Abstract
An extensive, highly diversified multigene family of novel immune-type receptor (nitr) genes has been defined in Danio rerio (zebrafish). The genes are predicted to encode type I transmembrane glycoproteins consisting of extracellular variable (V) and V-like C2 (V/C2) domains, a transmembrane region and a cytoplasmic tail. All of the genes examined encode immunoreceptor tyrosine-based inhibition motifs in the cytoplasmic tail. Radiation hybrid panel mapping and analysis of a deletion mutant line (b240) indicate that a minimum of ≈40 nitr genes are contiguous in the genome and span ≈0.6 Mb near the top of zebrafish linkage group 7. One flanking region of the nitr gene complex shares conserved synteny with a region of mouse chromosome 7, which shares conserved synteny with human 19q13.3-q13.4 that encodes the leukocyte receptor cluster. Antibody-induced crosslinking of Nitrs that have been introduced into a human natural killer cell line inhibits the phosphorylation of mitogen-activated protein kinase that is triggered by natural killer-sensitive tumor target cells. Nitrs likely represent intermediates in the evolution of the leukocyte receptor cluster.
Ever-increasing interest is being directed to the role of innate processes in the immune system (1). Particular attention has been given recently to the group of genes that includes the natural killer cell (NK) receptors that are encoded at the leukocyte receptor cluster (LRC) locus on human chromosome 19q13.4 and similar receptors on mouse chromosome 7. These genes, which include killer Ig-like receptors (KIR), paired Ig-like receptors, Ig-like transcripts (ILT) [leukocyte inhibitory receptors (LIR) and monocyte inhibitory receptors (MIR)], leukocyte-associated inhibitory receptor genes, and NKp46/MAR-1 (2–8), constitute a subset of the Ig gene superfamily (IgSF), and at least some members mediate innate recognition (9). The present understanding of LRC genes is based on studies in mammals; however, it is presumed that they arose early in vertebrate phylogeny.
On the basis of considerations of both structural specialization and genetic complexity, it is possible that the Ig and T cell antigen receptor (TCR) multigene families, which undergo segmental reorganization and mediate adaptive immunity, arose from innate immune precursor receptors that are not associated with somatic reorganization. The ability of the Ig and TCR subset of the IgSF to recognize a multitude of antigens relies on extensive diversity in their variable (V) regions and is achieved through site-specific rearrangement of V, diversity (D), and joining (J) segmental elements as well as through additional somatic variation and ultimately somatic hypermutation. Within the V region, diversity in both families of antigen receptors is concentrated in complementarity determining regions (CDRs); variation in CDR1 and CDR2 is encoded in the germ line, whereas variation in CDR3 originates somatically. No evidence has been found for structural features or somatic reorganization associated with V regions in known members of the LRC.
A third family of diversified V region-containing receptors, termed novel immune-type receptor (NITR) genes, was identified recently in the compact genome of the pufferfish (10). V regions of NITR genes in this species are organized in families, exhibit variation in CDR1 and CDR2, and like Ig and TCR genes encode J regions but neither rearrange nor appear to exhibit other forms of somatic variation (11). In an overall sense, NITRs are structurally similar to certain members of the LRC in that they possess two extracellular Ig domains, a transmembrane region and a cytoplasmic tail, which contains immunoreceptor tyrosine-based inhibition motifs (ITIMs) that function in negative signal transduction pathways. NITRs possess features of both adaptive and innate immune receptors, and their consideration is significant in addressing the evolution of immune function (12).
Despite the utility of the pufferfish genome in terms of the initial identification of NITR genes, this model system is limited in terms of further defining the genetics, developmental regulation, cell lineage-specific expression, and function of NITRs. To address these issues, we have identified and further characterized NITR orthologs, henceforth termed nitr, in the zebrafish (Danio rerio), a well defined developmental model for which increasing amounts of genetic linkage information is available (13–20) and for which complete genomic sequencing is in progress. The highly diverse nitr genes in this species can be categorized into at least four subfamilies, which are all encoded at a single genomic region, which is flanked by regions sharing conserved synteny with two separate loci in human and mouse. One region flanking the nitr gene complex shares conserved synteny with a region of mouse chromosome 7 that shares conserved synteny with human chromosome 19q13, which encodes the genes of the LRC. Like LRC orthologs found in higher vertebrates, Nitrs are able to transduce negative regulatory signals in response to receptor crosslinking.
Materials and Methods
General Methods.
DNA sequencing and the analysis of DNA sequences were carried out as described (10). cDNA libraries were constructed in λ ZAP Express and λ ZAP II (Stratagene) as described (21).
P1 Artificial Chromosome (PAC) Identification and Characterization.
An arrayed zebrafish genomic PAC library (22) was screened as described (23). PAC isolates were restriction digested and biotin end-labeled, followed by hybridization to gene-specific probes as described (24). Southern blotting used a standard protocol.
Chromosomal Mapping.
The T51 RH panel (Research Genetics, Huntsville, AL) (14) and the LN54 RH panel (15) (generous gift from M. Ekker) were used in radiation hybrid panel mapping. All mapping PCRs were done in duplicate.
Unless otherwise stated, mapping data were previously determined (25) or obtained by using the Mouse Genome Database (26) or locuslink software (27) (http://www.ncbi.nlm.nih.gov/LocusLink/). Mouse Tip30 has been identified as a sequence-tagged site (STS) (930601) and placed between D7Mit69 and D7Mit193 (http://www.ncbi.nlm.nih.gov/genome/sts/). Human TIP30 was mapped by ePCR (SHGC-17402) and placed between D11S1755 and D11S1165 with National Center for Biotechnology Information mapviewer software (http://www.ncbi.nlm.nih.gov/genome/sts/).
