Abstract
Induced pluripotent stem cells (iPSC) hold significant promise for advancing biomedical research. In the case of monogenic diseases, patient-iPSC and their derivatives contain the disease-causing mutation, suggesting the possibility of recapitulating salient disease features in vitro. Fanconi anemia (FA) is the most common inherited bone marrow failure syndrome. The etiology of bone marrow failure in FA remains largely unclear, but limited studies on patient bone marrow cells indicate cell intrinsic defects as causative. We examined the feasibility of modeling FA in a system based on hematopoietic differentiation of patient-specific iPSC. An informative iPSC-based model is predicated on the ability to derive disease-specific (uncorrected) patient iPSC that contain the disease-causing mutation, are pluripotent, maintain a normal karyotype and are capable of hematopoietic differentiation. Careful analysis of hematopoietic differentiation of such iPSC holds the promise of uncovering new insights into bone marrow failure and may enable high-throughput screening with the goal of identifying compounds that ameliorate hematopoietic failure. Ultimately, genetic correction, molecular characterization and successful engraftment of iPSC-derived cells may provide an attractive alternative to current hematopoietic stem cell-targeted gene therapy in some monogenic diseases, including FA.
Keywords: Fanconi anemia, bone marrow failure, drug discovery, hematopoietic differentiation, induced pluripotent stem cells
Introduction
The discovery of novel therapies is often supported by pre-clinical models that recapitulate salient disease features. While highly penetrant monogenic diseases are generally amenable to this approach, the pre-clinical models available for the study of the inherited bone marrow failure syndrome Fanconi anemia (FA) are limited. FA results from biallelic mutations in any of 15 genes, leading to characteristic developmental anomalies, cancer predisposition and near universal onset of bone marrow failure during childhood.1 Current tools for biomedical research include cell-free extracts for biochemical studies, immortalized patient cell lines, knockdown of the FA pathway in zebrafish2 and gene-targeted knockout mice. Individual murine genetic knockouts of Fanca, Fancc, Fangg and Fancd2 have been generated but do not develop bone marrow hypoplasia.3 Limited studies of murine knockout and human and FA hematopoietic progenitors have revealed an intrinsic hypersensitivity to the inhibitory cytokines (tumor necrosis factor α, interferon-gamma) and oxidative stress.4-7 However, the mechanisms underlying bone marrow failure remain elusive, and there are currently no effective pharmacologic treatments that can halt the progression of the disease.
Direct reprogramming represents a novel approach to obtaining patient-specific stem cells. Because of their virtually unlimited replicative capacity and clonability, induced pluripotent stem cells (iPSC) can provide adequate material for sophisticated molecular analysis. In addition, large quantities of otherwise limited differentiated cells, such as hematopoietic progenitor cells, can be generated ex vivo (reviewed in ref. 8). A growing number of reports indicate the possibility of eliciting disease-relevant phenotypes in iPSC-derived cells. Conceptually, direct reprogramming of somatic cells results in iPSC lines harboring the patient mutation. In the case of monogenic disorders, iPSC-derived cells are obligate carriers of the patient mutation, and cell types afflicted by the disease can therefore be expected to display a disease-relevant phenotype. This paradigm is illustrated by a growing number of neuronal, muscular and hematopoietic diseases that have been recapitulated in iPSC-based models (reviewed in ref. 9)
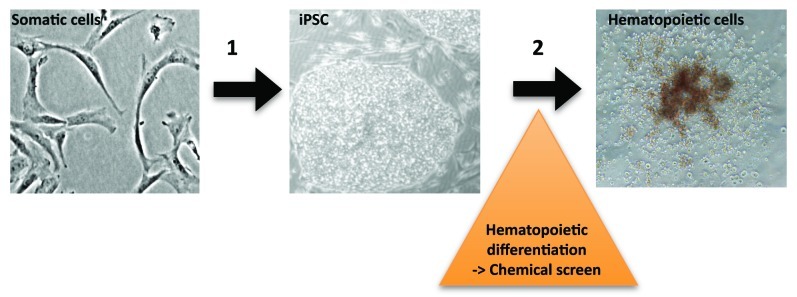
Given the strong penetrance of bone marrow failure in FA, Tulpule and colleagues reasoned that human embryonic stem cells (hESC) rendered FA-deficient by a RNA interference (RNAi) knockdown of FANCA or FANCD2 would display deficits in hematopoietic differentiation in vitro. Indeed, directed differentiation of FANCD2 (and to a lesser degree FANCA) deficient hESC, resulted in measurable decreases of CD45+ cells, reduced numbers of hematopoietic progenitor colonies and reduced expression levels of hematopoietic-specific genes, demonstrating that perturbation of the FA biochemical pathway in human pluripotent stem cells causes measurable defects in hematopoietic differentiation.10 Based on the hypothesis that defects of blood formation in vitro may provide insights into critical processes occurring in vivo, we reasoned that human FA iPSC can provide a platform for dissecting disease-specific cellular and molecular perturbations of hematopoietic differentiation. In a second step, such a system may enable high-throughput screening of chemical libraries with the goal of identifying compounds that may ameliorate hematopoietic failure (Fig. 1).
