roughly two-fold (Figure 2C), as was the reciprocal recovery (sgs1-r1 by Rpa70, Supplementary Figure S2). This suggests that the deleted domain indeed mediates Sgs1–RPA interaction in vivo, although other contacts may support interactions in the context of the holo-RPA complex. Indeed, residual binding could be explained by the interaction detected between Sgs1 and Rpa32 (Supplementary Figure S1B), through a site unaffected by the sgs1-r1 mutation, or by an indirect interaction of sgs1-r1–13Myc and Rpa70–3HA to DNA.
To ensure that the sgs1-r1 protein retained helicase activity, despite the reduced interaction with RPA, we tested the ability of sgs1-r1 to support growth in a srs2 null background. Rothstein and colleagues have shown that the helicase activity of Sgs1 is essential for growth in the absence of the Srs2 helicase (Weinstein and Rothstein, 2008). Tetrad analysis confirms that spores containing helicase-dead Sgs1 (K706R or sgs1-hd) in combination with srs2Δ showed almost no growth, while the sgs1-r1 srs2Δ double mutant spores grew normally (Figure 2D). We conclude that sgs1-r1 retains helicase activity, consistent with its weak suppression of top3Δ slow growth (Supplementary Figure S3).
We next tested whether the sgs1-r1 allele yields the same levels of sensitivity to DNA damage as sgs1Δ in drop assays. This assay monitors the capacity of cells to repair DNA damage induced by different genotoxic drugs. Previous work has shown that sgs1Δ is sensitive to low concentrations of the replication fork inhibitor HU and the alkylating agent MMS, and that this phenotype is dramatically enhanced when sgs1Δ is coupled to the S phase-specific Mec1 mutant, mec1-100 (Cobb et al, 2005). As described previously, we observed that both sgs1Δ and the helicase-dead mutant sgs1-hd are sensitive to HU and MMS, and show synthetic sensitivity when combined with the mec1-100 allele (Figure 2E). This was not observed for the sgs1-r1 mutant, although it did show a mild sensitivity to high concentrations of MMS (0.033% MMS, Figure 2E). Surprisingly, no additive effects were seen when sgs1-r1 mec1-100 cells were compared with the single mec1-100 mutant, suggesting that either sgs1-r1 acts on the same pathway as mec1-100 or it simply does not affect survival during persistent replicative stress (Figure 2E).
The Sgs1 helicase function, but not its R1 domain, stabilizes DNA pol α on HU
Given that fork restart after prolonged exposure to HU requires different activities than does growth under persistent damage, we next arrested S-phase cells in 0.2 M HU for 2–6 h, and quantitatively measured cell survival after plating on HU-free YPAD (Figure 3A). Similar to the lack of sensitivity observed on HU-containing plates, we found that sgs1-r1 did not confer sensitivity to this acute HU treatment. Unlike the sgs1Δ mec1-100 or sgs1-hd mec1-100 double mutants (Figure 3A), the sgs1-r1 mec1-100 combination showed no additive or synergistic effects. This suggested that strong RPA-Sgs1 binding is not crucial for fork recovery after HU-induced replication fork arrest.
Figure 3.
sgs1-r1 does not destabilize DNA pol α from HU-stalled replication forks in contrast to a helicase-deficient sgs1 mutant. (A) Recovery from replication fork stalling was monitored as colony outgrowth of cells which were synchronized in G1 by α-factor arrest and released into S phase in the presence of 0.2 M HU for the indicated times. Strains used were GA-1981 (WT), GA-180 (WT rad5-535), GA-5457 (sgs1Δ), GA-4978 (mec1-100), GA-4967 (sgs1Δ mec1-100), GA-5076 (sgs1-r1), GA-4504 (sgs1-r1 mec1-100 rad5-535), GA-5445 (sgs1-hd) and GA-5447 (sgs1-hd mec1-100). 0 min after release indicates G1 phase. (B, C) ChIP was performed on synchronized cultures released into S phase in the presence of 0.2 M HU. 3HA-tagged DNA pol α was precipitated with monoclonal anti-HA antibody (F-7) coupled to Dynabeads. Strains used were GA-4973 (WT), GA-4974 (mec1-100), GA-5055 (sgs1-r1), GA-5075 (sgs1-r1 mec1-100), GA-5449 (sgs1-hd) and GA-5451 (sgs1-hd mec1-100). The ChIP data for sgs1Δ and sgs1Δ mec1-100 from Cobb et al (2005) are shown for comparison (indicated by the dashed lines). The relative enrichment for ARS607 or ARS522 was obtained by normalizing the absolute enrichment at ARS607 or ARS522 for each time point to the absolute enrichment at a locus 14 kb away from ARS607. Mutant strains are indicated using the same colour code as in (A). (D) ChIP was performed as described above using strains GA-4973 (WT), GA-6266 (sgs1-r1-hd), GA-6260 (sgs1-r1-hd mec1-100) and GA-4975 (sgs1Δ mec1-100). (E) Primers (grey bars) used for ChIP that amplify the genomic regions corresponding to the early firing origin ARS607, a region 14 kb away from ARS607 (+14 kb) and the late firing origin ARS522 are shown.
We next tested the effects of sgs1-r1 on DNA pol α association at replication forks arrested on HU. We performed ChIP for DNA pol α after synchronizing single and double mutants in G1 and releasing them into S phase in the presence of 0.2 M HU. Over 1 h, the abundance of DNA-bound DNA pol α was quantified by real-time PCR analysis of the recovered DNA, using primers that amplify the early firing origin ARS607 and the late firing origin ARS522 (formerly ARS501; Figure 3E). As a negative control, a primer set placed 14 kb away from ARS607 was analysed and used to normalize the absolute enrichment at ARS607 or ARS522. As previously reported (Cobb et al, 2005), sgs1Δ or mec1-100 cells yield lower recoveries of DNA pol α at HU-arrested replication forks (Figure 3B, black solid and stippled lines). This effect is additive when both mutations are combined, resulting in complete loss of DNA pol α from the stalled fork (Figure 3B, blue stippled line). The sgs1-r1 mutant showed a much weaker displacement of DNA pol α than sgs1Δ, and was not additive when combined with the mec1-100 mutation (Figure 3B, second panel). Similar results were obtained, when we scored DNA pol α association at two other early firing origins, ARS306 and ARS606, while ARS603.5, which fires in mid-S phase, displayed a pattern similar to the late origin ARS522 (Supplementary Figure S4).
Because HU treatment activates the intra-S phase checkpoint response, late firing origins like ARS522 are repressed, and show no DNA pol α in wild-type cells (Figure 3C, black line). However, due to the compromised intra-S-phase checkpoint in mec1-100 cells, DNA pol α is detected at the late firing origin ARS522, by 40 min on HU (Figure 3C, grey line). Interestingly, loss of Sgs1 combined with mec1-100 reduces polymerase recovery at this late origin firing, or else partially maintains repression. However, the sgs1-r1 mutation alone did not promote late origin firing, and was largely epistatic with mec1-100 for late origin activation (Figure 3C, second panel). Similar effects were observed at the mid-to-late firing origin ARS603.5 (Supplementary Figure S4). We conclude that deletion of a major Sgs1–RPA interaction domain has little or no effect on DNA pol α binding at HU-arrested forks, consistent with the colony outgrowth assay in Figure 3A.
Given that the R1 deletion did not affect polymerase binding, nor act synergistically with mec1-100 like sgs1Δ, we examined if Sgs1 helicase activity is responsible for these functions. By performing ChIP in the sgs1-hd (K706R) and the sgs1-hd mec1-100 double mutant (Figure 3B and C, third panel), we scored a significant destabilization of DNA pol α at the HU-stalled fork, comparable to sgs1Δ. This shows that the enzymatic activity of Sgs1 helicase is indeed critical for DNA pol α stabilization. However, the sgs1-hd mutation was not additive with mec1-100; the double mutant showed the same level of residual DNA pol α recovery as sgs1-hd alone (Figure 3B). Thus, whereas sgs1-hd and mec1-100 are synergistic in the recovery assay (Figure 3A), they are not with respect to polymerase stability. The sgs1Δ and mec1-100 defects, on the other hand, are additive in both assays (Cobb et al, 2005).