Recombinant Nitr3 Expression Constructs, Vaccinia Viral Delivery, NITR Crosslinking, and Detection of Active Mitogen-Activated Protein Kinase (MAPK).
A plasmid construct (pcDNA3.1) expressing nitr3.1 was derived in which the translational start site was modified to increase expression (28), and a FLAG tag (29) was introduced carboxyl-terminal to the predicted leader cleavage site and amino-terminal of the V region. Mutations were introduced in the ITIM sequence of nitr3.1 using the Transformer Site-Directed Mutagenesis kit (CLONTECH). Recombinant vaccinia viruses encoding nitr sequences were constructed by using the pSC11 vector in combination with the WR strain of vaccinia; CD56-expressing vaccinia was used as a control. Recombinant vaccinia-treated NK92 cells expressing different nitr3.1 constructs (or mock infected) were incubated for 4 h at 37°C and then analyzed for surface expression of the receptors by flow cytometry with a mouse α-FLAG M2 monoclonal antibody (Sigma). Goat α-mouse IgG Fc was used to stimulate the treated cells. Incubation with Raji tumor cells was for 5 min at 37°C. An α-active MAPK/ERK antibody was used for Western analysis.
Results and Discussion
nitr Gene Subfamilies Are Present in Zebrafish.
Initial efforts to identify nitr genes in zebrafish either by cross-species DNA hybridization or by direct amplification using short primer PCR (21) were unsuccessful. An alternative strategy based on the use of touch-down PCR (30) and degenerate primers, including CODE-HOP primer design (31) with a giant Danio (Danio aequipinnatus) spleen cDNA template, resulted in the amplification of DNA fragments corresponding to the carboxyl-terminal Ig domains of nitr1 (clone 1483) and nitr2 (clone 1889), which are 60% identical to one another. The giant Danio nitr1 and nitr2 probes were hybridized to an arrayed zebrafish PAC library (22). The nitr3 and nitr4 sequence families were identified through genomic sequencing of subclones of nitr1+/nitr2+ PACs. Extensive sequencing of nitr+ PACs was used to identify sequence diversity.
Screening of zebrafish kidney and spleen cDNA libraries with nitr probes yielded nitr1.1, nitr 3.1, and nitr 4.1 cDNAs (Fig. 1). A PCR priming strategy, which was based on degenerate primers complementing the 5′ and 3′ nitr exons, with zebrafish blood and intestine cDNA templates resulted in the identification of (i) several nitr1, nitr2, nitr3, and nitr4 cDNAs, (ii) a potential mRNA splice variant of nitr4, which encodes 19 additional amino acids between the V and V-like C2 (V/C2) domains, and (iii) nitr3r (nitr3-related), a variant of nitr3, which lacks a V/C2 domain (Fig. 1). The recent identification of a homologous nitr gene lacking a V/C2 domain, in another bony fish model system, suggests that nitr genes constitute a more structurally complex range of receptors than recognized initially (N. A. Hawke, J.A.Y., N. Miller, and G.W.L., unpublished observation).
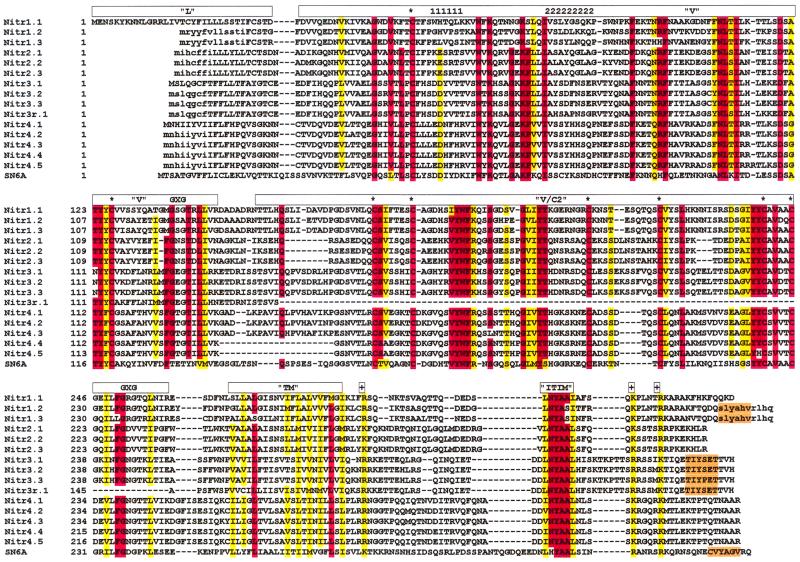
Figure 1.
Sequence identity patterns in Nitrs. clustalw alignment of predicted translation products from zebrafish nitr cDNAs and the representative pufferfish cDNA, SN6A. Leader (L), V, V/C2, transmembrane (TM), and ITIM sequences are indicated above the alignment. A second putative ITIM in Nitr1, Nitr3, and SN6A is highlighted in orange. Identical residues are enclosed in red. Residues that are in similar functional groups (defined in ref. 10) are boxed in yellow. Conserved cysteines are indicated with an asterisk. The glycine bulges of the J domains (GXG) are indicated. Conserved charged residues are indicated (+). CDR1 and CDR2 are indicated by “1111111” and “222222222”, respectively. Residues shown in lowercase letters are predicted from genomic sequence. Note that Nitr4 transcripts encode long (Nitr4.1, Nitr4.2, and Nitr4.3) or short (Nitr4.4 and Nitr4.5) forms, which differ by 19 amino acids between the V and V/C2 exons. The nucleotide identity between members within each nitr subfamily ranges from 80% to 99%.
