Abstract
Autophagy is an intracellular trafficking pathway sequestering cytoplasm and delivering excess and damaged cargo to the vacuole for degradation. The Atg1/ULK1 kinase is an essential component of the core autophagy machinery possibly activated by binding to Atg13 upon starvation. Indeed, we found that Atg13 directly binds Atg1, and specific Atg13 mutations abolishing this interaction interfere with Atg1 function in vivo. Surprisingly, Atg13 binding to Atg1 is constitutive and not altered by nutrient conditions or treatment with the Target of rapamycin complex 1 (TORC1)-inhibitor rapamycin. We identify Atg8 as a novel regulator of Atg1/ULK1, which directly binds Atg1/ULK1 in a LC3-interaction region (LIR)-dependent manner. Molecular analysis revealed that Atg13 and Atg8 cooperate at different steps to regulate Atg1 function. Atg8 targets Atg1/ULK1 to autophagosomes, where it may promote autophagosome maturation and/or fusion with vacuoles/lysosomes. Moreover, Atg8 binding triggers vacuolar degradation of the Atg1–Atg13 complex in yeast, thereby coupling Atg1 activity to autophagic flux. Together, these findings define a conserved step in autophagy regulation in yeast and mammals and expand the known functions of LIR-dependent Atg8 targets to include spatial regulation of the Atg1/ULK1 kinase.
Keywords: Atg1-ULK1 kinase, Atg8, autophagy, Cvt pathway, LIR motif
Introduction
Autophagy is an important cellular mechanism to eliminate excess or damaged proteins, protein complexes and organelles. This conserved process serves to ensure cellular homeostasis, and plays crucial roles during nutrient starvation and the cellular response to stress conditions, as well as in embryonic development and the defense against several human pathogens. Not surprisingly, defects in autophagy pathways have been associated with numerous human pathologies, including infectious diseases, neurodegenerative disorders and cancer. Despite these fundamental functions, the regulation and coordination of the steps required for cargo selection and transport to the cellular degradation site remain poorly understood.
Autophagy involves the engulfment of cargo by double-membraned autophagosomes, which then fuse with the lysosome/vacuole, where their content is degraded. Both non-selective ‘bulk’ (macro)autophagy and selective autophagy of specific proteins or organelles have been described (Kraft et al, 2009, 2010). In addition, yeast cells use the related cytoplasm-to-vacuole targeting (Cvt) pathway to fulfill a biosynthetic function by selectively delivering to the vacuole three of its resident enzymes, aminopeptidase 1 (Ape1), α-mannosidase (Ams1) and aspartyl aminopeptidase (Ape4) (Harding et al, 1995; Hutchins and Klionsky, 2001; Yuga et al, 2011). Genetic analysis in yeast has identified over 35 mostly conserved components that are required for different steps of autophagy called Atg1 to Atg36 (Suzuki and Ohsumi, 2007; Motley et al, 2012). Although most of these components are common to autophagy and the Cvt pathway, autophagy- and Cvt-specific genes also exist (Inoue and Klionsky, 2010).
The cellular signals that induce or restrict autophagy, and the mechanisms that spatially and temporally act to ensure cargo selection remain poorly understood. Atg1 is a conserved serine-threonine kinase that is required for both selective and bulk autophagy pathways, and epistasis analysis revealed that Atg1 functions upstream of cargo selection and autophagosome formation. Atg1 assembles into a large complex composed of several proteins including Atg13 (Scott et al, 2000). Atg1 activity is regulated by the Target of rapamycin (TOR) kinase, which regulates cell growth and autophagy as part of the TOR complex 1 (TORC1) in response to the nutrient availability (Chan et al, 2009; Hosokawa et al, 2009a; Kamada et al, 2009). Both Atg1 and Atg13 are highly phosphorylated under nutrient-rich conditions, and Atg13 phosphorylation is partly mediated by TORC1 (Kamada et al, 2009). Atg13 is rapidly dephosphorylated upon starvation in both yeast and mammalian cells (Scott et al, 2000; Hosokawa et al, 2009b; Yeh et al, 2010), which might be part of the regulatory mechanism for autophagy induction. In yeast, Atg13 phosphorylation is thought to reduce its binding affinity for Atg1, which in turn prevents Atg1 kinase activity under nutrient-rich conditions. In contrast, in flies and mammals, Atg1/ULK1 and Atg13 constitutively bind to each other, implying that Atg1 regulation may predominantly be mediated by phosphorylation rather than by complex formation (Hara et al, 2008; Chan et al, 2009).
The ubiquitin-like protein Atg8 recently emerged as a critical component that plays a dual role in autophagy regulation. On the one hand, Atg8 is conjugated to phosphatidylethanolamine (PE) and recruited to nascent autophagosomes, where it promotes autophagosome expansion (Mizushima et al, 1998; Ichimura et al, 2000) and appears to trigger fusion of autophagosomal membranes (Nakatogawa et al, 2007). On the other hand, a critical role of Atg8 in cargo selection has recently been discovered, where Atg8 binds to a family of cargo-recruiting receptors thereby directing them to autophagosomes. For example, Ape1 interacts with the Cvt-specific cargo receptor Atg19, which also binds to Atg8 (reviewed in Xie and Klionsky, 2007). Likewise, Ams1 binds to the adaptor Atg34, which through a separate domain also interacts with Atg8 (Watanabe et al, 2010).
Whereas yeast express a single Atg8 protein, mammals encode at least eight different isoforms, which are grouped into two families: three MAP1 light chain 3 proteins (LC3A (two splice variants), B and C) and four γ-aminobutyrate receptor-associated protein (GABARAP) and GABARAP-like proteins (ATG8L/GEC-1/GABARAPL1, GATE-16/GABARAPL2 and GABARAPL3) (Xin et al, 2001; He et al, 2003). Although LC3 is believed to be the main Atg8 protein involved in starvation-induced autophagy (Kabeya et al, 2004), GABARAP, GABARAPL2 and GABARAPL1 are indispensable for autophagy function (Weidberg et al, 2010). In mammals, several adaptors that bind to Atg8 family proteins and to specific cargo have been described (Kraft et al, 2010). These include p62 and Nbr1, which are involved in the autophagic degradation of ubiquitinated aggregates (Pankiv et al, 2007; Kirkin et al, 2009). Both proteins bind LC3 through a conserved hydrophobic WXXL/I motif called the LC3-interaction region (LIR), and a similar motif (AIM for Atg8-interacting motif) has been identified in yeast Atg19 and Atg34 for their interaction with Atg8 (Noda et al, 2010; Suzuki et al, 2010). Supported by a comprehensive LC3-interaction network (Behrends et al, 2010), the available data suggest that a large number of LIR domain-containing proteins may function as receptors, which specifically interact with different cargo and promote their engulfment into autophagosomes.
We were interested in how selective autophagy pathways are regulated, and thus investigate the mechanism of Atg1 activation. Here, we identified specific mutations in the positive regulator Atg13 that abolished its interaction with Atg1. Surprisingly, while the interaction of Atg13 and Atg1 was important for full kinase activity, it was neither essential for autophagy induction nor regulated by nutrient starvation, implying that additional regulators of Atg1 exist. Indeed, we found that Atg1 interacts with Atg8 in a LIR-dependent manner and travels with autophagosomes to the vacuole, leading to downregulation of the Atg1 complex. This interaction is conserved and maintained in mammals as we show that ULK1 interacts with LC3 and GABARAP family members. Importantly, in vivo analysis revealed that Atg13 and Atg8 cooperate at different steps to regulate Atg1 function in autophagy and the Cvt pathway. Together, these findings define new steps in autophagy regulation and expand the known functions of Atg8-interacting proteins to include spatial regulation of the Atg1 kinase complex in yeast and mammals.