Figure 1. In vitro blood formation of Fanconi anemia induced pluripotent stem cells. (1) Direct reprogramming of human FA fibroblasts yields disease-specific iPSC containing patient gene mutations. (2) Directed differentiation of iPSC results in hematopoietic progenitor cells, enabling disease modeling and chemical screens.
In 2009, Raya and colleagues reported the failure of six FA patient samples to undergo direct reprogramming in 28 attempts. FA iPSC could only be obtained if the somatic cells were first corrected by transgenic expression of the wild type FA cDNA in the somatic cells, suggesting that the FA biochemical pathway is critical for the derivation and maintenance of pluripotent stem cells.11 We recently demonstrated that reprogramming activated the FA pathway and resulted in increased double-strand DNA breaks and senescence in cells defective in the FA pathway. Consistent with an important role of the FA pathway in the reprogramming process, the reprogramming efficiency of Fanca-/- tail-tip fibroblasts was 10-fold decreased as compared with wild type littermate controls, indicating that somatic FA cells are resistant but not refractory to reprogramming. In our hands, attempts at reprogramming seven FA patient fibroblast samples under standard conditions (21% oxygen tension) with retroviral vectors expressing the four reprogramming factors (OCT4, SOX2, KLF4, c-MYC) failed to yield any iPSC. However, utilizing optimized reprogramming conditions including hypoxia (5% O2) and a lentiviral vector co-expressing all four reprogramming factors,12 we were able to derive both uncorrected and corrected patient-specific iPSC lines from two patients belonging to the FA-A and FA-C complementation group, respectively13. Utilizing these uncorrected, disease-specific human FA iSPC lines, we sought to evaluate the feasibility of disease modeling.
Results
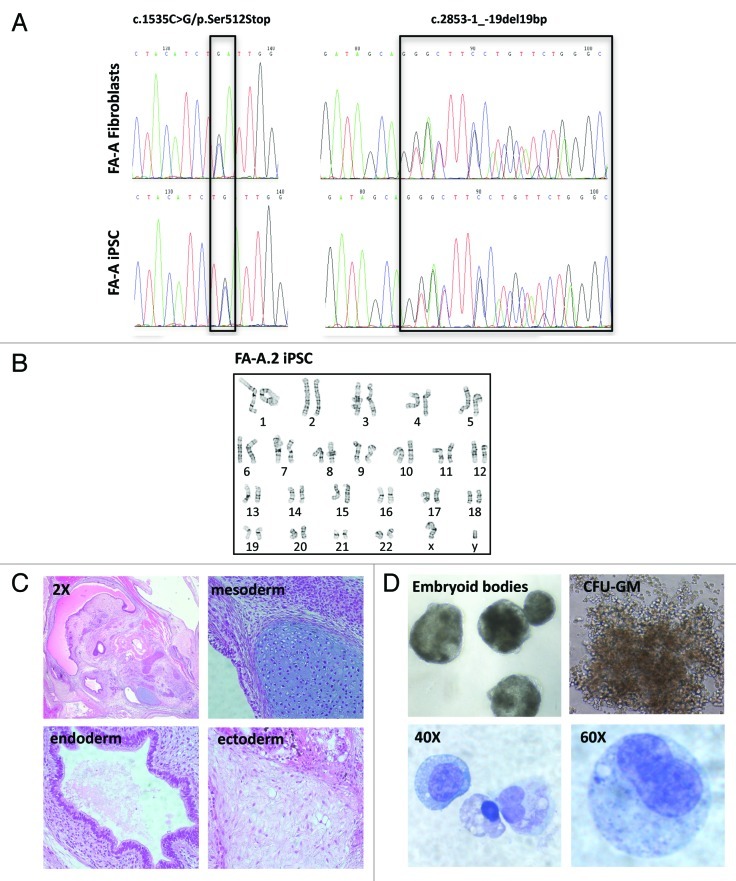
Somatic mosaicism may occur in up to 25% of FA patients.14-17 Given the significantly reduced reprogramming efficiency of somatic FA cells, we evaluated the possibility that reprogramming might have occurred in a selected subset of somatic cells that had undergone reversion of the FANCA gene mutation. DNA sequencing of the patient fibroblasts and the resultant iPSC clones revealed that the patient sample contained biallelic mutations in the FANCA gene and is heterozygous for c.2853–1_-19del19bp, inherited paternally, and c.1535C > G/p.Ser512Stop, inherited maternally. Analysis of the corresponding iPSC line confirmed the presence of the same disease-causing mutations, effectively ruling out a reversion event (Fig. 2A).
