This work examines the role of CYC2 clade genes in floral asymmetry in Primulina heterotricha, finding that CYC1C and CYC1D positively auto- and cross-regulate both themselves and each other in a double positive autoregulatory feedback loop, a regulatory mechanism that may be conserved in other clades with zygomorphic flowers.
Abstract
Members of the CYCLOIDEA2 (CYC2) clade of the TEOSINTE BRANCHED1, CYCLOIDEA, and PCF transcription factor genes are widely involved in controlling floral zygomorphy, a key innovation in angiosperm evolution, depending on their persistently asymmetric expression in the corresponding floral domains. However, it is unclear how this asymmetric expression is maintained throughout floral development. Selecting Primulina heterotricha as a model, we examined the expression and function of two CYC2 genes, CYC1C and CYC1D. We analyzed the role of their promoters in protein–DNA interactions and transcription activation using electrophoresis mobility shift assays, chromatin immunoprecipitation, and transient gene expression assays. We find that CYC1C and CYC1D positively autoregulate themselves and cross-regulate each other. Our results reveal a double positive autoregulatory feedback loop, evolved for a pair of CYC2 genes to maintain their expression in developing flowers. Further comparative genome analyses, together with the available expression and function data of CYC2 genes in the core eudicots, suggest that this mechanism might have led to the independent origins of floral zygomorphy, which are associated with plant–insect coevolution and the adaptive radiation of angiosperms.
INTRODUCTION
How phenotypic novelties originate is a central question for evolutionary developmental biologists. Floral zygomorphy, which is bilateral floral symmetry with floral organs of different shapes or sizes arranged with one plane of symmetry (as opposed to actinomorphic or radial floral symmetry), is one key innovation associated, at least in part, with the explosive radiation of angiosperms because it promotes the coevolution between angiosperms and insects. This reciprocal animal–plant relationship is well developed and widespread among zygomorphic plants and anthophilous insects, which live on flowers (Dilcher, 1979, 2000; Cubas, 2004; Busch and Zachgo, 2009; Specht and Bartlett, 2009). Paleontological and phylogenetic studies have shown that floral zygomorphy arose independently several times from actinomorphic ancestors and expanded to many large and successful angiosperm clades that have adaptive advantages attributed to an enhanced preference by specific insects; these clades include the Fabaceae, Lamiales, Asteraceae, and Orchidaceae (Donoghue et al., 1998; Endress, 1999; Dilcher, 2000; Cubas, 2004; Busch and Zachgo, 2009; Jabbour et al., 2009; Specht and Bartlett, 2009). However, it is still a key challenge to decipher the molecular bases for the independent events leading to establishment of zygomorphy. Morphologically, the shift from actinomorphy to zygomorphy requires a symmetry-breaking event. Theoretically, a locally strong effect on cell proliferation and growth would drive the shift from the existing symmetry to an alternative one. In angiosperms, especially in the core eudicots, most zygomorphic flowers have an apomorphic feature of reduced or enlarged dorsal petals, in many cases with the abortion of the dorsal stamen (Donoghue et al., 1998; Endress, 1999). Therefore, regulatory genes determining floral dorsal identity would be potential candidates for the genes underlying the evolutionary origin of floral zygomorphy.
In snapdragon (Antirrhinum majus), a model organism in the Lamiales, the CYCLOIDEA (CYC) gene and its paralog DICHOTOMA (DICH) play a crucial role in floral zygomorphy via their dorsal identity function (i.e., determining the fate of dorsal floral organs in the second and third whorls) (Luo et al., 1996, 1999). CYC and DICH encode proteins belonging to the plant-specific TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (TCP) family of transcription factors (Cubas et al., 1999). Based on the TCP domain, members of the TCP family are classified into PCF (TCP-P or class I) and CYC/TB1 (TCP-C or class II) subfamilies (Cubas et al., 1999; Navaud et al., 2007), and the latter is further divided into CIN and ECE lineages (Howarth and Donoghue, 2006). Phylogenetic analyses show that two major duplication events within the ECE lineage took place just before the radiation of core eudicots; these duplication events gave rise to three clades, CYC1, CYC2, and CYC3 (Howarth and Donoghue, 2006). Increasing evidence indicates that CYC2 clade genes, specific for the core eudicots, were repeatedly recruited to function in the control of floral zygomorphy based on their strong, dorsoventrally asymmetric expression (Luo et al., 1996, 1999; Feng et al., 2006; Busch and Zachgo, 2007, 2009; Broholm et al., 2008; Gao et al., 2008; Kim et al., 2008; Wang et al., 2008, 2010; Preston and Hileman, 2009; Song et al., 2009; Zhang et al., 2010; Howarth et al., 2011). Nevertheless, as a linkage between genotype and phenotype, the maintenance of expression of a gene that controls a key developmental process would be crucial in generating phenotypic effects (Crews and Pearson, 2009). This expression maintenance, together with the tissue- or organ-specific expression location defined by trans- or cis-activities, would result in a specific phenotype. Evidence from the backpetals mutant in snapdragon shows that a cis-regulatory region ∼4.2 kb upstream of the start codon would determine the dorsal location of CYC gene expression by repressing its transcription in the lateral and ventral regions (Luo et al., 1999). Therefore, it is important to understand how the maintenance of expression of CYC2 clade genes has been achieved, as co-option of its dorsal-specific location could be functionally sufficient to generate morphological zygomorphy. Furthermore, transient or weak expression of CYC2 clade genes in young floral or floral organ meristems in the ancestral actinomorphic lineages usually has no or very weak morphological effects (Cubas et al., 2001; Zhang et al., 2010; Busch et al., 2012). Therefore, the regulatory changes required to maintain and strengthen CYC2 expression, co-opted along with the regulatory mechanisms that produce dorsal-specific CYC2 expression, may represent a major step in the evolutionary origin of floral zygomorphy.
The TCP domain characteristic of TCP transcription factors is essential and sufficient for binding to GC-rich DNA motifs (GGNCCCAC and GTGGNCCC) (Kosugi and Ohashi, 2002; Aggarwal et al., 2010; Viola et al., 2012). Similar motifs have been identified as cis-acting elements in many plant genes, such as cell cycle– and protein synthesis–related genes, seed germination– and bud outgrowth–associated genes, and regulatory genes (reviewed in Martín-Trillo and Cubas, 2010). However, it is still poorly understood how TCP gene activities are controlled, especially at the transcriptional level (Martín-Trillo and Cubas, 2010). Transcription factors interacting with target DNA of their own genes is one of the primary mechanisms by which their expression is modulated and maintained (Dowell, 2010). Therefore, it is a key issue whether the expression maintenance of CYC2 clade genes in zygomorphic clades involves autoregulation.
Lamiales is a major angiosperm clade where most species have zygomorphic flowers. As the basal-most group in the Lamiales, the family Gesneriaceae has diverse forms of zygomorphy relating to floral organ differentiation early in the order (Cubas, 2004). Recent studies show that CYC2 clade genes in Gesneriaceae play key roles in establishment of zygomorphy, depending on their dorsoventrally asymmetric expression (Du and Wang, 2008; Gao et al., 2008; Song et al., 2009). One zygomorphic member of the Gesneriaceae, Primulina heterotricha (formerly known as Chirita heterotricha; Wang et al., 2011) is characterized by a zygomorphic corolla with the abortion of both dorsal and lateral stamens. Accordingly, two P. heterotricha CYC2 genes, CYC1C and CYC1D, are expressed in the corresponding floral organs from early to late stages (Gao et al., 2008). Therefore, P. heterotricha represents an ideal candidate to further explore the possible regulatory mechanisms for the expression maintenance of CYC2 clade genes.
In this study, we selected P. heterotricha as our model plant to examine the expression and function of two CYC1 genes and further analyze the function of their promoters. We found that the CYC binding site located in the promoter of either CYC1C or CYC1D was functionally bound by both CYC1C and CYC1D proteins and was required for activating a β-glucuronidase (GUS) reporter gene in transient gene expression assays. The results indicate that CYC1C and CYC1D have evolved a double positive autoregulatory feedback loop to maintain their expression in the corresponding floral domains. These findings, along with the putative CYC binding sites found in different zygomorphic lineages and comparative analyses of CYC2 gene expression and function in the core eudicots, provide an interpretation for CYC2 clade genes’ wide involvement in the control of floral zygomorphy and shed light on the evolutionary mechanism underlying the independent origins and subsequent rapid diversification of the major zygomorphic lineages in angiosperms.
RESULTS
Floral Development in P. heterotricha
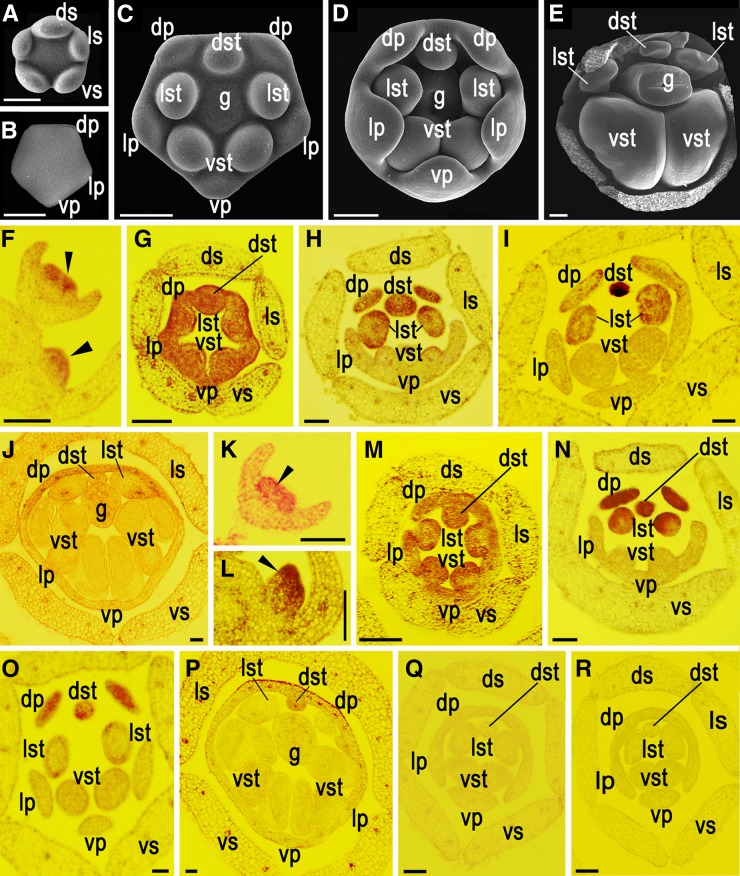
To provide a morphological basis for interpretation of gene expression patterns, we first investigated the development of P. heterotricha flowers using scanning electron microscopy. The five sepals emerge much earlier than other floral organs (Figure 1A). Inside the sepal whorl, the five petals initiate simultaneously, immediately followed by five stamens, without clear differences along the dorsoventral axis (Figures 1B and 1C). In early floral development, the petal and stamen primordia are nearly equal in size, with two dorsal petal and one dorsal stamen primordia slightly smaller relative to the lateral and ventral ones (Figure 1C). Gradually, both the dorsal and lateral stamens are suppressed in development at different levels, while the ventral ones continue to grow and further differentiate into filaments and anthers, and the dorsal petals are retarded in growth relative to the lateral and ventral ones (Figures 1D and 1E). At last, the mature flowers of P. heterotricha are formed, with two small dorsal petals and aborted dorsal and lateral stamens. Also, one dorsal staminode is barely visible and two lateral ones have only short filaments plus sterile anthers (see Supplemental Figure 1 online).
