Abstract
Inositol phosphates are implicated in the regulation of autophagy; however, the exact role of each inositol phosphate species is unclear. In this study, we systematically analyzed the highly conserved inositol polyphosphate synthesis pathway in S. cerevisiae for its role in regulating autophagy. Using yeast mutants that harbored a deletion in each of the genes within the inositol polyphosphate synthesis pathway, we found that deletion of KCS1, and to a lesser degree IPK2, led to a defect in autophagy. KCS1 encodes an inositol hexakisphosphate/heptakisposphate kinase that synthesizes 5-IP7 and IP8; and IPK2 encodes an inositol polyphosphate multikinase required for synthesis of IP4 and IP5. We characterized the kcs1Δ mutant strain in detail. The kcs1Δ yeast exhibited reduced autophagic flux, which might be caused by both the reduction in autophagosome number and autophagosome size as observed under nitrogen starvation. The autophagy defect in kcs1Δ strain was associated with mislocalization of the phagophore assembly site (PAS) and a defect in Atg18 release from the vacuole membrane under nitrogen deprivation conditions. Interestingly, formation of autophagosome-like vesicles was commonly observed to originate from the plasma membrane in the kcs1Δ strain. Our results indicate that lack of KCS1 interferes with proper localization of the PAS, leads to reduction of autophagosome formation, and causes the formation of autophagosome-like structure in abnormal subcellular locations.
Keywords: Kcs1, S. cerevisiae, autophagy, electron microscopy, fluorescence microscopy, inositol polyphosphate
Introduction
Inositol polyphosphates are a unique class of highly conserved molecules that have multiple functions within both the yeast and mammalian cells. These molecules can act as signaling components.1 Inositol polyphosphates can regulate protein localization by binding to proteins containing PH-domains as shown during the inhibition of AKT1 activation or during chemotaxis.2-4 They also play a role in endocytosis, apoptosis and telomere elongation.5-9 In addition, the high-energy phosphates can phosphorylate proteins without the involvement of protein kinases.10,11 The inositol polyphosphate synthesis pathway begins with inositol 1,4,5 triphosphate (Ins(1,4,5)P3), a product generated from phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) by phospholipase C.12,13 The sequential phosphorylation of Ins(1,4,5)P3 by the enzyme Arg82/Ipk2 results in inositol tetrakisphosphate (IP4) and inositol pentakisphosphate (IP5).14,15 The pathway continues with additional phosphates being placed on IP5 by Ipk1, resulting in formation of IP6. IP6 is further phosphorylated by Kcs1 to form 5-IP7 (pyrophosphate on the 5 position) or by a different inositol hexakisphosphate kinase Vip1 to form e-IP7 (where “e” denotes a pyrophosphate on an “equivalent” ring position at the 1/3 or 4/6 but not the 5 or 2 positions).15 The pathway is summarized in Figure 1A.
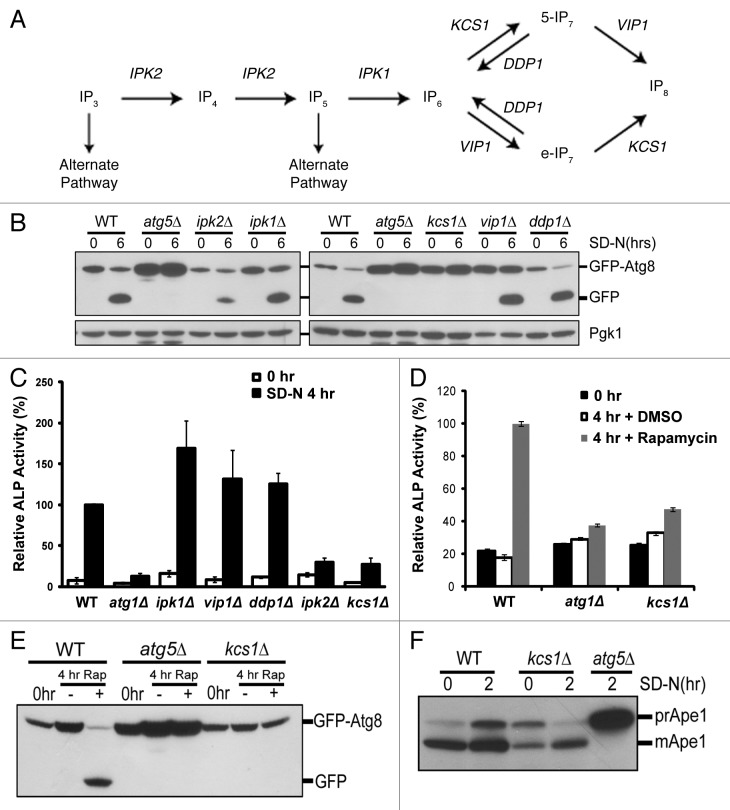
Figure 1. Deletion of genes required for generation of inositol polyphosphates reduces autophagic degradation under nitrogen starvation. (A) The inositol polyphosphate pathway. (B) Immunoblotting assay for GFP-Atg8 processing upon nitrogen deprivation. The inositol polyphosphate mutant strains (ipk2Δ, ipk1Δ, kcs1Δ, vip1Δ, ddp1Δ) as well as control wild-type and atg5Δ strains were transformed with the pCuGFP-Atg8(416) expression plasmid. Cells were grown to mid-log phase in minimal medium (0 h) followed by nitrogen starvation with synthetic dextrose medium minus nitrogen for six h (6 h SD-N). Mid-log phase cells (0 h) or cells that underwent six-hour nitrogen deprivation (6 h SD-N) were analyzed for vacuolar processing of the GFP fusion protein using immunoblotting analysis with an antibody against GFP. The levels of Pgk1 served as loading controls. (C) Vacuolar alkaline phosphatase (ALP) assay to quantify autophagy activity upon nitrogen starvation. Wild-type (WT) and inositol polyphosphate mutant strains expressing Pho8Δ60 were grown in YPD and shifted to SD-N for 4 h. Samples were collected and protein extracts were assayed for alkaline phosphatase activity. The value of SD-N 4 h in the wild-type strain was set to 100% (D) Vacuolar alkaline phosphatase assay to quantify autophagy activity upon rapamycin treatment. Wild-type (WT), atg1Δ (negative control) and the kcs1Δ cells expressing Pho8Δ60 were grown in YPD to early log phase (0 h) and treated with rapamycin (0.2 μg/ml, dissolved in DMSO) for 4 h. Samples were collected and protein extracts were assayed for phosphatase activity. The value for the wild-type strain was set to 100%. (E) Immunoblotting assay for GFP-Atg8 processing upon rapamycin (Rap) treatment. Wild-type (WT), atg5Δ (negative control), and the kcs1Δ mutant transformed with pCuGFP-Atg8(416) were grown to early log phase (0 h) in minimal medium and treated with the TOR inhibitor rapamycin (0.2 μg/ml, dissolved in DMSO) for 4 h. Strains were analyzed for vacuolar processing of the GFP fusion protein using an antibody against GFP. (F) Immunoblotting assay for Ape1 processing. Wild-type, kcs1Δ and atg5Δ cells were grown to mid log phase and starved for 2 h. Samples were collected and protein extracts were analyzed by western analysis with the Ape1 antibody. These data are representative results from three independent experiments.