Conserved Features of Nitrs.
An alignment of the predicted peptide sequence of representative members of the Nitr subfamilies and the prototypic NITR from pufferfish is shown in Fig. 1. The overall identities between cDNA sequences of prototypic members of the four families of nitr genes range from 51% to 54%. Eliminating the pufferfish sequence (SN6A) from the comparison set does not substantially change the overall identity profile. The FGXG peptide motif, a conserved feature of Ig and TCR J regions (32) as well as pufferfish NITRs (10), is found in the V domains of Nitr3 and Nitr4 in zebrafish; a GXG motif is present in the V domains of Nitr1. An FGXG motif is present in the V/C2 domains of D. rerio Nitr1, Nitr3, and Nitr4.
A single positive-charged residue is located adjacent to the transmembrane region in the cytoplasmic tail in all four zebrafish Nitrs as well as in SN6A in pufferfish. The function of this residue is unknown, but its position differs from that of the intramembrane-charged residues of TCRs and activating receptors of the LRC (9, 33). The overall extent of intergenic relatedness between the cytoplasmic regions of Nitrs is reduced markedly relative to that seen in the V, V/C2, and the transmembrane regions; each zebrafish Nitr possesses one of five different ITIM consensus sequences; a second putative ITIM-related sequence is located more carboxyl-terminal in Nitr1 and Nitr3. However, in genes encoded by the LRC, the majority of inhibitory function is associated with the amino-terminal, consensus ITIM (9). The absolute conservation of two positively charged residues in the Nitrs, which are carboxyl-terminal to the consensus ITIM, is potentially significant.
A Complex Multigenic Family Encodes Nitrs.
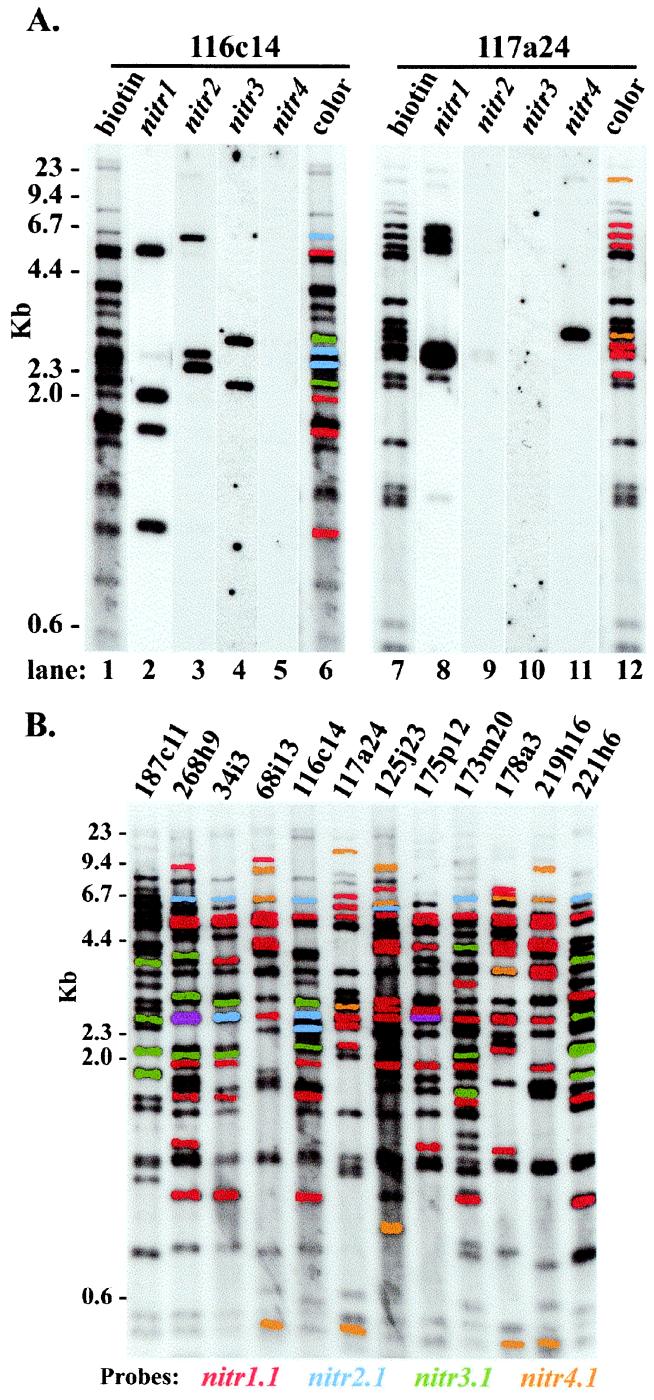
To further characterize the nitr multigene family, 12 of the 24 nitr PACs were selected on the basis of different patterns of restriction fragments and were hybridized with probes complementing the four subfamilies of nitr genes. The probe-specific hybridization patterns for two representative PACs are shown in Fig. 2A. Comparison of these patterns to 10 additional PACs is illustrated in Fig. 2B. Collectively, the hybridization patterns are most consistent with a minimal estimate of ≈22 nitr1, four nitr2, nine nitr3 (including nitr3r.1), and three nitr4 genes. Subcloning and sequencing of various representative nitr+ PACs indicate that the leader, V, V/C2, transmembrane, and two cytoplasmic regions are encoded in separate exons, distinguishing these genes from the pufferfish orthologs, which lack an intron between V and V/C2 (10). Absolute sequence identities between both zebrafish and pufferfish NITRs and other immune receptors, including LRC family members, are not adequate for a meaningful molecular phylogenetic comparison.