Figure 1.
Flower Development and Tissue-Specific Expression of CYC1C and CYC1D in Developing P. heterotricha Flowers.
(A) to (E) Morphological development of P. heterotricha flowers as revealed by scanning electron microscopy. Five sepal primordia initiate much earlier than other floral organs (A). Five petal primordia are becoming visible inside the sepal whorl (removed) (B) immediately followed by five stamen primordia (data not shown) without clear sequence along the dorsoventral axis. During early stage, the five petal and stamen primordia are of roughly equal size in their respective whorls, with the dorsal petal and stamen primordia slightly smaller relative to the lateral and ventral ones (C). The differences in size between the dorsal and lateral/ventral floral organs (including petals and stamens) become manifested as they grow and enlarge (D). As floral organ development advances further, one dorsal and two lateral stamens are suppressed in development, while the ventral ones continue growth and further differentiate into filaments and anthers (E).
(F) to (J) RNA in situ hybridization with the antisense CYC1C probe. Its mRNA is first detected across the apex of young floral meristems (arrows) (F). Its transcripts are evenly distributed in all five petals and stamens as petal and stamen primordia emerge (G). As petals and stamens expand in size, its transcripts are mainly distributed in two dorsal petals and one dorsal and two lateral stamens, while becoming undetectable in the lateral and ventral petals and two ventral stamens (H). Its transcripts continue to accumulate in the two lateral staminodes in addition to the dorsal petals and staminode as ventral anthers begin to widen (I). Weak expression signal is detected in the dorsal petals and staminode and lateral staminodes as pollen sacs of ventral stamens develop (J).
(K) to (P) RNA in situ hybridization with the CYC1D antisense probe. CYC1D shows a similar expression pattern to CYC1C ([F] to [H]) before floral organ development advance further ([K] to [N]). CYC1D is strongly expressed in the dorsal petals and staminode with rapid reduction in lateral stamens as ventral anthers begin to swell (O). Weak expression signal is only detected in the dorsal petals and staminode as ventral pollen sacs develop (P).
(Q) and (R) As negative controls, the CYC1C (Q) and CYC1D (R) sense probes detect no signal of expression in floral tissues.
ds/ls/vs, dorsal/lateral/ventral sepals; dp/lp/vp, dorsal/lateral/ventral petals; dst/lst/vst, dorsal/lateral/ventral stamens; g, gynoecium. Bars = 100 μm.
Expression Analyses of CYC1C and CYC1D in Developing P. heterotricha Flowers
Our previous RT-PCR results show that CYC2A and CYC2B are only transiently expressed in the very young inflorescences, but CYC1C and CYC1D are transcribed in both young inflorescences and developing flowers with specific expression in the dorsal/lateral floral organs in P. heterotricha (Gao et al., 2008). To explore how floral zygomorphy establishment depends upon specific CYC1 expression in P. heterotricha, here, we further examined their expression patterns in developing flowers by RNA in situ hybridization. Both CYC1C and CYC1D transcripts were first detected across the apex of young floral meristems (Figures 1F, 1K, and 1L). As petal and stamen primordia emerged, the transcripts were evenly distributed in all five petals and stamens, consistent with the equal sizes of floral organs in each whorl (Figures 1G and 1M). However, when dorsal/lateral and ventral organs (including petals and stamens) differentiated in size, strong CYC1C and CYC1D expression was only detected in two dorsal petals and one dorsal and two lateral staminodes (Figures 1H and 1N). Meanwhile, their expression signals soon weakened and became undetectable in the lateral and ventral petals and the ventral stamens (Figures 1H and 1N). As floral organ development advanced further, the CYC1C and CYC1D expression patterns differentiated slightly. When the ventral stamen anthers began to swell, strong CYC1C expression was detected in both the dorsal petals and staminode and the lateral staminodes, while CYC1D transcripts mainly accumulated in the dorsal petals and staminode with weak expression in the lateral staminodes (Figures 1I and 1O). Both CYC1C and CYC1D expression signals weakened and gradually became undetectable as pollen sacs of two ventral stamens developed (Figures 1J and 1P).
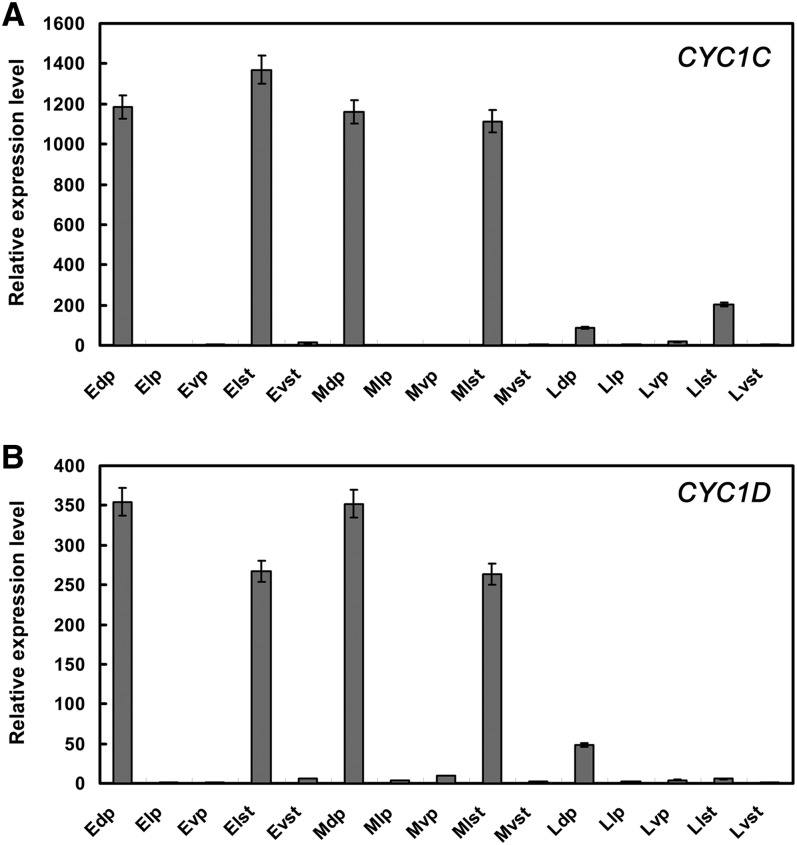
Real-time quantitative PCR (qPCR) was further conducted to check the expression of CYC1C and CYC1D in dissected floral organs (including the dorsal/lateral/ventral petal lobes and the lateral/ventral stamens) divided into early (<1.5 cm long), middle (1.5 to 3 cm long), and late (3 to 4 cm long, at anthesis) stages, respectively. In early and middle stages, both CYC1C and CYC1D were strongly expressed in the dorsal petal lobes, as well as the lateral staminodes in a similar way (Figures 2A and 2B). Their expression differentiation was manifested at late stages, in which CYC1C transcripts were mainly detected in the lateral staminodes, while CYC1D was mainly expressed in the dorsal petals (Figures 2A and 2B). RNA in situ hybridization and real-time qPCR showed that both CYC1C and CYC1D exhibit persistently asymmetric expression patterns with expression expansion from the dorsal to lateral regions, especially the strong transcription of CYC1C in the lateral staminodes at late stage, correlated with the abortion of both dorsal and lateral stamens in P. heterotricha flowers.
Figure 2.
Expression of CYC1C and CYC1D in the Dissected Floral Organs of P. heterotricha Flowers by Real-Time qPCR.
Real-time qPCR expression analysis of CYC1C and CYC1D genes in dissected floral organs. The expression level of each gene in the sample with the lowest expression level was set to 1 after all samples were normalized to the ACTIN reference gene. The values shown (mean ± sd) are the average of three replicates. E/M/L, early/middle/late floral developmental stages; dp/lp/vp, dorsal/lateral/ventral petals; lst/vst, lateral/ventral stamens.
Analysis of the Peloric Flowers of P. heterotricha
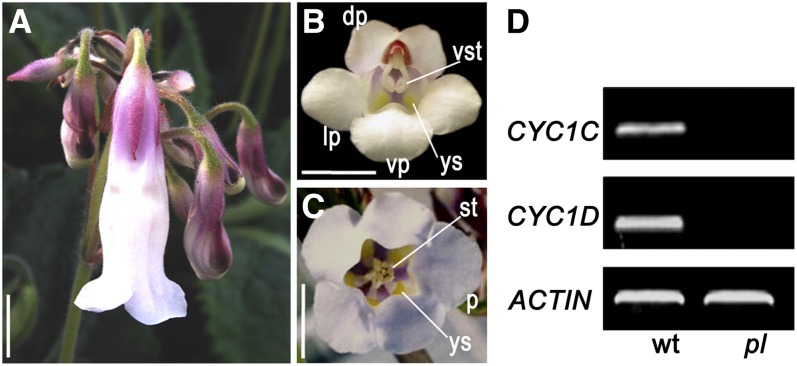
During cultivation of a large number of P. heterotricha plants in greenhouse, we identified a few plants with peloric flowers that consistently occur at the apex of the cymose inflorescences at a very low frequency (Figure 3A). In the peloric flowers, five petals of the same size and five fertile stamens equal in length correspond to the ventral petals and stamens of wild-type flowers, which are characterized by yellow spots at the joints between filaments and petals (Figures 3B and 3C). Apparently, all petals and stamens in the peloric flowers have adopted ventral identities, thus forming fully ventralized flowers. Consistent with this, neither CYC1C nor CYC1D transcription was detected in these flowers (Figure 3D), indicating that the silencing of both CYC1C and CYC1D genes gives rise to these peloric flowers, reminiscent of the snapdragon cyc dich double mutant, which has fully ventralized flowers (Luo et al., 1996). Even though it is unknown how CYC1 genes lose activity in the peloric flowers, the point is clearly demonstrated here that CYC1 genes are functionally essential to generate morphological zygomorphy in the flowers of P. heterotricha as CYC and DICH in snapdragon flowers (Luo et al., 1996, 1999).
Figure 3.
Analysis of Naturally Occurring Peloric Flowers of P. heterotricha.