Figure 2. Feasibility of deriving hematopoietic cells from uncorrected FA-A iPSC. (A) DNA sequencing chromatograms showing disease-causing compound heterozygous mutations in the FANCA gene, present in the patient fibroblasts and resultant iPSC. (B) Normal karyogram of the FA-A.2 iPSC line. (C) Teratoma derived from the FA-A.2 iPSC line (2X overview and 20X magnification). (D) Formation of embryoid bodies (EBs) from FA-A iPSC.2. Dissociated EBs yielded hematopoietic colony forming units (CFU); Cytology of hematopoietic cells derived from CFU (Wright-Giemsa stain).
Reprogramming has been shown to generate genotoxic stress, causing cell cycle arrest,18 cellular senescence13,19 and copy number variations.20 Given the significantly reduced reprogramming efficiency of somatic FA cells, it is conceivable that FA cells accumulate an increased burden of genomic damage prior to or during reprogramming, blocking damaged cells from becoming iPSC. Conversely, it is also possible that only a small subset of somatic FA cells that acquire mutations conferring a growth and survival advantage achieves pluripotency. Because of the increased susceptibility of FA cells to genomic stress and DNA damage, analysis of copy number variations or gene mutations by single nucleotide polymorphism array and whole-exome sequencing is likely to be informative with regard to changes occurring in the somatic cells vs. the resultant iPSC. We performed cytogenetic analysis of G-banded metaphase cells in two uncorrected human FA-A iPSC lines. One line (FA-A.1) contained cytogenetic abnormalities in all 20 metaphases that were analyzed, including trisomy 12, a translocation of unknown genetic material to the long-arm of chromosome 15, and an abnormal Y-chromome (data not shown). The second line (FA-A.2) did not show any clonal cytogentic aberrations, instead revealing a normal 46, XY karyotype in 18 of 20 metaphases (Fig. 2B). We therefore elected to conduct subsequent experiments utilizing the FA-A.2 iPSC line.
To assess whether FA-A.2 iPSC is pluripotent, we injected this iPSC line intramuscularly into non-obese diabetic/severe combined immunodeficient (NOD/SCID) mice. Histologic analysis of the cystic tumors that arose after nine weeks demonstrated well-differentiated teratomas containing endo-, meso- and ectodermal elements (Fig. 2C). We next evaluated the ability of FA-A.2 iPSC to undergo directed differentiation and form hematopoietic cells. Embryoid bodies were formed in the presence of hematopoietic cytokines as previously described.21 After 16 d, the embryoid bodies were dissociated and seeded into cytokine-containg semisolid methylcellulose. We observed the formation of granulocyte, macrophage and mixed (GM) colonies. Cytologic analysis of individual colonies confirmed the presence of hematopoietic cells (Fig. 2D). Taken together, these data indicate that modeling of FA hematopoietic differentiation in a human iPSC-based system is feasible and warrants detailed studies involving a panel of individual patient iPSC lines.