Figure 2.
Characterization of nitr-containing P1 artificial chromosomes. (A) Zebrafish nitr PAC clones 116c14 and 117a24 were digested with HindIII, 5′ end-labeled with biotin, electrophoresed, and transferred to a nylon membrane. Restriction fragments were identified by a streptavidin–enzyme-linked chemiluminescent assay (lanes 1 and 7). The membrane was stripped and hybridized sequentially with probes complementing the four different subfamilies of nitr genes (lanes 2–5 and 8–11). The fragments that hybridized to each probe are digitally color-coded on the original image (lanes 6 and 12); color-coding can be discerned from coding in B. (B) Similar analyses were performed for the 12 PACs that exhibit the greatest number of banding differences and are presumed to correspond to different regions of the extended nitr gene complex. Purple bands reflect hybridization with both nitr1 and nitr3 probes. Color-coding corresponding to different nitr subfamilies is indicated at the bottom of B and also corresponds to lanes 6 and 12 in A.
Nitrs Are Encoded in an Extended Chromosomal Region of Linkage Group 7.
STSs for the nitr encoding PACs 116c14, 173m20, 117a24, and 219h16 were mapped to linkage group 7 (LG7) with the T51 RH panel (14); 116c14, 173m20, and 117a24 also were mapped to LG7 with the LN54 RH panel (15) (Fig. 3A). The zebrafish deficiency strain b240 has deleted a large region of LG7, including markers that flank the nitr region (34). Genomic DNA from b240 was amplified along with wild-type control DNA using various STS primer sets that are specific to the nitr region. The lack of product formation from the genome of b240 with various nitr-linked STSs is consistent with nitrs occupying a single region on LG7 (Fig. 3B). The b240 deficiency line is embryonic lethal and thus not informative from a physiological standpoint (A. Fritz, personal communication).
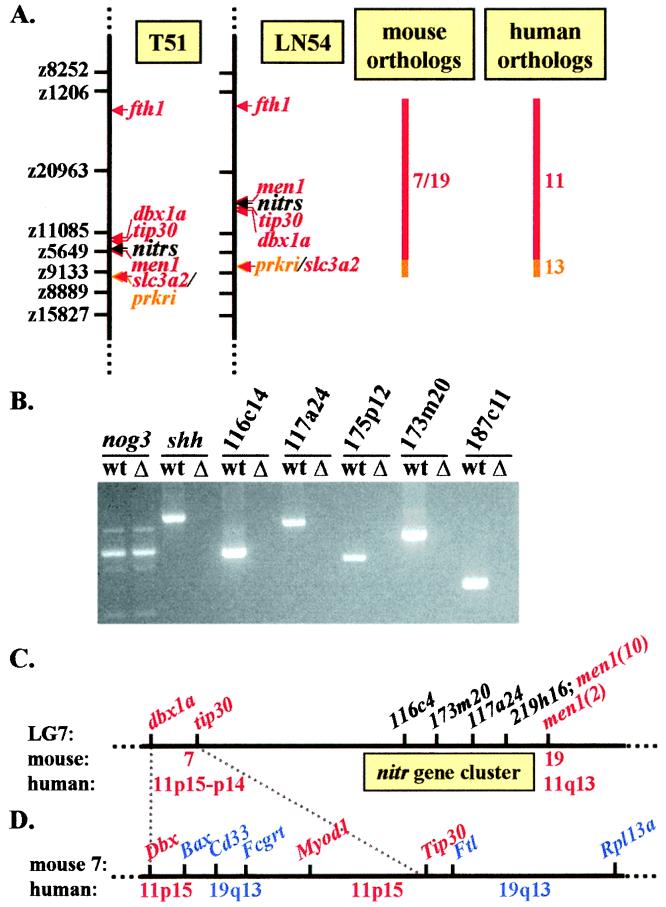
Figure 3.
Characterization of the nitr genomic region. (A) Intronic STSs from nitr-encoding PACs were mapped to LG7 with the T51 RH panel (14) and the LN54 RH panel (15). The regions of shared synteny between LG7 and the human and mouse genomes are shown. Mapping positions of genes shown have been published (25) except for tip30, which was identified for this study. (B) PCR analysis of nitr STSs in wild type (wt) and the b240 deficiency strain zebrafish (Δ). The positive control is noggin (nog3) (50), which is retained, and the negative control is sonic hedgehog (shh), which is deleted in b240 (A. Fritz, personal communication). Zebrafish strain b240 has lost a large region of LG7 and does not encode sequences from PACs 116c14, 117a24, 175p12, 173m20, and 187c11, strongly supporting the hypothesis that all of the nitr genes are located on LG7. (C) The nitr gene complex on LG7 is flanked by the men1 and tip30 genes in both RH panels. Exon 10 of the men1 gene has been mapped to one end of the nitr gene complex in both RH panels. Exon 2 of the men1 gene has been mapped 33 centiRays distal to the nitr genes on the T51 RH panel. A nitr-encoding PAC was identified that also encodes the 3′ end of the men1 gene placing the nitr gene complex within 120 kb of men1. The figure shown is not to scale and merges position data from both RH panels. Corresponding regions of the mouse and human genomes are shown below the line. (D) Multiple genes from the Tip30/Dbx region on mouse chromosome 7 are shown. Corresponding regions of the human genome are shown below the line. Note that this region of mouse chromosome 7 shares conserved synteny with human 11p15 and 19q13.