(A) A ventralized peloric flower occurs at the apex of a cymose inflorescence.
(B) A mature wild-type flower.
(C) A ventralized peloric flower.
(D) RT-PCR results show strong expression of CYC1C and CYC1D in the wild-type (wt) flowers but no expression signal in the peloric (pl) ones.
dp/lp/vp, dorsal/lateral/ventral petals; p, petal; st, stamen; vst, ventral stamen; ys, yellow spots. Bars = 1 cm.
Function Analysis of CYC1C in Transgenic Arabidopsis Plants
To further investigate the function of CYC1 genes, we constructed the 35S:CYC1C plasmid, using Primulina CYC1C, and studied the effect of CYC1C constitutive expression in Arabidopsis thaliana plants. We obtained 22 plants with visible phenotype (i.e., smaller flowers compared with the wild-type ones) out of 48 independent T1 transgenic lines.
Several representative T1 plants with obvious phenotypes were selfed to obtain T2 populations to confirm the heritability of phenotypes observed in T1 plants. In T2 plants, the typical phenotypes included dwarf plants, smaller leaves and flowers, and delayed flowering time compared with wild-type plants (Figure 4A; see Supplemental Figures 2 and 3 online). In addition, overexpression of CYC1C obviously enhanced the outgrowth of lateral branches (see Supplemental Figure 4 online), similar to the transgenic Arabidopsis plants overexpressing Iberis amara TCP1 (Busch and Zachgo, 2007). Furthermore, T2 plants with a strong phenotype showed abnormal growth during both vegetative and reproductive developmental stages. In early stages, T2 transgenic seedlings had smaller cotyledons and poorly developed roots compared with the wild-type ones (see Supplemental Figure 2A online). During late stage, CYC1C overexpression continued to repress the vegetative growth of Arabidopsis with reduced leaf sizes (see Supplemental Figures 2B to 2D online). During reproductive development, CYC1C overexpression usually produced dwarfish plants with smaller flowers (Figures 4A and 4B). Furthermore, all floral organs, including petals, stamens, and carpels, were respectively smaller than those of the wild type (see Supplemental Figures 5A to 5C online). Therefore, different from the snapdragon CYC gene, which promotes Arabidopsis petal growth, both the CYC1C gene from P. heterotricha and the TCP1 gene from I. amara retard Arabidopsis petal growth (Costa et al., 2005; Busch and Zachgo, 2007). Subsequent RT-PCR results showed that strong CYC1C expression signal was detected in both the leaves and flowers of transgenic T2 plants with no transcript in those of the wild-type ones (Figure 4C; see Supplemental Figure 2E online).
Figure 4.
Constitutive Expression of P. heterotricha CYC1C Affects Arabidopsis Reproductive Development.
(A) The wild-type (left) and representative T2 plants overexpressing CYC1C (right).
(B) T2 transgenic plants often produce smaller flowers (right) compared with the wild-type ones (left).
(C) RT-PCR results show strong expression of CYC1C in the flowers of T2 transgenic (tg) plants with no transcription signal in the wild-type (wt) ones.
Bars = 5 cm in (A) and 2 mm in (B).
To determine whether the reduction of petal area was achieved via an effect on cell proliferation or expansion, we examined the adaxial epidermal layer of mature petals from both the transgenic and wild-type plants by scanning electron microscopy. The results showed that the petal cell sizes of transgenic T2 plants were obviously smaller than those of the wild-type ones (see Supplemental Figures 6A and 6B online), indicating that CYC1C reduced the petal area of Arabidopsis plants probably mainly by affecting cell growth, a similar mechanism to the snapdragon CYC gene even though the latter has an opposite effect on Arabidopsis petal cell expansion (Costa et al., 2005).
In Vitro Protein–DNA Interaction Analyses
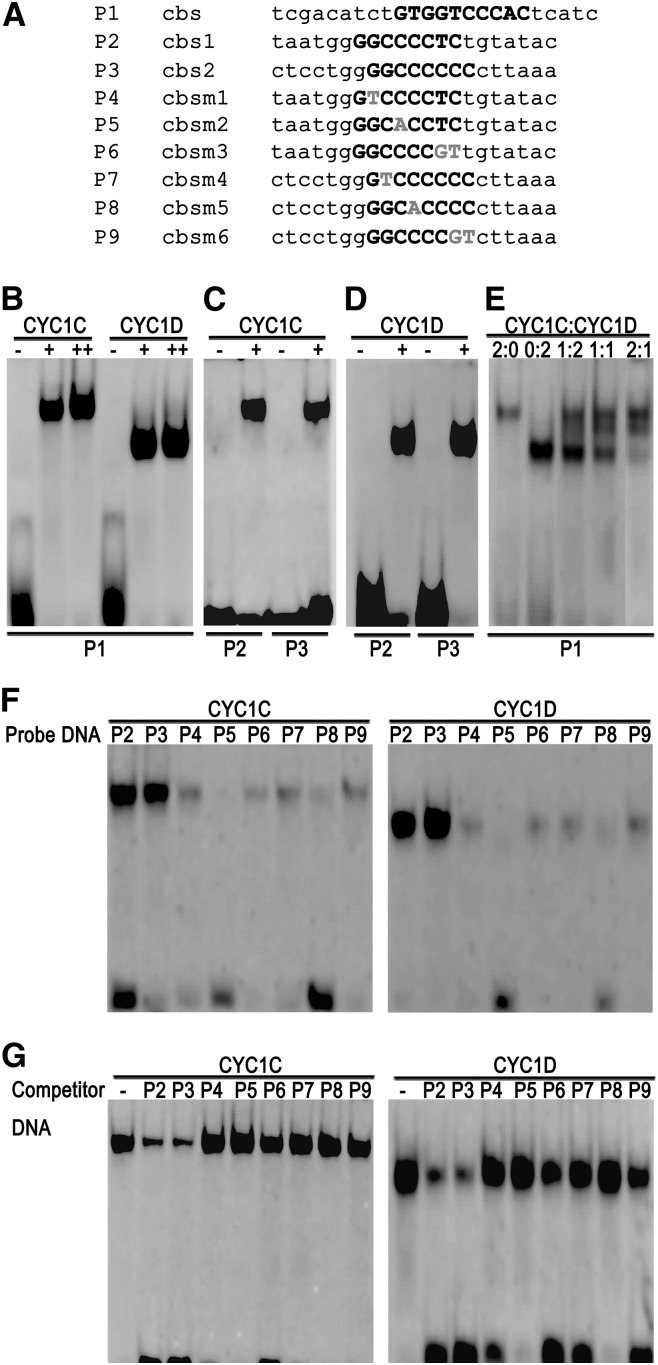
The promoters of both CYC1C and CYC1D were recently shown to contain sequences matching the consensus CYC binding site (GGNCCCNC) (Yang et al., 2010; see Supplemental Figures 7A and 7B online). To elucidate whether these sequences are functional or not, we performed electrophoresis mobility shift assays (EMSAs) to analyze the protein–DNA interaction in vitro.
At first, the purified recombinant CYC1C and CYC1D proteins were incubated with the 24-bp double-stranded oligonucleotide probe cbs, which contained the overlapping consensus DNA binding sites (Figure 5A) for both class I and class II TCP factors (Kosugi and Ohashi, 2002; Costa et al., 2005). Our results showed that the addition of either CYC1C or CYC1D resulted in the formation of a specific retarded band in a concentration-dependent manner (Figure 5B), indicating that CYC1 proteins have a conserved DNA binding activity like the rice (Oryza sativa) PCF and snapdragon CYC proteins (Kosugi and Ohashi, 2002; Costa et al., 2005). Subsequently, a 21-bp double-stranded oligonucleotide probe cbs1, containing one copy of the CYC binding site from the CYC1C promoter (Figure 5A), was incubated with CYC1C protein. As shown in Figure 5C (left), the probe cbs1 could be retarded by the recombinant CYC1C, indicating that CYC1C could bind to its own promoter in vitro. In addition, CYC1C could interact with the CYC1D promoter as it retarded the electrophoretic mobility of the probe cbs2, the CYC binding site from the CYC1D promoter (Figure 5C, right). Similar to CYC1C, CYC1D could also bind to the promoters of both its own and CYC1C genes (Figure 5D). CYC1C and CYC1D have similar DNA binding activities probably owing to the identical amino acid sequences in their TCP domains (see Supplemental Figure 8 online), which are essential and sufficient for interacting with target DNA (Aggarwal et al., 2010).
Figure 5.
In Vitro Protein–DNA Interaction by EMSA Assays.
(A) Oligonucleotide probe sequences used for EMSA assays. On the basis of the sequences of cbs1 and cbs2, one or two mutated nucleotides were introduced. Bold capital letters represent the consensus sequences, and gray letters indicate mutated nucleotides.
(B) to (D) Retardation of the probes cbs (B), cbs1 (C), and cbs2 (D) by recombinant CYC1C and CYC1D.
(E) CYC1C and CYC1D bind to the probe cbs by forming both homo- and heterodimers.
(F) Mutant probes cbsm1–6 have no or very low binding activity to CYC1C (left) and CYC1D (right) proteins.
(G) The unlabeled oligonucleotides cbs1 and cbs2, rather than cbsm1–6, strongly affect the interaction between the cbs1 probe and the CYC1C (left) or CYC1D (right) protein. −, +, and ++ represent no, onefold, and twofold amounts of recombinant proteins added, respectively.
To determine whether CYC1C and CYC1D bind to target sequences by forming a heterodimer, we added CYC1D protein into the reaction mixture after CYC1C had incubated with the cbs probe at room temperature for 15 min. The mixture was incubated for an additional 30 min. Figure 5E shows that CYC1C and CYC1D could bind to target sequences by forming both homo- and heterodimers, reminiscent of the rice PCF proteins (Kosugi and Ohashi, 2002).
We further confirmed the specificity of the binding reaction by introducing several independently mutated nucleotides into the probes cbs1 and cbs2, respectively (Figure 5A). The results showed that the cbs1 and cbs2 probes could be retarded by both CYC1C and CYC1D, but the mutated probes (cbsm1–6) had no or very low activity of interacting with the CYC1 proteins (Figure 5F). In subsequent competition assays, we added 125-fold excess of unlabeled oligonucleotides cbs1, cbs2, and cbsm1–6 into the binding reaction mixture in addition to the labeled probes cbs1 and cbs2. Our results showed that the addition of the unlabeled oligonucleotides cbs1 and cbs2, rather than cbsm1–6, strongly affected the formation of the specific retarded bands (Figure 5G), further confirming the specificity of the interaction between CYC1 proteins and their own promoters.