Results
Binding of Atg13 promotes Atg1 kinase activity, but is not regulated by nutrient starvation
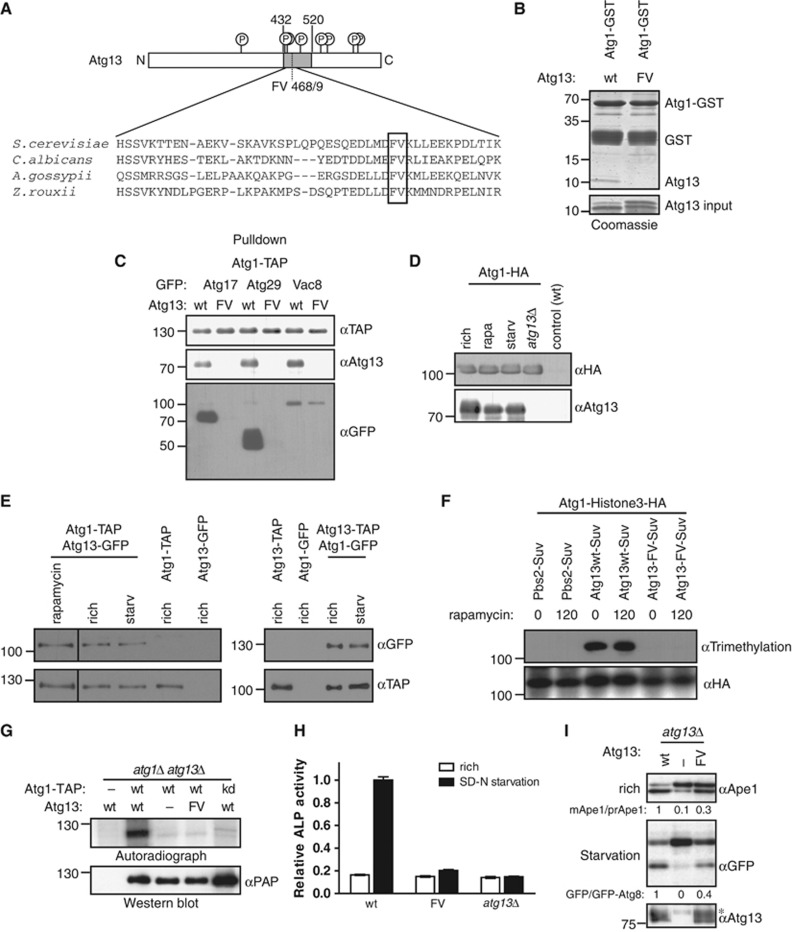
Available evidence suggests that Atg1 is activated by its binding partner Atg13, and this interaction is thought to be regulated by nutrient availability (Kamada et al, 2000). To examine the functional significance of the Atg1–Atg13 interaction, we analysed the Atg1-binding region in Atg13 with the aim to identify mutations that specifically abolish Atg1 binding. Previous work identified that residues 432–520 in yeast Atg13 are involved in Atg1 binding (Kamada et al, 2000), and sequence alignments showed several highly conserved residues in this region (Figure 1A). Indeed, mutational analysis followed by a yeast two-hybrid interaction study revealed that mutations of phenylalanine 468 and valine 469 to alanine residues (Atg13-FV) abolished Atg13 binding to Atg1, whereas mutation of F468 alone had a partial effect (Supplementary Figure S1A). Other mutations in the Atg1 binding domain had no impact on the interaction of Atg1 with Atg13, suggesting that the interaction is specifically mediated by F468 and V469 (Supplementary Figure S1A). This interaction is direct as Atg1 and the binding domain of Atg13 purified from E. coli bind to each other in vitro in an F468- and V469-dependent manner with almost 1:1 stoichiometry (Figure 1B).
Figure 1.
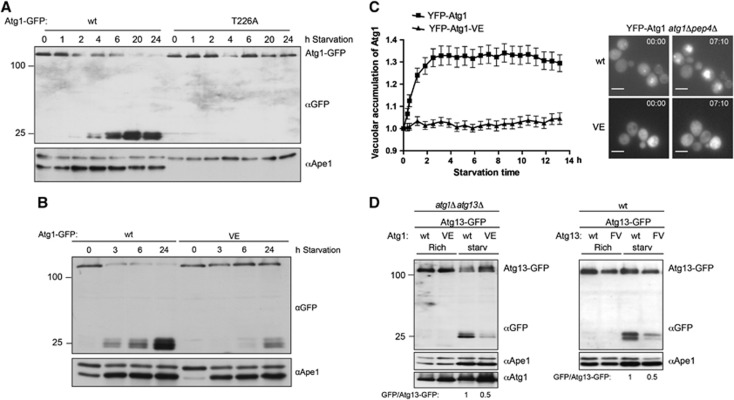
Atg13 binding to Atg1 promotes Cvt and autophagy function, but is not regulated by starvation conditions. (A) Schematic representation of budding yeast Atg13 and alignment of the Atg1-binding region with homologues from different yeast species. The residues required for Atg1 binding are marked with a grey box. Circled ‘P’ mark known phosphorylation sites on Atg13. N: amino-terminus; C: carboxy-terminus. (B) A fragment encompassing amino acids 432–520 of Atg13 (wt) or the corresponding FV mutant (FV) was expressed in E. coli and incubated in excess with an immobilized GST-tagged fragment of Atg1 (amino acids 501–897) purified from E. coli (GST-Atg1). Input (5.5%) and bound proteins were analysed by coomassie blue staining, which stains proteins proportional to their size. The size of marker proteins (in kDa) is indicated on the left. (C) ATG1-TAP atg13Δ cells containing endogenously GFP-tagged Atg17, Atg29 or Vac8 and wild-type (wt) or the Atg13 FV-mutant (F468A; V469A) were grown to mid log phase. Atg1 was immunoprecipitated and its association with Atg13 and the GFP-tagged proteins was analysed by immunoblotting. Extract inputs are shown in Supplementary Figure S1B. (D) atg1Δ and atg1Δatg13Δ cells expressing HA-tagged Atg1 and wild-type (wt) cells containing an empty plasmid were grown to mid log phase (rich), and then treated with rapamycin (rapa) or starved for 4 h in SD-N medium (starv). Atg1 was immunoprecipitated and its association with Atg13 analysed by immunoblotting. Note that phosphorylated Atg13 isolated from cells grown in rich medium migrates slightly slower on SDS–PAGE, and the extent corresponds to the shift typically observed under our gel conditions. (E) Yeast cells containing endogenously tagged Atg1-TAP with and without endogenously tagged Atg13-GFP, or conversely cells containing endogenously tagged Atg13-TAP and Atg1-GFP expressed from a centromeric plasmid were grown to mid log phase (rich) and treated with rapamycin (rapa) or starved for 4 h in SD-N medium (starv). Atg1-TAP and Atg13-TAP were affinity purified and their association with GFP-tagged proteins was analysed by western blotting. (F) Histone3-HA tagged Atg1 was expressed together with wild-type (wt) or the Atg13-FV mutant fused to Suv methylase (Suv) in atg1Δatg13Δ cells, or for control together with Pbs2-Suv in atg1Δ cells. Logarithmically growing cultures were treated with rapamycin (rapa) for 0 or 120 min, and methylation was monitored by preparing cell extracts and western blotting with an anti-trimethylation-specific antibody. Note that Suv-, GFP- or TAP-tagged Atg13 are fully functional, however, do not show the characteristic phosphorylation-induced reduced mobility observed with the untagged protein under our gel conditions. (G) atg1Δatg13Δ cells containing TAP-tagged wild-type Atg1 (wt) or the Atg1 kinase-dead (kd) K54A mutant and wild-type Atg13, the Atg13-FV mutant or an empty plasmid were grown to mid log phase. Atg1 was immunoprecipitated and its autophosphorylation activity was measured by autoradiography in vitro. (H) pho8Δ60pho13Δatg13Δ cells expressing wild-type Atg13 (wt), the Atg13-FV mutant (FV) or an empty plasmid were grown to mid log phase and starved for 4 h in SD-N medium. Pho8Δ60-specific alkaline phosphatase (ALP) activity (nmol/min/mg) was measured in three independent experiments as described in ‘Materials and methods’, and plotted as relative ALP activity with standard deviation (s.d.) compared to the wild-type controls. (I) atg13Δ cells expressing GFP-Atg8 and containing wild-type Atg13, the Atg13-FV mutant or an empty plasmid were grown to mid log phase with and without starvation for 4 h in SD-N. Processing of endogenous Ape1 and cleavage of GFP-Atg8 was analysed by western blotting, and quantified by calculating the ratio of cleaved versus uncleaved Ape1 or GFP-Atg8, respectively. The asterisk marks a non-specific band detected by the Atg13 antibody. Figure source data can be found with the Supplementary data.
Co-immunoprecipitation experiments confirmed that Atg13-FV was unable to bind Atg1 (Figure 1C). Interestingly, its association with Atg11, Atg17 and Atg29 was also abolished, while the interaction with Vac8, a putative complex member, was only slightly reduced (Figure 1C; Supplementary Figure S1B). We concluded that F468 and V469 are required for the ability of Atg13 to directly interact with Atg1 and stabilize the Atg1 kinase complex.
To compare the interaction of Atg1 with Atg13 under autophagic and non-autophagic conditions, we purified HA- or TAP-tagged Atg1 from yeast before and after rapamycin treatment as well as upon nitrogen starvation, and monitored co-precipitation of Atg13 by western blotting and silver staining (Figure 1D and E; Supplementary Figure S1C and D). As expected, Atg13 readily co-precipitated with Atg1, but this interaction did not change under different growth conditions, irrespective whether the affinity-purified complex was eluted from the beads with sample buffer or TEV protease cleavage (Supplementary Figure S1D). Similar results were obtained when conversely Atg13-TAP was purified and analysed for Atg1 binding (Figure 1E, right panel). Importantly, expression of all fusion proteins rescued the Cvt and autophagy defects of the corresponding deletion strains, excluding the possibility that the tags interfere with their function (Supplementary Figure S1E). Moreover, the stable Atg1–Atg13 interaction was not caused by artificial binding during extract preparation, as post-growth mixing of differentially tagged Atg1 and Atg13 cultures did not result in co-precipitation of the two proteins (Supplementary Figure S1F). Finally, floatation experiments revealed that the majority of Atg1 was not lipid associated after extract preparation, implying that the interaction was not indirectly caused by association with vesicles under these conditions (Supplementary Figure S1G).
To analyse the regulation of the Atg1–Atg13 interaction in vivo, we used a novel interaction assay in yeast (Zuzuarregui et al, 2012). This in vivo protein-proximity assay is based on fusing a histone lysine methyltransferase domain to the bait protein and its substrate, histone H3, to the prey protein. Upon binding, the prey is stably methylated in vivo, which can subsequently be detected by cell lysis and immunoblotting using methylation-specific antibodies. Importantly, this assay confirmed that the Atg1–Atg13 interaction is mediated by F468 and V469 in Atg13, and the complex is stable in different growth conditions in vivo (Figure 1F). Taken together, these results suggest that Atg1 and Atg13 constitutively interact in vivo, irrespective of nutrient availability.