To see if a strain that carries both the R1 deletion and helicase deficiency in Sgs1 (sgs1-r1-hd) shows even stronger polymerase destabilization, we tested this strain alone and in combination with mec1-100. However, sgs1-r1-hd mec1-100 cells resemble sgs1-r1-hd for polymerase ChIP, with neither completely displacing DNA pol α from ARS607 (Figure 3D). We conclude that the helicase function of Sgs1, and not its interaction with RPA, is crucial for stabilizing DNA pol α at stalled replication forks. We note that other Sgs1 functions may contribute to lagging strand replisome retention, given that sgs1-r1-hd mec1-100 cells still retain low levels of polymerase on HU-stalled forks. Consistently, sgs1-r1-hd mutants grew better than sgs1Δ cells on 10 mM HU, although both display synergistic lethality with mec1-100 (Supplementary Figure S5).
The sgs1-r1 mutant is a separation-of-function mutant that does not cause synthetic defects with replication mutants
To shed light on the function of the R1 domain within Sgs1, we next compared the behaviour of sgs1Δ and sgs1-r1 in a synthetic lethal screen against a sublibrary of yeast deletion strains. Previously, Collins et al (2007) had sorted 743 S. cerevisiae genes using hierarchical clustering according to the similarity of their genetic interaction profiles, which allowed them to distinguish functionally multi-protein complexes and to group different protein complexes into pathways. We performed a similar epistasis analysis (epistasis miniarray profile, E-MAP), comparing the interaction profiles of sgs1Δ and sgs1-r1 cells with 1536 genes involved in various chromatin functions (Guénole et al, under review). For this, we created four sets of double mutants in two independent experiments using a high-throughput setup (Schuldiner et al, 2006). Genetic interactions were quantified by calculating scores based on colony sizes of double mutants (Collins et al, 2006). Threshold levels of genetic interaction scores equal or lower than −2 and equal or higher than +2 were used to define negative (synthetically sick, blue) or positive (epistatic or suppressive, yellow) genetic interactors. In Figure 4A, we list genes showing genetic interaction with either sgs1Δ or sgs1-r1 in two independent experiments (bold), and genes that were previously shown to be synthetically lethal with sgs1Δ (in red; Ooi et al, 2003; Tong et al, 2004), along with other genes falling within the relevant functional clusters (Collins et al, 2007).
Figure 4.
sgs1-r1 does not interact genetically with lagging strand replication machinery. (A) Genetic interaction profiles of sgs1Δ and sgs1-r1 were determined by crossing with a strain library comprising 1536 knockout alleles (Guénole et al, under review) and measuring growth of resulting double mutants as described in Materials and methods. Genetic interaction scores (GIS) of selected double mutants obtained from two independent experiments are depicted by colour (positive (yellow)=epistatic/suppressive, negative (blue)=synthetically sick). Genes were sorted into functional groups as performed earlier (Collins et al, 2007). Bold represents the mutant genes with GIS above threshold (−2 for negative or +2 for positive interactions) for double mutants with sgs1Δ and/or sgs1-r1 in two independent screens. Red represents gene deletions previously reported as synthetically lethal with sgs1Δ (Tong et al, 2001). Genes marked with ‡ were previously reported to be a target of checkpoint kinases (Chen et al, 2010). Mus81 (*) was reported to be a target of S. pombe Cds1 (S. cerevisiae Rad53) (Kai et al, 2005). The † indicates genes that showed synthetic lethality with sgs1-r1, while o indicates no synthetic lethal interaction. Arrows indicated mutants further analysed in (B). (B) Left: a representative picture of 10-fold serial dilutions of the following strains plated onto YPAD and YPAD with 50 mM HU: GA-1981 (WT), GA-5457 (sgs1Δ), GA-5076 (sgs1-r1), GA-7549 (dia2Δ), GA-7540 (dia2Δ sgs1Δ), GA-7542 (dia2Δ sgs1-r1), GA-7548 (rad27Δ), GA-7530 (rad27Δ sgs1Δ), GA-7532 (rad27Δ sgs1-r1), GA-7498 (pol32Δ), GA-7499 (pol32Δ sgs1-r1) GA-7494 (mus81Δ) and GA-6326 (mus81Δ sgs1-r1). Right: the number of colonies scored on 50 mM HU plates normalized by colony number on YPAD plates. Error bars indicate standard deviation of three independent experiments. (C, D) ChIP of 13Myc-tagged RPA or 3HA-tagged Ddc2 using strains GA-5525 (WT) and GA-5365 (sgs1-r1) as described in Figure 3.
In the E-MAP, the sgs1-r1 mutant displayed a distinct interaction profile from either that of sgs1Δ or wild-type cells (Figure 4A). There are a few clear conclusions from this analysis. Importantly, sgs1-r1 did not display negative interaction with replication mutants. In particular, sgs1-r1 shows mildly improved growth with strains that lack enzymes involved in lagging strand synthesis, such as Rad27, Pol32 and RNAseH2 (Qiu et al, 1999; Arudchandran et al, 2000; Ii and Brill, 2005), whereas the sgs1Δ mutant showed synthetic sickness with these strains, even under non-damaging conditions. Neither query strain scored negatively with genes involved in homologous recombination, perhaps because the screen was performed under non-damaging conditions. Nonetheless, several interactions stand out as having negative synthetic effects with sgs1-r1, namely mus81Δ and wss1Δ. Mus81 works together with Mms4 as a Holliday junction resolvase that is thought to work together with Sgs1–Top3–Rmi1 to process homologous recombination repair (HRR) intermediates that occur behind replication forks (for review, Hickson and Mankouri, 2011). Wss1 is a SUMO peptidase associated with Slx5/Slx8, the SUMO-dependent Ubiquitin ligase required for telomere-telomere recombination and recovery from fork collapse (Mullen et al, 2010; Nagai et al, 2011).
We confirmed the strongly negative interaction of sgs1-r1 with mus81Δ by generating the double mutant independently. The slow growth phenotype persisted and cells were hypersensitive to HU (Figure 4B), while the mus81Δ sgs1Δ strain was synthetic lethal (data not shown and Tong et al, 2001). This implicates the R1 domain of Sgs1 in the resolution of, or recovery from, recombination events at the fork, particularly those that might form in the absence of Mus81-Mms4. We note, however, that sgs1-r1 does not aggravate the HRR pathway per se (Figure 4A, including rad51Δ, Supplementary Figure S6B). Based on the E-MAP data, we suspected that slx4Δ, slx5Δ, slx8Δ and mre11Δ might also be synthetic lethal with sgs1-r1, since they failed to yield E-MAP results. We crossed strains bearing these null alleles with sgs1-r1 and again tested synthetic lethality. We confirmed that slx4Δ, slx5Δ and slx8Δ are synthetic lethal with sgs1-r1, whereas mre11Δ is not (Figure 4A; data not shown). This result suggests that the R1 domain of Sgs1 confers an essential function when cells lack the activity for Slx5/Slx8 SUMO-dependent ubiquitin ligase or the Slx4-Slx1 structure-specific endonuclease. These results are fully consistent with the synthetic lethality observed in the absence of Wss1, which forms a complex with Slx5/Slx8, or of Mus81, which together with Mms4 helps resolve strand-exchange events, in parallel to the Slx4-Slx1 complex (Muñoz-Galván et al, 2012). In contrast, even though the loss of topoisomerase I should enhance torsional stress and promote strand exchange, we saw only a minor negative interaction between sgs1-r1 and top1Δ, by E-MAP or in drop assays on HU (Supplementary Figure S6A).
We extended the E-MAP observations to conditions of replicative stress by testing different mutant combinations with sgs1-r1 or sgs1Δ on plates containing HU (Figure 4B). Consistently, we saw that combinations of sgs1-r1 with genes that function in lagging strand synthesis (pol32Δ, rad27Δ) do not show negative interactions even on HU, while the corresponding double mutants with sgs1Δ showed either synthetic lethality or synthetic slow growth (Figure 4A and B). Similarly, there was no synthetic defect when sgs1-r1 was combined with the deletion mutant of the origin and replication fork associated F-box protein Dia2 (Mimura et al, 2009; Morohashi et al, 2009), either with or without HU (Figure 4A and B). Consistent with the ChIP data, we conclude that the sgs1-r1 mutation does not destabilize the stalled replication fork. However, sgs1-r1 is important when cells are deficient for functional complexes that promote replication fork recovery through exchange or invasion of the sister chromatid.