In Vivo Protein–DNA Interaction Analyses
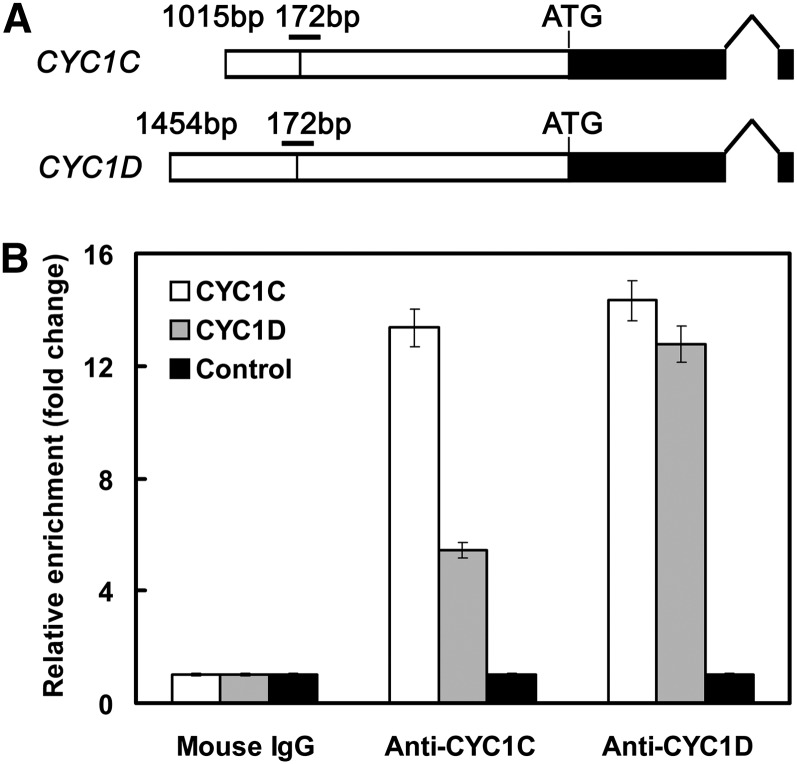
Chromatin immunoprecipitation (ChIP) assays were further conducted to analyze the protein–DNA interaction in vivo. Specific peptides were used to prepare anti-CYC1C and anti-CYC1D antibodies in rabbits. The antibodies were used to precipitate the cross-linked chromatin DNA isolated from P. heterotricha young flower buds. Real-time qPCR was conducted to determine whether CYC1C and CYC1D are direct targets of their own or each other’s products using gene-specific primers. As a negative control, a fragment of CYC1C located at ∼1.5 kb downstream of the CYC binding site was amplified with gene-specific primers. Our results showed that both CYC1C and CYC1D promoters were specifically enriched by anti-CYC1C and anti-CYC1D antibodies compared with the negative control (Figure 6), indicating that either CYC1C or CYC1D could bind to both their own and each other’s promoters in vivo. Therefore, as outlined above, EMSA and ChIP assays show that both CYC1C and CYC1D genes are direct targets of their own and each other’s products, and they might auto- and cross-regulate to maintain their expression throughout flower development by forming both homo- and heterodimers.
Figure 6.
Enrichment of CYC1C and CYC1D Promoters by Anti-CYC1C and Anti-CYC1D Antibodies in ChIP Assays.
(A) The structure of P. heterotricha CYC1 genes. The white boxes represent sequences upstream of the start codon, with vertical lines indicating the position of the CYC binding sites. The horizontal bars above the binding sites indicate the fragments amplified in ChIP-qPCR experiments. The black boxes represent the coding regions.
(B) Enrichment of either the CYC1C or CYC1D promoter by both anti-CYC1C and anti-CYC1D antibodies. The coding sequence of CYC1C located at ∼1.5 kb downstream of the CYC binding site is amplified as a negative control. The data (mean ± sd) are determined from at least three fully independent experiments.
Function Analysis of the CYC Binding Site Using a GUS Reporter Gene
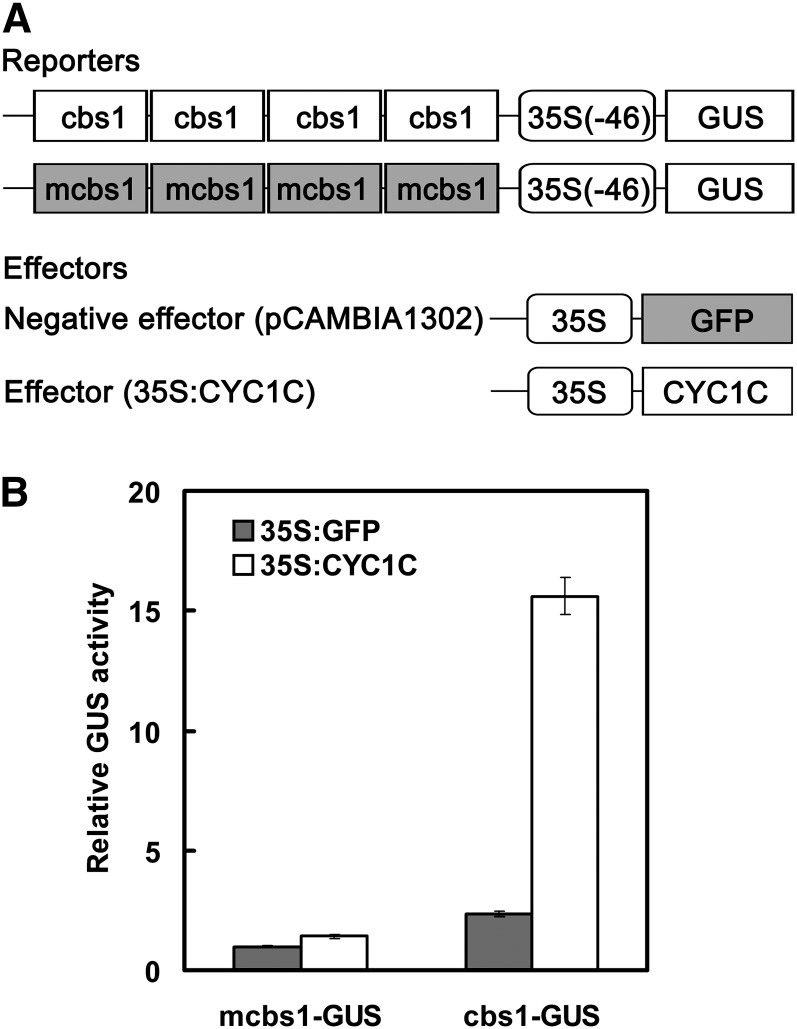
Transient gene expression assays were performed to examine the activity of the CYC binding site in activating a GUS reporter gene. The 35S(-46)-cbs1:GUS and 35S(-46)-mcbs1:GUS reporter plasmids were constructed by inserting a tetramer of the CYC binding site from the CYC1C promoter, either wild-type (cbs1) or mutated (mcbs1), into the upstream of the 35S minimal promoter, and introduced into Arabidopsis mesophyll protoplasts with the cotransfection of the 35S:CYC1C effector plasmid or the pCAMBIA1302 control vector. The results showed that while the transfection of the 35S(-46)-cbs1:GUS plasmid resulted in about a 10-fold increase in the GUS activity relative to the mutated construct in the presence of the effector plasmid, no obvious difference was observed between the wild-type and mutated constructs when the control vector was cotransfected (Figure 7). Apparently, the cbs1 sequence has a role in activating the GUS reporter by interacting with CYC1C.
Figure 7.
Activity of the CYC Binding Site in Activating a GUS Reporter Gene in Transient Gene Expression Assays.
(A) The GUS reporter gene plasmids are constructed by inserting tetramer of the CYC binding sites in the CYC1C promoter, either wild-type (cbs1) or mutated (mcbs1), into the upstream of 35S minimal promoter. The pCAMBIA1302 and 35S:CYC1C plasmids are used as a negative control and an effector plasmid, respectively.
(B) The sequence cbs1 activates the GUS reporter gene in the presence of CYC1C. The GUS activity is examined with 4-methylumbelliferyl-β-d-glucuronide. The data shown (mean ± sd) are the average of three experiments.
Identification of Putative CYC Binding Sites in Upstream Sequences of CYC2 Clade Genes from Zygomorphic Lineages in the Core Eudicots
Since the independent origins of floral zygomorphy have been widely considered to involve the repeated recruitment of CYC2 clade genes in several major zygomorphic lineages of the core eudicots, we next examined the upstream sequences of these genes from other zygomorphic lineages to see if they also contain the potential CYC binding site. We identified four CYC-like genes, namely, Glyma08g28690, Glyma13g07480, Glyma19g05910, and Glyma10g39140, from the soybean (Glycine max; Fabaceae, Rosids) genome and one CYC-like gene each from the genomes of Medicago truncatula (Fabaceae, Rosids) and Mimulus guttatus (Phrymaceae, Asterids). In addition, we identified one CYC-like gene each from the genomes of some ancestral actinomorphic lineages, including a TCP1-like gene from Brassica rapa (Brassicaceae), a CYC-like gene from grape (Vitis vinifera; Vitaceae), and a TCP7 gene from tomato (Solanum lycopersicum; Solanaceae), in addition to the TCP1 gene from the model plant Arabidopsis (Cubas et al., 2001). Phylogenetic analyses based on the protein sequences exclusive of the regions between TCP and R domains (see Supplemental Data Set 1 online) showed that all the above-mentioned genes, together with snapdragon CYC/DICH (Luo et al., 1996, 1999), Lotus japonicus CYC1/2/3 genes (Feng et al., 2006), and the P. heterotricha CYC1C/1D genes (this study), were grouped into the CYC2 clade with a high bootstrap value (82%; see Supplemental Figure 9 online).
Subsequently, we analyzed 3-kb upstream sequences of the start codons of these genes. The results showed that CYC-like genes from those zygomorphic species, including soybean, M. truncatula, and M. guttatus, contained at least one putative CYC binding site (Table 1). On the contrary, no such site was found in the corresponding regions of CYC-like genes from those ancestral actinomorphic species. The above results indicate that the presence of CYC binding site in CYC2 clade genes is positively correlated with the formation of zygomorphic flowers.
Table 1. Putative CYC Binding Sites Found in Upstream Sequences of CYC2 Clade Genes.
| Speciesa | Gene | CYC Binding Siteb | Reference or Source | Family (Subclass) |
|---|---|---|---|---|
| Arabidopsis | TCP1 | No | Cubas et al. (2001); NCBI | Brassicaceae (Rosids) |
| B. rapa | TCP1-like gene | No | NCBI | Brassicaceae (Rosids) |
| Grape | LOC100256607 | No | NCBI | Vitaceae (Rosids) |
| Tomato | TCP7 | No | NCBI | Solanaceae (Asterids) |
| Soybean | Glyma08g28690 | cttGACCCACtat (−1603) | PlantGDB | Fabaceae (Rosids) |
| tttGACCCTCtct (−468) | ||||
| Glyma13g07480 | ggtGACCCACaaa (−1198) | |||
| Glyma19g05910 | ggtGACCCACaaa (−1252) | |||
| Glyma10g39140 | tttGGCCCCAaag (−2284) | |||
| M. truncatula | CYC1A | tcaGGGCCCTgcc (−1901) | NCBI | Fabaceae (Rosids) |
| M. guttatus | CYC-like gene | cccGGCCCTCttc (−1894) | NCBI | Phrymaceae (Asterids) |
| aatGGGGCCCgtt (−1673) | ||||
| ctcGGCCCCCgcc (−401) | ||||
| P. heterotricha | CYC1C | tggGGCCCCTCtgt (−942) tggGGCCCCCCctt (−960) | Gao et al. (2008); this study | Gesneriaceae (Asterids) |
| CYC1D |
The top half is actinomorphic species, and the bottom half is zygomorphic species.