To examine the impact of Atg13-FV on Atg1 kinase activity, we immunoprecipitated TAP-tagged wild-type (wt) and mutant Atg1 complexes and monitored Atg1 autophosphorylation using in vitro kinase assays in the presence of radioactive γ-32P-ATP (Figure 1G). As expected, Atg1 activity was abolished when analysing a kinase-dead Atg1 mutant (Atg1-kd) (Kamada et al, 2000). Interestingly, the Atg13-FV mutant resulted in a similar loss of Atg1 autophosphorylation as in atg13Δ cells, demonstrating that Atg13 binding to Atg1 indeed promotes its kinase activation (Figure 1G). To probe the functional relevance of Atg13 binding to Atg1, we compared autophagic activity in wild-type and Atg13-FV mutant cells. First, we performed pho8Δ60 assays (Noda et al, 1995; Klionsky et al, 2007), which quantify the activity of the amino-terminally truncated alkaline phosphatase Pho8 (Pho8Δ60) that is activated in the vacuole after its delivery by autophagy. As expected, Pho8Δ60 activity in wild-type cells was increased after 4 h of nitrogen starvation, while no such activity was measured in atg13Δ cells. Interestingly, Pho8Δ60 activity increased by <10% in atg13-fv mutant cells, indicating that Atg13 binding to Atg1 is required in vivo for efficient autophagy (Figure 1H; Supplementary Figure S2C).
To corroborate these results, we examined Cvt activity by measuring Ape1 processing in nutrient-rich conditions (Klionsky et al, 1992), and followed the transport of GFP-Atg8 into the vacuole upon starvation. Due to the high stability of the GFP fold, the GFP moiety remains stable after cleavage in the vacuole and can be monitored as free GFP by western blotting (Meiling-Wesse et al, 2002). Surprisingly, whereas deletion of Atg13 resulted in an almost complete loss of Atg8 cleavage and Cvt activity, the Atg13-FV mutant only showed a partial reduction, although the mutant and wild-type proteins were expressed at comparable levels (Figure 1I; Supplementary Figure S2C). Other mutations in the Atg1 binding domain did not alter Cvt activity (Supplementary Figure S2A), implying that the defect in the Cvt pathway and autophagic activity is caused by loss of Atg13 binding rather than indirect defects caused by protein misfolding. Consistent with this notion, the partial loss of Cvt and autophagic activity was not due to mislocalization of the Atg13-FV mutant, as Atg13-FV was recruited to the PAS (Supplementary Figure S2B). A stronger defect in the pho8Δ60 assay compared to Ape1 and GFP-Atg8 processing has previously been observed, and might reflect a defect in autophagosome capacity (Cheong et al, 2005). Taken together, we conclude that Atg13 binding to Atg1 is important for efficient Cvt pathway and autophagy function, however, Atg13 might have an additional role in autophagy progression that is independent of Atg1 binding. Importantly, these data further imply that Atg1 requires additional activator(s) in vivo.
Atg1/ULK1 binds to Atg8 in an LIR-dependent manner
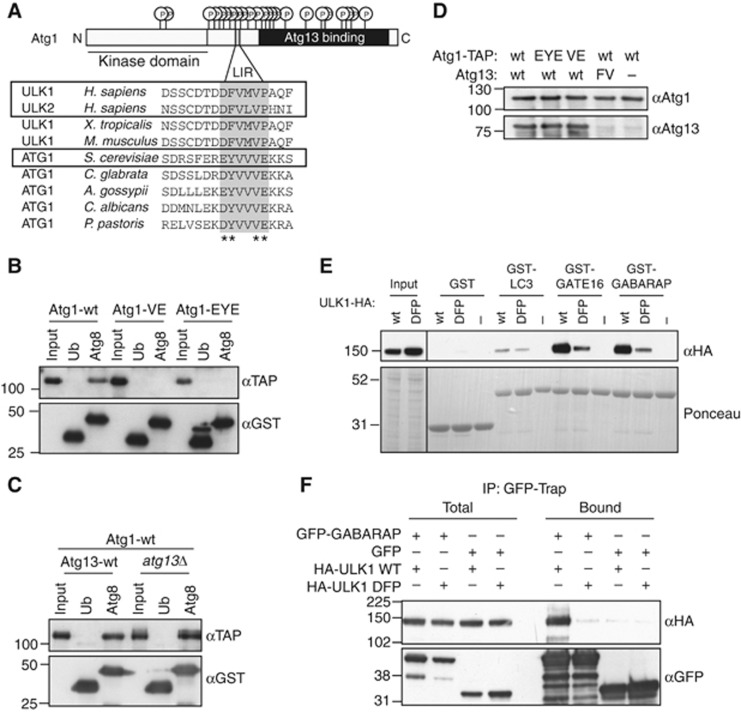
As recent work identified the mammalian Atg1 homologue ULK1 as a possible interactor of Atg8/LC3 (Behrends et al, 2010), we hypothesized that Atg8 may be involved in Atg1 regulation. A thorough bioinformatic analysis identified a conserved phenylalanine/tyrosine-X-X-valine (F/YXXV) sequence present in both ULK1 and ULK2 as well as yeast Atg1 (Figure 2A), which fulfills the criteria for a functional LIR motif. To investigate if yeast Atg1 indeed binds to Atg8 through this putative LIR motif, we performed in vitro interaction studies. As shown in Figure 2B, Atg1 strongly bound to Atg8 but not to ubiquitin. Importantly, mutation of valine 432 and glutamate 433 (Atg1-VE) or glutamate 428, phenylalanine 429 and glutamate 433 (Atg1-EYE) in the predicted LIR motif of Atg1 completely abolished this binding, suggesting that the interaction of Atg1 with Atg8 is mediated in a LIR-dependent manner. This interaction is independent of Atg13, as binding of Atg1 to Atg8 persisted in cells lacking Atg13 (Figure 2C). Moreover, as expected from the non-overlapping binding sites (Figure 2A), the Atg1-LIR mutants efficiently bound Atg13, implying that the LIR motif in Atg1 is not required for binding to Atg13 (Figure 2D; Supplementary Figure S3A).
Figure 2.
Atg1 binds Atg8 in a LIR-dependent manner. (A) Schematic representation of budding yeast Atg1 and sequence alignment of its putative LIR motif with Atg1 homologues from various species. Conserved residues of the predicted LIR motif are indicated with grey boxes, residues targeted by mutagenesis are marked with asterisks (*). The Atg1 kinase domain is underlined and the Atg13-binding region is shaded in black. Circled ‘P’ mark known phosphorylation sites on Atg1. N: amino-terminus; C: carboxy-terminus. (B) GST-Atg8 or GST-Ubiquitin (Ub) was purified from E. coli and bound to GSH beads. Wild-type Atg1-TAP or the VE and EYE mutants were purified from yeast, incubated with the immobilized GST proteins and bound proteins analysed by western blotting. (C) GST-Atg8 and GST-Ubiquitin were immobilized as described in (B), and probed for their ability to bind Atg1-TAP isolated from wild-type or atg13Δ cells. (D) atg1Δatg13Δ cells expressing as indicated wild-type Atg1-TAP or the VE or EYE mutants, and wild-type Atg13 or the Atg13-FV mutant were grown to mid log phase. Atg1 was immunoprecipitated and its association with Atg13 was determined by western blotting. (E) GST, GST-LC3B, GST-GATE16 and GST-GABARAP were purified from E. coli and bound to beads. HEK293 cell lysates prepared from cells expressing wild-type (wt) HA-ULK1 or the DFP mutant were incubated with the beads, and bound proteins analysed by western blotting. (F) HEK293 cells were co-transfected with GFP or GFP-GABARAP along with either wild-type HA-ULK1 or the DFP mutant. GFP and GFP-GABARAP were immobilized on GFP-Trap resin and HA-ULK1 binding was analysed by western blotting. Figure source data can be found with the Supplementary data.
Interestingly, the region encompassing the conserved LIR motif in human ULK1 was previously shown to bind GABARAP, GATE-16, and weakly to LC3 (Okazaki et al, 2000). To corroborate these results, we mutated aspartate 356, phenylalanine 357 and proline 361 in the predicted LIR motif of ULK1 to alanine (DFP) and tested if wild-type or the ULK1-DFP mutant could bind the Atg8 homologues LC3B, GATE-16 and GABARAP immobilized on beads. ULK1 was efficiently retained on GABARAP- and GATE16-coated beads, and also bound to LC3, though to a lower extent. These interactions were substantially reduced in the DFP mutant (Figure 2E), implying that they are mediated by this conserved LIR motif. To investigate this interaction in vivo, HEK293 cells were co-transfected with GFP or GFP-GABARAP together with either wild-type HA-ULK1 or the DFP mutant. GFP and GFP-GABARAP were immobilized on GFP-Trap resin, and binding proteins were analysed by immunoblotting. Consistent with our previous observations, both endogenous and HA-tagged ULK1 bound to GFP-GABARAP in vivo, and this interaction required a functional LIR motif in ULK1 (Figure 2F; Supplementary Figure S3B). Together, these data identify Atg1/ULK1 as a conserved Atg8 interactor, which directly binds to Atg8 in yeast and mammals.