Experimental Procedures
Patient-specific fibroblasts were obtained after informed consent under a protocol approved by the Boston Children’s Hospital Institutional Review Board.
Teratoma formation and karyotype analysis.
Teratomas were assessed by injecting 3 x106 iPSCs intramuscularly into NOD/SCID mice (NOD.CB17-Prkdcscid/J strain; The Jackson Laboratory). Teratomas were dissected after 9 weeks, fixed in 10% paraformaldehyde, embedded in paraffin, and 4-μm sections were stained with H&E (Rodent Histopathology Core, Dana-Farber Cancer Institute /Harvard Cancer Center). Images were obtained using a Nikon Eclipse 90i microscope. Cytogenetic analysis of g-banded metaphases was performed by Cell Line Genetics.
Hematopoietic differentiation.
Hematopoietic colony-forming activity of human iPSC lines was assayed as described previously.21 Briefly, embryoid bodies were dissociated after 16 d, and 1x104 or 3x104 cells were seeded in methylcellulose containing recombinant cytokines (MethoCult H4434; StemCell Technologies). Individual CFU colonies were picked, and cytospins were stained with the Wright-Giemsa stain method. Photographs were taken using a Nikon Eclipse TS100 microscope.
DNA sequence analysis.
DNA was extracted from the patient fibroblasts, and the resultant iPSC lines and PCR were amplified by primers flanking the mutations that were previously observed in this family. PCR fragments were sequenced on an ABI prism sequencer and interpreted with Sequencher sequence analysis software.
Discussion
Bone marrow failure remains the primary cause of morbidity and mortality in FA and occurs nearly universally, with a median age of onset of seven years. By age 40, 90% of FA patients develop bone marrow failure.22 The mechanisms underlying bone marrow failure remain unclear, and there are currently no pharmacologic treatments with long-term efficacy. In vitro myeloid and erythroid colony growth of bone marrow and peripheral blood cells from FA patients is decreased, suggesting the contribution of an intrinsic cellular defect to the bone marrow failure.23 However, studies of primary human FA bone marrow cells are limited, and knockout mouse models do not recapitulate the human phenotype. For instance, these mice do not develop bone marrow failure.3
Hematopoietic differentiation of hESC in the embryoid body (EB) system recapitulates many important aspects of early hematopoietic development.21,24,25 Tulpule et al. recently demonstrated that knockdown of FA genes in hESC results in decreased blood formation in vitro, including significantly decreased numbers of CD45+ cells, decreased expression of hematopoietic genes and decreased hematopoietic colonies in hematopoietic growth factor-containing semisolid methylcellulose. These defects were rescued by transgenic expression of the complementing FA cDNA,10 thus proving, in principle, that human pluripotent stem cells with defects in the FA pathway display a disease-specific phenotype. Other recent examples of disease modeling include iPSC-derived cardiomyocytes from patients with long QT syndrome and neuronal cells derived from patients with the neurodegenerative Rett syndrome. The former showed characteristic electrophysiologic alterations that were exacerbated by treatment with catecholamines and attenuated by β blockade,26 while the latter showed fewer synapses and altered electrophysiology. The defects in Rett syndrome neurons were rescued genetically by expression of the wild type MeCP2 gene and chemically by exposure to gentamycin.27
We recently demonstrated that uncorrected, disease-specific human FA iPSC can be obtained under optimized conditions, albeit at a decreased efficiency.13 Importantly, we showed here that FA iPSC are pluripotent, capable of maintaining a normal karyotype, forming embryoid bodies and yielding hematopoietic cells upon directed differentiation. Contrary to hESC that represent generic cell lines unrelated to patients with a specific disease of interest, human FA iPSC contain patient-specific mutations. Given that FA is a monogenic disease with strong penetration, we speculate that FA iPSC will demonstrate a measurable hematopoietic phenotype in vitro. Such a model system, based on expandable, disease- and patient-specific stem cells (rather than generic human ES cell lines) will enable biologic studies that were previously limited due to a paucity of primary FA hematopoietic bone marrow cells. As indicated in Figure 1, this model system may provide the opportunity to screen chemical libraries for compounds with biologic effects on FA hematopoiesis. Conceivable results of such a screen include molecules that specifically enhance FA blood formation vs. molecules that demonstrate the opposite effect. Any compound resulting in either effect holds promise for gaining new insights into pathways and mechanisms that are important in FA hematopoiesis and bone marrow failure. In the long-term, screening of large unbiased chemical libraries may facilitate the discovery of pharmacologic agents with clinical efficacy in bone marrow failure patients.