Given that the R1 domain of Sgs1 binds RPA, we asked whether the RPA levels at the stalled replication fork might be altered in the sgs1-r1 mutant. However, levels of Myc-tagged RPA at the early origin ARS607 were not changed in sgs1-r1 cells synchronously arrested in S phase by 0.2 M HU (Figure 4C). RPA was undetectable at the late firing origin ARS522 (Supplementary Figure S7A). This argues that although RPA–Sgs1 interaction is diminished in sgs1-r1 cells, it neither affects RPA deposition at stalled replication forks, nor the amount of ssDNA generated. Synthetic lethality might also arise through an inability to recruit or activate Mec1-Ddc2. To test this, we performed ChIP for the essential Mec1 cofactor, Ddc2, in both wild-type and sgs1-r1 cells (Figure 4D; Supplementary Figure S7B). Again, we detected no change in the level of Mec1-Ddc2 recruitment in the R1 mutant. We conclude that the R1 domain functions, but on a pathway that is essential in the absence of Mus81, Slx4/Slx1 or the Slx5/Slx8/Wss1 ubiquitin conjugating complex.
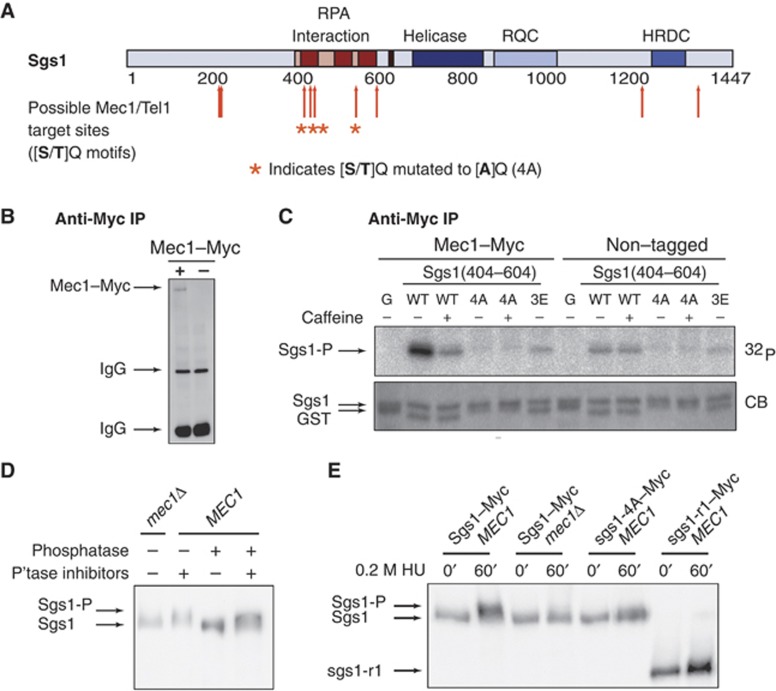
The Sgs1 R1 domain is phosphorylated by Mec1 in vitro and in vivo
Earlier work had implicated a function of Sgs1 in intra-S phase checkpoint activation, independent of its enzymatic activity (Frei and Gasser, 2000; Bjergbaek et al, 2005). We therefore investigated whether the R1 domain of Sgs1 might be implicated in checkpoint activation, or be a target of Mec1-Ddc2 phosphorylation. Activation of the intra-S phase checkpoint by Mec1 occurs through the phosphorylation of numerous targets containing SQ/TQ motifs. Previous studies report that Mec1 substrates often contain several closely spaced motifs, which are also referred to as SQ/TQ cluster domains (SCDs; Traven and Heierhorst, 2005). Such SCDs are defined by at least three SQ/TQ motifs within 100 aa with additional sites within another 100 aa. The human Sgs1 homologues BLM and WRN contain such clusters and are phosphorylated by ATR/ATRIP (Pichierri et al, 2003; Davies et al, 2004; Rao et al, 2005), the human homologue of Mec1-Ddc2.
Sequence analysis revealed nine SQ or TQ sites within Sgs1, of which four are clustered in the R1 region (T451Q452, S470Q471, S482Q483, T585Q586) with a fifth nearby (S628Q629; Figure 5A). To test if this domain of Sgs1 was indeed a target for Mec1 in vitro, we expressed Sgs1 aa 404–604 in bacteria and challenged it with kinase immunoprecipitated from a yeast strain expressing an epitope-tagged Mec1. A parallel precipitate from a yeast strain lacking the 18Myc tag was used as a control (Figure 5B and C). The Sgs1(404–604) domain is a robust substrate for Mec1–Myc phosphorylation (Figure 5C). We could significantly reduce the phosphate incorporation into the domain by adding the Mec1/Tel1 inhibitor caffeine to the in-vitro reaction, while the background phosphorylation in a non-tagged strain was not caffeine sensitive.
Figure 5.
Mec1 targets the RPA-interacting site on Sgs1 in vitro and in vivo. (A) Schematic representation of Sgs1 with its functional domains. Mec1/Tel1 consensus target sites are indicated by red arrows, while asterisks tag phospho-acceptor sites mutated to A or E. (B) Exponentially growing GA-1456 (Mec1–18Myc) and GA-426 (non-tagged) cells were exposed for 1 h to 0.1% MMS. After cell lysis, Mec1–18Myc was precipitated using anti-Myc coupled Sepharose beads and analysed by western blot. (C) Kinase assay using Mec1–18Myc or control immunoprecipitates (non-tagged lysates). Sgs1 peptides (aa 404–604) were used as substrates in the presence of γ-32P-ATP and analysed by gel electrophoresis and autoradiography. Where indicated, 30 mM caffeine was added to the reaction to inhibit Mec1. Mutant Sgs1 peptides were as follows: 4A=Sgs1(404–604-T451A-S470A-S482A-T585A) or 3E=Sgs1(404–604-T451E-S470E-S482E) 32P autoradiography and Coomassie Brilliant Blue (CB) staining are shown. (D) Exponentially growing GA-6402 (Sgs1–13Myc) cells were exposed for 2 h to 0.2 M HU. Native extracts were treated with phosphatase and/or phosphatase inhibitors as described in Materials and methods and subjected to western blot analysis using anti-Myc antibody. A denatured extract of GA-6403 (Sgs1–13Myc mec1Δ) cells treated as described in (E) was loaded as a control. (E) Cells were synchronized in G1 by α-factor arrest and released into S phase in the presence of 0.2 M HU for 60 min. Protein extracts were analysed by western blot using anti-Myc antibody. Strains used were GA-6402 (Sgs1–13Myc), GA-6403 (Sgs1–13Myc mec1Δ), GA-7485 (sgs1-4A–13Myc) and GA-5313 (sgs1-r1–13Myc).
We next mutated the SQ/TQ motifs in this region and purified substrates with either alanine or glutamate substitutions at the relevant phospho-acceptor sites, generating sgs1-4A (Sgs1(404–604-T451A-S470A-S482A-T585A)) and sgs1-3E (Sgs1(404–604-T451E-S470E-S482E). These mutations efficiently abolished the modification of Sgs1 by precipitated Mec1–18Myc (Figure 5C). As the sgs1-3E mutant was also sufficient to abrogate Sgs1 phosphorylation in vitro, it is likely that Mec1-Ddc2 targets T451, S470 and/or S482.
To investigate whether Sgs1 is phosphorylated in vivo, we examined extracts from both wild-type and mec1Δ sml1Δ strains expressing Sgs1–13Myc, after growth for 2 h in 0.2 M HU. Indeed, in wild-type cells a diffuse and more slowly migrating form of Sgs1–13Myc was observed, that was not detected in cells lacking Mec1 (Figure 5D), or in wild-type cells not treated with HU (data not shown). Treatment of the cell lysate with phosphatase eliminated the presence of the slower migrating band, while the addition of both phosphatase and phosphatase inhibitors restored the signal. We conclude that the slower migrating band of Sgs1 reflects a phosphorylated form, generation of which requires Mec1 kinase.