Intracellular degradation of cytoplasm, proteins and organelles through the lysosome is governed by a process known as autophagy, which is hallmarked by formation of double-membrane vesicles known as autophagosomes. Cellular components are engulfed in this vesicle and are delivered to the vacuole/lysosome for deconstruction. The process of autophagy can be broken into six unique phases. These phases include: initiation, expansion of a double membrane, closure of the double membrane to form the autophagosome, trafficking toward the vacuole, docking and fusion with the vacuole, and breakdown.16,17 These steps are governed by a set of four core complexes. The four complexes include: (1) The initiation complex of Atg1, Atg13 and Atg17; (2) the PtdIns3K complex Vps34, Vps15, Atg6 and Atg14; (3) the ubiquitin-like protein conjugation systems of Atg12–Atg5-Atg16 and Atg8; and finally (4) the cycling system of Atg9.18 In the yeast system, multiple signaling pathways converge on the initiation complex of the autophagy pathway. These pathways include TOR,19 Pho85,20,21 Ras/PKA,22 Sch9,23 Snf1,21 Sic1 and Gcn2.24 Each of these pathways is controlled by a different set of molecules that represent the energy and nutrient status of the cell. Phosphatidylinositol 3-phosphate (PtdIns3P) is an essential component of autophagy; failure to generate it in vps34Δ yeast completely abolishes autophagic activity.25 PtdIns3P has been shown to be necessary for the double-membrane expansion in addition to the proper localization of lipid binding domain proteins Atg18 and Atg2.26-28 Improper localization of Atg18 leads to deficient autophagy.27-29 A few inositol polyphosphate species have been reported to be involved, or inferred, in regulating autophagy.30,31 Ins(1,4,5)P3, which is the initial molecule for the inositol polyphosphate synthesis pathway, has been shown to inhibit autophagy. Using inhibitors of prolyl oligopeptidase or adding myo-inositol to increase Ins(1,4,5)P3 levels caused a reduction in autophagy activity.32 In addition, it was documented in a yeast genomic screen for mitophagy mutants that yeast containing a deletion within the ARG82/IPK2 locus failed to induce mitophagy, a selective form of autophagy.33 More recently, it has been documented in mammalian cells that siRNA knockdown of Ins(P)6Ks decrease autophagy while overexpression increases autophagy.30 However, a systematical study on the function of each of the inositol polyphosphates on autophagy has not been performed.
In yeast, the entire inositol polyphosphate synthesis pathway has been identified (Fig. 1A). This allows us to systematically determine the function of each gene on autophagy. In the present study, we characterized the complete set of deletion mutant strains in the inositol polyphosphate synthesis pathway. We identified two genes, KCS1 and IPK2, which are important for normal autophagy activation under nitrogen deprivation. The kcs1Δ strain was further characterized in detail. To our knowledge, this is the first time a complete genetic analysis on the inositol polyphosphate pathway has been conducted to determine its role in governing autophagy in yeast.
Results
Deletion of KCS1 or IPK2 leads to defect in autophagic degradation
To determine the possible role of the various inositol polyphosphates in autophagy regulation, we initially screened through the entire yeast inositol polyphosphate synthesis pathway by analyzing deletion mutants that are defective for inositol polyphosphate production. The yeast strains ipk2Δ, ipk1Δ, kcs1Δ, vip1Δ and ddp1Δ were transformed with the GFP-Atg8 fusion construct and analyzed for general autophagic flux by monitoring the processing of GFP-Atg8 during nitrogen starvation induced autophagy.34 During autophagy activation, Atg8 is cleaved, conjugated to phosphatidylethanolamine, and translocated to the membrane of autophagosomes. GFP-Atg8 is part of the inner membrane of the completed autophagosome. Upon translocation and fusion with the vacuole, GFP-Atg8 is degraded. Since the GFP molecule is more resistant to vacuolar proteases, the accumulation of GFP reflects autophagic flux. Yeast were grown to mid-log phase in minimal media and shifted to starvation conditions (SD-N) for 6 h. As shown in Figure 1B, as compared with the wild-type strain, the ipk2Δ strain had reduced autophagy and kcs1Δ had no measurable autophagy. The other inositol polyphosphate pathway mutants (ipk1Δ, vip1Δ, ddp1Δ) had levels of autophagy similar to that of wild-type strain.
To further quantify the autophagy levels, we analyzed vacuolar alkaline phosphatase (ALP) activity of the mutants in the Pho8Δ60 background. Under normal conditions the vacuolar alkaline phosphatase is inactive in the cytoplasm and only becomes active upon delivery to the vacuole by autophagy.35 The activity of the alkaline phosphatase correlates with the level of autophagic activity. Yeast mutants harboring gene deletions within the inositol polyphosphate synthesis pathway were generated in the Pho8Δ60 background. The cells were grown to mid-log phase in YPD, and shifted to SD-N medium for 4 h to induce autophagy (Fig. 1C). Consistent with the GFP-Atg8 processing assay, both the ipk2Δ and kcs1Δ mutant strains had reduced levels of autophagic activity, which was similar to the autophagy defective atg1Δ strain. All of the other mutants (ipk1Δ, vip1Δ, ddp1Δ) showed normal to slightly increased levels of autophagic activity. Moreover, the treatment of rapamycin also failed to induce autophagy in the kcs1Δ mutant strain analyzed by ALP assays and by GFP-Atg8 processing assays (Fig. 1D and E). In addition, we also analyzed the function of the Cvt pathway using aminopeptidase I processing as the readout. As shown in Figure 1F, the kcs1Δ strain has a partial defect in processing Ape1 under nonstarvation conditions, indicating a partial defect in the Cvt pathway. It is not as severe as the atg5Δ autophagy mutant, which has no processing at all even under starvation conditions. In addition, the processing of Ape1 appears to be restored under starvation conditions in the kcs1Δ strain. These results indicate that KCS1 and to a lesser degree IPK2 are important for autophagy activation in response to nitrogen starvation. In the following studies, we characterized the kcs1Δ mutant in more detail.