The sequences with capital letters represent the consensus CYC binding sites. The 3-kb upstream region of respective gene is analyzed in this study. The number in the parentheses represents the position of each site relative to the start codon of respective gene.
NCBI, National Center for Biotechnology Information.
DISCUSSION
CYC1C and CYC1D Are Crucial in Controlling Floral Zygomorphy in P. heterotricha
In the zygomorphic flowers of P. heterotricha, the characteristic size reduction of the two dorsal petals and the arrest of one dorsal and two lateral stamens is accompanied by the expression of CYC1C and CYC1D in the dorsal petals and both the dorsal and lateral stamens. Both P. heterotricha CYC1 and snapdragon CYC belong to CYC2 clade of TCP genes, which are widely conserved in floral zygomorphy patterning in the core eudicots (Luo et al., 1996, 1999; Feng et al., 2006; Busch and Zachgo, 2007, 2009; Broholm et al., 2008; Gao et al., 2008; Kim et al., 2008; Wang et al., 2008, 2010; Song et al., 2009; Zhang et al., 2010; Howarth et al., 2011). In snapdragon, floral zygomorphy is established mainly through the CYC/DICH dorsal identity function in controlling the fate of dorsal organs, namely, promoting dorsal petal growth and arresting dorsal stamen development (Luo et al., 1996, 1999). Similar to CYC, L. japonicus CYC1/2 and pea (Pisum sativum) CYC1/2 in the Fabaceae specifically promote the growth of the single dorsal petal (Feng et al., 2006; Wang et al., 2008, 2010). However, in I. amara, a close relative of Arabidopsis, TCP1 dorsal-specific expression inhibits the growth of two dorsal petals (Busch and Zachgo, 2007). The downregulation or loss of CYC2 function usually gives rise to ventralized peloric flowers (Luo et al., 1996, 1999; Feng et al., 2006; Busch and Zachgo, 2007). This is also true in P. heterotricha, as CYC1C and CYC1D are silenced in the ventralized peloric flowers. Further considering that overexpression of P. heterotricha CYC1C inhibits petal cell expansion in transgenic Arabidopsis plants, we suggest that floral zygomorphy in P. heterotricha might be established through CYC1 function in repressing the development of dorsal and lateral stamens and retarding the growth of two dorsal petals, probably mainly by inhibiting cell growth. The sterilization of two lateral stamens comes from CYC1 evolving a new domain, probably through a regulatory modification, to carry out the same role as in the dorsal floral region just like the CYC1C gene in Opithandra dinghushanensis (Gesneriaceae) and the CYC1/2 genes in Mohavea confertiflora, a close relative of snapdragon (Hileman et al., 2003; Hileman and Cubas, 2009; Song et al., 2009).
A Double Positive Autoregulatory Feedback Loop Accounting for Expression Maintenance of CYC1C and CYC1D
It was hypothesized that the expression maintenance of the snapdragon CYC gene might have arisen through evolution of an autoregulatory loop and its promoter could contain sequences matching the consensus CYC binding sites, but the TCP1 gene, with very early and transient expression in Arabidopsis actinomorphic flowers, does not contain such a site in its promoter (Costa et al., 2005). However, it is still open to question whether CYC can regulate itself because no direct evidence to date shows that its promoter contains such a site (Martín-Trillo and Cubas, 2010). We previously reported that the promoters of P. heterotricha CYC1C and CYC1D contain putative CYC binding sites (Yang et al., 2010). The results herein reveal that these sites are all functionally bound by both their own and each other’s proteins in both EMSA and ChIP assays and are required for activating a GUS reporter gene by interacting with the CYC1 proteins in transient gene expression assays. Also, mutated sites lose the binding activity and fail to activate transcription. Apparently, CYC1 genes have achieved persistently asymmetric expression in the corresponding floral domains, at least in part, by acquiring an autoregulation loop (Figure 8). By contrast, no CYC binding site is found in CYC2 clade genes from the ancestral actinomorphic lineages both in Rosids and Asterids, such as TCP1-like gene from B. rapa and TCP1 from Arabidopsis in Brassicaceae, CYC-like gene from grape in Vitaceae, and especially TCP7 from tomato, an actinomorphic species in Solanaceae that has a sister relationship with the Lamiales, which include P. heterotricha (Table 1). Therefore, our results identify an autoregulatory loop required for the maintenance of expression of CYC2 clade genes in angiosperms.
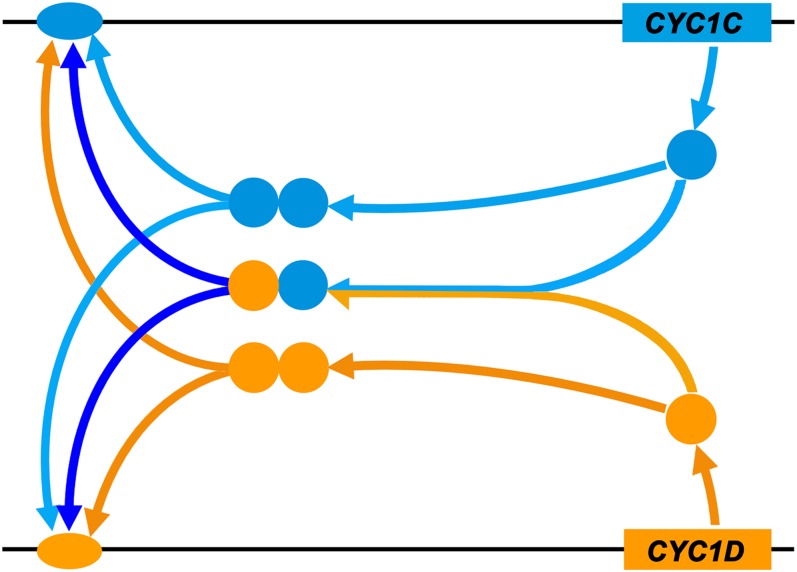
Figure 8.
A Model Showing the Double Positive Autoregulatory Feedback Loop Evolved in the P. heterotricha CYC1 Genes.
After expression initiation, CYC1C (blue box) and CYC1D (orange box) genes maintain expression throughout floral development via a double positive autoregulatory feedback loop by forming both homo- and heterodimers. The blue and orange circles represent the CYC1C and CYC1D proteins, and the blue and orange ovals represent the CYC binding sites found in the CYC1C and CYC1D promoters, respectively.
Autoregulation, a transcriptional strategy in which a given transcription factor binds to its own promoter to either activate or repress its own transcription, is employed by a wide variety of eukaryotes to refine and maintain transcription levels after induction of gene expression by transient extracellular signals (Bateman, 1998; Crews and Pearson, 2009). The occurrence of an autoregulatory loop may be positively correlated with the developmental or physiological importance of a transcription factor, and master developmental regulators that control large number of genes are usually autoregulated as their expression levels must be tightly controlled (Crews and Pearson, 2009). A positive autoregulatory loop is a general mechanism to amplify and maintain gene expression that arises from transient inputs to create morphological and physiological effects during development (Crews and Pearson, 2009). The autoregulation of P. heterotricha CYC1 should be a positive loop, a common feature of developmental transcription factors in animals and plants (Crews and Pearson, 2009).
Furthermore, developmental regulatory networks in animals and plants often use positive autoregulatory feedback loops of two transcription factors that regulate both themselves and each other (i.e., the double positive autoregulatory feedback loops) to promote bistability and thereby trigger threshold-dependent genetic switches (Alon, 2007; Kaufmann et al., 2010). Classical examples include Myocyte enhancing factor2 and Twist, which function in Drosophila melanogaster embryonic muscle development (Crews and Pearson, 2009), and DEFICIENS and GLOBOSA, which specify petal and stamen identities in snapdragon (Schwarz-Sommer et al., 1992). In our study, P. heterotricha CYC1C protein binds to both its own promoter and the CYC1D promoter, and vice versa, indicating that a double positive autoregulatory feedback loop has also been employed by P. heterotricha CYC1 genes to maintain their asymmetric expression in P. heterotricha flowers (Figure 8). Considering their slight expression differentiation during late stages of flower development, in which CYC1C is still strongly expressed in both the dorsal and lateral regions while CYC1D is mainly transcribed in the dorsal organs, we suggest that the abortion of two lateral stamens at late stages is probably mainly determined by CYC1C, which functions in repressing cell expansion, relying on a single positive autoregulation loop by forming homodimers. A dosage effect of CYC2 gene function is also demonstrated in the difference between the dorsal and lateral stamens in both morphology and CYC1 expression in P. heterotricha, which has also been shown in previous studies (Song et al., 2009; Busch et al., 2012).
Evolutionary Significance of the Autoregulatory Loops in CYC2 Clade Genes Relating to the Origin of Floral Zygomorphy in Angiosperms
Parallel to the sudden emergence of zygomorphic flowers ∼60 million years after the origin of angiosperms, advanced anthophilous insects dramatically diversified after the mid-Cretaceous, suggesting coevolution between angiosperms and insects (Grimaldi, 1999; Crepet, 2000; Dilcher, 2000). It is important to know why such close animal–plant relationships had not been established during the long period of angiosperm origin (across 100 million years, as suggested in Stuessy [2004]) and 60 million years of slow evolutionary process of early angiosperms (Crepet, 2000; Dilcher, 2000). Additionally, what developmental and genetic bases were driving the sudden origin of floral zygomorphy that triggered the establishment of animal–plant coevolution?