On a cautionary note, the value of any iPSC-based model hinges on the assumption that in vitro phenotypes are representative of in vivo pathophysiology. While bone marrow failure is a universal outcome in FA patients, inter-patient variability does exist with regard to the age of onset, rapidity of progression and severity of the disease.22 Another important consideration is clonal variability, the phenomenon that certain pluripotent cell lines are more amenable to directed differentiation than others, despite the lack of genotypic differences. Finally, recent papers indicate that early passage iPSC partially retain an “epigenetic memory” of the cell of origin, favoring their differentiation along lineages related to the donor cell while restricting alternative cell fates.28 These aspects warrant careful consideration in the development of an iPSC-based disease model. To minimize experimental variability, it will be crucial to establish iPSC lines from comparable somatic cell types and conduct reprogramming using identical methodology. In the context of bone marrow failure, there may be some value in utilizing a defined somatic hematopoietic cell (e.g., lymphocytes or CD34+ cells) as a starting population, facilitating sample collections form patients via simple phlebotomy and offering the hypothetical advantage of yielding a more pronounced hematopoietic phenotype.
As a means of overcoming clonal variability, the ideal model system should include patient-specific iPSC that have been conditionally gene-corrected, enabling a comparison of isogenic iPSC that are distinct only with regard to the expression of a specific FA protein. Undoubtedly, human pluripotent stem cells hold great potential for advancing biomedical research and developing new therapies. While the long-term goal of regenerative medicine remains compelling but futuristic, it is likely that iPSC-based disease models will yield important new insights in critical areas, including bone marrow failure.
Acknowledgments
We would like to thank the Dana Farber/Harvard Cancer Center Rodent Histopathology Core for assisting with teratoma histology. We appreciate the assistance with DNA sequence analysis provided by Bai-Lin Wu, Va Lip and Yiping Shen of the Genetics Diagnostic Lab at Boston Children’s Hospital. Grant support: Supported by a fellowship grant from the St. Baldrick’s Foundation (L.M.), Harvard Stem Cell Institute (L.M.), Hood Foundation (L.M.), and NIH1RC4DK090913–01 (D.A.W.).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21109
References
- 1.Soulier J. Fanconi anemia. Hematology Am Soc Hematol Educ Program. 2011;2011:492–7. doi: 10.1182/asheducation-2011.1.492. [DOI] [PubMed] [Google Scholar]
- 2.Liu TX, Howlett NG, Deng M, Langenau DM, Hsu K, Rhodes J, et al. Knockdown of zebrafish Fancd2 causes developmental abnormalities via p53-dependent apoptosis. Dev Cell. 2003;5:903–14. doi: 10.1016/S1534-5807(03)00339-3. [DOI] [PubMed] [Google Scholar]
- 3.Parmar K, D’Andrea A, Niedernhofer LJ. Mouse models of Fanconi anemia. Mutat Res. 2009;668:133–40. doi: 10.1016/j.mrfmmm.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Yang Y, Yuan J, Hong P, Freie B, Orazi A, et al. Continuous in vivo infusion of interferon-gamma (IFN-gamma) preferentially reduces myeloid progenitor numbers and enhances engraftment of syngeneic wild-type cells in Fancc-/- mice. Blood. 2004;104:1204–9. doi: 10.1182/blood-2004-03-1094. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Sejas DP, Zhang X, Qiu Y, Nattamai KJ, Rani R, et al. TNF-alpha induces leukemic clonal evolution ex vivo in Fanconi anemia group C murine stem cells. J Clin Invest. 2007;117:3283–95. doi: 10.1172/JCI31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joenje H, Arwert F, Eriksson AW, de Koning H, Oostra AB. Oxygen-dependence of chromosomal aberrations in Fanconi’s anaemia. Nature. 1981;290:142–3. doi: 10.1038/290142a0. [DOI] [PubMed] [Google Scholar]