We next asked whether Mec1 indeed targets the R1 region of Sgs1, by expressing Myc-tagged versions of wild-type Sgs1, and mutant forms bearing either the four S/T mutations (sgs1-4A–13Myc) or a deletion of the entire R1 domain (sgs1-r1–13Myc). We examined the shift in mobility in cells that were first synchronized in G1 and released into medium containing 0.2 M HU. A strain lacking Mec1 kinase (mec1Δ sml1Δ) was used as a control (Figure 5E). Sgs1–13Myc did not show a shift in G1-arrested MEC1 cells, while the slower migrating, phosphorylated form was evident in cells exposed to HU (Figure 5E). The phosphorylation of Sgs1 was lost in mec1Δ sml1Δ cells exposed to HU, and importantly, also greatly diminished in both the sgs1-4A–13Myc and sgs1-r1–13Myc mutants (Figure 5E). We detected a residual shift in the sgs1-4A–13Myc and sgs1-r1–13Myc, which may be explained by phosphorylation outside of the R1 domain. Indeed, our own LC-MS/MS data and data of (Bodenmiller et al, 2008, 2010) indicate that Sgs1 may be phosphorylated either on residue 1221/1222/1223 (Supplementary Table S2) or on residues 348, 1268 or in peptide aa 605–619. These Sgs1 phospho-acceptor sites are not typical Mec1 target sites as they lack the SQ/TQ consensus, and they may reflect phosphorylation by a kinase downstream of Mec1. Important to note is that the confirmed in-vitro Mec1 target sites within the R1 domain (T451 and/or S470) are found within a large tryptic cleavage fragment that could not be detected in our MS analysis (47 aa; see Supplementary Table S2), even when combined chymotryptic and tryptic digestions were used (data not shown). In contrast, S482 is in a smaller peptide that is not phosphorylated in vivo.
Earlier work from our laboratory showed that Sgs1 interacts directly with the FHA1 domain of the major Mec1 target and checkpoint kinase, Rad53 (Bjergbaek et al, 2005). The FHA1 domain of Rad53 is a phosphopeptide binding module that has a key role for both intra-S checkpoint activation and late origin control (Pike et al, 2004a). To determine whether the Mec1-modified R1 domain of Sgs1 is the site of Rad53 interaction, we first employed Y2H assays. We detect a strong interaction between the core Sgs1 fragment (290–1180) and the FHA1 domain of Rad53, which is entirely lost upon deletion of aa 404–604 (sgs1-r1; Figure 6A). The Sgs1 subdomain aa 292–661 was sufficient to mediate binding to the Rad53 FHA1 domain, and again, deletion of aa 404–604 ablated interaction with Rad53 FHA1 (Figure 6A). We also tested a non-phosphorylatable sgs1-4A substitution within the smaller Sgs1 fragment, which gave a 50% drop in β-galactosidase signal, suggesting that phosphorylation is important for high binding affinity, although contacts to flanking residues may contribute to the interaction (Figure 6A). Given that Y2H assays can detect indirect interactions, we decided to turn to a more quantitative binding assay to evaluate the importance of the Mec1 phosphorylation sites on the Sgs1-Rad53 interaction.
Figure 6.
Sgs1 phosphorylation promotes Sgs1-Rad53 interaction. (A) Y2H analysis of Rad53-FHA1 fused to the B42-AD and Sgs1 fragments fused to LexA-DBD, as in Figure 1. Asterisks indicate phospho-acceptor sites mutated to alanine. (B) ITC assay of the FHA1 domain of Rad53, Rad53(22–162), and two Sgs1(446–456) peptides, Sgs1 (DDLDPTQDTDY), encompassing either phosphorylated T451Q452 (left panel) or non-phosphorylated T451Q452 (right panel). Parameters as in Figure 1E. (C) Co-IP of 13Myc-tagged Sgs1 (GA-7487), sgs1-4A (GA-7483) or non-tagged Sgs1 (GA-7467) with 9PK-tagged Rad53. Cells were released from G1 (α-factor arrest) for 60 min in the presence of 0.2 M HU and lysed for Dynabead-IP using monoclonal anti-Myc. Western blots were probed with anti-Myc for Sgs1 or sgs1-r1 and anti-PK for Rad53. An anti-PK-cross-reacting 250 kDa protein served as loading control. (D) Quantification of three independent Sgs1-Rad53 co-IP experiments. The unspecific binding (no tag control) signal was subtracted from Rad53-9PK signals in Sgs1- and sgs1-4A IPs, and normalized to Sgs1–13Myc or sgs1-4A–13Myc signals in the IP fractions. Error bars indicate standard deviation. (E) Rad53 activation in GA-1981 (WT), GA-5321 (rad24Δ), GA-5932 (sgs1-4A), GA-5934 (sgs1-4A rad24Δ), GA-5076 (sgs1-r1), GA-5324 (sgs1-r1 rad24Δ), GA-1761 (sgs1Δ) and GA-2056 (sgs1Δ rad24Δ). Cells were synchronized in G1 by α-factor arrest and released for 60 min into 0.2 M HU before denatured extract preparation as done previously (Pike et al, 2003). Rad53 phosphorylation (Rad53-P) was monitored by western blot with anti-Rad53. (F) Ten-fold serial dilutions of (WT), GA-5321 (rad24Δ), GA-5932 (sgs1-4A), GA-5934 (sgs1-4A rad24Δ), GA-5076 (sgs1-r1), GA-5324 (sgs1-r1 rad24Δ), GA-5457 (sgs1Δ) and GA-2056 (sgs1Δ rad24Δ) on YPAD ±50 mM HU, 0.03% MMS, or 40 μg/ml Zeocin.
Using ITC with the FHA1 domain and short Sgs1 peptides that encompass phosphorylated T451, S470 or S482 (Sgs1(446–456), Sgs1(466–475) and Sgs1(478–487) respectively), we analysed the interaction between these peptides and the minimal FHA1 domain (aa 22–162) comparing phosphorylated and non-phosphorylated acceptor sites. Remarkably, only the Sgs1(446–456) peptide carrying a phosphorylated T451Q452, and not its unphosphorylated counterpart, bound the FHA1 domain of Rad53 (Rad53(22–162) significantly (Figure 6B). The phosphorylated Sgs1(446–456) peptide had a dissociation constant KD=21±0.2 μM for the FHA1 domain, a value that is similar to that observed between Rpa70(3–133) and Sgs1(404–560) (Figure 1F). Two other peptides bearing phosphor-serine (S470 in Sgs1(466–475) and S482 in Sgs1(478–487)) did not show significant binding to the Rad53 FHA1 domain by ITC (Supplementary Figure S8), consistent with the demonstrated preference of FHA1 for p-Thr over p-Ser (Durocher et al, 2000; Pennell et al, 2010).
To determine if the phosphorylation of Sgs1 by Mec1 on these residues also mediates Rad53 interaction in vivo, we performed co-IP experiments with strains expressing Rad53-9PK and either Sgs1–13Myc or sgs1-4A–13Myc. A strain lacking the Sgs1–Myc tag was used as a control. Cells were synchronized in G1 and released into 0.2 M HU for 60 min, prior to lysis in buffer with phosphatase inhibitors. Precipitation by anti-Myc antibody, and subsequent western blotting with anti-PK or anti-Myc antibodies, showed that Rad53-9PK and Sgs1–13Myc precipitate efficiently as a complex (Figure 6C and D). The amount of Rad53-9PK recovered with sgs1-4A–13Myc was >2-fold less than with wild-type Sgs1 (Figure 6C and D). We conclude that Sgs1 interacts with Rad53 in a phosphorylation-dependent manner, requiring modification on SQ/TQ acceptor sites within its R1 domain. The RPA-interaction site on Sgs1 would thus be targeted by Mec1 after HU-induced replication fork stalling, enabling phospho-T451 interaction with Rad53.
sgs1-r1 cells display a defect in Rad53 activation in a rad24 background
Previous studies have shown that Sgs1 and Rad24 act on parallel pathways to activate Rad53 when cells are exposed to HU during S phase (Frei and Gasser, 2000; Bjergbaek et al, 2005), yet how Sgs1 leads to Rad53 activation is unclear. Since both the helicase activity of Sgs1 and its interaction with Top3 are dispensable for Rad53 activation (Bjergbaek et al, 2005), we next examined whether the sgs1-r1 and/or sgs1-4A mutations, which lack the Mec1-dependent phosphorylation sites and show reduced binding to Rad53 in Y2H, ITC and in-vivo assays, have an effect on checkpoint activation.