Comparative genome analyses have revealed at least two whole-genome duplication (WGD) events during the early evolution of angiosperms: The first occurred before the monocot–eudicot split and the second predated the divergence of major core eudicot lineages (De Bodt et al., 2005; Soltis et al., 2007; Van de Peer et al., 2009a, 2009b; Proost et al., 2011). Following these WGD events, the genomes of early angiosperms became greatly enlarged with a dramatically increased number of genes. These genes, especially those associated with transcription regulation and development, further underwent functional redundancy, subfunctionalization, and neofunctionalization, accompanied by novel regulatory changes (De Bodt et al., 2005; Soltis et al., 2007; Van de Peer et al., 2009a, 2009b). For example, linked with the two WGD events, the MADS box floral organ identity genes seem to have undergone duplication bursts respectively close to the origin of angiosperms and in the ancestor of the core eudicots (De Bodt et al., 2005; Soltis et al., 2007). The ECE lineage of the TCP gene family, specific to angiosperms, also underwent two major duplication events just before the radiation of the core eudicots that gave rise to the CYC1, CYC2, and CYC3 clades (Howarth and Donoghue, 2006). The CYC2 clade might have further experienced frequent duplications that gave birth to multiple copies of CYC2 genes in most members of zygomorphic clades within the Rosids and Asterids (Howarth and Donoghue, 2006). Of these, there is usually a pair of CYC2 clade genes involved in floral zygomorphy establishment redundantly, at least partially, with other copies having no or transient expression signals, such as CYC and DICH in snapdragon (Luo et al., 1996, 1999), CYC1 and CYC2 in M. confertiflora (Hileman et al., 2003), CYC1 and CYC2 in L. japonicus (Feng et al., 2006; Wang et al., 2010), CYC1 and CYC2 in pea (Wang et al., 2008), CYC1A and CYC1B in Lupinus nanus (Fabaceae) (Citerne et al., 2006), CYC1C and CYC2A in O. dinghushanensis (Song et al., 2009), CYC2A and CYC2B in Byrsonima crassifolia (Malpighiaceae) (Zhang et al., 2010), and CYC2A and CYC2B in Lonicera (Caprifoliaceae) (Howarth et al., 2011). A central question is whether the double positive autoregulatory feedback loops have also been employed by these genes in different zygomorphic clades. It seems very likely because we find putative CYC binding sites in the upstream sequences of CYC2 clade genes from M. guttatus of Asterids and G. max and M. truncatula of Rosids (Table 1). In addition, we detect putative TCP binding sites in the upstream sequences of rice REP and TB1 genes, as well as the maize (Zea mays) TB1 gene (see Supplemental Table 1 online), indicating that these ECE genes involved in regulating axillary organs in the monocots (Doebley et al., 1997; Yuan et al., 2009) could also be regulated by autoregulatory loops, a possibility that deserves further study.
The independent evolution of morphological similarities is widespread in both animals and plants and usually involves repeated recruitment of the same kind of genes in independent lineages, such as Yellow genes for pigmentation traits in flies (Prud’homme et al., 2006, 2007; Werner et al., 2010), Pitx1 genes for pelvic reduction in sticklebacks (Shapiro et al., 2006), and RPL (shl) genes for fruit shattering in Arabidopsis and rice (Arnaud et al., 2011). Natural selection usually biases these genes, especially certain regulatory regions that behave as genomic hotspots for phenotypic evolution (Papa et al., 2008; Arnaud et al., 2011). Given that the same CYC binding sites are found in the zygomorphic lineages of both Asterids and Rosids, with no such site detected in their respective ancestral actinomorphic lineages, and the establishment of floral zygomorphy frequently involves the dorsal-specific activities of a pair of CYC2 genes, we suggest that the double positive autoregulatory feedback loops revealed in Lamiales might have become established independently in the Asterids and Rosids, in accordance with the independent origins of several major zygomorphic lineages in the core eudicots (Donoghue et al., 1998; Cubas, 2004; Jabbour et al., 2009). A growing amount of evidence shows that many polyploids experience extensive and rapid genomic alterations even within the first few generations, and changes in the architecture of gene regulatory networks and in particular functional changes within cis-regulatory elements are frequently associated with evolutionary innovations (Adams and Wendel, 2005; Moore and Purugganan, 2005; Soltis, 2005; Prud’homme et al., 2006, 2007; Wagner, 2008). Therefore, this neofunctionalization (i.e., the double positive autoregulatory feedback loops underlain by a key regulatory change in CYC2 clade genes) might have been subsequent to the second WGD event in angiosperms that provided novel opportunities for the evolutionary success of the core eudicots. With a possible exception in the family Brassicaceae, it is likely that a single CYC2 gene can function to establish a perfect floral zygomorphy, such as TCP1 in I. amara flowers (Busch and Zachgo, 2007; Busch et al., 2012). It would be interesting to know whether the CYC2 genes in Brassicaceae have employed a single positive autoregulatory loop to maintain their expression with different regulatory and evolutionary mechanisms underlying the diverse forms of floral symmetry, such as dorsal/ventral dosage effect of TCP1-like genes as suggested by Busch et al. (2012).
It is suggested that the establishment and maintenance of Hox gene asymmetric expression, relying on auto- and cross-regulation, might have been associated with the evolutionary switch from the primary oral–aboral axis to the elaborated anterior–posterior axis of Metazoans and subsequent rapid diversification of the bilaterian lineages, thus implicated in the Cambrian explosion ∼520 million years ago (Gellon and McGinnis, 1998; Knoll and Carroll, 1999; Valentine et al., 1999; Pearson et al., 2005). The double positive autoregulatory feedback loop as a novel mechanism maintaining and strengthening CYC2 gene expression in later developmental stages, co-opted with the regulation for their dorsal-specific location evolved previously or subsequently, might have driven the shift of floral symmetry from actinomorphy to zygomorphy. This shift further promoted the establishment of plant–insect coevolution, which, as Dilcher (1979) suggested, once established, underwent an explosive radiation in both angiosperms and insects. The genetic control of floral zygomorphy unraveled in snapdragon is a major breakthrough in advancing our understanding of the formation of zygomorphy. The findings herein suggest that the gain of double positive autoregulatory feedback loops is responsible for CYC2 gene expression maintenance that, as a linkage between molecular evolution and morphological macroevolution, might be associated with the independent origins and subsequent rapid diversification of the major zygomorphic lineages in angiosperms. This novel genetic mechanism makes Darwin’s abominable mystery of angiosperm explosive radiation (Darwin and Seeward, 1903) somewhat less of a mystery, on which further studies of the regulatory region of CYC2 clade genes in the Rosids would shed new light.
Over the past decade, comparative genetic studies have shown that the multiple origins of a novel trait are usually built using old genes wired in new ways, largely through the modification of ancestral cis-regulatory elements (Gompel et al., 2005; Prud’homme et al., 2006, 2007; Wray, 2007; Monteiro and Podlaha, 2009; Werner et al., 2010). Further studies will be required to understand whether the double positive autoregulatory feedback loops of CYC2 clade genes have evolved from the modification of ancestral cis-regulatory elements that preexisted in their actinomorphic ancestors or generation of novel elements. In addition, eukaryotic organisms are usually characterized using complex regulatory networks in the form of a hierarchy, in which a specific autoregulatory loop, together with other regulatory network motifs, can be organized into larger network structures (Lee et al., 2002). The dorsal identity function of CYC2 clade genes in patterning floral zygomorphy necessarily requires the coupling of at least two components (i.e., the expression maintenance by employing an autoregulatory loop and the expression location by evolving certain dorsal-specific cis-regulatory elements). Zygomorphy evolved independently several times from actinomorphic ancestors and is also frequently lost in the major zygomorphic lineages in angiosperms (Donoghue et al., 1998; Citerne et al., 2006; Zhou et al., 2008; Pang et al., 2010; Zhang et al., 2010). Therefore, it would be important to understand whether or how these secondary shifts are associated with a possible uncoupling of the two components, including a secondary change of the autoregulation loop and the regulatory alteration for CYC2 expression location, in the CYC2 gene regulatory pathway. Further studies employing an integrative approach to bridge phylogeny, genetics, development, and ecology would greatly improve our understanding of how the autoregulatory loops of CYC2 clade genes are co-opted into complex regulatory networks in the establishment of floral zygomorphy and how autoregulatory circuits change during adaptive evolution and diversification of floral zygomorphy in angiosperms, especially in the core eudicots.
METHODS
Plant Materials
Primulina heterotricha plants were transplanted from the field and cultivated in the greenhouse of the Botanical Garden, Institute of Botany, Chinese Academy of Sciences, Beijing. Arabidopsis thaliana ecotype Columbia-0 plants used in this study were grown to flowering stage under 16-h-light (150 to 200 µmol m−2 s−1, 23°C) and 8-h-dark (20°C) conditions.
Scanning Electron Microscopy and RNA in Situ Hybridization
P. heterotricha floral buds of different developmental stages were collected and fixed immediately. Their morphology was examined using a Hitachi S-4800 scanning electron microscope as described (Zhou et al., 2008).
Total RNA was extracted from P. heterotricha young flower buds using an SV Total RNA Isolation System (Promega). cDNA was synthesized using a RevertAid H Minus First-Strand cDNA synthesis kit (Fermentas). The CYC1C and CYC1D fragments amplified with PhCYC1C-F1/R1 and PhCYC1D-F1/R1 (see Supplemental Table 2 online) were used as templates to prepare the digoxygenin-labeled probes. Floral tissues were fixed, sectioned, and hybridized to the probes as described (Zhou et al., 2008).
Real-Time qPCR and RT-PCR
For real-time qPCR, wild-type flowers of early (<1.5 cm long), middle (1.5 to 3 cm long), and late (3 to 4 cm long, at anthesis) stages were dissected into dorsal/lateral/ventral petal lobes and lateral/ventral stamens, respectively. PhCYC1C-F2/R2 and PhCYC1D-F2/R2 were respectively used to amplify CYC1C and CYC1D, and ACTIN was amplified as a reference gene using PhACTIN-F/R (see Supplemental Table 2 online). The SYBR Premix Ex Taq (TaKaRa) was used to perform real-time qPCR with ROX as a reference dye using a Stratagene Mx3000P real-time system (Agilent Technologies). The relative expression levels were determined by normalizing the PCR threshold cycle number of each gene with that of ACTIN gene. The expression level of each gene in the sample with the lowest expression level was set to 1, and the data were the average of three replicates.
Naturally occurring peloric flowers were collected whole for RT-PCR. The RT-PCR conditions were as follows: initial denaturation at 94°C for 3 min; 25 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s; final extension at 72°C for 10 min. For normalization, ACTIN was amplified with 22 cycles. The amplified products were separated on a 1.5% agarose gel and visualized using a bioimaging system (Gene Tools Program; Syngene).
Transgenic Analysis
Full-length P. heterotricha CYC1C was amplified with PhCYC1C-F4/R4 (see Supplemental Table 2 online) and inserted into the NcoI-SpeI sites of the binary pCAMBIA1302 vector. The resulting construct was introduced into Arabidopsis plants by Agrobacterium tumefaciens LBA4404-mediated floral dipping method (Clough and Bent, 1998). Transgenic seedlings were selected on half-strength of Murashige and Skoog medium containing 25 µg/mL Hygromycin and 250 µg/mL Cefotaxime. T1 plants with obvious phenotype were selfed to obtain T2 populations. RT-PCR was performed to detect CYC1C expression in both flowers and leaves from at least three independent T2 plants. As a reference gene, Arabidopsis EF1α was amplified with 22 cycles using AtEF1α-F/R. For scanning electron microscopy, the adaxial epidermal petal cells of wild-type and transgenic plants were collected and fixed.