- 7.Dufour C, Corcione A, Svahn J, Haupt R, Poggi V, Béka’ssy AN, et al. TNF-alpha and IFN-gamma are overexpressed in the bone marrow of Fanconi anemia patients and TNF-alpha suppresses erythropoiesis in vitro. Blood. 2003;102:2053–9. doi: 10.1182/blood-2003-01-0114. [DOI] [PubMed] [Google Scholar]
- 8.Müller LU, Daley GQ, Williams DA. Upping the ante: recent advances in direct reprogramming. Mol Ther. 2009;17:947–53. doi: 10.1038/mt.2009.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherry AB, Daley GQ. Reprogramming cellular identity for regenerative medicine. Cell. 2012;148:1110–22. doi: 10.1016/j.cell.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tulpule A, Lensch MW, Miller JD, Austin K, D’Andrea A, Schlaeger TM, et al. Knockdown of Fanconi anemia genes in human embryonic stem cells reveals early developmental defects in the hematopoietic lineage. Blood. 2010;115:3453–62. doi: 10.1182/blood-2009-10-246694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raya A, Rodríguez-Pizà I, Guenechea G, Vassena R, Navarro S, Barrero MJ, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–9. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warlich E, Kuehle J, Cantz T, Brugman MH, Maetzig T, Galla M, et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 2011;19:782–9. doi: 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller LU, Milsom MD, Harris CE, Vyas R, Brumme KM, Parmar K, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119:5449–57. doi: 10.1182/blood-2012-02-408674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soulier J, Leblanc T, Larghero J, Dastot H, Shimamura A, Guardiola P, et al. Detection of somatic mosaicism and classification of Fanconi anemia patients by analysis of the FA/BRCA pathway. Blood. 2005;105:1329–36. doi: 10.1182/blood-2004-05-1852. [DOI] [PubMed] [Google Scholar]
- 15.Gross M, Hanenberg H, Lobitz S, Friedl R, Herterich S, Dietrich R, et al. Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self-correction. Cytogenet Genome Res. 2002;98:126–35. doi: 10.1159/000069805. [DOI] [PubMed] [Google Scholar]
- 16.Waisfisz Q, Morgan NV, Savino M, de Winter JP, van Berkel CG, Hoatlin ME, et al. Spontaneous functional correction of homozygous fanconi anaemia alleles reveals novel mechanistic basis for reverse mosaicism. Nat Genet. 1999;22:379–83. doi: 10.1038/11956. [DOI] [PubMed] [Google Scholar]
- 17.Lo Ten Foe JR, Kwee ML, Rooimans MA, Oostra AB, Veerman AJ, van Weel M, et al. Somatic mosaicism in Fanconi anemia: molecular basis and clinical significance. Eur J Hum Genet. 1997;5:137–48. [PubMed] [Google Scholar]
- 18.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–5. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–9. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 21.Chadwick K, Wang L, Li L, Menendez P, Murdoch B, Rouleau A, et al. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood. 2003;102:906–15. doi: 10.1182/blood-2003-03-0832. [DOI] [PubMed] [Google Scholar]
- 22.Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101:1249–56. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 23.Daneshbod-Skibba G, Martin J, Shahidi NT. Myeloid and erythroid colony growth in non-anaemic patients with Fanconi’s anaemia. Br J Haematol. 1980;44:33–8. doi: 10.1111/j.1365-2141.1980.tb01181.x. [DOI] [PubMed] [Google Scholar]
- 24.Zambidis ET, Peault B, Park TS, Bunz F, Civin CI. Hematopoietic differentiation of human embryonic stem cells progresses through sequential hematoendothelial, primitive, and definitive stages resembling human yolk sac development. Blood. 2005;106:860–70. doi: 10.1182/blood-2004-11-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kennedy M, D’Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109:2679–87. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 27.Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–39. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–90. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]