We released G1-synchronized cells into HU-containing media for 30 or 60 min and monitored Rad53 activation on SDS–PAGE. The hyper-phosphorylated, activated form of Rad53 was visualized on western blots as a band with delayed migration (Figure 6E). We tested the impact of either sgs1-r1 or sgs1-4A in both wild-type and rad24 mutant backgrounds, since Rad24/9-1-1 contributes to checkpoint activation through an alternative pathway in response to fork stalling (Frei and Gasser, 2000). Upon release from α factor arrest into HU, Rad53 became phosphorylated, not only in wild-type cells, but also in rad24Δ, sgs1-r1 or sgs1-4A single mutant cells, as well (Figure 6E). In the sgs1-r1 rad24Δ double mutant, however, the upshift of Rad53 was significantly reduced, as observed in the sgs1Δ rad24Δ mutant. Similarly, the sgs1-4A in combination with rad24Δ showed a strongly impaired upshift of Rad53. Since Mec1-Ddc2 recruitment is not altered in sgs1-r1 cells (Figure 4D), we conclude that the drop in Rad53 activation likely stems from impaired Rad53 recruitment by Sgs1.
Finally, we checked the effect of the Mec1 target sites in Sgs1 by combining the sgs1-4A mutant with mec1-100 or rad24Δ. In a drop assay on media containing HU, cells expressing a non-phosphorylatable sgs1-4A protein, behaved like sgs1-r1 cells: both showed wild-type sensitivity to HU and both were epistatic with mec1-100 (Figure 2E; Supplementary Figure S9A). Indeed, when we scored recovery from HU-induced arrest in S phase, the sgs1-4A mutant cells behaved like wild-type cells, even though the synergy of sgs1Δ with mec1-100 was readily reproducible (Supplementary Figure S9B). This was also true, when we combined sgs1-r1 and sgs1-4A mutant cells with rad24Δ cells in survival assays on HU (Figure 6F). However, when double mutants were scored on MMS, we found that sgs1-r1 is slightly additive with rad24Δ, and, both alone and in combination with rad24Δ, sgs1-r1 was as sensitive as either rad24Δ or sgs1Δ to Zeocin (Figure 6F). Surprisingly, sgs1-4A was not, suggesting that the R1 domain contributes more than just assisting checkpoint activation, which was reduced identically in sgs1-r1 and sgs1-4A mutants (Figure 6E). This is consistent with the E-MAP data which implicated the R1 domain in the resolution of strand-exchange intermediates that might arise as a result of double-strand breaks. The sensitivity of the sgs1-r1 mutation to Zeocin may reflect both Rad53 recruitment and a downstream function that is partially redundant with Mus81-Mms4 or Slx4-Slx1 (Figure 4A and B).
Discussion
RecQ helicases have been implicated in the maintenance of genome stability in multiple pathways, some of which involve the double cross-over resolving activity of Top3, while others do not (Cobb and Bjergbaek, 2006). Here, we show that the role of Sgs1 in preserving DNA pol α on the lagging strand at stalled replication forks is largely due to its helicase activity (Figure 3). Despite the strong affinity shown by Sgs1 for RPA, deletion of a major interaction site within Sgs1 for Rpa70 did not destabilize DNA pol α during HU arrest. On the other hand, we found that the Rpa70-binding domain also mediates interaction with the downstream checkpoint kinase Rad53 (Figure 5). Given that both in vitro and in vivo this interaction requires the phosphorylation of Sgs1 by the ATR homologue, Mec1, we propose that the Sgs1 phospho-R1-Rad53 interaction occurs only under conditions that activate Mec1-Ddc2, for example, at stalled replication forks.
Functional studies confirm the relevance of the interaction between the phosphorylated Sgs1 R1 domain with Rad53 FHA1, a domain of demonstrated importance for intra-S phase checkpoint activation (Boddy et al, 2000; Pike et al, 2004b; Smolka et al, 2006). Using appropriate mutants, we show that the R1 domain and the Mec1 phosphorylation sites within this domain contribute to Rad53 activation during fork arrest. This is most obvious in a rad24Δ background. Whether loss of Rad24 leads to a particular type of damage that requires Sgs1-mediated Rad53 recruitment, or whether it simply unmasks the dependence of the event on Sgs1, is unknown. However, our results suggest that the sgs1-r1 mutation neither impairs the generation of ssDNA nor Mec1-Ddc2 recruitment (Figure 4C and D); nonetheless, it affects Rad53 activation during fork arrest on HU (Figure 6E). We therefore suggest that the R1 domain of Sgs1, once modified by Mec1, recruits Rad53 to the complex of ssDNA, RPA, Ddc2-Mec1, to facilitate its activation. This demonstrates a non-enzymatic role of Sgs1 in the DNA damage response at stalled forks (Figure 7). Sgs1 clearly plays other roles in the pathways leading to replication fork recovery, namely, the helicase-dependent reversal of fold-back structures and the resolution of unproductive strand exchange events behind the fork (reviewed in Bernstein et al, 2010).
Figure 7.
Multiple roles for Sgs1 helicase at stalled replication forks: reversal of fold-back structures, polymerase stabilization and Rad53 recruitment. (A) Sgs1 helicase activity contributes to the retention of DNA polymerases at stalled replication forks possibly by reversal of fold-back or strand invasion structures between nascent or template strands. The checkpoint kinase Mec1-Ddc2 is recruited to ssDNA coated by RPA and phosphorylates Sgs1 in the R1 domain. The green hexameric ring is the MCM helicase. (B) Sgs1 phosphorylation may shift Sgs1 binding preference from RPA to the Rad53 FHA1 domain. This could recruit Rad53 into close proximity of Mec1-Ddc2 which is associated with stalled forks. Mec1-Ddc2 then phosphorylates Rad53, to activate it and initiate the intra-S phase checkpoint response.
Multiple binding sites in a domain upstream of the Sgs1 helicase domain
The domain we identify here in Sgs1 as both an RPA and Rad53 binding site is upstream of the helicase domain and overlaps partially with an acidic domain (aa 502–648) implicated in replication fork stability on MMS (Bernstein et al, 2009). We define both an RPA-binding region (aa 404–560) and a Rad53-binding motif (aa 446–456), and show by ITC, Y2H interaction and co-IP that the latter interaction is phosphorylation dependent, while the former is not.
Both the interaction sites and the Mec1 acceptor sites that we have mapped are distinct from the sgs1-D664Δ point mutant studied by Bernstein et al (2009), which partially mimics the larger deletion. Their AR2 deletion does not eliminate the Sgs1-FHA1 interaction site, but may have indirect effects, given that the mutant protein appeared to be unstable. Both the RPA and Rad53 binding activities of Sgs1 are also independent of the Siz1/Siz2 sumoylation site K621 in Sgs1, which remains intact in our mutants, but was removed by the AR2 deletion (SC Teng, personal communication). Nonetheless, the sgs1-r1 mutant shows similar phenotypes as sgs1-AR2Δ or sgs1-D664Δ with respect to HU and MMS sensitivity during continuous growth on plates. Both sets of mutations suppress top3Δ slow growth (Supplementary Figure S3), and thus retain Sgs1 helicase activity (Bernstein et al, 2009).
sgs1-hd mutants fail to stabilize the replisome at stalled forks
In our attempt to explore the importance of Sgs1–RPA interaction on lagging strand polymerases, we found instead that Sgs1 helicase activity is essential for the maintenance of DNA pol α at ARS607 (Figure 3). Additional removal of the R1 domain in the sgs1-r1-hd mutant did not aggravate this effect. This result supports earlier findings from our laboratory suggesting that sgs1-hd expression cannot complement sgs1Δ for DNA pol ε stability on HU (Cobb et al, 2003). We show by epistasis analysis that sgs1-r1 is a separation of function mutant, because sgs1-r1, unlike sgs1Δ, does not exhibit negative synthetic interaction when combined with replication mutants. Nonetheless, the R1 domain contributes to Rad53 activation on HU, and shows strong synergistic lethality with deletions of mus81, slx4, or with slx5 and/or slx8 (Figure 4).
