Abstract
Glycolipids presented by the major histocompatibility complex class I (MHC I) homolog CD1d are recognized by natural killer T (NKT) cells characterized by either a semi-invariant (type I or iNKT) or a relatively variable (type II) T cell receptor (TCR) repertoire. Here we describe the first structure of a type II NKT TCR complexed with CD1d-lysosulfatide (LSF). Both TCR α and β chains contacted the CD1d molecule with a diagonal footprint, typical of MHC-TCR interactions, while the antigen was recognized exclusively with a single TCR chain, similar to the iNKT TCR. Type II NKT cells, therefore, recognize CD1d-sulfatide complexes with a distinct recognition mechanism characterized by features of both iNKT cells as well as conventional peptide-reactive T cells.
Natural killer T (NKT) cells represent one of the most extensively studied non-conventional T lymphocyte lineages1,2. Characterization of this population has provided important insights related to the development and physiological role of innate-like T cells in immunity as well as a range of antigen recognition mechanisms employed by αβ TCRs. NKT cells reactive to CD1d-presented antigens can by divided in type I or iNKT cells (expressing a semi-invariant TCR characterized in mice by a conserved Vα14-Jα18 rearrangement paired with a limited repertoire of Vβ chains, mainly Vβ8.2, Vβ7 and Vβ2) and type II NKT cells (expressing a more variable TCR repertoire). Lipid antigens bind to the CD1d groove with their hydrophobic portion occupying the two main groove pockets (named A′ and F′) and the polar portion of the antigen exposed to the solvent3. While the iNKT TCR can recognize both α-linked and β-linked glycolipids by forcing the antigens in a conserved conformation4–7, type II NKT cells do not respond to α-linked glycolipids. Crystal structures of the iNKT TCR complexed with several CD1d-antigens showed that the semi-invariant TCR docks with an unique mode parallel to the CD1d antigen binding groove, with most of the interface dominated by the germline-encoded α chain4,7,8. The lack of a universal type II NKT antigen, analogous to the potent iNKT ligand α-galactosylceramide (αGalCer)9, has so far hampered our understanding of the role of this population in normal as in pathological states, despite type II NKT cells constitute a substantial portion of the CD4+ T cells in MHC II-deficient mice10,11 and in human bone marrow12, liver13 and gut14. Nonetheless, type II NKT cells reactive to the widely expressed self antigen sulfatide15 were found to be involved in the suppression of tumor immunity (reviewed in Terabe et al.16), regulation of ischemic reperfusion17,18, experimental autoimmune encephalomyelitis15, concanavalin A-induced hepatitis19 and type 1 diabetes20. Importantly, the finding that administration of sulfatide protected mice from inflammatory diseases in the central nervous system and liver in a type II NKT cell-dependent manner15–20 suggests a potential for the development of novel agonists able to harness the immunomodulatory potential of this population. Sulfatide-reactive type II NKT cells use an oligoclonal repertoire with characteristics of both antigen-specific conventional and innate-like T cells21. However, it is not known how these features of type II NKT TCRs determine self-antigen recognition. We report here the 2.1 Å resolution crystal structure of a type II NKT TCR (Vα1-Jα26, Vβ16-Jβ2.1) isolated from the sulfatide-reactive, CD1d-restricted T cell hybridoma Hy19.3 (a subclone of the XV19 hybridoma)15,22,23, as well as the crystal structure of the ternary complex of the TCR bound to mouse CD1d-lysosulfatide (LSF) at a resolution of 3.5 Å. LSF, which lacks the fatty acid chain present on most sulfatide molecules, was previously identified as the most potent antigen recognized by Hy19.3 (refs. 22,23) and elevated LSF tissue concentrations are found in subjects with metachromatic leukodistrophy24. The structural and biochemical data presented here describe how type II NKT cells recognize antigens using a distinct recognition mechanism, providing a rationale for the TCR repertoire used by this population.
Results
The Hy19.3 TCR binds diagonally above the CD1d A′ pocket
To determine the structural basis of sulfatide recognition by type II NKT cells, we determined the crystal structure of the mouse CD1d-LSF-Hy19.3 TCR complex at a resolution of 3.5 Å (Fig. 1, Table 1). Two highly similar ternary complexes (RMSD of 0.7 Å over Cα atoms) were present in the asymmetric unit and the following analysis will be limited to one complex, unless otherwise stated. Electron density for both CD1d and the TCR at the complex interface was unambiguous (Supplementary Fig. 1). Strong density was present for the galactosyl moiety of LSF while relatively weak density was observed for the sphingosine acyl chain modeled in the F′ pocket and a spacer occupying the A′ pocket (modeled as palmitic acid, Supplementary Fig. 1), similarly to what has been observed for the single alkyl chain ligand lysophosphatidylcholine from a recently solved human CD1d-iNKT TCR complex25. The CD1d-TCR buried surface area (BSA) is ~910 Å2, slightly higher than the iNKT TCR (~760 Å2)8 but below the range of values observed for MHC-TCR complexes (1200–2400 Å2)26. The Hy19.3 TCR docked on the CD1d antigen binding groove with an almost perpendicular orientation, roughly centered above the A′ pocket (Fig. 1a) with its Vα domain contacting the α2 helix (residues 154–167) while the Vβ domain bound to the α1 helix (residues 68–79, Fig. 1b). This binding orientation is radically different from the iNKT TCR, which binds above the opposite CD1d F′ pocket parallel to the CD1d α-helices (Fig. 1c)8. The docking angle of the Hy19.3 TCR was calculated to be 74°, at the high end of the range of docking angles reported for MHC-TCR (21-70°)26. Therefore, glycolipid-reactive type I and type II NKT TCRs mark the extreme ends of binding orientations of conventional pMHC-reactive TCRs that typically bind diagonally above the antigen-presenting molecule (e.g. 2C-H-2Kb-dEV8, Fig. 1d)26,27. Interestingly, the only known TCR with a higher docking angle (84°), is specific for a myelin protein-derived antigen28. This TCR also docks above the N-terminal portion of the peptide corresponding to the A′ pocket (Supplementary Fig. 2) and it was proposed that this unusual binding mode allows escaping negative selection. Overall, the structure of the ternary complex revealed that the Hy19.3 TCR docks on CD1d with an almost perpendicular orientation resulting in a radically different footprint compared to a previously described iNKT TCR.