Inositol hexakisphosphate/heptakisphosphate kinase Kcs1 affects autophagosome biogenesis
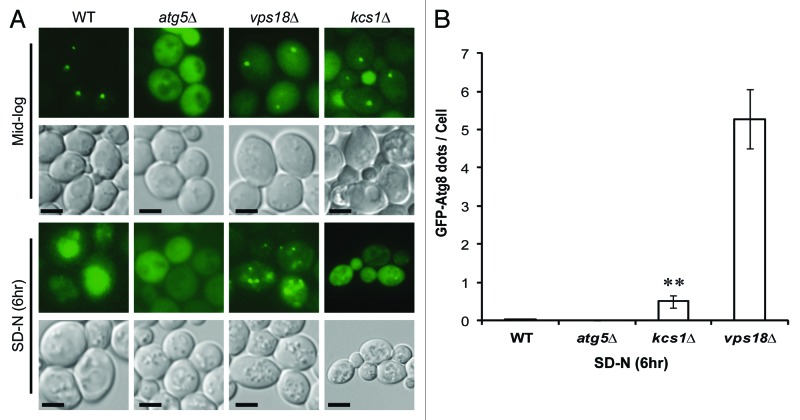
Both the GFP-Atg8 processing and the ALP assays indicate a defect in autophagy in the kcs1Δ mutant. To examine the possible cause for an autophagy defect, we first visualized the cells expressing GFP-Atg8 using fluorescent microscopy (Fig. 2). Yeast strains were grown to mid-log phase in minimal media and then shifted to SD-N media for 6 h. Under log phase growth, GFP-Atg8 in the wild-type strain accumulates at a perivacuolar site known as the phagophore assembly site (PAS), while GFP-Atg8 in the autophagy deficient strain atg5Δ failed to accumulate at the PAS and was dispersed in the cytosol.36 During mid-log phase, some of the kcs1Δ mutant cells did develop a single PAS. Under SD-N conditions, the wild-type strain accumulates GFP inside the vacuole, indicating efficient autophagy flux. As a negative control, the atg5Δ mutant failed to accumulate GFP inside the vacuole. Consistent with the autophagy defect observed by immunoblotting analysis in Figure 1, the kcs1Δ mutant had drastically reduced GFP accumulation inside their vacuoles, indicating reduction of autophagy flux. A defect in vacuolar accumulation (autophagy flux) of GFP may due to deficiency in autophagosome formation, or defect in autophagosome trafficking or fusion with vacuole. To differentiate the different causes of autophagy defect, we compared the kcs1Δ mutant to the vps18Δ mutant, which has a highly fragmented vacuole and is known to have autophagosome trafficking/vacuolar fusion defect but not autophagosome biogenesis defect.37 As shown in Figure 2A and quantified in Figure 2B, the vps18Δ mutant cells have dramatically increased GFP-Atg8 puncta in cytosol under nitrogen starvation conditions, indicating accumulation of autophagosomes. Despite the fact that the kcs1Δ mutant also has a slight vacuole morphology change, kcs1Δ mutant cells accumulated less than 10% of cytoplasmic GFP-Atg8 puncta as found in the vps18Δ mutant cells (Fig. 2A and B) under the same conditions. This result suggests that the defect in autophagy flux in the kcs1Δ mutant cells is not a simple consequence of vacuolar defect, which usually leads to increase of autophagosome accumulation in cytosol. Instead, the result suggests that KCS1 deletion may have impact on autophagosome formation.
Figure 2. Deletion of KCS1 affects autophagosome biogenesis analyzed by fluorescent microscopy. (A) Fluorescent microscopic analysis of mid-log phase cells (Mid-log) and the six-hour nitrogen deprived cells (SD-N) from the wild-type and kcs1Δ cells transformed with pCuGFP-Atg8(416). The atg5Δ and vps18Δ mutants were used as negative controls. Scale bar: 1.6 μm. (B) Accumulation of cytoplasmic GFP-Atg8 puncta in wild-type (WT), atg5Δ, kcs1Δ and vps18Δ cells after growing in SD-N medium for 6 h. **p < 0.01. The data are representative results from three independent experiments.
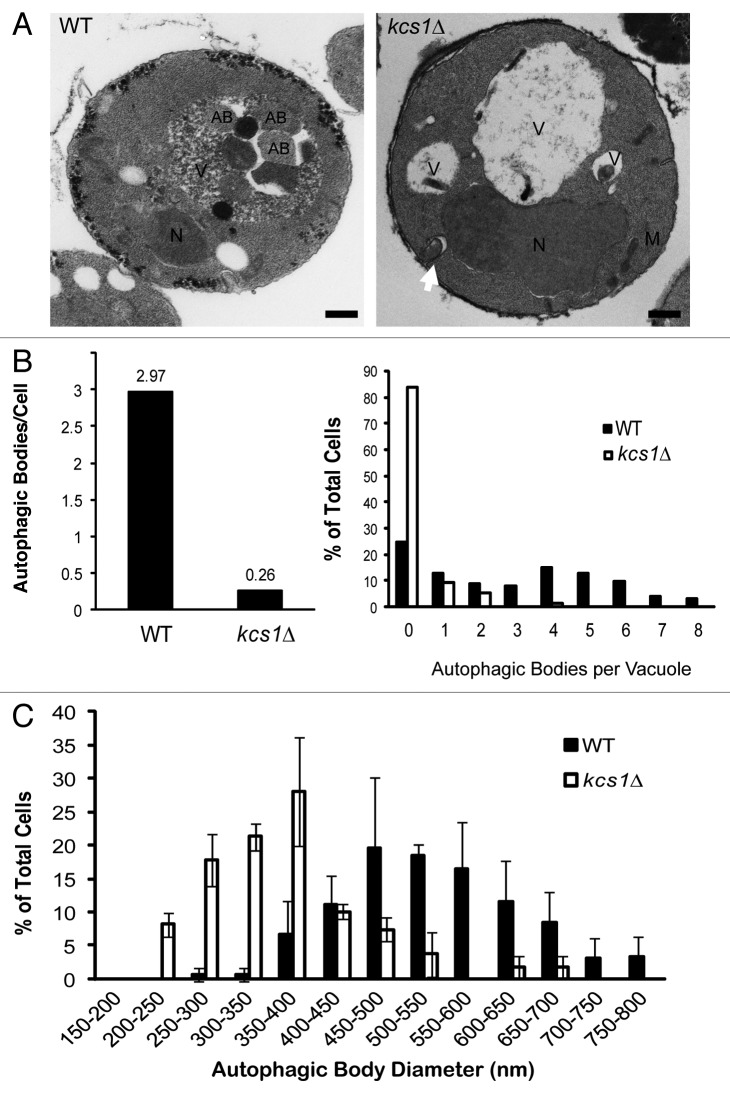
We performed electron microscopy to directly examine the possible autophagosome formation defect in kcs1Δ mutant cells (Fig. 3). Yeast strains were grown to mid-log phase in YPD, and then shifted to SD-N media in the presence of PMSF for 4 h in order to prevent the breakdown of autophagic bodies in the vacuole. As shown in Figure 3A, wild-type cells typically contained multiple autophagic bodies inside the vacuole, indicating proper formation and delivery of autophagosomes to the vacuole. In contrast, most kcs1Δ mutant cells failed to accumulate autophagic bodies in the vacuole. Consistent with the fluorescent microscopy results shown in Figure 2, the kcs1Δ mutant cells contained many fewer cytoplasmic autophagosomes when compared with the VPS18 deficient cells as described before.37 Moreover, some of the autophagosomes were untypical and appeared to originate from plasma membrane in kcs1Δ mutant, which were further characterized later. We analyzed the electron microscopic pictures quantitatively as previously described.38 As shown in Figure 3B, the kcs1Δ strain had far fewer autophagic bodies in the vacuole as compared with the wild-type cells. In addition, the size of the autophagic body found in the mutant yeast was also smaller than those in wild-type cells (Fig. 3C). Together, the results strongly suggest that a defect in autophagosome biogenesis has contributed to the autophagy defect observed in the kcs1Δ mutant cells.