These results argue that Sgs1 probably contributes to replication polymerase stability by a dissolution of fold-back structures (i.e., upstream of Mec1 activation) and/or resolution of strand exchange behind the fork (Bennett et al, 1999; Bjergbaek et al, 2005; Ashton and Hickson, 2010). Holliday junction dissolution is especially important for restart of a collapsed fork after DNA damage, which occurs after persistent exposure with MMS or HU in drop assays. Consistently, both Sgs1 helicase activity and its Top3 interaction contribute to DNA repair and survival during prolonged exposure to HU or MMS. This may explain the synthetic lethality observed between sgs1-r1 and mus81Δ and slx4Δ in the E-MAP analysis, since strand exchange is thought to enable the fork restart that is necessary when lesions block leading or lagging strand elongation (Lambert et al, 2010; Muñoz-Galván et al, 2012). Given that these pathways may be controlled by Slx5-Slx8 ubiquitination and subsequent degradation of a factor blocking strand exchange, all E-MAP results are consistent with a contribution of the R1 domain of Sgs1 to this pathway.
What, then, is the importance of the Sgs1–RPA interaction? Whereas RPA has been shown to stimulate RecQ helicase activity in human cells (Shen et al, 2003; Machwe et al, 2006; Sowd et al, 2009; Yodh et al, 2009), it is not clear that RPA truly promotes unwinding. Direct interaction of RPA with RecQ helicases may alter enzymatic activity through a conformational change or switch in the oligomeric state of the RecQ helicase. It is noteworthy that RPA also counteracts the annealing activity of the RecQ helicase by stabilizing the ssDNA that occurs from unwinding (Bachrati and Hickson, 2008). Different Sgs1–RPA disruption mutants or further analysis of the relevant Rpa70 OB fold will be necessary to identify the exact role of Sgs1–RPA interaction for helicase activity and replication fork integrity.
Sgs1 R1 domain phosphorylation by Mec1-Ddc2 aids Rad53 activation
The most novel aspect of our study is to show how Sgs1 contributes to Rad53 activation during conditions that arrest replication forks. We propose that phosphorylated Sgs1 interacts with the FHA1 domain of Rad53 to promote Rad53 recruitment to sites of damage. We found that Sgs1 is a target of Mec1 in vitro and in vivo and we map the phosphorylation sites to the Rad53-interaction domain. Importantly, ITC studies show that the binding of Sgs1(446–456) to Rad53 FHA1 is phospho-threonine dependent, while Sgs1 binding to RPA through the same domain is not. We confirm that mutation of the key Mec1 acceptor sites alters Rad53 activation, Sgs1–Rad53 interaction, and deletion of the domain confers sensitivity to Zeocin. We do not know if phosphorylation of the Sgs1 R1 domain also facilitates Rad53 recruitment to DSBs, but we consider it likely.
An additional fork-associated adaptor protein, Mrc1, has been implicated as a bridge between Mec1-Ddc2 and Rad53 activation at stalled replication forks (Alcasabas et al, 2001; Osborn and Elledge, 2003; Crabbé et al, 2010). Indeed, in-vitro checkpoint reconstitution assays have shown that Mrc1 can facilitate the phosphorylation and activation of Rad53 probably by promoting Mec1–Rad53 interaction (Chen and Zhou, 2009). A recent study demonstrated that Mec1- but not Rad53-dependent phosphorylation of Mrc1 is necessary for the establishment of a positive feedback loop that leads to Mec1 stabilization at stalled replication forks (Naylor et al, 2009). Thus, Mrc1 may act prior to Sgs1 function ensuring Mec1-Ddc2 recruitment to stalled replication forks, which in turn is necessary for Sgs1 phosphorylation (Naylor et al, 2009). While Mrc1 and Sgs1 are epistatic for Rad53 activation on HU, they nonetheless show strong synergistic defects for stalled fork recovery and DNA polymerase stability on HU (Bjergbaek et al, 2005). This argues that Mrc1, like Sgs1, serves multiple roles at stalled forks. One possible scenario for the synergy observed between Sgs1 and Mrc1 would be that they act preferentially on different strands (the leading versus the lagging strand). Indeed, Mrc1 has been shown to function together with leading strand polymerase pol ε (Lou et al, 2008).
As in yeast, the human RecQ helicase BLM has been shown to be a target of ATR in a region N-terminal of its helicase domain (Davies et al, 2004; Rao et al, 2005). Interestingly, the N-terminal region of BLM (aa 1–477) including the ATR-target sites (T99Q100, T122Q123) binds hRPA in vitro (Davies et al, 2004, 2007). In contrast to Sgs1, BLM is not a constitutive component of the replisome, but is recruited from PML bodies to sites of stalled replication forks in response to HU (Sengupta et al, 2003). This re-localization requires ATR-dependent phosphorylation of BLM (Davalos et al, 2004). It has been suggested that ATR phosphorylation of BLM is required for recovery from HU-mediated replication fork stalling, but not for the recruitment of BLM to damaged forks nor for the suppression of sister chromatid exchanges with hTOPO IIIα-hRMI1-hRMI2 (Davies et al, 2004; Wu, 2007). Davies et al (2007) demonstrated that ATR-dependent phosphorylation of BLM is required for efficient replication fork resumption and repression of new origin firing after aphidicolin treatment. Furthermore, and similar to our findings, the presence of BLM at stalled replication forks is required for robust intra-S phase checkpoint activation in human cells (Franchitto and Pichierri, 2002; Davalos and Campisi, 2003). Nonetheless, the mechanisms through which mammalian ATR and BLM work together to maintain replisomes at stalled replication forks and activate the intra-S phase checkpoint are unknown. A conserved, damage-specific assembly of a RecQ homologue (BLM and/or WRN) with a downstream checkpoint kinase is an attractive hypothesis.
Materials and methods
Yeast strains and plasmids
S. cerevisiae strains (Supplementary Table S1) were derived from W303-1A (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100). If not stated otherwise, all strains were cultured at 30°C in YPAD media. The sgs1-r1 allele was generated using pop-in/pop-out mutagenesis as previously described (Tam et al, 2007). For Y2H analyses, fragments of Sgs1, Rpa70 and Rpa32 were fused in frame to the B42 activator domain in the pJG46 or the lexA DNA binding domain in the pGAL-lexA vector (Bjergbaek et al, 2005). Y2H was performed as described (Bjergbaek et al, 2005), except that both bait and prey were under GALUAS control. EGY191 cells (GA-1211) containing the lacZ reporter pSH18-34, the bait and the prey were glucose depleted, then 2% galactose added to the exponentially growing culture to induce expression of the fusion proteins. The quantitative β-galactosidase assay for permeabilized cells (Adams et al, 1997) was performed on four independent transformants in at least two independent experiments for each data point. Expression of the fusion proteins was confirmed by western blot analysis (data not shown). β-Galactosidase units are defined as OD420/(OD600 × dilution × time(min)).
Survival and drop assays
For liquid survival assays, overnight cultures were diluted to OD600=0.15 and grown for 3 h, then synchronized with α-factor in G1 and released into YPAD containing 0.2 M HU. At the indicated time points, relevant dilutions were plated onto fresh YPAD plates and colonies were counted after 3–4 days. Survival is defined as the fraction of colonies compared to the untreated control (0 h) normalized to the survival of WT cells for each time point. Drop tests used overnight cultures diluted to OD600=0.5 with 2 μl drops of 10-fold dilutions plated on YPAD or the appropriate selective medium with indicated concentrations of damaging agents.
ChIP analysis, co-IP and kinase assays
ChIP experiments were performed as described (Cobb et al, 2005) using monoclonal anti-HA (F-7, Santa Cruz) to precipitate HA-tagged DNA pol α or Ddc2, and anti-Myc (9E10) to precipitate Myc-tagged Rpa70. Details are in Supplementary Methods. For each time point, the relative enrichment for ARS607 or ARS522 was obtained by normalizing the absolute enrichment at ARS607 or ARS522 to the absolute enrichment 14 kb away from ARS607.
Co-IP was performed essentially as described in Bjergbaek et al (2005), except that two additional rounds of washing with 100 mM Tris pH 8.0, 500 mM NaCl, 1 mM EDTA, 0.5% NP-40 and 0.5% sodium deoxycholate were performed. Note that the use of more stringent washing conditions most probably explains why the Sgs1-Rad53 interaction was not dependent on checkpoint induction in the previous study (Bjergbaek et al, 2005). Mec1 immunoprecipitation and kinase assays are described in Supplementary Methods.