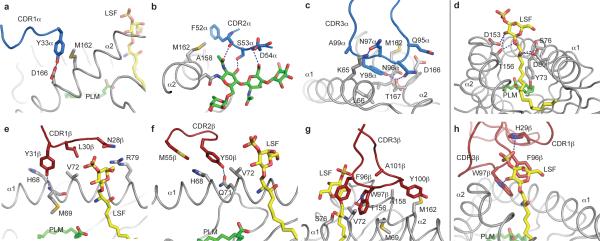
Figure 1. Overall structure and docking orientation of the Hy19.3 TCR on the CD1d-LSF complex.
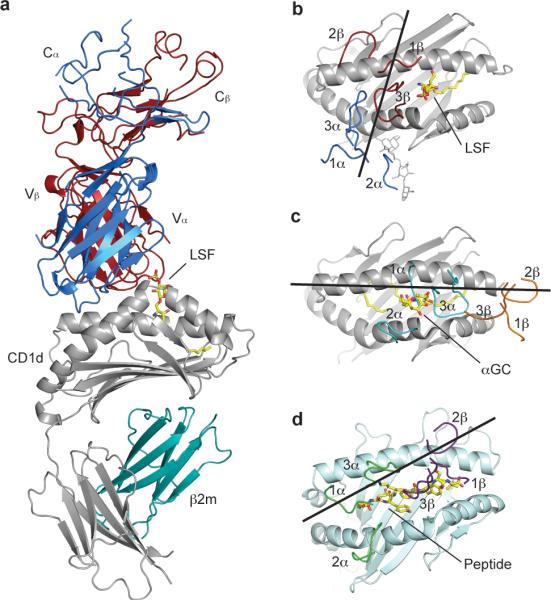
a, Crystal structure of the mouse CD1d-LSF-Hy19.3 TCR with CD1d in grey, β2-microglobulin in aqua, TCR α chain in blue and TCR β chain in dark red. The ligand is shown in yellow. b-d, TCR footprint and binding orientation observed for the CD1d-LSF-Hy19.3 TCR complex (b), the CD1d-αGalCer-iNKT TCR complex (c, PDB code 3HE6) and an MHCI-peptide-TCR complex (d, PDB code 2CKB). The lines represent the vector connecting the centroids of the conserved disulphide bond present in each V domain. Note how the Hy19.3 TCR docks on CD1d with an almost perpendicular orientation (b), in stark contrast to the parallel docking of the iNKT TCR on the same antigen-presenting molecule (c).
Table 1.
Data collection and refinement statistics (molecular replacement) One crystal was used for each structure. Values in parentheses are for highest-resolution shell.
| mCD1d-LSF- Hy19.3 TCR | Hy19.3 TCR | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 98.51,126.99,104.35 | 73.22,101.53,134.49 |
| α, β,.γ (°) | 90.00, 110.5, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 92.3-3.5 (3.69-3.50) | 49.0-2.1 (2.15-2.1) |
| Rmerge (%) | 24.0 (81.4) | 7.9 (56.3) |
| Rmeas (%) | 25.5 (86.5) | 9.0 (64.3) |
| Rp.i.m. (%) | 8.7 (29.2) | 4.2 (30.3) |
| Average I / σ I | 8.3 (2.8) | 11.5 (2.5) |
| Completeness (%) | 99.9 (99.9) | 99.9 (99.9) |
| Redundancy | 8.3 (8.5) | 4.3 (4.2) |
| Refinement | ||
| Resolution (Å) | 92.3-3.5 | 38.0-2.1 |
| No. reflections | 29051 | 56586 |
| Rwork / Rfree | 0.210 / 0.266 | 0.188 / 0.227 |
| Ramachandran plot (%) | ||
| Favored | 91.3 | 97.6 |
| Allowed | 98.9 | 99.9 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.007 |
| Bond angles (°) | 1.11 | 1.04 |
CDR1, CDR2 contacts and the type II NKT cell repertoire
Both TCR chains made similar contributions to the interface with CD1d in terms of BSA (α chain, 49%, β chain 51%). In contrast to the iNKT TCR, all six complementarity-determining region (CDR) loops contacted the CD1d-LSF complex (Fig. 2a-h). Specific residues on the germline-encoded CDR1 and CDR2 loops bound CD1d, with CDR1β also contacting the antigen. In particular, residues Tyr33α on CDR1α, together with Phe52α, Ser53α and Asp54α on CDR2α interacted with the α2 helix and a N-linked oligosaccharide (Asn165, Fig. 2a,b) while Tyr31β on CDR1β and Tyr50β on CDR2β contacted residues on the α1 helix (Fig. 2e,f). These loops contacted CD1 in correspondence of two “hotspots” previously identified on the MHC protein surface (corresponding to Met69 on the α1 helix and Ala158 on the α2 helix), and two of the three residues equivalent to the “restriction triad” always contacted by MHC-reactive TCRs2 were also contacted by the Hy19.3 TCR (His68, Val72, Supplementary Table 1), resulting in the diagonal, albeit shifted, docking mode typical of MHC-TCR interactions29. Notably, among the murine Vβ gene segments, only Vβ16, Vβ8.1 and Vβ8.3 possessed the combination of critical residues employed to contact CD1d-LSF (His30, Tyr31, Tyr50) while only Vα1, Vα3, Vα5 and Vα8 possess the combination Tyr33, Phe or Tyr52 contacting the α2 helix. Interestingly, these genes represent the majority of the V segments identified in type II NKT cells11 and in particular among sulfatide-reactive ones21, suggesting a conserved docking mode for type II NKT TCRs. Thus, the requirement for specific contacts between the CDR1 and CDR2 loops and the CD1d α1 and α2 helices observed for the Hy19.3 TCR provides an explanation for the TCR repertoire used by type II NKT cells.