Figure 3. Deletion of KCS1 affects autophagosome biogenesis analyzed by electron microscopy. (A) Representative electron microscopic pictures of wild-type (WT) and kcs1Δ mutant cells after a 4 h incubation in SD-N medium in the presence of 1 mM PMSF. N, nucleus; AB, autophagic body; M, mitochondria; V, vacuole. Scale bars: 500 nm. Arrow pointing to an untypical autophagosome. (B) Quantification of the number of autophagic bodies per cell and per vacuole. Total cells analyzed: wild-type (n = 125) and kcs1Δ (n = 129). (C) Size distribution of autophagic body in wild-type and kcs1Δ cells. Size of autophagic bodies (diameter in nm) were measured using the ImageJ software.
Loss of Kcs1 disrupts proper localization of the phagophore assembly site
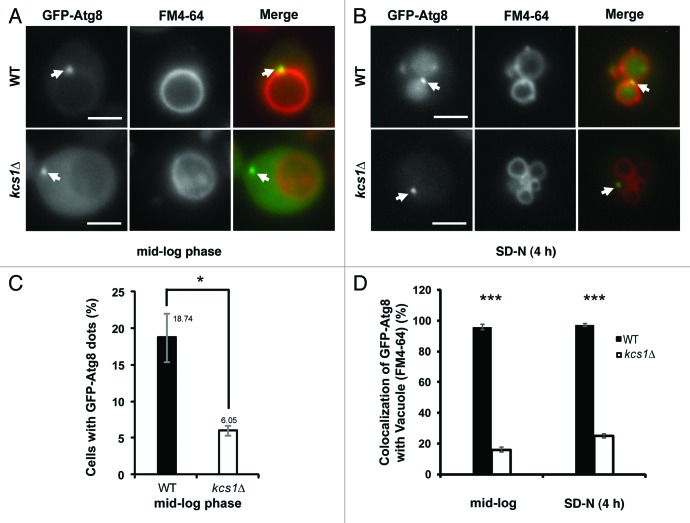
Failure to generate wild-type levels of autophagosomes and autophagosomes of correct size led us to analyze the location of some autophagy proteins. Disruption of the proper localization of Atg8 has been documented to cause smaller autophagosome formation and a reduction in autophagic flux.38 We analyzed the GFP-Atg8 localization in wild-type and kcs1Δ cells. In mid-log phase, it is well documented that wild-type yeast cells contain a single PAS that is enriched with Atg8p and is always associated with vacuolar membrane (Fig. 4A). In any given time, the GFP-Atg8 containing PAS puncta could be observed in about 20% of mid-log wild-type cells (Fig. 4C). Similarly, the kcs1Δ cells in mid-log growth phase also contained a single GFP-Atg8 punctum (Fig. 4A). However, the population of kcs1Δ cells that exhibited a single GFP-Atg8 punctum reduced dramatically as compared with wild-type cells (Fig. 4C, 18.74% vs 6.05%, p < 0.05). In the rest of kcs1Δ cells, GFP-Atg8 showed a diffused distribution pattern in cytosol. This result suggests that deletion of KCS1 might lead to reduction of PAS formation.
Figure 4. Loss of Kcs1 affects localization of phagophore assembly site. Fluorescence microscopy pictures of wild-type or kcs1Δ cells in (A) mid-log growth phase or (B) 4 h SD-N nitrogen starvation. Wild-type or kcs1Δ cells transformed with pCuGFP-Atg8(416) were either grown to mid-log phase (A), or further followed by 4 h SD-N nitrogen starvation (B). Cells were then stained with FM 4–64 to label vacuolar membrane. Arrows indicate GFP-Atg8 puncta. Scale bar: 2.5 μm. (C) Percentage of cells containing GFP-Atg8 puncta in mid-log growth phase. Approximately 400 cells were analyzed. *p < 0.05. (D) Quantification of colocalization between GFP-Atg8 puncta and vacuolar membrane in either wild-type or kcs1Δ cells under mid-log phase or nitrogen starvation condition as indicated. The data are representative results from three independent experiments. ***p < 0.001.
Strikingly, while in wild-type cells all the GFP-Atg8 containing PAS were associated with the FM 4–64 laden vacuolar membrane, the single GFP-Atg8 puncta in the kcs1Δ cells were usually not associated with vacuolar membrane (Fig. 4A and D). Among the kcs1Δ cells where a single GFP-Atg8 punctum was observed in mid-log phase, over 80% of them did not show a colocalization between GFP-Atg8 and vacuole. This observation strongly suggests that the function of KCS1 is required for the correct localization of PAS onto the vacuolar membrane.
In addition, we analyzed the GFP-Atg8 localization under nitrogen starvation. After shifting to SD-N medium, GFP distribution in wild-type cells was primarily localized inside vacuole in a diffused pattern, indicating efficient autophagic flux into vacuole. In a small percentage of cells where a single or multiple GFP-Atg8 containing puncta could be observed, they were all colocalized with vacuolar membrane (Fig. 4B and D). In contrast, in about 80% kcs1Δ cells, the GFP-Atg8 containing puncta were not associated with vacuoles under starvation condition (Fig. 4B and D). It is worth noting that the GFP-Atg8 containing structure was not specifically associated with plasma membrane (labeled with mRFP-Snc139) in the kcs1Δ cells either, as shown in Figure S1.