The interaction of Atg8 with Atg1 is functionally relevant in vivo
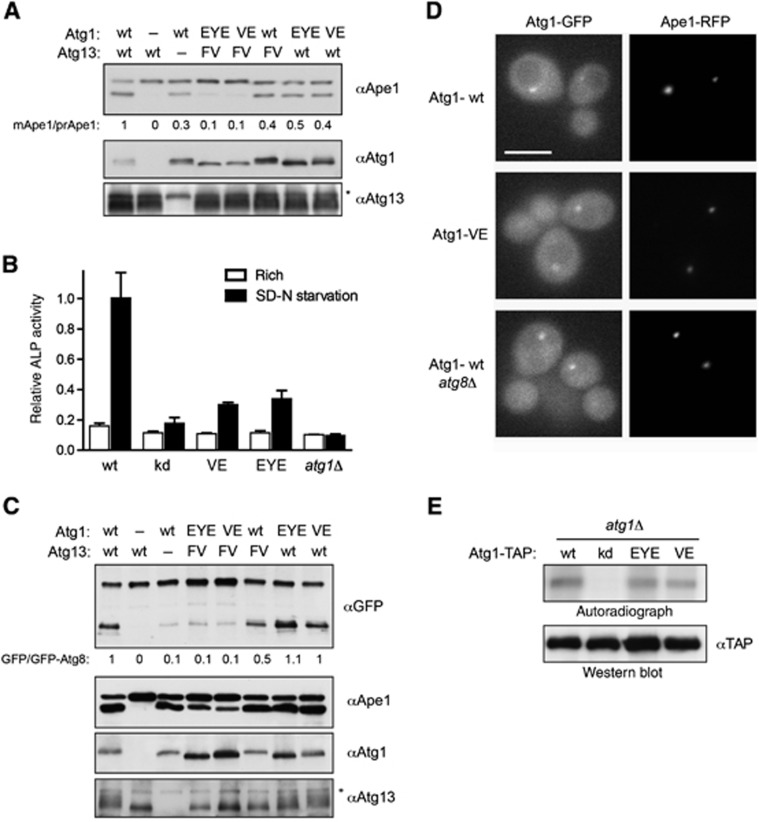
To test whether the interaction of Atg1 with Atg8 is important for its function in vivo, we compared the activity of wild-type and Atg1 mutant cells in the Cvt pathway and autophagy. Interestingly, cells expressing Atg1-EYE or Atg1-VE were partially defective for Cvt activity, although both mutant proteins were expressed at comparable levels to wild-type Atg1. Quantification revealed that vacuolar processing of Ape1 at steady state was reduced by ∼60% (Figure 3A; Supplementary Figure S3C). Using the pho8Δ60 assay, atg1-EYE and atg1-VE mutant cells showed little Pho8Δ60 activity after 4 h starvation compared to wild-type controls (Figure 3B; Supplementary Figure S2C), indicating that the LIR motif in Atg1 is required for both Cvt pathway and autophagy function.
Figure 3.
The LIR domain in Atg1 is functionally important in vivo. (A) atg1Δatg13Δ cells containing an empty control plasmid, wild-type or the indicated Atg1 mutants, and Atg13 wild-type, the Atg13-FV mutant or an empty plasmid were grown to mid log phase. Processing of endogenous Ape1 was analysed by western blotting, and quantified as described in the legend of Figure 1I. Expression of the different proteins was controlled by immunoblotting with antibodies against Atg1 or Atg13. The asterisk marks a non-specific band detected by the Atg13 antibody. (B) pho8Δ60pho13Δatg1Δ cells expressing wild-type (wt), kinase-dead (kd), Atg1-VE (VE) or -EYE (EYE) mutants or an empty plasmid were grown to mid log phase and starved for 4 h in SD-N medium. Pho8Δ60-specific alkaline phosphatase (ALP) activity (nmol/min/mg) was measured in three independent experiments as described in ‘Materials and methods’, and plotted as relative ALP activity with standard deviation (s.d.) compared to the wild-type controls. (C) Cells as in (A) expressing GFP-Atg8 were grown to mid log phase with starvation for 4 h in SD-N medium. The processing of GFP-Atg8 was analysed and quantified as in Figure 1I. (D) Exponentially growing atg1Δ and atg1Δatg8Δ strains expressing the PAS marker Ape1-RFP and either GFP-tagged wild-type Atg1 or the Atg1-VE mutant were examined by fluorescence microscopy. Bar=5 μm. (E) atg1Δ cells containing TAP-tagged wild-type or the indicated Atg1-mutants were grown to mid log phase. Atg1 was immunoprecipitated and its in vitro autophosphorylation activity was measured by autoradiography. The kinase-dead (kd) Atg1-mutant K54A serves as a negative control. Figure source data can be found with the Supplementary data.
In contrast, no obvious defects were observed when analysing autophagy by monitoring GFP-Atg8 cleavage or Ape1 processing upon nutrient starvation (Figure 3C; Supplementary Figure S3C and D), indicating that Atg8 binding to Atg1 may affect later steps in autophagy (Cheong et al, 2005). Indeed, fluorescence microscopy revealed that the Cvt defect of the Atg1-EYE and Atg1-VE mutants was not caused by a defect in their recruitment to the PAS, which occurs in an Atg8-independent manner (Figure 3D). Moreover, interfering with binding of Atg1 to Atg8 did not alter its kinase activity in vitro (Figure 3E). Thus, although the interaction of Atg1 and Atg8 is functionally important in vivo, Atg8 binding is not required for kinase activity or PAS localization of Atg1.
To examine whether the Atg1-LIR and Atg13-FV mutants affect the same or different aspects of Atg1 function, we analysed the double mutants for defects in autophagic pathways. Interestingly, whereas the Atg1 and Atg13 single mutants showed a partial reduction in Cvt activity (Figures 1I and 3A), the double mutant was almost completely impaired in Ape1 processing (Figure 3A). Likewise, although Atg1-LIR mutants did not show obvious defects in GFP-Atg8 processing (Figure 3C), the double mutants with Atg13-FV resulted in a strong autophagy defect (Figure 3C). Together, these data demonstrate that Atg8 and Atg13 mutually act on Atg1 to regulate its function in vivo, possibly at different steps of the autophagy pathway.
Atg1/ULK1 associates with autophagosomes in an LIR-dependent manner
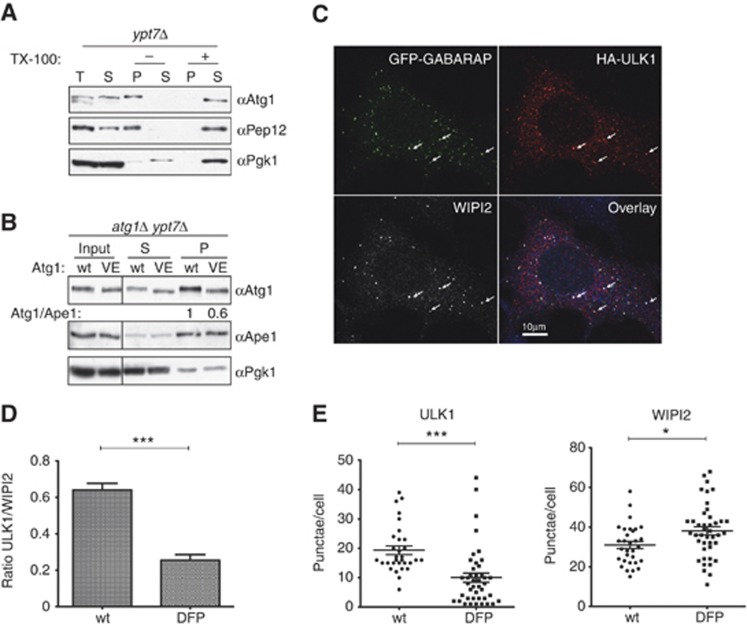
To test whether Atg8 may recruit Atg1/ULK1 to autophagosomes, we first separated yeast cell lysates into cytoplasmic (supernatant) and membrane (pellet) fractions, and analysed the presence of Atg1 by immunoblotting. To enrich for autophagosomes, we used starved ypt7Δ cells, which are defective for efficient fusion of autophagosomes with the vacuolar membrane. Interestingly, Atg1 was enriched in Triton X-100-sensitive membrane fractions, together with the endosomal membrane protein Pep12, indicating that Atg1 associates with membranes (Figure 4A; Supplementary Figure S3E). Membrane-associated Atg1 was resistant to proteolytic cleavage by proteinase K, indicating that part of Atg1 may be protected in completed autophagosomes (Supplementary Figure S3E). Importantly, membrane association of Atg1 depended on a functional LIR motif, as the Atg1-VE mutant showed a decreased fractionation with membranes (Figure 4B). Together, this biochemical analysis suggests that binding of Atg1 to Atg8 recruits Atg1 to membranes, which most likely represent autophagosomes.
Figure 4.
Atg1/ULK1 localize to autophagosomes in yeast and mammals. (A) Exponentially growing ypt7Δ cells were starved in SD-N medium for 4 h, lysed and the extract separated into a cytoplasmic (S) and a 5000 g membrane pellet fraction. The pellet was treated with (+) or without (−) TX-100, centrifuged and the supernatant (S) and pellet (P) fractions analysed by western blotting with anti-Atg1, anti-Pep12 and anti-Pgk1 antibodies. (B) Exponentially growing ypt7Δatg1Δ cells expressing wild-type Atg1 or the Atg1-VE mutant protein were starved in SD-N for 4 h, lysed and the extract separated into cytoplasmic (S) and membrane (P) fractions. The fractions were analysed by western blotting with anti-Atg1, anti-Pgk1 and anti-Ape1 antibodies, and the ratio of Atg1 to Ape1 in the pellet fractions was quantified and indicated as a ratio compared to wild-type cells. (C) HEK293 cells were co-transfected with GFP-GABARAP and HA-ULK1, starved for 2 h, and subsequently immunostained for HA (red) and WIPI2 (white). The arrows mark putative autophagosomes that stain positive for all three markers. Bar=10 μm. (D) HEK293 cells stably expressing GFP-LC3 were transfected with wild-type HA-ULK1 or the DFP mutant, and immunostained for HA and WIPI2 after 2 h starvation. HA-ULK1 and WIPI2 spot numbers were counted using Imaris software in over 70 cells (from one out of two experiments) and plotted as the ratio of ULK1 to WIPI2 puncta. ***Indicates a statistically significant P-value of <0.0001. One of the two independent experiments is shown. (E) Quantification of HA-ULK1 and WIPI2 spot numbers in HEK293 cells expressing wild-type HA-ULK1 or the DFP mutant. Cells were treated as above, and the spot number counted using Imaris software. A representative immunofluorescence image is shown in Supplementary Figure S3F. *** and * indicate P-values of <0.0001 or 0.0158, respectively. Figure source data can be found with the Supplementary data.