Figure 2. The CD1d-antigen-TCR interface.
a-c, Contacts between the TCR α chain CDR loops (in blue) and CD1d (grey). d, Contacts between CD1d (grey) and LSF (yellow). e-g, Contacts between CD1d (grey) and the TCR β chain CDR loops (dark red). h, Recognition of LSF (yellow) by the Hy19.3 TCR CDR1β and CDR3β loops (dark red). The spacer lipid in the A’ pocket (PLM) is shown in green. b, The N-linked glycosylation (green) at Asn165 of CD1d is shown contacting the CDR2α loop. Polar contacts are shown as dashed blue lines.
Both CDR3α and CDR3β are critical for complex formation
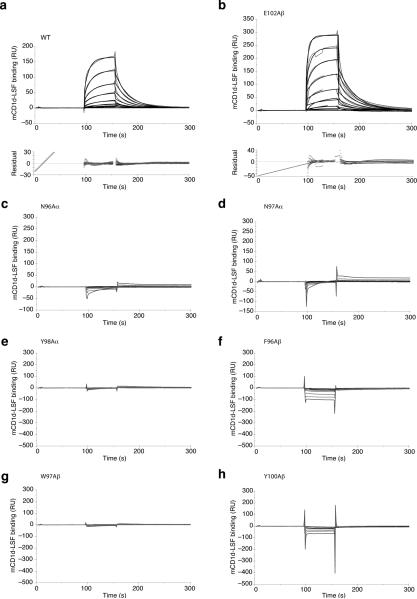
The Hy19.3 TCR bound to mCD1d-LSF with micromolar affinity, as measured by surface plasmon resonance (SPR, Fig. 3a). The binding was characterized by relatively slow on–rate and fast off-rates, not dissimilar to what reported for weak iNKT antigens and MHC-TCR interactions30. Since the majority of the interface contacts in the ternary complex were mediated by the non-germline encoded CDR3 loops (CDR3α and CDR3β account for 34% and 32% of total BSA respectively), we next determined whether mutation of residues on these two loops affected TCR binding. Consistent with a pivotal role of the CDR3 loops at the CD1d-TCR interface, mutation to alanine of any of the residues at the tip of these two CDR loops (N96Aα, N97Aα, Y98Aα, F96Aβ, W97Aβ, Y100Aβ) resulted in the abrogation of binding, as measured by surface plasmon resonance, while a control mutant (E102Aβ) was not affected (Fig. 3b-h). This result is reminiscent of what has been observed for MHC-reactive TCRs, where both CDR3 loops play a critical role in complex formation, but contrasts with the iNKT TCR, where the CDR3β is thought to only modulate the affinity of the TCR for the CD1-antigen complex26,31.
Figure 3. Effect of TCR mutations on binding affinity as measured by SPR.
a Binding response of increasing concentrations (0.3-22 μm) of mCD1d-LSF complexes to immobilized WT TCR as measured by surface plasmon resonance. The response shown is reference-substracted. Binding of mCD1d-lysosulfatide is shown with the calculated fit as black lines and the corresponding residuals in the plot below. The diagonal line at the early time points in the residual plot is the result of a known software bug and it should be ignored. Kd WT = 5.98 ± 4.73 μM, ka WT = 6.0 ± 1.3 × 103 M-1 s-1, kd WT = 0.033 ± 0.006 s-1. b-h Binding response of increasing concentrations (0.3-36 μm) of mCD1d-LSF complexes to immobilized CDR3α and CDR3β mutant TCRs. Note how all the mutants but the control mutation at position 102 of CDR3β were unable to elicit measurable responses. Kd E102Aβ = 6.65 ± 3.18 μM, ka E102Aβ = 4.9 ± 2.0 × 103 M-1 s-1, kd E102Aβ = 0.029 ± 0.003 s-1. Values represent average and standard deviation of at least two independent measurements.
Interestingly, the Hy19.3 TCR CDR3α loop contacted exclusively the CD1d surface (Fig. 2c) while the CDR3β loop contacted both CD1d and the antigen (Fig. 2g). A QNNY amino acid motif on CDR3α (Gln95, Asn96, Asn97, Tyr98) bound to the top of the A′ roof of CD1d, contacting both helices via polar and van der Waals contacts (Supplementary Table 1). The central NN motif observed on CDR3α is present on a sizeable proportion of sulfatide-reactive type II NKT TCRs21 (Supplementary Fig. 3). Moreover, the CDR3α length of this TCR (10 aa) is similar to the highly restricted length (8-9 aa) of this loop observed in Vα3 and Vα1 sulfatide-reactive TCRs21 and other type II NKT cells11. Therefore, both CDR3α and CDR3β play a critical role in the formation of the ternary complex, with the interaction between CD1d and CDR3α resulting in the oligoclonal nature of the CDR3α loops observed in type II NKT cells.