Rad53 and Sgs1 phosphorylation and mass spectroscopic analysis
Conditions used to monitor Rad53 or Sgs1 phosphorylation by western blot and by semi-quantitative mass spectroscopy are described in Supplementary Methods. In brief, peptides were separated by nano-HPLC (Agilent 1100 nanoLC system, Agilent Technologies) coupled to an LTQ Orbitrap Velos hybrid mass spectrometer (Thermo Scientific) using a top 15 DDA method and an LC-MSMS method containing m/z values of predicted phosphopeptides. Peptides were identified by searching SwissProt database (version 2010-09) with Mascot 2.3.0 (Matrix Science). Data were compiled and evaluated with Scaffold 3.4.3 and Scaffold PTM 1.1.3 (Proteome Software).
Protein purification and ITC
The Rad53(22–162) and Rpa70(1–133) and Rpa70(3–133) constructs inserted into a pET15 derived vector, containing a TEV protease cleavable His6-tag, and Sgs1 constructs (404–485, 404–560) were amplified and inserted into a pET15 derived vector, containing a thrombin protease cleavable His6 tag. All were expressed in E. coli BL21 cells and purified by metal chelate affinity (His-Select Nickel Affinity Gel, Sigma-Aldrich), anion-exchange (Resource 15 Q, GE Healthcare), and gel-filtration chromatography (Superdex S-200, GE Healthcare). ITC experiments were conducted with a MicroCal VP-ITC calorimeter as described in Supplementary Methods.
Genetic interactions and sequence analysis
GA-6998 (sgs1-r1) and yHA429 (sgs1Δ) cells together with 33 other query strains (data to be published elsewhere) were crossed with a library of 1536 mutants (Guénole et al, under review) as described (Schuldiner et al, 2006), except that four sets of double mutants were created in two independent experiments. Results and the analysis of the library by pooled TAG amplification are described in Supplementary Methods. Genetic interaction scores (S scores) were calculated using the E-MAP toolbox (Collins et al, 2006). The analysis of sequence motifs in Sgs1 is described in Supplementary Methods.
Supplementary Material
Acknowledgments
The Gasser laboratory acknowledges the help from T Aust, J Eichenberger, S Schuierer and D Hoepfner of the Novartis Institutes for Biomedical Research (Basel, Switzerland); H Vlaming from the van Leeuwen laboratory; and M Rebhan, D Gaidatzis and R Sack (FMI). Experimental assistance and discussion were contributed by Y Moriyama (Kyoto University), P Maillard (U of Geneva), S Kueng and other Gasser laboratory members. The mutant sgs1-hd was a gift from R Rothstein (Columbia U, NY). Funding from the Swiss Cancer League, the Novartis Research Foundation, the Swiss National Science Foundation and the EU ITR Image DDR is gratefully acknowledged.
Author contributions: AMH performed experiments, contributed to writing and assembled figures; NH performed experiments, contributed to writing and assembled figures; KS advised, performed experiments, contributed to writing and figures; BLP advised, performed experiments and contributed to writing; PA and MV purified proteins and cooperated on biochemical analyses; SMR performed and analysed ITC experiments; NHT advised MV, PA, and contributed to writing; HvA advised AG who generated strains used in the E-MAP analysis, and FvL hosted NH for same; SMG advised, interpreted results, planned experiments, contributed to writing, figures and funding of the project.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams A, Gottschling DE, Kaiser CA, Stearns T (1997) Methods in Yeast Genetics New York: Cold Spring Harbor Laboratory Press, [Google Scholar]
- Aguilera A, Gomez-Gonzalez B (2008) Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ (2001) Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol 3: 958–965 [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Stout AM, Bell SP (1999) Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci USA 96: 9130–9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arudchandran A, Cerritelli S, Narimatsu S, Itaya M, Shin DY, Shimada Y, Crouch RJ (2000) The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells 5: 789–802 [DOI] [PubMed] [Google Scholar]
- Ashton TM, Hickson ID (2010) Yeast as a model system to study RecQ helicase function. DNA Repair 9: 303–314 [DOI] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID (2008) RecQ helicases: guardian angels of the DNA replication fork. Chromosoma 117: 219–233 [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Keck JL, Wang JC (1999) Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J Mol Biol 289: 235–248 [DOI] [PubMed] [Google Scholar]
- Bennett RJ, Noirot-Gros MF, Wang JC (2000) Interaction between yeast sgs1 helicase and DNA topoisomerase III. J Biol Chem 275: 26898–26905 [DOI] [PubMed] [Google Scholar]
- Bernstein KA, Gangloff S, Rothstein R (2010) The RecQ DNA helicases in DNA repair. Annu Rev Genet 44: 393–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein KA, Shor E, Sunjevaric I, Fumasoni M, Burgess RC, Foiani M, Branzei D, Rothstein R (2009) Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J 28: 915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz SK, Sheehan AM, Wold MS (2004) Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 3: 1015–1024 [DOI] [PubMed] [Google Scholar]
- Bjergbaek L, Cobb JA, Tsai-Pflugfelder M, Gasser SM (2005) Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J 24: 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Lopez-Girona A, Shanahan P, Interthal H, Heyer WD, Russell P (2000) Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol Cell Biol 20: 8758–8766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R (2008) PhosphoPep—a database of protein phosphorylation sites in model organisms. Nat Biotechnol 26: 1339–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, Brusniak MY, Roschitzki B, Zhang C, Shokat KM, Schlapbach R, Colman-Lerner A, Nolan GP, Nesvizhskii AI, Peter M, Loewith R et al. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal 3: rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brush GS, Morrow DM, Hieter P, Kelly TJ (1996) The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci USA 93: 15075–15080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer GA, Boone C, Brown GW (2005) RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J 24: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Brill SJ (2007) Binding and activation of DNA topoisomerase III by the Rmi1 subunit. J Biol Chem 282: 28971–28979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Albuquerque CP, Liang J, Suhandynata RT, Zhou H (2010) A proteome-wide analysis of kinase-substrate network in the DNA damage response. J Biol Chem 285: 12803–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Zhou H (2009) Reconstitution of Rad53 activation by Mec1 through adaptor protein Mrc1. J Biol Chem 284: 18593–18604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L (2006) RecQ helicases: lessons from model organisms. Nucleic Acids Res 34: 4106–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM (2005) Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS (2006) A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol 7: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbé L, Thomas A, Pantesco V, De Vos J, Pasero P, Lengronne A (2010) Analysis of replication profiles reveals key role of RFC-Ctf18 in yeast replication stress response. Nat Struct Mol Biol 17: 1391–1397 [DOI] [PubMed] [Google Scholar]
- Davalos AR, Campisi J (2003) Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks. J Cell Biol 162: 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos AR, Kaminker P, Hansen RK, Campisi J (2004) ATR and ATM-dependent movement of BLM helicase during replication stress ensures optimal ATM activation and 53BP1 focus formation. Cell Cycle 3: 1579–1586 [DOI] [PubMed] [Google Scholar]
- Davies SL, North PS, Dart A, Lakin ND, Hickson ID (2004) Phosphorylation of the Bloom’s syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol 24: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SL, North PS, Hickson ID (2007) Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol 14: 677–679 [DOI] [PubMed] [Google Scholar]
- De Piccoli G, Katou Y, Itoh T, Nakato R, Shirahige K, Labib K (2012) Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol Cell 45: 696–704 [DOI] [PubMed] [Google Scholar]
- Dubrana K, van Attikum H, Hediger F, Gasser SM (2007) The processing of double-strand breaks and binding of single-strand-binding proteins RPA and Rad51 modulate the formation of ATR-kinase foci in yeast. J Cell Sci 120: 4209–4220 [DOI] [PubMed] [Google Scholar]
- Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB (2000) The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell 6: 1169–1182 [DOI] [PubMed] [Google Scholar]
- Franchitto A, Pichierri P (2002) Bloom’s syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J Cell Biol 157: 19–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C, Gasser SM (2000) The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev 14: 81–96 [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Kaliraman V, Brill SJ (2001) Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J Biol Chem 276: 8848–8855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel AM, Pike BL, Gasser SM (2009) ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol 21: 237–244 [DOI] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R (1994) The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol 14: 8391–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guénole A, Srivas R, Vreeken K, Wang S, Krogan NJ, Ideker T, Van Attikum H Dissection of DNA damage response pathways using a multi-conditional genetic interaction map. (Submitted). [DOI] [PMC free article] [PubMed]