The nitr gene complex spans at least 10 centiRays on the T51 RH panel, which corresponds to ≈600 kb (14). Both RH panels place the nitr gene complex between men1 and expressed sequence tag (fb15b04). A cDNA corresponding to this expressed sequence tag was identified and shown to be ≈63% identical to mouse and human TIP30 (data not shown) and is designated tip30 (Fig. 3C) (35). Mapping assignments on both RH panels place one end of the nitr gene complex at the same position as the 3′ end of the men1 gene. A single zebrafish PAC was identified (150L22), which encodes the 3′ end of men1 and at least one nitr gene, physically linking these loci within ≈110 kb (data not shown). Human MEN1 is found on chromosome 11q13, whereas the human orthologs corresponding to the other side of the nitr gene complex, DBX and TIP30, are found on 11p15-p14 (25). The involvement of 11q13 in translocations in multiple malignancies suggests that it is not stable (36), and the fact that it shares conserved synteny with two loci in mouse argues that the nitr/men1 linkage observed in zebrafish may not be shared across phyla. In any case, no genes have been identified in the 11q13 or 11p15-p14 regions that resemble nitr genes.
Dbx and Tip30 map to 22.8 cM and 24.5 cM, respectively, on mouse chromosome 7. The human orthologs of the genes that map between mouse Dbx and Tip30 are encoded on either 11p15 or chromosome 19, of which most map to 19q13.3-q13.4, which encodes the LRC (Fig. 3D). Taken together, these findings suggest a distant evolutionary link between the nitr gene complex and the LRC. These data should be interpreted with caution as additional genes may be found to map between the nitr gene complex and the dbxla/tip30 locus.
Finally, it has been proposed that zebrafish LG7 is derived from an ancient chromosome that fragmented to become the modern human chromosomes 11, 15, and 19q (37); the only multigene transmembrane Ig family found on these human chromosomes includes the genes at 19q13.3–19q13.4, further supporting that the nitr gene complex is evolutionarily linked to the human LRC.
Recombinant Nitr3 Transduces Negative Regulatory Signals in Mammalian NK Cells.
As discussed above, despite the presence of a V region, the Nitrs share a number of overall structural features with the inhibitory members of the LRC locus, such as KIRs. Upon ligand binding and subsequent tyrosine phosphorylation, KIR ITIMs can associate with the protein-tyrosine phosphatases SHP-1 and SHP-2 (9). SHP proteins are responsible for dephosphorylating secretory effector substrates involved in the NK lytic pathway. One key effector molecule is MAPK/ERK, which critically regulates NK function toward the tumor target (38–40). Expression of a KIR in NK cells has been shown to down-regulate MAPK activation by tumor ligand, leading to loss of NK function (41).
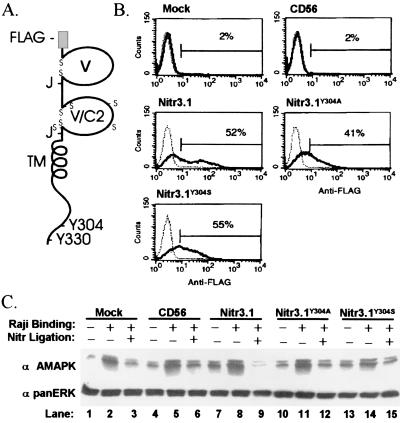
FLAG tag-containing Nitr3.1 (Fig. 4A) as well as Nitr3.1 mutants, in which the tyrosine in the ITIM motif (Tyr-304) is substituted with alanine (Nitr3.1Y304A) or serine (Nitr3.1Y304S), were used to infect NK92 cells (Fig. 4B). Addition of Raji tumor target cells to NK92 cells expressing mock, CD56 (an unrelated protein), Nitr3.1, or the two Nitr3.1 mutants induced active MAPK within 5 min (Fig. 4C, lanes 2, 5, 8, 11, and 14). The activation of MAPK by Raji tumor cells was blocked by crosslinking of Nitr3.1 (Fig. 4C, lane 9), whereas crosslinking of Nitr3.1 with mutated ITIMs had little blocking effect (Fig. 4C, lanes 12 and 15). The results indicate that Nitrs, at the density present in the transfected NK92 cells, can function as negative regulators of MAPK and that this function is controlled at the level of the ITIM motif.
Figure 4.
Inhibition of MAPK activation in NK cells. (A) Predicted protein structure of recombinant Nitr3.1; tyrosines 304 and 330 are indicated as Y304 and Y330, respectively. (B) Human NK92 cells, mock-infected or infected with vaccinia virus carrying CD56, FLAG-tagged Nitr3.1, FLAG-tagged mutant Nitr3.1Y304A, or FLAG-tagged Nitr3.1Y304S, were analyzed for surface expression of the receptors by flow cytometry. (C) The same NK92 cells were then incubated with Raji tumor cells before lysis and Western analysis for active MAPK. The membrane was stripped and probed for panERK to verify equal loading; triplicate determinations verified the findings.