Mutation of the A′ pocket abolishes Hy19.3 TCR binding
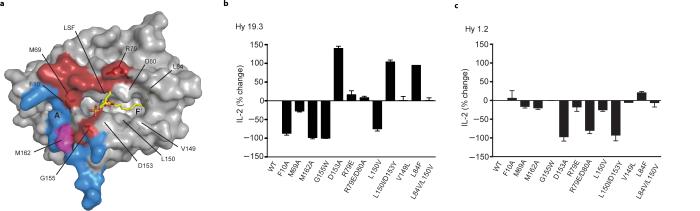
Mutation of selected residues on CD1d (Fig. 4a) showed that perturbation of the area around the A′ pocket (Phe10Ala, Met69Ala, Met162Ala) results in the disruption of the CD1d-Hy19.3 TCR interaction, as measured by an antigen presentation assay (Fig. 4b). Those mutations, however, do not greatly affect the binding of the iNKT TCR that sits on the opposite side of CD1d4,7,8, above the F′ pocket (Fig. 4c). Interestingly, mutation of Asp153 (which is crucial for the binding of iNKT antigens) to alanine or tyrosine resulted in increased stimulation of the type II NKT hybridoma (Fig. 4b), suggesting that this residue is not involved in specific interactions with the antigen but rather impairs TCR recognition. As expected, residues around the F′ pocket were not crucial for Hy19.3 TCR binding, except for Leu150. However, as this residue is not close to the Hy19.3 TCR, the alanine mutation likely affected LSF binding to CD1d. The presence of polymorphisms at the CD1d residue 162 between laboratory and wild-type mouse strains have been previously shown to strongly affect type II NKT cell recognition of CD1d-antigen complexes32. In the ternary complex structure described here, the side chain of Met162 was “capped” by the aromatic residues Tyr33α, Phe52α and Tyr100β (Supplementary Fig. 4), providing an explanation why modifications at this position can have direct consequences on the binding of the TCR. Interestingly, we also observed some degree of cross-species reactivity between human CD1d molecules and the Hy19.3 TCR, although to a lesser extent compared to the iNKT TCR (Supplementary Fig. 5).
Figure 4. Mutation of CD1d residues at the interface affects activation of the Hy19.3 hybridoma.
a Localization of the CD1d mutated residues at the CD1d-TCR interface. CD1d is shown as grey surface with the residues comprising the α or β chain footprint on CD1d shown in blue and dark red, respectively. The only shared residue between the two footprints (Met162) is shown in magenta. LSF is shown in yellow. b-c Relative changes in the amount of IL-2 release, a measure of hybridoma activation, by the Hy19.3 line (b) or the iNKT hybridoma line Hy1.2 (c) in a coated plate assay when stimulated with different CD1d mutants loaded with either LSF (2 μg/ml, Hy19.3) or αGalCer (0.5 μg/ml, Hy1.2). The antigen concentration used represents the optimal concentration determined by antigen titration. The results are representative of at least five independent experiments.
The Hy19.3 TCR recognizes LSF exclusively with the β chain
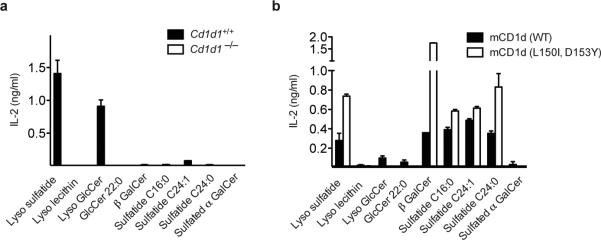
The side chain of His29β on CDR1β contacted the sulfate group of LSF, while the bulky side chains Phe96β and Trp97β on CDR3β “pinned” the galactosyl residue against Asp153 on the CD1d α2 helix (Fig. 2h), explaining the preference of this TCR for the extended ligand conformation typical of β-linked glycolipids. This rather nonspecific binding mode suggests that modifications at the 4’ and 6’ positions of the hexose moiety are likely to be tolerated. Moreover, comparison of the binding orientation of LSF and the closely related cis-tetracosenoyl sulfatide that we had previously crystallized bound to mCD1d in absence of any TCR (Supplementary Fig. 6)33 shows a striking similarity further suggesting that this TCR is able to bind several CD1d-ligand complexes with a conserved mode of recognition23. Interestingly, LSF was shifted by approximately 3 Å toward the F’ pocket, likely as a result of the presence of the bulky Trp97 on CDR3β (Supplementary Fig. 6). Furthermore, while the lyso isoforms of sulfatide and glucosylceramide (GlcCer) are the predominant antigenic lipids able to stimulate Hy19.3 when presented by splenocytes (Fig. 5a), several dual-alkyl chain β-linked glycolipids, such as different forms of sulfatide (with C16:0, C24:0 and C24:1 acyl chains) and β-GalCer, were also recognized when presented by plate bound recombinant CD1d (Fig. 5b). Mutation of Asp153 and Leu150 generally enhanced the antigenicity of most lipids (Fig. 5b). The lack of antigenicity observed for lysolecithin (also known as lysophospatidylcholine) is likely the result of the drastically different polar moiety (a phosphorylcholine group) of this lipid compared to the other antigens carrying hexose sugars. Moreover, the differences in the antigen presentation between plate bound CD1d and antigen-presenting cells (APCs) suggests that APCs either process antigens into an inactive form or prevent their presentation at the cell surface, consistent with the reported requirement of mCD1d trafficking to the lysosomal compartment for type II NKT cell activation22,34.