Production of Recombinant Proteins and EMSA Assays
The CYC1C and CYC1D fragments amplified with PhCYC1C-F3/R3 and PhCYC1D-F3/R3 (see Supplemental Table 2 online), respectively, were digested and inserted into the pET30a vector (Novagen). The recombinant CYC1C and CYC1D proteins were expressed in BL21 Escherichia coli cells and purified from the soluble fractions using the His SpinTrap kit (GE Healthcare).
The DIG gel shift kit (Roche) was used to perform EMSA. In brief, the digoxygenin-labeled DNA probes were generated using the recombinant terminal transferase (Roche). The binding reaction was performed in a volume of 10 μL containing 0.8 ng of probe, 1× binding buffer [20 mM HEPES-KOH, pH 7.6, 1 mM EDTA, 10 mM (NH4)2 SO4, 1 mM DTT, Tween 20, 0.2% (w/v), and 30 mM KCl], 100 ng of salmon sperm DNA, 50 ng of poly L-Lys, and 20 to 200 ng of purified recombinant proteins. The mixture was incubated at room temperature for 15 min and loaded on the 6% native polyacrylamide gel. To check whether CYC1C and CYC1D could form heterodimers, CYC1D protein was added into the reaction mixture after CYC1C had incubated with the cbs probe at room temperature for 15 min. The mixture was incubated for another 30 min. Electrophoresis was conducted at 12 V/cm for 40 min with 0.25× TBE (22.25 mM Tris, 22.25 mM boric acid, and 0.5 mM EDTA, pH 8.0) buffer at 4°C. The probes were detected according to the manufacturers’ instructions using a Gene Gnome Syngene Bio imaging system (Gene Company).
ChIP Assays
The gene-specific peptides (CYC1C, CKLNESRNMNNLFE; and CYC1D, GSNLNPSVPIQRNC) were used to prepare anti-CYC1C and anti-CYC1D antibodies in rabbits (Genomics). The EpiQuik Plant ChIP kit (Epigentek) was used to perform the ChIP assays. In brief, 0.8 to 1.0 g of young P. heterotricha floral buds was fixed with 20 mL of 1.0% formaldehyde by vacuuming for 10 min. The chromatin DNA was extracted and sheared to 200- to 1000-bp fragments by sonicating. One hundred microliters of the sheared DNA was immunoprecipitated with 3 to 5 µg purified antibodies for 90 min at 50 to 100 rpm at room temperature. In addition, 1 μL of normal mouse IgG (Epigentek) was used as a negative control. DNA fragments that specifically associated with CYC1 were released, purified, and used as templates for real-time qPCR using primers PhCYC1C-F5/R5 and PhCYC1D-F4/R4 (see Supplemental Table 2 online). As a negative control, the coding region of CYC1C, which is ∼1.5 kb from the CYC binding site, was amplified using primers PhCYC1C-F6/R6 (see Supplemental Table 2 online).
Transient Gene Expression Assays
A fragment extending from −46 to +6 of the cauliflower mosaic virus 35S promoter was synthesized and inserted into the HindIII-XbaI sites of the binary pBI121 vector to create a 35S(-46)-GUS construct. Then, a double-stranded synthetic oligonucleotide comprising a tetramer of the CYC binding site in the CYC1C promoter, either wild-type (5′-GCTAATGGGGCCCCTCTGTATAC-3′) or mutated (5′-GCTAATGGGTCAACTCTGTATAC-3′), was inserted into the HindIII site of the 35S(-46)-GUS construct. The obtained GUS reporter constructs were introduced into Arabidopsis mesophyll protoplasts by the polyethylene glycol–mediated transfection method. The pCAMBIA1302 vector and the 35S:CYC1C construct were cotransfected into the protoplasts as a negative control and an effector plasmid, respectively. The protoplasts were cultured at room temperature for 48 h in the dark, and the GUS activity was determined with 4-methylumbelliferyl-β-d-glucuronide.
Comparative Analyses of the Upstream Sequences of Related CYC2 Clade or Other ECE Linage Genes
We downloaded the genomic DNA sequences of related CYC2 clade or other ECE linage genes comprising the full-length coding regions and 3-kb upstream sequences of the start codon from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) or PlantGDB (http://www.plantgdb.org/) database using the BLAST search tools. In brief, Lotus japonicus CYC1/2/3 (Feng et al., 2006) were used as queries to retrieve CYC2-like genes from the genome of soybean (Glycine max) and Medicago truncatula, the At TCP1 gene (Cubas et al., 2001) was used to blast the genomes of Brassica rapa and grape (Vitis vinifera), and the Am CYC gene (Luo et al., 1996) was used to search for CYC-like genes from the genomes of Mimulus guttatus and tomato (Solanum lycopersicum).
Phylogenetic analyses were conducted based on the protein sequences exclusive of the regions between TCP and R domains of Ph CYC1C and Ph CYC1D genes (this study), CYC and DICH genes from snapdragon (Antirrhinum majus; Luo et al., 1996, 1999), At TCP1/2/4/12/16 genes from A. thaliana (Martín-Trillo and Cubas, 2010), and PCF1 and PCF2 from rice (Oryza sativa; Kosugi and Ohashi, 1997), in addition to the above-mentioned CYC2 clade genes. The sequences were first aligned using ClustalX 1.81 (Thompson et al., 1997) and further manually refined. The neighbor-joining method with p-distance was performed using MEGA 4 (Tamura et al., 2007). The bootstrap values were calculated for 1000 replicates.
DNAMAN 5.2.2 software (Lynnon Biosoft) was used to search for the putative CYC or TCP binding sites in the upstream sequences of these genes using the core sequence GGNCCC of the consensus DNA binding sites for both class I and class II TCP factors (Kosugi and Ohashi, 2002) as a query.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Am CYC, Y16313; Am DICH, AF199465; At TCP1, NM_001084312; At TCP2, NM_001084938; At TCP4, NM_001202967; At TCP12, NM_105554; At TCP16, NM_114384; At TCP18, NM_112741; Br TCP1, AC189200; Vvi LOC100256607, XM_002279676; Mg CYC, AC182570; Mt CYC1A, XM_003637000; Sl TCP7, NM_001246868; Lj CYC1, DQ202475; Lj CYC2, DQ202476; Lj CYC3, DQ202477; PCF1, DE7260; PCF2, D87261; Ph CYC1C, JX020500; Ph CYC1D, JX020501; Ph CYC2A, JX020502; and Ph CYC2B, JX020503.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mature Flowers of P. heterotricha.
Supplemental Figure 2. Overexpression of P. heterotricha CYC1C Represses the Vegetative Growth of Arabidopsis T2 Plants.
Supplemental Figure 3. P. heterotricha CYC1C Constitutive Expression Delays the Flowering Time of Arabidopsis T2 Plants.
Supplemental Figure 4. P. heterotricha CYC1C Overexpression Enhances the Outgrowth of Lateral Branches in Arabidopsis T2 Plants.
Supplemental Figure 5. Arabidopsis Plants Overexpressing P. heterotricha CYC1C Produce Smaller Flowers than Wild-Type Arabidopsis.
Supplemental Figure 6. P. heterotricha CYC1C Overexpression Reduces Arabidopsis Petal Areas Mainly by Reducing Petal Cell Sizes.
Supplemental Figure 7. The P. heterotricha CYC1C and CYC1D Promoters Contain Sequences Matching the Consensus CYC binding Sites.
Supplemental Figure 8. Alignment of Protein Sequences of P. heterotricha CYC and Snapdragon CYC.
Supplemental Figure 9. The Neighbor-Joining Tree of CYC2 Clade Genes Using MEGA 4.
Supplemental Table 1. The Putative TCP binding Sites Found in Other ECE Genes in the Monocots.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Alignment of Protein Sequences of CYC2 Clade Genes Used in the Phylogenetic Analysis.
Supplementary Material
Acknowledgments
We thank Tao Sang, James F. Smith, Song Ge, Chao-Ying He, and Sabine Zachgo for their constructive comments and language improvements on this article. This work was funded by the National Natural Science Foundation of China (Grants 30990240 and 31170198).
AUTHOR CONTRIBUTIONS
Y.-Z.W. initiated, designed, and supervised the research and wrote the article. X.Y. designed and performed the research, analyzed the data, and wrote the article. H.-B.P. and B.-L.L. were involved in performing the transgene experiments and transient gene expression assays. Z.-J.Q. was involved in performing the RNA in situ hybridization experiments. Q.G. was involved in analyzing the peloric flowers. L.W. and Y.D were involved in performing the scanning electron microscopy experiments.