Nitrs Are a Unique Subset of the IgSF.
Nitrs possess a number of unique features that distinguish them from other members of the IgSF. Specifically, the combination of V and V/C2 extracellular domains is comparable to both CD22 (33) and CTX/CTH/CTM (42), which possess V and C2 domains but represent single copy genes. Two extracellular Ig domains, a transmembrane and an ITIM-containing cytoplasmic tail found in Nitrs, also are present in KIRs, ILTs, FcγRIIB, gp49B1, CDW150, as well as CD33 (a member of the siglec family) (9, 43). The KIRs, ILTs, and CD33 represent multigene families; however, the unusual nature of the amino-terminal extracellular domain in CD33 (33) makes it a less attractive potential ortholog of a Nitr than do either KIRs or ILTs. The presence of V and J sequences as well as the overall predicted structures of Nitrs resembles TCRs (33); however, TCRs possess a C1-type extracellular domain, undergo recombination, and lack ITIMs. Whereas the variation in the structure of the amino-terminal exons distinguish the nitrs from the LRC genes, the overall genomic organization and general features of protein architecture of the nitrs most resemble the mammalian KIRs and ILT (LIR/MIR).
Evolutionary Significance of the Nitrs.
Ig, TCR, MHC I, and MHC II as well as other genes that function in immune recognition have been identified in zebrafish in forms resembling their mammalian orthologs (44, 45). The organization of the nitr gene complex, as well as the general architecture of predicted Nitr proteins (notwithstanding the differences noted above) and their inhibitory function, is most like the gene products of the mammalian LRC. It could be argued that the differences in structures of the extracellular domains found in NITRs distinguish them from LRC genes; however, these regions in certain members of the LRC differ as extensively from genes such as KIRs as do the NITRs.
Large-scale genomic sequencing offers the greatest promise for identifying genetic features that may shed light on the patterns of divergence of multigene families. Investigations of shared synteny between mammals and zebrafish have established that certain chromosomal orders have been maintained over the long period since these species shared a common ancestor. Based on the mapping conducted here, it seems likely that putative human nitr orthologs would be found on chromosomes 11 or 19q. The current resolution of the available zebrafish RH panels falls short of allowing an unequivocal assignment of absolute synteny; however, the mapping data are consistent with a potential relatedness between the LRC and nitr genes. If the Nitrs are distant orthologs of contemporary LRC molecules, their high level of variation well exceeds that described for individual LRC gene families in mammals (3) but certainly does not exceed the variation between the LRC gene families. The relationships between nitr genes and various LRC genes could be better defined by identifying (and mapping) additional LRC orthologs in bony fish; however, even this type of finding is confounded by the extensive sequence variation that has been demonstrated for some LRC multigene families between closely related species (primates) (46) and two haplotypes from a single individual (3).
If the nitr genes are LRC orthologs, are they highly derived representatives or do they reflect the nature of the common progenitor from which the rearranging antigen binding receptors and contemporary LRC gene family evolved? One argument in favor of the latter hypothesis comes in the generally accepted view that the precursor molecule from which Ig and TCR genes evolved encoded contiguous V and J regions and was the target for RAG-mediated transposition events (47, 48). The identity of the J-like regions in the Nitrs in both pufferfish (10) and the orthologous forms described here emphasizes their common phylogenetic origin and apparent significance. Despite the obvious complexity of reconstructing a molecular phylogeny for the highly diversified LRCs, a model can be proposed in which an nitr precursor sustained the loss of cytoplasmic ITIM-containing exons, acquired a C1 extracellular domain, and became a target for this critical transposition event, which ultimately gave rise to a rearranging MHC-independent TCR-like system (the origin of which at present only can be traced to the same jawed vertebrate ancestors). Irrespective of the origins of the V association, it is notable that the extraordinary variation between the extracellular domains of LRC members (including the V-containing Nitrs) is countered by the conserved features of the transmembrane and cytoplasmic regions. It is possible that the nitr genes and various inhibitory LRC receptors reflect the association of different extramembrane domains with a common transmembrane structure and inhibitory signal transduction mechanism.
Further resolution of the role of Nitrs in immune function will be facilitated by continuing studies of their functional properties as well as by identifying potential ligands, determining their expression patterns during development, using hematopoietic mutant animals, and refining methods to search for short sequence motifs (49) in the rapidly resolving genomic sequence databases of higher vertebrates. Notwithstanding their obvious significance from a phylogenetic perspective, the nitr genes may prove critical in understanding one aspect of the divergence of adaptive and innate function.
Acknowledgments
We thank Dr. C. Amemiya for zebrafish PACs, Dr. M. Ekker for the LN54 RH panel, Dr. R. Haire for cDNA libraries, Dr. H.G. Klingeman for the NK92 cell line, and Drs. S. Chandrasekarappa and G. Otto (Terry Fox Laboratory, Vancouver, BC, Canada) for sharing information about the zebrafish men1 locus before publication. The b240 genomic DNA and the shh and nog3 primers were a generous gift from Drs. A. Fritz (Emory University, Atlanta, GA) and M. Halpern (Carnegie Institution of Washington, Baltimore, MD), who were supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant R01 DK55390-01. This work was supported by National Institutes of Health Grant R37 AI23338 and a grant from The Pediatric Cancer Foundation (to G.W.L.). J.A.Y. was supported by a fellowship from the H. Lee Moffitt Cancer Center and Research Institute and by National Institutes of Health Grant F32 GM20231.