Figure 5. Recognition of self-lipids by the Hy19.3 TCR in a plate-bound assay.
a A typical NKT cell stimulation assay in the presence of APC (splenocytes) from either wild-type (Cd1d1+/+) or CD1d-knockout mice (Cd1d1-/-). A concentration of 2.5 μg/ml of each antigen was used. b APC-free CD1d-coated plate assay using recombinant WT mCD1d and the mutant L150I,D153Y mCD1d. The data are representative of three independent experiments.
Flexibility in the Hy19.3 TCR and binding thermodynamics
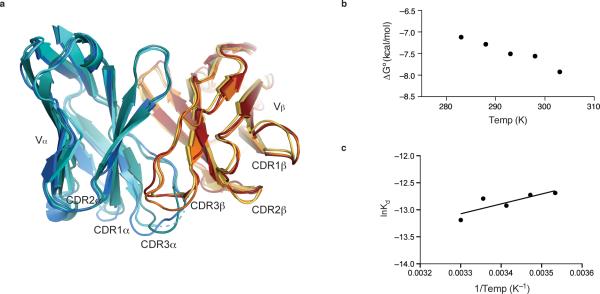
We also determined the structure of the Hy19.3 TCR alone at 2.1 Å resolution (Table 1) and compared it to the structure of the same TCR in the ternary complex. Superposition of the two uncomplexed TCR structures present in the asymmetric unit with the TCR derived the ternary complex revealed flexibility within the CDR1α, CDR2β and CDR3 loops (Fig. 6a), potentially allowing the TCR to recognize an even broader spectrum of antigens through fine positioning of the CDR footprint, as has been reported for some MHC-reactive TCRs2. This finding is in contrast to the iNKT TCR which binds to CD1d-antigens without significant conformational changes in its CDR loops4,8. Also in contrast to the iNKT TCR35, we observed that the binding affinity of the Hy19.3 TCR is temperature dependent (Fig. 6b, Supplementary Fig. 7). Interestingly, van't Hoff analysis of the binding data (Fig. 6c) suggests that the complex formation was entropically driven, possibly as a result of the hydrophobic interactions created at the CD1-TCR interface and/or water displacement (although the low resolution of the ternary complex did not allow to us to model water molecules), similar to what has been reported for the HLA-B8-FLR-L13 TCR complex36.
Figure 6. Flexibility in the CDR loops of the Hy19.3 TCR.
a, Superposition of the complexed and uncomplexed TCR structures showing the V domain of α (complexed in blue, uncomplexed in cyan and aqua) and β (complexed in dark red, uncomplexed in orange and yellow) chains. Flexibility is observed for CDR1α, CDR2β and most notably both CDR3 loops. The disordered portion of the CDR3α loop in one of the uncomplexed TCRs is shown as a dashed line. b Plot of free energy (ΔG) values against temperature. The values represent the average of two independent experiments. c van't Hoff analysis of Hy19.3 TCR binding to mCD1d-LSF. The enthalpic and entropic energetic contributions to the complex formation were estimated by linear regression of data from a van't Hoff plot yielding ΔH = 3.63 ± 3.58 kcal/mol and TΔS@25°C = 11.28 ± 3.65 kcal/mol. The binding of the Hy19.3 TCR is entropically driven, possibly as a result water displacement and/or the formation of hydrophobic interactions upon complex formation.
Discussion
The CD1d-LSF-Hy19.3 TCR structure reported here provides the first insight into lipid antigen recognition by a non-invariant NKT TCR. Surprisingly, the Hy19.3 TCR binding mode combines features of both iNKT cells, as well as MHC-restricted T cells, resulting in a unique antigen recognition mechanism among αβ TCRs. On one hand, in the type II NKT ternary complex both TCR α and β chain contact the CD1d molecule with a diagonal footprint, typical of MHC-TCR interactions26. For pMHC-reactive T cells, optimal T cell activation occurs when the co-receptor is engaged, which correlates with a limited range of TCR docking angles37. However, as NKT cells can be double negative for CD4 and CD8 co-receptor expression1, the TCR docking angle on CD1d is not influenced by the engagement of co-receptors, and as such, its significance in T cell activation remains unclear. On the other hand, the Hy19.3 TCR recognized the antigen exclusively with only one TCR chain, similar to the iNKT TCR3, while MHC-reactive TCRs require both CDR3 loops to contact the antigen26. However, unlike the iNKT TCR, which contacts the ligand with its α chain, the type II NKT TCR bound lysosulfatide solely through the TCRβ chain, suggesting that the latter chain is responsible for the fine antigen specificity of the TCR. Consistent with this notion, oligoclonal Vβ gene usage and variability in the length and sequence of the CDR3β loop have been observed in sulfatide-reactive NKT cells21. The role of the CDR1β and His29 in particular appeared less critical, as antigens lacking a sulfate group such as β-GalCer and lysoGlcCer could still be recognized by this TCR. The conservation of key CDR1 and CDR2 interacting residues between the lysosulfatide-reactive TCR and other sulfatide reactive TCRs21 strongly suggests a shared recognition logic for this class of self-antigens that could extend to a substantial proportion of type II NKT cells11. Moreover, the finding that the CDR3α loop was not directly involved in antigen recognition but instead recognized the CD1d surface provides an explanation for the restricted length and conserved motifs observed on the CDR3α loop of most sulfatide reactive and type II NKT TCRs. We hypothesize that TCR α chain rearrangement is not a result of positive selection by a self-antigen, as is the case for the iNKT cells, but is rather driven by the binding to the non-polymorphic CD1d molecule, consistent with the presence of sulfatide-reactive type II NKT cells in mice deficient in sulfatide15,21–23. Collectively, our data suggests that the immune system developed two radically different strategies enabling the “non”-polymorphic CD1d to bind at least two different types of NKT TCRs. The finding that a substantial proportion of type II NKT cells responds to the myelin-derived glycolipid sulfatide15 was pivotal in determining the role of these cells in normal as in pathological processes. Given the growing recognition of the role sulfatide-reactive type II NKT cells play in the suppression of tumor immunity and regulation of autoimmune diseases15–20, with the consequent therapeutic potential, the structure described here opens the way to the rational development of new ligands with immunomodulatory potential.