Loss of Kcs1 is associated with a defect in the release of Atg18 from vacuolar membrane upon nitrogen deprivation
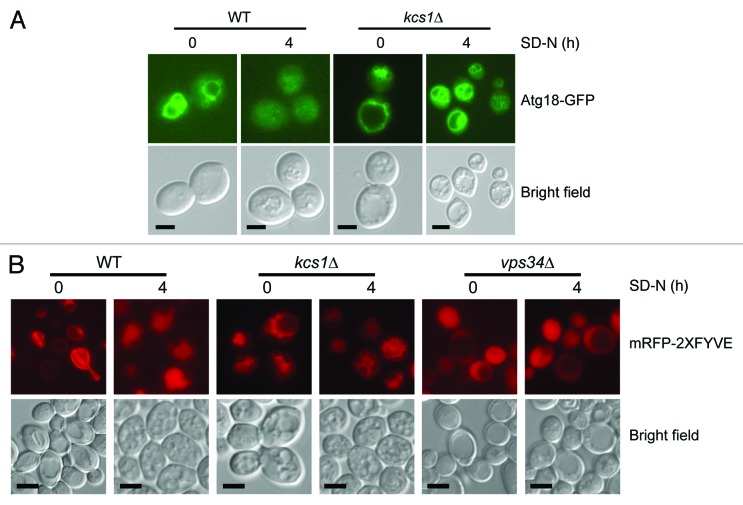
Mislocalization of Atg8 has been documented in a number of autophagy mutants, particularly in the atg21 and atg18 mutants.40,41 In addition, disruption of the lipid binding domains of Atg18 and Atg21 has been shown to mislocalize Atg8 and reduce autophagy.28,29 Since one of the roles of inositol polyphosphates is to bind to lipid binding domains of proteins, we extended our analysis to study the localization of Atg18. We generated wild-type and kcs1Δ mutant yeast strains in which Atg18-GFP is expressed under Atg18 endogenous promoter. We analyzed each of the strains under the fluorescent microscope during mid-log phase and up to 4 h of starvation (Fig. 5A). Under mid-log phase growth, Atg18-GFP was localized to the vacuolar membrane in both the wild-type and kcs1Δ mutant. When the wild-type cells were starved for up to 4 h, Atg18-GFP was almost completely released from the vacuole membrane, similar to what has been observed by others.42 However, the rate of Atg18-GFP releasing from vacuolar membrane was much slower in the kcs1Δ strain. After 4 h of starvation, significant amount of Atg18-GFP was still associated with vacuolar membrane in the mutant cells (Fig. 5A and quantification in Table 1).
Figure 5. Loss of Kcs1 affects Atg18-GFP and mRFP-2XFYVE translocation. (A) Intracellular translocation of Atg18 upon nitrogen starvation. Wild-type and kcs1Δ cells were transformed with an Atg18-GFP plasmid and grown to mid-log phase in minimal media minus URA. Cells were then nitrogen starved up to 4 h in SD-N and analyzed by fluorescence microscopy. (B) Intracellular PtdIns3P distribution analyzed by mRFP-2XFYVE binding. Wild-type and kcs1Δ cells were transformed with mRFP-2XFYVE plasmid and grown to mid log phase in minimal media minus URA. Cells were then nitrogen starved up to 4 h in SD-N and analyzed by fluorescence microscopy. Fluorescent pictures are representative images from two independent results. Scale bars: 1.6 μm.
Table 1. Ratiometric measurement of Atg18-GFP signal present on the vacuolar membrane vs. that of the cytosol in mid-log and SD-N 4hr growth condition.
| Strain | Growth condition | GFP signal ratio (vacuolar membrane/cytosolic GFP) | t testb (p value) |
|---|---|---|---|
| Wta |
mid-log |
2.49 ± 0.23 |
< 0.005 |
| SD-N 4 h |
1.44 ± 0.06 |
||
| kcs1Δa | mid-log |
2.36 ± 0.25 |
0.312 |
| SD-N 4 h | 2.70 ± 0.21 |
a WT cells and kcs1Δ cells were transformed with Atg18-GFP. bTwo-tailed Student’s t test (n = 15)
The translocation of Atg18 from the vacuolar membrane to autophagic membrane is mediated by its ability to bind PtdIns3P.42 To determine if loss of Kcs1 affects PtdIns3P distribution, we also analyzed the localization of a well-documented PtdIns3P binding protein marker mRFP-2XFYVE.26 This marker is known to bind to the vacuolar membrane under log phase conditions and translocate to the vacuolar lumen during autophagy induction due to its recruitment onto the autophagosome membrane.26 We transformed wild-type and kcs1Δ cells with the expression construct of mRFP-2XFYVE and analyzed its location under mid-log and starvation conditions. Consistent with what has been described in the literature, wild-type cells had mRFP staining on the vacuole membrane during mid-log phase growth and diffused mRFP staining inside the vacuole under starvation conditions (Fig. 5B). However, in the kcs1Δ strain mRFP-2XFYVE did not release from the vacuole membrane during starvation conditions (Fig. 5B). Together, these results indicate that lack of Kcs1 is associated with a defect in the translocation of Atg18 from vacuolar membrane to autophagic membrane under nitrogen deprivation conditions. This phenotype is associated with a similar defect of releasing of PtdIns3P from vacuolar membrane under those conditions. It remains to be determined whether the defect of Atg18 and PtdIns3P localization is the cause of autophagy defect in the kcs1Δ cells or the consequence of autophagy flux defect in these kcs1Δ cells.