Abbreviations
- CDR
complementarity determining region
- IgSF
Ig gene superfamily
- ILT
Ig-like transcripts
- ITIM
immunoreceptor tyrosine-based inhibition motif
- KIR
killer Ig-like receptors
- LRC
leukocyte receptor cluster
- MAPK
mitogen-activated protein kinase
- NK
natural killer
- nitr
novel immune-type receptor
- STS
sequence-tagged site
- TCR
T cell antigen receptor
- PAC
P1 artificial chromosome
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF318390 (clone 1483), AF318391 (clone 1889), AF318392 (nitr1.1), AF318393 (nitr1.2), AF318394 (nitr1.3), AF318395 (nitr2.1), AF318396 (nitr2.2), AF318397 (nitr2.3), AF318398 (nitr3.1), AF318399 (nitr3.2), AF318400 (nitr3.3), AF318401 (nitr3r.1), AF318402 (nitr4.1), AF318403 (nitr4.2), AF318404 (nitr4.3), AF318405 (nitr4.4), AF318406 (nitr4.5), and AF329850 (tip30)]. The map position and annotation of the described genes have been deposited in the ZFIN database (51) (http://zfin.org/ZFIN/).
References
- 1.Medzhitov R, Janeway C A J. Cell. 1997;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 2.Wende H, Colonna M, Ziegler A, Volz A. Mamm Genome. 1999;10:154–160. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 3.Wilson M J, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Proc Natl Acad Sci USA. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. . (First Published April 18, 2000, 10.1073/pnas.080588597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubagawa H, Cooper M D, Chen C C, Ho L H, Alley T L, Hurez V, Tun T, Uehara T, Shimada T, Burrows P D. Curr Top Microbiol Immunol. 1999;244:137–149. doi: 10.1007/978-3-642-58537-1_12. [DOI] [PubMed] [Google Scholar]
- 5.Moretta L, Biassoni R, Bottino C, Mingari M C, Moretta A. Immunol Today. 2000;21:420–422. doi: 10.1016/s0167-5699(00)01673-x. [DOI] [PubMed] [Google Scholar]
- 6.Ravetch J V, Lanier L L. Science. 2000;290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 7.Biassoni R, Pessino A, Bottino C, Pende D, Moretta L, Moretta A. Eur J Immunol. 1999;29:1014–1020. doi: 10.1002/(SICI)1521-4141(199903)29:03<1014::AID-IMMU1014>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 8.Colonna M, Moretta A, Vely F, Vivier E. Immunol Today. 2000;21:428–431. doi: 10.1016/s0167-5699(00)01697-2. [DOI] [PubMed] [Google Scholar]
- 9.Blery M, Olcese L, Vivier E. Hum Immunol. 2000;61:51–64. doi: 10.1016/s0198-8859(99)00157-3. [DOI] [PubMed] [Google Scholar]
- 10.Strong S J, Mueller M G, Litman R T, Hawke N A, Haire R N, Miracle A L, Rast J P, Amemiya C T, Litman G W. Proc Natl Acad Sci USA. 1999;96:15080–15085. doi: 10.1073/pnas.96.26.15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoder J A, Litman G W. Curr Top Microbiol Immunol. 2000;248:271–284. doi: 10.1007/978-3-642-59674-2_12. [DOI] [PubMed] [Google Scholar]
- 12.Barclay A N. Proc Natl Acad Sci USA. 1999;96:14672–14674. doi: 10.1073/pnas.96.26.14672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dooley K, Zon L I. Curr Opin Genet Dev. 2000;10:252–256. doi: 10.1016/s0959-437x(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 14.Geisler R, Rauch G-J, Baier H, van Beber F, Bross L, Dekens M P S, Finger K, Fricke C, Gates M A, Geiger H, et al. Nat Genet. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- 15.Hukriede N A, Joly L, Tsang M, Miles J, Tellis P, Epstein J A, Barbazuk W B, Li F N, Paw B, Postlethwait J H, et al. Proc Natl Acad Sci USA. 1999;96:9745–9750. doi: 10.1073/pnas.96.17.9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbazuk W B, Korf I, Kadavi C, Heyen J, Tate S, Wun E, Bedell J A, McPherson J D, Johnson S L. Genome Res. 2000;10:1351–1358. doi: 10.1101/gr.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimoda N, Knapik E W, Ziniti J, Sim C, Yamada E, Kaplan S, Jackson D, de Sauvage F, Jacob H, Fishman M C. Genomics. 1999;58:219–232. doi: 10.1006/geno.1999.5824. [DOI] [PubMed] [Google Scholar]
- 18.Gates M A, Kim L, Egan E S, Cardozo T, Sirotkin H I, Dougan S T, Lashkari D, Abagyan R, Schier A F, Talbot W S. Genome Res. 1999;9:334–347. [PubMed] [Google Scholar]
- 19.Woods I G, Kelly P D, Chu F, Ngo-Hazelett P, Yan Y-L, Huang H, Postlethwait J H, Talbot W S. Genome Res. 2000;10:1903–1914. doi: 10.1101/gr.10.12.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ton C, Hwang D M, Dempsey A A, Tang H-C, Yoon J, Lim M, Mably J D, Fishman M C, Liew C-C. Genome Res. 2000;10:1915–1927. doi: 10.1101/gr.10.12.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rast J P, Litman G W. Proc Natl Acad Sci USA. 1994;91:9248–9252. doi: 10.1073/pnas.91.20.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amemiya C T, Zon L I. Genomics. 1999;58:211–213. doi: 10.1006/geno.1999.5827. [DOI] [PubMed] [Google Scholar]