Methods
Cloning, expression and purification of the Hy19.3 TCR and mouse CD1d
Total RNA was isolated from 5 × 106 hybridoma cells using the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. First-strand cDNA synthesis of 5′RACE and standard PCR (second strand reaction) were performed with a previously described protocol39. The PCR products were cloned into the pGEM-T easy vector (Promega) for sequencing and then subcloned in Escherichia coli expression vectors containing the human constant domains of either the α or β chain (pET22b+ for Vα1-Jα26, pET30a+ for the Vβ16-Jα2.1 gene segment) as previously described for the iNKT TCR40. Fully glycosylated mCD1d protein was expressed and purified as described previously40. The α and β chains of the Hy19.3 TCR were expressed separately in E. coli BL21(DE3) cells as inclusion bodies, extracted with 6M guanidine-HCl in 50 mM Tris-HCl pH 7.0, 5 mM EDTA, 2 mM DTT and refolded by mixing 32 mg of α and 48 mg of β chain in presence of 1 mM DTT. The mixture was then added dropwise to 1 L of refolding buffer under stirring at 4°C (50 mM Tris-HCl, 0.4 M arginine, 5 M urea, 2 mM EDTA, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, 0.2 mM PMSF, pH 8 at 25°C). After 16 h, 32 mg of both α and β chains were added to the refolding buffer and the mixture stirred for an additional 8-10 h. The mixture was then dialyzed against 10 mM Tris-HCl, 0.1 M urea, pH 8.0 for ~16 h, followed by dialysis against 10 mM Tris-HCl, pH 8.0 for 24 h. DEAE sepharose beads (GE Healthcare, 3 ml of settled resin) were added to refolding solution for 2–4 h before being collected on an Econo column (Bio-Rad Laboratories). The refolded TCR was eluted with 100-150 mM NaCl in 10 mM Tris pH 8.0 and further purified by anion-exchange chromatography (MonoQ 5/50 GL, GE Healthcare in 10 mM Tris pH 8.0) using a linear gradient of NaCl (0-300 mM). The fractions containing the TCR were purified by gel filtration chromatography (Superdex S200 10/300 GL, GE Healthcare, in 50 mM HEPES, 150 mM NaCl, pH 7.5) and concentrated to 5 mg/ml for further analysis.
Lipid loading and complex formation
Lysosulfatide (Matreya) was dissolved in DMSO at 4 mg/ml. Purified mouse CD1d was incubated with lysosulfatide (3-6x molar excess) for 16 h at 25°C before further purification by gel filtration chromatography (Superdex S200 10/300 GL, GE Healtcare, in 50 mM HEPES, 150 mM NaCl, pH 7.5) to remove the excess lipid. The CD1d-LSF complex and the Hy19.3 TCR (1:1 molar ratio, both proteins at a concentration of ~5 mg/ml) were then mixed and incubated for 1 h at 25°C to promote complex formation.
Crystallization and data collection
Crystals of the TCR were grown at 23 °C by sitting drop vapor diffusion mixing 0.1 μl protein (5 mg/ml) with 0.1 μl precipitant (18% polyethylene glycol 3350, 0.2 M ammonium citrate dibasic). Crystals of the ternary complex were grown over several weeks at 4 °C by sitting-drop vapour diffusion while mixing 1 μl protein (~5 mg/ml) with 1 μl precipitant (11% polyethylene glycol 4000, 4% tacsimate pH 6). The crystals were flash-cooled at 100 K in a solution containing the mother liquor and 20% glycerol. Diffraction data were collected at the Stanford Synchrotron Radiation Lightsource beamline 7.1 (Hy19.3 TCR) or at the Advanced Light Source beamline 5.0.3 (ternary complex). The Hy19.3 TCR crystals belong to space group P212121 with cell parameters a=73.2 Å; b=101.6 Å; c=134.5 Å. The mCD1d-LSF-Hy19.3 TCR crystal belongs to space group P21 with cell parameters a=98.5 Å; b=127.0 Å; c=104.4 Å β=110.5°. The data were processed with iMOSFLM and SCALA in the CCP4 suite38 (Table 1).