To substantiate these data, we next asked whether ULK1 binding to autophagosomes is dependent on the LIR motif in mammalian cells. Indeed, upon amino-acid starvation, HA-tagged ULK1 expressed in HEK293 cells at low levels was found on punctate structures identified by labelling with WIPI2, the mammalian homologue of the PI3P-binding protein Atg18 (Polson et al, 2010), and GFP-GABARAP or GFP-LC3 (Figure 4C; Supplementary Figure S3F). These structures most likely represent autophagosomes, but may also include phagophores and omegasomes. The number of ULK1-positive structures was significantly reduced in the DFP mutant of ULK1, suggesting that efficient recruitment of ULK1 to autophagosomes requires its interaction with mammalian Atg8 (Figure 4D and E). Interestingly, the total number of WIPI2 spots was increased after expression of the ULK1-DFP mutant (Figure 4E), implying that the WIPI2-positive autophagosomes or autophagosome precursors were stalled at an early stage during autophagy. Together, these results demonstrate that Atg8 recruits Atg1/ULK1 to autophagosome-like structures in yeast and mammals, which appears to be functionally important for autophagosome formation and/or maturation.
Autophagy induces vacuolar degradation of the Atg1–Atg13 complex
While analysing the levels of yeast Atg1 during nutrient limiting conditions, we observed that its level decreased upon starvation. This decrease required vacuolar proteases, as Atg1 was stabilized in pep4Δ cells defective for vacuolar degradation (data not shown). To analyse Atg1 degradation in more detail, we followed Atg1-GFP levels during starvation by western blotting. Interestingly, upon starvation yeast Atg1-GFP was transported to the vacuole releasing free GFP, representing proteolytic digestion of this protein in the vacuole similar to that of GFP-Atg8 (Figure 5A and B; Supplementary Figure S3G). This transport depends on Atg1 kinase activity, and on a functional autophagy pathway, as the kinase-dead Atg1-T226A mutant as well as wild-type Atg1 in atg8Δ cells failed to be degraded in the vacuole (Figure 5A; Supplementary Figure S3G). Moreover, even though autophagic transport of GFP-Atg8 to the vacuole is still functional in atg1-VE mutant cells, the Atg1-VE mutant protein was not transported to the vacuole upon starvation (Figure 5B). To corroborate these data, we used time-lapse microscopy to follow the selective transport of YFP-Atg1 in cells lacking the major vacuolar protease Pep4. Vacuolar translocation was quantified by comparing the ratio of cytoplasmic to vacuolar YFP-Atg1 after induction of autophagy by nitrogen starvation. Indeed, YFP-Atg1 strongly accumulated in the vacuole upon starvation in a time-dependent manner (Figure 5C; Supplementary Figure S3H). In contrast, even after 13 h of nitrogen deprivation, hardly any vacuolar enrichment was observed when analysing the YFP-Atg1-VE mutant, demonstrating that Atg8 binding is required for selective transport of Atg1 to the vacuole.
Figure 5.
Atg1 travels with autophagosomes to the vacuole during starvation. (A) atg1Δ cells containing either wild-type Atg1 or the kinase-dead Atg1-T226A mutant tagged with GFP were grown to mid log phase followed by starvation for 24 h in SD-N. Samples were taken at indicated time points and GFP cleavage was analysed by western blotting. Processing of endogenous Ape1 under starvation conditions monitors the autophagic flux. (B) atg1Δ cells containing either GFP-tagged wild-type Atg1 or the Atg1-VE mutant were grown and analysed as in (A). (C) atg1Δpep4Δ cells containing YFP-tagged wild-type Atg1 or the Atg1-VE mutant were grown to log phase and starved in SD-N. Cells were monitored by time-lapse microscopy and the vacuolar accumulation of Atg1 was quantified as described in ‘Materials and methods’. Data are plotted as the mean±s.e.m. from at least 90 quantified cells. Additional images are shown in Supplementary Figure S3H. Bar=5 μm. (D) atg1Δatg13Δ cells expressing GFP-tagged wild-type Atg13, and either wild-type Atg1 or the Atg1-VE mutant (left panel) and wild-type cells containing Atg13-GFP and either wild-type Atg13 or Atg13-FV were grown to mid log phase followed by starvation for 6 h in SD-N media. GFP cleavage was analysed by western blotting. Figure source data can be found with the Supplementary data.
Similarly, Atg13 levels were also decreased in starved cells, suggesting that Atg1 and its activator Atg13 travel with autophagosomes (Figure 5D). Interestingly, the vacuolar turnover of Atg13 also depends on the interaction between Atg1 and Atg8, as a significant decrease in Atg13-GFP processing was observed when the protein was expressed in the atg1-VE mutant (Figure 5D, left panel). Even in a situation where autophagy is functional, the Atg13-FV mutant protein showed slower degradation (Figure 5D, right panel), confirming that the interaction of Atg1 with Atg13 is required for this process.
Taken together, these results suggest that the Atg1–Atg13 kinase complex associates with autophagosomes in a selective and Atg8-dependent manner, resulting in its transport to and degradation in the vacuole.
Discussion
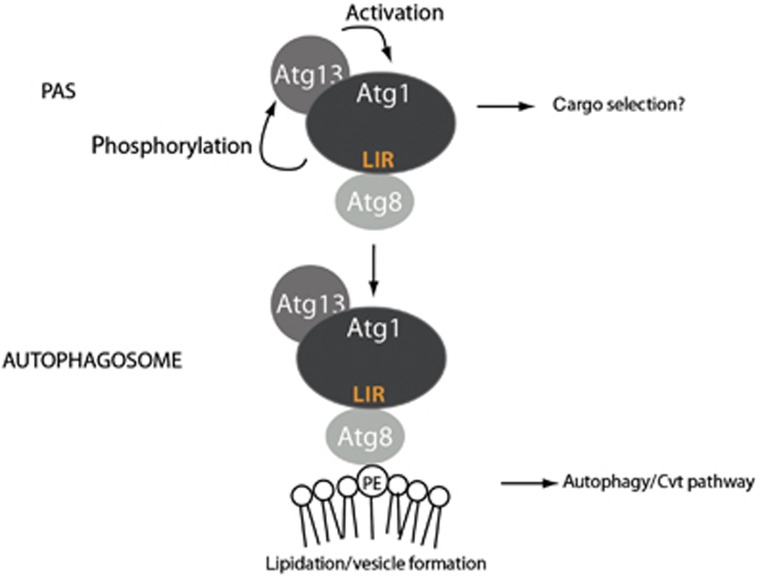
Here, we analysed the regulation of the Atg1–Atg13 complex in autophagy. Surprisingly, while Atg13 promotes Atg1 kinase activity, this interaction is neither regulated by starvation conditions nor essential for autophagy induction. However, Atg13 cooperates with Atg8 to regulate Atg1 function in vivo. Atg8 directly binds Atg1 through a conserved LIR motif, and this interaction targets the Atg1–Atg13 complex to autophagosomes and ultimately results in its vacuolar degradation. Together, these results revealed a conserved mechanism for targeting Atg1/ULK1 to autophagosomes, and may suggest a novel function to balance autophagic flux during limiting nutrient conditions (Figure 6).
Figure 6.
Model for the cooperation of Atg13 and Atg8 on Atg1 regulation. Atg13 constitutively binds Atg1. Upon starvation, Atg1 is activated by autophosphorylation and in turn phosphorylates Atg13, which further increases its kinase activity. Atg1 binds to Atg8 via its LIR motif, which triggers its removal from the PAS and mediates its association with autophagosomes. Atg1 on autophagosomes may phosphorylate unknown substrates involved in autophagosome maturation. After autophagosomal fusion, the Atg1–Atg13 complex is degraded in the vacuole/lysosome, thereby limiting autophagic flux during starvation.