- 23.Amemiya C T, Ota T, Litman G W. In: Construction of P1 Artificial Chromosome (PAC) Libraries from Lower Vertebrates. Lai E, Birren B, editors. New York: Academic; 1996. pp. 223–256. [Google Scholar]
- 24.Ota T, Amemiya C T. Genet Anal. 1996;12:173–178. [PubMed] [Google Scholar]
- 25.Yoder J A, Litman G W. Gene. 2000;261:235–242. doi: 10.1016/s0378-1119(00)00503-5. [DOI] [PubMed] [Google Scholar]
- 26.Blake J A, Eppig J T, Richardson J E, Bult C J, Kadin J A Mouse Genome Database Group. Nucleic Acids Res. 2001;29:91–94. doi: 10.1093/nar/29.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruitt K D, Maglott D R. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak M. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 29.Chubet R G, Brizzard B L. BioTechniques. 1996;20:136–141. doi: 10.2144/96201pf01. [DOI] [PubMed] [Google Scholar]
- 30.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose T M, Schultz E R, Henikoff J G, Pietrokovski S, McCallum C M, Henikoff S. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chothia C, Novotny J, Bruccoleri R, Karplus M. J Mol Biol. 1985;186:651–663. doi: 10.1016/0022-2836(85)90137-8. [DOI] [PubMed] [Google Scholar]
- 33.Barclay A N, Brown M H, Law S K A, McKnight A J, Tomlinson M G, van der Merwe P A. The Leucocyte Antigen FactsBook. San Diego: Academic; 1997. [Google Scholar]
- 34.Fritz A, Rozowski M, Walker C, Westerfield M. Genetics. 1996;144:1735–1745. doi: 10.1093/genetics/144.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao H, Tao Y, Greenblatt J, Roeder R G. Proc Natl Acad Sci USA. 1998;95:2146–2151. doi: 10.1073/pnas.95.5.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong K F. Cancer Genet Cytogenet. 1999;113:93–95. doi: 10.1016/s0165-4608(98)00285-4. [DOI] [PubMed] [Google Scholar]
- 37.Postlethwait J H, Woods I G, Ngo-Hazelett P, Yan Y-L, Kelly P D, Chu F, Huang H, Hill-Force A, Talbot W S. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- 38.Henkart P A. Immunity. 1994;1:343–346. doi: 10.1016/1074-7613(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 39.Wei S, Gamero A M, Liu J H, Daulton A A, Valkov N I, Trapani J A, Larner A C, Weber M J, Djeu J Y. J Exp Med. 1998;187:1753–1765. doi: 10.1084/jem.187.11.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei S, Gilvary D L, Corliss S, Sebti S, Sun J, Straus D B, Leibson P J, Trapani J A, Hamilton A D, Weber M J, Djeu J Y. J Immunol. 2000;165:3811–3819. doi: 10.4049/jimmunol.165.7.3811. [DOI] [PubMed] [Google Scholar]
- 41.Zingoni A, Palmieri G, Morrone S, Carretero M, Lopex-Botel M, Piccoli M, Frati L, Santoni A. J Immunol. 2000;30:644–651. doi: 10.1002/1521-4141(200002)30:2<644::AID-IMMU644>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 42.Chretien I, Marcuz A, Courtet M, Katevuo K, Vainio O, Heath J K, White S J, Du Pasquier L. Eur J Immunol. 1998;28:4094–4104. doi: 10.1002/(SICI)1521-4141(199812)28:12<4094::AID-IMMU4094>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 43.Angata T, Varki A. J Biol Chem. 2000;275:22127–22135. doi: 10.1074/jbc.M002775200. [DOI] [PubMed] [Google Scholar]
- 44.Haire R N, Rast J P, Litman R T, Litman G W. Immunogenetics. 2000;51:915–923. doi: 10.1007/s002510000229. [DOI] [PubMed] [Google Scholar]
- 45.Bingulac-Popovic J, Figueroa F, Sato A, Talbot W S, Johnson S L, Gates M, Postlethwait J H, Klein J. Immunogenetics. 1997;46:129–134. doi: 10.1007/s002510050251. [DOI] [PubMed] [Google Scholar]
- 46.Khakoo S I, Rajalingam R, Shum B P, Weidenbach K, Flodin L, Muir D G, Canavez F, Cooper S L, Valiante N M, Lanier L L, Parham P. Immunity. 2000;12:687–698. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- 47.Agrawal A, Eastman Q M, Schatz D G. Nature (London) 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 48.Hiom K, Melek M, Gellert M. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 49.Hawke N A, Yoder J A, Litman G W. Immunogenetics. 1999;50:124–133. doi: 10.1007/s002510050588. [DOI] [PubMed] [Google Scholar]
- 50.Bauer H, Meier A, Hild M, Stachel S, Economides A, Hazelett D, Harland R M, Hammerschmidt M. Dev Biol. 1998;204:488–507. doi: 10.1006/dbio.1998.9003. [DOI] [PubMed] [Google Scholar]
- 51.Sprague J, Doerry E, Douglas S, Westerfield M ZFIN Group. Nucleic Acids Res. 2001;29:87–90. doi: 10.1093/nar/29.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]