Structure determination and refinement
The two structures were solved by molecular replacement using PHASER41. The structure of an iNKT TCR (PDB ID 2Q86) with its CDR loops removed was used as a template for the Hy19.3 TCR data, yielding two molecules in the asymmetric unit. For the ternary complex, a search for mouse CD1d (PDB ID 3ILQ with the ligand, water molecules and oligosaccharides removed) was completed first, yielding two CD1d molecules. A search performed using the Hy19.3 uncomplexed TCR structure with its CDR loops removed resulted in two TCR molecules in the asymmetric unit, for a total of two highly similar ternary complexes in the asymmetric unit (RMSD of 0.7 Å on Cα atoms). For both structures, refinement was carried out with REFMAC38 (the final cycles of the uncomplexed Hy19.3 TCR refinement were performed with the software Phenix42) applying both TLS and NCS restraints, intercalated with cycles of manual building in COOT43. The final Hy19.3 TCR structure was refined to 2.1 Å with a final R/Rfree of 18.8/22.7% (Ramachandran allowed/favored residues 97.6/99.9%) while the final ternary complex structure was refined to 3.5 Å with a final R/Rfree of 21.0/26.6% (Ramachandran allowed/favored residues 91.3/98.9%). Refinements statistics are presented in Table 1.
Mutant generation
The CD1d mutants used here were previously described44,45. Hy19.3 TCR mutants were generated by site-mutagenesis using the Quick Change II kit (Stratagene, Agilent Technologies) with the primers designed on the Quick Change Primer Design server. The proteins were expressed and purified as the wild-type proteins.
Antigen presentation assays
CD1d mutants were tested using a CD1d-coated plate assay essentially as previously described15,22. For the APC assay, irradiated splenocytes from C57BL/6 and Cd1d1-/- mice were used. Synthetic and semi-synthetic lipids were acquired from Matreya and as described earlier15,21,22,33.
Surface Plasmon Resonance measurements
SPR kinetics measurements were performed on a Biacore3000 by immobilizing biotinylated Hy19.3 TCR (WT or mutant variants carrying a C-terminal birA tag on the β chain) on a CAP chip and flowing increasing concentrations of purified mCD1d-LSF at 30 μl/min in 10 mM HEPES pH7.5, 150 mM NaCl, 3 mM EDTA. For the thermodynamics studies, 8 μM mCD1d-LSF was injected over immobilized WT TCR, covering a range of temperatures from 10 °C to 30 °C, in 5 °C increments. Kinetic parameters were calculated a simple Langmuir 1:1 model in the BIA evaluation software version 4.1. Enthalpic and entropic contributions to the binding were estimated by van't Hoff analysis. Data points were fitted with a linear regression in GraphPad Prism.
Supplementary Material
Acknowledgments
We thank the Stanford Synchrotron Radiation Lightsource (SSRL, beamline 7-1) and the Advanced Light Source (ALS, beamline 5.0.3) for their support during remote data collection. D.M.Z is supported by NIH grant AI074952. V.K. is supported by NIH grant CA100660 and JDRF and MSNRI. We thank M. Kronenberg (LIAI) for providing some of the CD1d mutants, R. Stanfield (TSRI) for the scripts to measure docking angles and N. Vu and J. Nourblin (LIAI) for their help during cloning and protein expression.
Footnotes
Author contributions
E.G. generated the TCR constructs, purified and crystallized the proteins, determined the structures and performed SPR experiments. I.M. performed the antigen presentation assays. J.W. and P.I. provided assistance with mutant generation and protein purification. T.T.M sequenced the TCR. E.G., V.K. and D.M.Z. analyzed the data and wrote the manuscript.
Accession codes
The nucleotide sequence of the Vα and Vβ domains of the Hy19.3 TCR have been deposited in the DDB/GenBank/EBI Data Bank with accession numbers BankIt1529027 and BankIt1529045. Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession codes 4ELK and 4ELM.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Godfrey DI, Macdonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what's in a name? Nat. Rev. Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 2.Godfrey DI, Rossjohn J, Mccluskey J. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity. 2008;28:304–314. doi: 10.1016/j.immuni.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Zajonc D, Wilson IA. Architecture of CD1 proteins. Curr. Top. Microbiol. Immunol. 2007;314:27–50. doi: 10.1007/978-3-540-69511-0_2. [DOI] [PubMed] [Google Scholar]
- 4.Borg NA, et al. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 5.Pellicci DG, et al. Recognition of β-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat. Immunol. 2011;12:827–834. doi: 10.1038/ni.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu ED, Girardi E, Wang J, Zajonc D. Cutting Edge: Structural Basis for the Recognition of {beta}-Linked Glycolipid Antigens by Invariant NKT Cells. J. Immunol. 2011;187:2079–2083. doi: 10.4049/jimmunol.1101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, et al. The Vα14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J. Exp. Med. 2010;207:2383–2393. doi: 10.1084/jem.20101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellicci DG, et al. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 10.Cardell S, et al. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. J. Exp. Med. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park SH, et al. The mouse CD1d-restricted repertoire is dominated by a few autoreactive T cell receptor families. J. Exp. Med. 2001;193:893–904. doi: 10.1084/jem.193.8.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Exley MA, et al. A major fraction of human bone marrow lymphocytes are Th2-like CD1d-reactive T cells that can suppress mixed lymphocyte responses. J. Immunol. 2001;167:5531–5534. doi: 10.4049/jimmunol.167.10.5531. [DOI] [PubMed] [Google Scholar]
- 13.Exley MA, et al. Cutting edge: Compartmentalization of Th1-like noninvariant CD1d-reactive T cells in hepatitis C virus-infected liver. J. Immunol. 2002;168:1519–1523. doi: 10.4049/jimmunol.168.4.1519. [DOI] [PubMed] [Google Scholar]