Atg13 promotes Atg1 activity, but forms a stable complex with Atg1 regardless of the nutrient conditions
While Atg1 is required for autophagy induction from yeast to mammals, different activation mechanisms have been proposed. Our biochemical and mutational analysis in yeast confirmed that the interaction of Atg13 and Atg1 is direct and promotes Atg1 kinase activity in vivo. The autophagy defects of Atg13 mutants unable to interact with Atg1 are however less pronounced when compared to cells deleted for Atg13, indicating that Atg13 may have functions outside of the Atg1 complex. In yeast, Atg13 promotes autophosphorylation and dimerization of Atg1 (Yeh et al, 2011), and TORC1-mediated phosphorylation of Atg13 is thought to prevent its interaction with Atg1. In contrast, fly and mammalian Atg13 constitutively bind to the Atg1 homologues dAtg1 and ULK1, respectively, suggesting that in these organisms the complex is activated by other means such as phosphorylation or the association of additional factors (Hara et al, 2008; Chan et al, 2009; Alers et al, 2011). Surprisingly, we did not detect any significant difference in the ability of yeast Atg13 to bind to the Atg1 complex by comparing the complex formation in cells grown under rich or nitrogen-starved conditions, or after inhibition of the TOR kinase by rapamycin. Consistent with this notion, the size of the Atg1 complex does not change with its activation status, as the kinase-inactive Atg1-T226AS230A shows a similar sedimentation behaviour in sucrose gradients as the wild-type Atg1 complex (Kijanska et al, 2010). Finally, Atg13 mutants unable to interact with Atg1 exhibit a Cvt defect, implying that the association of Atg13 and Atg1 is functionally important under conditions where TORC1 is active. Together, these results strongly suggest that like in other organisms, activation of the yeast Atg1 kinase is not mediated by regulated binding of Atg13. While it remains to be determined whether proteins other than Atg13 join the complex in a nutrient-dependent manner, regulated phosphorylation of Atg1 on conserved residues in its activation loop is essential for Atg1 activity in vitro and in vivo (Kijanska et al, 2010; Yeh et al, 2010). Likewise, the activation loop is phosphorylated in mammalian ULK1, and phosphorylation of this site is required for its activation (Bach et al, 2011). Interestingly, further sites in mammalian ULK1 are phosphorylated by AMP-activated protein kinase (AMPK), suggesting that glucose starvation also activates autophagy through phosphorylation (Egan et al, 2011). Similarly, PKA-dependent phosphorylation and activation of yeast Atg1 has been observed (Stephan et al, 2009). Finally, the phosphorylation status of Atg13 is regulated by TORC1, suggesting that these sites are relevant for its nutrient-dependent regulation in vivo. Taken together, these results suggest that like in higher eukaryotes, activation of yeast Atg1 requires phosphorylation of Atg1 and most likely Atg13, but is not regulated by binding of Atg1 and Atg13.
Atg8 recruits Atg1/ULK1 to autophagosomes and promotes Atg1 degradation in the vacuole
Interestingly, our results demonstrate that Atg8 directly interacts with Atg1 through a conserved LIR motif, which is not required for Atg13 binding. Mutations that specifically abolish Atg8 binding decrease the ability of Atg1 to promote progression of the Cvt pathway, and sensitize cells for autophagy defects, implying that Atg13 and Atg8 independently promote Atg1 function in vivo. In contrast to Atg13, Atg8 binding is dispensable for Atg1 kinase activity, implying that binding of Atg1 to Atg8 and Atg13 may affect different aspects of Atg1 function, and most likely at different steps of the pathway. The LIR motif of yeast Atg1 is conserved in human ULK1, where it mediates binding to the Atg8-like proteins GABARAP and GATE-16, which are known to regulate later steps in autophagosome formation (Weidberg et al, 2010). It has previously been observed that ULK1 associates with membranes (Chan et al, 2009), and our results indicate that the recruitment of ULK1 to autophagosome-like structures is dependent, at least in part, on its interaction with GABARAP/GATE16. While the precise molecular mechanism leading to accumulation of WIPI2 punctae in ULK1-DFP expressing cells remains to be determined, it may reflect a requirement for ULK1 in the function of GABARAP/GATE16 during late stages of autophagy (Weidberg et al, 2010). Likewise, binding of Atg8 to Atg1 is required for its recruitment to membranes and vacuolar degradation in yeast, and the Atg1–Atg13 complex has also been visualized on autophagosomes in plants (Suttangkakul et al, 2011). These results suggest that Atg1/ULK1 may play a conserved role in cargo engulfment or autophagosome formation, or their maturation and delivery to the vacuole. For example, Atg1/ULK1 may phosphorylate and thereby activate factors involved in closure of autophagosomes or vacuolar fusion. Interestingly, yeast Atg1 mutants unable to bind Atg8 exhibit defects in the Cvt pathway, but did not significantly affect autophagy progression when measured with the GFP-Atg8 cleavage assay. Because Cvt vesicles are generally smaller than autophagosomes, it is possible that higher local Atg1 activity is needed at late steps, and thus autophagy defects of the Atg1-LIR mutants become only apparent when the pathway is already sensitized. This view is consistent with Atg1 mutants with reduced kinase activity, which affect the Cvt pathway with minimal defects in autophagy. Alternatively, the Atg8–Atg1 complex may specifically be important for selective autophagy pathways that require special mechanisms to select and engulf different cargo. It is possible that these special steps may require localized Atg1/ULK1 activity, which is dispensable for starvation-induced bulk autophagy.
Irrespective of the precise molecular function, our results demonstrate that Atg1–Atg13 complexes tethered to autophagosomes are subsequently degraded in the vacuole. Interestingly, this Atg8-dependent degradation mechanism efficiently downregulates activated Atg1–Atg13 complexes, and may thus prevent excessive autophagic degradation by coupling autophagy flux to the availability of amino acids and energy required to newly synthesize Atg1 and Atg13. Since starvation-regulated signalling pathways such as TORC1 not only induce autophagy but also inhibit protein translation, these mechanisms are expected to cooperate to adjust autophagy flux to the physiological needs. Such a negative feedback loop has previously been proposed based on genetic experiments in flies, where TORC1 activates the Atg1 kinase, which in turn feeds back on TORC1 to fine-tune the activity of the pathway (Chang and Neufeld, 2009).
Atg8 plays multiple roles in autophagy by interacting with different effectors involved in regulating the basic machinery and cargo adaptors
Atg8-family members are essential components of the autophagy machinery, which are activated by the covalent attachment of a PE moiety that anchors the proteins to phagophore and autophagosome membrane. Recent results revealed that Atg8-like proteins regulate several steps in selective and non-selective autophagy pathways. For example, yeast Atg8 and its mammalian LC3 homologues are involved in elongating the phagophore membrane required to ultimately engulf the cargo. Furthermore, the GABARAP/GATE-16 subfamily members are essential for autophagosome closure (Weidberg et al, 2010). Finally, the conjugation of yeast Atg8 to PE is required for the hemifusion of membranes in yeast (Nakatogawa et al, 2007), and Atg8-like proteins may directly promote expansion and fusion of autophagosomes through their amino-terminal domains (Weidberg et al, 2011).
On the other hand, specific binding partners of Atg8/LC3 have recently been identified, which interact through defined LIR motifs on the target proteins and a LIR-interacting surface on Atg8/LC3. In some cases, these LIR domain-containing proteins function as specificity adaptors for the selective degradation of certain cargo by autophagy. For example, the LC3-adaptor p62 is not required for general autophagy, but targets ubiquitinated protein aggregates for lysosomal destruction. Likewise, NDP52 and optineurin link LC3 to intracellular pathogens, thereby selectively removing them from the cell by autophagy (Thurston et al, 2009; Wild et al, 2011). Thus, these results suggest that LC3 plays a direct role in the selection and engulfment of specific cargo for autophagy, and many LIR-containing proteins are thought to function in this process. However, LIR-dependent interactions of Atg8 with autophagy core proteins that may serve a regulatory function have also been reported. Both Atg3 in yeast and Atg4B in mammals interact with Atg8/LC3 (Satoo et al, 2009; Yamaguchi et al, 2010), and binding of Atg8 to the LIR motif of Atg3 is required for its efficient lipidation in the Cvt pathway. Similarly, our results expand the known functions of Atg8 and strengthen the notion that Atg8-interacting targets may not only serve as receptors for cargo recruitment but also directly regulate multiple components of the general autophagy machinery. It will thus be important to carefully analyse LIR-containing proteins whether they affect general processes or are specifically implicated in vacuolar/lysosomal degradation in selective autophagy pathways.
Materials and methods
Yeast strains, growth conditions and antibodies
Yeast strains are listed in Supplementary Table S1. Yeast cells were grown in synthetic medium (SD: 0.17% yeast nitrogen base, 0.5% ammonium sulphate, 2% glucose, amino acids as required). For starvation induction, cells of 0.5–0.8 OD600 were washed and resuspended in starvation medium (SD-N: 0.17% yeast nitrogen base without amino acids, 2% glucose) for 4 h, unless stated otherwise.
The following antibodies were used in this study: mouse monoclonal anti-GFP antibody (Roche), rabbit polyclonal PAP antibody (Sigma), rabbit polyclonal anti-TAP (Open Biosystems), mouse monoclonal anti-GST antibody (Sigma), mouse anti-Pep12 antibody (Molecular Probes), rabbit polyclonal anti-HA antibody (Sigma), rat anti-HA (Roche), rabbit polyclonal anti-Caveolin1 antibody (Santa Cruz), mouse anti-trimethylation-specific antibody (Zuzuarregui et al, 2012) and mouse anti-WIPI2 (Polson et al, 2010). The rabbit polyclonal anti-Ape1, anti-Atg1 and anti-Atg13 antibodies were kindly provided by D Klionsky, University of Michigan, and the rabbit polyclonal anti-Calnexin antibody was a generous gift from A Helenius, ETH Zurich.
Plasmid construction and two-hybrid analysis
Plasmids are listed in Supplementary Table S2. Site-directed mutagenesis was performed by QuikChange mutagenesis (Stratagene), and verified by sequencing. ATG13 with its endogenous promoter and terminator (698 and 371 bases, respectively) was amplified from genomic DNA and ligated into the pRS316 vector (Sikorski and Hieter, 1989) using the XbaI and HindIII restriction sites. TAP and GFP fusions of ATG1 were generated by ligating PCR-amplified TAP and GFP tags via PstI-SalI to the C-terminus of ATG1 expressed under its endogenous promoter (726 bases), cloned into pRS315. The pRS313-GFP-Atg8 plasmid was constructed by subcloning from pRS316-GFP-Atg8 plasmid via XhoI-SacI sites. Yeast two-hybrid experiments were performed as described (Kijanska et al, 2010). ATG13, ATG1 and the indicated mutants were subcloned into the yeast two-hybrid vectors pGAD and pLEX, respectively.