Loss of Kcs1 leads to occasional autophagosome formation from the plasma membrane
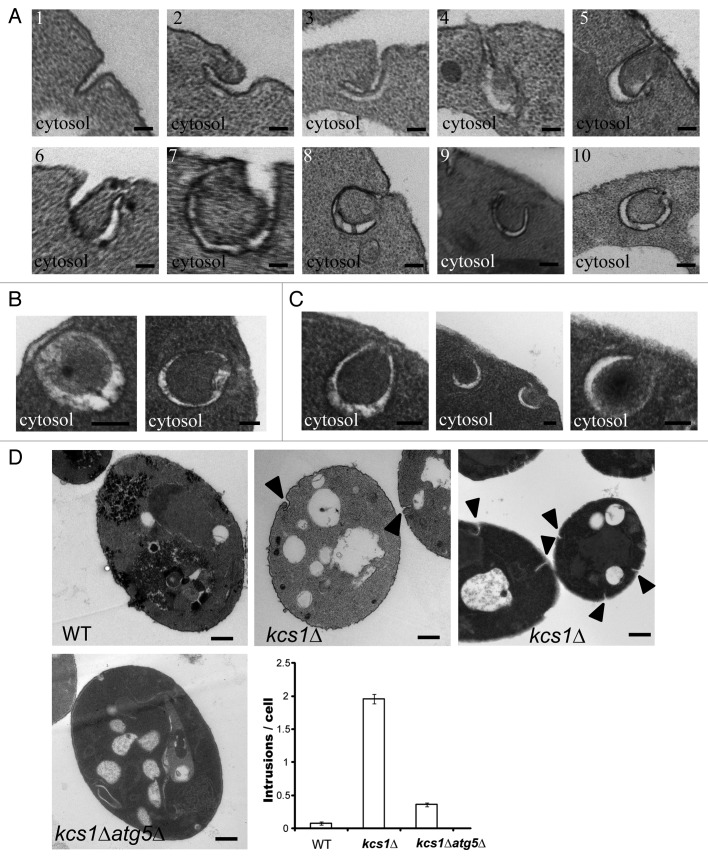
During the electron microscopy experiments, we frequently noticed a number of membrane intrusions located at the plasma membrane of the kcs1Δ mutants (Fig. 6A). Initially the intrusions resembled that of endocytosis; however, careful examination revealed that these membrane intrusions began to curve in a particular fashion that gave the resemblance to nascent autophagosome (Fig. 6A, pictures 2–5). As the membrane wrapped into a circle, cytoplasm was engulfed into the double-membrane vesicle (Fig. 6A, pictures 4–8). The plasma membrane was then seen to reseal itself and thus release the open autophagosome structure into the cytoplasm. Eventually the two ends of the membrane would come into close proximity and thus fuse in order to close (Fig. 6A, pictures 9 and 10). Detachment of the autophagosome-like structure from the plasma membrane resulted in two autophagosome-like species, sealed autophagosome-like structures (Fig. 6B) and unsealed/opened structures (Fig. 6C). The double-membrane vesicles formed from plasma membrane were structurally indistinguishable from those autophagosomes formed in cytosol of wild-type cells. This result suggests that inactivation of KCS1 may cause occasional formation of autophagosome-like double-membrane structure from plasma membrane.
Figure 6. Formation of double-membrane, autophagosome-like structures from cytoplasmic membrane in kcs1Δ cells. Electron microscopic analysis of kcs1Δ cells upon nitrogen deprivation. (A) Pictures show the different stages in the process of the formation of double-membrane, autophagosome-like vesicles from plasma membrane. Each picture (1 to 10) represents a different stage of the double-membrane, autophagosome-like structure formation in different cells. (B) Pictures represent typical sealed autophagosome-like vesicles in proximity to plasma membrane. (C) Pictures represent typical autophagosome-like vesicles in proximity to plasma membrane that have not completely closed. (D) EM pictures represent typical wild-type, kcs1Δ or kcs1Δ atg5Δ mutant cells after SD-N 4 h starvation. The arrowheads show intrusions on the plasma membrane. The plot shows quantification of the number of intrusions at the plasma membrane in wild-type, kcs1Δ or kcs1Δ atg5Δ mutant cells after 4 h SD-N nitrogen starvation. A total of 50 cells from each strain were analyzed. Scale bars: (A–C) 100 nm; (D) 500 nm.
To determine if the formation of the double-membrane vesicles originated from the plasma membrane are related to autophagosomes, we generated a KCS1 and ATG5 double deletion strain (kcs1Δ atg5Δ). The wild-type, kcs1Δ and kcs1Δ atg5Δ cells were subject to nitrogen starvation and they were subsequently analyzed by electron microscopy. As shown in Figure 6D, the kcs1Δ cells exhibited significantly more “crackles” or intrusions on plasma membrane. The intrusions on plasma membrane were quantified in these strains. The number of intrusions formed in the kcs1Δ atg5Δ mutant strain dramatically reduced as compared with the kcs1Δ strain. In addition, barely any autophagosome-like structures derived from plasma membrane was observed in kcs1Δ atg5Δ cells. These results support that the double-membrane vesicles derived from plasma membrane in the kcs1Δ mutant cells are most likely related to autophagosomes.
Discussion
Recent data in the mammalian system indicated the possibility that inositol polyphosphates may regulate the process of autophagy.30 In the present study, we used yeast as the model system to systemically determine the possible role of each enzyme involved in the synthesis of inositol polyphosphates in autophagy induced by nitrogen deprivation. The study showed that deletion of only two genes, KCS1 and IPK2, has impact on autophagy. KCS1 deletion led to a reduction in autophagosome formation and autophagy flux, which was associated with a reduction of the number of cells containing PAS and mislocalization of PAS. In addition, the kcs1Δ cells exhibited impaired Atg18 release during nitrogen starvation. Together, these observations strongly support that normal Kcs1 function is required for the correct localization of PAS for normal autophagosome formation under nitrogen starvation.
One unexpected and intriguing observation in the kcs1Δ mutant cells by electron microscopy study is that autophagosome-like, double-membrane vesicles were assembled from the plasma membrane. A dramatic increase in plasma membrane intrusion was observed in the kcs1Δ mutant strain. These plasma membrane intrusions and the autophagosome-like, double-membrane vesicles originated from plasma membrane diminish in the KCS1 and ATG5 double knockout strain. These results strongly support that the plasma membrane-derived double-membrane vesicles are most likely an autophagosome-related structure. Indeed, it has been previously described that autophagosomes can originate from the plasma membrane.43,44 However, this phenomenon is rarely observed in wild-type cells under the same conditions, indicating KCS1 deletion was a trigger for the formation of the double-membrane vesicles from plasma membrane. One simple explanation is that loss of Kcs1 leads to mislocalization of PAS and effectively eliminates the normal and main location (near vacuolar membrane) of autophagosome formation. Occasionally, the autophagic machinery (PAS) may by chance associate with cytoplasm membrane, the largest membrane reservoir in a cell, leading to autophagosome assembly at this location.
The exact molecular mechanism by which KCS1 deletion affects PAS localization is not clear. First, we do not know mislocalization of which PAS component protein causes the mislocalization of PAS. Although we have observed the abnormal translocation of Atg18-GFP under nitrogen deprivation, its localization under log-phase condition appears normal. Testing the localization of each individual PAS component protein will help answer the question. Second, we do not know which inositol phosphate species are involved in causing the mislocation of PAS. It may result from lack of production of 5-IP7, which may be required for regulating autophagy protein localization upon nitrogen starvation. Alternatively, it may be a result of the accumulation of IP6 or other inositol polyphosphate intermediates, which may interfere with the normal cellular localization of autophagy proteins. A direct measurement of the various inositol polyphosphates in the wild-type and mutant cells and a direct evaluation of the effect of the individual inositol polyphosphates on PAS localization would provide valuable insight into the question.