- 14.Fuss IJ, et al. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J. Clin. Invest. 2004;113:1490–1497. doi: 10.1172/JCI19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jahng A, et al. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. J. Exp. Med. 2004;199:947–957. doi: 10.1084/jem.20031389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terabe M, Berzofsky JA. NKT cells in immunoregulation of tumor immunity: a new immunoregulatory axis. Trends Immunol. 2007;28:491–496. doi: 10.1016/j.it.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Arrenberg P, Maricic I, Kumar V. Sulfatide-mediated activation of type II natural killer T cells prevents hepatic ischemic reperfusion injury in mice. Gastroenterology. 2011;140:646–655. doi: 10.1053/j.gastro.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang SH, et al. Sulfatide-reactive natural killer T cells abrogate ischemia-reperfusion injury. J. Am. Soc. Nephrol. 2011;22:1305–1314. doi: 10.1681/ASN.2010080815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halder RC, Aguilera C, Maricic I, Kumar V. Type II NKT cell-mediated anergy induction in type I NKT cells prevents inflammatory liver disease. J. Clin. Invest. 2007;117:2302–2312. doi: 10.1172/JCI31602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian L, et al. NKT cells stimulated by long fatty acyl chain sulfatides protect against type 1 diabetes in nonobese diabetic mice. PLoS One. doi: 10.1371/journal.pone.0037771. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arrenberg P, Halder R, Dai Y, Maricic I, Kumar V. Oligoclonality and innate-like features in the TCR repertoire of type II NKT cells reactive to a beta-linked self-glycolipid. Proc. Natl. Acad. Sci. USA. 2010;107:10984–10989. doi: 10.1073/pnas.1000576107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy KC, et al. Involvement of secretory and endosomal compartments in presentation of an exogenous self-glycolipid to type II NKT cells. J. Immunol. 2008;180:2942–2950. doi: 10.4049/jimmunol.180.5.2942. [DOI] [PubMed] [Google Scholar]
- 23.Blomqvist M, et al. Multiple tissue-specific isoforms of sulfatide activate CD1d-restricted type II NKT cells. Eur. J. Immunol. 2009;39:1726–1735. doi: 10.1002/eji.200839001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toda K, et al. Lysosulfatide (sulfogalactosylsphingosine) accumulation in tissues from patients with metachromatic leukodystrophy. J. Neurochem. 1990;55:1585–1591. doi: 10.1111/j.1471-4159.1990.tb04942.x. [DOI] [PubMed] [Google Scholar]
- 25.López-Sagaseta J, Sibener LV, Kung JE, Gumperz J, Adams EJ. Lysophospholipid presentation by CD1d and recognition by a human Natural Killer T-cell receptor. EMBO J. 2012;24:1–13. doi: 10.1038/emboj.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 27.Garcia KC, et al. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 28.Hahn M, Nicholson MJ, Pyrdol J, Wucherpfennig KW. Unconventional topology of self peptide-major histocompatibility complex binding by a human autoimmune T cell receptor. Nat. Immunol. 2005;6:490–496. doi: 10.1038/ni1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu. Rev. Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joyce S, Girardi E, Zajonc D. NKT cell ligand recognition logic: molecular basis for a synaptic duet and transmission of inflammatory effectors. J. Immunol. 2011;187:1081–1089. doi: 10.4049/jimmunol.1001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matulis G, et al. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PLoS Biol. 2010;8:e1000402. doi: 10.1371/journal.pbio.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmer MI, et al. Polymorphisms in CD1d affect antigen presentation and the activation of CD1d-restricted T cells. Proc. Natl. Acad. Sci. USA. 2009;106:1909–1914. doi: 10.1073/pnas.0808476106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zajonc D, et al. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J. Exp. Med. 2005;202:1517–1526. doi: 10.1084/jem.20051625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin JH, et al. Mutation of a Positively Charged Cytoplasmic Motif within CD1d Results in Multiple Defects in Antigen Presentation to NKT Cells. J. Immunol. 2012;188:2235–2243. doi: 10.4049/jimmunol.1100236. [DOI] [PubMed] [Google Scholar]
- 35.Cantu C, Benlagha K, Savage PB, Bendelac A, Teyton L. The paradox of immune molecular recognition of alpha-galactosylceramide: low affinity, low specificity for CD1d, high affinity for alpha beta TCRs. J. Immunol. 2003;170:4673–4682. doi: 10.4049/jimmunol.170.9.4673. [DOI] [PubMed] [Google Scholar]
- 36.Ely LK, et al. Disparate thermodynamics governing T cell receptor-MHC-I interactions implicate extrinsic factors in guiding MHC restriction. Proc. Natl. Acad. Sci. USA. 2006;103:6641–6646. doi: 10.1073/pnas.0600743103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams JJ, et al. T Cell Receptor Signaling Is Limited by Docking Geometry to Peptide-Major Histocompatibility Complex. Immunity. 2011;35:681–693. doi: 10.1016/j.immuni.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu ED, et al. Structural basis for the recognition of C20:2-αGalCer by the invariant natural killer T cell receptor-like antibody L363. J. Biol. Chem. 2012;287:1269–1278. doi: 10.1074/jbc.M111.308783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, et al. Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proc. Natl. Acad. Sci. USA. 2010;107:1535–1540. doi: 10.1073/pnas.0909479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burdin N, et al. Structural requirements for antigen presentation by mouse CD1. Proc. Natl. Acad. Sci. USA. 2000;97:10156–10161. doi: 10.1073/pnas.97.18.10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Girardi E, et al. Unique Interplay between Sugar and Lipid in Determining the Antigenic Potency of Bacterial Antigens for NKT Cells. PLoS Biol. 2011;9:e1001189. doi: 10.1371/journal.pbio.1001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.