Yeast cell extract preparation, immunoprecipitations, and the pho8Δ60 assay
Protein extraction with TCA and immunoblotting was carried out as described previously (Kijanska et al, 2010). To prepare extract under non-denaturing conditions, cells were grown in selective medium to OD600 of 0.5–0.8, harvested by centrifugation and washed in PBS with 2% glucose. Cells were then resuspended in a pellet volume of lysis buffer (PBS, 10% glycerol, 0.5% Tween-20, 1 mM NaF, 1 mM PMSF, 1 mM Na3VO4, protease inhibitor cocktail; Roche) and frozen in droplets in liquid nitrogen. After cell disruption with a freezer mill (6770, Spex, USA), extracts were cleared by centrifugation at 5000 g for 5 min, followed by two centrifugations at 10 000 g for 15 min. Note that in contrast to spheroplasting, freezer milling disrupts organelle-enclosed proteins.
HA immunoprecipitations were carried out as described (Kijanska et al, 2010). For protein A immunoprecipitations, Dyna epoxy magnetic beads (Invitrogen) were coupled with rabbit IgG according to manufacturer’s protocol and used instead of HA agarose.
The pho8Δ60 assay was performed as described (Klionsky, 2007). To ensure the linear range of the enzymatic reaction, the assay was performed with two different sample concentrations.
Cell fractionation and proteinase protection assay
Yeast cells starved for 4 h in SD-N were washed and incubated with 2-OD/ml DTT buffer (10 mM DTT, 10 mM Tris pH 9.4; 15 min, 30°C), resuspended in 10-OD/ml SP buffer (1 M sorbitol, 20 mM PIPES pH 6.8) and spheroplasted by Zymolyase-20T (MP Biomedicals) treatment. Spheroplasts were collected by centrifugation (2000 g for 5 min) and gently lysed with osmotic lysis buffer (200 mM sorbitol, 20 mM PIPES pH 6.8, 5 mM MgCl2, ‘input’) to preserve organelle-enclosed particles. After two 500 g centrifugation steps to remove unbroken cells, the resulting total lysate (‘T’) was further separated into a 5000 g supernatant (‘S’) and a pellet fraction (‘P’). The efficiency of fractionation and distribution of Atg1 was determined by immunoblot analysis using anti-Pgk1, anti-Ape1 and anti-Atg1 antibodies. Membrane association of Atg1 was controlled by treating the pellet fraction for 30 min with 0.2 % Triton X-100 in the presence of the complete protease inhibitor cocktail (Roche) and 0.1 mM PMSF, followed by a 5000 g centrifugation step to separate the supernatant (‘S’) and insoluble pellet fractions. For proteinase protection assays, the 5000 g pellet was resuspended in osmotic lysis buffer with or without proteinase K (50 μg/ml, AppliChem) and 0.2% Triton X-100. After 30 min incubation on ice, samples were precipitated with TCA and analysed by western blotting with anti-Ape1, anti-Atg1 and anti-Pep12 (Molecular Probes) antibodies.
Kinase assays
In all, 10 mg of cleared yeast extract was incubated with 50 μl IgG-coupled magnetic beads (Dynabeads, Invitrogen) for 1 h at 4°C, with rotation. The beads were washed 4 × in IP buffer (100 mM Tris–HCl pH 7.5, 300 mM NaCl, 10 mM EDTA, 10 mM EGTA, 1% NP-40, 10 mM Na3VO4, 10 mM NaF, 1 mM PMSF, protease inhibitor cocktail; Roche), and twice in kinase buffer (25 mM MOPS pH 7.5, 1 mM EGTA, 10 mM Na3VO4, 15 mM MgCl2). Kinase reactions were performed in a total volume of 11 μl containing 10 μCi γATP for 20 min at 30°C. Reactions were stopped with 10 μl urea loading buffer and analysed by SDS–PAGE and phosphor-imaging.
Floatation analysis
Floatation in sucrose gradients was performed according to Lingwood and Simons, (2007). In brief, cleared yeast or HeLa cell extracts were adjusted to 40% sucrose and layered under a 5–35% sucrose gradient in TNE (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 2 mM EDTA, protease inhibitor cocktail; Roche). The gradients were centrifuged at 200 000 g for 18 h in a SW41 rotor in a Beckman Coulter Optima TLX ultracentrifuge. In all, 1 ml fractions were collected and precipitated with TCA. Note that the bottom 4–5 fractions contain soluble proteins as calnexin, whereas lipid-associated complexes float in higher fractions (caveolin).
Atg8-binding assay and co-immunoprecipitation experiments
TAP-tagged proteins were immunoprecipitated overnight at 4°C from yeast cell extracts prepared under native conditions using IgG-coupled sepharose beads (GE Healthcare), washed and then cleaved with TEV protease for 3–6 h at 4°C or 2 h at 16°C. The TEV eluates were incubated with 3–4 μg of GST-Atg8 or GST-ubiquitin purified from E. coli. Bound proteins were analysed with rabbit polyclonal anti-TAP (Thermo Scientific) and mouse monoclonal anti-GST antibody (Sigma) antibodies. For co-immunoprecipitation experiments, 20 mg of cleared yeast extract was incubated with 30 μl anti-HA agarose beads (Sigma) or magnetic Dynabeads (Invitrogen) coupled to rabbit IgG on a rotating wheel for 1 h at 4°C. The beads were washed 6 × in lysis buffer and bound proteins eluted with 70 μl urea loading buffer.
Purification of mammalian Atg8 proteins and mammalian binding assays
GST, GST-LC3B, GST-GABARAP or GST-GATE-16 proteins were purified from E. coli on a Glutathione Sepharose 4B resin. In all, 33 μg of each protein was bound to Glutathione Sepharose 4B beads in PBS containing protease inhibitors (Roche). Cell lysates prepared from HEK293 cells transfected with HA-ULK1 wild-type or HA-ULK1 containing mutations D356A, F357A, P361A in TNTE buffer (20 mM Tris pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.3% Triton X-100) supplemented with Complete protease inhibitor cocktail were incubated with the GST-protein bound beads for 2 h followed by extensive washing in TNTE. Bound protein was eluted with 2 × sample buffer and analysed by SDS–PAGE and western blotting.
Co-immunoprecipitation of mammalian proteins
HEK293 cells were transfected using lipofectamine 2000 (Invitrogen) with GFP-GABARAP, or co-transfected with GFP-GABARAP and either wild-type HA-ULK1 or the DFP mutant. As a negative control, cells were transfected as above using GFP instead of GFP-GABARAP. The following day cells were washed once with cold PBS and lysed in TNTE buffer (20 mM Tris pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.3% Triton X-100) supplemented with Complete protease inhibitor cocktail. GFP-GABARAP and GFP proteins were immune-precipitated by incubating with GFP-Trap beads (ChromoTek) for 1 h at 4°C, followed by two washing steps with TNTE and an additional wash with 50 mM Tris 7.4. Beads were then added with 30 μl of 2 × sample buffer (125 mM Tris 6.8, 6% SDS, 20% glycerol, 6.66% β-mercaptoethanol, 0.2% bromophenol blue) and boiled at 100°C for 10 min. Binding of endogenous ULK1 or overexpressed HA-ULK1 was analysed by western blot using rabbit anti-ULK1 (Santa Cruz) or mouse anti-HA (Covance), respectively.
Purification and in vitro binding assays of Atg1 and Atg13 fragments
Amino acids 501–897 of Atg1 and amino acids 432–520 of either wild-type or the Atg13-FV mutant were cloned into pGEX4T1 resulting in N-terminal GST fusion proteins. Proteins were purified from E. coli on Glutathione Sepharose 4B resin, and the Atg13 proteins cleaved with thrombin in 1 × PBS pH 7.4. An excess of cleaved Atg13 was incubated in the presence of 4 mM pefabloc with GST-Atg1 immobilized on Glutathione Sepharose 4B beads. After extensive washing, bound proteins were eluted with sample buffer and analysed by SDS–PAGE and coomassie staining. The ratio of Atg13 bound to GST-Atg1 was quantified using ImageJ software and normalized by the size of the respective proteins. The ratio quantified from the coomassie stained gel of Atg13 to GST-Atg1 was 0.11, whereas the size ratio is 0.13. Therefore, the binding ratio is roughly 1:1.
In vivo methylation-interaction assay
Atg1 was C-terminally tagged with Histone3-HA and the C-terminus of wild-type and the Atg13-FV mutant with the Suv methylase domain. Both proteins were expressed from their endogenous promoters on a CEN plasmid in an atg1Δatg13Δ yeast strain and tested for functionality by analysing Ape1 processing. The M-track methylation assay was performed as described (Zuzuarregui et al, 2012).