Glossary
- GUS
β-glucuronidase
- qPCR
quantitative PCR
- EMSA
electrophoresis mobility shift assay
- ChIP
chromatin immunoprecipitation
- WGD
whole-genome duplication
References
- Adams K.L., Wendel J.F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141 [DOI] [PubMed] [Google Scholar]
- Aggarwal P., Das Gupta M., Joseph A.P., Chatterjee N., Srinivasan N., Nath U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. (2007). Network motifs: Theory and experimental approaches. Nat. Rev. Genet. 8: 450–461 [DOI] [PubMed] [Google Scholar]
- Arnaud N., Lawrenson T., Østergaard L., Sablowski R. (2011). The same regulatory point mutation changed seed-dispersal structures in evolution and domestication. Curr. Biol. 21: 1215–1219 [DOI] [PubMed] [Google Scholar]
- Bateman E. (1998). Autoregulation of eukaryotic transcription factors. Prog. Nucleic Acid Res. Mol. Biol. 60: 133–168 [DOI] [PubMed] [Google Scholar]
- Broholm S.K., Tähtiharju S., Laitinen R.A.E., Albert V.A., Teeri T.H., Elomaa P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. USA 105: 9117–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A., Horn S., Mühlhausen A., Mummenhoff K., Zachgo S. (2012). Corolla monosymmetry: Evolution of a morphological novelty in the Brassicaceae family. Mol. Biol. Evol. 29: 1241–1254 [DOI] [PubMed] [Google Scholar]
- Busch A., Zachgo S. (2007). Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc. Natl. Acad. Sci. USA 104: 16714–16719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A., Zachgo S. (2009). Flower symmetry evolution: Towards understanding the abominable mystery of angiosperm radiation. Bioessays 31: 1181–1190 [DOI] [PubMed] [Google Scholar]
- Citerne H.L., Pennington R.T., Cronk Q.C.B. (2006). An apparent reversal in floral symmetry in the legume Cadia is a homeotic transformation. Proc. Natl. Acad. Sci. USA 103: 12017–12020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Costa M.M.R., Fox S., Hanna A.I., Baxter C., Coen E. (2005). Evolution of regulatory interactions controlling floral asymmetry. Development 132: 5093–5101 [DOI] [PubMed] [Google Scholar]
- Crepet W.L. (2000). Progress in understanding angiosperm history, success, and relationships: Darwin’s abominably “perplexing phenomenon”. Proc. Natl. Acad. Sci. USA 97: 12939–12941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews S.T., Pearson J.C. (2009). Transcriptional autoregulation in development. Curr. Biol. 19: R241–R246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P. (2004). Floral zygomorphy, the recurring evolution of a successful trait. Bioessays 26: 1175–1184 [DOI] [PubMed] [Google Scholar]
- Cubas P., Coen E., Zapater J.M.M. (2001). Ancient asymmetries in the evolution of flowers. Curr. Biol. 11: 1050–1052 [DOI] [PubMed] [Google Scholar]
- Cubas P., Lauter N., Doebley J., Coen E. (1999). The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Darwin F., Seeward A.C. (1903). More Letters of Charles Darwin: A Record of His Work in a Series of Hitherto Unpublished Papers, Vol. II. (London: John Murray)
- De Bodt S., Maere S., Van de Peer Y. (2005). Genome duplication and the origin of angiosperms. Trends Ecol. Evol. (Amst.) 20: 591–597 [DOI] [PubMed] [Google Scholar]
- Dilcher D. (1979). Early angiosperm reproduction: an introductory report. Rev. Palaeobot. Palynol. 27: 291–328 [Google Scholar]
- Dilcher D. (2000). Toward a new synthesis: Major evolutionary trends in the angiosperm fossil record. Proc. Natl. Acad. Sci. USA 97: 7030–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Donoghue M.J., Ree R.H., Baum D.A. (1998). Phylogeny and the evolution of flower symmetry in the Asteridae. Trends Plant Sci. 3: 311–317 [Google Scholar]
- Dowell R.D. (2010). Transcription factor binding variation in the evolution of gene regulation. Trends Genet. 26: 468–475 [DOI] [PubMed] [Google Scholar]
- Du Z.-Y., Wang Y.-Z. (2008). Significance of RT-PCR expression patterns of CYC-like genes in Oreocharis benthamii (Gesneriaceae). J. Syst. Evol. 46: 23–31 [Google Scholar]
- Endress P.K. (1999). Symmetry in flowers: Diversity and evolution. Int. J. Plant Sci. 160: S3–S23 [DOI] [PubMed] [Google Scholar]
- Feng X., et al. (2006). Control of petal shape and floral zygomorphy in Lotus japonicus. Proc. Natl. Acad. Sci. USA 103: 4970–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Tao J.-H., Yan D., Wang Y.-Z., Li Z.-Y. (2008). Expression differentiation of CYC-like floral symmetry genes correlated with their protein sequence divergence in Chirita heterotricha (Gesneriaceae). Dev. Genes Evol. 218: 341–351 [DOI] [PubMed] [Google Scholar]
- Gellon G., McGinnis W. (1998). Shaping animal body plans in development and evolution by modulation of Hox expression patterns. Bioessays 20: 116–125 [DOI] [PubMed] [Google Scholar]
- Gompel N., Prud’homme B., Wittkopp P.J., Kassner V.A., Carroll S.B. (2005). Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487 [DOI] [PubMed] [Google Scholar]
- Grimaldi D. (1999). The co-radiations of pollinating insects and angiosperms in the Cretaceous. Ann. Mo. Bot. Gard. 86: 373–406 [Google Scholar]
- Hileman L.C., Cubas P. (2009). An expanded evolutionary role for flower symmetry genes. J. Biol. 8: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman L.C., Kramer E.M., Baum D.A. (2003). Differential regulation of symmetry genes and the evolution of floral morphologies. Proc. Natl. Acad. Sci. USA 100: 12814–12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth D.G., Donoghue M.J. (2006). Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. USA 103: 9101–9106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth D.G., Martins T., Chimney E., Donoghue M.J. (2011). Diversification of CYCLOIDEA expression in the evolution of bilateral flower symmetry in Caprifoliaceae and Lonicera (Dipsacales). Ann. Bot. (Lond.) 107: 1521–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour F., Nadot S., Damerval C. (2009). Evolution of floral symmetry: A state of the art. C. R. Biologies 332: 219–231 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Pajoro A., Angenent G.C. (2010). Regulation of transcription in plants: Mechanisms controlling developmental switches. Nat. Rev. Genet. 11: 830–842 [DOI] [PubMed] [Google Scholar]
- Kim M., Cui M.-L., Cubas P., Gillies A., Lee K., Chapman M.A., Abbott R.J., Coen E. (2008). Regulatory genes control a key morphological and ecological trait transferred between species. Science 322: 1116–1119 [DOI] [PubMed] [Google Scholar]
- Knoll A.H., Carroll S.B. (1999). Early animal evolution: Emerging views from comparative biology and geology. Science 284: 2129–2137 [DOI] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Lee T.I., et al. (2002). Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Copsey L., Vincent C., Clark J., Coen E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99: 367–376 [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L., Coen E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M., Cubas P. (2010). TCP genes: A family snapshot ten years later. Trends Plant Sci. 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Monteiro A., Podlaha O. (2009). Wings, horns, and butterfly eyespots: How do complex traits evolve? PLoS Biol. 7: e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R.C., Purugganan M.D. (2005). The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8: 122–128 [DOI] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Hervé C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65: 23–33 [DOI] [PubMed] [Google Scholar]
- Pang H.-B., Sun Q.-W., He S.-Z., Wang Y.-Z. (2010). Expression pattern of CYC-like genes relating to a dorsalized actinomorphic flower in Tengia (Gesneriaceae). J. Syst. Evol. 48: 309–317 [Google Scholar]
- Papa R., Martin A., Reed R.D. (2008). Genomic hotspots of adaptation in butterfly wing pattern evolution. Curr. Opin. Genet. Dev. 18: 559–564 [DOI] [PubMed] [Google Scholar]
- Pearson J.C., Lemons D., McGinnis W. (2005). Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6: 893–904 [DOI] [PubMed] [Google Scholar]
- Preston J.C., Hileman L.C. (2009). Developmental genetics of floral symmetry evolution. Trends Plant Sci. 14: 147–154 [DOI] [PubMed] [Google Scholar]
- Proost S., Pattyn P., Gerats T., Van de Peer Y. (2011). Journey through the past: 150 million years of plant genome evolution. Plant J. 66: 58–65 [DOI] [PubMed] [Google Scholar]
- Prud’homme B., Gompel N., Carroll S.B. (2007). Emerging principles of regulatory evolution. Proc. Natl. Acad. Sci. USA 104 (Suppl. 1): 8605–8612
- Prud’homme B., Gompel N., Rokas A., Kassner V.A., Williams T.M., Yeh S.-D., True J.R., Carroll S.B. (2006). Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440: 1050–1053 [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Hue I., Huijser P., Flor P.J., Hansen R., Tetens F., Lönnig W.-E., Saedler H., Sommer H. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: Evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M.D., Bell M.A., Kingsley D.M. (2006). Parallel genetic origins of pelvic reduction in vertebrates. Proc. Natl. Acad. Sci. USA 103: 13753–13758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E., Ma H., Frohlich M.W., Soltis P.S., Albert V.A., Oppenheimer D.G., Altman N.S., dePamphilis C., Leebens-Mack J. (2007). The floral genome: an evolutionary history of gene duplication and shifting patterns of gene expression. Trends Plant Sci. 12: 358–367 [DOI] [PubMed] [Google Scholar]
- Soltis P.S. (2005). Ancient and recent polyploidy in angiosperms. New Phytol. 166: 5–8 [DOI] [PubMed] [Google Scholar]
- Song C.-F., Lin Q.-B., Liang R.-H., Wang Y.-Z. (2009). Expressions of ECE-CYC2 clade genes relating to abortion of both dorsal and ventral stamens in Opithandra (Gesneriaceae). BMC Evol. Biol. 9: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht C.D., Bartlett M.E. (2009). Flower evolution: The origin and subsequent diversification of the angiosperm flower. Annu. Rev. Ecol. Evol. Syst. 40: 217–243 [Google Scholar]
- Stuessy T.F. (2004). A transitional-combinational theory for the origin of angiosperms. Taxon 53: 3–16 [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine J.W., Jablonski D., Erwin D.H. (1999). Fossils, molecules and embryos: new perspectives on the Cambrian explosion. Development 126: 851–859 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., Fawcett J.A., Proost S., Sterck L., Vandepoele K. (2009a). The flowering world: A tale of duplications. Trends Plant Sci. 14: 680–688 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y., Maere S., Meyer A. (2009b). The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10: 725–732 [DOI] [PubMed] [Google Scholar]
- Viola I.L., Reinheimer R., Ripoll R., Manassero N.G., Gonzalez D.H. (2012). Determinants of the DNA binding specificity of class I and class II TCP transcription factors. J. Biol. Chem. 287: 347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. (2008). Gene duplications, robustness and evolutionary innovations. Bioessays 30: 367–373 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang Y., Luo D. (2010). LjCYC genes constitute floral dorsoventral asymmetry in Lotus japonicus. J. Integr. Plant Biol. 52: 959–970 [DOI] [PubMed] [Google Scholar]
- Wang Y.-Z., Mao R.-B., Liu Y., Li J.-M., Dong Y., Li Z.-Y., Smith J.F. (2011). Phylogenetic reconstruction of Chirita and allies (Gesneriaceae) with taxonomic treatments. J. Syst. Evol. 49: 50–64 [Google Scholar]
- Wang Z., et al. (2008). Genetic control of floral zygomorphy in pea (Pisum sativum L.). Proc. Natl. Acad. Sci. USA 105: 10414–10419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Koshikawa S., Williams T.M., Carroll S.B. (2010). Generation of a novel wing colour pattern by the Wingless morphogen. Nature 464: 1143–1148 [DOI] [PubMed] [Google Scholar]
- Wray G.A. (2007). The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8: 206–216 [DOI] [PubMed] [Google Scholar]
- Yang X., Cui H., Yuan Z.-L., Wang Y.-Z. (2010). Significance of consensus CYC-binding sites found in the promoters of both ChCYC and ChRAD genes in Chirita heterotricha (Gesneriaceae). J. Syst. Evol. 48: 249–256 [Google Scholar]
- Yuan Z., Gao S., Xue D.-W., Luo D., Li L.-T., Ding S.-Y., Yao X., Wilson Z.A., Qian Q., Zhang D.-B. (2009). RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 149: 235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Kramer E.M., Davis C.C. (2010). Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proc. Natl. Acad. Sci. USA 107: 6388–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.-R., Wang Y.-Z., Smith J.F., Chen R.J. (2008). Altered expression patterns of TCP and MYB genes relating to the floral developmental transition from initial zygomorphy to actinomorphy in Bournea (Gesneriaceae). New Phytol. 178: 532–543 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.