We cannot rule out the possibility that the autophagy defect is a secondary effect caused by the vacuolar defect observed in kcs1Δ cells, although this scenario is unlikely because of the following reasons. First, other deletion strains with a vacuolar defect, such as vps18Δ as shown in this study and other VPS genes (e.g., VAM3) shown in the literature,37,45 do not exhibit autophagosome biogenesis defect. Instead, they have increased autophagosome accumulation in the cytosol because of impairment of autophagosome trafficking. Second, the kcs1Δ cells exhibited a specific phenotype, namely mislocalization of PAS, which is again unique to kcs1Δ cells but not observed in other strains that have vacuole phenotypes. Future studies to identify the specific inositol polyphosphate that is responsible for the phenotypical defect and a direct evaluation of this molecule on autophagy as well as vacuolar function would yield a definitive conclusion.
In summary, we have systematically investigated the individual inositol polyphosphate synthesis pathway components for their possible involvement in regulating autophagy. This study reveals a specific interaction between the two pathways and shows that KCS1 is critical for correct PAS localization and autophagy activation under nitrogen deprivation. Future studies on how Kcs1 may regulate the autophagic proteins are warranted to dissect the detail interactions. In addition, the unexpected observation that autophagosome-like, double-membrane vesicle assembles from cytoplasm membrane in the kcs1Δ cells may provide a novel platform to investigate the molecular mechanism governing the membrane dynamic during autophagosome biogenesis.
Materials and Methods
Yeast strains and growth conditions
The yeast strains used in this study were from the diploid knockout library (BY4743: MATa/α his3Δ1/his3Δ1 leu2Δ0 /leu2Δ0 lys2Δ0/LYS2 MET15/met15Δ0 ura3Δ0 /ura3Δ0) created by the Saccharomyces Genome Deletion Project and purchased from Open Biosystems. Yeast cultures were grown to an OD600 of 0.5–0.7 (mid-log) in minimal media (MM, 0.175% yeast nitrogen base without amino acids and ammonium sulfate, casamino acids minus appropriate amino acids + 40 mg/ml adenine, 2% ammonium sulfate, pH 5.5) supplemented with 2% glucose. Upon reaching the desired growth phase, cells were submitted to amino acid and nitrogen starvation (SD-N, 0.175% yeast nitrogen base without amino acids and ammonium sulfate). Cells were washed twice with ddH2O and resuspended in SD-N media supplemented with 2% glucose. Cells were then incubated for 0 to 6 h depending on experiments. For electron microscopy, yeast strains were grown in YPD (1% yeast extract, 2% peptone, 2% dextrose) to mid-log phase. All incubations took place at 30°C and cultures were shaken at 250 rpm. Haploid kcs1Δ atg5Δ strains were made in BY4741 and BY4742 de novo. By mating haploid kcs1Δ atg5Δ strains of BY4741 and BY4742, kcs1Δ atg5Δ diploid strain was generated and selected in SC-lysine medium with α factor. The kcs1Δ atg5Δ diploid strain was confirmed by PCR and GFP-Atg8 vacuolar processing immunoblotting assay.
Plasmids
Yeast strains were transformed with pCuGFP-Atg8(415) or pCuGFP-Atg8(416);46 pAtg18-GFP(416), which has a pRS416 backbone and expresses using an endogenous Atg18 promoter;28 mRFP-2XFYVE (pB88-A2), which has a pRS416 backbone and is expressed by the ADH1 promoter;47 mRFP-SNC1 (pKT1563), which has a pRS416 backbone and is under TPI1 promoter. Transformation was conducted using the Lithium Acetate/ssDNA/PEG 3500 protocol.48
Immunoblotting
Yeast cultures were grown and starved as described above. At each time point, protein was isolated as described previously.49 Briefly, 2.5 OD600 of cells were spun down and resuspended in 0.1M NaOH for 5 min. Cells were again pelleted, resuspended in SDS lysis buffer (0.06 M TRIS-HCl pH 6.8, 5% glycerol, 2% SDS, 4% β-mercaptoethanol, 0.0025% bromophenol blue), and boiled for 5 min at 95°C. Supernatant were collected as protein extract samples. Immunoblotting was performed by using an Invitrogen XCell SureLock Mini-Cell apparatus and the appropriate percentage Tris-Glycine Gel (Invitrogen). Standard SDS-PAGE protocol was conducted, proteins were transferred onto a PVDF membrane and immunoblotting was performed using a standard protocol with a mouse monoclonal antibody against GFP (clone B-2 from Santa Cruz Biotechnology Inc., sc-9996) and a mouse monoclonal against toward Pgk1 (Clone 22C5 from Molecular Probes/Invitrogen) for the loading controls.
Fluorescence microscopy
Yeast cells were visualized by placing ~3 × 105 cells on a microscope slide containing an agar slab (2% low melt agarose). A coverslip was then placed on top of the slab and the cells were visualized using a Zeiss Axioplan 2 microscope with a 100× oil immersion lens. Pictures were taken using the AxioCam (Zeiss) and the OpenLab Software. GFP was visualized using the Chroma Technology Filter Em535/Ex480 (41001). mRFP-2XFYVE was visualized using the Chroma Technology Filter Em640/Ex575 (41043). Pictures were edited using Adobe Photoshop CS3 software.
Quantification of vacuolar fluorescence intensity
Fluorescence intensity of the green fluorescent protein (GFP; FITC) of unprocessed TIFF images was quantified by using ImageJ 1.43 (NIH, Bethesda, MD). Lines were drawn from the cytosol to the vacuole lumen; lines started around the midpoint of the cytosol and were typically 30–40 pixels long with a 5-pixel width line. Line positioning across the vacuole was random, but puncta were avoided. Fluorescence intensity values were exported to Excel 2007 (Microsoft, Redmond, WA) where the background was subtracted. Note that the contrast and the brightness of the images shown in Figure 5 have been adjusted after acquiring line plot profiles.
Electron microscopy
Yeast cultures were grown and starved as described above and analyzed after 4 h of starvation. PMSF was added at a final concentration of 1 mM. Approximately ~2 × 108 cells were harvested on a 0.45-μm filter apparatus, washed with 0.1 M cacodylate (pH 6.8), fixed in 2.5% gluteraldehyde/4% paraformaldehyde in 0.1 M cacodylate buffer. After Zymolyase digestion, the cells were then post-fixed in buffered 2% osmium tetroxide, dehydrated in a graded series of ethanol and embedded in SPURR resin. 90-nm thin sections were cut on a Reichert ultracut E microtome and picked up on copper grids. The grids were stained with ethanoic uranyl acetate and lead citrate.