- Hickson ID, Mankouri HW (2011) Processing of homologous recombination repair intermediates by the Sgs1-Top3-Rmi1 and Mus81-Mms4 complexes. Cell Cycle 10: 3078–3085 [DOI] [PubMed] [Google Scholar]
- Ii M, Brill SJ (2005) Roles of SGS1, MUS81, and RAD51 in the repair of lagging-strand replication defects in Saccharomyces cerevisiae. Curr Genet 48: 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai M, Boddy MN, Russell P, Wang TS (2005) Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes Dev 19: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Mizuno K, Blaisonneau J, Martineau S, Chanet R, Freon K, Murray JM, Carr AM, Baldacci G (2010) Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol Cell 39: 346–359 [DOI] [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M (2005) Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev1 9: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H, Komata M, Katou Y, Guan Z, Reis CC, Budd M, Shirahige K, Campbell JL (2008) Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol Cell 32: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucca C, Vanoli F, Cotta-Ramusino C, Pellicioli A, Liberi G, Haber J, Foiani M (2004) Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 23: 1206–1213 [DOI] [PubMed] [Google Scholar]
- Machwe A, Lozada EM, Xiao L, Orren DK (2006) Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol Biol 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J, Niedziela-Majka A, Burgers PM (2006) The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell 24: 891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Ashton TM, Hickson ID (2011) Holliday junction-containing DNA structures persist in cells lacking Sgs1 or Top3 following exposure to DNA damage. Proc Natl Acad Sci USA 108: 4944–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, Cohen J, Toczyski DP (2001) Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev 15: 2809–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S, Komata M, Kishi T, Shirahige K, Kamura T (2009) SCF(Dia2) regulates DNA replication forks during S-phase in budding yeast. EMBO J 28: 3693–3705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Nam EA, Cortez D (2008) Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci USA 105: 18730–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi H, Maculins T, Labib K (2009) The amino-terminal TPR domain of Dia2 tethers SCF(Dia2) to the replisome progression complex. Curr Biol 19: 1943–1949 [DOI] [PubMed] [Google Scholar]
- Mullen JR, Chen CF, Brill SJ (2010) Wss1 is a SUMO-dependent isopeptidase that interacts genetically with the Slx5-Slx8 SUMO-targeted ubiquitin ligase. Mol Cell Biol 30: 3737–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ (2005) Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol 25: 4476–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Galván S, Tous C, Blanco MG, Schwartz EK, Ehmsen KT, West SC, Heyer WD, Aguilera A (2012) Distinct roles of Mus81, Yen1, Slx1-Slx4, and Rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol Cell Biol 32: 1592–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Kolodner RD (2002) Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 99: 4500–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S, Davoodi N, Gasser SM (2011) Nuclear organization in genome stability: SUMO connections. Cell Res 21: 474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM (2008) Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem 283: 35853–35859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM (2011) Cell-cycle-specific activators of the Mec1/ATR checkpoint kinase. Biochem Soc Trans 39: 600–605 [DOI] [PubMed] [Google Scholar]
- Naylor ML, Li JM, Osborn AJ, Elledge SJ (2009) Mrc1 phosphorylation in response to DNA replication stress is required for Mec1 accumulation at the stalled fork. Proc Natl Acad Sci USA 106: 12765–12770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SL, Shoemaker DD, Boeke JD (2003) DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat Genet 35: 277–286 [DOI] [PubMed] [Google Scholar]
- Osborn AJ, Elledge SJ (2003) Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev 17: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V, Clerici M, Scotti M, Lucchini G, Longhese MP (2001) Characterization of mec1 kinase-deficient mutants and of new hypomorphic mec1 alleles impairing subsets of the DNA damage response pathway. Mol Cell Biol 21: 3913–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell S, Westcott S, Ortiz-Lombardia M, Patel D, Li J, Nott TJ, Mohammed D, Buxton RS, Yaffe MB, Verma C, Smerdon SJ (2010) Structural and functional analysis of phosphothreonine-dependent FHA domain interactions. Structure 18: 1587–1595 [DOI] [PubMed] [Google Scholar]
- Pichierri P, Rosselli F, Franchitto A (2003) Werner’s syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene 22: 1491–1500 [DOI] [PubMed] [Google Scholar]
- Pike BL, Tenis N, Heierhorst J (2004a) Rad53 kinase activation-independent replication checkpoint function of the N-terminal forkhead-associated (FHA1) domain. J Biol Chem 279: 39636–39644 [DOI] [PubMed] [Google Scholar]
- Pike BL, Yongkiettrakul S, Tsai MD, Heierhorst J (2003) Diverse but overlapping functions of the two forkhead-associated (FHA) domains in Rad53 checkpoint kinase activation. J Biol Chem 278: 30421–30424 [DOI] [PubMed] [Google Scholar]
- Pike BL, Yongkiettrakul S, Tsai MD, Heierhorst J (2004b) Mdt1, a novel Rad53 FHA1 domain-interacting protein, modulates DNA damage tolerance and G(2)/M cell cycle progression in Saccharomyces cerevisiae. Mol Cell Biol 24: 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirzio LM, Pichierri P, Bignami M, Franchitto A (2008) Werner syndrome helicase activity is essential in maintaining fragile site stability. J Cell Biol 180: 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Qian Y, Frank P, Wintersberger U, Shen B (1999) Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol Cell Biol 19: 8361–8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VA, Fan AM, Meng L, Doe CF, North PS, Hickson ID, Pommier Y (2005) Phosphorylation of BLM, dissociation from topoisomerase IIIalpha, and colocalization with gamma-H2AX after topoisomerase I-induced replication damage. Mol Cell Biol 25: 8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP (2002) Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol Cell 9: 857–869 [DOI] [PubMed] [Google Scholar]
- Schmidt KH, Kolodner RD (2006) Suppression of spontaneous genome rearrangements in yeast DNA helicase mutants. Proc Natl Acad Sci USA 103: 18196–18201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Weissman JS, Krogan NJ (2006) Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods 40: 344–352 [DOI] [PubMed] [Google Scholar]
- Segurado M, Diffley JF (2008) Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev 22: 1816–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Linke SP, Pedeux R, Yang Q, Farnsworth J, Garfield SH, Valerie K, Shay JW, Ellis NA, Wasylyk B, Harris CC (2003) BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J 22: 1210–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JC, Lao Y, Kamath-Loeb A, Wold MS, Loeb LA (2003) The N-terminal domain of the large subunit of human replication protein A binds to Werner syndrome protein and stimulates helicase activity. Mech Ageing Dev 124: 921–930 [DOI] [PubMed] [Google Scholar]
- Smolka MB, Chen SH, Maddox PS, Enserink JM, Albuquerque CP, Wei XX, Desai A, Kolodner RD, Zhou H (2006) An FHA domain-mediated protein interaction network of Rad53 reveals its role in polarized cell growth. J Cell Biol 175: 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Sowd G, Wang H, Pretto D, Chazin WJ, Opresko PL (2009) Replication protein a stimulates the werner syndrome protein branch migration activity. J Biol Chem 284: 34682–34691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam AT, Pike BL, Hammet A, Heierhorst J (2007) Telomere-related functions of yeast KU in the repair of bleomycin-induced DNA damage. Biochem Biophys Res Commun 357: 800–803 [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L et al. (2004) Global mapping of the yeast genetic interaction network. Science 303: 808–813 [DOI] [PubMed] [Google Scholar]
- Tourriere H, Pasero P (2007) Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 6: 900–913 [DOI] [PubMed] [Google Scholar]
- Traven A, Heierhorst J (2005) SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. Bioessays 27: 397–407 [DOI] [PubMed] [Google Scholar]
- Watt PM, Hickson ID, Borts RH, Louis EJ (1996) SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J, Rothstein R (2008) The genetic consequences of ablating helicase activity and the Top3 interaction domain of Sgs1. DNA Repair (Amst) 7: 558–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L (2007) Role of the BLM helicase in replication fork management. DNA Repair (Amst) 6: 936–944 [DOI] [PubMed] [Google Scholar]
- Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T (2009) BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO J 28: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
- Zou Y, Liu Y, Wu X, Shell SM (2006) Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J Cell Physiol 208: 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.