Immunofluorescence
HEK293 cells stably expressing GFP-LC3 were generated as previously described (Kochl et al, 2006), grown on coverslips and transfected with wild-type HA-ULK1 or the DFP mutant using the Fugene transfection reagent (Promega). To analyse co-localization of HA-ULK1 with GFP-GABARAP, HEK293 cells were co-transfected with GFP-GABARAP and HA-ULK1 using the same method. Autophagy was induced the next day by incubating the cells for 2 h in Earls Balanced Saline Solution (EBSS). Cells were then fixed in 4% paraformaldehyde, quenched with 50 mM NH4Cl for 10 min and permeabilized with 0.2% Triton X-100 for 3 min. Blocking was done in 5% BSA for 1 h, followed by 1 h incubation with rat anti-HA (Roche #11867423001) and mouse anti-WIPI2 (Polson et al, 2010) antibodies, three times washing in PBS and incubation with secondary antibodies (Goat anti-Rat 555, Donkey anti-Mouse 647; Invitrogen) for 1 h. Cells were then washed three times with PBS and mounted on glass slides using mowiol (Calbiochem). Images were acquired with an LSM 510 laser scanning confocal microscope equipped with a × 63 oil immersion objective (Carl Zeiss MicroImaging Inc). Quantifications of HA-ULK1 and WIPI2 spots were done with blinded samples using the spot function in the Imaris image analysis software, keeping the same threshold for all analysed cells.
Protein localization in yeast and quantitative live-cell imaging
Yeast cells expressing fluorescently tagged proteins were grown in liquid media to OD600 of 0.6–0.8, and starved for 4 h in SD-N. Images were acquired with required excitation and emission filters (GFP excitation: 470±40 nm, GFP emission: 525±50 nm, RFP excitation: 572±35 nm, RFP emission: 632±60 nm, YFP excitation: 500±20 nm, YFP emission: 535±30 nm). Quantitative live-cell imaging of starved cells was performed in a microfluidic cell culture chamber (Y4, CellASIC Corp, USA) coupled to an automated controller (ONIX, CellASIC Corp), allowing rapid SD to SD-N media exchange. Cells in exponential phase were loaded into the microfluidic chamber (height=4–5 μm), maintained in a fixed position (Lee et al, 2008) and followed by fluorescence microscopy during the N-starvation. Images of yeast cells in the microfluidic chamber were acquired on an inverted fluorescence microscope (Eclipse Ti, Nikon Instruments) using an oil immersion objective lens (CFI Plan Apo 60 × , Nikon) in a temperature incubator set at 30°C. The hardware-based focusing system (Perfect Focus System (PFS), Nikon Instruments) stably maintained the focus during the whole experiment. Images were automatically analysed with MATLAB routines, using cell segmentation based on defocused and focused transmission images to define the cell boundary (Gordon et al, 2007). Standard deviation (s.d.) of normalized average intensity in of YFP in single cells was assessed to quantify vacuolar accumulation of Atg1, and normalized by comparing the initial value of each cell.
Supplementary Material
Acknowledgments
We thank Larissa Wilhelm, Dan Klionsky, Egon Ogris and Zvulun Elazar for plasmids and antibodies; Serge Pelet for advice on microscopy; Fulvio Reggiori for sharing unpublished results; members of the Peter laboratory for helpful discussions; and Alicia Smith, Reinhard Dechant and Fulvio Reggiori for critical reading of the manuscript. CK is supported by a Marie-Heim Vögtlin fellowship from the Swiss National Science Foundation (SNF) and a ‘Vienna Research Groups for Young Investigators’ grant from the Vienna Science and Technology Fund (WWTF). MK is a member of the Molecular Life Science PhD program (MLS), and MP and SSL of the Competence Center for Systems Physiology and Metabolic Diseases (SPMD). SAT and EK are supported by Cancer Research UK. Work in the Peter laboratory is supported by the European Research Council (ERC), the 7th EU framework project UNICELLSYS, and by grants from SystemsX.ch, the SNF and the ETH Zürich, respectively.
Author contributions: CK, MK, EK, ES, SL, GS, IS, AB, IH and MV performed the experiments. CK, MK, EK, ES, SL, GA, KH, ST and MP participated in the experimental design. CK and MP wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alers S, Loffler AS, Paasch F, Dieterle AM, Keppeler H, Lauber K, Campbell DG, Fehrenbacher B, Schaller M, Wesselborg S, Stork B (2011) Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction. Autophagy 7: 1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach M, Larance M, James DE, Ramm G (2011) The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem J 440: 283–291 [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW (2010) Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Longatti A, McKnight NC, Tooze SA (2009) Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol 29: 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Neufeld TP (2009) An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell 20: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang CW, Klionsky DJ (2005) Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell 16: 3438–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ (2011) Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A, Colman-Lerner A, Chin TE, Benjamin KR, Yu RC, Brent R (2007) Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat Methods 4: 175–181 [DOI] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ (1995) Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol 131: 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Dang Y, Dai F, Guo Z, Wu J, She X, Pei Y, Chen Y, Ling W, Wu C, Zhao S, Liu JO, Yu L (2003) Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem 278: 29278–29287 [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N (2009a) Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 20: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N (2009b) Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 5: 973–979 [DOI] [PubMed] [Google Scholar]
- Hutchins MU, Klionsky DJ (2001) Vacuolar localization of oligomeric alpha-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem 276: 20491–20498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Inoue Y, Klionsky DJ (2010) Regulation of macroautophagy in Saccharomyces cerevisiae. Semin Cell Dev Biol 21: 664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 117: 2805–2812 [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150: 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y (2009) Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 30: 1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijanska M, Dohnal I, Reiter W, Kaspar S, Stoffel I, Ammerer G, Kraft C, Peter M (2010) Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy 6: 1168–1178 [DOI] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Overvatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33: 505–516 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ (2007) Monitoring autophagy in yeast: the Pho8Delta60 assay. Methods Mol Biol 390: 363–371 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Seglen PO (2007) Methods for monitoring autophagy from yeast to human. Autophagy 3: 181–206 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cueva R, Yaver DS (1992) Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol 119: 287–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochl R, Hu XW, Chan EY, Tooze SA (2006) Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic 7: 129–145 [DOI] [PubMed] [Google Scholar]
- Kraft C, Peter M, Hofmann K (2010) Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol 12: 836–841 [DOI] [PubMed] [Google Scholar]
- Kraft C, Reggiori F, Peter M (2009) Selective types of autophagy in yeast. Biochim Biophys Acta 1793: 1404–1412 [DOI] [PubMed] [Google Scholar]
- Lee PJ, Helman NC, Lim WA, Hung PJ (2008) A microfluidic system for dynamic yeast cell imaging. Biotechniques 44: 91–95 [DOI] [PubMed] [Google Scholar]
- Lingwood D, Simons K (2007) Detergent resistance as a tool in membrane research. Nat Protoc 2: 2159–2165 [DOI] [PubMed] [Google Scholar]
- Meiling-Wesse K, Barth H, Thumm M (2002) Ccz1p/Aut11p/Cvt16p is essential for autophagy and the cvt pathway. FEBS Lett 526: 71–76 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y (1998) A protein conjugation system essential for autophagy. Nature 395: 395–398 [DOI] [PubMed] [Google Scholar]
- Motley AM, Nuttall JM, Hettema EH (2012) Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J 31: 2852–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130: 165–178 [DOI] [PubMed] [Google Scholar]
- Noda NN, Ohsumi Y, Inagaki F (2010) Atg8-family interacting motif crucial for selective autophagy. FEBS Lett 584: 1379–1385 [DOI] [PubMed] [Google Scholar]
- Noda T, Matsuura A, Wada Y, Ohsumi Y (1995) Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun 210: 126–132 [DOI] [PubMed] [Google Scholar]
- Okazaki N, Yan J, Yuasa S, Ueno T, Kominami E, Masuho Y, Koga H, Muramatsu M (2000) Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Brain Res Mol Brain Res 85: 1–12 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA (2010) Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6: 506–522 [DOI] [PubMed] [Google Scholar]
- Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F (2009) The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J 28: 1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Nice DC 3rd, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ (2000) Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem 275: 25840–25849 [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK (2009) The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA 106: 17049–17054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttangkakul A, Li F, Chung T, Vierstra RD (2011) The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kondo C, Morimoto M, Ohsumi Y (2010) Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J Biol Chem 285: 30019–30025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Ohsumi Y (2007) Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett 581: 2156–2161 [DOI] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F (2009) The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 10: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Noda NN, Kumeta H, Suzuki K, Ohsumi Y, Inagaki F (2010) Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34. J Biol Chem 285: 30026–30033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shpilka T, Shvets E, Abada A, Shimron F, Elazar Z (2011) LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell 20: 444–454 [DOI] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z (2010) LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 29: 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, Dotsch V, Bumann D, Dikic I (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333: 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ (2007) Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Xin Y, Yu L, Chen Z, Zheng L, Fu Q, Jiang J, Zhang P, Gong R, Zhao S (2001) Cloning, expression patterns, and chromosome localization of three human and two mouse homologues of GABA(A) receptor-associated protein. Genomics 74: 408–413 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Noda NN, Nakatogawa H, Kumeta H, Ohsumi Y, Inagaki F (2010) Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J Biol Chem 285: 29599–29607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YY, Shah KH, Herman PK (2011) An Atg13 protein-mediated self-association of the Atg1 protein kinase is important for the induction of autophagy. J Biol Chem 286: 28931–28939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh YY, Wrasman K, Herman PK (2010) Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics 185: 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuga M, Gomi K, Klionsky DJ, Shintani T (2011) Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J Biol Chem 286: 13704–13713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuzuarregui A, Kupka T, Bhatt B, Dohnal I, Mudrak I, Friedmann C, Schuchner S, Frohner IE, Ammerer G, Ogris E (2012) M-Track: detecting short-lived protein-protein interactions in vivo. Nat Methods 9: 594–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.