Alkaline phosphatase assay
Yeast strains were generated by standard PCR knockout techniques50 using the wild-type background (WLY176, SEY6210 MATα his3-Δ200 leu2–3,112 lys2–801 trp1-Δ901 ura3–52 suc2-Δ9 GAL pho13Δ pho8Δ60).51 The assay was conducted as previously described.35
Supplementary Material
Acknowledgments
We are very grateful to Dr. Daniel Klionsky for the CuGFP-Atg8 (pRS416) construct, Atg18-GFP and the alkaline phosphatase mutant (WLY176), Dr. Scott Emr for the mRFP-2XFYVE construct, Dr. Yoshinori Ohsumi for the Ape1 antibody and Dr. Tanaka for the pKT1563 plasmid (pRS416 mRFP-SNC1). The work is supported by 1R01CA116088 and 1R01AG030081.
Glossary
Abbreviations:
- IP6
inositol hexakisphosphate
- IP7
inositol heptakisphosphate
- IP4
inositol tetrakisphosphate
- IP5
inositol pentakisphosphate
- PtdIns(4,5)P2
phosphatidylinositol 4,5-bisphosphate
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/20681
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/20681
References
- 1.Lee Y-S, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–12. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, et al. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3,4,5)P3 interactions. Cell. 2003;114:559–72. doi: 10.1016/S0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 3.Falasca M, Chiozzotto D, Godage HY, Mazzoletti M, Riley AM, Previdi S, et al. A novel inhibitor of the PI3K/Akt pathway based on the structure of inositol 1,3,4,5,6-pentakisphosphate. Br J Cancer. 2010;102:104–14. doi: 10.1038/sj.bjc.6605408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty A, Koldobskiy MA, Bello NT, Maxwell M, Potter JJ, Juluri KR, et al. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubois E, Scherens B, Vierendeels F, Ho MMW, Messenguy F, Shears SB. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J Biol Chem. 2002;277:23755–63. doi: 10.1074/jbc.M202206200. [DOI] [PubMed] [Google Scholar]
- 6.Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc Natl Acad Sci U S A. 2002;99:14206–11. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-beta in ovarian carcinoma cells. J Biol Chem. 2001;276:24965–70. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc Natl Acad Sci U S A. 2005;102:1911–4. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280:4264–9. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- 10.Bhandari R, Saiardi A, Ahmadibeni Y, Snowman AM, Resnick AC, Kristiansen TZ, et al. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc Natl Acad Sci U S A. 2007;104:15305–10. doi: 10.1073/pnas.0707338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saiardi A, Bhandari R, Resnick AC, Snowman AM, Snyder SH. Phosphorylation of proteins by inositol pyrophosphates. Science. 2004;306:2101–5. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- 12.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–9. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 13.York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 14.Saiardi A, Caffrey JJ, Snyder SH, Shears SB. Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett. 2000;468:28–32. doi: 10.1016/S0014-5793(00)01194-7. [DOI] [PubMed] [Google Scholar]
- 15.Auesukaree C, Tochio H, Shirakawa M, Kaneko Y, Harashima S. Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J Biol Chem. 2005;280:25127–33. doi: 10.1074/jbc.M414579200. [DOI] [PubMed] [Google Scholar]
- 16.Wang C-W, Klionsky DJ. The molecular mechanism of autophagy. Mol Med. 2003;9:65–76. [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–61. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–6. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z, Geng J, Yen W-L, Wang K, Klionsky DJ. Positive or negative roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae. Mol Cell. 2010;38:250–64. doi: 10.1016/j.molcel.2010.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Wilson WA, Fujino MA, Roach PJ. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol Cell Biol. 2001;21:5742–52. doi: 10.1128/MCB.21.17.5742-5752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004;279:20663–71. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:4180–9. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tallóczy Z, Jiang W, Virgin HW, 4th, Leib DA, Scheuner D, Kaufman RJ, et al. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A. 2002;99:190–5. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–30. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obara K, Noda T, Niimi K, Ohsumi Y. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells. 2008;13:537–47. doi: 10.1111/j.1365-2443.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- 27.Obara K, Sekito T, Niimi K, Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem. 2008;283:23972–80. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair U, Cao Y, Xie Z, Klionsky DJ. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J Biol Chem. 2010;285:11476–88. doi: 10.1074/jbc.M109.080374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krick R, Tolstrup J, Appelles A, Henke S, Thumm M. The relevance of the phosphatidylinositolphosphat-binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy. FEBS Lett. 2006;580:4632–8. doi: 10.1016/j.febslet.2006.07.041. [DOI] [PubMed] [Google Scholar]
- 30.Nagata E, Saiardi A, Tsukamoto H, Satoh T, Itoh Y, Itoh J, et al. Inositol hexakisphosphate kinases promote autophagy. Int J Biochem Cell Biol. 2010;42:2065–71. doi: 10.1016/j.biocel.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar S, Rubinsztein DC. Inositol and IP3 levels regulate autophagy: biology and therapeutic speculations. Autophagy. 2006;2:132–4. doi: 10.4161/auto.2387. [DOI] [PubMed] [Google Scholar]
- 32.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–11. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanki T, Wang K, Klionsky DJ. A genomic screen for yeast mutants defective in mitophagy. Autophagy. 2010;6:278–80. doi: 10.4161/auto.6.2.10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–94. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–32. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–18. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 37.Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–27. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen W-L, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101–14. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakane H, Yamamoto T, Tanaka K. The functional relationship between the Cdc50p-Drs2p putative aminophospholipid translocase and the Arf GAP Gcs1p in vesicle formation in the retrieval pathway from yeast early endosomes to the TGN. Cell Struct Funct. 2006;31:87–108. doi: 10.1247/csf.06021. [DOI] [PubMed] [Google Scholar]
- 40.Meiling-Wesse K, Barth H, Voss C, Eskelinen E-L, Epple UD, Thumm M. Atg21 is required for effective recruitment of Atg8 to the preautophagosomal structure during the Cvt pathway. J Biol Chem. 2004;279:37741–50. doi: 10.1074/jbc.M401066200. [DOI] [PubMed] [Google Scholar]
- 41.Strømhaug PE, Reggiori F, Guan J, Wang C-W, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell. 2004;15:3553–66. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obara K, Ohsumi Y. PtdIns 3-Kinase Orchestrates Autophagosome Formation in Yeast. J Lipids. 2011;2011:498768. doi: 10.1155/2011/498768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuervo AM. The plasma membrane brings autophagosomes to life. Nat Cell Biol. 2010;12:735–7. doi: 10.1038/ncb0810-735. [DOI] [PubMed] [Google Scholar]
- 44.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12:747–57. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–29. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim J, Huang W-P, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rue SM, Mattei S, Saksena S, Emr SD. Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol Biol Cell. 2008;19:475–84. doi: 10.1091/mbc.E07-07-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gietz RD, Schiestl RH. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:35–7. doi: 10.1038/nprot.2007.14. [DOI] [PubMed] [Google Scholar]
- 49.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–60. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 50.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–30. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20:4730–8. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.